Abstract
People living with HIV are more exposed to the adverse health effects of the worldwide COVID-19 pandemic. The pandemic's health and social repercussions may promote drug abuse and inadequate HIV management among this demographic. The coronavirus pandemic of 2019 (COVID-19) has caused unprecedented disruption worldwide in people's lives and health care. When the COVID-19 epidemic was identified, people with HIV faced significant obstacles and hurdles to achieving optimal care results. The viral spike protein (S-Protein) and the cognate host cell receptor angiotensin-converting enzyme 2 (ACE2) are both realistic and appropriate intervention targets. Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin have anti-HIV effects. Our computational biology study investigated that these compounds all had interaction binding scores related to S protein of coronavirus of −9.0 kcal /mol, −7.1 kcal /mol, −9.1 kcal /mol, and −10.3 kcal/mol/mol, respectively. A combination of plant-derived anti-HIV compounds like protease inhibitors and nucleoside analogs, which are commonly used to treat HIV infection, might be explored in clinical trials for the treatment of COVID-19.
Keywords: HIV, Covid-19, ACE2, S protein, Calanolides A, Holy Basil, Kuwanon-L, Patentiflorin A
Abbreviations: ACE2, Angiotensin-converting enzyme-2; HIV, Human Immunodeficiency Virus; AIDS, Acquired immunodeficiency syndrome; NETs, neutrophil extracellular traps; CD4, Cluster of Differentiation 4; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; AZT, Azidothymidine; NRTIs, nucleoside analog reverse transcriptase inhibitor; NNTRIs, Non-nucleoside analogs transcriptase reverse inhibitor; RT, Reverse Transcriptase; IN, Integrase; HAART, Highly active antiretroviral therapy, ART, Antiretroviral therapy
Background
Immunodeficiency is the source of hereditary malignancy, whereby the hosts cannot fight against the disease-causing pathogen. We are prone to getting infections quickly if our body's immune system doesn't generate enough white blood cells to combat external threats [25]. Immune function loss due to different agents’ exposure, such as HIV, is the product of an acquired immunodeficiency syndrome (AIDS) that has become pandemic in size and almost invariably leads to death in some populations. The two most common diseases, HIV and the coronavirus are currently fighting worldwide.
In the United States of America, AIDS was first documented in 1981 [7]. Transfusion of HIV-infected blood in homosexual and heterosexual sex is widely utilized. In many situations, the appearance of AIDS symptoms may take a long time after HIV infection. But with a coronavirus, it takes 2–14 days for someone to be infected with the virus, and Coronavirus transmission comprises droplets of the cough, sneezing, or breathing of the individual [24]. In Wuhan, China, in December 2019, a group of individuals suffering from pneumonia with unknown etiology was found. The pathogen is a novelty of the coronavirus, termed a severe acute respiratory syndrome, called the coronavirus 2 (SARS-CoV-2).
Over millennia, sexuality has evolved from a stigmatized topic to one openly discussed in many cultures worldwide. One of the world's most significant health and social concerns is AIDS, which has affected millions of people worldwide. All infections begin with someone who has already been exposed. New infections develop when infected individuals fail to take the appropriate precautions or when protective services are inadequately structured.
Human behavior has a direct impact on the spread of HIV. As a result, it is influenced by both biological and socio-cultural factors [32]. Illiterate and unaware people are in danger of engaging in high-risk behavior. Similarly, COVID-19 affects all sections of society and is most damaging to the most disadvantaged people. It continues to afflict various communities, such as those who live in poverty, the elderly and the disabled, and young and indigenous populations.
In both conditions, “othering” and maltreatment are frequent discrimination components among those with either COVID-19 or HIV. Confident people see these two disorders negatively due to their Sociodemographic features, microaggressions, discriminating attitudes, and social disconnection. Their stance on the two diseases, HIV and COVID-19, is that people, who know that they have this virus but do not make efforts to stop them from transmitting it, should commit a criminal offense.
Hypothesis
The apparent reduction of HIV-related risk for developing severe COVID-19 may be attributed to the protection offered by Antiretroviral therapy (ART). The presence of anti-HIV compounds derived from plants, such as protease inhibitors and nucleoside analogs, suppresses CoV-2 replication. An ART drug that shows a modest but significant effect on CoV-2 has now been compared to another bioactive anti-viral medication. Drug design is an integrated developing discipline that portends an era of ‘tailored drugs’. It entails investigating biologically active substances' effects based on molecular interactions in molecular structure or physicochemical characteristics.
The drug is usually a small organic molecule that activates or inhibits the action of a biomolecule such as protein, providing a therapeutic benefit to the patient. Drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and, therefore, bind to it [40].
A biomolecular target (usually a protein or a nucleic acid) is a key molecule engaged in a metabolic or signalling pathway linked to a specific disease state or pathology and the infectivity or survival of a microbial pathogen [41], [42].
Potential therapeutic targets do not have to be disease-causing, but they must be disease-modifying. Small compounds may be tailored to boost or inhibit the target function in a disease-modifying pathway in some situations [43].
In recent years, integrating computational models and predictions with experimental validation has enabled more effective investigations of the structure, function, and regulation of key therapeutic targets [37]. Computational biology and bioinformatics have the potential to transform not just the way drugs are developed but also to speed up the drug development process and reduce costs [38]. For example, molecular docking has been widely used in recent decades as a quick and inexpensive technology in academic and industry contexts [39].
Needless to mention that computational study minimizes biological waste and research period and is cost-effective. In this context, we have employed computational approaches to design natural compound-based therapeutic interventions against COVID-19.
Support of hypothesis, a brief overview of HIV and its protein
HIV is presently classified into two types: HIV-type 1 (HIV-1) and HIV-type 2 (HIV-2) (HIV-2). The retrovirus genome comprises two identical copies of single-stranded RNA molecules [11], and it is distinguished by the presence of structural genes gag, pol, and env. The genomes of HIV-1 and HIV-2 viruses differ in their organization; however, the essential structure (the existence of the three structural genes, gag, pol, and env) is the same for all retroviruses. In reality, the HIV-1 and HIV-2 genomes contain a complicated mix of additional regulatory/accessory genes in addition to these three. Virologically, HIV-1 and HIV-2 particles are the same. Samudrala and Jenwitheesuk predict HIV-1 protease inhibitor resistance on a molecular level [46]. Chaudry et al., 2020 use computational approaches to study the inhibitory impact of selected phytochemicals against HIV-2 protease [48].
Like other retroviruses, the gag gene encodes the core (p24, p7, and p6) and matrix (p17) structural proteins, whereas the env gene encodes the viral envelope glycoproteins gp120 and gp41, which recognize cell surface receptors. The pol gene encodes for viral replication enzymes such as reverse transcriptase, which transforms viral RNA into DNA, integrase, which inserts viral DNA into host chromosomal DNA (the provirus); and protease, which cleaves huge Gag and Pol protein precursors into their components. Integrase (IN), a critical enzyme in the human immunodeficiency virus type 1 (HIV-1) life cycle, helps the integration of viral DNA to host DNA, making it a potential target for anti-HIV medication development. Zhou et al. (2021) used machine learning to classify and design HIV-1 Integrase inhibitors [47]. HIV particles have a diameter of 100 nm and are surrounded by a lipoprotein-rich membrane. Each membrane of a viral particle contains glycoprotein heterodimers formed by the binding of trimers of the external surface glycoprotein gp120 to the transmembrane spanning glycoprotein gp41. Since the connection between gp120 and gp41 is not covalent, gp120 can be released spontaneously and be detected in HIV-infected persons' blood and lymphatic tissue. The virus may integrate different proteins into the infected cell membrane, such as HLA class I and II proteins or adhesion proteins like ICAM-1, which may increase adherence to additional target cells during budding. The interior of the viral lipoprotein membrane is attached by a matrix protein (p17). Capsids composed of polymers of the core antigen (p24) are contained in the membrane and matrix proteins of the virus. In addition to a nucleoprotein, HIV RNA carries the enzymes reverse transcriptase, integrase, and protease [13]. Another accessory/regulatory gene in HIV viruses is essential in controlling viral replication [9]. Specifically, the Tat gene produces a protein (Tat) made very early after infection. This protein stimulates HIV gene expression.For appropriate messenger and genomic RNA to arrive in the cytoplasm. The function of the other auxiliary HIV proteins is less well defined; the Vpr protein is thought to be involved in cell cycle arrest.
Vpx is also necessary for reverse transcription to enter the nucleus of non-dividing cells, including macrophages, what HIV-2 does through this protein. The Vpu gene encodes for a protein necessary for viral particle release, while the VIF gene encodes for an intriguing protein (Vif) that increases the infectiousness of progeny viruses. In addition to its functions in signal communication, the Nef protein downregulates the CD4 receptor on the cell surface, allowing the late stages of viral replication to allow budding. Genetically, HIV belongs to the Lentivirus genus within the Retroviridae family. Lentivirus infections often have chronic symptomology, with a protracted period of clinical latency, persistent viral replication, and central nervous system involvement. Both viruses can produce AIDS, albeit central nervous system dysfunction may be more common in HIV-2 infection [21]. Furthermore, HIV-2 appears to be less virulent than HIV-1, and infection progression to AIDS takes longer [33].
In the development of anti-HIV drugs, computational approaches are vital. Various computational approaches, such as virtual screening, QSAR, molecular docking, and homology modelling, have been used in the discovery of anti-HIV drugs in a significant number of studies [44], [45].
A brief overview of SARS-CoV-2 and its protein
Coronavirus particles resemble spheres and have a diameter of 80–120 nm. An envelope (E), membrane (M), nucleocapsid (N), and spike (S) protein are components of this virus family [22]. The S protein of coronavirus develops large protrusions from its surface to resemble a crown; for this reason, it's called Coronavirus [35]. A host cell releases its offspring virions via the S and HE proteins (transmembrane proteins). Virus releases can be achieved by coupling the M and E proteins to form homopentamers/dimers. The N protein facilitates a viral genome's replication, RNA packing, and immune system evasion. Numerous chemicals and processes modify this general strategy, such as glycosylation, palmitoylation, phosphorylation, etc. [31]. Coronaviruses rapidly mutate and spread from one person to another. It comprises a single strand of positive-sense RNA and measures around 27–32 kb. Many strains exist, but A2, A3, A3I, B1, B4, and A1A are the most significant. Coronavirus strains A2A and A3I make up the majority of infections in Indian patients. In addition, the A2A strain is found all over the world. The SARS-CoV-2 virus first propagates in the upper respiratory tract to spread to the lungs. A mutation and genetic diversity in the pathogen also cause the common cold, oral/nasopharyngeal diseases, and CNS inflammation [18]. Recently, Khare et al. (2020) proposed that natural compounds might be explored as target-specific treatment agents against COVID-19 infection using silico analysis [49].
The distinction between HIV and SARS-CoV-2
These two viruses are phylogenetically different. HIV infects T cells that are CD4+; an envelope of HIV protein attaches to the CD4 antigen and infects monocytes and other CD4 cells [15]. Coronavirus infects alveolar cells, intestinal epithelial cells, endothelium cells, kidney cells, monocytes, and neuroepithelial cells via binding to the ACE-2 spike protein.
Persons get diagnosed with AIDS when their number of CD4 cells falls below 200 cells per mm, and those diagnosed with Covid-19 have a heading of more than 1.4 au/ml [6]. AIDS may be exceedingly contagious and highly viral as a coronavirus. There are generally fewer odds of survival for persons with AIDS and COVID-19 without treatment.
Similarities between HIV and SARS-CoV-2
HIV and coronavirus have an RNA genome integrated as a DNA copy into the human genome. These two viruses raise cytokine levels; SARS-CoV-2 increases cytokine levels by a similar amount. As a result of HIV infection, the production of cytokines is a long-term process that causes chronic inflammation to occur. Acute cytokine production is associated with clinical symptoms of COVID-19.
HIV-infected people infected with COVID19: Symptoms and treatment
The risk posed by COVID-19 to people with HIV varies among different sources. For example, studies have hypothesized that patients with HIV may be at a reduced risk for COVID-19 complications since HIV antiretroviral drugs might inhibit coronaviruses, such as SARS-CoV-2. Conversely, others have had the opposite experience due to the immunosuppression of HIV patients [29].
Consequently, Benkovic et al. reported four cases of HIV infection with COVID-19 on Long Island, New York, USA. High CD4 T cell counts in HIV-infected individuals are due to their HIV medications. Out of four patients, only one required hospitalization for further complications related to influenza A. Several incidents suggest that HIV-infected patients can be treated for COVID-19 when the disease is uncomplicated [1]. COVID‐19 has also been observed in five patients with HIV in Barcelona, Spain, four of whom were on antiretroviral therapy when admitted; and Common symptoms included fever and cough. The four patients who responded well to treatment have been discharged, except for one who needed extracorporeal support [2].
As reported by Jiaxiang Chen et al., a 24 year old HIV-infected man with a history of living HIV infection was admitted to Guizhou Provincial People's Hospital with a 1‐day history of fever (37.8 °C) and dry cough. No abnormalities were found in the leukocyte count, lymphocyte count, C‐reactive protein, erythrocyte sedimentation rate, or flow cytometry. A CT scan of the middle and lower lobes of the right lung revealed multiple patches of shadows with unclear borders in the subpleural regions and the involvement of adjacent terlobar pleura [5]. Ultimately he was diagnosed with non-severe COVID-19 pneumonia after real-time reverse transcription-polymerase chain reaction (RT-PCR) tests were performed on oropharyngeal swabs. After being diagnosed with HIV 2 years ago, the patient received antiretroviral therapy (tenofovir 0.3 g, lamivudine 0.3 g, favirenz 0.6 g) to suppress his viral load. No prior history of drug use or blood transfusions was reported, and he did not disclose his sexual history. His treatment included lopinavir/ritonavir (300/75 mg, bid) combined with interferon inhalation (5 µg, bid) after a diagnosis of COVID-19. In the meantime, tenofovir, lamivudine, and efavirenz were given orally. The symptoms improved during treatment, and a chest CT on day 7 of hospitalization showed that pulmonary lesions decreased and were partially absorbed on day 12. CT imaging of the chest showed that the pulmonary lesions substantially resolved after 15 days of treatment, leaving only a few residual foci of fibrosis. Ultimately, the patient was discharged from the hospital and transferred to Guizhou Provincial Staff Hospital for observation after three RT-PCR tests on the pharyngeal swab specimens (interval greater than 24 h) were negative. Moreover, the nasopharyngeal swabs were submitted for testing to Guizhou Provincial Center for Disease Control and Guizhou Provincial People's Hospital Laboratory Department to detect SARS‐CoV‐2 RNA. PCR tests conducted on days 14 and 28 both produced negative results. COVID‐19 disease is characterized by fever, dry cough, and fatigue as its main clinical symptoms [34]. The immune system of patients with impaired immune function makes them more likely to develop severe acute respiratory distress syndrome and even lead to death [14]. Although this HIV-infected case presented mild symptoms, laboratory examinations detected no abnormalities. Moreover, the symptoms improved after a short treatment period, with a successful outcome. There may have been a connection between the quick absorption of lesions and ART before the infection with SARS-CoV-2. Tenofovir is effective as part of an ART treatment against the SARS-CoV-2 by binding the RNA polymerase [8].
Thus, people with HIV who receive ART for COVID-19 pneumonia may experience moderate symptoms and faster recovery than the general population. Consequently, HIV-infected patients who receive anti-HIV treatment and do not have any other complications from unrelated infections are likely to be cured of COVID‐19. Findings from the study have brought a bit of hope to HIV-infected patients who were apprehensive of complications and death if infected with the novel virus.
Research Hypothesis
The current trend in drug design is to develop new clinically effective agents that have structural modifications of target biomolecules. Lead is a prototype compound with the desired biological or pharmacological activity but may have many undesirable characteristics, like high toxicity, other biological activity, and insolubility or metabolism problems.
Such organic leads, once identified, are easy to exploit. This process is relatively straightforward. The real test lies in identifying these lead bioactive compounds on the basic skeleton. Conformational changes occur when a drug molecule interacts with the biomolecules. The activity of a drug can be correlated to its structure in terms of the contribution of its structure and functional group to the lipophilicity, and electronic and steric features of the drug skeleton.
Possible treatment/ the relevance of medicine
No adequate treatment for these two immunodeficiency diseases has been developed so far. The following are the several alternative techniques for treating AIDS: 1) vaccination for AIDS and 2) medicines for reverse symptoms of AIDS. The blocking of 1) reverse transcriptase, 2) viral protease can be developed for AIDS. The reverse transcriptase was the first effective medication targeted; zidovudine (AZT) [28], an inhibitor of nucleoside analog reverse transcriptase (NRTIs). Similarly, in AIDS treatments, that are a Nonnucleoside analog inhibitor of transcriptase reverse (NNRTI) and a protease inhibitor; thus, many anti-viral therapists are utilized [3]. These NRTIs and NNRTIs bind to HIV reverse transcriptase and inhibit its activity. To transform its RNA to DNA, HIV requires reverse transcriptase. HIV cannot replicate by inhibiting reverse transcriptase and reverse transcription. The current AIDS treatment consists of two nucleoside analogs and one protease inhibitor, the most aggressive antiretroviral therapy (HAART) [19]. HAART has been very successful in decreasing the viral blood load and lowering mortality rates. anti-viral treatment may reduce the length of the disease in specific individuals and reduce consequences because the COVID-19 causing coronavirus is new, with little evidence of special anti-viral. Some anti-viral medications used for treating Covid-19 include 1) Remdesivir, 2) Dexamethasone, and 3) Hydroxychloroquine, which reduces the viral load to blood below the level of detection. In some cases, such anti-viral drugs are successful; their long-term usage causes various adverse effects, and in treated patients, mutant viruses emerge.
Natural compounds can be utilized as an anti-viral medicine therapy to combat the side effects [20]. In drug development, the study of natural compounds is not a recent idea. Many of the best medicinal therapies were identified originally in nature. Viral suppression can be a natural product-based compound. Early in the history of HIV research, several beneficial compounds on plants were researched and are still studied today.
These new compounds can lead to an increase in suppressive function, such as antiretroviral treatment (ART) [17]. Calanolides A and B are plant-based antiretroviral drugs from Calophyllum, a tree from the families of mangosteens found in the tropical rainforest of Malaysia[16]; the herb tulsi, or holy basil, is traditionally used in Indian Ayurvedic medicine for treating a wide range of ailments while promoting overall health, Tulsi leaves contain esters and amides that have anti-RT activity in vitro [27], kuwanon-L, derived from Morus nigra black mulberry tree [10], [23], Patentiflorin A isolated from the Justica gendarussa plant of Vietnam [36]. All of these plant-based anti-HIV viral drugs have reverse transcriptase inhibitor activity, as shown in Table 1 . It suppresses HIV replication and can impede coronavirus activity by inhibiting reverse transcriptase and reverse transcription, as shown in Fig. 1 [4]. Further, we have investigated the binding energy of these drugs with S protein (Table 2 ) to study its impact on coronavirus. The binding affinities of S protein with various drugs are evaluated using the molecular docking software Auto Dock Tools 1.5.6 [30]. The canonical SMILES ids of multiple medications are obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). CHIMERA 1.11.2 program is used for the conversion of 2D to 3D structures of drugs [26]. To identify the binding site of S protein, various parameters such as binding affinity, receptor-interacting atom, receptor pocket atom, receptor-ligand interaction site, atomic contact energy (ACE), and side amino acid residues have been studied.
Table 1.
The source, structure, and anti-HIV activity of the bioactive compounds.
| Natural Product | Active Compounds | Structure | Anti-HIV Activity | Reference |
|---|---|---|---|---|
Calophyllumlanigerum
|
Calanolides A | 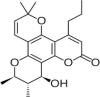 |
NNRT Inhibitor | (Kashman, Y. et al., 1992) |
Morusnigra (Black mulberry)
|
Kuwanon L | 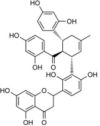 |
RT Inhibitor | (Esposito, F. et al., 2015; Martini, R. et al., 2017) |
Justicagendarussa
|
Patentiflorin A |  |
RT and IN Inhibitor | (Zhang, H. J. et al., 2017) |
Ocimumtenuiflorum
|
Holy Basil |  |
RT Inhibitor | (Sonar, V.P. et al., 2017) |
Nonnucleoside Reverse Transcriptase (NNRT); Reverse Transcriptase (RT); Integrase (IN).
Fig. 1.
HIV replication is inhibited by a natural substance. Compounds generated from natural products have been investigated to target different phases in HIV replication. For example, Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A inhibit HIV RT, preventing HIV genomic RNA from reverse-transcribing into proviral DNA. In addition to anti-RT action, Kuwanon-L has anti-IN activity, which hinders HIV proviral DNA integration into the host genome.
Table 2.
Calanolides A, Holy Basil, Kuwanon L and Patentiflorin A docking research for S-protein.
| Bioactive compounds | 2D Interaction | Binding Energy in Kcal/Mole |
|---|---|---|
| S Protein-Calanolide A |  |
−9.0 |
| S Protein- Holy Basil | 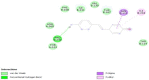 |
−7.1 |
| S Protein- Kuwanon L | 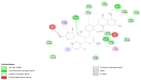 |
−9.1 |
| S Protein- Patentiflorin A | 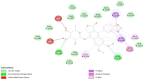 |
−10.3 |
Interaction of anti-HIV drugs (Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A) with the coronavirus S protein (which binds to human cell ACE2 and allows it to enter the cell) at the molecular level was investigated (Table 2). The binding modes of Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A with S Protein were investigated through Auto Dock Vina 1.1.2. The binding energy of S Protein with Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A scored to be − 9.0 kcal/mol, − 7.1 kcal/mol, − 9.1 kcal/mol, and − 10.3 kcal/mol, respectively (Table − 2). The binding energies of the compounds listed above indicate that they have a high binding affinity towards S-protein, with Patentiflorin-A having the highest binding affinity.
Results obtained from the anti-HIV active compounds bind preferably to sites of S Protein which are crucial in host cell binding [12]. Thus, the present computational studies suggest possible prevention of the coronavirus by using Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A, widely used anti-HIV active compounds. This inhibitory machinery of blocking the binding of host cell receptors to viruses and inhibiting the cellular entry of viral protein could be an effective therapeutic target, as evident from an array of computational studies. These compounds are already widely available on the market as antiretroviral drugs in nucleotide analogs. Compounds with these bioactive properties have been extensively used for HIV-infected patients, primarily due to the limited duration of COVID-19 treatment, and these compounds have different targets and may reduce SARS-CoV-2 viral load more rapidly. However, this needs to be experimentally validated before translational intervention.
Conclusions
Here we conclude that a plant-based anti-HIV viral medication used to treat AIDS may be less likely to contract SARS-CoV-2. However, Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A can be regarded as potential anti-HIV viral medicines against SARS CoV2, according to the results of this computational investigation. anti-HIV compounds bind to S Protein on the coronavirus surface, which binds to ACE2, allowing the coronavirus into human cells. When anti-HIV medicines interact with the S-protein, the coronavirus is unable to bind to the human cell receptor ACE2. They all have excellent interactions with coronavirus surface protein (S Protein). This inhibitory machinery of blocking the binding of host cell receptors to viruses and inhibiting the cellular entry of viral protein could be an effective therapeutic target, as evident from an array of computational studies. However, this needs to be experimentally validated before translational intervention. In the future, we may explore the bioactive anti-viral substance that can be found in unique natural product sources through our intensive study; more research could be undertaken on the discovery and continuing efforts to develop revolutionary treatment techniques of purified active compounds from natural sources.
Consent statement/Ethical approval: Not required.
Funding Statement: Funding was not received for this study.
Authors’ Contributions: The review was conceptualized, and DD contributed to study design, and writing the manuscript. ABJ contributed to Data Analysis and computational investigation. SP and AKDR corrected the manuscript. All authors have read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to the Chettinad Academy of Research and Education (CARE), Chettinad Hospital and Research Institute (CHRI), Chennai, India, and the Faculty of Medicine, University of Oslo, Norway.
References
- 1.Benkovic S., Michelle K., Eric S. 4 cases: HIV and SARS-CoV-2 co-infection in patients from Long Island. New York J Med Virol. 2020 doi: 10.1002/jmv.26029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco J.L., Ambrosioni J., Garcia F., Martínez E., Soriano A., Mallolas J., et al. COVID-19 in patients with HIV: clinical case series. The lancet HIV. 2020;7(5):e314–e316. doi: 10.1016/S2352-3018(20)30111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown T.T., Li X., Cole S.R., Kingsley L.A., Palella F.J., Riddler S.A., et al. Cumulative exposure to nucleoside analog reverse transcriptase inhibitors is associated with insulin resistance markers in the Multicenter AIDS Cohort Study. Aids. 2005;19(13):1375–1383. doi: 10.1097/01.aids.0000181011.62385.91. [DOI] [PubMed] [Google Scholar]
- 4.Cary DC, Peterlin BM. Natural products and HIV/AIDS. AIDS research and human retroviruses. 2018; 34(1):31-8. Doi: 10.1089/aid.2017.0232. [DOI] [PMC free article] [PubMed]
- 5.Chen J, Cheng X, Wang R, Zeng X. Computed Tomography Imaging of an HIV-infected Patient with Coronavirus Disease 2019 (COVID-19). J Med Virol; 2020. https://doi.org/10.1002/jmv.25879. [DOI] [PMC free article] [PubMed]
- 6.Donders F, Lonnée-Hoffmann R, Tsiakalos A, Mendling W, Martinez de Oliveira J, Judlin P, Xue F, Donders GG, COVID I, Workgroup G. ISIDOG recommendations concerning COVID-19 and pregnancy. Diagnostics. 2020;(4):243. https://doi.org/10.3390/diagnostics10040243. [DOI] [PMC free article] [PubMed]
- 7.Donovan M.C. Social constructions of people with AIDS: Target populations and United States policy, 1981–1990. Rev Policy Res. 1993;3–4:3–29. doi: 10.1111/j.1541-1338.1993.tb00548.x. [DOI] [Google Scholar]
- 8.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS‐CoV‐2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci; 2020:117592253. Doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed]
- 9.Emerman M., Malim M.H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280(5371):1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 10.Esposito F., Tintori C., Martini R., Christ F., Debyser Z., Ferrarese R., et al. Kuwanon-L as a new allosteric HIV-1 integrase inhibitor: molecular modeling and biological evaluation. ChemBioChem. 2015;16(17):2507–2512. doi: 10.1002/cbic.201500385. [DOI] [PubMed] [Google Scholar]
- 11.Fanales-Belasio E., Raimondo M., Suligoi B., Buttò S. HIV virology and pathogenetic mechanisms of infection: a brief overview. Annalidell'Istitutosuperiore di sanita. 2010;46:5–14. doi: 10.4415/ANN_10_01_02. [DOI] [PubMed] [Google Scholar]
- 12.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J MedChem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 13.Gelderblom H.R., Özel M., Pauli G. Morphogenesis and morphology of HIV structure-function relations. Arch Virol. 1989;106(1):1–3. doi: 10.1007/BF01311033. [DOI] [PubMed] [Google Scholar]
- 14.Guan W.-J., Ni Z.-y., Hu Y.u., Liang W.-H., Ou C.-Q., He J.-X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser P., Joos B., Niederöst B., Weber R., Günthard H.F., Fischer M. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol. 2007;81(18):9693–9706. doi: 10.1128/JVI.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashman Y., Gustafson K.R., Fuller R.W., Cardellina J.H., 2nd, McMahon J.B., Currens M.J., et al. The calanolides, a novel HIV-inhibitory class of coumarin derivatives from the tropical rainforest tree, Calophyllum lanigerum. J MedChem. 1992;35(15):2735–2743. doi: 10.1021/jm00093a004. [DOI] [PubMed] [Google Scholar]
- 17.Katlama C., Deeks S.G., Autran B., Martinez-Picado J., van Lunzen J., Rouzioux C., et al. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. The Lancet. 2013;381(9883):2109–2117. doi: 10.1016/S0140-6736(13)60104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Coronavirus Dis. 2019;COVID-19:2020:23. doi: 10.1007/978-981-15-4814-7. [DOI] [Google Scholar]
- 19.Kumari G.K., Singh R. Anti-HIV drug development: structural features and limitations of present day drugs and future challenges in the successful HIV/AIDS treatment. Curr Pharm Des. 2013;19(10):1767–1783. doi: 10.2174/13816128113199990295. [DOI] [PubMed] [Google Scholar]
- 20.Li Z., Li L.J., Sun Y., Li J. Identification of natural compounds with anti-hepatitis B virus activity from Rheum palmatum L. ethanol extract. Chemotherapy. 2007;53(5):320–326. doi: 10.1159/000107690. [DOI] [PubMed] [Google Scholar]
- 21.Lucas S.B., Hounnou A., Peacock C., Beaumel A., Djomand G., N’Gbichi J.-M., et al. The mortality and pathology of HIV infection in a west African city. AIDS (London, England) 1993;7(12):1569–1579. doi: 10.1097/00002030-199312000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Mallick R., Duttaroy A.K. Origin and Structural Biology of Novel Coronavirus (SARS-CoV-2) Adv Exp Med Biol. 2021;1352:1–13. doi: 10.1007/978-3-030-85109-5_1. [DOI] [PubMed] [Google Scholar]
- 23.Martini R., Esposito F., Corona A., Ferrarese R., Ceresola E.R., Visconti L., et al. Natural Product Kuwanon-L Inhibits HIV-1 Replication through Multiple Target Binding. ChemBioChem. 2017;18(4):374–377. doi: 10.1002/cbic.201600592. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson F. (2020). Infectious diseases: the role of the healthcare professional. In Clinical forensic medicine (pp. 343-392). Springer, Cham. Doi: 10.1007/978-3-030-29462-5_10.
- 25.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat RevImmunol. 2018;18(2):134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 26.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Sonar V.P., Corona A., Distinto S., Maccioni E., Meleddu R., Fois B., et al. Natural product-inspired esters and amides of ferulic and caffeic acid as dual inhibitors of HIV-1 reverse transcriptase. Eur J Med Chem. 2017;130:248–260. doi: 10.1016/j.ejmech.2017.02.054. [DOI] [PubMed] [Google Scholar]
- 28.Thomas J., Ruggiero A., Paxton W.A., Pollakis G. Measuring the success of HIV-1 cure strategies. Front Cell Infect Microbiol. 2020;10:134. doi: 10.3389/fcimb.2020.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian C., Tang L., Wu J., Li W., Ming X., Zhou H., et al. An HIV-infected patient with coronavirus disease 2019 has a favourable prognosis: a case report. Ann Palliat Med. 2021;10(5):5808–5812. doi: 10.21037/apm-20-576. [DOI] [PubMed] [Google Scholar]
- 30.Trott O., Olson A.J. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White A. Men and COVID-19: the aftermath. Postgrad Med. 2020;132(sup4):18–27. doi: 10.1080/00325481.2020.1823760. [DOI] [PubMed] [Google Scholar]
- 33.Whittle H., Morris J., Todd J., Corrah T., Sabally S., Bangali J., et al. HIV-2-infected patients survive longer than HIV-1-infected patients. AIDS (London, England) 1994;8(11):1617–1620. doi: 10.1097/00002030-199411000-00015. [DOI] [PubMed] [Google Scholar]
- 34.Wu J., Wu X., Zeng W., Guo D., Fang Z., Chen L., et al. Chest CT findings in patients with corona virus disease 2019 and its relationship with clinical features. Invest Radiol. 2020;55(5):257–261. doi: 10.1097/RLI.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183(3):730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H.J., Rumschlag-Booms E., Guan Y.F., Wang D.Y., Liu K.L., Li W.F., et al. Potent inhibitor of drug-resistant HIV-1 strains identified from the medicinal plant Justicia gendarussa. J Nat Prod. 2017;80(6):1798–1807. doi: 10.1021/acs.jnatprod.7b00004. [DOI] [PubMed] [Google Scholar]
- 37.Zheng M, Zhao J, Cui C, Fu Z, Li X, Liu X, Ding X, Tan X, Li F, Luo X, Chen K. Computational chemical biology and drug design: Facilitating protein structure, function, and modulation studies. Medicinal research reviews. 2018 May;38(3):914-50.Doi: 10.1002/med.21483. [DOI] [PubMed]
- 38.Ambesi-Impiombato A., Bernardo D. Computational biology and drug discovery: From single-target to network drugs. Curr Bioinform. 2006 Jan 1;1(1):3–13. doi: 10.2174/157489306775330598. [DOI] [Google Scholar]
- 39.Torres PH, Sodero AC, Jofily P, Silva-Jr FP. Key topics in molecular docking for drug design. Int J Mol Sci; 2019 Jan;20(18):4574.Doi: 10.3390/ijms20184574. [DOI] [PMC free article] [PubMed]
- 40.Cavalla D., Oerton E., Bender A. Drug Repurpos Rev. 2017 doi: 10.1016/B978-0-12-409547-2.12283-8. [DOI] [Google Scholar]
- 41.Schenone M., Dančík V., Wagner B.K., Clemons P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013 Apr;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh S, Malik BK, Sharma DK. Molecular drug targets and structure based drug design: A holistic approach. Bioinformation. 2006;1(8):314. Doi: 10.6026/97320630001314. [DOI] [PMC free article] [PubMed]
- 43.Minikel E.V., Zhao H.T., Le J., O’Moore J., Pitstick R., Graffam S., et al. Prion protein lowering is a disease-modifying therapy across prion disease stages, strains and endpoints. Nucleic Acids Res. 2020;48(19):10615–10631. doi: 10.1093/nar/gkaa616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu W.-G., Zhang X., Yuan J.-F. Anti-HIV drug development through computational methods. AAPS J. 2014;16(4):674–680. doi: 10.1208/s12248-014-9604-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordes F., Kaiser R., Selbig J. Bioinformatics approach to predicting HIV drug resistance. Exp Rev Molecul Diagnost. 2006;6(2):207–215. doi: 10.1586/14737159.6.2.207. [DOI] [PubMed] [Google Scholar]
- 46.Jenwitheesuk E., Samudrala R. Prediction of HIV-1 protease inhibitor resistance using a protein–inhibitor flexible docking approach. Antiviral Therapy. 2005 Jan;10(1):157–166. doi: 10.1177/135965350501000115. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J., Hao J., Peng L., Duan H., Luo Q., Yan H., et al. Classification and Design of HIV-1 Integrase Inhibitors Based on Machine Learning. Comput Math Methods Med. 2021;2021:1–11. doi: 10.1155/2021/5559338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaudry S.N., Hussain W., Rasool N. Inhibitory role of selective phytochemicals against HIV-2 protease: a study of molecular docking, ADMET and DFT computations. Int J Computat Biol Drug Design. 2020;13(4):390–414. [Google Scholar]
- 49.Khare P., Sahu U., Pandey S.C., Samant M. Current approaches for target-specific drug discovery using natural compounds against SARS-CoV-2 infection. Virus Res. 2020 Dec;1(290) doi: 10.1016/j.virusres.2020.198169. [DOI] [PMC free article] [PubMed] [Google Scholar]



