Abstract
Purpose
Our objective was to develop a tool promoting early detection of COVID-19 cases by focusing epidemiological investigations and PCR examinations during a period of limited testing capabilities.
Methods
We developed an algorithm for analyzing medical records recorded by healthcare providers in the Israeli Defense Forces. The algorithm utilized textual analysis to detect patients presenting with suspicious symptoms and was tested among 92 randomly selected units. Detection of a potential cluster of patients in a unit prompted a focused epidemiological investigation aided by data provided by the algorithm.
Results
During a month of follow up, the algorithm has flagged 17 of the units for investigation. The subsequent epidemiological investigations led to the testing of 78 persons and the detection of eight cases in four clusters that were previously gone unnoticed. The resulting positive test rate of 10.25% was five time higher than the IDF average at the time of the study. No cases of COVID-19 in the examined units were missed by the algorithm.
Conclusions
This study depicts the successful development and large scale deployment of a textual analysis based algorithm for early detection of COVID-19 cases, demonstrating the potential of natural language processing of medical text as a tool for promoting public health.
Keywords: COVID-19, Outbreak detection, Natural Language Processing
1. Introduction
The COVID-19 pandemic poses a significant challenge to healthcare systems worldwide potentially overwhelming local resources [1]. As a result, many countries around the world implemented measures to prevent its spread through lockdowns and contact tracing [2,3].
The first cases of COVID −19 infections were detected in Israel during late February 2020 resulting in the first wave of the pandemic during March and April [4]. Early and aggressive measures were utilized by the Israeli authorities to prevent the disease spread and were effective in stopping the first wave of infections by early May [5]. These efforts were hindered but by the lack of sufficient testing kits, leaving many potentially infected person untested. Thus, early detection of new outbreaks, an immensely important part in controlling the disease spread, was delayed [6].
The Israeli Defense Forces (IDF) provide health care services for its enlisted personnel, practically serving as a health maintenance organization (HMO). During the first wave of the COVID-19 outbreak in Israel a team of physicians and data scientists were assembled to tackle challenges posed by the new epidemic among the enlisted population.
This article depicts the development and testing of a computational tool for analyzing electronic medical records (EMR) that employs Natural Language Processing tools(NLP) to examine free text written by physicians to detect potential COVID-19 cases and guide focused epidemiological investigations and testing.
2. Methods
2.1. Study population
The study population included all military personnel that were on active duty throughout the months of March and April of 2020 in 92 units of an approximate size of a company (80–150 persons).
2.2. Healthcare providers
IDF personnel are treated exclusively by the medical corps’ staff consisting of physicians, nurses, and medics. Additionally, during the COVID-19 pandemic a designated call center was built to address issues regarding potential contact with infected individuals or for reporting new onset of symptoms. All healthcare staff, including the call center personnel, are required to record any visit or call in the EMR. The designed system had access to records by all healthcare providers.
2.3. Data collection
Data was extracted from all visits conducted by physicians, nurses, medics and emergency call center personnel that were recorded in the IDF Electronic Medical Records(EMRs) starting on February 1st. The records contain three text fields – patient-recounted medical history, physical examination and treatment plan. Due to significant variation in the way healthcare workers filled these fields, they were combined into a single body of text for analysis. Additionally, fever measurements that were recorded during visits were extracted from pre-specified data fields of the EMR.
2.4. Symptom detection
The constructed algorithm made use of textual analysis to scan the EMR text for suspected COVID-19 cases. The search focused on the most common symptoms presented by COVID-19 patients: fever and cough [7].
The text analysis was done in a rule based manner. First, the text was examined for verbs and nouns depicting each symptom, for example “cough” and “coughed”. The three nearby words to the symptom declaration were examined for the presence of commonly used negation terms that would rule it out as a positive occurrence. This is similar to looking for “no fever” or “without fever” in the English language. This approach, while simple, proved highly effective due to the tendency of IDF medical personnel to repeat the wording and sentence formulation when reporting symptoms. Additional corrections were made for cases in which several symptoms were recorded as negative together (i.e. “the patient presents without fever or cough”).
Fever was not always recorded in the intended field and additional fever measurements were commonly described in the text. To extract these measurement, numerical values between 34 and 43 were searched in a window of five words before and after the term “fever”. The numerical range is highly specific for body temperature fitting no other vital signs values that have higher values, while most common blood test results have lower values. Additional corrections were made to avoid common instructions that did not represent actual fever measurement, for example, “return to the clinic if your fever rises above 38″. Our algorithm used the sub-febrile cutoff of 37.5 °C to flag the visit as containing a symptom of fever.
2.5. Definition of suspected infection cluster
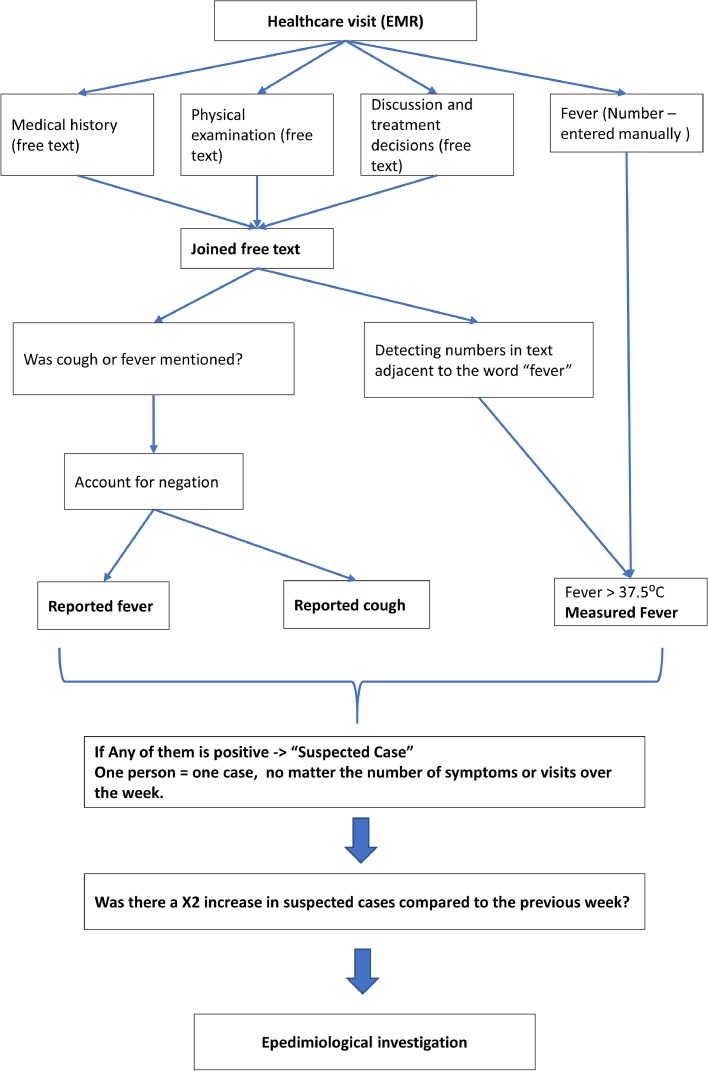
A person suspected for having COVID-19 or a “potential case” was defined as a person suffering from cough or fever and these symptoms were recorded by a healthcare professional in the EMR. Each person was counted once no matter how many symptoms or how many visits they had during the measurement time-window of a week. The results were compared to the previous week, with a suspected cluster defined as a twofold increase in unique suspected cases among the unit's registered personnel. The system's workflow is presented in Fig. 1 .
Fig. 1.
The algorithm workflow.
2.6. Algorithm evaluation
After internal validation the algorithm was evaluated in two stages. First, the algorithm was examined in its ability to detect cough and fever symptoms in EMR records. The performance of the algorithm was compared to a manual survey conducted by physicians of the medical corps of the IDF independently to our study.
Second, the algorithm was examined in its ability to assist in discovering new COVID-19 outbreaks. This was accomplished prospectively by monitoring 92 randomly selected units over a period of one month, and alerting the epidemiological branch of the IDF medical corps when the algorithm detected a possible outbreak. It is important to note that the epidemiological effort conducted as a result of the alert, while guided by the results of the algorithm, was independent of our study and no author of this study participated in the investigation.
2.6.1. Ethics statement
The article was approved by the IDF medical corps IRB
3. Results
3.1. Symptom detection
Manual monitoring by physicians was conducted regarding nine units during a period of one month and detected 32 persons who reported cough and 27 who reported fever. Our algorithm was able to detect all cases reported manually as well as three more cases of cough and four more cases of fever. To confirm the additional detected symptomatic cases, we contacted the physicians who conducted the original manual survey and ask them to re-examine the cases in question. The repeated evaluation found that all the additional symptom detected by our algorithm were correctly labeled, and the human error was attributed to the hastiness of the original survey.
3.2. Detection of new COVID-19 outbreaks
The algorithm was next tested for its ability to assist in discovering new Covid-19 cases. To achieve this goal, 92 randomly selected units were followed for a period of one month.
During the follow-up period, a twofold or more increase in suspected cases were detected in 17 of the units examined. The results were subsequently reported to the epidemiology branch of the IDF medical corps. The following epidemiological investigations used the data provided by the algorithm but were not limited to it. The investigation identified and quarantined 78 suspected patients who underwent polymerase chain reaction(PCR) testing for COVID-19. The test results diagnosed eight COVID-19 patients (10.25% of tests) in four clusters. The specificity for cluster detection was 81.5% and the positive predictive value (PPV) was 23.5%. Thus resulting in false positive rate of 76.5% for disease clusters.
We compare the testing results achieved using our algorithm to testing conducted using the standard survey of the IDF medical corps for COVID-19 cases during this period. This standard survey was conducted based on symptoms and exposure history to known sick patients and each PCR test was approved by a public health officer. The symptoms used for this survey included cough, shortness of breath, sore throat, fever > 38 °C and loss of taste and smell [8].
Our method achieved a positive test rate of 10.25% compared to 2.25% using the standard IDF survey (P value < 0.001), the comparison is presented in Table 1 . It should be noted that asymptomatic and mildly symptomatic persons were not tested for COVID-19 during this period due to shortage of PCR tests, thus, the sensitivity of both methods cannot be fully assessed and compared.
Table 1.
Testing results using the study algorithm compared to the standard IDF COVID-19 survey method.
| IDF standard survey | The study algorithm | |
|---|---|---|
| Tested | 5231 | 78 |
| Positive tests | 118 | 8 |
| Positive test rate | 2.25% | 10.25% |
During the study period the persons included in the study continued to receive the same medical treatment as any other soldiers in IDF and could undergo COVID-19 testing as part of the standard survey. However, all persons who were tested positive in the units included in the study were detected using our algorithm.
4. Discussion
In this study, we described the construction and evaluation of a free-text analyzing algorithm that through examination of electronic medical records on a large scale was instrumental in detecting new COVID-19 outbreaks. The algorithm assisted testing resulted in a five-time higher positive test rate during a period of scarce testing capabilities.
Covid-19 disease posed a significant challenge for health care systems around the globe. With no effective treatment or vaccine available during the initial phases of the pandemic, the emphasis was put on containment. The tools available for governments were crude, including large scale lockdowns and cumbersome contact tracing systems [9]. Another obstacle faced during the first wave of the pandemic, was the shortage of sufficient testing capabilities [10]. To help mitigate these challenges our group was tasked with devising a monitoring system for potential COVID-19 outbreaks through the use of electronic medical records.
The IDF has been a pioneer in adopting electronic health records in Israel, resulting in today's highly digitalized healthcare system of the medical corps that enabled this study. EMRs provide an indispensable resource for the treating physician and have been shown to improve clinical outcomes [11] and provide an efficient platform for research [12]. This study adds to the growing use of EMRs for surveillance and monitoring with the goal of improving public health [13,14].
Manual collection of data and monitoring the spread of a disease tends to be a slow and mistake ridden process [15], giving rise to various automation attempts. Previous notable examples include the use of EMR surveillance of bronchiolitis in the emergency department as an early warning for increased winter cases [16] and detection of local clusters of Legionnaires' disease [17]. Automated monitoring proved crucial in the face of a fast spreading COVID-19 infection [18,19].
Free medical text, while harder to analyze compared to structured data, holds a promise of vast information as patients’ symptoms and sings on physical examination. Previous attempts to predict and monitor infectious disease outbreaks using free text analysis used mainly social media or search data with the notable example of Google Flu system [20,21]. These approaches were later deemed unreliable due to various artifacts and distortions that are inherent when dealing with free-text generated by the general public [22].
Medical text written by trained health care professional is likely to be better structured and less biased, mitigating the set-backs that plagued previously mentioned systems [22]. During the COVID-19 pandemic natural language processing and medical text mining have been used to improve disease registration of patients with undocumented positive PCR results [23] and provided a comprehensive research tool regarding the disease symptoms and progression using the COVID-19 SignSym tool [24]. Additionally, previous work has shown that COVID-19 like symptoms, detected from EMR free text correlate well with the trend in positive PCR results [25].
Our system advanced previous attempts by deploying a large-scale monitoring system encompassing a large population that was widespread geographically. The system is based on extracting COVID-19 like symptoms from free medical text emphasizing simplicity to allow early deployment. It also took advantage of the military social dynamic where intra-unit is much more common than inter-unit contact, therefore clusters of infections were well defined within units. Another advantage of our system was its design as a decision support tool, both focusing and supplying information for the subsequent epidemiological investigation. While human judgment was still part of the process the augmentation and support of our system culminated in a significant increase in positive test rate compared to testing according to human judgment alone that was conducted beforehand.
Similar civilian circumstances could arise in a tightly connected local communities and education systems where clusters of COVID-19 infections could be detected and prompt action can be taken to prevent further spread [26]. Similarly, tools for symptom monitoring can be used for the detection of local environmental exposures (e.g., water or air pollution) that might go unnoticed until a critical mass of cases amount [27].
This study has several limitations. First, the study was conducted during the first wave of the pandemic in Israel when the rate of COVID-19 cases was low. However, despite this limitation the study showed an acceptable PPV even in such low prevalence situation. Second, the algorithm was designed to detect clusters of patients presenting symptoms that can be caused by COVID-19 but also by other respiratory infections. To mitigate the personal cost of a false positive results an additional stage of manual investigation determined which patients should be tested or quarantined. Third, the sensitivity of the algorithm cannot be fully assessed as a general screening was not performed and mildly symptomatic or a-symptomatic cases could have been missed.
In summary, this study demonstrated the use of a natural language processing tool for early detection of possible COVID-19 patient clusters by analyzing medical free text stored in EMRs. Thus, provided an automatic, online monitoring system that successfully guided testing and epidemiological investigation for early detection of infection cases.
Funding sources
This research did not receive any funding from agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Miller I.F., Becker A.D., Grenfell B.T., et al. Disease and healthcare burden of COVID-19 in the United States. Nat. Med. 2020;26:1212–1217. doi: 10.1038/s41591-020-0952-y. [DOI] [PubMed] [Google Scholar]
- 2.Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020;395:1225–1228. doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie J., Tong Z., Guan X., et al. Critical care crisis and some recommendations during the COVID-19 epidemic in China. Intensive Care Med. 2020:6–9. doi: 10.1007/s00134-020-05979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leshem E., Afek A., Kreiss Y. Buying time with COVID-19 outbreak response, Israel. Emerg. Infect. Dis. 2020;26:2251. doi: 10.3201/EID2609.201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossman H., Keshet A., Shilo S., et al. A framework for identifying regional outbreak and spread of COVID-19 from one-minute population-wide surveys. Nat. Med. 2020 doi: 10.1038/s41591-020-0857-9. Published Online First: April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu H.Y., Englund J.A., Starita L.M., et al. Early detection of Covid-19 through a citywide pandemic surveillance platform. N. Engl. J. Med. 2020;383:185–187. doi: 10.1056/nejmc2008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang F., Deng L., Zhang L., et al. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19) J. Gen. Intern. Med. 2020;2019:1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitecki M., Taran B., Ketko I., et al. Self-reported symptoms in healthy young adults to predict potential coronavirus disease 2019. Clin. Microbiol. Infect. 2021;27:618–623. doi: 10.1016/J.CMI.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretzschmar M.E., Rozhnova G., Bootsma M.C.J., et al. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health. 2020;5:e452–e459. doi: 10.1016/S2468-2667(20)30157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yelin I., Aharony N., Tamar E.S., et al. Evaluation of COVID-19 RT-qPCR test in multi sample pools. Clin. Infect. Dis. 2020;71:2073–2078. doi: 10.1093/CID/CIAA531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menachemi N., Collum T.H. Benefits and drawbacks of electronic health record systems. Risk Manag. Healthc. Policy. 2011;4:47–55. doi: 10.2147/RMHP.S12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen P.B., Jensen L.J., Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 2012;13:395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 13.Birkhead G.S., Klompas M., Shah N.R. Uses of electronic health records for public health surveillance to advance public health. Annu. Rev. Public Health. 2015;36:345–359. doi: 10.1146/annurev-publhealth-031914-122747. [DOI] [PubMed] [Google Scholar]
- 14.Friedman D.J., Parrish R.G., Ross D.A. Electronic health records and US public health: current realities and future promise. Am. J. Public Health. 2013;103:1560–1567. doi: 10.2105/AJPH.2013.301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal S., Chowell G., Simonsen L., et al. Big data for infectious disease surveillance and modeling. J. Infect. Dis. 2016;214:S375–S379. doi: 10.1093/infdis/jiw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes H.E., Morbey R., Hughes T.C., et al. Emergency department syndromic surveillance providing early warning of seasonal respiratory activity in England. Epidemiol. Infect. 2016;144:1052–1064. doi: 10.1017/S0950268815002125. [DOI] [PubMed] [Google Scholar]
- 17.Wijngaard C.C.V.D., Asten L.V., Pelt W.V., et al. Syndromic surveillance for local outbreaks of lower-respiratory infections: would it work? PLoS One. 2010;5:e10406. doi: 10.1371/JOURNAL.PONE.0010406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C.J., Ng C.Y., Brook R.H. Response to COVID-19 in Taiwan: big data analytics, new technology, and proactive testing. JAMA J. Am. Med. Assoc. 2020;323:1341–1342. doi: 10.1001/jama.2020.3151. [DOI] [PubMed] [Google Scholar]
- 19.Ting D.S.W., Carin L., Dzau V., et al. Digital technology and COVID-19. Nat. Med. 2020;26:459–461. doi: 10.1038/s41591-020-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginsberg J., Mohebbi M.H., Patel R.S., et al. Detecting influenza epidemics using search engine query data. Natwork. 2009;457:1012–1014. doi: 10.1038/nature07634. 2009 4577232. [DOI] [PubMed] [Google Scholar]
- 21.Dugas A.F., Jalalpour M., Gel Y., et al. Influenza forecasting with Google flu trends. PLoS One. 2013;8:e56176. doi: 10.1371/journal.pone.0056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazer D., Kennedy R., King G., et al. The parable of Google flu: traps in big data analysis. Science. 2014;343:1203–1205. doi: 10.1126/science.1248506. (80-.) [DOI] [PubMed] [Google Scholar]
- 23.Chapman A.B., Peterson K.S., Turano A., et al. A natural language processing system for national COVID-19 surveillance in the US department of veterans affairs. 2020. https://www.va.gov/health/ (accessed 25 Sep 2021).
- 24.Wang J., Abu-el-Rub N., Gray J., et al. COVID-19 SignSym: a fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model. J. Am. Med. Inform. Assoc. 2021;28:1275–1283. doi: 10.1093/JAMIA/OCAB015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo J.T.H., Dinu V., Bernal W., et al. Real-time clinician text feeds from electronic health records. npj Digit Med. 2021;4:1–3. doi: 10.1038/s41746-021-00406-7. 2021 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aherfi S., Gautret P., Chaudet H., et al. Clusters of COVID-19 associated with Purim celebration in the Jewish community in Marseille, France, March 2020. Int. J. Infect. Dis. 2020;100:88–94. doi: 10.1016/j.ijid.2020.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoshino T., Hoshino A., Nishino J. Relationship between environment factors and the number of outpatient visits at a clinic for nonallergic rhinitis in Japan, extracted from electronic medical records. Eur. J. Med. Res. 2015;20:1–17. doi: 10.1186/S40001-015-0151-3. 2015 201. [DOI] [PMC free article] [PubMed] [Google Scholar]