Abstract
Synthetic Genomics is the construction of viruses, bacteria, and eukaryotic cells with synthetic genomes. It involves two basic processes: synthesis of complete genomes or chromosomes and booting up of those synthetic nucleic acids to make viruses or living cells. The first Synthetic Genomics efforts resulted in the construction of viruses. This led to a revolution in viral reverse genetics and improvements in vaccine design and manufacture. The first bacterium with a synthetic genome led to construction of a minimal bacterial cell and recoded Escherichia coli strains able to incorporate multiple non-standard amino acids in proteins and resistant to phage infection. Further advances led to a yeast strain with a synthetic genome and new approaches for animal and plant artificial chromosomes. On the horizon there are dramatic advances in DNA synthesis that will enable extraordinary new opportunities in medicine, industry, agriculture, and research.
ETOC:
Synthetic Genomics is a branch of synthetic biology that constructs viruses, bacteria and eukaryotic cells with synthetic genomes. Venter and colleagues review the foundations, advances, challenges and future prospects of Synthetic Genomics.
Introduction
The term genomics is attributed to Jackson Laboratory scientist Tom Roderick. Reportedly Roderick and some of his colleagues were brainstorming over beer to come up with a name for a new journal about the study of genomes (McKusick and Ruddle, 1987). In 2005, a consortium including J. Craig Venter and Nobel Laureate Hamilton O. Smith founded a company they called Synthetic Genomics, Inc. that would merge genomics and synthetic biology to address problems in industry and medicine. The term Synthetic Genomics is now in wide use to refer to a branch of synthetic biology that constructs viruses, bacteria, and eukaryotic cells with synthetic genomes. It utilizes two basic processes: design and synthesis of complete genomes or chromosomes and booting up of those synthetic nucleic acids to make viruses or living cells (Zhang et al., 2020). Nucleic acid synthesis technology advances since the first tRNA gene was synthesized in 1972 have resulted in a steady, exponential improvements in the size of synthetic DNA that could be made and the accuracy of the sequence. Remarkably, one can now purchase synthetic DNAs encoding whole bacterial genomes comprised of more than a million basepairs. The processes for installing synthetic viral, bacterial or eukaryotic genomes (or chromosomes) in viral capsids or living cells so that the resulting viruses and cells manifest the phenotypes encoded in the new synthetic genomes has advanced less linearly. This is largely because each viral or cellular species requires a different approach for this booting up process. In our review we will present advances in both elements of Synthetic Genomics focusing on viruses, bacteria, and eukaryotes in that order. Additionally, we will introduce new Synthetic Genomics technologies that can lead to still more efficient DNA synthesis of much larger genomes than have been done previously and that are enabling pig to human organ transplant from pigs whose genomes have been altered to make the xenotransplantation work.
Synthetic Viruses
Given that infectious diseases with the periodic likelihood of pandemics remain a major worldwide problem, there is a desperate need to develop and distribute vaccines more quickly. While vaccines come in different forms, including live attenuated, inactivated, virus-vectored and subunit among others (Lee et al., 2018), nucleic acid-based vaccines have recently gained prominence and renewed enthusiasm due to the resounding success of the mRNA vaccines that were developed in almost record time for COVID-19 (Golob et al., 2021). Nucleic acid vaccines are attractive choices due to their potential to be safe, effective and economical. In addition, both DNA and RNA vaccines can induce humoral and cellular immune responses, generally showing greater ability to induce T-cell responses than other noninfectious vaccine platforms (Qin et al., 2021). A major advantage of nucleic acid vaccines [including DNA, non-amplifying RNA and self-amplifying RNA (SAM)] is that they are simple to construct since they only need to express the gene encoding the antigen. While they are not without some drawbacks, such as limited immunogenicity in vivo (DNA vaccines) or instability (RNA vaccines), the ease and cost of manufacturing nucleic acid vaccines provide a strong case for them (Qin et al., 2021).
While many factors, including mode of delivery, formulation, presence of adjuvants, etc., are important for an effective recombinant vaccine, in this review, we will highlight the contribution of Synthetic Genomics in accelerating the development of recombinant vaccines, from design to efficient and speedy generation of the constructs and recovery of vaccine candidates. For the other factors, we direct the reader to recent reviews (Gary and Weiner, 2020; Lee et al., 2018; Qin et al., 2021; Sandbrink and Shattock, 2020). Below, we describe some key studies that laid the groundwork to develop recombinant vaccines for viruses, in particular for RNA viruses, from a Synthetic Genomics perspective which includes development of reverse genetics for viruses, design/synthesis of genes or full-length viral genomes and how these are incorporated to developing synthetic recombinant vaccines.
Even before improvements in DNA synthesis resulted in the production of viruses using only nucleic acid sequences in the early 2000s, virologists were designing and constructing viral genomes to test hypotheses about viral pathogenesis and to develop attenuated strains to serve as vaccines. Viral reverse genetics were important for vaccine development and had been development for both positive-strand and negative-strand RNA viruses (Enami et al., 1990; Pattnaik and Wertz, 1990; Wertz et al., 1998). However, a major advance using Synthetic Genomics to facilitate viral reverse genetics was the generation of DNA copies of viral genomes from synthetic oligonucleotides. It is widely assumed that this was first done in 2002 when Eckard Wimmer and colleagues produced poliovirus from a synthetic DNA molecule encoding the 7.9 kb poliovirus genome after nearly two years of effort (Cello et al., 2002). However, 18 months earlier, Charles Rice and colleagues constructed an 8,001 base pair (bp) DNA copy of a hepatitis C virus (HCV) sub-genomic replicon from oligonucleotides in less than 6 months. This accomplishment was overshadowed by the report in the same paper of the first robust, cell-based system for genetic and functional analyses of HCV replication that was enabled by the synthetic HCV replicon (Blight et al., 2000). For their process, initially, sub-genomic cDNAs spanning 600 to 750 bases in length were assembled in a stepwise PCR assay with 10 to 12 gel-purified 60–80 base oligonucleotides with complementary overlaps of 16 nucleotides. These molecules were cloned in plasmids, sequence verified and then assembled to construct the 8 kilobase (kb) replicon.
The next major advance in viral genome synthesis was a radical increase in the speed at which such DNA synthesis projects could be completed. In 2003, we used a pool of 259 overlapping gel purified oligonucleotides to synthesize 5,386 bp bacteriophage φX174 DNA genome in less than two weeks. As with the previous genome syntheses of viral genomes, the process utilized sequential ligation and polymerase cycling reactions to accomplish the assembly. The main differences between our approach and what was done previously by the Wimmer and Rice teams were the single step construction of the entire >5 kb DNA without cloning and sequence verification of sub-genomic 400–600 base-pair DNAs (Smith et al., 2003). While this process was not the same as the process our team used to assemble the sub-genomic segments used to build complete bacterial genomes (Gibson et al., 2008a), it set the stage for development the genome assembly methods we used to create bacteria with chemically synthesized genomes (Gibson et al., 2010; Hutchison et al., 2016).
The above studies demonstrated the synthesis of virus genomes smaller than 10 kb. However, in 2005, the Drew Endy group reported the refactoring of the bacteriophage T7 genome to physically separate and enable unique manipulation of primary genetic elements to facilitate modeling and functional scientific research (Chan et al., 2005). In doing so, they replaced approximately the left quarter of the 40 kb T7 genome with constructed synthetic DNA that contained separate individually assigned functional elements. The resulting chimeric phage was viable and subsequent analysis confirmed that the individual parts could be independently manipulated (Chan et al., 2005). Although, in this case, the T7 genome was not entirely synthetic, this research was seminal in that it brought systematic engineering principles to biology and the concept of redesign and building anew to biological systems in support of rational scientific discovery.
Subsequently, in 2008, researchers led by Mark Denison and Ralph Baric extended the capacity to synthesize full-length viral genomes to about 30 kb when they reported the rational design, synthesis and recovery of a recombinant bat SARS-like coronavirus [SCoV] (Becker et al., 2008). Importantly, this study showed that Synthetic Genomics can be used to recover non-cultivable viruses since prior to this study, no bat coronavirus had been successfully grown in culture or animals. In their work, the team first established a putative consensus Bat-SCoV from the available Bat-SCoV sequences but used the defined and functional 5’ UTR and transcriptional regulatory sequences from SARS-CoV-1 due to incomplete 5’ UTR sequences of BAT-SCoVs. The team then designed cDNA fragments with junctions precisely aligned to the existing SARS-CoV-1 reverse genetics system and obtained them commercially. The cDNA fragments were assembled into a full-length cDNA, transcribed in vitro to yield genomic RNA and then transfected into host cells to recover virus. Although with this iteration, recombinant Bat-SCoV was recovered, infectious recombinant virus was successfully obtained when the Bat-SCoV receptor-binding domain (RBD) was replaced with that of SARS-CoV-1 (Becker et al., 2008). While this study can be considered controversial due to the gain-of-function, it did demonstrate the power of Synthetic Genomics to aid rapid public health responses to emerging virus threats. As an example, more recently, a team led by Volker Thiel and Joerg Jores developed a reverse genetics system for the pandemic-causing SARS-CoV-2 by assembling a full-length cDNA genome from commercially synthesized DNA fragments using a Synthetic Genomics approach that was developed for the synthetic cell (Gibson et al., 2010) and extended to large DNA viruses (Oldfield et al., 2017; Vashee et al., 2017), transcribing in vitro to generate genomic RNA and then recovering infectious virus (Thi Nhu Thao et al., 2020). Impressively, the team was able to recover an infectious clone about a month after the first SARS-CoV-2 genome sequence was reported using the Synthetic Genomics platform.
By 2013, advanced protocols for synthetic DNA production enabled our team’s efforts to speed up production of virus to be used in vaccines for rapid response to pandemics (Dormitzer et al., 2013). The first example of these efforts was provided by us at the JCVI together with researchers from Novartis Vaccine and Diagnostics (NV&D), Synthetic Genomics Vaccines Inc. (SGVI) and the Biomedical Advanced Research and Development Authority (BARDA), U.S. Department of Health and Human Services, who collaborated to develop a rapid process for synthetic vaccine influenza virus production. The team addressed three major technical hurdles to more rapid and reliable pandemic responses: the speed of synthesizing DNA cassettes to drive production of influenza RNA genome segments, the accuracy of rapid gene synthesis and the yield of hemagglutinin protein (HA) from vaccine viruses. First, using a method pioneered by the JCVI’s Dan Gibson, uncloned synthetic linear DNA copies of viral genome segments encoding the immunogenic 1.7 kb HA and 1.5 kb neuraminidase (NA) viral genome segments were synthesized in less than a day in a process that included oligonucleotide design and synthesis (Figure 1A). This approach was different from previous viral genome synthesis projects in several regards. One, oligonucleotide synthesis from some foundries was now of sufficient quality that no gel purification was necessary as was the case in previous viral genome syntheses (Blight et al., 2000; Cello et al., 2002; Smith et al., 2003). Our team found that there were fewer sequence errors if the oligonucleotides were designed to include the entire sequences of both DNA strands. Thus, unlike in previous efforts where the overlapping oligonucleotides tiled across the whole design sequence with gaps between the oligonucleotides on the same strand, here there were no gaps. The assembly process involved isothermal assembly, and polymerase chain reaction (PCR) amplification similar to previous described synthetic efforts. Two, at the next step, any error-containing DNA was removed enzymatically by treating melted and reannealed DNA with a commercially available error correction kit that excises areas of base mismatch in double-stranded DNA molecules before another round of PCR amplification (Dormitzer et al., 2013).
Figure 1. Synthetic Gene Segment Assembly with Error Correction and Rescue of Synthetic Influenza Viruses from a Panel of Backbones.
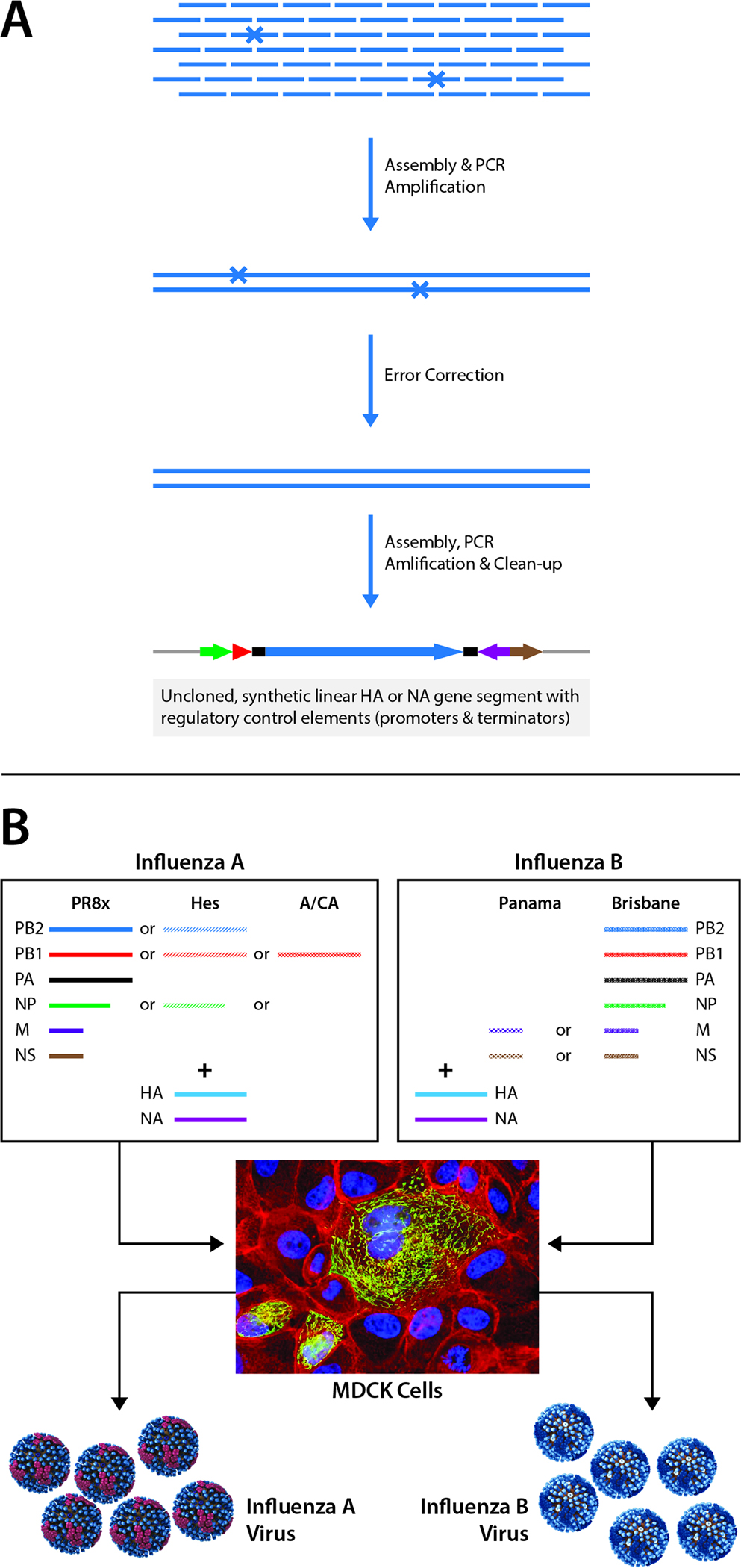
A. Schematic diagram of the assembly procedure. Error correction reduced the rate from 1 error per 1328 bp to 1 error per 9589. X, sites of oligonucleotide errors. Blue arrow, HA or NA coding sequence; gray, plasmid backbone sequence; green arrow, CMV promoter; purple arrow, human pol I promoter; red arrow, murine pol I terminator; brown arrow, pol II terminator; black rectangle, UTR. B. Schematic diagram of the rescue of synthetic influenza viruses from multiple backbones for types A and B influenza strains. PR8x, derivative of a PR8 strain adapted over 5 passages for growth in MDCK cells; Hes, A/Hessen/105/2007 (H1N1); A/CA, A/California/7/2009 (H1N1); Brisbane, B/Brisbane/60/2008 (Victoria lineage); Panama, B/Panama/45/1990 (Yamagata lineage). The photograph of MDCK cells was made by and used with permission from Benjamin Sievers, JCVI.
Then, the team improved the speed and efficiency of virus rescue by first, using a manufacturing qualified MDCK 33016PF cell line for both seed generation and vaccine antigen production and second, by identifying individual optimized backbones (sets of genome segments encoding influenza virus proteins other than HA and neuraminidase (NA)) for influenza type A and B strains (Figure 1B). Furthermore, as a proof-of-concept test of this synthetic system’s first iteration, our team generated a synthetic vaccine virus in a simulated pandemic response. For this, BARDA personnel not involved in the project provided us with unidentified, partial HA and NA genome segment sequences. The given sequences, which included complete coding regions but incomplete UTRs, mimicked information likely to be available early in a pandemic. Sequence analysis of the HA segment revealed that it was closely related to a low-pathogenicity North American avian H7N3 virus whereas the NA segment was closely related to a low-pathogenicity North American avian H10N9 virus. In addition, we reconstructed the HA and NA UTRs by alignment of each sequence with high quality full-length H7 HA and N9 NA genome segments, respectively. Using the synthetic vaccine virus system, the team generated seed viruses of multiple HA or NA variants with multiple backbones in 5 days from the start of oligonucleotide design. Our team further validated the synthetic vaccine virus system by generating a variety of influenza strains, including seasonal H3N2 (Dormitzer et al., 2013). Thus, this study demonstrated that simultaneous rescue of multiple variants is faster and more easily accomplished with this synthetic approach than standard plasmid mutagenesis approaches, which should lead to a much faster end to a pandemic through rapid vaccine responses.
The researchers from NV&D and SGVI then teamed up to reduce the time further, potentially to days after the discovery of a new virus, for the first availability of a vaccine candidate. In this study, the team was able to generate a vaccine candidate in eight days for the H7N9 influenza outbreak in Shanghai, China in 2013 (Hekele et al., 2013). To accomplish this, the team again used the gene assembly and error correction approach described above to synthesize the H7 HA coding sequence after the China Center for Disease Control and Prevention posted the HA and NA gene coding sequences of the outbreak H7N9 strain on the Global Initiative for Sharing All Influenza Data system. However, rather than generate virus, the team placed the synthetic HA coding sequence into their SAM vaccine platform [a synthetic self-amplifying mRNA, delivered by a synthetic lipid nanoparticle (LNP)] DNA template that contained elements for self-amplification and expression of the H7 HA (Geall et al., 2012). mRNA was then produced in vitro by T7 RNA polymerase and transfected into BHK7 to demonstrate expression of influenza H7 HA. Furthermore, after two immunizations with the H7/LNP SAM RNA vaccine, mice produced HA inhibition titers considered protective as well as virus neutralizing titers (Hekele et al., 2013). Thus, this study demonstrated that fully Synthetic Genomics vaccine technologies may provide unmatched speed of response to reduce the impact of pandemics or novel emerging viruses.
Finally, in 2018, David Evans and colleagues constructed an infectious horsepox virus vaccine from chemically synthesized DNA fragments (Noyce et al., 2018). In their work, they transfected ten synthesized overlapping 10–30 kb DNA fragments obtained from a commercial company together with Vaccinia virus terminal sequences into host cells infected with a helper virus, Shope fibroma virus, where they were recombined into a live synthetic chimeric horsepox virus. The resulting virus produced smaller plaques, less extracellular virus and was less virulent in mice than Vaccinia virus while providing vaccine protection against a lethal challenge (Noyce et al., 2018). However, this study was controversial due to potential dual use, access and benefit sharing issues (Rourke et al., 2020). Regardless, this study demonstrated the possibility to generate virtually any live virus from sequence alone.
In summary, Synthetic Genomics has already proven useful in helping to develop the next generation of vaccines due to the capacity to rapidly design and construct not only synthetic genes but also complete viral genomes. This capacity has facilitated the development of higher throughput production of genes for recombinant, subunit and nucleic acid vaccines as well as viral reverse genetics systems to quickly understand their biology and facilitate vaccine or therapeutic development. With continued reduction in the cost of nucleic acid synthesis and further advances to increasing speed and scale of synthesis, we expect that the field will significantly contribute to an even faster response to emerging infectious diseases and potential pandemics. Unfortunately, the synthetic virus/vaccine platform has not yet led to commercial success. However, with the recent full approval by the U. S. Food and Drug Administration of Pfizer-BioNTech COVID-19 Vaccine (on August 23, 2021, marketed as Comirnaty) and the Moderna COVID-19 Vaccine (on January 31, 2022, marketed as Spikevax) for 16 and 18 years of age, respectively and older, we can expect more vaccines made using the synthetic virus/vaccine platform to be available in the market.
Prokaryotic genome sequencing led to construction of bacteria with synthetic genomes
When the J. Craig Venter Institute (JCVI) undertook the goal of sequencing the first cellular genome in history (Fleischmann et al., 1995), the primary goal was to see how the life of the cell could be understood based on its gene content. The team spent tremendous effort to annotate every gene and pathway of Mycoplasma genitalium, but it soon became clear that there were so many genes of unknown function that this goal was unreachable at that time. Because this was the first sequenced genome in history there was clearly no other genomes to compare with. We thought if we had at least one more genome that we might be able to make more gene identifications and have a clearer understanding of how the genome coded for life. We chose to sequence the smallest known cellular genome, thinking it would have fewer non-essential genes, which would aid in understanding the first genome. Thus, in 1995 a second genome was sequenced, that of M. genitalium, which had the smallest known genome of any species capable of independent growth (Fraser et al., 1995). Again, it was disappointing that the second genome did not have many genes that overlapped with the first genome and that it also had a significant percentage of genes of unknown function. We felt that two major approaches were needed going forward. The most obvious was that the number of sequenced genomes needed to be increased by orders of magnitude, the DOE agreed and began funding multiple genome sequences from diverse organisms including the first archaea. It became clear that it would be a multiple decade approach with this new field of comparative genomics to yield a clear understanding of life at the genome level.
The JCVI team decided that the best way to try to understand life, at the genome level, was to try to synthesize a genome chemically and attempt to reconstitute life using this synthetic genome. Building a minimal bacterial cell to facilitate basic studies of living cells had been a goal of biologists ever since Max Delbruck’s Phage School in the 1930s, but never before had the technology to make such a cell been available (Glass et al., 2017).
The team at the JCVI launched its Synthetic Genomics efforts to construct a minimal bacterial cell in late 2002. By then, methods for DNA synthesis had advanced such that <10 kb viral genomes had already been synthesized and booted up to produce virus (Blight et al., 2000; Cello et al., 2002). Those early 21st century efforts marked the dawn of the field of Synthetic Genomics, which is the construction of viruses, bacteria, and eukaryotic cells with synthetic genomes. It involves two basic processes: synthesis of complete genomes or chromosomes and booting up of those synthetic nucleic acids to make viruses or living cells.
Using the molecular biology and microbiology technologies of the early 1990’s, construction of a bacterial cell with a synthetic genome would have been impossible. Driven by the ambition to construct a minimal bacterial cell that could be used to investigate the first principles of cellular life, the JCVI’s Synthetic Genomics efforts led to the landmark synthetic biology tool building accomplishments that enabled construction of the first “synthetic organism”, JCVI-syn1.0 in 2010 (Gibson et al., 2010) and the construction of the first minimal bacterial cell, JCVI-syn3.0 in 2016 (Hutchison et al., 2016). Guided by the writings of Harold Morowitz, the JCVI team elected to return to Mycoplasma species as starting points for building minimal cells (Morowitz, 1984). The same features that made Mycoplasma genitalium an appealing candidate for the early whole-genome sequencing efforts described above, again made it appealing as a basis for a minimal synthetic cell. It seemed reasonable to assume that the M. genitalium genome had near the minimum number of genes required for cellular life. However, it was found that more than 100 of the 525 M. genitalium genes could be disrupted by transposon insertion without affecting viability (Glass et al., 2006; Hutchison et al., 1999). At that time mycoplasmas were largely genetically intractable, so stepwise deletion of non-essential genes would be very time-consuming and laborious. Thus it was decided to adopt a Synthetic Genomics approach to minimizing the mycoplasma genome.
In 2002 two efforts aimed at eventually enabling design, construction, and booting up of a minimized M. genitalium genome were initiated at JCVI (Marshall, 2002). One team developed improved DNA synthesis methods that would be capable of constructing a 583 kb synthetic M. genitalium genome. The other team sought to devise a plan to boot up the synthetic genome.
As mentioned in the Synthetic Viruses section above, in 2003 a protocol was developed that enabled rapid synthesis of a ~5 kb φX174 bacteriophage genome from synthetic oligonucleotides (Smith et al., 2003). Within a few years, presumably in part due to that study, synthesis of DNA molecules up to 5 kb had become an affordable commodity.
A synthetic M. genitalium genome was built starting with ~5 kb cassettes. These were assembled in five stages using a combination of in vitro enzymatic joining methods and in vivo recombination in yeast cells. Clones of intermediate products were sequence verified as assembly proceeded. While synthesizing a complete M. genitalium genome was not achieved without difficulties, the processes employed relied on technologies that had been proven to work to produce smaller synthetic DNAs (Kodumal et al., 2004; Smith et al., 2003). There were no such precedents to guide the search for a method to boot up the synthetic M. genitalium genome. The plan was to install the M. genitalium genome containing a tetracycline resistance marker in a Mycoplasma pneumoniae cell, and then after these two genome cells had time to divide, only cells containing the M. genitalium genome would survive antibiotic treatment. M. pneumoniae is closely related to M. genitalium. All but a couple of non-essential M. genitalium genes have orthologous counterparts in the M. pneumoniae genome. So it appeared reasonable to assume that the M. genitalium genome would likely function in the cytoplasm of its close relative. However, efforts to boot up isolated M. genitalium genomes were all unsuccessful.
Because of the difficulties involved with working with M. genitalium (it takes up to six weeks to form colonies and those colonies can only be visualized using a microscope), JCVI scientist Carole Lartigue attempted to develop the needed technology using a more tractable set of closely related Mycoplasma species. After two years of experimentation, she was able to install an isolated Mycoplasma mycoides genome in a Mycoplasma capricolum cell. After antibiotic selection, she recovered cells containing only complete M. mycoides genomes. M. mycoides and M. capricolum are very closely related species that have ~1 mb genomes and that grow rapidly to produce 1 mm diameter colonies in 2–3 days. This faster growth greatly accelerated the Lartigue’s pace of experimentation. Lartigue called this process genome transplantation (Lartigue et al., 2007). It in many ways is similar to a chemical transformation of E. coli. The transplantation reaction involves treating the M. capricolum recipient cell with CaCl2 and polyethylene glycol, Figure 2.
Figure 2. Genome Transplantation.
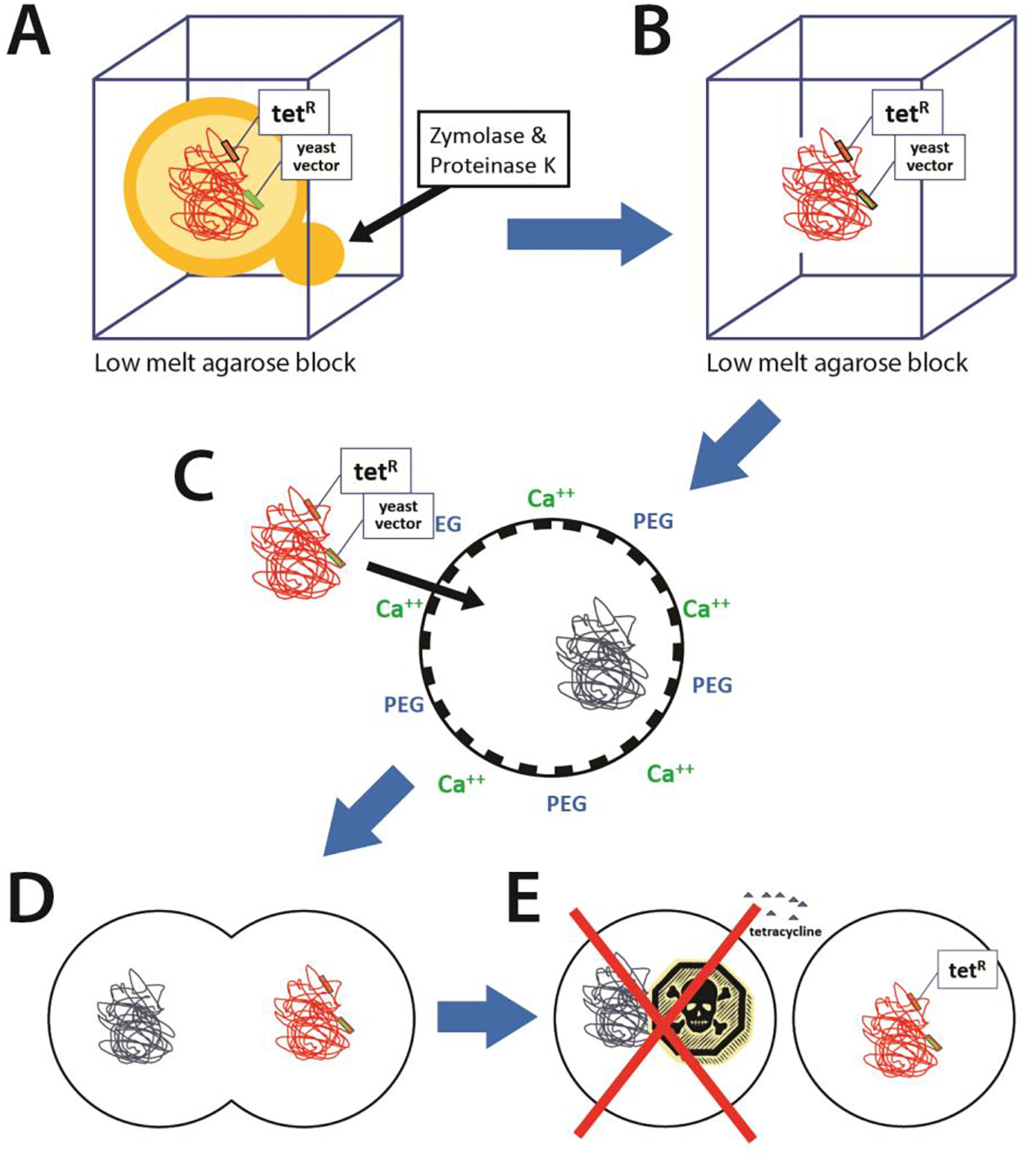
A. Yeast cells or bacterial cells containing the donor genome to be transplanted are encased in low melt agarose blocks. Yeast cells are spheroplasted using zymolase and digested with proteinase K. B. This leaves the donor genome inside caverns in the agarose, and not sheared during the purification The agarose is melted to gently retrieve the DNA. C. The donor DNA (red) and M. capricolum cells are mixed with polyethylene glycol (PEG) to increase recipient cell membrane fluidity and CaCl2, to mask the DNA charge, resulting in the donor genome entering the recipient cell (at very low frequency). D. The transiently diploid cells are transferred to growth media and begin to grow and divide. E. After several hours, the cells are treated with tetracycline. Only the cells with the synthetic donor genome containing a tetracycline resistance marker survive.
Two years later Lartigue et al. (2009) reported genome transplantation of an M. mycoides genome that was cloned as a yeast centromeric plasmid into an M. capricolum cell whose single restriction enzyme gene had been disrupted. Because M. mycoides encodes the same restriction enzyme gene as M. capricolum, when isolated M. mycoides genomes were transplanted into M. capricolum, CCATC sites were methylated and unaffected. To transplant M. mycoides genomes isolated from yeast, it was essential that the M. capricolum restriction enzyme be inactivated (Lartigue et al., 2009). Unfortunately, intense efforts to adapt the M. mycoides-M. capricolum genome transplantation technique to install the M. genitalium genome via interspecies transfer into M. pneumoniae or intraspecies transfer into a different strain of M. genitalium were not successful. In fact, genome transplantation has only worked for a subgroup of mycoplasma called the mycoides cluster. Why this is so is not entirely clear. One known issue is the treatment of recipient cells with calcium chloride as is done for the mycoplasma genome transplantation activates surface associated nucleases in many other bacteria. Efforts are now underway at the JCVI to develop genome transplantation for non-mycoplasma species. It is unlikely that the JCVI team was so lucky as to choose to work with the only bacteria capable of this useful approach.
Because it was possible to transplant a M. mycoides genome cloned as a yeast centromeric plasmid and because there were reliable protocols for genome assembly, in 2009 it was decided to set aside the M. genitalium effort and instead to synthesize and install an M. mycoides genome. Learned lessons and improvements in DNA assembly technology made synthesis of the 1.079 Mb M. mycoides genome required only a fraction of one person’s time for a few months relative to the years required to develop the methods for and complete the M. genitalium assembly. The genome was assembled in three stages by transformation and homologous recombination in yeast from 1078 1 kb DNA cassettes. The synthetic genome differed from the wildtype M. mycoides genome in that it lacked genes encoding a glycerol transporter associated with pathogenesis, and it contained a four watermark sequences so that the synthetic genome could readily be distinguished from a wildtype genome. The genome was transplanted to produce the first cell with a chemically synthesized genome. That “synthetic“ bacterium was called JCVI-syn1.0 (Gibson et al., 2010).
To create JCVI-syn1.0, the JCVI developed three key Synthetic Genomics technologies (Figure 3). First, while construction of synthetic DNA molecules larger than 10 kb was possible before the JCVI complete genome synthesis efforts (Kodumal et al., 2004; Smith et al., 2003), those methods were not practical for rapid construction DNAs larger than 100 kb. Gibson Assembly for in vitro assembly of DNA molecules was a critical advance that led to synthetic genomes and has become one of the basic methods in synthetic biology (Gibson et al., 2008a; Gibson et al., 2009). Second, while in vitro DNA assembly of whole genomes is theoretically possible, it was construction of complete bacterial genomes as yeast centromeric plasmids that enabled both yeast recombination-based assembly of whole genomes and accumulation of sufficient amounts of synthetic genome that would be needed to boot up those genomes (Gibson et al., 2008a; Gibson et al., 2008b). Finally, genome transplantation installed the complete synthetic genomes, isolated from yeast, in a suitable recipient cell so that the new genome commandeered the cellular machinery to produce “synthetic” cells with the genotype and phenotype of the synthetic genome (Lartigue et al., 2009).
Figure 3. Three technologies critical to the construction of the first bacterium with a synthetic genome.
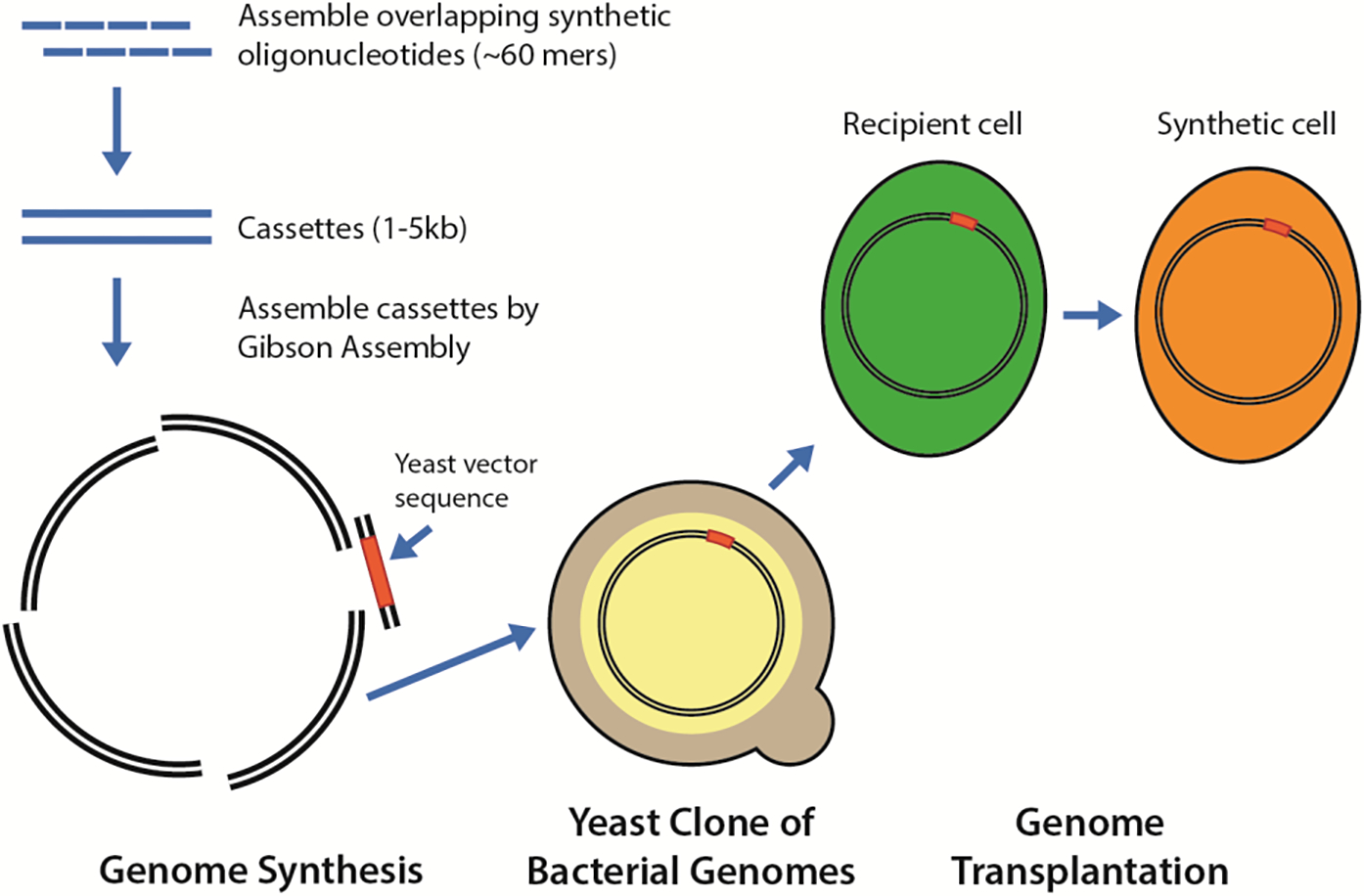
These Synthetic Genomics technologies were developed by the JCVI to enable construction of bacteria with chemically synthesized genomes. Prior to the synthesis of the M. genitalium genome in 2008 (Gibson et al., 2008a), DNA synthesis was used to produce molecules only as large as 32 kb; however the process was slow and inefficient. The genome synthesis technology developed by the JCVI greatly accelerated the process as well as enabling the in vitro synthesis of much larger DNA molecules. Yeast cloning of bacterial genomes was developed both for the final assembly of large overlapping sub-genomic DNA molecules that were transformed into yeast along with a 3–5 kb yeast vector sequence as yeast centromeric plasmids. This enabled parking the synthetic genome in yeast cells so that amount of bacterial genomic DNA needed for genome transplantation could be produced from large amounts of those yeast. Genome transplantation as depicted in Figure 2, boots up the synthetic genome isolated from yeast by installing it in a suitable bacterial recipient cell so that the new genome commandeers the recipient cell to produce a new cell with the genotype and phenotype of the synthetic genome.
The next step in the effort to construct a minimal bacterial cell was to perform transposon bombardment to identify the essential and non-essential genes in M. mycoides. This showed that about half of the M. mycoides genes were non-essential. Based on those findings, a reduced genome was designed and synthesized in eight overlapping segments just as in constructing JCVI-syn1.0. Eight different versions of the M. mycoides genome were then constructed, with each being comprised of a different reduced genome segment and seven wildtype segments. These were all transplanted and all resulted in viable transplants. This indicated that no essential genes had been deleted from any of the segments. A similar synthetic genome segment validation step was employed using genomes made from synthetic and natural one eighth genome segments in the construction of JCVI-syn1.0. Such validation steps are required in all similar large genome design and construction projects to localize potential design or synthesis flaws. While each of the eight reduced genome segments was individually viable, the fully minimized genome did not yield any successful transplants, indicating that pairs of non-essential genes located on different segments of the genome that both encode proteins that performed the same essential function, i.e. synthetic lethals had likely been deleted. Additional rounds of transposon bombardment on partially minimized genomes guided design of a new reduced genome that could be transplanted. First, JCVI-syn2.0 was produced, which has a 576 kb genome and encodes 475 protein coding genes. A final cycle of transposon bombardment and genome design resulted in construction of JCVI-syn3.0, which has a 531 kb genome and encodes 438 genes (Hutchison et al., 2016). That genome is smaller than the smallest known naturally occurring genome for an independently replicating bacterium. Currently the JCVI-syn3.0 minimal bacterial cell is used as a chassis both by the JCVI and more than fifty other research groups to investigate the fundamental aspects of cell biology.
Synthetic Escherichia coli genomes
The first successful effort using synthetic biology technology for grand scale redesign of the Escherichia coli genome was achieved by a team led by George Church and Farren Isaacs that recoded the genome to eliminate all the UAG stop codons (Lajoie et al., 2013). The resulting genomically recoded organism was thus amenable to the incorporation of non-standard amino acids and also showed resistance to phage infection. Rather than synthesize a new genome as was done by the Venter team to produce JCVI-syn1.0, this team employed multiplex automated genome engineering (MAGE) (Wang et al., 2009) and conjugative assembly genome engineering (CAGE) (Isaacs et al., 2011) to recode their genome. This effort paved the way for more ambitious E. coli recoding efforts described below.
Jason Chin’s Cambridge University team constructed a semi-synthetic 3.97 Mb E. coli genome, called Syn61, that was recoded to free up three codons that could be used to produce proteins containing as many as three non-standard amino acids (Fredens et al., 2019). Rather than construct an entire genome and then install it in a suitable recipient cell as was done by the JCVI, here ~100 kb synthetic recoded segments were substituted for natural equivalent regions using processes involving iterative programmed recombination called REXER and GENESIS (Wang et al., 2016). That process was repeated more than 35 times using a sophisticated marker exchange process, and iterative segment exchange (Wang et al., 2016). This project leveraged earlier work done by Chin and others that developed systems to utilize non-standard amino acids (Chin, 2017; Wang et al., 2016). As was used in the earlier mycoplasma synthetic genome projects, the modular, 100 kb at a time enabled identification of any design flaws. The synthetic 100 kb segments were constructed as bacterial artificial chromosomes in a two-step process that assembled in commercially synthesized DNA sub-genomic cassettes in yeast (Fredens et al., 2019).
A similar effort to construct an E. coli strain with a synthetic, recoded genome is ongoing in George Church’s Harvard University lab. They constructed a 3.97 Mb synthetic genome that utilized only 57 codons where seven codons were replaced with synonymous alternatives (for all but 13 of 2229 genes. Not only did the Church team free up seven codons to enable use of non-standard amino acids, but they also made the cell, which they call “rE.coli-57”, resistant to phage infection or alteration by horizontal import of new genes. As would be expected, some a small number of efforts at recoding some genes were unsuccessful and required troubleshooting. The ten failures the team reported are instructive for future genome design projects (Ostrov et al., 2016).
A synthetic Caulobacter crescentus genome
Eidgenössische Technische Hochschule (ETH) Zürich synthetic biologist Beat Christen recoded the essential genes of Caulobacter crescentus to produce a new genome called Caulobacter ethensis 2.0 (Venetz et al., 2019). C. crescentus is an important model organism for understanding bacterial cell cycles (McAdams and Shapiro, 2009). The 786 kb synthetic C. entensis-2.0 genome encodes 676 genes. Approximately 133,000 base substitutions were made in the genome to alter ~124,000 codons. One goal of this effort was to reduce the number of sequence elements that could interfere with genome synthesis such as high GC regions, direct repeats, hairpins, homopolymers and restriction sites. The number of synthesis constraints went from 7,014 in C. crescentus to 301 in C. ethensis-2.0. The recoding also eliminated three rare codons (TTG, TTA, TAG). The recoding to facilitate genome synthesis resulted in successful manufacture of 235 out of 236 3–4 kb DNA fragments, and only the one fragment needing custom synthesis. While this recoding likely facilitated genome synthesis, and maintained protein amino acid sequences, it also erased genetic information that may affect gene expression and other critical elements. In merodiploid studies in C. crescentus where plasmids containing segments of the C. ethensis-2.0 genome were expressed, ~20% of the genes had lower capacity than natural counterparts to support viability (Venetz et al., 2019). The findings here offer important lessons about the design of future synthetic genomes. This study offered no plans to boot up the C. entensis-2.0 genome (Venetz et al., 2019).
Synthetic Genomics using eukaryotes: Yeast 2.0
The other most widely known example of Synthetic Genomics resulting in organisms with synthetic genomes is the Synthetic Yeast Genome Project, Sc2.0, more commonly known as the Yeast-2.0 project. Members of this international consortium of 21 institutions designed and built synthetic versions of all 16 chromosomes of Saccharomyces cerevisiae (Zhao et al., 2022). Absent from the redesigned synthetic chromosomes are extra copies of tRNA genes, introns, and transposons, which result in more efficient chromosome synthesis during stepwise reconstruction of the DNA molecules using homologous recombination as well as providing a chassis to probe the role of mobile elements on yeast biology. Other changes include TAG/TAA stop codon replacement to enable use of non-standard amino acids and insertion of numerous loxPsym sites to enable genome scrambling that enables inducible evolution and genome reduction (Pretorius and Boeke, 2018; Zhao et al., 2022). Collectively, the already completed yeast-2.0 chromosomes are being used to investigate numerous questions about yeast and eukaryotic chromosome function and evolution. A series of publications reporting both the synthesis of 6.5 of the 16 yeast chromosomes and important findings about both yeast and eukaryotic chromosome biology gleaned from the organisms with the Sc2.0 chromosomes are in the literature with more papers describing the other chromosomes expected. (Annaluru et al., 2014; Dymond et al., 2011; Mitchell et al., 2017; Richardson et al., 2017; Shen et al., 2017; Wu et al., 2017; Xie et al., 2017; Zhang et al., 2017). In 2022, the Sc2.0 consortium reported the construction of a yeast strain encoding all 6.5 of those synthetic chromosomes from previously reported strains single synthetic chromosome using a technique they called endoreduplication intercross. Working with this strain, the Yeast-2.0 team discovered unknown interactions between synthetic chromosomes linking transcriptional regulation, inositol metabolism and tRNASerCGA abundance (Zhao et al., 2022).
In sum, the Yeast 2.0 project will produce a cell with ~8% less DNA and a remarkable ~1.1 Mb of alterations relative to wild type S. cerevisiae. This grand scale genome engineering not only maintained the fitness of the organism, it also encoded new features in the yeast genome, such as the genome SCRaMbLE (Synthetic Chromosome Rearrangement and Modification by LoxPsym-mediated Evolution), which enables genome restricting via inducible evolution (Dymond and Boeke, 2012), that are facilitating the understanding of both yeast and eukaryotic biology as well (Richardson et al., 2017; Zhang et al., 2020). A planned Yeast-3.0 project would address questions of how many genes can be deleted and how optimal is the current gene organization (Dai et al., 2020). The methods developed for grand scale yeast chromosome engineering by this massive project will be vital to future efforts to build synthetic or artificial chromosomes for higher eukaryotes first in yeast before booting them up in their intended host. This will be discussed further below.
Why are there so few bacterial and eukaryotic Synthetic Genomics efforts?
Based on the reaction in the scientific community and beyond to the construction by the JCVI of bacteria with synthetic genomes, i.e. JCVI-syn1.0 in 2010 and minimal cell JCVI-3.0 in 2016, and to the Yeast 2.0 project, a wave of building microbes with synthetic genomes that might address a variety of basic research and applied problems was predicted (Cohan, 2010; Elowitz and Lim, 2010; Fritz et al., 2010; Service, 2016). That has not happened, yet at least; although, there are reports of three other bacterial genome synthesis projects.
The reasons for this come down to need and cost. The JCVI’s mycoplasma efforts were driven by a need to construct a minimal bacterial cell using naturally near minimal organisms for which few genetic tools existed. The Synthetic Genomics approach was the best option. The Cambridge University and Harvard University E. coli projects rebuilt the most widely used bacterium in the biotechnology industry so that the recoded E. coli could be used for production of proteins containing multiple non-standard amino acids and so that the organisms would be resistant to bacteriophage infection. The utility and value of the E. coli strains with synthetic genomes is obvious. The goal of the ETH Zürich Caulobacter effort was to develop methods that facilitate synthesis of genomes and other large DNA molecules using a widely used model organism. All of these bacterial Synthetic Genomics were slow, expensive efforts. The same is true for the Yeast 2.0 effort. There could be commercial entities that without publicizing the work constructed or are constructing bacteria, yeasts or algae with synthetic genomes to produce molecules that could not be made otherwise. On the other hand, there have been numerous publications describing computational approaches to design recoded and minimal genomes as well as laboratory approaches that may facilitate grand scale genome reconstruction and methods to replace large sections of native bacterial genomes with synthetic versions (Krishnakumar et al., 2014; Kuznetsov et al., 2017; Lamoureux et al., 2020; Lau et al., 2017; Libicher et al., 2020; Rees-Garbutt et al., 2020; Yoneji et al., 2021).
It may be that until the cost of microbial genome synthesis significantly decreases and methods to boot up synthetic genomes become as rapid as the genome transplantation process, production of bacteria, yeasts, fungi, and algae with synthetic genomes will be rare. One can imagine a time when genome synthesis costs are so low that a researcher might computationally design thousands or millions of bacterial genomes in an effort to build an organism optimized for a specific purpose and a DNA foundry would synthesize and deliver them in a few days. These would be booted up using a high throughput version of genome transplantation and the organisms best able to perform a desired task isolated.
The cost of DNA synthesis continues to decline (see the next section of this review). For making synthetic bacteria the likely bottleneck to achieving such a future is our current inability to boot up a complete bacterial genome for any species that is not a mycoplasma. The methods used to build the recoded E. coli genomes by iteratively exchanging as many as fifty synthetic sub-genomic DNA molecules with their natural counterparts would not be amenable to high throughput efforts. Thus the requirement for a high throughput approach to boot up a complete bacterial genomes. Grand scale engineering of eukaryotic microbial genomes, such as was done for yeast, will not likely need to swap in new altered ~50 kb synthetic DNA chromosomal regions for native sequences as was done for E. coli. With rare exceptions, eukaryotic genomes are comprised of multiple chromosomes. Methods that would enable installation of new synthetic chromosomes into eukaryotic microbes via transformation and/or conjugation have been demonstrated for several species. Once a new chromosome or chromosomes are installed, CRISPR methods could be used to destroy the native chromosomes the new synthetic ones are replacing.
Synthetic Genomics using eukaryotes: Animal & plant synthetic artificial chromosomes
As a sort of follow up to the Yeast 2.0 project, a cohort of synthetic biologists founded the Genome Project Write, which will focus on what is coming up next in Synthetic Genomics. Among its ambitions is driving a reduction in the costs of synthesizing and booting up large genomes from higher eukaryotes by 1000X by 2029 (Boeke et al., 2016). At present, it is not possible to synthesize and boot up large chromosomes of higher eukaryotes; however, Synthetic Genomics is having an impact on eukaryotes in work with synthetic mammalian (including human) and plant artificial chromosomes.
Mammalian artificial chromosomes could be used to build cell-based anticancer therapeutics (Kouprina et al., 2018) or alter the genomes of animals so that they produce humanized organs or pharmaceuticals. Plant artificial chromosomes could enable improved foods or plants with new metabolic pathways requiring large numbers of added genes (Birchler and Swyers, 2020; Dawe, 2020; Jakubiec et al., 2021). Until very recently, most artificial chromosome studies in higher eukaryotes were top down approaches where small natural chromosomes were whittled down using telomere-associated chromosome fragmentation in cells that would produce mini-chromosomes, via homologous recombination, that contain a natural centromere. For human artificial chromosomes (HACs), this approach has been used on at least five of the smallest chromosomes to produce linear HACs from 0.5 kb to 10 Mb. These are mitotically stable so long as the HAC size stays above ~300 kb (Kouprina et al., 2018).
More recently, bottom up approaches to create fully synthetic mammalian artificial chromosomes are making advances. Critical issues for this technology are the construction of functional synthetic centromeres and enabling Mendelian inheritance of the synthetic chromosomes via meiotic transmission. HACs with synthetic centromeres containing alphoid DNA arrays (repetitive tandem repeats in centromeric DNA) harboring binding sites for the DNA sequence-specific binding protein CENP-B, which serve as centromeres, have been created primarily in the laboratory of Vladimir Larionov and Natalay Kouprina at the United States National Cancer Institute (Kouprina et al., 2018; Lee et al., 2021; Sinenko et al., 2018). In a different approach to constructing synthetic HACs, Larionov and Ben Black at the University of Pennsylvania used what they termed a “CENP-A nucleosome seeding strategy” to create mitotically stable HACs that did not contain repetitive centromeric DNA characteristic of both natural eukaryotic chromosomes and the previously described Larionov lab HACs (Logsdon et al., 2019; Yang et al., 2020). Both the Larionov and Black approaches involve construction of the HACs either as yeast or bacterial artificial chromosomes.
Meiotic transmission of synthetic chromosomes is perhaps a more difficult problem. We are not aware of any attempts to create synthetic mammalian chromosomes that truly function as chromosomes in meiosis. Studies using whittled down natural plant chromosomes have shown that small chromosomes do not pair properly in meiosis (Birchler et al., 2016; Han et al., 2007). A possible solution for this is to design chromosomes in a way to promote recombination of sister chromatids (Dawe, 2020).
How large a chromosome is needed and can be synthesized? At present most of mammalian and plant artificial chromosome projects that do not add new DNA sequences to existing chromosomes build DNAs that are smaller than 1 Mb. Given that for many years, the yeast artificial chromosome (YAC) community suspected the maximum possible YAC size might be around two million base pairs. This was based on the largest reported YAC being only 2.3 Mb (Marschall et al., 1999). That concern, which could have made construction of animal and plant artificial chromosomes larger than a few megabases was recently eliminated. In 2018 a yeast strain with a single 12 Mb chromosome comprised of all 16 linear yeast chromosomes was reported (Shao et al., 2018). This suggests that yeast could be used to construct mammalian and plant artificial chromosomes comprised of a tiny amount of yeast sequence and perhaps hundreds of higher eukaryotic genes could be constructed in yeast.
These shuttle chromosomes that might be larger than 10 Mb could then be gently transferred, so as not break the artificial chromosome, to higher eukaryotes by fusing yeast spheroplasts with the target cell using polyethylene glycol (PEG) to mediate the fusion (Brown et al., 2017; Brown and Glass, 2020). Indeed, transfer of yeast artificial chromosomes to mammalian cells using PEG has been used for decades, but the process was very inefficient. The JCVI’s David Brown improved the efficiency of the transfer more than 100X by using colchicine to arrest the mammalian cell cycle at a point when the nuclear membrane was degraded. Thus the DNA does not have to traverse any cell membranes during the transfer process (Brown et al., 2017). These sort of advances may make it much more practical to construct plant or mammalian artificial chromosomes as larger as 10 Mb in yeast and then move them into target cells without shearing the chromosomes by pipetting.
Agricultural research is growing more interested in employing plant artificial chromosomes (often called mini-chromosomes) to add functions to plants (Birchler and Swyers, 2020; Dawe, 2020; Gaeta et al., 2012; Jakubiec et al., 2021). As with mammalian artificial chromosomes, efforts in plants employ partially synthetic approaches whereby new DNA is added to small existing natural chromosomes that have been whittled down to produce mini-chromosomes. More relevant to this review are fully synthetic approaches. The most important work involves nuclear chromosomes; although, in some cases, this involves replacing chloroplast genomes with new synthetic genomes (Frangedakis et al., 2021). Many of the approaches being developed for mammalian chromosomes are also in use with plants. The problems of building and booting up synthetic centromeres and enabling meiotic artificial chromosome inheritance are similar. While it is reasonable to construct synthetic plant chromosomes in yeast as is being done in mammalian systems, the difficulty in transferring large DNA molecules across plant cell walls has slowed progress.
As more groups are focusing on development of animal and plant artificial chromosomes as a method for grand scale alteration of the genetic and metabolic capacities of organisms, progress will accelerate. Artificial chromosomes smaller than 1 Mb are often problematic because of instability; however it seems clear that chromosomes much larger than that can be constructed in yeast and transferred to higher eukaryotic cells. Construction of synthetic centromeres is advancing and will likely continue. Mendelian inheritance of artificial chromosomes will be a challenging problem in part because of our limited understanding of what sequences and chromatin structures are critical to the effective inheritance of chromosomes. Plus there is much to learn about turning a naked DNA molecule that has a natural chromosome sequence into a functional chromosome. Once again, our capacity to synthesize megabase plus sized DNA molecules exceeds our ability to design eukaryotic chromosomes that will do what we want. While there is a long way to go, ongoing work by many groups using animal and plant synthetic artificial chromosomes is generating knowledge about chromosome biology in higher eukaryotes that will move science towards more effective grand scale engineering to solve human problems.
Emerging technologies that could revolutionize biology: Advances in DNA synthesis
The minimal bacterial cell project was a multi-year project largely due to the rate limitations of DNA synthesis. The JCVI could only make one megabase genome at a time to test the insertion or deletion of genes or gene cassettes for viability. If they could have made ten or one hundred different chromosomes, they would have reached the correct answer to what was needed for a viable cell much faster. New technology has been slow in coming but recent breakthroughs using semiconductor chips may provide the key to the future of the synthetic genome field. Avery Digital Data is one of the developers of this new technology using semiconductor chips for DNA synthesis. The JCVI and Avery Digital Data have plans to synthesize a complete eucaryote genome. This scale of Synthetic Genomics would not be feasible without this new technology.
Economical synthesis of very large numbers of oligonucleotides is fundamental for searching genome variation space in synthetic biology applications. To extend synthesis scales to the practical generation of billions to trillions of oligonucleotides, one emerging approach is to transfer the classical phosphoramidite synthesis chemistry onto a highly scalable semiconductor chip device—- ideally a standard complementary metal oxide semiconductor (CMOS) chip, fabricated in existing commercial foundries—having up to billions of independent oligo synthesis sites on a single chip. Early work by Ed Southern demonstrated the ability to drive DNA synthesis with electrochemical reactions (Egeland and Southern, 2005). Recent work by Microsoft has shown this approach can be scaled to the extremes of nanoscale electrodes, for future DNA data storage applications (Nguyen et al., 2021). The latest work in this area, by Avery Digital Data and Drew Hall’s Biosensors & Bioelectronics Group at the University of California at San Diego, demonstrates a fully integrated platform for scalable DNA synthesis in bioengineering, consisting of a scalable CMOS chip device, combined with a CMOS-compatible electrochemistry (Merriman, 2022). This platform uses electrochemical acid generation on micro-electrodes, from a novel hydroquinone solution, to provide a localized acid concentration that drives the deprotection reaction step of the phosphoramidite synthesis chemistry (Figure 4A). This low-voltage chemistry is compatible with deployment on electronic CMOS chips (Hall et al., 2022). Each chip would contain more than 100 million oligonucleotide synthesis sites and each site can support synthesis of ~100,000 copies of a specified oligonucleotide (Figure 4B). Synthesis of up to 100-mers has been demonstrated (Figure 4C). At high volume production, circa 2022, the cost of each oligonucleotide synthesis chip would be approximately $25 (Merriman, 2022). Thus, this first step of putting DNA synthesis “on chip” provides a path to greatly enhanced scales of oligo production—and ultimately gene/genome assembly—for synthetic biology.
Figure 4. DNA synthesis by electrochemistry.
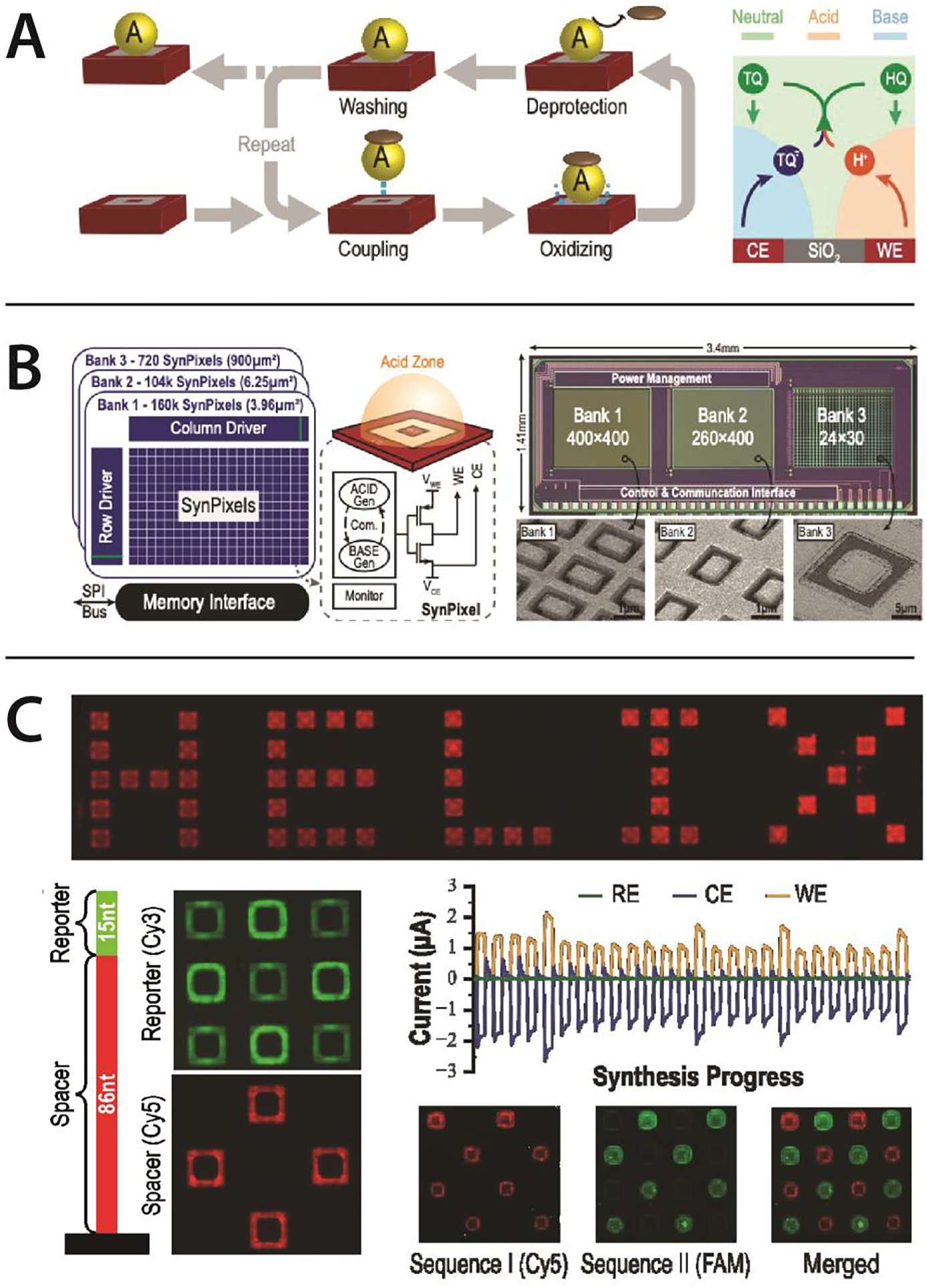
(A) (Left) Cycle of synthesis, illustrated adding a phosphoramidite to a site: deprotection is driven by localized acid generation at the site. (Right) Localized Acid for deprotection, achieved by decomposition of Hydroquinone (HQ) to release H+ acid at the local Working Electrode (WE), and active removal of acid by recombination with a cognate base, oxidized tetrachloro-1,4-benzoquinone (TQ) generated at the local Counter Electrode (CE). (B) DNA synthesis chip. (Left) A CMOS chip device to drive on-chip DNA synthesis. The chip has three sub-arrays of synthesis pixels (SynPixels) of different sizes to illustrate scalability: Banks 1—3 have pixels with footprint (in microns) 2 × 2, 2 × 3 and 30 × 30 respectively. Each array is controlled by row and column driver circuits, which program the pixels for activation, and provide connection to peripheral current monitoring circuits to monitor the electrochemical processes. (Middle) Voltage control of the central Working Electrode (WE) for acid generation and surrounding Counter Electrode (CE) for base generation, is controlled by a transistor switch circuit. (Right) Annotated microscopic image of the CMOS chip die, showing chip size and subarray dimensions. Insets show electron microscope images of the central Platinum WE and common peripheral CE. (C) On-chip scalable 100-mer synthesis. (Upper) Example of localized, controlled synthesis of oligos on the pixel array, spelling out “HELIX”, with synthesis visualized via a fluorescent microscope image of the synthesized oligos labeled by hybridizing to a fluorescently labeled complementary oligo. (Lower Left) Structure of the 100-mer oligo: 86 nucleotide (nt) poly-T and 15 nt complex sequence. Signals from red (Cy5) and green (FAM) labelling oligos hybridized to these segments are shown. Oligo synthesis is seen to be primarily in the annular silicon surface area between the central platinum electrode surrounding platinum counter electrode. (Lower Right) Example of checkerboard pattern synthesis of two different 15 nt sequences, illustrating the ability to sequence independent sequences at each site, along with the current versus time observed during the 30 cycles of synthesis, showing the net electrochemical currents drawn on the array by the Working Electrodes (WE) and Counter Electrodes (CE) (the information and images are through personal communication with Barry Merriman of Avery Digital Data).
For decades there has only been one method for synthesis of DNA, the phosphoramidite nucleoside method. While widely used, it has drawbacks such as a maximum effective synthesis of only ~200 base oligonucleotides and the production of hazardous wastes. Non-templated enzymatic synthesis of oligonucleotides using terminal deoxynucleotidyl transferase (TdT), which is the only known polymerase whose predominant activity is to add deoxynucleotide triphosphates (dNTPs) indiscriminately to the 3′ end of single-stranded DNA, has been proposed as an alternative conventional phosphoramidite chemistry. Enzymatic DNA synthesis offers several advantages over chemical synthesis. Longer oligonucleotides can likely be synthesized because of both the high specificity of enzymatic reactions and the mild biological conditions under which the aqueous polymerase reactions take place. This should reduce the formation of reactive side products that can lead to DNA damage like depurination. Enzymatic reactions will not generate hazardous wastes. Unfortunately a variety of technical issues, such as an inability of TdT to synthesize even small DNA hairpins, has kept enzymatic DNA synthesis from realization. Recently, a team led by Nathan Hillson and Jay Keasling has achieved two breakthroughs that may lead to a competitive enzymatic DNA synthesis technology. First, in their reaction scheme, they conjugate each TdT molecule to a single dNTP. The TdT then adds the dNTP to the 3’ end of an existing DNA primer. The TdT remains linked to the growing DNA stand so that the end of the DNA is inaccessible to other TdT-dNTP conjugates. At the end of that extension step the linkage between the TdT and newly added nucleotide is cleaved by the addition of β-mercaptoethanol thus allowing the next extension reaction. Keasling and Hillson’s team demonstrated that the TdT-dNTP conjugates can add a new nucleotide to the 3’ end of primer every 10–20 seconds, and that the reaction can be used to generate 10 base oligonucleotides (Palluk et al., 2018). The other advance worked around the problem of TdT polymerization being inhibited by DNA hairpins. To do this they optimized the divalent cation cofactor concentrations in the polymerization reaction and they remodeled the TdT to make it more thermostable so that the polymerization reactions could take place at higher temperatures where the hairpins would be less of an issue. These improvements, when combined with the aforementioned TdT-dNTP conjugate method, enabled dTTP addition onto the 3’ end of an 8 basepair guanine-cytosine hairpin (Barthel et al., 2020). In sum these advances make enzymatic non-templated oligonucleotide synthesis seem plausible for the first time in decades and may eventually lead to enzymatic oligonucleotide synthesizers.
Emerging technologies that could revolutionize biology: Humanized pig genome and organ xenotransplantation.
One of the potentially most important medical breakthroughs came earlier this year when a 57-year-old male patient received a genetically-modified pig heart transplant at the University of Maryland Medical Center (UMMC) (Reardon, 2022). The surgery was a “world-first” and deemed the patient’s only chance for survival after he was declared unsuitable for a human donor transplant or an artificial heart pump. (Figure 5). The recipient lived for two months, which is twice as long as the first human to human heart transplant (Rabin, 2022). To make this pig to human xenotransplantation possible, ten genetic changes were made in the pig genome including addition of human genes, and knockouts and alterations of pig genes. Two weeks after the pig heart transplant, the first pig kidney transplants were performed (Porrett et al., 2022). As this work is perfected the human impact will be enormous as there are over 100,000 patients in the USA on the transplant wait list.
Figure 5. World’s first pig to human heart transplant performed on January 7, 2022.
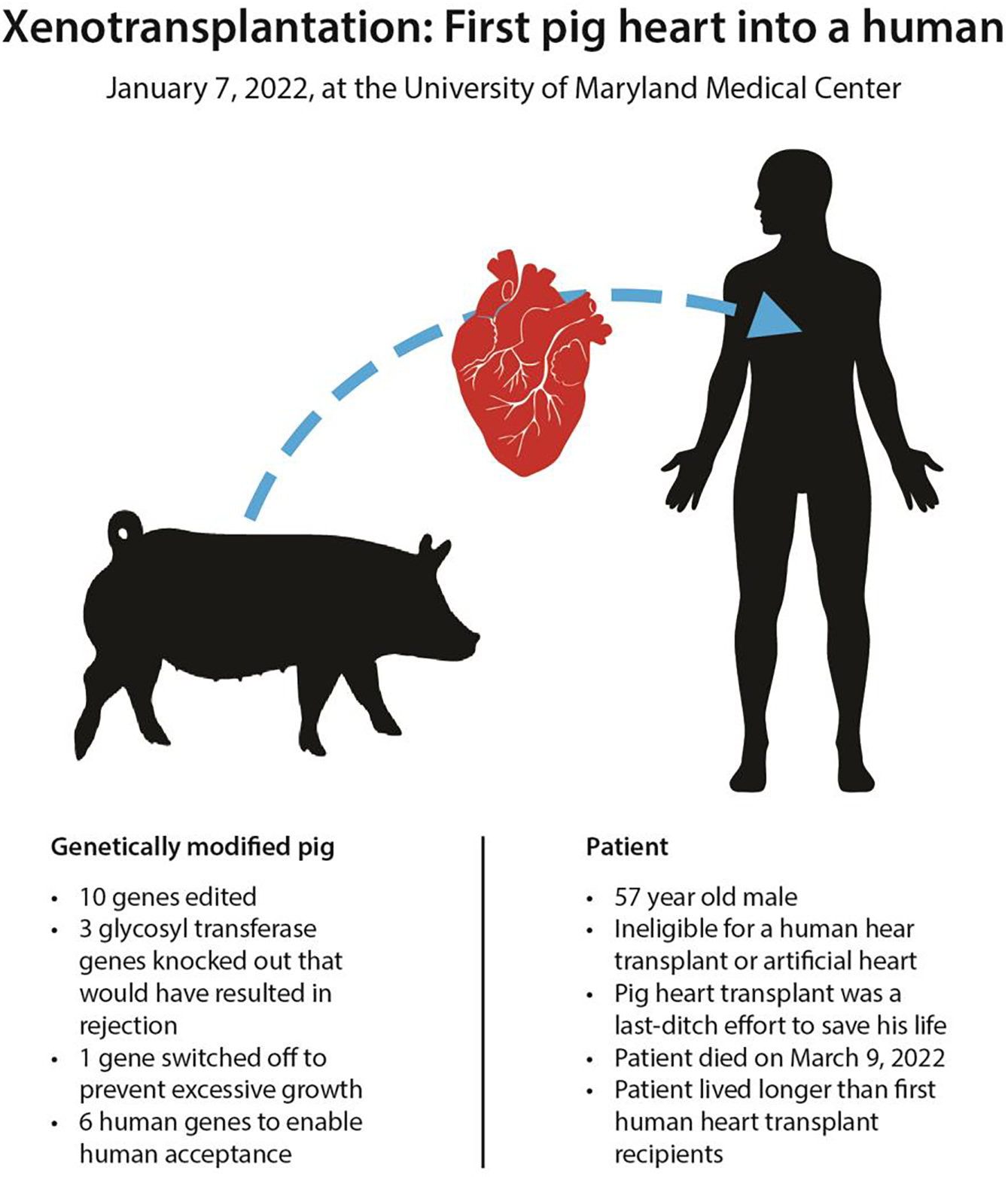
The patient, Robert Bennett was not a candidate for a human heart transplant or an artificial heart. The xenotransplantation was a compassionate-use case. Bennett lived for two months.
Xenotransplantation has long been a dream of medicine. Xenotransplantation experiments were described as early as the seventeenth century. Post-World War II xenotransplantation efforts shifted to the use of organs from primates in human patients. In the 1960s, experiments by various teams showed that while it was technically possible to transplant animal organs into humans, there were still too many clinical challenges at that time for the approach to be viable. Significant advancements have been made in recent years in understanding the molecular mechanisms of xeno-rejection responses (Cooper, 2012; Stevens, 2020). These have been made possible due to the advances in Synthetic Genomics and fundamental genomics, which while essential are only a small part of the overall transplantation process.
The immune system is very complex and our response to foreign tissues is far from simple. For example unlike in humans where vascular endothelium expresses the ABH blood group antigens, the pig’s vascular endothelium expresses a galactose oligosaccharide, galactose-α(1,3)-galactose (Gal). The presence of Gal in the pig and its absence in humans, has proved a major challenge due to the generation of anti-Gal antibodies, which cause acute rejection of the pig organ (Phelps et al., 2003). Additionally, the N-glycolyl neuraminic acid (Neu5Gc or Hanganutziu-Deicher antigen) is also a major pig xenoantigen, given that humans have Nue5Gc antibodies. These are two of three (unlinked) glycosyl transferases that have been deleted from the pig genome to reduce immune reactions to porcine tissue (Tector et al., 2020).
A second way of reducing human antibody binding to pig antigens is to provide the pig with increased resistance to human complement-mediated injury. This has been achieved by inserting into the pig genes one or more human complement-regulatory proteins, such as CD55 or CD46. The combination of GTKO and expression of CD46 and/or CD55 has made hyperacute rejection a rarity in experimental organ xenotransplantation studies. While this sounds straight forward it was far from simple. CRISPRs for example have many off-target effects so while you think you are targeting one gene you may be targeting many. Also random integration of human genes into the pig genome could cause major biological disruptions. As a result, the genetics team at Synthetic Genomics, Inc. lead by Sean Stevens who together with Martine Rothblatt and Craig Venter took a unique approach (this effort was later transferred to United Therapeutics, Inc.). They started with a highly accurate human genome and added a new diploid pig genome from the line used for the transplantation. The pig genome was sequenced to very high coverage to ensure accuracy. The accurate genome allowed for highly specific targeted gene knockouts and for the insertion of cassettes of human genes in specific sites without disrupting genome functions (Stevens, 2020).
Biosafety and bioethical concerns.
Bioethical and biosecurity issues have been part of the Synthetic Genomics field from the outset. There have been multiple committees, boards and review teams discussing essentially every possible aspect of issues associated with synthetic DNA applications (Addressing Biosecurity Concerns Related to the Synthesis of Select Agents, 2006; Synthetic biology - Ethical considerations, 2010; Biodefense in the Age of Synthetic Biology, 2018; Capurro et al., 2010; Carter et al., 2014; Cho et al., 1999; Craig et al., 2022; Garfinkel et al., 2007; Heavey, 2017; Issues and of, 2015; Kaebnick and Murray, 2013; National Academies of Sciences, 2017; Relman, 2009). Some of these reports offer action items and proposed regulation while others just raise the issues. Of greatest concern to the authors of this review are biosecurity issues and environmental release of synthetic organisms and viruses.
The ability to de novo synthesize DNA and assemble mega-base size constructs is clearly in the category of dual use technology. Clearly any virus including the large pox viruses that have been sequenced can be regenerated by DNA synthesis as can most bacterial pathogens. It is not therefore illogical to try to limit the access to DNA synthesis to legitimate researchers. Most reputable DNA synthesis companies screen all orders against the “A” list of pathogen agents. When Synthetic Genomics DNA, INC. (now Codex DNA) designed its DNA assembly robot multiple levels of security were built into the machine which block users from assembling non-approved pathogens (Boles et al., 2017). Oligonucleotides are provided by custom order in sealed cassettes that the machine can detect any alteration and will not proceed with assembly. None of these measures are fool proof as the reagents for benchtop assembly are readily available. Meaningful regulation is lacking due in no small part to the lack of knowledge and understanding of this field and its potential for good and harm. It is noteworthy that the United States Health and Human Services Department has proposed new policies on synthetic DNA that will lower the risk of dangerous toxins, viruses or bacteria being synthesized for nefarious intent by expanding current guidance to include a requirement for synthetic DNA providers to screen oligonucleotides for sequences of concern (Screening Framework Guidance for Providers of Synthetic Double-Stranded DNA, 2022).
We are strongly opposed to any environmental release of synthetic organisms and even sharing between laboratories needs to be done carefully. With the first synthetic genome/cell we introduced the watermarking of synthetic DNA with a code that allows for the entire English alphabet and standard punctuation (Gibson et al., 2010). We included authors names and institutions so that no one would confuse our cell with a naturally existing organism. Other measures to prevent escape of synthetic organisms from the laboratory could be engineering the organism to need metabolites not found in nature. Such simple measures as well as biological kill switches can prevent unintended environmental consequences from occurring.
Conclusions:
Synthetic genomics is still a young field that is still seeing a limited number of efforts. However synthetic viruses have already altered vaccine design and production starting with the synthetic flu virus and leading to rapid development of RNA based Covid vaccines. Synthetic cells will hopefully become more feasible with new DNA synthetic approaches. If thousands of versions can be made and tested simultaneously the field will move forward at least an order of magnitude faster. We have yet to see an actual synthetic DNA version of a eukaryote cell. With all the regulation at the gene and genome level design will depend heavily on trial and error and the ability to test rapidly multiple versions.
Genome design and construction could lead to a new industrial revolution for food and chemical production. It will be key to develop cellular mechanisms to limit the viability of synthetic organisms to laboratory and production facilities. Similarly, alteration of the genetic code of synthetic organisms can eliminate concerns that potentially dangerous genes in the synthetic strain will be horizontally transferred to natural organisms and expressed. Water marking the genetic code should be a requirement for any synthetic organisms to avoid confusion of evolution analysis.
Synthetic genomes are already saving lives through new vaccines and now humanized pig organs for transplantation. The future of this field will clearly be exciting to see unfold.
ACKNOWLEDGMENTS
The authors thank Matt LaPointe for assistance preparing the manuscript figures. S. V. was supported by the National Institutes of Health (Grant numbers R01AI137365, R03AI146632) and the International Development Research Centre (Grant number 109212). J.I.G. and C.A.H. were supported by the National Science Foundation (MCB 1840320 and MCB 1818344). J.C.V., J.I.G., C.A.H. and S.V. were supported by the J. Craig Venter Institute.
Footnotes
DECLARATION OF INTERESTS
- US20150344837A1 Minimal bacterial genome
- US20210254046A1 Generation of synthetic Genomes
- US20160177322A1 Methods for cloning and manipulating genomes
- WO2011109031A1 Methods for cloning and manipulating genomes
- US20140274806A1 Influenza virus reassortment
- AU2009214435C1 Methods for in vitro joining and combinatorial assembly of nucleic acid molecules
- US20180340165A Method of nucleic acid cassette assembly
- AU2018201085B2 Digital to biological converter
- US9434974B2 Installation of genomes or partial genomes into cells or cell-like systems
- EP2207899B1 Assembly of large nucleic acids
- WO2008144192A9 Methods of genome installation in a recipient host cell
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Addressing Biosecurity Concerns Related to the Synthesis of Select Agents. (2006). https://osp.od.nih.gov/wp-content/uploads/2013/06/Final_NSABB_Report_on_Synthetic_Genomics.pdf.
- Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, et al. (2014). Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58. 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel S, Palluk S, Hillson NJ, Keasling JD, and Arlow DH (2020). Enhancing Terminal Deoxynucleotidyl Transferase Activity on Substrates with 3’ Terminal Structures for Enzymatic De Novo DNA Synthesis. Genes (Basel) 11. 10.3390/genes11010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, Pickles RJ, Corti D, Johnston RE, Baric RS, and Denison MR (2008). Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A 105, 19944–19949. 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biodefense in the Age of Synthetic Biology. (2018). (National Academies Press; (US: )). 10.17226/24890. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Graham ND, Swyers NC, Cody JP, and McCaw ME (2016). Plant minichromosomes. Curr Opin Biotechnol 37, 135–142. 10.1016/j.copbio.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Birchler JA, and Swyers NC (2020). Engineered minichromosomes in plants. Exp Cell Res 388, 111852. 10.1016/j.yexcr.2020.111852. [DOI] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, and Rice CM (2000). Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y, Carlson R, Chakravarti A, Cornish VW, Holt L, et al. (2016). GENOME ENGINEERING. The Genome Project-Write. Science 353, 126–127. 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- Boles KS, Kannan K, Gill J, Felderman M, Gouvis H, Hubby B, Kamrud KI, Venter JC, and Gibson DG (2017). Digital-to-biological converter for on-demand production of biologics. Nature biotechnology 35, 672–675. 10.1038/nbt.3859. [DOI] [PubMed] [Google Scholar]
- Brown DM, Chan YA, Desai PJ, Grzesik P, Oldfield LM, Vashee S, Way JC, Silver PA, and Glass JI (2017). Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic acids research 45, e50. 10.1093/nar/gkw1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, and Glass JI (2020). Technology used to build and transfer mammalian chromosomes. Exp Cell Res, 111851. 10.1016/j.yexcr.2020.111851. [DOI] [PubMed] [Google Scholar]
- Capurro R, Kinderlerer J, Martinho de Silva P, and Rosell PP (2010). Ethics of Synthetic Biology. European Group on Ethics in Science and New Technologies to the European Commission. https://www.coe.int/t/dg3/healthbioethic/cometh/ege/20091118%20finalSB%20_2_%20MP.pdf
- Carter SR, Rodemeyer M, Garfinkel MS, and Friedman RM (2014). Synthetic Biology and the U.S. Biotechnology Regulatory System: Challenges and Options. J. Craig Venter Institute. https://www.jcvi.org/sites/default/files/assets/projects/synthetic-biology-and-the-us-regulatory-system/full-report.pdf. [Google Scholar]
- Cello J, Paul AV, and Wimmer E (2002). Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297, 1016–1018. 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- Chan LY, Kosuri S, and Endy D (2005). Refactoring bacteriophage T7. Molecular systems biology 1, 2005 0018. 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW (2017). Expanding and reprogramming the genetic code. Nature 550, 53–60. 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- Cho MK, Magnus D, Caplan AL, and McGee D (1999). Policy forum: genetics. Ethical considerations in synthesizing a minimal genome. Science 286, 2087, 2089–2090. [DOI] [PubMed] [Google Scholar]
- Cohan FM (2010). Synthetic biology: now that we’re creators, what should we create? Curr Biol 20, R675–677. 10.1016/j.cub.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Cooper DK (2012). A brief history of cross-species organ transplantation. Proc (Bayl Univ Med Cent) 25, 49–57. 10.1080/08998280.2012.11928783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig W, Sara R, and Moronta-Barrios F (2022). Synthetic Biology. https://www.cbd.int/doc/publications/cbd-ts-100-en.pdf.
- Dai J, Boeke JD, Luo Z, Jiang S, and Cai Y (2020). Sc3.0: revamping and minimizing the yeast genome. Genome Biol 21, 205. 10.1186/s13059-020-02130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RK (2020). Charting the path to fully synthetic plant chromosomes. Exp Cell Res 390, 111951. 10.1016/j.yexcr.2020.111951. [DOI] [PubMed] [Google Scholar]
- Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, Alperovich N, Barro M, Brown DM, Craig S, et al. (2013). Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Science translational medicine 5, 185ra168. 10.1126/scitranslmed.3006368. [DOI] [PubMed] [Google Scholar]
- Dymond J, and Boeke J (2012). The Saccharomyces cerevisiae SCRaMbLE system and genome minimization. Bioeng Bugs 3, 168–171. 10.4161/bbug.19543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, Blake WJ, Schwerzmann JW, Dai J, Lindstrom DL, et al. (2011). Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477, 471–476. 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeland RD, and Southern EM (2005). Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication. Nucleic acids research 33, e125. 10.1093/nar/gni117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M, and Lim WA (2010). Build life to understand it. Nature 468, 889–890. 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami M, Luytjes W, Krystal M, and Palese P (1990). Introduction of site-specific mutations into the genome of influenza virus. Proceedings of the National Academy of Sciences of the United States of America 87, 3802–3805. 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, and et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512. [DOI] [PubMed] [Google Scholar]
- Frangedakis E, Guzman-Chavez F, Rebmann M, Markel K, Yu Y, Perraki A, Tse SW, Liu Y, Rever J, Sauret-Gueto S, et al. (2021). Construction of DNA Tools for Hyperexpression in Marchantia Chloroplasts. ACS Synth Biol 10, 1651–1666. 10.1021/acssynbio.0c00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. (1995). The minimal gene complement of Mycoplasma genitalium. Science 270, 397–403. [DOI] [PubMed] [Google Scholar]
- Fredens J, Wang K, de la Torre D, Funke LFH, Robertson WE, Christova Y, Chia T, Schmied WH, Dunkelmann DL, Beranek V, et al. (2019). Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518. 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz BR, Timmerman LE, Daringer NM, Leonard JN, and Jewett MC (2010). Biology by design: from top to bottom and back. J Biomed Biotechnol 2010, 232016. 10.1155/2010/232016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Masonbrink RE, Krishnaswamy L, Zhao C, and Birchler JA (2012). Synthetic chromosome platforms in plants. Annu Rev Plant Biol 63, 307–330. 10.1146/annurev-arplant-042110-103924. [DOI] [PubMed] [Google Scholar]
- Garfinkel MS, Endy D, Epstein GL, and Friedman RM (2007). Synthetic genomics | options for governance. Biosecur Bioterror 5, 359–362. 10.1089/bsp.2007.0923. [DOI] [PubMed] [Google Scholar]
- Gary EN, and Weiner DB (2020). DNA vaccines: prime time is now. Curr Opin Immunol 65, 21–27. 10.1016/j.coi.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, Cu Y, Beard CW, Brito LA, Krucker T, et al. (2012). Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A 109, 14604–14609. 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. (2008a). Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220. 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, and Hutchison CA 3rd (2008b). One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proceedings of the National Academy of Sciences of the United States of America 105, 20404–20409. 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. (2010). Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56. 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA 3rd, Smith HO, and Venter JC (2006). Essential genes of a minimal bacterium. Proceedings of the National Academy of Sciences of the United States of America 103, 425–430. 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Merryman C, Wise KS, Hutchison CA 3rd, and Smith HO (2017). Minimal Cells-Real and Imagined. Cold Spring Harb Perspect Biol 9. 10.1101/cshperspect.a023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golob JL, Lugogo N, Lauring AS, and Lok AS (2021). SARS-CoV-2 vaccines: a triumph of science and collaboration. JCI Insight 6. 10.1172/jci.insight.149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Manabhan NA, Choi C, Zheng L, Pan PP, Fuller CW, Padayatti PP, Gardner C, Gebhardt D, Majzik Z, et al. (2022). A CMOS Molecular Electronics Chip for Single-Molecule Biosensing. 2022 IEEE International Solid- State Circuits Conference (ISSCC), 1–3. 10.1109/ISSCC42614.2022.9731770. [DOI] [Google Scholar]
- Han F, Gao Z, Yu W, and Birchler JA (2007). Minichromosome analysis of chromosome pairing, disjunction, and sister chromatid cohesion in maize. Plant Cell 19, 3853–3863. 10.1105/tpc.107.055905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavey P (2017). Synthetic Biology: The Response of the Commission of the (Catholic) Bishops’ Conferences of the European Community. Camb Q Healthc Ethics 26, 257–266. 10.1017/S0963180116000852. [DOI] [PubMed] [Google Scholar]
- Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, Otten GR, Brazzoli M, Buccato S, Bonci A, et al. (2013). Rapidly produced SAM((R)) vaccine against H7N9 influenza is immunogenic in mice. Emerg Microbes Infect 2, e52. 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, et al. (2016). Design and synthesis of a minimal bacterial genome. Science 351, aad6253. 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, and Venter JC (1999). Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. (2011). Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333, 348–353. 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presidential Commission for the Study of Bioethical Issues. (2015). New Directions the Ethics of Synthetic Biology and Emerging Technologies (CreateSpace Independent Publishing Platform; ). https://bioethicsarchive.georgetown.edu/pcsbi/synthetic-biology-report.html [Google Scholar]
- Jakubiec A, Sarokina A, Choinard S, Vlad F, Malcuit I, and Sorokin AP (2021). Replicating minichromosomes as a new tool for plastid genome engineering. Nat Plants 7, 932–941. 10.1038/s41477-021-00940-y. [DOI] [PubMed] [Google Scholar]
- Kaebnick GE, and Murray TH, eds. (2013). Synthetic Biology and Morality: Artificial Life and the Bounds of Nature (MIT Press; ). 10.7551/mitpress/9780262019392.001.0001. [DOI] [Google Scholar]
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, and Santi DV (2004). Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proceedings of the National Academy of Sciences of the United States of America 101, 15573–15578. 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Petrov N, Molina O, Liskovykh M, Pesenti E, Ohzeki JI, Masumoto H, Earnshaw WC, and Larionov V (2018). Human Artificial Chromosome with Regulated Centromere: A Tool for Genome and Cancer Studies. ACS synthetic biology 7, 1974–1989. 10.1021/acssynbio.8b00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Grose C, Haft DH, Zaveri J, Alperovich N, Gibson DG, Merryman C, and Glass JI (2014). Simultaneous non-contiguous deletions using large synthetic DNA and site-specific recombinases. Nucleic acids research 42, e111. 1 10.1093/nar/gku509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov G, Goodman DB, Filsinger GT, Landon M, Rohland N, Aach J, Lajoie MJ, and Church GM (2017). Optimizing complex phenotypes through model-guided multiplex genome engineering. Genome Biol 18, 100. 10.1186/s13059-017-1217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, et al. (2013). Genomically recoded organisms expand biological functions. Science 342, 357–360. 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux CR, Choudhary KS, King ZA, Sandberg TE, Gao Y, Sastry AV, Phaneuf PV, Choe D, Cho BK, and Palsson BO (2020). The Bitome: digitized genomic features reveal fundamental genome organization. Nucleic Acids Res 48, 10157–10163. 10.1093/nar/gkaa774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA 3rd, Smith HO, and Venter JC (2007). Genome transplantation in bacteria: changing one species to another. Science 317, 632–638. 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- Lartigue C, Vashee S, Algire MA, Chuang RY, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, et al. (2009). Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325, 1693–1696. 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- Lau YH, Stirling F, Kuo J, Karrenbelt MAP, Chan YA, Riesselman A, Horton CA, Schafer E, Lips D, Weinstock MT, et al. (2017). Large-scale recoding of a bacterial genome by iterative recombineering of synthetic DNA. Nucleic acids research 45, 6971–6980. 10.1093/nar/gkx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Arun Kumar S, Jhan YY, and Bishop CJ (2018). Engineering DNA vaccines against infectious diseases. Acta Biomater 80, 31–47. 10.1016/j.actbio.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NCO, Petrov NS, Larionov V, and Kouprina N (2021). Assembly of Multiple Full-Size Genes or Genomic DNA Fragments on Human Artificial Chromosomes Using the Iterative Integration System. Curr Protoc 1, e316. 10.1002/cpz1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libicher K, Hornberger R, Heymann M, and Mutschler H (2020). In vitro self-replication and multicistronic expression of large synthetic genomes. Nat Commun 11, 904. 10.1038/s41467-020-14694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, and Black BE (2019). Human Artificial Chromosomes that Bypass Centromeric DNA. Cell 178, 624–639 e619. 10.1016/j.cell.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall P, Malik N, and Larin Z (1999). Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine. Gene therapy 6, 1634–1637. 10.1038/sj.gt.3300975. [DOI] [PubMed] [Google Scholar]
- Marshall E (2002). Genetics. Venter gets down to life’s basics. Science 298, 1701. 10.1126/science.298.5599.1701. [DOI] [PubMed] [Google Scholar]
- McAdams HH, and Shapiro L (2009). System-level design of bacterial cell cycle control. FEBS Lett 583, 3984–3991. 10.1016/j.febslet.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA, and Ruddle FH (1987). Toward a complete map of the human genome. Genomics 1, 103–106. 10.1016/0888-7543(87)90001-2. [DOI] [PubMed] [Google Scholar]
- Mitchell LA, Wang A, Stracquadanio G, Kuang Z, Wang X, Yang K, Richardson S, Martin JA, Zhao Y, Walker R, et al. (2017). Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science 355. 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- Morowitz HJ (1984). The completeness of molecular biology. Israel journal of medical sciences 20, 750–753. [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. (2017). Preparing for Future Products of Biotechnology (National Academies Press; (US: )). 10.17226/24605. [DOI] [PubMed] [Google Scholar]
- Nguyen BH, Takahashi CN, Gupta G, Smith JA, Rouse R, Berndt P, Yekhanin S, Ward DP, Ang SD, Garvan P, et al. (2021). Scaling DNA data storage with nanoscale electrode wells. Science advances 7, eabi6714. 10.1126/sciadv.abi6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce RS, Lederman S, and Evans DH (2018). Construction of an infectious horsepox virus vaccine from chemically synthesized DNA fragments. PLoS One 13, e0188453. 10.1371/journal.pone.0188453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield LM, Grzesik P, Voorhies AA, Alperovich N, MacMath D, Najera CD, Chandra DS, Prasad S, Noskov VN, Montague MG, et al. (2017). Genome-wide engineering of an infectious clone of herpes simplex virus type 1 using synthetic genomics assembly methods. Proceedings of the National Academy of Sciences of the United States of America 114, E8885–E8894. 10.1073/pnas.1700534114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, Zhou M, Singh K, Napolitano MG, Moosburner M, et al. (2016). Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822. 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- Palluk S, Arlow DH, de Rond T, Barthel S, Kang JS, Bector R, Baghdassarian HM, Truong AN, Kim PW, Singh AK, et al. (2018). De novo DNA synthesis using polymerase-nucleotide conjugates. Nature biotechnology 36, 645–650. 10.1038/nbt.4173. [DOI] [PubMed] [Google Scholar]
- Pattnaik AK, and Wertz GW (1990). Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol 64, 2948–2957. 10.1128/JVI.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CJ, Koike C, Vaught TD, Boone J, Wells KD, Chen SH, Ball S, Specht SM, Polejaeva IA, Monahan JA, et al. (2003). Production of alpha 1,3-galactosyltransferase-deficient pigs. Science 299, 411–414. 10.1126/science.1078942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrett PM, Orandi BJ, Kumar V, Houp J, Anderson D, Cozette Killian A, Hauptfeld-Dolejsek V, Martin DE, Macedon S, Budd N, et al. (2022). First clinical-grade porcine kidney xenotransplant using a human decedent model. Am J Transplant 22, 1037–1053. 10.1111/ajt.16930. [DOI] [PubMed] [Google Scholar]
- Pretorius IS, and Boeke JD (2018). Yeast 2.0-connecting the dots in the construction of the world’s first functional synthetic eukaryotic genome. FEMS Yeast Res 18. 10.1093/femsyr/foy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Xia F, Chen H, Cui B, Feng Y, Zhang P, Chen J, and Luo M (2021). A Guide to Nucleic Acid Vaccines in the Prevention and Treatment of Infectious Diseases and Cancers: From Basic Principles to Current Applications. Front Cell Dev Biol 9, 633776. 10.3389/fcell.2021.633776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin RC (2022). Patient in Groundbreaking Heart Transplant Dies. New York Times, 03/10/2022. [Google Scholar]
- Reardon S (2022). First pig-to-human heart transplant: what can scientists learn? Nature 601, 305–306. 10.1038/d41586-022-00111-9. [DOI] [PubMed] [Google Scholar]
- Rees-Garbutt J, Chalkley O, Landon S, Purcell O, Marucci L, and Grierson C (2020). Designing minimal genomes using whole-cell models. Nat Commun 11, 836. 10.1038/s41467-020-14545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D (2009). Addressing Biosecurity Concerns Related to Synthetic Biology. https://osp.od.nih.gov/wp-content/uploads/Relman-NSABB_Draft_Report_on_Synthetic_Biology.pdf.
- Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, Lee D, Huang CL, Chandrasegaran S, Cai Y, et al. (2017). Design of a synthetic yeast genome. Science 355, 1040–1044. 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- Rourke MF, Phelan A, and Lawson C (2020). Access and benefit-sharing following the synthesis of horsepox virus. Nat Biotechnol 38, 537–539. 10.1038/s41587-020-0518-z. [DOI] [PubMed] [Google Scholar]
- Sandbrink JB, and Shattock RJ (2020). RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front Immunol 11, 608460. 10.3389/fimmu.2020.608460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screening Framework Guidance for Providers of Synthetic Double-Stranded DNA. (2022). Federal Register, 5. https://www.phe.gov/preparedness/legal/guidance/syndna/documents/syndna-guidance.pdf [Google Scholar]
- Service RF (2016). SYNTHETIC BIOLOGY. Synthetic microbe has fewest genes, but many mysteries. Science 351, 1380–1381. 10.1126/science.351.6280.1380. [DOI] [PubMed] [Google Scholar]
- Shao Y, Lu N, Wu Z, Cai C, Wang S, Zhang LL, Zhou F, Xiao S, Liu L, Zeng X, et al. (2018). Creating a functional single-chromosome yeast. Nature 560, 331–335. 10.1038/s41586-018-0382-x. [DOI] [PubMed] [Google Scholar]
- Shen Y, Wang Y, Chen T, Gao F, Gong J, Abramczyk D, Walker R, Zhao H, Chen S, Liu W, et al. (2017). Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 355. 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinenko SA, Skvortsova EV, Liskovykh MA, Ponomartsev SV, Kuzmin AA, Khudiakov AA, Malashicheva AB, Alenina N, Larionov V, Kouprina N, and Tomilin AN (2018). Transfer of Synthetic Human Chromosome into Human Induced Pluripotent Stem Cells for Biomedical Applications. Cells 7. 10.3390/cells7120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA 3rd, Pfannkoch C, and Venter JC (2003). Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America 100, 15440–15445. 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S (2020). Genome Engineering for Xenotransplantation (IntechOpen; ). 10.5772/intechopen.84782. [DOI] [Google Scholar]
- Synthetic biology - Ethical considerations. (2010). https://www.ekah.admin.ch/inhalte/ekah-dateien/dokumentation/publikationen/e-Synthetische_Bio_Broschuere.pdf.
- Tector AJ, Mosser M, Tector M, and Bach JM (2020). The Possible Role of Anti-Neu5Gc as an Obstacle in Xenotransplantation. Front Immunol 11, 622. 10.3389/fimmu.2020.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Nhu Thao T, Labroussaa F, Ebert N, V’Kovski P, Stalder H, Portmann J, Kelly J, Steiner S, Holwerda M, Kratzel A, et al. (2020). Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582, 561–565. 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Vashee S, Stockwell TB, Alperovich N, Denisova EA, Gibson DG, Cady KC, Miller K, Kannan K, Malouli D, Crawford LB, et al. (2017). Cloning, Assembly, and Modification of the Primary Human Cytomegalovirus Isolate Toledo by Yeast-Based Transformation-Associated Recombination. mSphere 2. 10.1128/mSphereDirect.00331-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetz JE, Del Medico L, Wolfle A, Schachle P, Bucher Y, Appert D, Tschan F, Flores-Tinoco CE, van Kooten M, Guennoun R, et al. (2019). Chemical synthesis rewriting of a bacterial genome to achieve design flexibility and biological functionality. Proceedings of the National Academy of Sciences of the United States of America 116, 8070–8079. 10.1073/pnas.1818259116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, and Church GM (2009). Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898. 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Fredens J, Brunner SF, Kim SH, Chia T, and Chin JW (2016). Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64. 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz GW, Perepelitsa VP, and Ball LA (1998). Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proceedings of the National Academy of Sciences of the United States of America 95, 3501–3506. 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li BZ, Zhao M, Mitchell LA, Xie ZX, Lin QH, Wang X, Xiao WH, Wang Y, Zhou X, et al. (2017). Bug mapping and fitness testing of chemically synthesized chromosome X. Science 355. 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZX, Li BZ, Mitchell LA, Wu Y, Qi X, Jin Z, Jia B, Wang X, Zeng BX, Liu HM, et al. (2017). “Perfect” designer chromosome V and behavior of a ring derivative. Science 355. 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- Yang Y, Lampson MA, and Black BE (2020). Centromere identity and function put to use: construction and transfer of mammalian artificial chromosomes to animal models. Essays Biochem 64, 185–192. 10.1042/EBC20190071. [DOI] [PubMed] [Google Scholar]
- Yoneji T, Fujita H, Mukai T, and Su’etsugu M (2021). Grand scale genome manipulation via chromosome swapping in Escherichia coli programmed by three one megabase chromosomes. Nucleic Acids Res 49, 8407–8418. 10.1093/nar/gkab298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mitchell LA, Bader JS, and Boeke JD (2020). Synthetic Genomes. Annual review of biochemistry 89, 77–101. 10.1146/annurev-biochem-013118-110704. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhao G, Luo Z, Lin Y, Wang L, Guo Y, Wang A, Jiang S, Jiang Q, Gong J, et al. (2017). Engineering the ribosomal DNA in a megabase synthetic chromosome. Science 355. 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Coelho C, Hughes AL, Lazar-Stefanita L, Yang S, Brooks AN, Walker RSK, Zhang W, Lauer S, Hernandez C, et al. (2022). Debugging and consolidating multiple synthetic chromosomes reveals combinatorial genetic interactions. bioRxiv, 2022.2004.2011.486913. 10.1101/2022.04.11.486913. [DOI] [PubMed] [Google Scholar]


