Abstract
Flavonoids are phenolic compounds naturally found in plants and commonly consumed in diets. Herein, flavonoids were sequentially evaluated by a comparative in silico study associated with systematic literature search. This was followed by an in vitro study and enzyme inhibition assays against vital SARS‐CoV‐2 proteins including spike (S) protein, main protease (Mpro), RNA‐dependent RNA‐polymerase (RdRp), and human transmembrane serine protease (TMPRSS2). The results obtained revealed 10 flavonoids with potential antiviral activity. Out of them, silibinin showed promising selectivity index against SARS‐CoV‐2 in vitro. Screening against S protein discloses the highest inhibition activity of silibinin. Mapping the activity of silibinin indicated its excellent binding inhibition activity against SARS‐CoV‐2 S protein, Mpro and RdRP at IC50 0.029, 0.021, and 0.042 μM, respectively, while it showed no inhibition activity against TMPRSS2 at its IC50(SARS‐CoV‐2). Silibinin was tested safe on human mammalian cells at >7‐fold its IC50(SARS‐CoV‐2). Additionally, silibinin exhibited >90% virucidal activity at 0.031 μM. Comparative molecular docking (MD) showed that silibinin possesses the highest binding affinity to S protein and RdRP at −7.78 and −7.15 kcal/mol, respectively. MDs showed that silibinin exhibited stable interaction with key amino acids of SARS‐CoV‐2 targets. Collectively, silibinin, an FDA‐approved drug, can significantly interfere with SARS‐CoV‐2 entry and replication through multi‐targeting activity.
Keywords: enzyme inhibition, flavonoids, in silico study, in vitro evaluation, SARS‐CoV‐2, silibinin
Abbreviations
- ACE2
angiotensin converting enzyme II
- ADME
absorption, distribution, metabolism, and excretion of a drug molecule
- BSA
bovine serum albumin
- COVID‐19
coronavirus disease‐2019
- DMEM
Dulbecco's modified Eagle's medium
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EUA
emergency use authorization
- FBS
fetal bovine serum
- FDA
Food and Drug Administration
- H‐bond
hydrogen bond
- IC50
the concentration of drug required for 50% inhibition
- LPS
lipopolysacccharides
- MDs
molecular dynamic simulation
- Mpro
main protease
- PBS
phosphate buffered saline
- PDB
protein data bank
- PDBQT
protein data bank, partial charge (Q), & atom type (T)
- PLS
partial least square
- QSAR
quantitative structure–activity relationship
- RCSB
research collaboratory for structural bioinformatics
- RdRp
RNA dependent RNA polymerase
- RMSD
root mean square deviation
- S protein
spike protein
- SARS‐CoV‐2
severe acute respiratory syndrome
- SP
standard precision
- TMPRSS2
type 2 transmembrane serine protease
1. INTRODUCTION
SARS‐CoV‐2 is an emerging single stranded, positive‐sense RNA virus (Yang et al., 2020). By March 11th 2020, the World Health Organization (WHO) considered SARS‐CoV‐2 infection as a pandemic disease, named COVID‐19, due to the dramatic increase in the number of deaths (Riva et al., 2020). Despite the presence of several potential targets for the inhibition of SARS‐CoV‐2 (Gil et al., 2020), U.S. FDA has granted the emergency use authorization (EUA) to remdesivir against SARS‐CoV‐2 infection (Wang et al., 2020). Similarly, molnupiravir and paxlovid were granted EUA to manage the COVID‐19 hospitalized patients (Wen et al., 2022). Paxlovid is a combination therapy of nirmatrelvir and ritonavir. Both showed a significant reduced rate of mortality and severity (Wen et al., 2022).While the emergence of new virus variants is a serious health problem, the discovery of novel drugs effective against SARS‐CoV‐2 that can overcome the virus resistance mechanisms is urgent.
Flavonoids, on the other hand, are naturally occurring polyphenolic compounds with potential antiviral (Kim, Leem, Lee, & Kim, 2020), and immunomodulatory (Lalani & Poh, 2020) activities. The antiviral activity of flavonoids has been identified since 1940s. The mechanism of action mainly involves the inhibition of various essential enzymes associated with the virus life cycle (Coelho et al., 2018). For example, the potential role of quercetin and its derivatives against SARS‐CoV2 infection have been highlighted in several studies (Di Petrillo, Orrù, Fais, & Fantini, 2021). Quercetin has potential antioxidant, antiviral and anti‐inflammatory activities and showed promising binding affinity towards SARS‐CoV‐2 proteins (Derosa, Maffioli, D'Angelo, & Di Pierro, 2021). Quercetin has also been reported as potent inhibitor against SARS‐CoV‐2 Mpro at Ki 7 μM (Abian et al., 2020). On the other hand, rutin, a glycosylated analogue of quercetin, showed potent inhibition against SARS‐CoV‐2 Mpro at K i 11 μM, while its l‐arginine derivative was not able to inhibit the viral replication at the cellular level, despite its enhanced solubility (Sancineto et al., 2021). The literature is rich in various in silico studies that suggest the potential importance of flavonoids as potent antiviral against SARS‐CoV‐2 infection. Molecular docking studies indicated the potential binding of naringin and hesperetin against ACE‐2, consequently they can control SARS‐CoV‐2 entry (Cheng et al., 2020). The binding efficiency of herbacetin, isobavachalcone, quercetin 3‐β‐d‐glucoside and helichrysetin flavonoids against MERS‐CoV 3C‐like protease (3CLpro) were confirmed by fluorescence resonance energy transfer (FRET) and tryptophan‐based fluorescence methods (Jo, Kim, Kim, Shin, & Kim, 2019). Naringin also showed significant inhibition activity on the expression of proinflammatory cytokines in raw macrophage cell line (Cheng et al., 2020). Furthermore, the immunomodulatory role of flavonoids especially epicatechin, epigallocatechin gallate, hesperidin, naringenin, quercetin, rutin, luteolin, baicalin, diosmin, genistein, biochanin A, and silymarin in COVID‐19‐associated cytokine storm suggested their potential as phytotherapeutics against SARS‐CoV‐2 infection (Gour, Manhas, Bag, Gorain, & Nandi, 2021).
Flavonoids are recognized as safe phytochemicals, and hence they are intensively used as food and health supplements especially for upper respiratory tract infection and immune‐related diseases (Somerville, Braakhuis, & Hopkins, 2016). Remarkably, a significant antiviral activity of flavonoids against SARS‐CoV following in vitro and in vivo studies was reported (Jo, Kim, Shin, & Kim, 2020). Flavonoids also showed excellent therapeutic activity as antiviral when taken in a combination therapy such as the synergistic effect of apigenin with acyclovir (Mucsi, Gyulai, & Beladi, 1992). Flavonoids attract the interest as candidate against SARS‐CoV‐2 infection. However, the lack of supportive experimental validations prohibits their implementation against SARS‐CoV‐2. Accordingly, all accumulated evidence questioned how far the flavonoids would be proven as beneficial treatment against SARS‐CoV‐2. On the other hand, the higher cost and long timeline required for the development of new antiviral drug draw the attention for drug repurposing particularly those are FDA‐approved. Obviously, there is an urgent need to elucidate the crucial role of flavonoids and verify their mechanisms of action, as well as the adequate application against SARS‐CoV‐2 infection.
Several in silico‐based studies showed the potential importance of variable flavonoids against SARS‐CoV‐2. However, careful comparative studies along with experimental validation were very limited. Thus, in this study, we propose that a comparative in silico study accompanied with systematic evaluation of the published data can narrow down these reported flavonoids to a shortlist with potential anti‐SARS‐CoV‐2. Consequently, this list can be explored experimentally through in vitro study. The ones with promising activity can be further mapped against vital viral target proteins in order to select a flavonoid with superior and multi‐targeting activity against SARS‐CoV‐2. Selecting a single compound with promising activity against multiple targets can increase the probability to overcome the developed resistance by viruses including SARS‐CoV‐2.
2. MATERIALS AND METHODS
2.1. Materials
Flavonoids were purchased from Sigma‐Aldrich Company, Germany. Flavonoids employed in this study are eriodictyol (Cat#: 94258, 95%), hesperetin (Cat#: H4125, 95%), fisetin (Cat#: PHL82542, 98%), kaempferol (Cat#: 60010, 90%), morin (Cat#: M4008, 98%), myricetin (Cat#: M6760, 96%), luteolin (Cat#: L9283, 98%), daidzein (Cat#: D7802, 98%), genistein (Cat#: G6649, 98%), and silibinin (Cat#: S0417, 98%). Mammalian cells including Vero‐E6 cells and normal human fibroblast cell line (HDF, 106‐05A) were purchased from Sigma‐Aldrich. HDF was cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F‐12 (DMEM/F‐12) (Sigma‐Aldrich), supplemented with FBS (Sigma‐Aldrich), and penicillin–streptomycin (Sigma‐Aldrich).
2.2. Computational studies
All computational work was carried out using Schrödinger suite 12.7 available at www.Schrödinger.com and using Maestro graphical user interface software.
2.2.1. Protein and ligand preparation
The 3D crystal structures of SARS‐CoV‐2 S protein (PDB ID: 6VSB), Mpro (PDB ID: 6LU7), RdRp (PDB ID: 7BV2), and TMPRSS2 (PDB ID: 2OQ5) enzymes were downloaded from the protein data bank (https://www.rcsb.org/). The proteins were prepared and refined using the Protein Preparation Wizard (Sastry, Adzhigirey, Day, Annabhimoju, & Sherman, 2013). Crystallographic water molecules that beyond 5 Å were removed. All the missing hydrogen atoms were added at appropriate ionization pH and the tautomerization state were assigned. Next, the refining of protein structures was performed and the water molecules with <3 hydrogen bonds to non‐waters were deleted. Finally, the energy minimization was performed using OPLS‐4 to relieve the steric clashes (Harder et al., 2016). The 2D structures of the generated library were converted to 3D structures using LigPrep, Schrodinger (Giardina et al., 2020). Hydrogen atoms were added, and the salt ions were removed. The most probable ionization states were calculated at appropriate ionization pH using the Epik module (Greenwood, Calkins, Sullivan, & Shelley, 2010; Shelley et al., 2007). During the ligand preparation, specified chirality of the 3D crystal structure was retained. The subsequent energy minimization of each structure was carried out using OPLS4 force field (Harder et al., 2016) and filtered through a relative energy tool to exclude the high energy structures from the given input. Besides, any errors in the ligands were eradicated in order to enhance the accuracy of the molecular docking (Ganai, Abdullah, Rashid, & Altaf, 2017).
2.2.2. Grid generation and molecular docking
The ligand in the crystal structure of RdRp and TMPRSS2 enzymes was used for grid generation. A grid box was generated at the centroid of the active site for docking studies, and the active site was defined around the ligand crystal structure. Since the crystal structure of S protein with its ligand is not available, a sitemap analysis was used for the identification of the predictable binding site. The binding site that showed the highest site score and Dscore was used for receptor grid generation with Glide.
Molecular docking was performed within the catalytic pocket site of the proteins using standard precision (SP) mode of Grid using Glide (Friesner et al., 2004; Halgren et al., 2004). The Docking method validation was carried out using the crystal structure of TMPRSS2 (PDB: 2OQ5) with its co‐crystallized ligand. It was prepared using the protein preparation wizard followed by separation of ligand and protein. The ligand was then re‐docked against the protein using standard precision (SP) flexible docking protocol, and the RMSD was calculated between the crystal structure ligand pose and re‐docked pose. The accuracy of the docking procedure was determined by the low RMSD value predicted by the Glide scoring function resembles the validity of the docking.
2.2.3. Molecular dynamic simulation MDs
The top‐ranked ligand‐enzyme complexes were selected for MDs using the Desmond software (Chow et al., 2008). The Desmond system was built using the TIP4P water model (Zeiske, Stafford, Friesner, & Palmer III, 2013). The orthorhombic water box that showed the boundary conditions for setting up the shape and size was also generated and then predefined the simple point charge (SPC) solvent model, OPLS3 as a force field was selected. Sodium/chloride ions were added for neutralization by maintaining the salt concentration of 0.15 M (Na+ and Cl−). The built system was minimized to relax a model system to a minimum local energy. The minimized system was used for performing the MD simulation for 100 ns using NPT (constant temperature, constant number of atoms, and constant pressure ensemble) class at 300 K temperature and 1.01325 bar pressure. The results from MD simulation were detailed information like protein and ligand root mean square deviation (RMSD), root mean square fluctuation (RMSF), and ligand interaction profile were generated from the simulation trajectory of ligand‐enzyme complexes (Hamdy et al., 2021).
2.2.4. Pharmacophore model and field‐3D‐QSAR
The pharmacophore hypothesis of flavonoid was generated with Phase Schrödinger. The prepared structures were imported with their respective pIC50. The flavonoids with pIC50 > 5 were assigned as active ligands, while those <3.7 were assigned inactive, and the rest were assigned as moderately active. A pharmacophore model was created using shape screening‐based alignments and a default set of six chemical features including hydrogen‐bond acceptor (A), hydrogen‐bond donor (D), hydrophobic (H), negative ionizable (N), positive ionizable (P), and aromatic ring (R). The resulting hypotheses were scored and ranked using vector, volume, sites, survival, and survival inactive scores.
The correlation between the 3D structures of flavonoids and their biological activity was built using a predictive 3D QSAR model and Phase Schrödinger. The model was generated by applying Partial Least Square (PLS) regression statistics and by keeping a grid spacing of 1 Å. The number of PLS factor included in the development of model was two. The pharmacophore model was validated by its accuracy in predicting the training set of ligands activity. The validation of the model based on the cross‐validation coefficient (Q 2 = 0.672) and the regression coefficient for the training set was 0.86, which showed relevance of the model.
2.2.5. Pharmacokinetics and drug‐likeness filter and target prediction
ADME properties of flavonoids were predicted using Swiss‐ADME server (Daina, Michielin, & Zoete, 2017), and flavonoids in a SMILES format. Two different filters including Lipinski's rule of five and Ghose filter were used for flavonoids selection.
2.3. In vitro inhibition assay of flavonoids against SARS‐CoV‐2
The antiviral activity of flavonoids was performed as previously described (Mostafa et al., 2020). SARS‐CoV‐2 strain NRC‐03‐nhCoV isolated in Egypt and deposited in GSAID (Accession # EPI_ISL_430820) (Kandeil et al., 2020) was used. Vero‐E6 cells (passage #11) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% Fetal Bovine Serum (FBS) (Invitrogen, Waltham, Massachusetts, USA) and 1% penicillin/streptomycin antibiotic mixture at 37°C, 5% CO2 for 24 h. In 96‐well tissue culture plates, 2.4 × 104 Vero‐E6 cells/well were incubated overnight at a humidified 37°C incubator under 5% CO2. The virus was adsorbed on the cell monolayers and further overlaid with 50 μl DMEM containing different concentrations of the compounds. Flavonoids were assigned random numbers and have been tested blindly. The plates were then incubated at 37°C, 5% CO2 for 72 h. The cells were then fixed with 4% paraformaldehyde (100 μl) for 20 min and stained with 0.1% crystal violet (CV) for 15 min at room temperature. CV was then dissolved in absolute methanol (100 μl/well) and the produced color was measured at 570 nm using Anthos Zenyth 200rt plate reader (Anthos Labtec Instruments, Heerhugowaard, Netherlands). The half maximal viral inhibitory (IC50) concentrations of the compounds were measured as previously described (Mostafa et al., 2020). The cytotoxicity of various concentrations compared to the untreated cells was determined using nonlinear regression analysis by plotting log inhibitor concentrations versus normalized responses.
2.4. Virucidal effect of silibinin
SARS‐CoV‐2 strain was treated with silibinin at 15 μg/ml and incubated for 30 min at room temperature. Vehicle, employed as negative control, was subjected to the same incubation condition. The mixture is then diluted three times (10‐fold) to reach 10−3 of the original viral titers. The diluted control virus and silibinin‐treated virus were then inoculated in duplicates to cultured monolayers of Vero‐E6 cells in 6 well plates and titrated using plaque infectivity assay as previously described (Mostafa et al., 2020).
2.5. Cell viability assay
The cytotoxic activities of flavonoids were assessed blindly using MTT (3‐[4, 5‐dimethylthiazolyl‐2]‐2, 5‐diphenyltetrazolium bromide) assay according to Soliman, Alhamidi, et al. (2020) and Soliman, Saeed, et al. (2020). Briefly, in 96‐well plates, normal human fibroblast cell line (HDF, passage #6, 106‐05A, Sigma‐Aldrich) were seeded at 4000 cells/50 μl and incubated for 24 h at 37°C, and 5% CO2 with the compounds at different concentrations. A 20 μl of sterile filtered MTT reagent in PBS (5 mg/ml) was added to each well and incubated for 4 h. DMSO (200 μl/well) was added, and the developed purple color was measured using Multiskan Go machine (spectrophotometer) at 570 nm. Each experiment was repeated 6 times. Cell viability percentage was calculated using the following formula,
2.6. SARS‐CoV‐2 S protein inhibition assay
SARS‐CoV‐2 S protein inhibition assay was performed using SARS‐CoV‐2 Spike: ACE‐2 inhibitor Screening Assay Kit (CAT # 79331, BPS Bioscience, San Diego, CA, USA) following the supplier protocol. Briefly, in 96 flat‐bottom well plate, 50 μl of SAS‐CoV‐2 S protein (1 μg/ml PBS) was added and incubated at 4°C overnight. Next day, supernatant was decanted followed by several washings with supplied washing buffer. The plates were then blocked for 1 h. This was followed by the addition of 10 μl of the tested flavonoids at 15 μg/ml compared to remdesivir at its in vitro IC50 (2.5 μg/ml) and the plates were incubated for 1 h at room temperature. Then, 20 μl ACE‐2‐His‐Tag protein (2.5 ng/ml) was added and incubated for additional 1 h at room temperature. The supernatants were then removed, and the plates were washed several times, followed by the addition of 100 μl blocking buffer (supplied with the kit) for 10 min. For detection, 100 μl anti‐His‐HRP was added and incubated for 1 h, then the plates were washed and blocked for 10 min. This was followed by the addition of 100 μl mixture of ECL substrate A and B (1:1) and the chemiluminescence was measured using microtiter‐plate reader (Synergy H1, Biotek Ltd, Winoosk, VT, USA). The reaction without inhibitors was employed as positive control, while the reaction without inhibitors and ACE‐2‐His‐Tag protein was used as negative control (blank). To measure the IC50, different concentrations (3.87, 7.75, 15.5, 31, and 62 nM) of the inhibitors were used and the inhibition activity was evaluated following the same procedure. The inhibitory activity of flavonoids was plotted against the logarithm of the inhibitor concentrations to calculate the IC50.
2.7. SARS‐CoV‐2 main protease (Mpro, 3CLpro ) inhibition assay
Mpro assay was performed employing 3CL Protease (3CLpro), Untagged (SARS‐CoV‐2) Assay Kit (CAT # 78042‐1, BPS Bioscience) following the supplier protocol with minor modifications. Initially, 2.5 μl of the inhibitors at concentrations (3.87, 7.75, 15.5, 31, and 62 nM) were incubated for 1 h at room temperature with 10 μl 3CLpro enzyme (1.5 ng/μl) in a reaction buffer (20 mM Tris–HCl pH 7.3, 100 mM NaCl, 1 mM EDTA, 0.01% BSA, and 1 mM 1,4‐dithio‐d, l‐threitol (DTT). This was followed by the addition of 12.5 μl of 80 mM 3CLpro substrate (Dabcyl KTSAVLQSGFRKME‐Edans fluorogenic substrate). The reaction was incubated for 1 h at room temperature in dark, then the fluorescence intensity was measured by a microtiter plate‐reader (Synergy H1, Biotek Ltd) at an emission and excitation wavelengths 460 and 360 nm, respectively. Cysteine protease covalent inhibitor (GC376) provided by the supplier was used as positive control at 50 μg/ml according to Fu et al. (2020), while the reaction without inhibitor and 3CLpro was used as negative control.
2.8. SARS‐CoV‐2 RNA‐dependent RNA polymerase (RdRp) inhibition assay
SARS‐CoV‐2 RdRp inhibition assay was performed using viral RNA‐dependent RNA polymerase assay Kit (Cat # VRT500K, Profoldin, Hudson, MA, USA) following the supplier protocol with modification according to Yanira et al. 2019 (Sáez‐Álvarez, Arias, del Águila, & Agudo, 2019). Initially, 300 ng (RdRp/NSP7/NSP8, Cat # 100839, BPS Bioscience) was added to different concentrations (3.87, 7.75, 15.5, 31, and 62 nM) of the tested inhibitors in 10 μl reaction buffer (200 mM Tris–HCl, pH 8, 500 mM NH4Cl, 80 mM Mg[OAc]2, 0.05% Tritone X‐100) for 30 min at 30°C. The reaction was initiated by the addition of 5 μl 3 mM MnCl2, 0.5 μl of 0.4 mg/ml RNA template, 0.5 μl of 50 mM NTPs (ATP, UTP, GTP, CTP) and 34 μl of the reaction buffer. The reaction was incubated for 1 h at 37°C, and then stopped by adding 50 μl SYTO‐9 fluorescence dye. The detection was performed by measuring the fluorescence developed due to the binding of fluorescence dye with dsRNA using microtiter‐plate reader (Synergy H1, Biotek Ltd) using excitation and emission filters at 485 and 520 nm, respectively. For positive control, remdesivir was employed at 2.5 μg/ml, while the reactions without inhibitors was used as negative control.
2.9. Human TMPRSS2 fluorogenic assay
TMPRSS2 fluorogenic assay was carried out using TMPRSS2 fluorogenic assay kit (CAT # 78083, BPS Bioscience) following the supplier protocol. Briefly, 10 μl silibinin (3 mg/ml) was incubated with 30 μl TMPRSS2 (5 ng/μl) for 30 min at room temperature. Following incubation, 10 μl TMPRSS2 substrate (50 μM) was added, and the fluorescence intensity was measured in dark by a microtiter plate‐reader (Synergy H1, Biotek Ltd) at an emission and excitation wavelengths 383 and 455 nm, respectively. Camostat mesylate (10 μM) was used as positive control (Hoffmann et al., 2021), while the reaction without inhibitor and enzyme was employed as negative control.
2.10. Statistical analysis
The data were collected and graphed using GraphPad Prism (5.04, GraphPad Inc., La Jolla, CA, USA). The enzyme inhibition and cytotoxic activities of the compounds were analysed by one‐way analysis of variance (ANOVA) using Bonferroni's multiple comparisons test. p < 0.05 was considered as significant. The data display the mean ± SEM of 3–6 replicas. The best practice in natural products pharmacological research has been taken into account (Heinrich et al., 2020; Izzo et al., 2020) including the purity and activity of the tested compounds, their mechanism of action, their activity in comparison to the vehicle as a negative control and clinically approved drug as a positive control, the appropriate concentrations were also reported for further pharmaceutical development, and the safety of the compounds was also mentioned.
3. RESULTS
3.1. Rational selection of candidate flavonoids with potential anti‐SARS‐CoV‐2 activity
The rational procedure for the selection of flavonoids with potential activity against SARS‐CoV‐2 was summarized in Figure 1. SARS‐CoV‐2 S protein was selected as a target since it can provide a potential strategy to inhibit the viral binding and cell entry. Flavonoids database with ~500 compounds was downloaded from PubChem and filtered using PAINS filter. Lipinski rule of 5 was used as a second filter to reject those showed 3 or more violations (Table S1). Literature search using keywords such as flavonoids and antiviral filtered >50 articles that narrow down the selection of candidates that have been previously reported with antiviral activity and good binding affinity to essential viral proteins (Table S1). Molecular docking of 45 flavonoids was performed to select the flavonoids with high binding affinity to SARS‐CoV‐2 S protein. The cut off binding score value was set at −6 kcal/mol (Figure 2 and Table S1). The top candidates were then mapped against other vital SARS‐CoV‐2 targets to select the most promising candidate flavonoid (Figure 2). Eventually, the process resulted in the selection of 10 flavonoids, which were purchased for further experimental validation against SARS‐CoV‐2 and its vital target proteins (Figure 3).
FIGURE 1.
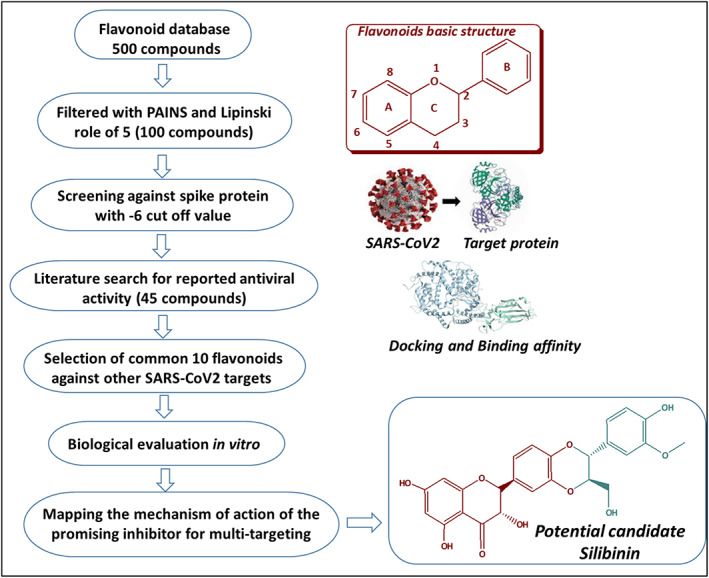
Schematic representation for the rational selection of flavonoids with potential activity against SARS‐CoV‐2
FIGURE 2.
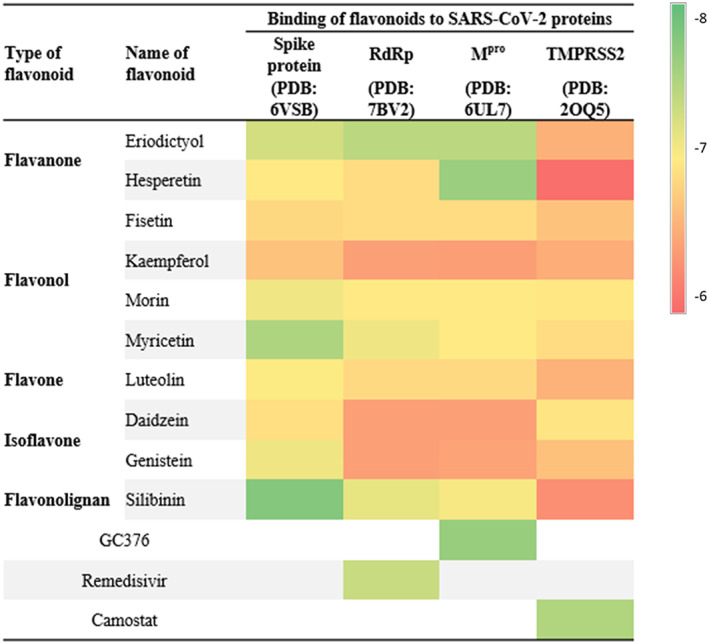
Heatmap representing the levels of binding affinity of flavonoids against vital SARS‐CoV‐2 protein targets. Relative binding affinity levels were displayed as colors ranging from red to green as shown in the key
FIGURE 3.
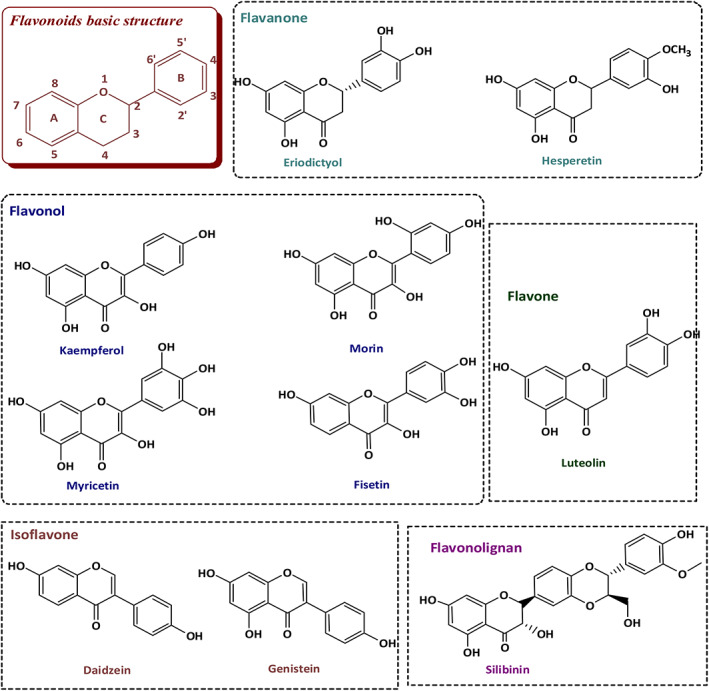
Chemical structures of selected flavonoids
The filtered flavonoids were further classified according to the structure to select the flavonoid with optimal anti‐SARS‐CoV‐2 activity for further future lead optimization. These included flavonoids in which the B ring is linked in position 3 to the C ring (isoflavones) such as daidzein and genistein, and flavonoids in which the B ring is linked in position 2 to the C ring (typical flavonoids) (Figure 3). The flavonoids from the second type were further classified based on (i) the absence of (OH) at position 3 and the absence of 2,3‐double bond (flavanone) such as eriodyctiol and hesperetin, (ii) the absence of (OH) at position 3 and the presence of 2,3‐double bond (flavone) such as luteolin, (iii) the presence of (OH) at position 3 and the presence of 2,3‐double bond (flavonol) such as fisetin, kaempferol, morin, and myricetin. Furthermore, the classification depended on the number, and position of (OH) group on the B ring (Figure 3). They also classified according to the blockage of OH group on the B ring. This includes a single blocking by forming a methoxy ether such as hesperetin or blocking both (OH) groups by oxidative coupling with coniferyl alcohol (flavonolignan) such as silibinin (Figure 3).
3.2. Identification of S protein binding site using sitemap analysis
To determine the S protein best binding site for molecular docking, a sitemap analysis was performed. The best docking sites were determined based on the site score and Dscore (Table 1). Among these sites, site 3 with the highest Dscore of 1.076 and site score of 1.006 were selected for receptor‐grid generation and subsequent docking studies. Molecular docking study indicated the superiority and potential multi‐targeting activity of eriodictyol, myricetin, hesperetin and silibinin against SARS‐CoV‐2.Our molecular docking study against SARS‐CoV‐2 S protein (PDB: 6VSB) revealed that all selected flavonoids showed moderate binding affinity close to the cut off value ranged from – 6.68 to −7.78 kcal/mol (Figure 2 and Table S1). The binding affinity of the selected flavonoids was further mapped against Mpro (PDB: 6UL7), RdRp (PDB: 7BV2), and human TMPRSS2 (PDB: 2OQ5). Eriodictyol showed high binding affinity at −7.28, −7.43, and −7.43 towards S protein, RdRp and Mpro, respectively. Hesperetin showed high binding affinity against Mpro at −7.64 kcal/mol similar to the Mpro inhibitor GC‐376 (Figure 2 and Table S1). Myricetin showed significant binding affinity at −7.51, and −7.1 kcal/mol towards S and RdRp, respectively. Silibinin showed also significant binding affinity at −7.78, −7.15, and −7.05 kcal/mol against S, RdRp, and Mpro, respectively. This in silico study indicated that eriodictyol, myricetin, hesperetin and silibinin out of the selected flavonoids showed potential binding affinity to multiple SARS‐CoV‐2 targets.
TABLE 1.
Sitemap analysis of S protein
| Title | SiteScore | Size | Dscore |
|---|---|---|---|
| sitemap_spikep_site_3 | 1.067 | 898 | 1.006 |
| sitemap_spikep_site_1 | 1.06 | 932 | 1.049 |
| sitemap_spikep_site_4 | 1.037 | 657 | 1.055 |
| sitemap_spikep_site_2 | 1.022 | 916 | 1.004 |
| sitemap_spikep_site_5 | 1.009 | 353 | 0.97 |
3.3. ADME indicated the drug‐likeness of the selected flavonoids
The selected flavonoids were examined with the Lipinski rule of 5 including hydrogen bond donors (HBD) <5, hydrogen bond acceptors (HBA) <10, octanol–water partition coefficient (Log P) <5, and molecular mass (Mwt) <500 (Table S1). Flavonoids that showed three or more violations were rejected since they do not fulfil the criteria of drug‐likeliness. The passing criteria were set at rotatable bonds ≤10, topological polar surface area (TPSA) ≤ 140, and rotatable bonds ≥6. The compounds that follow Lipinski rule were considered promising drug candidates. Further, a good gastrointestinal absorption with good bioavailability, and good solubility (log s < 5) was employed (Table S1). All the selected flavonoids were complied with the Lipinski rule of five. Although the in silico study including ADME indicated the drug‐likeness of all selected flavonoids, molecular docking favored eriodictyol, myricetin, hesperetin and silibinin because of their potential binding affinity with multiple viral targets.
3.4. Out of the selected flavonoid candidates, eriodictyol, myricetin, and silibinin showed significant anti‐SARS‐CoV‐2 in vitro
The in vitro anti‐SARS‐CoV‐2 activity of flavonoids under study was screened in comparison to remdesivir as positive control and DMSO as negative control. The results obtained revealed that only eriodictyol, myricetin, and silibinin showed high selectivity index for antiviral activity relative to Vero‐E6 cells toxicity with CC50 values higher than the IC50 values by 2‐, 2.5‐, and 1.5‐fold, respectively (Table 2 and Figure 4).The pharmacophore hypothesis also indicated that the best model with the higher survival score of 5.04 was AADRR (Figure 5a), which consisted of two acceptors, one donor and two aromatic rings. These chemical features existed in silibinin, myricetin, and eriodictyol (Figure 5b) alongside their good alignment to the pharmacophore model, indicating their importance against SARS‐CoV‐2. On the other hand, the good alignment of silibinin as the most active flavonoid (Figure 5c) was compared to the bad alignment of daidzein (Figure 5d), which illustrated their different activity profiles.
TABLE 2.
Flavonoids IC50 and CC50 values in μM
| Name of flavonoid | CC50 | IC50 |
|---|---|---|
| Eriodictyol | 42.2 ± 0.6 | 22.3 ± 1.2 |
| Hesperetin | 53.8 ± 1.0 | 465.8 ± 1.4 |
| Fisetin | 60.9 ± 0.7 | 315.6 ± 1.1 |
| Kaempferol | 68.3 ± 0.5 | 182.5 ± 0.9 |
| Morin | 115.1 ± 0.8 | 527.2 ± 1.1 |
| Myricetin | 49.0 ± 0.6 | 19.11 ± 0.5 |
| Luteolin | 32.2 ± 0.5 | 212.6 ± 0.6 |
| Daidzein | 48.5 ± 0.8 | 655.9 ± 0.9 |
| Silibinin | 34.3 ± 0.5 | 31.2 ± 0.5 |
FIGURE 4.
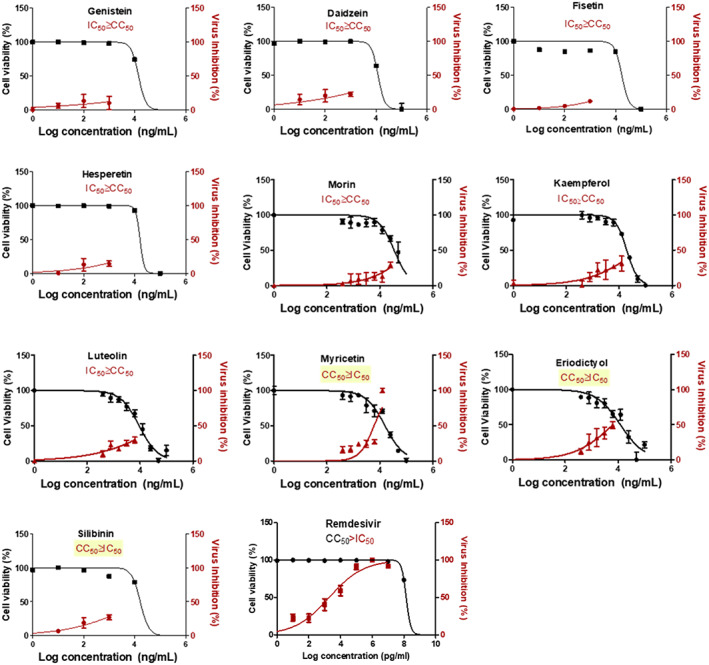
In vitro antiviral activity of flavonoids under study. The antiviral activity of the compounds was screened on SARS‐CoV‐2 co‐cultured with Vero‐E6 cells. Values of inhibitory concentration 50% (IC50) on viral cells and cytotoxic concentration 50% (CC50) on Vero cells were calculated using nonlinear regression analysis. The data display the mean of cell viability percentage ± SEM of 4 replicates
FIGURE 5.
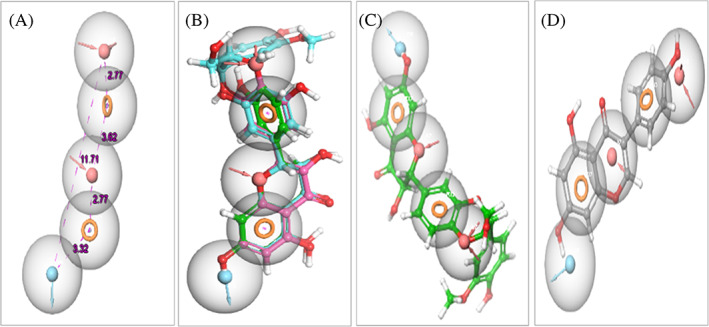
Pharmacophore features of flavonoids required for the anti‐SARS‐CoV‐2 activity. (a) Common pharmacophore generation sites with distance in Å with two acceptors [pink sphere with two arrows], two aromatic rings [dark yellow circle], and one donor [blue sphere with arrow]. (b) Alignment of all active flavonoids including silibinin, myricetin and eriodictyol to the pharmacophore. (c) The good alignment of the active silibinin to the pharmacophore. (d) The bad alignment of inactive daidzein to the pharmacophore
3.5. Eriodictyol, myricetin, and silibinin are safe on normal human cells
The cytotoxic activity of all tested flavonoids was performed on normal human fibroblast cells (HDF) using MTT assay. Although most of the compounds were tested safe on human cells, eriodictyol, myricetin, and silibinin showed ~100% cell viability at 17‐, 8.5‐, and seven‐fold their IC50(SARS‐CoV‐2), respectively, compared to 65% for remdesivir (Figure 6).
FIGURE 6.
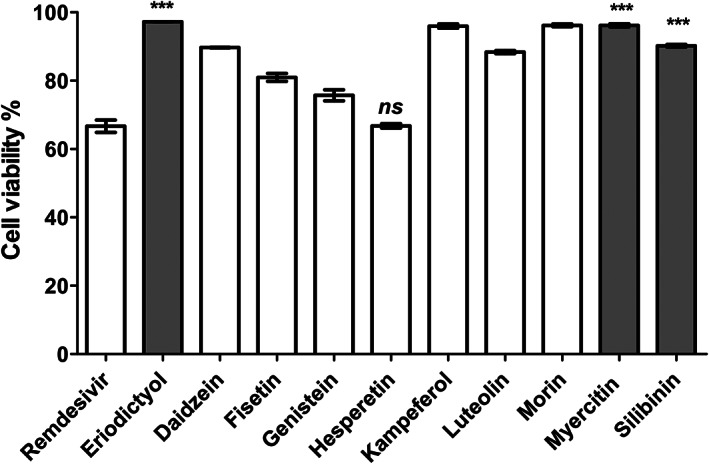
Cytotoxic activities of flavonoids understudy compared to remdesivir, employed as positive control drug. The cytotoxic activity of the compounds was tested on normal human cells at 100 μg/ml. The data was analyzed using one‐way ANOVA and statistical significance was calculated with Dunnett's multiple comparisons test and significance level indicated by asterisks (*p < 0.05; **p < 0.01: ***p < 0.001; ****p < 0.0001). The data display the mean of cell viability percentage ± SEM of 3 replicates
3.6. Silibinin at its IC50 (SARS‐CoV‐2) showed promising inhibition activity against SARS‐CoV‐2 S protein, Mpro and RdRp, but not against human TMPRSS2
Following the in vitro SARS‐CoV‐2 inhibition and cell viability assays, eriodictyol, myricetin, and silibinin were selected for further screening against SARS‐CoV‐2 S protein. Our data indicated that only silibinin showed potent inhibition activity at 15 μg/ml (~52% ± 3.0) against SARS‐CoV‐2 S protein, while eriodictyol and myricetin showed lower inhibition activity at double their IC50 value and with inhibition percentages ~22 ± 0.03 and 31 ± 0.69, respectively, compared to 4% ± 0.02 for remdesivir at its IC50 value (Figure 7). Following the primary screening, silibinin IC50 was determined by evaluating the inhibition efficacy in a dose–response curve. The data obtained showed that silibinin IC50 was 0.029 ± 0.004 μM (Figure 8a). The inhibition activity of silibinin was further tested against SARS‐CoV‐2 Mpro and RdRp. Interestingly, silibinin showed potent inhibition activity against Mpro (Figure 8b) and RdRp (Figure 8c) at IC50 0.021 ± 0.003 and 0.042 ± 0.004 μM, respectively. The inhibition activity of silibinin against RdRp was ~35% less than remdesivir (Figure 8d). Silibinin did not inhibit TMPRSS2 at the identified IC50(SARS‐CoV‐2) concentration. Additionally, silibinin exhibited >90% virucidal effect, indicating its direct inactivation effect on the virion (Figure 9a,b).
FIGURE 7.
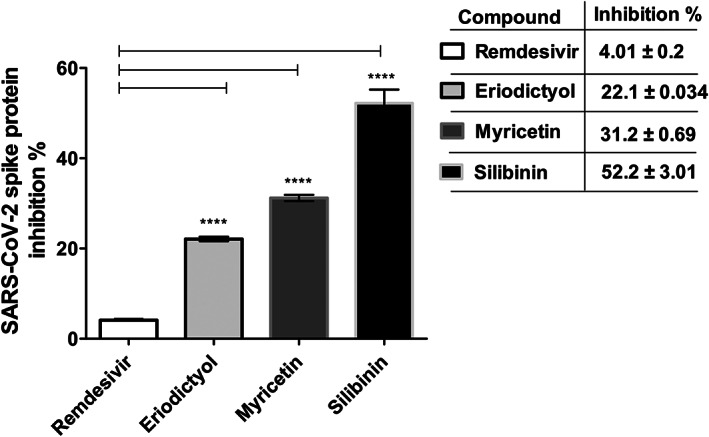
Inhibition percentage of selected flavonoids against SARS‐CoV‐2 S protein compared to remdesivir. The inhibition percentage of eriodictyol, myricetin, and silibinin flavonoids to the binding of S protein to ACE‐2 receptor was screened at 15 μg/ml compared to remdesivir at its IC50 (2.5 μg/ml). The data was analyzed using one‐way ANOVA and statistical significance was calculated with Dunnett's multiple comparisons test and significance level indicated by asterisks (*p < 0.05; **p < 0.01: ***p < 0.001; ****p < 0.0001). The data display the mean ± SEM of three independent replicates. p‐value <0.05 was considered as significant
FIGURE 8.
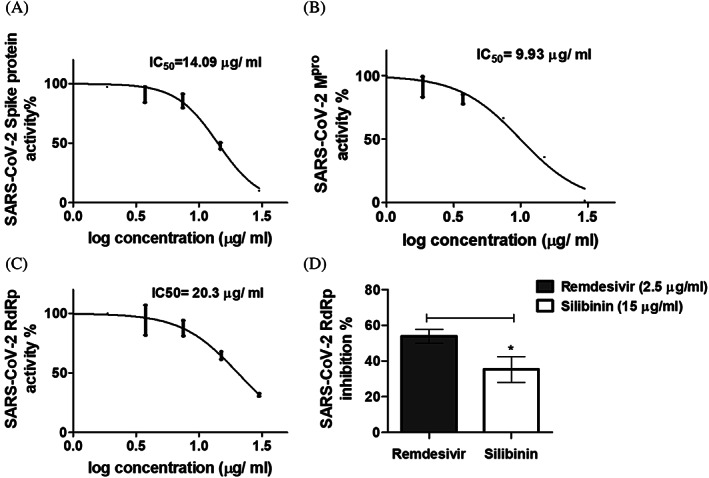
Inhibition percentage of silibinin against selected critical SARS‐CoV‐2 targets. (a) IC50 calculation of silibinin against SARS‐CoV‐2 S protein. (b) IC50 calculation of silibinin against SARS‐CoV‐2 Mpro enzyme. (c) IC50 calculation of silibinin against SARS‐CoV‐2 RdRp. (d) Inhibition percentage of silibinin on SARS‐CoV‐2 RdRp compared to remdesivir. The data was analyzed using Unpaired t test with Welch's correction and significance level indicated by asterisks. The data display the mean of the percentage of the enzyme inhibition ± SEM of 3 replicates
FIGURE 9.
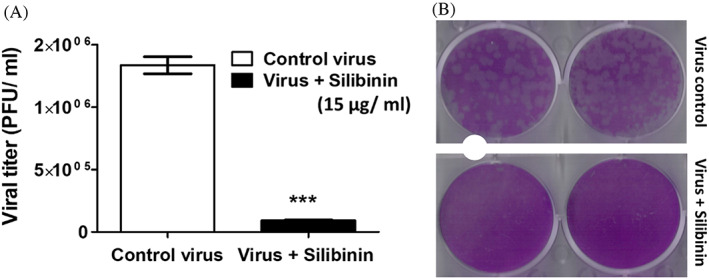
Silibinin exhibited strong virucidal effect on SARS‐CoV‐2 strain NRC‐03‐nhCoV. The diluted pre‐incubated control virus and silibinin‐treated virus were cultured with Vero E6 cells using plaque infectivity assay. (a) Viral titer of virus control (untreated) and silibinin‐treated virus. (b) Plaque reduction by treatment with silibinin. The data was analyzed using Unpaired t test with Welch's correction and significance level indicated by asterisks. The data display the mean of the percentage of virucidal activity of silibinin ± SEM of 4 replicates
3.7. Molecular docking and ADME analysis indicated the superiority of silibinin against SARS‐CoV‐2 target proteins
The interactions of silibinin as the best candidate from the experimental analysis with the target proteins compared to native ligand were further studied by Glide standard precision mode. Silibinin showed several interactions with S protein (PDB: 7VSB) amino acid residues. Silibinin formed 5 H‐bonds with Asn‐907, Lys‐1038, Ile‐909, Glu‐1092, and Tyr‐904, alongside 3 aromatic H‐bonds with Asn‐907, Gly‐904, and Tyr‐904, in addition to hydrophobic interactions with Tyr‐904, Asn‐907, Gly‐910, Gly‐908, Lys‐1038, Glu‐1092, and Tyr‐904 that stabilize the complex (Table 3, Figure 10a).
TABLE 3.
Molecular modeling of silibinin within the binding active site of SARS‐CoV‐2 S protein
| Compound | Moiety | Interaction | Amino acid residue |
|---|---|---|---|
| Silibinin | OH | H‐bond | Asn‐907 |
| CH2OH | H‐bond | Lys‐1038 | |
| OH | H‐bond | Ile‐909 | |
| OH | H‐bond | Glu‐1092 | |
| 1,4‐dioxane | H‐bond | Tyr‐904 | |
| CH | Aromatic H‐bond | Asn‐907 | |
| Phenyl | Aromatic H‐bond | Gly‐904 | |
| 1,4‐dioxane | Aromatic H‐bond | Tyr‐904 | |
| Phenyl | Hydrophobic bond | Gly‐908, Gly‐910, Asn‐907 and Glu‐1092, Asn‐907 and Tyr‐904 | |
| Phenyl | Hydrophobic bond | Lys‐1038 | |
| CH | Hydrophobic bond |
FIGURE 10.
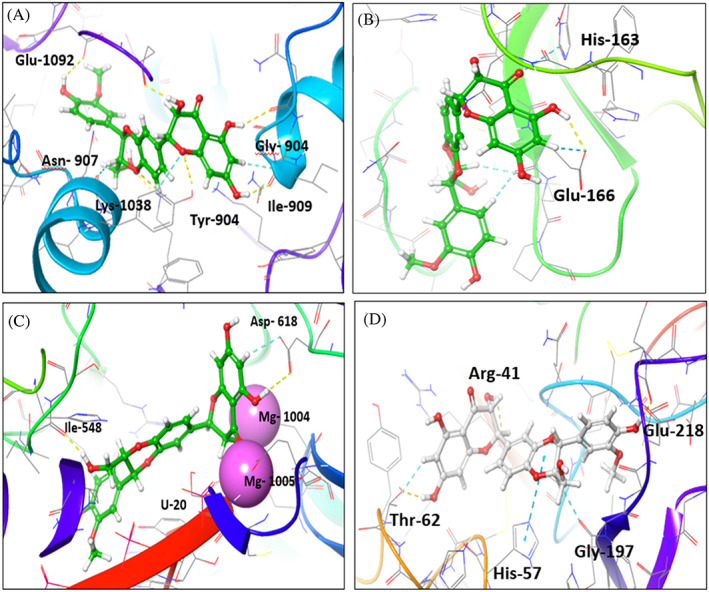
Interaction of silibinin with SARS‐CoV‐2 target proteins. (a–d) Interaction of silibinin within the active site of viral proteins. (a) Interaction of silibinin with SARS‐CoV‐2 S protein (PDB:6VSB). (b) Interaction of silibinin with SARS‐CoV‐2 Mpro enzyme (PDB: 6LU7). (c) Interaction of silibinin with SARS‐CoV‐2 RdRp enzyme (PDB:7BV2). (d) Interaction of silibinin with TMPRSS2 enzyme (PDB:2OQ5)
The interactions of silibinin with RdRp (PDB: 7VB2) compared to remdesivir, the native ligand, were evaluated (Table 4). The silibinin showed three H‐bonds with U‐20, Asp‐618, and Ile‐548, while remdesivir showed one H‐bond with U‐20. Both showed two metal bond interactions with Mg‐1004, and Mg‐1005. Silibinin showed aromatic H‐bond interaction with Asp‐618 and hydrophobic interactions with Arg‐836, Asp‐618, Ser‐814, Glu‐811, Lys‐545, Lys‐551, Ala‐547, Asp‐760, and Ile‐548 (Figure 10b). Remdesivir showed pi‐cation interaction with Arg‐555 and hydrophobic interaction with Asp‐761, and Arg‐555.The interaction of silibinin with SARS‐CoV‐2 Mpro enzyme (PDB:6LU7) showed H‐bond with Glu‐166, pi‐pi stacking interaction with His‐163, and Glu‐166 and hydrophobic interaction with Gly‐143, Arg‐188, Phe‐140, Ser‐144, Asn‐142, Leu‐167, Pro‐168, Thr‐190, Ala‐191, Gln‐189, and Glu‐166 amino acids (Table 5, Figure 10c). Native ligand showed 3 H‐bonds with Gly‐143, Glu‐166, and catalytic amino acid Cys‐145, in addition to hydrophobic interaction with Met‐49, Met‐165, His‐163, and Gln‐189 (Table 5). Although, silibinin showed good interaction with TMPRSS2 (Table 6, Figure 10d), it did not show inhibition at its IC50 (SARS‐CoV‐2) value. Therefore, we turn our attention to examine the mechanism of inhibition at the IC50 concentration of silibinin.
TABLE 4.
Molecular modeling of silibinin within the binding active site of SARS‐CoV‐2 RdRp compared to the native ligand
| Compound | Moiety | Interaction | Amino acid residue |
|---|---|---|---|
| Silibinin | OH | H‐bond | U‐20 |
| OH | Metal bond | Mg‐1004 | |
| CO | Metal bond | Mg‐1005 | |
| OH | H‐bond | Asp‐618 | |
| CH2OH | H‐bond | Ile‐548 | |
| CH | Aromatic H‐bond hydrophobic bond | Asp‐618 | |
| Phenyl ring | Hydrophobic bond | Lys‐545, Glu‐811, Asp‐618 and Ser‐814 | |
| CH | Hydrophobic bond | Ile‐548, Lys‐551, ala‐547 and Asp‐760 | |
| OCH3 | Arg‐836 | ||
| Native ligand | OH | H‐bond | U‐20 |
| Phosphate OH | Metal bond | Mg‐1004 | |
| Phosphate OH | Metal bond | Mg‐1005 | |
| Pyrrole | Pi‐cation | Arg‐555 | |
| Phenyl | Hydrophobic bond | Arg‐555 and Asp‐761 | |
| NH2 | Hydrophobic bond | Thr‐687 |
TABLE 5.
Molecular modeling of silibinin within the binding active site of SARS‐CoV‐2 Mpro compared to the native ligand
| Compound | Moiety | Interaction | Amino acid residue |
|---|---|---|---|
| Silibinin | OH | H‐bond | Glu‐166 |
| Phenyl ring | Pi‐pi stacking bond | Glu‐1166 | |
| Phenyl ring | Pi‐pi stacking bond | His‐163 | |
| Phenyl ring | Hydrophobic bond | Leu‐167, Glu‐166, Asn‐142 and Gln‐189 | |
| Phenyl CH | Hydrophobic bond | Gly‐143, Arg‐188, Thr‐190 and ala‐191 | |
| OH | Hydrophobic bond | Phe‐140, Ser‐144 and Pro‐168 | |
| Native ligand | NH2 | H‐bond | Gly‐143 |
| Carbonyl | H‐bond | Cys‐145 | |
| NH | H‐bond | Glu‐166 | |
| Hydrophobic bond | Met‐49, His‐163, Met‐165 and Gln‐189 |
TABLE 6.
Molecular modeling of silibinin within the binding active site of TMPRSS2 compared to the native ligand
| Compound | Moiety | Interaction | Amino acid residue |
|---|---|---|---|
| Silibinin | OH | H‐bond | Glu‐218 |
| OH | H‐bond | Arg‐41 | |
| OH | H‐bond | Thr‐62 | |
| Phenyl | Pi‐cation bond | Arg‐41 | |
| CH2OH | Pi‐cation bond | His‐57 and Gly‐197 | |
| Phenyl ring | Hydrophobic bond | Cys‐219, Gln‐192 and Thr‐63 | |
| Native ligand | NH | H‐bond | Asp‐189 |
| NH2 | H‐bond | Asp‐189 | |
| NH2 | H‐bond | Glu‐218 | |
| CH | Aromatic H‐bond | Glu‐216 and Gly‐216 | |
| Phenyl ring | Hydrophobic bond | Val‐213 | |
| Phenyl ring | Hydrophobic bond | Ala‐190 |
The bioavailability radar of silibinin was represented in Figure 11. ADME indicates that silibinin is in the conformity range and with acceptable pharmacokinetics parameters. The pink areas revealed the optimum range of properties including lipophilicity, flexibility, unsaturation, insolubility, polarity, and size.
FIGURE 11.
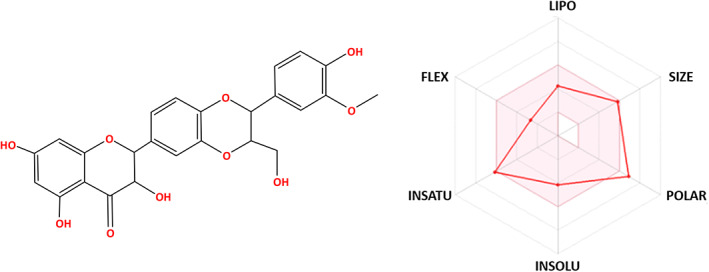
Bioavailability radar of silibinin. The pink area indicates the preferred properties range
3.8. Molecular dynamic simulation confirmed the stability of silibinin complex with SARS‐CoV‐2 target proteins
MD simulation study provides information about the ligand‐protein complex interaction that mimics its physiological condition. The inhibitory activity was related to the decrease in residues fluctuation within the pocket site. MDs was performed for silibinin with the top selected targeting proteins. The results were represented in Figure 12. MDs root mean square deviation (RMSD) plot showed stability of protein structure with RMSD value <3 Å, while the ligand RMSD range was from 5.2 to 7.3 Å, indicating its stable interaction (Figure 12a).
FIGURE 12.
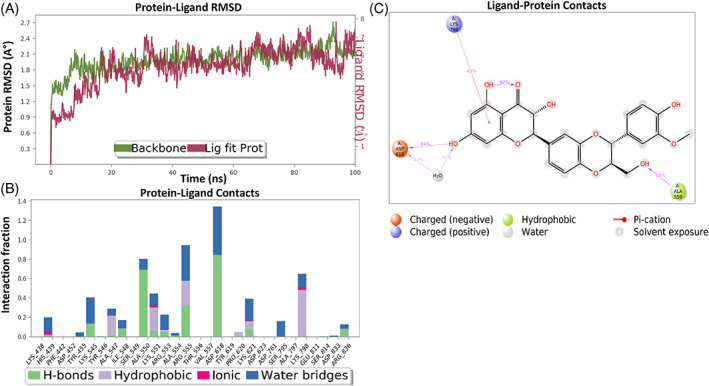
Interaction diagram of silibinin with RdRP observed during the molecular dynamic simulation. (a) Protein‐ligand interaction diagram. (b) Silibinin in contact with RdRP protein. (c) Schematic diagram of silibinin interaction with the amino acid residues of RdRP protein during MDs
Molecular dynamic simulation of silibinin with RdRp showed ligand contacts with the protein key amino acid residues. It showed H‐bond interaction with His‐438, Ala‐547, Ile‐548, Ala‐550, Lys‐551, Arg‐553, Ala‐554, Arg‐555, Asp‐618, Lys‐621, and Arg‐836, in addition to ionic bond interaction with His‐438, Lys‐551, Arg‐553, Lys‐621, and Lys‐798, in addition to hydrophobic interaction with Ala‐547, Lys‐551, Arg‐553, Arg‐555, Pro‐620, Lys‐621, and Lys‐798. The region made of Try‐619, Thr‐556, His‐164, and Val‐557 and Ser‐549, Asp‐452, and Phe‐442 residues was more flexible and with no interactions (Figure 12b). Silibinin‐protein complex showed similar interactions to the molecular docking interaction (Figure 12c). Silibinin exhibited hydrogen bond interactions between OH and Asp‐618, OH formed a water bridge with Asp‐618, phenyl ring formed pi‐cation bond with Lys‐798, while CH2OH displayed H‐bond interaction with Ala‐550 (Figure 12c).
MDs of silibinin with S protein (Figure 13) indicated a favorable ligand interaction with S protein amino acid residues. The complex showed fluctuation and reached stabilization after 30 ns with accepted RMSD range that revealed the complex stabilization (Figure 13a). MDs showed H‐bond interaction with chain C, Lys‐1038, Asn‐907, and Tyr‐904 as well as water bridge interaction with Lys‐1038, Trp‐886, Asn‐907, Tyr‐904, and Gln‐1036 along with hydrophobic contact with Lys‐1038, Trp‐886, and Tyr‐904 (Figure 13b). It also showed interaction with chain B via H‐bond interactions with Tyr‐904, Asn‐907, Gly‐910, Lys‐1038, and Arg‐1107, and hydrophobic contacts with Tyr‐904, and Lys‐1038, in addition to water bridge with Gly‐908, Ile‐909, Gly‐910, Val‐911, Gln‐1106, Tyr‐904, Asn‐907, Gln‐1036, and Asn‐1108 (Figure 13b). The OH of silibinin created water bridge with Gln‐1036, Lys‐1038, and Trp‐886, while the CH2OH exhibited water bridge with Asn‐907, and 1,4‐dioxane, and the O showed water bridge with Ile‐909 of chain C. OH also formed H‐bond with Lys‐1038, and Tyr‐906 (Figure 13c).
FIGURE 13.
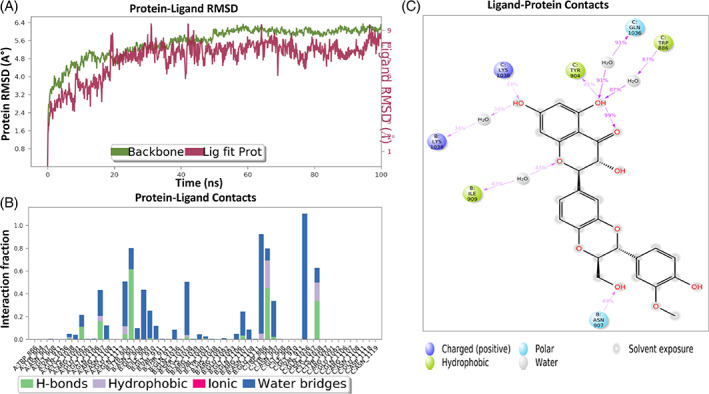
Interaction diagram of silibinin with S protein observed during the molecular dynamic simulation. (a) Protein‐ligand interaction diagram. (b) Silibinin in contact with S protein. (c) Schematic diagram of silibinin interaction with the amino acid residues of S protein during MDs
3.9. QSAR indicated the favorable pharmacophoric features of silibinin
To illustrate the effect of spatial arrangement on the activity of silibinin, contour plot analysis was performed. Silibinin showed the most significant favorable features as indicated in Figure 14. The positive contribution of steric, and electrostatic factors was indicated in green, and blue, respectively, while the negative one was shown as red.
FIGURE 14.
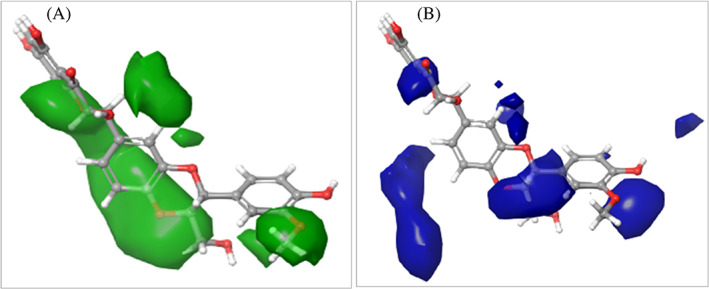
QSAR model visualized silibinin in the context of favorable and unfavorable factors. (a) Steric factor. (b) Electrostatic factors. Silibinin showed favorable binding features
4. DISCUSSION
The outbreak of the global pandemic caused by SARS‐CoV‐2 infection creates devastating social, economic, political, and health problems (Wang et al., 2020). SARS‐CoV‐2 is a coronavirus type with a lipid envelope and a positive‐sense single‐stranded RNA genome. The virion has structural and functional proteins that are essential for virion assembly and infection. These proteins include S protein, RdRp, and Mpro enzymes. Hence, the development of a therapeutic strategy that can target any of these proteins could combat SARS‐CoV‐2 and possibly other emergent new variants.
Plants natural products have been historically used in the prevention of many respiratory infections including viral infection. Recent extensive in silico studies using the natural products library indicated the potential activity of several natural products against COVID‐19. Withanone derived from Withania somnifera showed potential binding activity against TMPRSS2 (Kumar et al., 2020). Alkaloids such as 10‐hydroxyusambarensine, and cryptoquindoline, and terpenoids such as 6‐oxoisoiguesterin, and 22‐hydroxyhopan‐3‐one have been proposed as potential inhibitors against SARS‐CoV‐2 Mpro (Gyebi, Ogunro, Adegunloye, Ogunyemi, & Afolabi, 2021). Natural polyphenols including quercetin, naringenin, caffeine, oleuropein, ellagic acid, benzoic acid, resveratrol, and gallic acid showed potential inhibition activity against SARS‐CoV‐2 RdRp (El‐Aziz Abd, Mohamed, Awad, & El‐Sohaimy, 2020). Similarly, molecular docking indicated the potential activity of several flavonoids against SARS‐CoV‐2 targets (Alzaabi et al., 2021). Flavonoids are ubiquitous compounds of the plant kingdom and many have been approved as drugs or food supplements (Ahmad, Kaleem, Ahmed, & Shafiq, 2015; Yahia, García‐Solís, & Celis, 2019).
In this study, we showed that the flavonolignan silibinin out of 10 flavonoids with diverse structures including two flavanones, one flavone, four flavonols, and two isoflavones possesses significant and promising anti‐SARS‐CoV‐2 activity through in vitro assay and by targeting essential enzymes required for viral life cycle including Mpro, and RdRp. Silibinin showed also excellent binding activity to the viral S protein and hence inhibits the viral entry.
Our comparative in silico analysis of different flavonoids subclasses indicated the superior multi‐binding activity of the flavonolignan silibinin against SARS‐CoV‐2 vital proteins. This is consistent with several computational studies, which predict the formation of a stable complex between silibinin and SARS‐CoV‐2 S protein, its interaction with many residues of Mpro (Speciale et al., 2021), and the binding activity with SARS‐CoV‐2 RdRp (Bosch‐Barrera et al., 2020). The obtained in silico results were further confirmed in vitro. Silibinin, compared to other flavonoids subclasses, showed a promising selectivity index against SARS‐CoV‐2 incubated with Vero cells. Experimental validation of in vitro activity of silibinin against SARS‐CoV‐2 has never been investigated. For instance, silibinin shows excellent well‐tolerated antiviral activity against hepatitis C virus following intravenous injection particularly in patients not responding to treatment with interferon (Ferenci et al., 2008). Our obtained results were experimentally confirmed by investigating the enzyme binding activity of silibinin on vital SARS‐CoV‐2 proteins. In this study, silibinin exhibited excellent binding inhibition activity against SARS‐CoV‐2 S protein, Mpro and RdRp. MDs also confirmed the stability of silibinin complex with SARS‐CoV‐2 target proteins. However, as of our knowledge, there is no single report that tested the enzyme binding inhibition activity of silibinin with SARS‐CoV‐2 target proteins. Comparative in silico analysis accompanied with in vitro validation and enzyme inhibition assays of flavonoids representing different subclasses are considered as a powerful study to narrow down the selection of candidate compounds with promising activity, while the limitation of the QSAR model can be explained because of the few number of flavonoids identified with anti‐SARS‐CoV‐2 activity.
Generally, inhibition of SARS‐CoV‐2 can be achieved by targeting either the SARS‐CoV‐2 proteins or the host cell proteins that are important in viral infection (Shyr, Gorshkov, Chen, & Zheng, 2020). Targeting viral proteins have a major advantage over targeting the host cell targets as it provides higher specificity with minimal toxicity on humans. However, due to the frequent mutation that occurs in RNA virus, the emergence of drug resistance is a major challenge when developing new antiviral drugs. Therefore, it is essential to target more than one protein to overcome resistance mechanisms developed by the virus. Interestingly, silibinin showed excellent binding inhibition activity against three critical SARS‐CoV‐2 proteins including S protein, Mpro, and RdRp, which are essential for the virus entry (Zhu et al., 2020) and replication (Wu et al., 2020). On the other hand, silibinin did not show activity against TMPRSS2, despite its moderate binding affinity. This cannot exclude the potential binding inhibition activity of silibinin against TMPRSS2 since the examined concentrations were exceeded the in vitro IC50 by the double, whereas increasing the concentrations would increase the chance of toxicity. Therefore, it is not obvious to increase the concentration to reach the activity regardless of toxicity. It is important to prove the activity of the compound at its IC50, while avoiding any chance of toxicity.
Structural diversity of flavonoids including different electronic properties, hydrophobicity, and steric effects are closely correlated to their biological activity. Therefore, flavonoids are considered as an ideal candidate for structure activity‐related studies. The first screening revealed that silibinin showed the highest binding affinity to S protein, in addition to the potent in vitro antiviral activity. Further, the pharmacophore model discriminates the active and inactive flavonoids, explaining that spatial arrangement was crucial for the interaction and potential activity of silibinin, which has been confirmed by several computational studies as previously reported. The desirable structural features of silibinin and its good alignment indicates its superiority with the highest biological activity.
Recently, few studies highlighted the in vitro inhibition and enzyme inhibition efficacy of some flavonoids such as quercetin and its derivatives on SARS‐CoV‐2. For instance, rutin (quercetin‐3‐O‐rutinose) exhibited inhibition activity against Mpro with IC50 32 μM as reported by (Rizzuti et al., 2021). However, the obtained data was not validated on infected cells. Another recent study by (Mangiavacchi et al., 2021), showed that quercetin and its organoselenium derivative [8‐(p‐tolylselenyl) quercetin] blocked SARS‐CoV‐2 replication in infected cells at non‐toxic concentrations, and with IC50 192 μM and 8 μM, respectively. Quercetin and quercetin‐9 derivative exhibited inhibition activity against Mpro at IC50 21 μM and 2.2 μM, respectively. However, both compounds have not been screened against other SARS‐CoV‐2 proteins. By our study, silibinin is the only flavonoid tested on SARS‐CoV‐2 enzymes and on infected cells and showed multiple activity against SARS‐CoV‐2 S protein, Mpro and RdRp.
Silibinin also known as silybin, is a flavonolignan isolated from Silybum marianum (L.) Gaertn. (Asteraceae) (milk thistle) plant (Cheung, Gibbons, Johnson, & Nicol, 2010). Silibinin is the major active constituent of silymarin, a standardized extract of the milk thistle presents in the market under different trade names. Furthermore, following systematic administration, peak levels of silibinin were observed at 0.5–1 h in the liver, lung, lower bowel, pancreas, skin and prostate, indicating its excellent bioavailability and efficient targeting (Zhao & Agarwal, 1999). Silibinin like most flavonoids downregulates the NF‐κb pathway and pro‐inflammatory cytokine production (Giorgi, Peracoli, Peracoli, Witkin, & Bannwart‐Castro, 2012). Additionally, silibinin exhibits immunosuppressive and immunomodulatory activity by downregulating the secretion of pro‐inflammatory Th1 cytokines and upregulating the anti‐inflammatory Th2 cytokines (Min, Yoon, Kim, & Kim, 2007). This indicates the beneficial role of silibinin in COVID‐19 particularly in severe condition when cytokine storm is progressive.
Our study, which is a blind mapping study of selected flavonoids, indicated the multi‐targeting efficacy of silibinin against COVID‐19. Furthermore, in accordance with our safety study, silibinin confirms a wide safety profile and tolerability without interaction with immunosuppressive drugs (Rendina et al., 2014). Interestingly, a randomized placebo‐controlled trial to assess the clinical outcome in COVID‐19 pneumonia following administration of silymarin is currently in Phase III (Salem & Alfishawy, 2020). This study provided a tool to highlight the activity of untested flavonoids and to identify the features of their desirable structures for designing novel and more potent anti‐SARS‐CoV‐2 compounds.
5. CONCLUSIONS
Recently, flavonoids have gained lots of attention because of their potential activity against SARS‐CoV‐2, which is confirmed by several in silico studies. However, there is no single experimental validation as of our knowledge. Here, a comparative in silico study along with systematic evaluation of available literature were used to select a list of flavonoids with the highest scoring potential activity against SARS‐CoV‐2. This resulted in the selection of 10 flavonoids with diverse structures representing flavanone, flavone, flavonol, isoflavone, and flavonolignan. The selected flavonoids were then evaluated in vitro and by enzyme inhibition assays against vital SARS‐CoV‐2 target proteins. The results revealed that the flavonolignan silibinin possesses promising anti‐SARS‐CoV‐2 activity by multi‐targeting the virus itself, and its vital proteins including S protein, Mpro and RdRp, while maintaining a safety profile on normal human cells. Silibinin can be a subject of comparative study for further investigation in the future for the lead discovery of more potent and broader spectrum antiviral flavonolignans. Furthermore, silibinin and its flavonolignans analogues are U.S. FDA‐approved, giving an excellent opportunity for a future clinical study.
AUTHOR CONTRIBUTIONS
Rania Hamdy conducted the virtual screening, molecular docking, and develop the rational. Bahgat Fayed conducted the enzyme binding inhibition activity. Ahmed Mostafa and Noura M. Abo Shama conducted the in vitro activity. Rania Hamdy, Bahgat Fayed and Sameh S. M. Soliman wrote the first draft. Sameh S. M. Soliman designed the project, wrote the final draft, supervise the project, save the fund, and revise the manuscript. All data were generated in‐house, and no paper mill was used. Rania Hamdy, Ahmed Mostafa, Noura M. Abo Shama, Sameh S. M. Soliman, Bahgat Fayed agree to be accountable for all aspects of the work ensuring integrity and accuracy.
CONFLICT OF INTEREST
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Supporting information
TABLE S1 Pharmacokinetics and drug likeness of flavonoids
TABLE S2 Flavonoids with reported antiviral activity
TABLE S3 The binding affinity of flavonoids against vital SARS‐CoV‐2 protein targets
Hamdy, R. , Mostafa, A. , Abo Shama, N. M. , Soliman, S. S. M. , & Fayed, B. (2022). Comparative evaluation of flavonoids reveals the superiority and promising inhibition activity of silibinin against SARS‐CoV‐2. Phytotherapy Research, 36(7), 2921–2939. 10.1002/ptr.7486
Sameh S. M. Soliman and Bahgat Fayed contributed equally to this work.
Funding information This research has been funded by Sandooq Al‐Watan and University of Sharjah to SS under grant #133006 and by the Egyptian Academy of Scientific Research and Technology (ASRT) within the “Ideation Fund” program to AM under contract #7303
DATA AVAILABILITY STATEMENT
All data are available with the manuscript.
REFERENCES
- Abian, O. , Ortega‐Alarcon, D. , Jimenez‐Alesanco, A. , Ceballos‐Laita, L. , Vega, S. , Reyburn, H. T. , … Velazquez‐Campoy, A. (2020). Structural stability of SARS‐CoV‐2 3CLpro and identification of quercetin as an inhibitor by experimental screening. International Journal of Biological Macromolecules, 164, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, A. , Kaleem, M. , Ahmed, Z. , & Shafiq, H. (2015). Therapeutic potential of flavonoids and their mechanism of action against microbial and viral infections—A review. Food Research International, 77, 221–235. 10.1016/j.foodres.2015.06.021 [DOI] [Google Scholar]
- Alzaabi, M. M. , Hamdy, R. , Ashmawy, N. S. , Hamoda, A. M. , Alkhayat, F. , Khademi, N. N. , … Soliman, S. S. M. (2022). Flavonoids are promising safe therapy against COVID‐19. Phytochemistry Reviews, 21, 291–312. 10.1007/s11101-021-09759-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch‐Barrera, J. , Martin‐Castillo, B. , Buxó, M. , Brunet, J. , Encinar, J. A. , & Menendez, J. A. (2020). Silibinin and SARS‐CoV‐2: Dual targeting of host cytokine storm and virus replication machinery for clinical management of COVID‐19 patients. Journal of Clinical Medicine, 9(6), 1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Zheng, W. , Li, M. , Huang, J. , Bao, S. , Xu, Q. , & Ma, Z. (2020). Citrus fruits are rich in flavonoids for Immunoregulation and potential targeting ACE2. Preprint, 2020020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, C. W. , Gibbons, N. , Johnson, D. W. , & Nicol, D. L. (2010). Silibinin—A promising new treatment for cancer. Anti‐Cancer Agents in Medicinal Chemistry, 10(3), 186–195. 10.2174/1871520611009030186 [DOI] [PubMed] [Google Scholar]
- Chow, E. , Rendleman, C. A. , Bowers, K. J. , Dror, R. O. , Hughes, D. H. , Gullingsrud, J. , Sacerdoti, F. D. , & Shaw, D. E. (2008). Desmond performance on a cluster of multicore processors. DE Shaw Research Technical Report DESRES/TR–2008‐01. [Google Scholar]
- Coelho, G. R. , Figueiredo, C. A. , Negri, G. , Fernandes‐Silva, C. C. , Villar, K. D. S. , Badari, J. C. , … Mendonça, R. (2018). Antiviral activity of geopropolis extract from Scaptotrigona aff. postica against rubella virus. Food Research International, 7(6), 91–106. [Google Scholar]
- Daina, A. , Michielin, O. , & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug‐likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosa, G. , Maffioli, P. , D'Angelo, A. , & Di Pierro, F. (2021). A role for quercetin in coronavirus disease 2019 (COVID‐19). Phytotherapy Research, 35(3), 1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Petrillo, A. , Orrù, G. , Fais, A. , & Fantini, M. C. (2021). Quercetin and its derivates as antiviral potentials: A comprehensive review. Phytotherapy Research 36 (1), 266–278. 10.1002/ptr.7309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Aziz Abd, N. M. , Mohamed, G. S. , Awad, O. M. E. , & El‐Sohaimy, S. A. (2020). Inhibition of COVID‐19 RNA‐dependent RNA polymerase by natural bioactive compounds: Molecular docking analysis. Preprint. 10.21203/RS.3.RS-25850/V1 [DOI] [Google Scholar]
- Ferenci, P. , Scherzer, T. M. , Kerschner, H. , Rutter, K. , Beinhardt, S. , Hofer, H. , … Steindl‐Munda, P. (2008). Silibinin is a potent antiviral agent in patients with chronic hepatitis C not responding to Pegylated interferon/ribavirin therapy. Gastroenterology, 135(5), 1561–1567. 10.1053/j.gastro.2008.07.072 [DOI] [PubMed] [Google Scholar]
- Friesner, R. A. , Banks, J. L. , Murphy, R. B. , Halgren, T. A. , Klicic, J. J. , Mainz, D. T. , … Shenkin, P. S. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. [DOI] [PubMed] [Google Scholar]
- Fu, L. , Ye, F. , Feng, Y. , Yu, F. , Wang, Q. , Wu, Y. , … Gao, G. F. (2020). Both Boceprevir and GC376 efficaciously inhibit SARS‐CoV‐2 by targeting its main protease. Nature Communication, 11(1), 4417. 10.1038/s41467-020-18233-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai, S. A. , Abdullah, E. , Rashid, R. , & Altaf, M. (2017). Combinatorial in silico strategy towards identifying potential hotspots during inhibition of structurally identical HDAC1 and HDAC2 enzymes for effective chemotherapy against neurological disorders. Frontiers in Molecular Neuroscience, 10, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina, S. F. , Werner, D. S. , Pingle, M. , Feinberg, P. B. , Foreman, K. W. , Bergstrom, D. E. , … Barany, F. (2020). Novel, self‐assembling dimeric inhibitors of human β tryptase. Journal of Medicinal Chemistry, 63(6), 3004–3027. [DOI] [PubMed] [Google Scholar]
- Gil, C. , Ginex, T. , Maestro, I. , Nozal, V. , Barrado‐Gil, L. , Cuesta‐Geijo, M. Á. , … Martinez, A. (2020). COVID‐19: Drug targets and potential treatments. Journal of Medicinal Chemistry, 63(21), 12359–12386. [DOI] [PubMed] [Google Scholar]
- Giorgi, V. S. , Peracoli, M. T. , Peracoli, J. C. , Witkin, S. S. , & Bannwart‐Castro, C. F. (2012). Silibinin modulates the NF‐κb pathway and pro‐inflammatory cytokine production by mononuclear cells from preeclamptic women. Journal of Reproductive Immunology, 95(1–2), 67–72. 10.1016/j.jri.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Gour, A. , Manhas, D. , Bag, S. , Gorain, B. , & Nandi, U. (2021). Flavonoids as potential phytotherapeutics to combat cytokine storm in SARS‐CoV‐2. Phytotherapy Research, 35, 4258–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, J. R. , Calkins, D. , Sullivan, A. P. , & Shelley, J. C. (2010). Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug‐like molecules in aqueous solution. Journal of Computer‐Aided Molecular Design, 24(6), 591–604. [DOI] [PubMed] [Google Scholar]
- Gyebi, G. A. , Ogunro, O. B. , Adegunloye, A. P. , Ogunyemi, O. M. , & Afolabi, S. O. (2021). Potential inhibitors of coronavirus 3‐chymotrypsin‐like protease (3CL[pro]): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure & Dynamics, 39(9), 3396–3408. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren, T. A. , Murphy, R. B. , Friesner, R. A. , Beard, H. S. , Frye, L. L. , Pollard, W. T. , & Banks, J. L. (2004). Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry, 47(7), 1750–1759. [DOI] [PubMed] [Google Scholar]
- Hamdy, R. , Fayed, B. , Mostafa, A. , Shama, N. M. A. , Mahmoud, S. H. , Mehta, C. H. , … Soliman, S. S. M. (2021). Iterated virtual screening‐assisted antiviral and enzyme inhibition assays reveal the discovery of novel promising anti‐SARS‐CoV‐2 with dual activity. International Journal of Molecular Sciences, 22(16), 9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder, E. , Damm, W. , Maple, J. , Wu, C. , Reboul, M. , Xiang, J. Y. , … Friesner, R. A. (2016). OPLS3: A force field providing broad coverage of drug‐like small molecules and proteins. Journal of Chemical Theory and Computation, 12(1), 281–296. [DOI] [PubMed] [Google Scholar]
- Heinrich, M. , Appendino, G. , Efferth, T. , Fürst, R. , Izzo, A. A. , Kayser, O. , … Viljoen, A. (2020). Best practice in research—Overcoming common challenges in phytopharmacological research. Journal of Ethnopharmacology, 246, 112230. 10.1016/j.jep.2019.112230 [DOI] [PubMed] [Google Scholar]
- Hoffmann, M. , Hofmann‐Winkler, H. , Smith, J. C. , Krüger, N. , Sørensen, L. K. , Søgaard, O. S. , … Pöhlmann, S. (2020). Camostat mesylate inhibits SARS‐CoV‐2 activation by TMPRSS2‐related proteases and its metabolite GBPA exerts antiviral activity. The Lancet, 65, 103255. 10.1101/2020.08.05.237651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo, A. A. , Teixeira, M. , Alexander, S. P. , Cirino, G. , Docherty, J. R. , George, C. H. , … Panettieri, R. A. (2020). A practical guide for transparent reporting of research on natural products in the British Journal of Pharmacology: Reproducibility of natural product research. British Journal of Pharmacology, 177(10), 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, H. , Kim, S. , Shin, D. H. , & Kim, M. S. (2019). Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chemical Biology & Drug Design, 94(6), 2023–2030. 10.1111/cbdd.13604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, S. , Shin, D. H. , & Kim, M.‐S. (2020). Inhibition of SARS‐CoV 3CL protease by flavonoids. Journal of Enzyme Inhibition and Medicinal Chemistry, 35(1), 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeil, A. , Mostafa, A. , El‐Shesheny, R. , Shehata, M. , Roshdy, W. H. , Ahmed, S. S. , … Ali, M. A. (2020). Coding‐complete genome sequences of two SARS‐CoV‐2 isolates from Egypt. Microbiology Resource Announcements, 9(22), e00489‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. Y. , Leem, E. , Lee, J. M. , & Kim, S. R. (2020). Control of reactive oxygen species for the prevention of Parkinson's disease: The possible application of flavonoids. Antioxidants, 9(7), 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V. , Dhanjal, J. K. , Bhargava, P. , Kaul, A. , Wang, J. , Zhang, H. , … Sundar, D. (2022). Withanone and withaferin‐a are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS‐CoV‐2 into cells. Journal of Biomolecular Structure & Dynamics, 40, 1–13. 10.1080/07391102.2020.1775704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani, S. , & Poh, C. L. (2020). Flavonoids as antiviral agents for enterovirus A71 (EV‐A71). Viruses, 12(2), 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiavacchi, F. , Botwina, P. , Menichetti, E. , Bagnoli, L. , Rosati, O. , Marini, F. , … Kula‐Pacurar, A. (2021). Seleno‐functionalization of quercetin improves the non‐covalent inhibition of Mpro and its antiviral activity in cells against SARS‐CoV‐2. International Journal of Molecular Sciences, 22(13), 7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, K. , Yoon, W.‐K. , Kim, S. K. , & Kim, B.‐H. (2007). Immunosuppressive effect of silibinin in experimental autoimmune encephalomyelitis. Archives of Pharmacal Research, 30(10), 1265–1272. 10.1007/BF02980267 [DOI] [PubMed] [Google Scholar]
- Mostafa, A. , Kandeil, A. , Elshaier, Y. A. M. M. , Kutkat, O. , Moatasim, Y. , Rashad, A. A. , … GabAllah, M. (2020). FDA‐approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals, 13(12), 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucsi, I. , Gyulai, Z. , & Beladi, I. (1992). Combined effects of flavonoids and acyclovir against herpesviruses in cell cultures. Acta Microbiologica Hungarica, 39(2), 137–147. [PubMed] [Google Scholar]
- Rendina, M. , D'Amato, M. , Castellaneta, A. , Castellaneta, N. M. , Brambilla, N. , Giacovelli, G. , … di Leo, A. (2014). Antiviral activity and safety profile of silibinin in HCV patients with advanced fibrosis after liver transplantation: A randomized clinical trial. Transplant International, 27(7), 696–704. 10.1111/tri.12324 [DOI] [PubMed] [Google Scholar]
- Riva, L. , Yuan, S. , Yin, X. , Martin‐Sancho, L. , Matsunaga, N. , Pache, L. , … Hull, M. V. (2020). Discovery of SARS‐CoV‐2 antiviral drugs through large‐scale compound repurposing. Nature, 586(7827), 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuti, B. , Grande, F. , Conforti, F. , Jimenez‐Alesanco, A. , Ceballos‐Laita, L. , Ortega‐Alarcon, D. , … Velazquez‐Campoy, A. (2021). Rutin is a low micromolar inhibitor of SARS‐CoV‐2 main protease 3CLpro: Implications for drug design of quercetin analogs. Biomedicine, 9(4), 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez‐Álvarez, Y. , Arias, A. , del Águila, C. , & Agudo, R. (2019). Development of a fluorescence‐based method for the rapid determination of Zika virus polymerase activity and the screening of antiviral drugs. Scientific Reports, 9(1), 5397. 10.1038/s41598-019-41998-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, K. , & Alfishawy, M. (2020). Silymarin in COVID‐19 pneumonia (SCOPE). https://clinicaltrials.gov/ct2/show/record/NCT04394208 [Google Scholar]
- Sancineto, L. , Ostacolo, C. , Ortega‐Alarcon, D. , Jimenez‐Alesanco, A. , Ceballos‐Laita, L. , Vega, S. , … Dabrowska, A. (2021). l‐arginine improves solubility and ANTI SARS‐CoV‐2 Mpro activity of Rutin but not the antiviral activity in cells. Molecules (Basel, Switzerland), 26(19), 6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry, G. M. , Adzhigirey, M. , Day, T. , Annabhimoju, R. , & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer‐Aided Molecular Design, 27(3), 221–234. [DOI] [PubMed] [Google Scholar]
- Shelley, J. C. , Cholleti, A. , Frye, L. L. , Greenwood, J. R. , Timlin, M. R. , & Uchimaya, M. (2007). Epik: A software program for pK a prediction and protonation state generation for drug‐like molecules. Journal of Computer‐Aided Molecular Design, 21(12), 681–691. [DOI] [PubMed] [Google Scholar]
- Shyr, Z. A. , Gorshkov, K. , Chen, C. Z. , & Zheng, W. (2020). Drug discovery strategies for SARS‐CoV‐2. Journal of Pharmacology and Experimental Therapeutics, 375(1), 127–138. 10.1124/jpet.120.000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman, S. S. M. , Alhamidi, T. B. , Abdin, S. , Almehdi, A. M. , Semreen, M. H. , Alhumaidi, R. B. , … Omar, H. A. (2020). Effective targeting of breast cancer cells (MCF7) via novel biogenic synthesis of gold nanoparticles using cancer‐derived metabolites. PLoS ONE, 15(10), e0240156. 10.1371/journal.pone.0240156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman, S. S. M. , Saeed, B. Q. , Elseginy, S. A. , Al‐Marzooq, F. , Ahmady, I. M. , El‐Keblawy, A. A. , & Hamdy, R. (2020). Critical discovery and synthesis of novel antibacterial and resistance‐modifying agents inspired by plant phytochemical defense mechanisms. Chemico‐Biological Interaction, 333, 109318. 10.1016/j.cbi.2020.109318 [DOI] [PubMed] [Google Scholar]
- Somerville, V. S. , Braakhuis, A. J. , & Hopkins, W. G. (2016). Effect of flavonoids on upper respiratory tract infections and immune function: A systematic review and meta‐analysis. Advances in Nutrition, 7(3), 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speciale, A. , Muscarà, C. , Molonia, M. S. , Cimino, F. , Saija, A. , & Giofrè, S. V. (2021). Silibinin as potential tool against SARS‐CoV‐2: In silico spike receptor‐binding domain and main protease molecular docking analysis, and in vitro endothelial protective effects. Phytotherapy Research, 35, 1–10. 10.1002/ptr.7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Zhang, D. , du, G. , du, R. , Zhao, J. , Jin, Y. , … Wang, C. (2020). Remdesivir in adults with severe COVID‐19: A randomised, double‐blind, placebo‐controlled, multicentre trial. The Lancet, 395(10236), 1569–1578. 10.1016/S0140-6736(20)31022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, W. , Chen, C. , Tang, J. , Wang, C. , Zhou, M. , Cheng, Y. , … Mao, Q. (2022). Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID‐19: A meta‐analysis. Annals of Medicine, 54(1), 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Liu, Y. , Yang, Y. , Zhang, P. , Zhong, W. , Wang, Y. , … Li, H. (2020). Analysis of therapeutic targets for SARS‐CoV‐2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahia, E. M. , García‐Solís, P. , & Celis, M. E. M. (2019). Contribution of fruits and vegetables to human nutrition and health. In Yahia E. M. & Carrillo‐Lopez A. (Eds.), Postharvest physiology and biochemistry of fruits and vegetables (pp. 19–45). Woodhead Publishing. [Google Scholar]
- Yang, X. , Yu, Y. , Xu, J. , Shu, H. , Xia, J. , Liu, H. , … Shang, Y. (2020). Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: A single‐centered, retrospective, observational study. The Lancet Respiratory Medicine, 8(5), 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiske, T. , Stafford, K. A. , Friesner, R. A. , & Palmer III, A. G. (2013). Starting‐structure dependence of nanosecond timescale intersubstate transitions and reproducibility of MD‐derived order parameters. Proteins: Structure, Function, and Bioinformatics, 81(3), 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , & Agarwal, R. (1999). Tissue distribution of silibinin, the major active constituent of silymarin, in mice and its association with enhancement of phase II enzymes: Implications in cancer chemoprevention. Carcinogenesis, 20(11), 2101–2108. 10.1093/carcin/20.11.2101 [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Chen, C. Z. , Gorshkov, K. , Xu, M. , Lo, D. C. , & Zheng, W. (2020). RNA‐dependent RNA polymerase as a target for COVID‐19 drug discovery. SLAS, 25(10), 1141–1151. 10.1177/2472555220942123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Pharmacokinetics and drug likeness of flavonoids
TABLE S2 Flavonoids with reported antiviral activity
TABLE S3 The binding affinity of flavonoids against vital SARS‐CoV‐2 protein targets
Data Availability Statement
All data are available with the manuscript.


