Abstract
The ipl locus is a site for the preferential insertion of IS6110 and has been identified as an insertion sequence, IS1547, in its own right. Various deletions around the ipl locus of clinical isolates of Mycobacterium tuberculosis were identified, and these deletions ranged in length from several hundred base pairs up to several kilobase pairs. The most obvious feature shared by these deletions was the presence of an IS6110 copy at the deletion sites, which suggested two possible mechanisms for their occurrence, IS6110 transposition and homologous recombination. To clarify the mechanism, an investigation was conducted; the results suggest that although deletion transpositionally mediated by IS6110 was a possibility, homologous recombination was a more likely one. The implications of such chromosomal rearrangements for the evolution of M. tuberculosis, for IS6110-mediated mutagenesis, and for the development of genetic tools are discussed. The deletion of genomic DNA in isolates of M. tuberculosis has previously been noted at only a few sites. This study examined the deletional loss of genetic material at a new site and suggests that such losses may occur elsewhere too and may be more prevalent than was previously thought. Distinct from the study of laboratory-induced mutations, the detailed analysis of clinical isolates, in combination with knowledge of their evolutionary relationships to each other, gives us the opportunity to study mutational diversity in isolates that have survived in the human host and therefore offers a different perspective on the importance of particular genetic markers in pathogenesis.
The study of genome rearrangements in bacteria facilitates the development of genetic tools and the understanding of the mechanisms of genetic diversity of chromosomes. Genome rearrangements usually involve specific proteins and specific DNA sequences. Homologous recombination, which takes place between repeated DNA sequences, is one of the most important mechanisms for bacterial genome rearrangements. Any repeated DNA sequences on a chromosome can induce homologous recombination, but insertion sequences (ISs) are one of the most abundant of these (for reviews, see references 25 and 27).
Mycobacterium tuberculosis, which is responsible for more than 7 million new cases of human tuberculosis and about 3 million deaths annually (31), contains several different IS elements in its genome, one of which is IS6110. IS6110 is a member of the IS3 family of transposable elements and is 1,355 bp in length. From none to 25 copies of IS6110 are present in M. tuberculosis strains (16, 29). It ends in 28-bp imperfect inverted repeats designated IR-l and IR-r at the left and right ends, respectively (16, 20). IS6110 has several open reading frames (ORFs); ORFb is the longest (1,037 bp) and encodes a transposase which is transcribed in the direction from IR-l to IR-r (20). On transposition, 3 or 4 bp of the DNA sequence at the target site is duplicated at the ends of the new copy, the direct repeats (9, 12), a feature which is the same as in IS3 (32). IS6110 preferentially inserts into certain regions of the chromosome of M. tuberculosis, such as the ipl locus (9, 16).
Here we report the identification of various deletions around the ipl locus in clinical isolates of M. tuberculosis. This is the first report of naturally occurring chromosomal rearrangements of M. tuberculosis mediated by IS6110. The likely mechanisms of rearrangement and their implications are discussed.
MATERIALS AND METHODS
Bacterial isolates and DNA extraction.
We studied a total of 31 clinical isolates of M. tuberculosis, all from clinical specimens isolated in the Scottish Mycobacteria Reference Laboratory, Edinburgh, United Kingdom, and collected between 1990 and 1996; also, 10 “Beijing Family” strains isolated in Thailand and type strain H37Rv were studied. Culturing was in Middlebrook 7H9 medium in a 50-ml centrifuge tube at 37°C for about 4 weeks. After we determined that the cultures were free from other bacterial contamination, cells were harvested and heated to 80°C for 30 min and stored at −20°C prior to DNA extraction. DNA extraction was performed by a standard protocol (36).
Antibiotic susceptibility tests.
Drug sensitivity testing was conducted with a Bactec Radiometric System (Becton Dickinson, Paramus, N.J.).
Long PCR.
PCRs with 50-μl reaction mixtures were conducted with the Expand Long Template PCR System (Boehringer, Mannheim, Germany). Reaction mixtures contained 2 mM Tris-HCl, 10 mM KCl, 0.1 mM dithiothreitol, 0.01 mM EDTA, 0.05% (vol/vol) Tween 20, 0.05% (vol/vol) Nonidet P-40, 10% (vol/vol) glycerol, 350 mM (each) deoxynucleotide triphosphate, and 250 nM each primer. The reaction mixtures were subjected to 35 cycles of 1 min at 94°C, 1 min at 61°C, and 4 min at 72°C following denaturation of DNA for 3 min at 94°C; the program finished with 1 min at 68°C and 8 min at 72°C.
DNA sequencing.
DNA was sequenced with a model 377A automated DNA sequencer with a Prism-Ready Mix Kit based on Ampli-Taq CS and polymerase (Applied Biosystems, Inc., Warrington, United Kingdom). Each sequencing reaction mixture contained 150 μg of template DNA, a 3.2 pM concentration of one primer, and 8 μl of Prism-Ready Mix and were subjected to 25 cycles of denaturation (96°C, 30 s), annealing (50°C, 15 s), and extension (60°C, 4 min).
IS6110 restriction fragment length polymorphism (RFLP) analysis.
To make a digoxigenin (DIG)-labelled IS6110 DNA probe, 5 μl of Mycobacterium bovis BCG (Pasteur) DNA solution (10 μg/ml) was added to a PCR tube which contained 40 μl of PCR mixture (50 mM KCl, 10 mM Tris-HCl [pH 8.0], 1.5 mM MgCl2, 5% glycerol, 225 μM [each] primers P5 and P6 [17], 0.5 U of Taq polymerase). Five microliters of 10× DIG-dUTP–deoxynucleotide triphosphate labelling mixture (Boehringer) was added to the reaction mixture, which was then subjected to PCR at an annealing temperature of 65°C. PvuII-digested supercoiled DNA ladder (Gibco-BRL, Life Technologies Ltd., Paisley, United Kingdom) and HaeIII-digested φX174 DNA (Advanced Biotechnologies, London, United Kingdom) were DIG labelled by a randomly primed-DNA labelling method (Boehringer). The working probe solutions were prepared by diluting the DIG-labelled PCR product in standard hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1.0% blocking reagent for nucleic acid hybridization, 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate) to a concentration of 5 to 25 ng/ml.
Hybridization and detection were carried out according to the instructions of the manufacturer (Boehringer). Briefly, the blotted membrane was hybridized with the DIG-labelled probe at 68°C overnight in a hybridization oven (Hybaid Ltd., Middlesex, United Kingdom). The membrane was then washed, equilibrated, blocked, and incubated with a 1:10,000 solution of anti-DIG antibody–alkaline phosphates. Chemiluminescent substrate solution (1% CSPD [Boehringer], 100 mM Tris-HCl [pH 9.5], 50 mM MgCl2) was pipetted over the membrane, and the membrane was sealed in a plastic hybridization bag (after excess liquid was removed), incubated, and then exposed to X-ray films. The X-ray films were developed in a film processor (RPX-Omat model M6B; Kodak Diagnostic Imaging, New York, N.Y.). To reprobe a membrane, the previous probe was stripped off by incubation in 0.2 M NaOH solution.
Polymorphism analysis of codon 463 in katG.
PCR-based RFLP with primers K5 and K6 and endonucleotide restriction enzyme NciI (33) (Boehringer) was used in the polymorphism analysis of codon 463 in katG.
Primer oligonucleotide design and synthesis.
All primer oligonucleotides used in this study were designed with the software package OLIGO (version 5.0; National Bioscience Inc., Plymouth, Mass.), synthesized on an Applied Biosystems model 291 DNA synthesizer, and purified with OPC columns (Applied Biosystems). These primers are listed in Table 1 or previous publications. For primers INS1 and INS2, see reference 16; for IS1 and IS2, see reference 10; and for P3, P4, P5, P6, P7, and P8, see reference 11.
TABLE 1.
Some of the primers used in this study
| Primer | Sequence | Positions (reference) |
|---|---|---|
| INS3 | 5′-CGGAGACGGTGCGTAAGTG-3′ | nt 194–212 in IS6110 (1) |
| INS4 | 5′-TCGCGGTGGCCCTGATGAT-3′ | nt 400–418 in the complementary strand of IS6110 above |
| K5 | 5′-TCCCGTTGCGAGATACCTTGG-3′ | nt 3141–3161 in accession no. X68081 |
| K6 | 5′-CCGCCTTTGCTGCTTTCTCTA-3′ | nt 3633–3653 in the complementary strand of accession no. X68081 |
| Tf | 5′-TCAACCGCACCGACCGCTTGT-3′ | nt 3710383–3710403 in the genomic DNA sequence of M. tuberculosis H37Rv, Sanger Centre |
Sequence analysis with a computer.
Programs in the Genetics Computer Group package (version 8.1) used in this study were GAP (22), BESTFIT (30), and PUBLISH.
Nucleotide sequence accession numbers.
Fragments sequenced in this study have been deposited in the EMBL, GenBank, and DDBJ data banks under accession no. Y15749 and Y15805.
RESULTS AND DISCUSSION
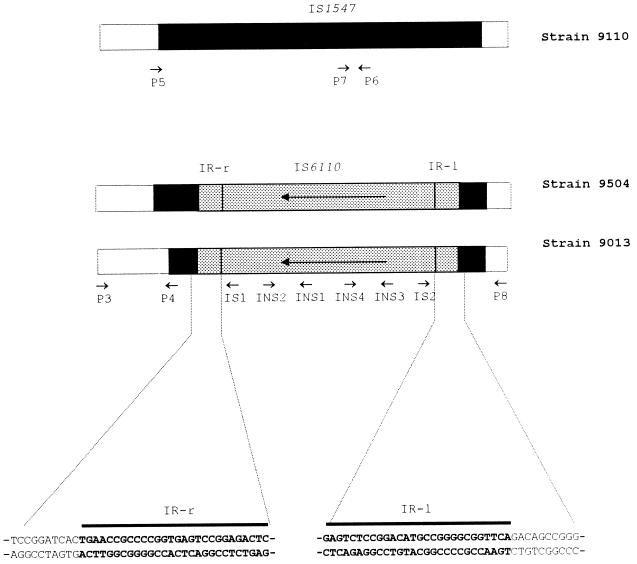
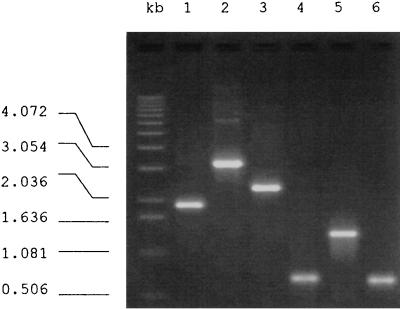
The ipl locus is a site of intensive insertion of IS6110, and this locus has been demonstrated to be part of IS1547. IS1547 is a member of the IS900 family of transposable elements and exists only in isolates of the M. tuberculosis complex, where it has two unique insertion sites on the chromosome (9, 11). One of these IS1547 copies is in the promoter region of hr-gr-lpdh (11; EMBL accession no. Y13470). The activity of IS6110 at this locus was investigated by long PCR with primers P3 and P8, which are located in the flanking DNA sequences of this IS1547 copy (see Fig. 2; Table 1). From the DNA sequence of this locus (EMBL accession no. Y13470) a PCR fragment of 545 bp would be predicted if this locus was free of IS1547 (i.e., hr-gr-lpdh), a PCR fragment of 1,895 bp would be predicted if IS1547 was present (hr-gr-lpdh::IS1547), and a PCR fragment of 3,250 bp would be predicted if IS6110 was also present (hr-gr-lpdh::IS1547::IS6110) (see Fig. 4). Unexpectedly, a PCR fragment of about 2.2 kb was obtained from M. tuberculosis clinical isolate 9013 (Fig. 1, lane 3), which is intermediate in size between those predicted from hr-gr-lpdh::IS1547 (Fig. 1, lane 1) and hr-gr-lpdh::IS1547::IS6110 (Fig. 1, lane 2).
FIG. 2.
Schematic illustration of locus hr-gr-lpdh::IS1547 of strains 9110, 9013, and 9504. The DNA sequences of the inverted repeats of the IS6110 copy in strain 9013 and their adjacent DNA sequences are detailed. The positions of the primers used in this study are illustrated with arrows.
FIG. 4.
Dendrogram of the 19 isolates and their IS6110 RFLP profiles with a scale of Dice coefficients. Two distinct subclusters (A and B) appear in the dendrogram.
FIG. 1.
PCR products from the hr-gr-lpdh::IS1547 loci in different isolates. Lanes 1 to 3, products of long PCR with primers P3 and P8 from isolate 9110 (hr-gr-lpdh::IS1547), isolate 41909 (hr-gr-lpdh::IS1547::IS6110), and isolate 9013, respectively; lanes 4 to 6, products of conventional PCR with the long PCR products from isolate 9013 as templates with primer pairs INS2 and INS3 (lane 4), P3 and INS1 (lane 5), and INS4 and P8 (lane 6).
The unusual nature of this locus in this isolate was confirmed by normal PCR with primers around this locus and within IS1547 and IS6110. First, the intactness of IS1547 in the fragment was determined by PCR with the IS1547 internal primers P4, P5, P6, and P7 in combination with the flanking sequence primers P3 and P8, i.e., P3 with P4, P5 with P6, and P7 with P8. Unexpectedly, none of these combinations yielded a product suggesting an incomplete IS1547 copy in the fragment. Second, as all previously known IS6110 copies in IS1547 are oriented with the IR-r towards the 3′ end of IS1547 (9), IS6110-internal primers INS1 and INS2 were combined with primers P3 and P8 (P3 with INS2, INS1 with P8) so that they would detect IS6110 inserted in this orientation. No PCR products, however, appeared. Neither of these experiments explained the unusual length of this fragment and left open the possibility that IS6110 was inserted in the opposite orientation. To test this possibility, the above-named primers were used in the opposite combination (P3 with INS1, INS2 with P8), and on this occasion products were obtained (Fig. 1). This IS6110 copy is then in the orientation opposite to that of the six IS6110 insertions at this locus observed previously in this laboratory (9). This clarification of the structure allowed the informed sequencing of the 2.2-kb fragment with primers IS1, IS2, INS3, and INS4 (Fig. 2; Table 1).
These sequencing results disclosed that the fragment was 2,174 bp in length and had several interesting features (Fig. 2). (i) It contained an IS6110 copy orientated with its IR-l towards the primer P8 side (Fig. 2). (ii) The IS6110 copy was intact and was 1,355 bp long (10, 11). (iii) The IR-r of the IS6110 copy was connected to nucleotide (nt) 103 of the IS1547 copy, and the IR-l of the IS6110 copy was connected to nt 1171 of the IS1547 copy (numbering from the beginning of IS1547, i.e., nt 1725 in EMBL accession no. Y13470), indicating the loss of a segment of 1,067 nt within the IS1547 copy. (iv) Most intriguingly, no target direct repeat sequences were found flanking this IS6110 copy (Fig. 2), a phenomenon not observed before at this locus (see below).
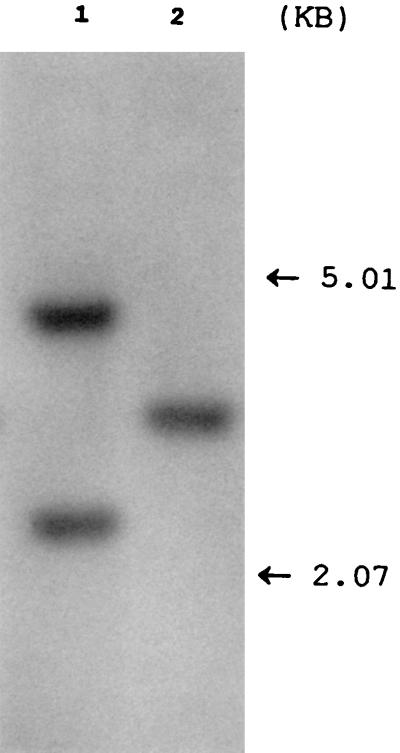
To test whether the loss of 1,067 bp was due to a local deletion or to other global genetic rearrangements, chromosomal DNA of isolate 9013 was subjected to a Southern blotting analysis. The probe was located in the DNA segment of IS1547 that was deleted in the copy of IS1547 in isolate 9013. The result showed that, unlike the majority of isolates of M. tuberculosis examined, which had two fragments which hybridized to the probe (see below), only one appeared in isolate 9013 (Fig. 3), suggesting that the loss of the DNA fragment in this isolate was due to local deletion rather than other global genetic rearrangements.
FIG. 3.
Autoradiograph of Southern blot of PvuII-digested chromosomal DNA of M. tuberculosis 41909 with intact IS1547 copies (lane 1) and isolate 9013 (lane 2) probed with the DIG-labelled IS1547 probe. In comparison with isolate 41909, isolate 9013 has lost one hybridized band due to the deletion of the DNA segment of IS1547 where the probe would hybridize.
To characterize the extent of this polymorphism in the population, this locus was intensively investigated with isolates closely related to isolate 9013. IS6110 RFLP-based dendrograms have been found to be quite efficient in grouping genetically closely related isolates of M. tuberculosis (10). We selected 19 isolates which formed a cluster in an RFLP-based dendrogram of about 700 isolates, and this cluster comprised two subclusters (A and B) which contained 11 and 8 isolates, respectively, with isolate 9013 being in subcluster A (Fig. 4). Long PCR and DNA sequencing of these isolates disclosed the following. (i) The isolates in subcluster A had the same locus structure as that in isolate 9013. (ii) Intriguingly, the isolates in subcluster B showed a PCR fragment of 2,348 bp with primers P3 and P8, which is longer than the fragment from isolate 9013. Sequencing of the PCR fragment from isolate 9504, which was representative of isolates in subcluster B, indicated that this isolate had nearly the same structure as that of isolate 9013; that is, there was an IS6110 insertion with the same orientation, with no target direct repeats flanking the IS6110 copy and the same flanking DNA sequence adjacent to the IR-l of the IS6110 copy. However, the IR-r flanking DNA sequence was different from that in isolate 9013, i.e., 911 bp was deleted from IS1547 (Fig. 2).
To clarify the evolutionary history of these two alleles, the relationship between isolates 9013 and 9504 was assessed with other markers. First, as presented above, these two isolates were quite closely linked in the IS6110 RFLP dendrogram, with about 70% Dice coefficient of similarity. They had 11 and 13 PvuII-digested fragments containing IS6110, respectively, with 9 of these being in common (Fig. 4). Second, both isolates were resistant to isoniazid but sensitive to rifampin, ethambutol, and pyrazinamide. Third, the examination of the useful evolutionary genetic polymorphism at codon 463 in the katG gene, either CTG (Leu) or CGG (Arg) (33), indicated that both of the isolates had the former allele. All these analyses indicated that these two isolates are closely related to each other and have a recent common ancestor.
Several mechanisms may explain the formation of the observed structures. First, site-specific recombination can generate DNA sequence deletions. One distinguishing feature of site-specific recombination is the involvement in specific DNA sites, as its name suggests (3, 19). If the structures observed in this study were due to site-specific recombination, this would imply that the IS1547 is a locus for site-specific recombination in the genome of M. tuberculosis. Clearly, this is unlikely, because the structures like those in strains 9310 and 9504 are the only examples found so far among more than 100 clinical isolates (9).
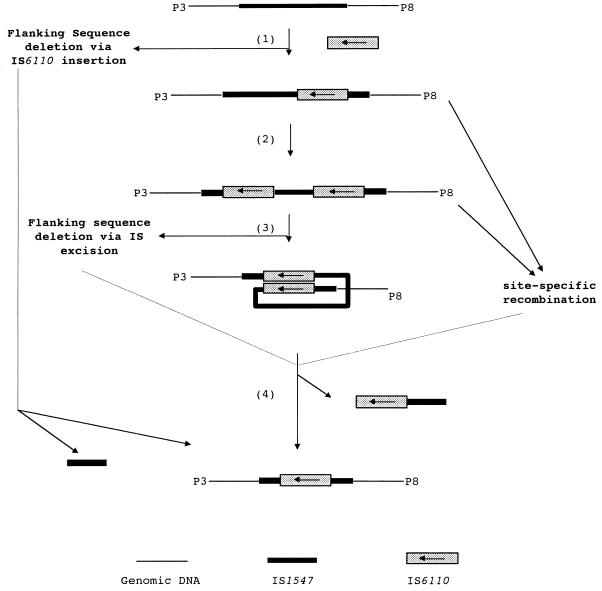
Second, IS6110 transposition may generate deletions of flanking DNA sequences on either integration into or transposition from a site. Generation of a target DNA deletion on the integration of transposable elements into a target site has been observed in bacteriophage Mu and Tn3 (24, 28). If this were the case after IS6110 transposition, then the different deletions seen in the two isolates would have been derived from separate insertion events. However, as isolates 9013 and 9504 are closely related and the IS6110 elements at the sites of the deletions have the same 3′-end flanking sequence, it is less likely that the two IS6110 insertions are separate insertion events which inserted in the same positions in the two strains and, subsequently, caused the different deletions. IS excision can also generate deletions, which may or may not include the loss of the IS copy, depending on the transposition mechanisms involved (26, 35). If so, then this generation of deletions must have involved a second IS6110 copy, since the isolates studied here still harbored an IS6110 copy at the site of the deletion, and the second IS6110 copies would have been located at the 5′ end of the sequence deleted in each of the strains (step 2 of Fig. 5). On excision of an IS6110 copy, the intervening sequence was also lost. However, it seems inconceivable that the endpoint of this deletion was right at the end of the other IS6110 copy since no target direct repeats were observed flanking the remaining IS6110 copy. The majority of IS6110 copies investigated so far either in the ipl locus or at other loci are delimited by direct repeats of 3 or 4 bp of DNA sequence (9, 16, 17). Of the transpositionally based deletion mechanisms, this leaves duplicative transposition at the donor site, in which flanking sequence adjacent to the donor IS copy was deleted while the donor IS remained in the place (35). If this were the case, an ancestor of both isolates would have harbored a single IS6110 copy located at the 3′ end of the deleted sequence and from this IS6110 deletions would have extended from the IR-r end of the element. Although possible here, this mechanism is less likely than the homologous-recombination mechanism proposed next. Above all, the frequency of such transpositionally mediated deletions is relatively low.
FIG. 5.
Models of the formation of the structures observed in isolates 9013 and 9504. Among the three possible mechanisms (IS6110 transposition, site-specific recombination, and homologous recombination) that may be responsible for the generation of the structures, homologous recombination is the most likely one and is detailed.
Third, homologous recombination may result in the genetic structures observed in these isolates. Any type of repeated sequence can provide regions of homology between which recombination can cause deletions (25, 27, 38), and bacterial IS elements are particularly well-documented examples (4, 8, 15, 23, 34). Homologous recombination by this mechanism in our isolates would be between two IS6110 elements, resulting in the deletion of the DNA segment between the two IS6110 elements and one of the IS6110 elements. The scenario would be as follows. An IS6110 insertion occurs in the IS1547 copy with its IR-l towards the flanking sequence, as the IS6110 IR-l did in isolates 9013 and 9504 (Fig. 5, step 1). After that, a second IS6110 copy inserts into the ipl locus in the same orientation as that of the first copy (Fig. 5, step 2); recombination between these two elements (Fig. 5, step 3), probably via a RecA-dependent system (see below), results in the deletions (Fig. 5, step 4). As observed in this study, as the insertions of the second IS6110 copies in isolates 9013 and 9504 were in different positions in the ipl, the consequential deletions were also different and, as a result, generated different structures for the loci.
This homologous-recombination-based mechanism is proposed as the most likely mechanism, and it is heavily based on the assumption that, sometime in evolutionary history, the recent common ancestor of these two isolates had two IS6110 insertions in the locus. In addition, the insertions should have had the same orientation, which is liable to generate deletions. A search for this expected ancestral isolate was carried out with our collection. Although we did not find this isolate, which is genetically closely related to isolates 9013 and 9504, we did find the same structures in strains B104 and B556 of the “Beijing Family,” which are genetically closely related to each other (37). Strain B104 carried an IS6110 in the ipl locus with the same orientation as that of the IS6110 copies in isolates 9013 and 9504, but strain B556 not only harbored this IS6110 copy but also carried a second IS6110 copy just downstream of the first IS6110 copy and with the same orientation. This result provided solid evidence for the mechanism proposed above.
RecA protein plays an important role in homologous recombination in promoting homologous pairing and strand exchange, especially where longer DNA sequences, typically greater than 1 kb, are involved (13, 27). The recA gene of M. tuberculosis has been cloned and sequenced, and its expression has been studied (6, 21). While this project was being conducted, the contiguous DNA sequence of the complete genome of M. tuberculosis H37Rv became available from the Sanger Centre (27a). Inspection of the IS1547 loci in the DNA sequence revealed that one of the two IS1547 copies, that ca. 3.7 Mb from oriC, harbored an IS6110 copy (ipl-1::IS6110) with a deletion to the left side of the IS6110 copy. It seems highly probable that this deletion was also mediated by homologous recombination between the IS6110 copy in IS1547 and a second copy at the end of the deleted flanking sequence 4.2 kb away. With the 1.3 kb of IS6110, this would make a total deletion of about 5.5 kb of DNA at this locus (Fig. 6).
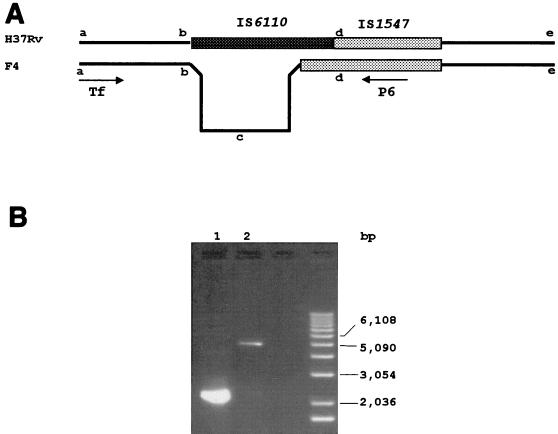
FIG. 6.
(A) Schematic illustration of strain H37Rv of M. tuberculosis in comparison with clinical isolate F4 of M. tuberculosis, showing strain H37Rv’s deletion. (B) PCR fragments from these two strains (lane 1, H37Rv, and lane 2, F4) with primers Tf and P6. The fragment in isolate F4 (i.e., segment of b-c-d) was confirmed to contain no IS6110 element by DNA sequencing (data not shown).
Studies of the genetics and pathogenesis of M. tuberculosis and development of a vaccine against tuberculosis largely rely on mutagenesis systems. Allelic exchange, a protocol to develop mutagenesis systems in which a foreign DNA fragment containing designed functions is introduced into the genome of a cell via homologous recombination, has been used with Escherichia coli and some mycobacterium species (7, 14). This protocol could be especially useful with M. tuberculosis owing to its characteristics of slow growth. Unfortunately, allelic exchange in M. tuberculosis was successful by using a long DNA fragment (>40 kb) as the recombination substrate but unsuccessful by using a short DNA fragment (<4 kb). One of the explanations for this is that short DNA sequences cannot induce a recombination system because of infrequent positioning (1, 2, 18). However, our findings suggest that a homologous DNA sequence of 1.3 kb (the length of IS6110) can promote a recombination system in M. tuberculosis.
The great abundance and varied distribution of IS6110 elements in the genome of M. tuberculosis in comparison to the restricted nucleotide polymorphism in the structural genes of M. tuberculosis has led to the suggestion that IS6110 transposition is a major force in generating chromosomal diversity in M. tuberculosis (29, 33). This study for the first time demonstrates IS6110-mediated deletions. The occurrence of these deletions in a transposable element (IS1547) does not imply that IS6110-mediated rearrangements cannot take place anywhere on the genome rather than in IS1547, because IS-mediated recombination does not require any IS-coded functions (34). It is thus reasonable to assume that IS6110-mediated homologous recombination duplications and/or IS6110-mediated homologous recombination inversions may also be possible in the genomes of M. tuberculosis. The genome of M. tuberculosis can contain up to 25 copies of IS6110 (34), 7 copies of IS1081 (5), and 2 copies of IS1547 (11); these IS elements, no doubt, can provide an abundance of opportunity for IS-mediated rearrangements and may have a significant influence on the evolution of M. tuberculosis.
ACKNOWLEDGMENTS
We thank A. Rayner and G. Harris at the Scottish Mycobacteria Reference Laboratory for bacteriological assistance, P. Carter, and K. Reay for DNA sequencing and synthesis of the oligonucleotide primers. DNA sequence analysis benefited from SEQNET, the SERC facility (Daresbury, United Kingdom).
This study was financially supported by the Department of Health, the Scottish Office; Chest, Heart and Stroke Scotland; and a Milner Scholarship from the University of Aberdeen.
Footnotes
Present address: Department of Biomedical Sciences, University of Bradford, Bradford, West Yorkshire, BD7 1DP, United Kingdom.
REFERENCES
- 1.Aldovini A, Husson R N, Young R A. The uraA locus and homologous recombination in Mycobacterium bovis BCG. J Bacteriol. 1993;175:7282–7289. doi: 10.1128/jb.175.22.7282-7289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardi A, Bernardi F. Site-specific deletions in the recombinant plasmid pSC101 containing the redB-ori region of phage lambda. Gene. 1981;13:103–109. doi: 10.1016/0378-1119(81)90047-0. [DOI] [PubMed] [Google Scholar]
- 4.Caspers P, Dalrymple B, Iida S, Arber W. IS30, a new insertion sequence of Escherichia coli K12. Mol Gen Genet. 1984;196:68–73. doi: 10.1007/BF00334094. [DOI] [PubMed] [Google Scholar]
- 5.Collins D M, Stephens D M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991;67:11–15. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- 6.Davis E O, Sedgwick S G, Colston M J. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J Bacteriol. 1991;173:5653–5662. doi: 10.1128/jb.173.18.5653-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ennis D G, Amundsen S K, Smith G R. Genetic functions promoting homologous recombination in Escherichia coli: a study of inversions in phage lambda. Genetics. 1987;115:11–24. doi: 10.1093/genetics/115.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Z, Morrison N, Watt B, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Z, Doig C, Morrison N, Watt B, Forbes K J. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex, and its association with IS6110. J Bacteriol. 1999;181:•••–•••. doi: 10.1128/jb.181.3.1021-1024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fomukong N G, Tang T H, al-Maamary S, Ibrahim W A, Ramayah S, Yates M, Zainuddin Z F, Dale J W. Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tubercle Lung Dis. 1994;75:435–440. doi: 10.1016/0962-8479(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 13.Galitski T, Roth J R. Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics. 1997;146:751–767. doi: 10.1093/genetics/146.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilhot C, Otal I, Van Rompaey I, Martin C, Gicquel B. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haack K R, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermans P W, van Soolingen D, Bik E M, de Haas P E, Dale J W, van Embden J D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermans P W, van Soolingen D, Dale J W, Schuitema A R, McAdam R A, Catty D, van Embden J D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalpana G V, Bloom B R, Jacobs W R., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci USA. 1991;88:5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leach D R F. Genetic recombination. Oxford, United Kingdom: Blackwell Science Ltd.; 1996. [Google Scholar]
- 20.McAdam R A, Hermans P W, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D, Dale J W. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 21.Movahedzadeh F, Colston M J, Davis E O. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J Bacteriol. 1997;179:3509–3518. doi: 10.1128/jb.179.11.3509-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 23.Noel K D, Ames G F. Evidence for a common mechanism for the insertion of the Tn10 transposon and for the generation of Tn10-stimulated deletions. Mol Gen Genet. 1978;166:217–223. doi: 10.1007/BF00285924. [DOI] [PubMed] [Google Scholar]
- 24.Pato M L. Bacteriophage Mu. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: ASM Press; 1989. pp. 23–52. [Google Scholar]
- 25.Petes T D, Hill C W. Recombination between repeated genes in microorganisms. Annu Rev Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- 26.Reif H J, Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137:17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- 27.Roth J R. Rearrangements of the bacterial chromosome formation and applications. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2256–2276. [Google Scholar]
- 27a.Sanger Centre Website. [Online.] Hinxton, Cambridge, United Kingdom: Sanger Centre; 1997, copyright date. http://www.sanger.ac.uk . [December 1997, last date accessed.] [Google Scholar]
- 28.Sherratt D. Tn3 and related transposable elements: site-specific recombination and transposition. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: ASM Press; 1989. pp. 163–184. [Google Scholar]
- 29.Small P M, van Embden J D. Molecular epidemiology of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: ASM Press; 1994. pp. 569–582. [Google Scholar]
- 30.Smith D, Waterman R. Adv. Appl Math. 1980;2:482–489. [Google Scholar]
- 31.Snider D E., Jr . Global burden of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: ASM Press; 1994. pp. 3–59. [Google Scholar]
- 32.Spielmann-Ryser J, Moser M, Kast P, Weber H. Factors determining the frequency of plasmid cointegrate formation mediated by insertion sequence IS3 from Escherichia coli. Mol Gen Genet. 1991;226:441–448. doi: 10.1007/BF00260657. [DOI] [PubMed] [Google Scholar]
- 33.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timmons M S, Lieb M, Deonier R C. Recombination between IS5 elements: requirement for homology and recombination functions. Genetics. 1986;113:797–810. doi: 10.1093/genetics/113.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turlan C, Chandler M. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 1995;14:5410–5421. doi: 10.1002/j.1460-2075.1995.tb00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Soolingen D, Qian L, de Haas P E, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Widom R L, Jarvis E D, LaFauci G, Rudner R. Instability of rRNA operons in Bacillus subtilis. J Bacteriol. 1988;170:605–610. doi: 10.1128/jb.170.2.605-610.1988. . (Erratum, 170:2003.) [DOI] [PMC free article] [PubMed] [Google Scholar]