Abstract
RbsC of Escherichia coli is the hydrophobic membrane component of ribose uptake system classified as the ATP-binding cassette transporter. To understand the structure and function of RbsC, its transmembrane topology was investigated by using 64 RbsC-PhoA fusions isolated either specifically or randomly. In order to confirm the cytoplasmic location of the short C-terminal region (5 amino acids), inside-out or right-side-out membrane vesicles were generated, and the C-terminal region was found to be digested by carboxypeptidase A only in inside-out vesicles. This result is consistent with the model, based on the results of alkaline phosphatase fusions, in which the protein traverses the membrane six times and the N and C termini are exposed to the cytoplasm.
The bacterial periplasmic binding protein-dependent ATP-binding cassette (ABC) transporters are versatile in substrate specificity, e.g., sugars, amino acids, oligopeptides, and ions, etc., and generally consist of three to five proteins that fall into three different functional categories (10). The first component is the periplasmic substrate binding protein, which has two separate domains with a substrate binding cleft in the middle (16). The second component, consisting of either one or two homologous proteins, is a hydrophobic membrane protein that possesses a short conserved hydrophilic segment, containing a consensus EAA-G motif, located at approximately 100 amino acids (aa) from the C terminus (22). The hydrophilic third component has a highly conserved ATP-binding motif (Walker A and B boxes) and the conserved helical domain for interaction with the EAA-G region of the membrane component (15).
Topologies of the inner membrane components of the binding protein transporters have been determined by gene fusion and/or proteolysis techniques. The OppB and OppC proteins involved in oligopeptide transport (17) and MalG for maltose transport (5) were found to have six transmembrane (TM) helices with both ends oriented toward the cytoplasm. The MalF protein (514 aa), interacting with MalG for transport, comprises eight membrane-spanning segments with both ends of the protein exposed to the cytoplasm (3). ProW involved in the uptake of glycine betaine/proline has been reported to contain seven membrane-spanning segments with the C terminus inside the cytoplasm and the large N-terminal extension (100 aa) located within the periplasmic space (7). HisQ (228 aa) and HisM (235 aa) have five TM helices with periplasmic and cytoplasmic N and C termini, respectively (13). However, of a group of permeases known as the AraH/RbsC superfamily (21), transporting sugars such as arabinose, ribose, methylgalactoside, xylose, and allose, none were characterized for their membrane structures.
Ribose is transported in Escherichia coli through the high-affinity, binding-protein-dependent system encoded by genes in the rbs region of the chromosome (1). The system is composed of the periplasmic ribose-binding protein (RbsB), the hydrophobic membrane protein (RbsC), and the hydrophilic ATP-binding protein (RbsA). Other AraH transporters (currently more than 20 are in the database) have similar organizations with periplasmic, membrane, and ATPase components (12, 14, 19, 23). Information on the structure of the membrane component is a key to an understanding of the mechanism of sugar transport. However, membrane topologies of AraH/RbsC homologs were not readily predicted by a theoretical analysis since overall hydrophobicities of their amino acid sequences are so broad as to suggest five to eight putative TM helices (Hofmann and Stoffel algorithm [11]). In order to determine the membrane structure of RbsC, we made a series of RbsC-PhoA fusions by using existing restriction enzyme sites, PCR mutagenesis, or exonucleolytic digestion (6, 20). The results indicate that the structure of RbsC comprises six TM helices and cytoplasmically located N and C termini.
RbsC-PhoA fusions generated specifically or randomly.
For creating PhoA fusions at chosen sites, we used restriction enzymes (48N and 316V) and PCR (93I, 117V, 120A, 123R, 126A, 129A, 152T, 199I, 264G, 313A, 317D, and 321Q). The sites were chosen to represent locations which are likely to be in the hydrophilic regions based on the hydropathy profile of the RbsC protein. The fusions were constructed in the pYP110 plasmid, a derivative of pACYC184 which contains the signal-sequence-less phoA gene (from the first amino acid of the mature part) preceded by a multicloning site. The expression of rbsC was driven by the promoter of the tetracycline resistance gene from pACYC184. The fusions were introduced into the BW14893 strain (27) lacking the phoA gene on the chromosome, and their phenotypes were tested on indicator plates containing 5-bromo-4-chloro-3-indolyl-phosphate-p-toluidine (X-P).
To obtain random RbsC-PhoA fusions, the rbsC gene on the plasmid (pYP113) was serially digested with exonuclease III and S1 nuclease and fused to phoA located on the same plasmid (25). The pYP113 plasmid contains both genes in the same orientation that allowed rejoining of the C-terminally deleted rbsC to phoA which remained intact after digestion with exonuclease III. The plasmid is an rbsC-containing derivative of pYP110 in which rbsC is expressed from the tetA promoter. Linearization of pYP113 generated a 5′ overhang downstream of rbsC which was digested by exonuclease III (9). Derivatives of pYP113 harboring the rbsC-phoA fusions were then introduced into the BW14893 strain and grown on Luria-Bertani medium containing X-P. One hundred twenty-five blue colonies among 2,464 transformants were isolated, and their plasmids were analyzed by restriction enzyme digestions. According to their sizes, the fusions were classified into three groups, among which 60 were sequenced. Fusions were found in 38 unique sites, which are located in the three periplasmic regions of the proposed structure (Table 1 and Fig. 1).
TABLE 1.
RbsC-PhoA fusions
| Fusion site | Residue with no.a (PhoA activityb) |
|---|---|
| N terminus | 2T (9.93) |
| TM helix I | 36L (242.84), 39N (168.21) |
| Periplasmic loop I | 48N (109.2),c 50L (251.16) |
| TM helix II | 62G (238.77), 64T (161.9), 65L (191.59), 69T (13.6), 77G (39.2) |
| Cytoplasmic loop I | 93I (28.34),d 100A (62.18), 113A (16.41), 117V (45.42),d 120A (16.72),d 123R (3.87)d |
| TM helix III | 126A (5.45),d 129A (4.52)d |
| Periplasmic loop II | 147G (295.11), 152T (189.24),d 155T (103.26), 159D (204.25), 160L (181.33), 162G (183.52), 163W (154.43), 167G (195.31), 168R (152.01) |
| TM helix IV | 171G (189.72), 173P (229.46), 177W (169.37), 178I (51), 180G (104.08), 181I (113.47), 184L (62.61), 186A (114.86), 187W (93.59) |
| Cytoplasmic loop II | 195L (10.75), 199I (5.77),d 209T (1.5) |
| TM helix V | 222I (1.35), 225S (1.12), 236I (207.12) |
| Periplasmic loop III | 250G (245.89), 255L (355.52), 263L (270.05), 264G (19.7),d 271G (158.61), 287F (156), 288L (279.06) |
| TM helix VI | 297V (248.91), 301Y (243.16), 302Q (199.34), 304I (230.33), 306K (266.89), 307A (254.92), 308V (214.52), 309V (169.66), 310I (231.48), 311L (233.15), 313A (34.12),d 316V (71.46)c |
| C terminus | 317D (16.25),d 320K (74.3), 321Q (8.75)d |
The amino acid residue at the fusion joint of RbsC is indicated. Most of the fusions were constructed by serial deletion with exonuclease III unless otherwise indicated.
Alkaline phosphatase activities of RbsC-PhoA fusions were measured by the method of Brickman and Beckwith (4). The values are averages of three or four separate experiments. Boldface indicates values of more than 75 U of enzyme activity.
The fusions were made by using unique restriction enzyme sites in rbsC.
The fusions were obtained at specific sites by PCR.
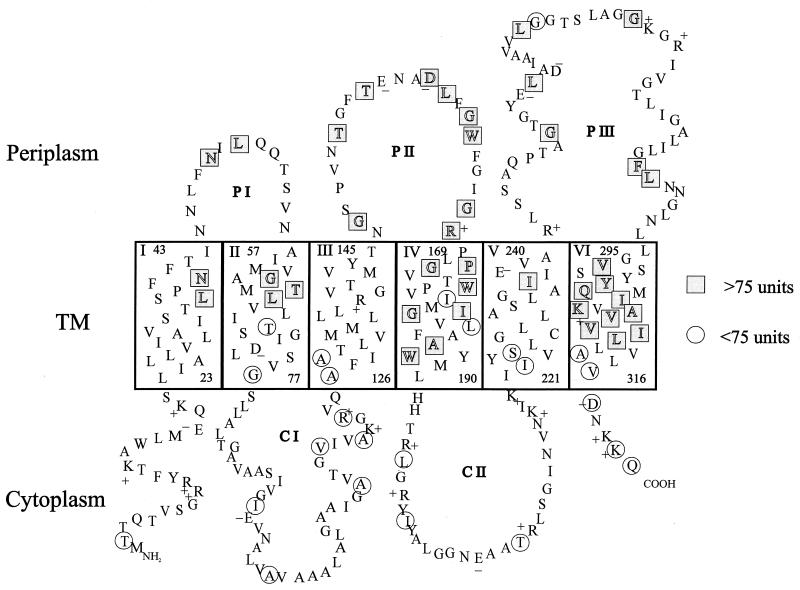
FIG. 1.
A proposed topology of RbsC. Residues in TM helices are arranged in an α-helical structure. Circles and rectangles indicate the residues of RbsC fused to PhoA. The periplasmic (P) and cytoplasmic (C) loops are designated. The charged residues are indicated with plus and minus signs.
For obtaining fusions in the cytoplasmic region, we first screened white and pale blue colonies that showed positive responses to PhoA immunoblotting, in order to avoid any out-of-frame construct. A total of 18 plasmids were isolated and subjected to DNA sequencing. Sixteen of them were found to be in frame with the phoA gene, giving rise to 12 fusions with unique sites (Table 1). These sites are located in the two cytoplasmic loops and the N- and C-terminal regions exposed to the cytoplasm (Fig. 1).
PhoA activities and stabilities of the fusion proteins.
Alkaline phosphatase activities of the RbsC-PhoA fusions shown in Table 1 ranged from 1.1 to 355 U. They are divided into two groups based on an arbitrary cutoff value of 75 U that can be used to predict their locations, i.e., more than 75 for periplasmic and less than 75 for cytoplasmic (8). As indicated in Fig. 1, it is likely that RbsC has six TM helices with three periplasmic and two cytoplasmic loops. Therefore, the N- and C-terminal regions of RbsC are located in the cytoplasm. Almost all the alkaline phosphatase fusions agreed with the six-TM model of RbsC. A possibility of RbsC having eight transmembrane regions, in which either CI or PIII forms an extra pair of TM peptides, has been carefully evaluated. However, it was deemed unlikely considering that the lengths of amino acids for the loops are barely enough to cover two TM spans and that the phoA fusions in the middle of the CI and PIII loops have distinctly low and high enzyme activities, respectively.
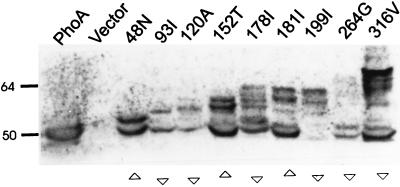
When steady-state levels of all the fusion products were analyzed by immunoblotting from whole-cell extracts, proteins corresponding to the RbsC-PhoA fusions and their degraded products were detected (only a few are shown in Fig. 2) with various intermediates and the PhoA moiety (47 kDa). In general, cytoplasmic fusions with lower PhoA activities exhibited reduced levels of intact or degraded PhoA proteins relative to periplasmic fusions (Fig. 2). This might be due to the fact that alkaline phosphatase located in the cytoplasm is often unable to fold correctly or to make its structure stable (20). The low alkaline phosphatase activity (Table 1) of the 264G fusion, predicted to be in a periplasmic loop, may be due to the instability of the fusion protein (6, 20). Indeed, the level of the protein was found (Fig. 2 and data not shown) to be lower than that of the other periplasmic fusions. On the other hand, the fusions in the C-terminal region, including 313A, 316V, 317D, 320K, and 321Q, produced fairly stable PhoA products (Fig. 2 and data not shown) despite their low alkaline phosphatase activities (ranging from 8.75 to 74.3 U). The reason might be that they have most of the membrane peptides without any truncated TM regions. It is noted that the fusions to inwardly directed TM peptides tended to exhibit high alkaline phosphatase activities (Fig. 1), which might be due to the fact that such fusions may reside in the periplasm (2).
FIG. 2.
Immunoblotting of RbsC-PhoA fusions. Equal amounts of solubilized cells containing RbsC-PhoA fusion proteins were resolved by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and immunoblotted with anti-alkaline phosphatase antiserum. Activities of alkaline phosphatase are designated underneath as more than 75 U (▵) or less than 75 U (▿). The positions of molecular mass standards (kilodaltons) are indicated.
Localization of the C terminus by carboxypeptidase A digestion.
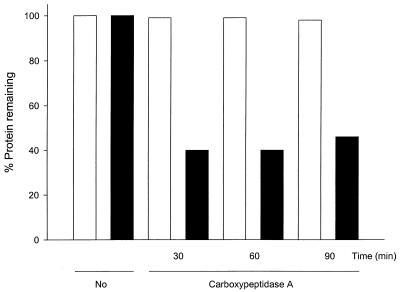
The location of the C terminus of RbsC was further confirmed by proteolysis. We constructed RbsC with a C-terminal His tag, which was functional when tested for sugar uptake and utilization (data not shown). The protein was expressed in cells under the IPTG (isopropyl-β-d-thiogalactopyranoside)-controlled promoter, and right-side-out vesicles and inside-out vesicles (IOV) were prepared as described elsewhere (13, 18). When right-side-out vesicles and IOV that contained the RbsC-His-tagged proteins were treated with carboxypeptidase A, the removal of the C-terminal histidines was observed only in IOV, confirming the location of the C-terminal region of RbsC in the cytoplasm (Fig. 3). However, the digestion was not complete even after treatment for 90 min. This appears to be due to an inaccessibility of the region to the enzyme, as reported for the N-terminal region of HisQ, which was not effectively digested by aminopeptidase (13). A similar approach employing proteolysis with vesicles or spheroplasts has proven useful in determining the topologies of ProW (7), HisQ, and HisM (13). The topology of OppB was confirmed by trypsin treatment for permeable and impermeable cytoplasmic membranes (17).
FIG. 3.
Carboxypeptidase A digestion of the C-terminal region of RbsC. Right-side-out vesicles and IOV with RbsC-His-tagged proteins were digested with carboxypeptidase A for various times (30, 60, and 90 min). The digestion products of vesicles were resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and immunoblotted with His tag conjugate. The immunoreactive bands were quantified with the ImageQuant program. The white and black bars indicate the percentages of the remaining immunoreactive bands of right-side-out vesicles and IOV, respectively.
Topology of RbsC and its implication for other AraH family members.
Our results suggest that RbsC is composed of six TM helices (TM I, II, III, IV, V, and VI), each with a size of 20 to 22 aa (Fig. 1). Two TM regions (TM I and IV) possess only neutral amino acids, while TM II, III, V, and VI each contain a charged amino acid (73D, 138R, 238E, and 306K, respectively). RbsC has three periplasmic loops, I (13 aa), II (23 aa), and III (54 aa). The cytoplasmic loops I (48 aa) and II (30 aa) also have +1 and +4 net charges, respectively (Fig. 1). The presence of the EAA-G motif in the cytoplasmic loop II is particularly notable, as it is conserved in all the membrane components of the ABC transporters and is supposed to interact with the cytoplasmic ATPase (RbsA [15]).
The N-terminal region of RbsC with a net charge of +3 (Fig. 1) is predicted to be located in the cytoplasm, which is consistent with the positive-inside rule (26), although the fusion at 2T with low alkaline phosphatase activity (9.93 U) may not be sufficient to prove its cytoplasmic location. The RbsC portion of the fusion is too short to contain a topogenic signal while the protein itself is stable, as seen on Western blots (data not shown). As a matter of fact, the evidence for the cytoplasmic location of the N terminus was already presented by the functionality of the RbsA-RbsC fusion in which the C terminus of the cytoplasmic RbsA is connected to the N terminus of RbsC (28). The short C-terminal region of RbsC has a net charge of +1 (Fig. 1), as is also predicted for AlsC (for allose), AraH (for arabinose), and MglC (for methyl-galactoside, all belonging to the AraH family) with positive net charges in the C-terminal regions.
As predicted for RbsC, other hydrophobic components belonging to the AraH superfamily may have six TM helices. In order to examine the structures of some AraH members that are close to RbsC, we made alkaline phosphatase fusions to the C-terminal regions of AlsC, AraH, MglC, and XylH. They exhibited low alkaline phosphatase activities (ranging from 2.47 to 24.45 U) with stable PhoA products (data not shown). Therefore, it is very likely that these and other AraH family members have structures very similar to that of RbsC. Some family members with unknown functions include YphD, YjfF, YtfT, YdeY, and YdeZ (from E. coli); TeuC1 and TeuC2 (from Rhizobium tropici); GguB (from Agrobacterium tumefaciens); and Y4mJ (from Rhizobium sp. strain NGR234), which are putative ABC-type sugar permeases with similarities to RbsC ranging from 22.8 to 33.5%. However, MalF, MalG, HisM, HisQ, OppB, OppC, and ProW in other families are more distant from RbsC, with 8.6 to 14.4% similarity. The majority of the inner membrane transport components other than the AraH members, are predicted to have six membrane-spanning segments with their amino and carboxy termini located in the cytoplasm (10).
In addition to their structural similarity, the AraH family members appear to have functional similarity. For example, it was found that the allose and xylose permeases can serve as a low-affinity transporter for ribose (14, 24), indicating that these hydrophobic components have substrate channels similar to those of RbsC. Interestingly, the ydeY-ydeZ (E. coli) and teuC1-teuC2 (R. tropici) pairs are each located in one operon. It seems plausible that they form heterodimers to serve as a functional permease, suggesting the possibility that RbsC forms a homodimer.
Acknowledgments
We thank W. Goebel for anti-PhoA antiserum, B. L. Wanner for the strain, H. Oh and S. Shin for the plasmids, and K. Park for computer analysis.
This work was supported by a grant, KOSEF 96-0401-01-01-3, from the Korea Science and Engineering Foundation and in part by the Creative Research Initiative Program.
REFERENCES
- 1.Bell A W, Buckel S D, Groarke J M, Hope J N, Kingsley D H, Hermodson M A. The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K12. J Biol Chem. 1986;261:7652–7658. [PubMed] [Google Scholar]
- 2.Boyd D, Beckwith J. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc Natl Acad Sci USA. 1989;86:9446–9450. doi: 10.1073/pnas.86.23.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 5.Dassa E, Muir S. Membrane topology of MalG, an inner membrane protein from the maltose transport system of Escherichia coli. Mol Microbiol. 1993;7:29–38. doi: 10.1111/j.1365-2958.1993.tb01094.x. [DOI] [PubMed] [Google Scholar]
- 6.Ginn S L, Brown M H, Skurray R A. Membrane topology of the metal-tetracycline/H+ antiporter TetA(K) from Staphylococcus aureus. J Bacteriol. 1997;179:3786–3789. doi: 10.1128/jb.179.11.3786-3789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol. 1996;178:5370–5381. doi: 10.1128/jb.178.18.5370-5381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagting A, Velde J, Poolman B, Konings W N. Membrane topology of the di- and tripeptide transport protein of Lactococcus lactis. Biochemistry. 1997;36:6777–6785. doi: 10.1021/bi963068t. [DOI] [PubMed] [Google Scholar]
- 9.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 10.Higgins C F. ABC transporters: from microorganism to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann K, Stoffel W. TM base—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166–170. [Google Scholar]
- 12.Hogg R W, Voelker C, Carlowitz I. Nucleotide sequence and analysis of the mgl operon of Escherichia coli K12. Mol Gen Genet. 1991;229:453–459. doi: 10.1007/BF00267469. [DOI] [PubMed] [Google Scholar]
- 13.Kerppola R E, Ames G F. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease: comparison with other members of the family. J Biol Chem. 1992;267:2329–2336. [PubMed] [Google Scholar]
- 14.Kim C, Song S, Park C. The d-allose operon of Escherichia coli K-12. J Bacteriol. 1997;179:7631–7637. doi: 10.1128/jb.179.24.7631-7637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mourez M, Hofnung M, Dassa E. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 1997;16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mowbray S L. Ribose and glucose-galactose receptors: competitors in bacterial chemotaxis. J Mol Biol. 1992;227:418–440. doi: 10.1016/0022-2836(92)90898-t. [DOI] [PubMed] [Google Scholar]
- 17.Pearce S R, Mimmack M L, Gallagher M P, Gileadi U, Hyde S C, Higgins C F. Membrane topology of the integral membrane components, OppB and OppC, of the oligopeptide permease of Salmonella typhimurium. Mol Microbiol. 1992;6:47–57. doi: 10.1111/j.1365-2958.1992.tb00836.x. [DOI] [PubMed] [Google Scholar]
- 18.Prossnitz E, Gee A, Ames G F. Reconstitution of the histidine periplasmic transport system in membrane vesicles: energy coupling and interaction between the binding protein and the membrane complex. J Biol Chem. 1989;264:5006–5014. [PubMed] [Google Scholar]
- 19.Rosenfeld S A, Tevis P E, Ho N W Y. Cloning and characterization of the xyl genes from Escherichia coli. Mol Gen Genet. 1984;194:410–415. doi: 10.1007/BF00425552. [DOI] [PubMed] [Google Scholar]
- 20.Sarsero J P, Pittard A J. Membrane topology analysis of Escherichia coli K-12 Mtr permease by alkaline phosphatase and β-galactosidase fusions. J Bacteriol. 1995;177:297–306. doi: 10.1128/jb.177.2.297-306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saurin W, Dassa E. Sequence relationships between integral inner membrane proteins of binding protein-dependent transport system: evolution by recurrent gene duplications. Protein Sci. 1994;3:325–344. doi: 10.1002/pro.5560030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saurin W, Koster W, Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 23.Scripture J B, Voelker C, Miller S, O’Donnell R T, Polgar L, Rade J, Horazdovsky B F, Hogg R W. High-affinity l-arabinose transport operon: nucleotide sequence and analysis of gene products. J Mol Biol. 1987;197:37–46. doi: 10.1016/0022-2836(87)90607-3. [DOI] [PubMed] [Google Scholar]
- 24.Song S, Park C. Utilization of d-ribose through d-xylose transporter. FEMS Microbiol Lett. 1998;163:255–261. doi: 10.1111/j.1574-6968.1998.tb13054.x. [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama J E, Mahmoodian S, Jacobson G R. Membrane topology analysis of Escherichia coli mannitol permease by using a nested-deletion method to create mtlA-phoA fusions. Proc Natl Acad Sci USA. 1991;88:9603–9607. doi: 10.1073/pnas.88.21.9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilmes-Riesenberg M R, Wanner B L. TnphoA and TnphoA′ elements for making and switching fusions for study of transcription, translation, and cell surface localization. J Bacteriol. 1992;174:4558–4575. doi: 10.1128/jb.174.14.4558-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaitseva J, Zhang H, Binnie R A, Hermodson M. The proteins encoded by the rbs operon of Escherichia coli. II. Use of chimeric protein constructs to isolate and characterize RbsC. Protein Sci. 1996;5:1100–1107. doi: 10.1002/pro.5560050612. [DOI] [PMC free article] [PubMed] [Google Scholar]