Abstract
Human nasal mucosa is susceptible to severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection and serves as a reservoir for viral replication before spreading to other organs (e.g. the lung and brain) and transmission to other individuals. Chronic rhinosinusitis (CRS) is a common respiratory tract disease and there is evidence suggesting that susceptibility to SARS‐CoV‐2 infection differs between the two known subtypes, eosinophilic CRS and non‐ECRS (NECRS). However, the mechanism of SARS‐CoV‐2 infection in the human nasal mucosa and its association with CRS has not been experimentally validated. In this study, we investigated whether the human nasal mucosa is susceptible to SARS‐CoV‐2 infection and how different endotypes of CRS impact on viral infection and progression. Primary human nasal mucosa tissue culture revealed highly efficient SARS‐CoV‐2 viral infection and production, with particularly high susceptibility in the NECRS group. The gene expression differences suggested that human nasal mucosa is highly susceptible to SARS‐CoV‐2 infection, presumably due to an increase in ACE2‐expressing cells and a deficiency in antiviral immune response, especially for NECRS. Importantly, patients with NECRS may be at a particularly high risk of viral infection and transmission, and therefore, close monitoring should be considered.
Keywords: ACE2, Chronic rhinosinusitis, Eosinophils Human nasal mucosa, SARS‐CoV‐2
Human nasal mucosa can be infected by SARS‐Cov‐2 because ACE2 is wildly expressed through the mucosa. We culture the tissues in vitro to mimic the infection progress. The results provide that non‐eosinophilic CRS (NECRS) tissue is more sensitive to SARS‐Cov‐2 infection than eosinophilic CRS (ECRS) tissue and healthy control.
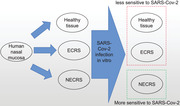
Introduction
SARS‐CoV‐2 was initially considered a viral pathogen of the lower respiratory tract because COVID‐19 is characterized by pneumonia, fever, and dry cough and appears to lack upper respiratory tract symptoms [1, 2]. However, it was quickly recognized that the upper respiratory system is also involved. Most infected individuals do not show symptoms or have mild‐to‐moderate symptoms and sensory dysfunction [3]. These patients are highly contagious with a high viral load of SARS‐CoV‐2 in nasal swab samples, suggesting that the upper respiratory tract, including the nasal cavity, is involved in disease progression and viral transmission.
As the main passway for inspired air, the nasal cavity not only acts as the front‐line respiratory defense mechanism but also serves as a primary site for respiratory pathogen infection and multiplication. Anatomically, the human nasal cavity is lined with a pseudostratified ciliated epithelium and is interspersed with tubulin+ mucus‐secreting goblet cells, differentiated from a basal layer of stem cells from the p63+ epithelium [4, 5]. Similar to other respiratory pathways, the human nasal mucosal epithelium, especially goblet and secretory cells, expresses high levels of ACE2 and TMPRSS2, key molecules in viral infection [6]. TMPRSS2 is widely expressed, while ACE2 expression is limited to specific organs and cell types. Hence, ACE2 expression is likely the main limiting factor for viral cell tropism [7, 8]. Emerging studies suggest that the human nasal mucosal epithelium is presumably susceptible to SARS‐CoV‐2, serving as a reservoir for viral replication before spreading to other organs, such as the lungs and central neural systems, and/or transmission to other individuals [9]. It is noteworthy that viruses in the nasal cavity may also escape vaccination and/or treatment with human neutralizing antibodies [10]. Considering SARS‐CoV‐2 has infected more than 508 million people worldwide, accounting for more than 6 million deaths, it is now imperative to comprehensively investigate SARS‐CoV‐2 infection in the human nasal cavity.
Chronic inflammation in the respiratory tract can compromise immunity, including the type I IFN response, and thus, increases susceptibility to viral infection [11, 12]. For patients with COVID‐19, pre‐existing asthma is associated with severe symptoms and/or mortality [13, 14, 15], and an upper respiratory tract infection and/or paranasal sinus predict worse disease symptoms and/or clinical outcomes [16]. Chronic rhinosinusitis (CRS) is a respiratory tract disease with a high prevalence, frequently with concomitant nasal polyps. CRS, characterized by inflammatory cell infiltration, can be subdivided into eosinophilic CRS (ECRS) and non‐ECRS (NECRS) [17]. ECRS is usually related to IL‐4‐ and IL‐13‐mediated Th2 responses and NECRS involves IFN‐related Th1 inflammation [18]. Clinical statistics suggested that the two types of CRS could have quite different susceptibilities to SARS‐CoV‐2, while NECRS may be at a higher risk with relatively severe symptoms [19, 20]. Further studies have shown that ACE2 expression was decreased in ECRS but increased in NECRS, presumably conferring a differential risk of infection between the two types, an assumption that merits experimental validation [9, 19, 20].
Considering the potential importance of the nasal mucosa in viral infection, intrapersonal spread, and interpersonal transmission, we investigated in detail whether the human nasal mucosa is susceptible to SARS‐CoV‐2 and how different types of CRS may impact infection.
Results
Demographic and clinical properties of patients with CRS
The clinical characteristics of the subjects are presented in Table 1. The groups were comparable in terms of age, female/male ratio, and nasal polyp status. The age of the patients ranged from 32 to 58 years, with a median age of 44.2 years. There were 34 males (56.7%) and 32 females (43.3%). As expected, a nasal mucosa and nasal polyp tissues evaluation revealed that patients with NECRS had substantially lower serum nasal nitric oxide (nNO) and total IgE levels and fewer infiltrating eosinophils than those of patients with ECRS (p < 0.01). Peripheral blood eosinophil cell (PBEC) counts were also significantly lower (p < 0.01).
Table 1.
Demographic characteristics and clinical properties of patients with chronic rhinitis
| Control | ECRS | NECRS | p‐value | |
|---|---|---|---|---|
| No. of subjects | 12 | 26 | 22 | / |
| Age (y) | 40.00 ± 8.29 | 45.00 ± 13.00 | 46.51 ± 10.64 | 0.64 |
| Sex (M/F) | 6/6 | 12/14 | 10/12 | / |
| Nasal polyps | 0 | 8 | 12 | / |
| nNO (ppb) | 394.00 ± 70.40 | 526.62 ± 84.00 | 180.27 ± 49.97 | <0. 01 |
| PBECs (1 × 109) | 0.25 ± 0.16 | 0.66 ± 0.11 | 0.55 ± 0.15 | <0. 01 |
| Serum total IgE (IU) | 135.00 ± 50.18 | 413.00 ± 46.00 | 112.26 ± 58.28 | <0. 01 |
| Local infiltrating eosinophilia (%) | 1.00 ± 0.74 | 32.05 ± 3.00 | 7.12 ± 3.30 | <0. 01 |
CRS, chronic rhinosinusitis; ECRS, eosinophilic CRS; IgE, immunoglobulin E; F, female; FeNO, fractional exhaled nitric oxide; Local infiltrating eosinophilia (%), eosinophils/total inflammatory cells; M, male; NECRS, non‐ECRS; PBEC, peripheral blood eosinophil cell. Data are presented as mean ± SD. Statistical significance was evaluated by one‐way ANOVA, chi‐squared test, and Tukey's test.
Expression and distribution of ACE2 and TMPRSS2 in the normal nasal mucosal epithelium
The results of a single‐cell RNA sequencing study were consistent with previous results [21, 22], ACE2 and TMPRSS2 were detected in the human nasal mucosa epithelium (Fig. 1A and B). ACE2 expression was mainly found in secretory cells and moderate in ciliated cells, suprabasal, and basal cells (Fig. 1A). ACE2 expression was also detected in cycling basal cells and glandular cells but was undetectable in most immune cells including CD4+ T cells, macrophages, and B cells. Conversely, TMPRSS2 was highly expressed in many cell types (Fig. 1A and B), including immune and stromal cells, suggesting that ACE2 is a key determinant of the types of susceptible cells in the human nasal mucosa.
Figure 1.
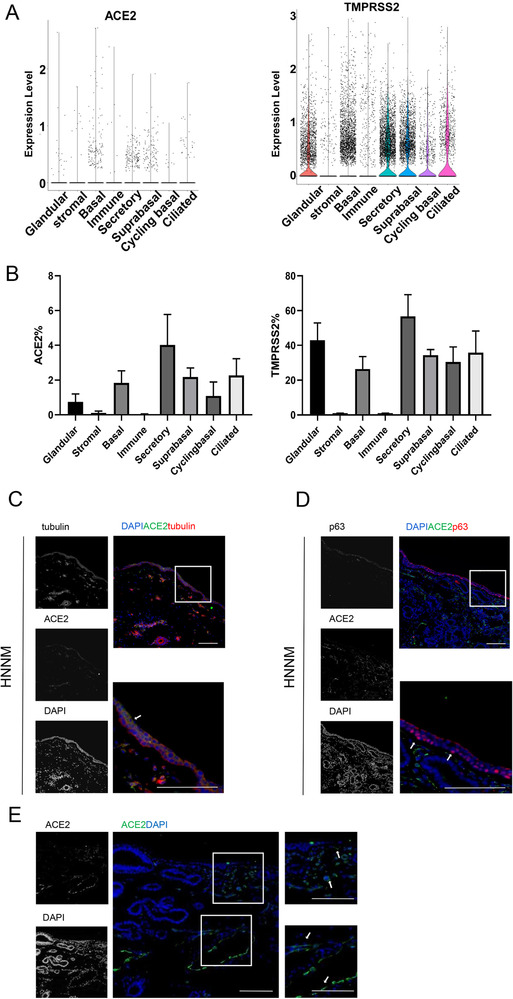
ACE2 expression pattern in human normal nasal mucosa tissues (HNNM). (A) ACE2 and TRPPSS expression across major cell types in HNNM. (B) Proportions of cells expressing ACE2 and/or TMPRSS2 across major cell types. Data are shown as mean ± SD and are pooled (n = 3) from three different samples. (C–D) ACE2 (green) expression in epithelial layer. (C) ACE2 (green) expression in tubulin+ cells (red) (including secretory cells, glandular cells, and ciliated cells). White arrows denote tubulin+ cells. (D) ACE2 (green) expression in p63+ basal cells (red). White arrows denote p63+ cells. (E) ACE2 (green) expression outside the epithelial layer. White arrows denote ACE2+ cells around blood vessels. (C‐E) One representative image from ten (C and E) or eight (D) independent samples is shown. Scale bar = 100 μm.
To further confirm the expression pattern of ACE2 in the human nasal mucosa epithelium, we performed immune costaining of ACE2 with tubulin, a marker for secretory cells, glandular cells, and ciliated cells, and p63, a marker for basal cells. ACE2 was expressed in the nasal mucosal epithelium (Fig. 1C and D). Similar to the results obtained by single‐cell RNA‐seq, ACE2 was highly expressed in tubulin‐positive cells (Fig. 1C). In contrast, p63‐positive basal cells expressed low levels of ACE2 (Fig. 1D). Furthermore, ACE2 was detected around blood vessels, presumably in pericytes and endothelial cells, but was not detected in most other tissues below the nasal mucosa (Fig. 1E).
Increased expression of ACE2 in NECRS
Then, we evaluated the expression dynamics of ACE2 in rhinosinusitis. Similar to the results for the normal human epithelium, ACE2 expression was highest in tubulin‐positive cells (Fig. 2A) in both ECRS and NECRS. ACE2 expression was also lower in p63‐positive basal cells (Fig. 2B). Interestingly, we noticed that the epithelial layers of tubulin and ACE2‐double positive cells, at least in the NECRS group, were considerably wider than those of the normal group, although the expression level of ACE2 was only slightly higher (Fig. 2C). In support of these observations, H&E staining clearly showed a significantly wider layer of epithelial cells in the NECRS group than in the control group (Fig. 2D).
Figure 2.
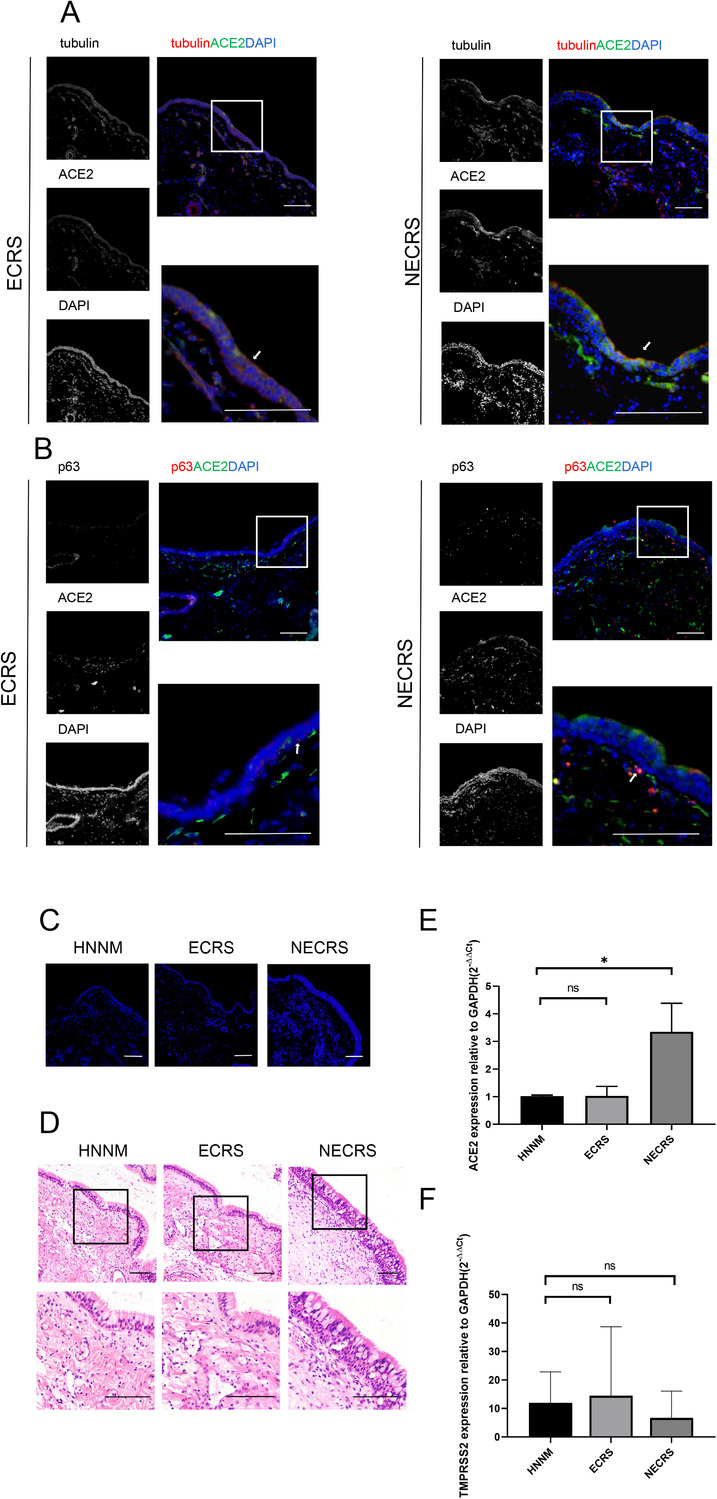
ACE2 expression dynamics in nasal mucosal tissues from patients with chronic rhinosinusitis (CRS). (A) ACE2 (green) expression in tubulin+ cells in both ECRS and NECRS tissues. (B) Expression of ACE2 (green) in p63+ basal cells (red). (C–D) DAPI and H&E staining show epithelial layers. (A–D) One representative image from five independent samples is shown for each type of tissues. Scale bar = 100 μm. (E–F) qRT‐PCR analysis show ACE2 expression (E) and TMPRSS2 expression (F) in NECRS group, control group, and ECRS group before infection. Data are shown as mean±SD of two to three samples for each type of the chronic sinusitis tissues and normal tissue. *p < 0.05; one‐way ANOVA.
To further quantify the differences in ACE2 and TMPRSS2 expression, we extracted total RNAs from the tissues and performed qRT‐PCR analyses. As shown in Fig. 2E, the level of ACE2 mRNA in the NECRS group was significantly higher than in the normal group (p < 0.05). The levels of ACE2 transcripts in ECRS were comparable to those of the normal group. No significant difference in the levels of TMPRSS2 transcripts was observed between the three groups (Fig. 2F).
Infection and replication of SARS‐CoV‐2 in the human nasal mucosa
Next, we established a primary tissue culture and infection system to evaluate the susceptibility of patients with different inflammation states (Fig. 3A) [23, 24]. Under the culture conditions used, primary nasal mucosa tissues can be maintained for longer than a week. As shown in Figure 3B, many vibrating cilia could be clearly observed in the culture on day 7. DAPI and H&E staining showed that the structure and integrity of the cells were maintained during the 7‐day in vitro culture period (Fig. 3C and D).
Figure 3.
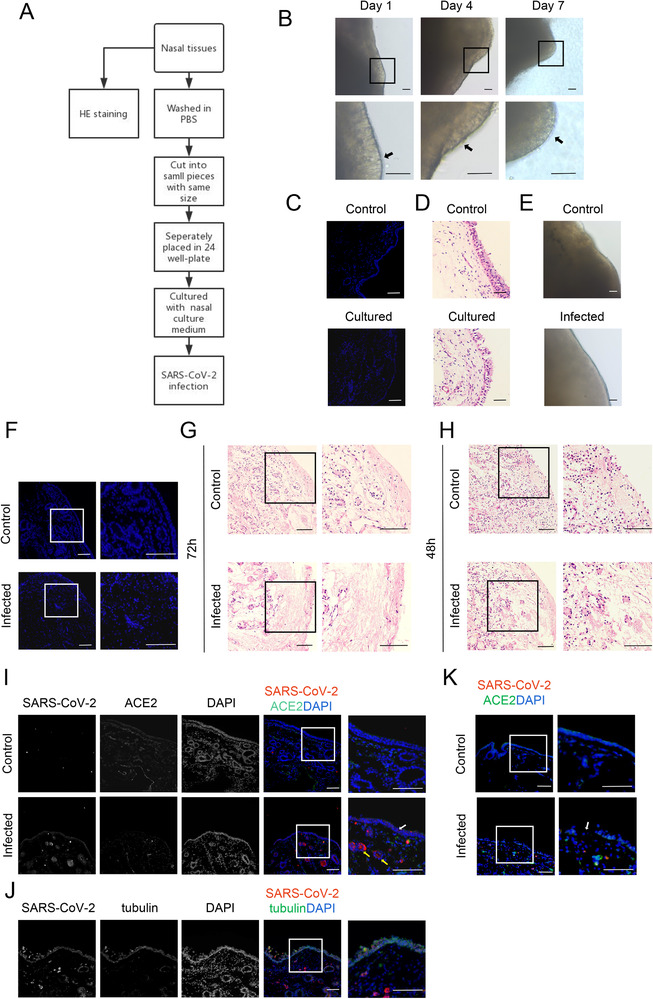
Human nasal mucosa tissue culture system and SARS‐CoV‐2 infection system. (A) Schematic diagram of nasal mucosa tissue culture and SARS‐CoV‐2 infection. (B) Phase contrast of nasal mucosa tissue cultured at days 1, 4, and 7. (C–D) DAPI and H&E staining of the structural integrity for nasal mucosa cultured on day 7. (E) Phase contrast of uninfected and infected tissues 72 h after viral infection. (F–G) DAPI and H&E staining show the structure of the nasal mucosa 72 h after viral infection. (H) H&E staining of nasal mucosa 48 h after viral infection. (I) SARS‐CoV‐2 N protein (red) in human nasal mucosa after 24 h of infection. White and yellow arrows denote viral infection in the epithelial layers and near the blood vessels. (J) SARS‐CoV‐2 N protein (red) and tubulin (green) in the human mucosal epithelium after 24 h of infection. (K) The structure of human nasal mucosa after 24 h of infection. White arrow denotes the broken epithelial layer. (B–K) One representative image from 20 (B) or 5 (C–K) independent samples is shown for each group. Scale bar = 100 μm, except in B and E (scale bar = 200 μm).
As expected, human nasal mucosa tissues were highly susceptible to SARS‐CoV‐2. As determined by light microscopy, both uninfected and infected tissues remained intact during the 72‐h infection period (Fig. 3E). Consistent with these observations, DAPI staining showed that uninfected control tissues remained intact (Fig. 3F). However, in striking contrast, DAPI staining clearly showed that the integrity of infected tissues was dramatically and globally compromised, well beyond the layers expressing ACE2 at 72 h (Fig. 3F). H&E staining revealed that most cells in infected tissues were absent, presumably due to direct viral infection and replication and indirect inflammation and stress/damage resulting from viral infection and replication (Fig. 3G). Nonetheless, tissues exposed to SARS‐CoV‐2 for 48 h remained grossly intact (Fig. 3H).
To determine the types of cells that were directly infected, we collected tissues at 24 and 48 h after viral infection. At 24 h, the infection was limited mainly to the epithelial layers (Fig. 3I), where tubulin and the double‐positive ACE2 cells are abundant (Fig. 3J). In the remaining tissues, infected cells were rarely seen, except around the blood vessels (Fig. 3I). However, at this early time point, tissues, including epithelial layers, remained intact (Fig. 3I). At 48 h, the infection was still limited to the epithelial layers and blood vessels. However, there were signs of structural deterioration of the tissues and the epithelial layers were no longer intact (Fig. 3K).
Increased susceptibility of NECRS to SARS‐CoV‐2
Finally, we examined the association between CRS and susceptibility to SARS‐CoV‐2. We subjected primary tissues from different disease states to SARS‐CoV‐2 infection. At 24 and 48 h after viral infection, we collected tissues for immunostaining and qRT‐PCR analyses.
Similar to the results for the control group, the nasal mucosal tissues in ECRS and NECRS were highly susceptible to SARS‐CoV‐2 (Fig. 4A). Infection was evident in tubulin‐positive epithelial cells (Fig. 4B). Importantly, we noticed significantly more infected cells in the NECRS group than in the other groups. At 24 h, when there were only scattered infected cells in the control and ECRS groups, almost the entire epithelial layer was infected in the NECRS group (Fig. 4A). A closer examination revealed that some parts of the infected epithelial layers showed thinning in the NECRS group, even at 24 h, while no signs of cell loss were detected in the other two groups at this early stage (Fig. 4C). This pattern became even more evident by hematoxylin staining, clearly showing that the nasal epithelial layers were considerably thinner in the NECRS group than in the uninfected group (Fig. 4D). There was also massive cell loss within the tissues (Fig. 4D). Conversely, the differences between infected and uninfected cells were negligible for the other two groups. In these groups, both the uninfected and infected tissues remained intact at this early stage.
Figure 4.
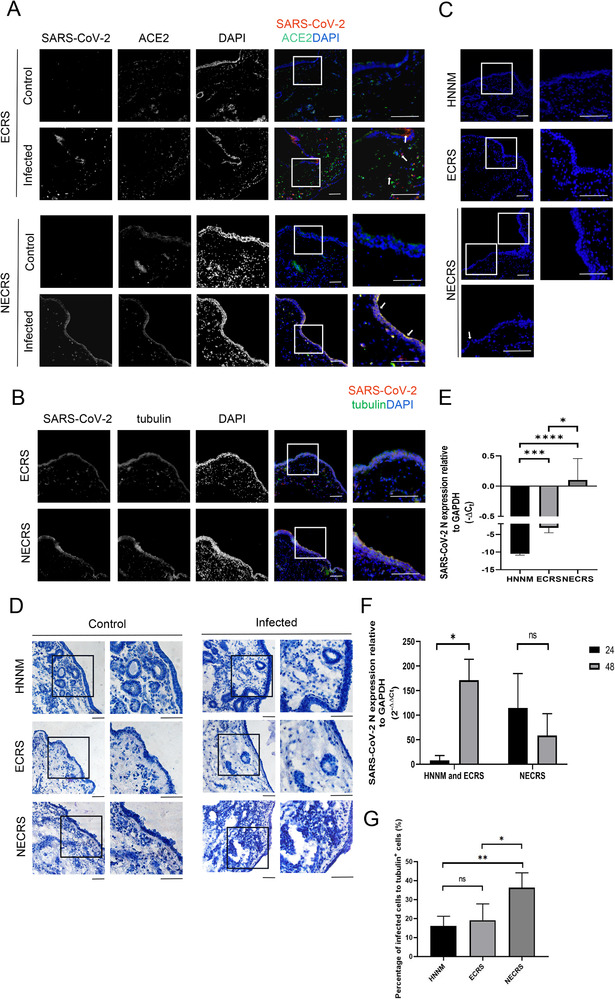
Increased susceptibility to SARS‐CoV‐2 in non‐ECRS (NECRS). (A) SARS‐CoV‐2 infection (red) in NECRS and ECRS nasal mucosal tissues. White arrows denote infected cells. (B) Viral infection (red) in tubulin+ cells (green). (C–D) DAPI and hematoxylin staining showing epithelial layer in NECRS after 24 h of infection. Some cavities visible within the infected NECRS nasal mucosal tissues in (D). (A–D) One representative image from a total of 30 (A–B) or 42 (C–D) independent samples (15 ECRS tissue samples, 15 NECRS tissue samples, and 12 healthy tissue samples) is shown for each group. Scale bar = 100 μm. (E) Viral susceptibility and production determined by qRT‐PCR at 24 h. (F) Comparation of viral infection and production at 24 and 48 h, as determined by qRT‐PCR. (E–F) Data are shown as mean ± SD of three to five group of samples from different patients for each type of the chronic sinusitis tissues and normal tissue.*p < 0.05,***p < 0.005,****p < 0.0005; one‐way ANOVA. (G) The percentage of infected cells to total cells according to immunofluorescence images (seven images of normal tissues, seven images of ECRS tissues, and seven images of NECRS tissues). Data are shown as mean ± SD. *p < 0.05, ***p < 0.005; one‐way ANOVA.
The above results strongly suggested that NECRS is related to increased susceptibility to SARS‐CoV‐2. To further confirm this finding, we prepared total RNAs from infected and uninfected samples and performed qRT‐PCR. At 24 h after viral infection, virus production in the NECRS group was significantly higher than that in the other two groups (Fig. 4E). At 48 h, SARS‐CoV‐2 production or titers increased dramatically in the ECRS and normal groups, suggesting continuous viral infection and replication over the course of time. However, instead of an increase, the titers in the NECRS group decreased substantially at 48 h (Fig. 4F). In some samples, viral titers at 48 h were several times lower than those at 24 h, presumably due to massive cell loss from viral infection and replication. Finally, we quantified the percentage of infected tubulin‐positive cells. At 24 h after viral infection, about 40% tubulin‐positive cells were already infected in the NECRS group, while only about 20% those were infected in the other two groups (Fig. 4G). Together, these results strongly suggest that NECRS tissues have increased susceptibility to SARS‐CoV‐2.
NECRS tissues showed little differences in gene expression after SARS‐CoV‐2 infection
To globally probe the mechanisms underlying the increased susceptibility in the NECRS group, we performed transcriptome analysis. As expected, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of ECRS and normal groups revealed that numerous pathways were differentially expressed before and 24 h after SARS‐Cov‐2 infection (Fig. 5A and B).The differentially regulated pathways were also similar between the groups, mainly including TNF signaling pathway, cytokine‐cytokine receptor interaction, and the chemokine signaling pathway. Strikingly, for the NECRS group, there was a very limited difference in gene expression before and after infection (Fig. 5C). But we did not find a significant difference in immune response genes between the NECRS and the ECRS groups after viral infection (Fig. 5D). These puzzling observations prompted us to compare the transcriptome between the NECRS group and the other two before infection. As expected, many differentially expressed genes between them were found, most enriched in immune and inflammatory processes such as viral protein interaction with cytokine and cytokine receptor, chemokine signaling pathway, neutrophil extracellular trap formation, NF‐kB signaling pathway, and/or TNF signaling (Fig. 5E and F).
Figure 5.
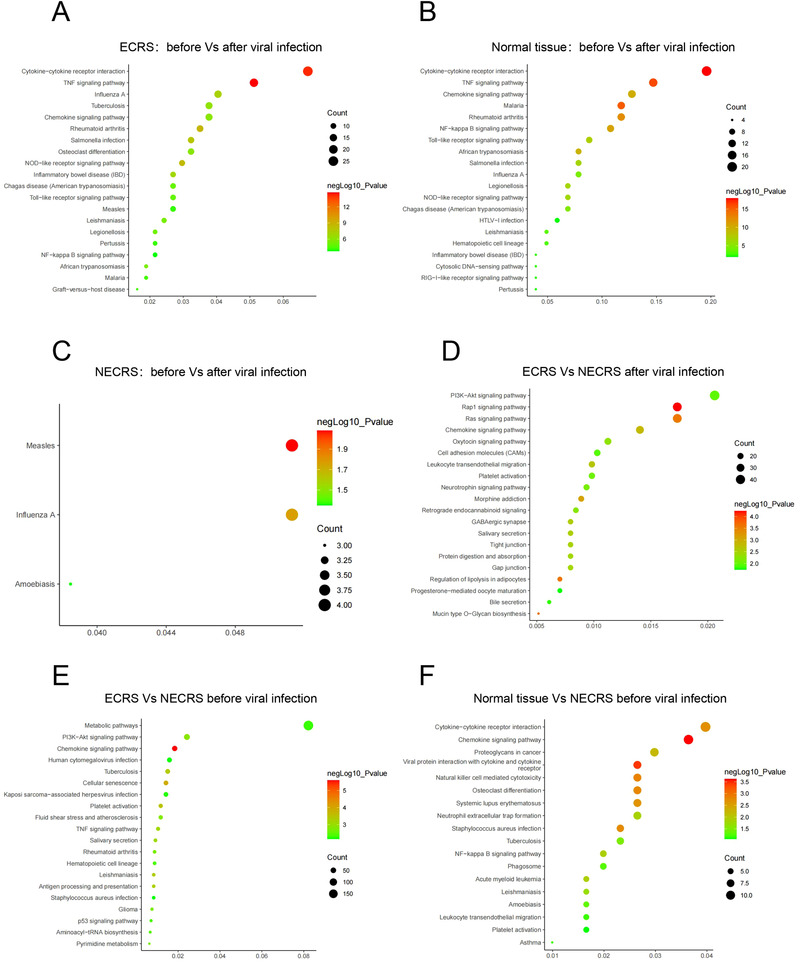
Differential gene signal pathway in infected nasal mucosa. (A) Differences in the gene signal pathway expression in ECRS tissues before and after the infection (n = 6). (B) Differences in the gene signal pathway expression in normal tissues before and after the infection (n = 6). (C) Differences in the gene signal pathway expression in NECRS tissues before and after the infection (n = 5). (D) Differences in the gene signal pathway expression in infected ECRS tissues (n = 6) and infected NECRS tissues (n = 5). (E) Differences in the gene signal pathway expression in ECRS tissues (n = 6) and NECRS tissues (n = 5) before infection. (F) Differences in the gene signal pathway expression in NECRS tissues (n = 6) and normal tissues (n = 6) before infection.
Discussion
The nasal cavity is not only the front‐line respiratory defense but also a major site of infection and reproduction by respiratory pathogens. Given the broad impact of the SARS‐COV‐2 pandemic and the central role of nasal infection in intra‐ and interpersonal viral spreading and transmission, it is of paramount importance to dissect the viral infection and reproduction in human nasal tissues. But such studies have been greatly hampered by the lack of human relevant clinical models. Therefore, it is imperative to develop a relatively easy and accessible in vitro model system that could closely recapture the characteristics of in vivo viral infection. In the present study, we have established an in vitro nasal mucosa tissue culture system, which can retain the physiological activity and immune function of the nasal mucosa. The system successfully supported the viral infection cycle and recaptured related immune responses. The ex vivo model developed herein represents a significant step forward, enabling functional studies and mechanistic investigations for implementable prevention strategies through nasal mucosa avenues.
Earlier studies reported that out of the two key molecules in SARS‐COV‐2 infection, TMPRSS2 is widely expressed and ACE2 expression is limited to specific cell types, often acting as a major limiting factor for viral cell tropism [23, 25]. Similarly, we have found that TMPRSS2 is broadly expressed in diverse cell types in the human nasal tissues. In contrast, ACE2 was only detected in a subset of nasal epithelium cells and presumably acted as a limiting factor for SARS‐COV2 infection. The selective expression of ACE2 in the nasal mucosa epithelium and the broad impact within 2 days of viral infection in our ex vivo culture system suggests that high ACE2‐expressing cells are not only susceptible to infection, but also may serve as a reservoir for SARS‐COV‐2 replication and amplification, leading to further infection. A number of studies have suggested that CRS, as a common nasal disease, might have an important impact on SARS‐CoV‐2 infection and transmission, thus, bearing importance in the ongoing pandemic [24, 26]. We observed an increased ACE2 expression in the NECRS nasal tissue, partially accounting for an increased SARS‐COV‐2 susceptibility as we and others have seen [9]. Our observation is also in line with earlier studies that have suggested that neutrophil inflammation and related cytokines, such as IFN‐γ and IL‐17, which are often associated with NECRS, could induce the expression of ACE2 and enhance viral infection [27]. Nonetheless, the ACE2 immunostaining patterns suggest that the significant expansion of ACE2‐positive cell layers in the NECRS tissue may matter more than the upregulation of ACE2 itself (Fig. 2). Utilizing the clinically relevant ex vivo model, we have showed that NECRS tissues had increased susceptibility to SARS‐Cov‐2, confirming earlier clinical observations [19, 20]. Our further transcriptomic analysis revealed a deficiency in immune response in NECRS tissue upon viral infection, presumably as a result of immune desensitization due to chronic inflammatory insults. This compromised antiviral response should also contribute to the increased susceptibility in the NECRS group. As such, patients with NECRS may be at higher risk both in terms of chances to be infected and disease severity once infected. As well, infected NECRS patients are likely to show faster and greater viral replication and, thus, stronger interpersonal transmission capacity.
Unlike NECRS, a rather different pattern may be expected for ECRS. Eosinophilic inflammation and related type 2 cytokines may downregulate the expression of ACE2 in human nasal mucosa tissues, exerting a protective effect on SARS‐CoV‐2 infection [20]. Similarly, because respiratory allergen exposure and the type 2 cytokine IL‐13 reduce the expression of the ACE2 gene in nasal and bronchial epithelial cells, patients with atopic asthma may have a reduced risk of developing diseases associated with COVID‐19 [19, 28]. Generally, in line with these earlier findings, our work with the ex vivo model showed that SARS‐CoV‐2 susceptibility in ECRS tissues was similar to that in the normal control ones.
In agreement with literature, we did not see a strong and timely antiviral IFN response in any of the three groups. Nonetheless, a dramatic induction of TNF signaling was evident in the ECRS and normal groups upon viral infection in our ex vivo model. TNF is one of the important cytokines in the acute phase of the inflammatory response and can trigger the activation of many pathways, including the NFkB and MAPK pathways, probably contributing to the viral defense response. TNF signaling could also trigger inflammatory cell death and result in tissue damage [29]. The failure to mount TNF signaling in the NECRS group is likely also resulted from chronic inflammation‐induced immune desensitization. In support of the speculation, a dampened immune response was already present in the NECRS tissue prior to viral infection, and as such, viral infection no longer promotes a strong antiviral immune response, rendering a milieu conducive to virus infection and contributing to the increased vulnerability of NECRS patients.
Finally, it is worth noting that the distribution of ECRS and NECRS is highly correlated with regions and races. Most patients with CRS in China are with ECRS but NECRS is more common in Europe [30, 31]. CRS is relatively common upper respiratory disease. Given the differential susceptibilities between the different types, our findings provide useful guidance for better management of the patients amid the pandemic.
In summary, we demonstrated that the human nasal mucosal epithelium contains abundant ACE2‐expressing cells and is highly susceptible to SARS‐CoV‐2. Our findings suggest that CRS, a common disease, is an important factor during the pandemic. In particular, the nasal mucosa epithelia from patients with NECRS contain more ACE2‐expressing cells and are highly susceptible to SARS‐CoV‐2 infection, increasing the risk of viral transmission [32]. As such, precautions and proper protective measures should be considered for patients with NECRS to protect the health of the patients and ensure public health safety. The ex vivo infection system that we have developed here also enables functional and mechanistic exploration directly with human samples and provides a great platform for drug testing and screening.
Materials and methods
Human nasal mucosal tissue samples
Sample collection was performed after obtaining Ethics Approval (Ethics Committee of Tongji Hospital) and informed consent from patients. Patients with CRS were diagnosed according to the European Position Paper on Rhinosinusitis and Nasal Polyps 2020 guidelines [33]. Patients who underwent septoplasty due to anatomical variation and had no other sinus diseases served as the control group. In total, 26 patients with ECRS, 22 patients with NECRS, and 12 healthy control subjects were enrolled at the Department of Otolaryngology of Tongji Hospital. Subjects with pregnancy, coagulopathy, fungal sinusitis, immunodeficiency, tumors, and/or infection with the SARS‐CoV‐2 virus were excluded. None of the patients received corticosteroids, antibiotics, or biological agents within 4 weeks before surgery. Nasal mucosa and nasal polyp tissues were collected during surgery from the middle nasal meatus and middle turbinate after obtaining informed consent. Necessary clinical data, such as age, sex, PBEC, total IgE, and nNO, were collected. Nasal tissues of the inferior or middle turbinate mucosa of healthy subjects were also collected as controls.
Pathological analysis was performed on a subset of tissue samples and the remaining were used for culture. The samples were washed with normal sterile saline immediately after isolation, transferred to sterile sample bottles, and stored temporarily in a 4°C refrigerator before subsequent analyses.
The studies involving human participants were reviewed and approved by the Research Ethics Committees of Tongji Hospital (K‐2021‐008). The patients or participants provided their written informed consent to participate in this study.
Single‐cell RNA‐seq analysis of ACE2 and TMPRSS2 in the human nasal mucosa epithelium
To examine the expression of ACE2 and TMPRSS2 in the human nasal mucosa, three normal nasal mucosa samples were chosen for single‐cell RNA sequencing analysis. Each sample was minced on ice, followed by enzymatic digestion. The supernatant was then centrifuged and removed without disturbing the cell pellet. The cell pellet was resuspended in 1 mL of RBC lysis buffer and filtered over 40‐μm cell strainers. The Cell Ranger software pipeline (version 3.1.0) provided by 10×Genomics was used to demultiplex cellular barcodes, map reads to the genome and transcriptome using the STAR aligner, and downsample reads as required to generate normalized aggregate data across samples, producing a matrix of gene counts versus cells. We processed the unique molecular identifier count matrix using the Seurat R package Seurat. The cells were then clustered according to a graph‐based clustering approach and visualized in two‐dimension using tSNE. The likelihood ratio test simultaneously tested for changes in mean expression and percentage of expressed cells was used to identify ACE2 and TMPRSS2 between groups.
Histological analysis
Nasal mucosal tissues were fixed in 4% paraformaldehyde (Fisher Scientific, Waltham, MA, USA) overnight. Dehydration and embedding in paraffin wax were performed after fixation. Sections of 8 μm were cut and stained with H&E. The presence of different types of inflammatory cells was evaluated by optical microscopy at ×400 magnification. For each patient, six nonoverlapping regions were randomly selected in each section and the absolute numbers and percentages of infiltrating inflammatory cells were quantified. Cell types mainly included eosinophils, neutrophils, plasma cells, and lymphocytes. The quantification was carried out by two independent pathologists blinded to the study design and clinical history of the patients. Tissues from patients with CRS were classified as ECRS or NECRS based on whether the percentage of infiltrating eosinophils exceeded 27% of total inflammatory cells [16, 34].
Human mucosal tissue culture
Human mucosal tissues were cultured in DMEM/F12 medium (Gibco, Waltham, MA, USA) with 500 ng/mL R‐spondin (CX83; Novoprotein, Summit, NJ, USA), 100 ng/mL Noggin (CB89; Novoprotein), 1×B27 (17504‐044; Gibco), 1× GlutaMAX 100 (A9921; Sigma, St. Louis, MO, USA), 1.25 mM N‐acetylcysteine (A9165; Sigma), nicotinamide (N0636; Sigma), 10 mM HEPES (15630080; Gibco), 25 ng/mL FGF7 (CH73; Novoprotein, Shanghai, China), 100 ng/mL FGF10 (CR11; Novoprotein, Shanghai, China), 5 μM Y‐27632 (HY‐10071; MCE, Princeton, NJ, USA), and 50 mg/mL Primocin (ant‐pm‐2; Invivogen, San Diego, CA, USA) at 37°C in 5% CO2. To monitor the integrity of the tissues, bright‐field images were obtained using a Nikon inverted microscope on days 1, 4, and 7.
SARS‐CoV‐2 virus amplification and infection
The SARS‐CoV‐2 virus (PubMed No: MT627325) was isolated, processed, and maintained at the ABSL‐3 laboratory of the Second Military Medical University. All infections were carried out in 24‐well plates with 1 ×105 viruses per well with the same batch of viruses. All experimental procedures involving live viruses were performed in the ABSL‐3 laboratory, with samples removed from the ABSL‐3 only after virus inactivation was completed. Uninfected samples were cultured at the same time as controls.
RNA extraction and quantitative PCR analysis of genes encoding the ACE2, TMPRSS2, and SARS‐CoV‐2 N proteins
Total RNA was extracted from uninfected and infected human nasal mucosa samples with TRIzol reagent (Invitrogen), following the manufacturer's instructions. Reverse transcription was performed using NovoScript Plus All‐in‐One 1st Strand cDNA Synthesis Supermix (GDNA Purge) (E047; Novoprotein, Shanghai, China). Quantitative real‐time PCR was performed with NovoStart SYBR Green Color qPCR Supermix (E168; Novoprotein, Shanghai, China) and QuantStudio 7 Flex. The primer sequences for the genes that encode the SARS‐CoV‐2 N protein, ACE2, and TMPRSS2 are included in Table 2. GAPDH was used as the internal control.
Table 2.
Primers
| Primers | Sequences |
|---|---|
| N‐protein‐R | CAGACATTTTGCTCTCAAGCTG |
| N‐protein‐F | GGGGAACTTCTCCTGCTAGAAT |
| ACE‐2‐R | GCTCTCTCCTTGGCCATGTT |
| ACE‐2‐F | ACACTGATGATGTTCAGACCTCC |
| GAPDH‐R | GAAGGCTGGGGCTCATTT |
| GAPDH‐F | CAGGAGGCATTGCTGATGAT |
| TMPRSS2‐R | CTCGGAGCATACTGAGGCA |
| TMPRSS2‐F | CAGTCTGAGCACATCTGTCCT |
RNA sequencing and gene expression analysis
RNA integrity was determined using an Agilent 2100 Bioanalyzer (Agilent; Palo Alto, CA, USA). The RNA quantity was determined using a NanoDrop (Thermo Fisher Scientific). Samples with a total RNA concentration greater than 50 ng/μL and OD260/OD280 between 1.8 and 2.1 were selected for RNA sequencing. In total, 12 normal tissue samples from four patients, 10 NECRS tissue samples from three patients, and 11 ECRS tissue samples from six patients were selected. The mRNA was purified using mRNA Capture Beads (VAHTS) and then fragmented and primed for cDNA synthesis using the VAHTS Universal V6 RNA‐seq Library Prep Kit according to the manufacturer's protocol. cDNA was converted to dsDNA using the kit reagents. The dsDNA was purified using VAHTS DNA clean beads. After adapter ligation, PCR was used to enrich DNA fragments with adapter molecules on both ends and to amplify the amount of DNA in the library. The resulting molecular libraries were pooled and sequenced on a NovaSeq 6000 system (Illumina Inc., San Diego, CA, USA). RNA sequencing was performed by Annoroad Gene Technology Inc. The KEGG and GO analyses were performed using online software (Morpheus, https://software.broadinstitute.org/morpheus).
Immunofluorescence staining
The control and infected human nasal mucosal tissues were fixed in 4% paraformaldehyde (Fisher Scientific) for 48 h at 4°C. The fixed tissues were then dehydrated in 20 and 30% sucrose solutions for 24 h before cryopreservation in opti‐mum cutting temperature compound (O.C.T. compounds) (4583; Sakura, Torrance, CA, USA). Sections of 12 μm were cut and transferred onto glass chips. Heat‐induced Epitope Retrieval was performed in a citrate buffer solution (pH 6.0) at 98°C for 15 min. After 20 min of permeabilization with 0.25% Triton X‐100 (Gibco), the samples were blocked for 2 h with immunostaining and primary antibody dilution solution (E674004; Sango Biotech, Shanghai, China) at room temperature. Primary antibodies were incubated at 4°C overnight, followed by a 2 h incubation with the secondary antibody at room temperature. Primary antibodies were as follows: anti‐ACE2 (10108‐T26; Sino Biological, Beijing, China), anti‐α‐tubulin (Invitrogen), anti‐p63/TP73L (AF1916; R&D Systems, Minneapolis, MN, USA), and the SARS‐CoV /SARS‐CoV‐2 N protein (MA5‐35943; Invitrogen). Images were obtained and analyzed using an Olympus BX53 microscope.
Statistical analyses
Statistical analyses were performed using SPSS v 20.0 (IBM Corp., Armonk, NY, USA). All data are expressed as mean ± SD. One‐way ANOVA was used to analyze differences, with the chi‐square test and Tukey's tests for multiple comparisons. p < 0.05 was considered statistically significant.
Conflict of interest
The authors have declared no financial or commercial conflict of interest.
Author contributions
Z.G., S.Y., P.Z., and D.D. conceived the idea, provided funds, and designed and supervised the study. Z.Z. H.P., L.J, and J.L. performed the experiments. K.F and Z.Z. collected the clinical samples. Z.Z. C.Z., S.J., L.W., and D.D analyzed the data and participated in the discussion. Z.G., S.Y., D.D., and Z.Z. wrote and revised the manuscript. All authors reviewed the manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202249805
Abbreviations
- CRS
chronic rhinosinusitis
- ECRS
eosinophilic CRS
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NECRS
non‐ECRS
- nNO
nasal nitric oxide
- PBEC
peripheral blood eosinophil cell
Acknowledgments
This work was supported by funds from the National Key R&D Program of China (No. 2019YFA0110300), the National Science Foundation of China (No. 81873689, 32070862, and 82173396), Clinical Research Plan of SHDC (No. SHDC2020CR4090), Clinical Science and Technology Innovation Project of SHDC (No. SHDC12019X07), and the Health Commission Advanced Technology Promotion Project of Shanghai City (No. 2019SY071).
Contributor Information
Dan Deng, Email: dengdan1234567@126.com.
Ping Zhao, Email: pnzhao@163.com.
Zhengliang Gao, Email: zhengliang_gao@tongji.edu.cn.
Shaoqing Yu, Email: yu_shaoqing@163.com.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , Zhao, X. et al., A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020. 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park, S. E. , Epidemiology, virology, and clinical features of severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2; Coronavirus Disease‐19). Clin. Exp. Pediatr. 2020. 63: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moein, S. T. , Hashemian, S. M. , Mansourafshar, B. , Khorram‐Tousi, A. , Tabarsi, P. and Doty, R. L. , Smell dysfunction: a biomarker for COVID‐19. Int. Forum Allergy Rhinol. 2020. 10: 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schleimer, R. P. , Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu. Rev. Pathol. 2017. 12: 331–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu, F. , Zhao, X. , Li, C. , Li, Y. , Yan, Y. , Shi, L. , Gordon, B. R. and Wang, D.‐Y. , Airway stem cells: review of potential impact on understanding of upper airway diseases. Laryngoscope 2012. 122: 1463–1469. [DOI] [PubMed] [Google Scholar]
- 6. Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. et al., SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020. 181: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sungnak, W. , Huang, N. , Bécavin, C. , Berg, M. , Queen, R. , Litvinukova, M. , Talavera‐López, C. et al., SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020. 26: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zou, X. , Chen, K. , Zou, J. , Han, P. , Hao, J. and Han, Z. , Single‐cell RNA‐seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019‐nCoV infection. Front. Med. 2020. 14: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang, M. , Bu, X. , Fang, G. , Luan, G. , Huang, Y. , Akdis, C. A. , Wang, C. et al., Distinct expression of SARS‐CoV‐2 receptor ACE2 correlates with endotypes of chronic rhinosinusitis with nasal polyps. Allergy 2021. 76: 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou, D. , Chan, J. F. , Zhou, B. , Zhou, R. , Li, S. , Shan, S. , Liu, L. et al., Robust SARS‐CoV‐2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021. 29: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Juhn, Y. J. , Risks for infection in patients with asthma (or other atopic conditions): is asthma more than a chronic airway disease? J. Allergy Clin. Immunol. 2014. 134: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edwards, M. R. , Strong, K. , Cameron, A. , Walton, R. P. , Jackson, D. J. and Johnston, S. L. , Viral infections in allergy and immunology: how allergic inflammation influences viral infections and illness. J. Allergy Clin. Immunol. 2017. 140: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhatraju, P. K. , Ghassemieh, B. J. , Nichols, M. , Kim, R. , Jerome, K. R. , Nalla, A. K. , Greninger, A. L. et al., Covid‐19 in critically ill patients in the Seattle region ‐ case series. N. Engl. J. Med. 2020. 382: 2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang, J. J. , Dong, X. , Cao, Y. Y. , Yuan, Y. D. , Yang, Y. B. , Yan, Y. Q. , Akdis, C. A. et al., Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy 2020. 75: 1730–1741. [DOI] [PubMed] [Google Scholar]
- 15. Li, X. , Xu, S. , Yu, M. , Wang, K. , Tao, Y. , Zhou, Y. , Shi, J. et al., Risk factors for severity and mortality in adult COVID‐19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020. 146: 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu, Z. , Hasegawa, K. , Ma, B. , Fujiogi, M. , Camargo, C. A. Jr. and Liang, L. , Association of obesity and its genetic predisposition with the risk of severe COVID‐19: analysis of population‐based cohort data. Metabolism 2020. 112: 154345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao, P. P. , Li, H. B. , Wang, B. F. , Wang, S. B. , You, X. J. , Cui, Y. H. , Wang, D.‐Y. et al., Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J. Allergy Clin. Immunol. 2009. 124: 478–484. [DOI] [PubMed] [Google Scholar]
- 18. Ouyang, Y. , Fan, E. , Li, Y. , Wang, X. and Zhang, L. , Clinical characteristics and expression of thymic stromal lymphopoetin in eosinophilic and non‐eosinophilic chronic rhinosinusitis. ORL J. Otorhinolaryngol. Relat. Spec. 2013. 75: 37–45. [DOI] [PubMed] [Google Scholar]
- 19. Yang, J. M. , Koh, H. Y. , Moon, S. Y. , Yoo, I. K. , Ha, E. K. , You, S. , Kim, S. Y. et al., Allergic disorders and susceptibility to and severity of COVID‐19: a nationwide cohort study. J. Allergy Clin. Immunol. 2020. 146: 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saheb Sharif‐Askari, F. , Saheb Sharif‐Askari, N. , Goel, S. , Fakhri, S. , Al‐Muhsen, S. , Hamid, Q. , Halwani, R. et al., Are patients with chronic rhinosinusitis with nasal polyps at a decreased risk of COVID‐19 infection? Int. Forum Allergy Rhinol. 2020. 10: 1182–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lukassen, S. , Chua, R. L. , Trefzer, T. , Kahn, N. C. , Schneider, M. A. , Muley, T. , Winter, H. et al., SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020. 39: e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bilinska, K. , Jakubowska, P. , Von Bartheld, C. S. , Butowt, R. , Expression of the SARS‐CoV‐2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem. Neurosci. 2020. 11: 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winkley, K. , Banerjee, D. , Bradley, T. , Koseva, B. , Cheung, W. A. , Selvarangan, R. , Pastinen, T. et al., Immune cell residency in the nasal mucosa may partially explain respiratory disease severity across the age range. Sci. Rep. 2021. 11: 15927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yao, Y. , Wang, H. and Liu, Z. , Expression of ACE2 in airways: Implication for COVID‐19 risk and disease management in patients with chronic inflammatory respiratory diseases. Clin. Exp. Allergy 2020. 50: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deprez, M. , Zaragosi, L. E. , Truchi, M. , Becavin, C. , Ruiz García, S. , Arguel, M. J. , Plaisant, M. et al., A single‐cell atlas of the human healthy airways. Am. J. Respir. Crit. Care Med. 2020. 202: 1636–1645. [DOI] [PubMed] [Google Scholar]
- 26. Wang, H. , Song, J. , Pan, L. , Yao, Y. , Deng, Y.‐K. , Wang, Z.‐C. , Liao, B. et al., The characterization of chronic rhinosinusitis in hospitalized patients with COVID‐19. J. Allergy Clin. Immunol. Pract. 2020. 8: 3597–3599.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hussman, J. P. , Cellular and molecular pathways of COVID‐19 and potential points of therapeutic intervention. Front. Pharmacol. 2020. 11: 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang, M. , Wang, C. , Zhang, L. , Inflammatory endotypes of CRSwNP and responses to COVID‐19. Curr. Opin. Allergy Clin. Immunol. 2021. 21: 8–15. [DOI] [PubMed] [Google Scholar]
- 29. Karki, R. , Sharma, B. R. , Tuladhar, S. , Williams, E. P. , Zalduondo, L. , Samir, P. , Zheng, M. et al., Synergism of TNF‐α and IFN‐γ triggers inflammatory cell death, tissue damage, and mortality in SARS‐CoV‐2 infection and cytokine shock syndromes. Cell 2021. 184(1): 149–168. https://pubmed.ncbi.nlm.nih.gov/33278357/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guan, W. J. , Zhong, N. S. , Clinical characteristics of Covid‐19 in China. Reply. N. Engl. J. Med. 2020. 382: 1861–1862. [DOI] [PubMed] [Google Scholar]
- 31. Bouayad, A. , Innate immune evasion by SARS‐CoV‐2: comparison with SARS‐CoV. Rev. Med. Virol. 2020. 30: 1–9. [DOI] [PubMed] [Google Scholar]
- 32. Goggin, R. K. , Bennett, C. A. , Bialasiewicz, S. , Vediappan, R. S. , Vreugde, S. , Wormald, P. J. and Psaltis, A. J. , The presence of virus significantly associates with chronic rhinosinusitis disease severity. Allergy 2019. 74: 1569–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fokkens, W. J. , Lund, V. J. , Hopkins, C. , Hellings, P. W. , Kern, R. , Reitsma, S. , Toppila‐Salmi, S. et al., European position paper on Rhinosinusitis and nasal polyps 2020. Rhinology 2020. 58: 1–464. [DOI] [PubMed] [Google Scholar]
- 34. Lou, H. , Zhang, N. , Bachert, C. and Zhang, L. , Highlights of eosinophilic chronic rhinosinusitis with nasal polyps in definition, prognosis, and advancement. Int. Forum Allergy Rhinol. 2018. 8, 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


