Abstract
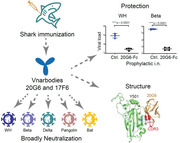
The identification of a novel class of shark‐derived single domain antibodies, named vnarbodies that show picomolar affinities binding to the receptor binding domain (RBD) of Wuhan and Alpha, Beta, Kappa, Delta, Delta‐plus, and Lambda variants, is reported. Vnarbody 20G6 and 17F6 have broad neutralizing activities against all these SARS‐CoV‐2 viruses as well as other sarbecoviruses, including Pangolin coronavirus and Bat coronavirus. Intranasal administration of 20G6 effectively protects mice from the challenges of SARS‐CoV‐2 Wuhan and Beta variants. 20G6 and 17F6 contain a unique “WXGY” motif in the complementary determining region 3 that binds to a hidden epitope on RBD, which is highly conserved in sarbecoviruses through a novel β‐sheet interaction. It is found that the S375F mutation on Omicron RBD disrupts the structure of β‐strand, thus impair the binding with 20G6. The study demonstrates that shark‐derived vnarbodies offer a prophylactic and therapeutic option against most SARS‐CoV‐2 variants and provide insights into antibody evasion by the Omicron variant.
Keywords: neutralization, receptor binding domain (RBD), SARS‐CoV‐2, variants of concerns (VOCs)
Shark‐derived vnarbodies 20G6 and 17F6 broadly neutralize SARS‐CoV‐2 Wuhan, Alpha, Beta, Kappa, Delta, Delta‐plus, Lambda variants, and sarbecovirus coronaviruses, but are unexpectedly ineffective for Omicron. Intranasal administration of 20G6 confers effective preventive and therapeutic effects in mice infected with SARS‐CoV‐2 Wuhan and Beta variant. Structural analysis elucidates the neutralizing mechanism and revealed the S375F mutation on Omicron receptor binding domain renders the evasion of neutralization by 20G6‐like antibodies.
1. Introduction
Since the outbreak of the COVID‐19 pandemic in December 2019,[ 1 , 2 ] about 5 billion people have been infected and more than 6 million deaths have been reported to World Health Organization as of April 9, 2022. Many SARS‐CoV‐2 variants appeared under selective pressure from host immunity with high infectivity and immune evasion, such as the Alpha (B.1.1.7),[ 3 , 4 ] Beta (B.1.351),[ 5 ] Delta (B.1.617.2),[ 6 ] Gamma (P.1)[ 7 ] and Omicron (B.1.1.529).[ 8 ] These variants brought considerable challenges to the prevention and treatment of SARS‐CoV‐2 infections. Effective and affordable preventive and therapeutic strategies are urgently needed. Understanding mechanisms of virus neutralization and the escape will expedite our effort in virus prevention and therapy.
In addition to the active immunity brought by vaccination, the passive immunity of neutralizing antibodies can play an important role in preventing and treating infectious diseases. The vaccine's protective effect is greatly reduced in people with weakened immune systems, such as elderly and people with immune‐compromised conditions. In this case, the role of passive immunity is significant. The convalescent serum has been used to treat COVID‐19 patients with a considerable effect but is limited by the scarcity of sources and possible side effects.[ 9 , 10 ]
Like other coronaviruses, SARS‐CoV‐2 entry into host cells is mediated by the homotrimeric spike (S) glycoprotein.[ 11 ] The spike is composed of two subunits, S1 and S2. S1 binds the host cell receptor angiotensin‐converting enzyme 2 (ACE2) by the receptor‐binding domain (RBD),[ 12 , 13 ] which serves as the primary target for neutralizing antibodies. S2 is responsible for fusing the virus with cellular membranes.[ 14 ] Many human‐derived SARS‐CoV‐2 neutralizing antibodies targeting S protein have been identified and developed, some of which have entered clinical trials.[ 15 ] However, the high production costs, large doses needed, and low‐temperature requirements for transportation and storage make it challenging to apply on a large scale cost‐effectively.
Single‐domain antibodies or variable domain of the heavy chain of HACbs (VHH) from camelids, having a smaller molecular weight (15 kD) than human antibodies (150 kD), are commonly termed nanobodies.[ 16 , 17 ] Nanobodies have shown great potential in biomedical applications, including cancer, infection, autoimmune disease, inflammation, and other diseases.[ 18 , 19 , 20 , 21 ] The first nanobody‐based medicine was approved to treat acquired thrombotic thrombocytopenic purpura (aTTP) in 2018.[ 22 ] Another class of single‐domain antibodies, namely variable new antigen receptor (VNAR), having a smaller molecular weight of 13 kD, was found in cartilaginous fish such as sharks, skates, and rays.[ 23 ] Single‐domain antibodies have advantages such as high affinity to target proteins, better thermal stability, small molecular weight, and low production cost in nonmammalian expression systems. One of the advantages of single‐domain antibodies is that they can bind to epitopes that are not reachable by traditional human antibodies because of their smaller size and structure.
A few camelid‐derived nanobodies against the RBD region of the SARS‐CoV‐2 Spike (S) protein have been reported recently.[ 24 , 25 , 26 , 27 ] We took a different approach by searching shark‐derived single domain antibodies, VNAR, which we termed it as vnarbodies to distinguish from camel‐derived nanobodies, for neutralizing SARS‐CoV‐2. Since shark blood and body fluids contain high concentration of urea (about 350 × 10−3 m),[ 28 ] shark‐derived vnarbodies may possess better physiochemical stability than human‐derived antibodies. We immunized bamboo sharks with SARS‐CoV‐2 Spike protein and constructed a phage display library to identify vnarbodies with picomolar binding affinities to RBD. A lead vnarbody, 20G6, was further accessed for its neutralizing activities against various SARS‐CoV‐2 variants and its efficacy in preventing and treating mice challenged with SARS‐CoV‐2. Structural biology analysis was carried out to understand the mechanism of neutralization and resistance.
2. Results
2.1. Identification of RBD Binding Vnarbodies from Phage Display Library Derived from Immunized Bamboo Shark (Chiloscyllium plagiosum)
We first immunized a bamboo shark with SARS‐CoV‐2 Spike protein (Figure 1A). Shark serum collected 10 days after a booster immunization contained RBD‐specific binding antibodies at the half‐maximal effective concentration (EC50) titer of 1:13740 (Figure S1A, Supporting Information). The immune serum could neutralize pseudotyped SARS‐CoV‐2 Wuhan at the half‐maximal neutralizing titer (NT50) of 1:10000 (Figure S1B, Supporting Information).
Figure 1.
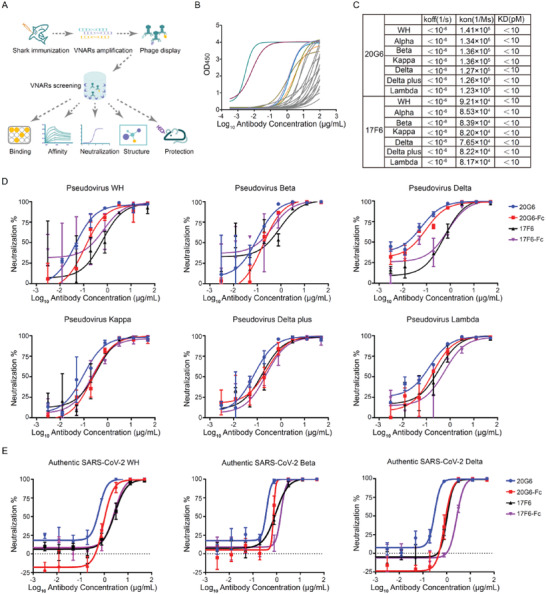
Identification and characterization of receptor binding domain (RBD) specific vnarbodies. A) Study design of this work. B) Enzyme‐linked immunosorbent assay (ELISA) measurement of 26 isolated vnarbodies against RBD of SARS‐CoV‐2 Wuhan. 17F6 and 20G6 (purple and green) show the best binding activities. C) The biolayer interferometry (BLI) binding kinetics of 20G6‐Fc and 17F6‐Fc to RBD of SARS‐CoV‐2 variants. D) The neutralization potency of 20G6 and 17F6 were calculated based on the pseudotyped SARS‐CoV‐2 neutralization assay (luciferase). Blue, red, black, and purple lines denote monovalent 20G6, bivalent 20G6 (20G6‐Fc), monovalent 17F6, and bivalent 17F6 (17F6‐Fc). Data represent as one of at least two independent experiments. The average neutralization percentage was shown for each data point (n = 3). E) The neutralization potency of 20G6 and 17F6 by SARS‐CoV‐2 focus reduction neutralization test (FRNT). The Wuhan (WH), Beta variant, and Delta variant SARS‐CoV‐2 virus were used. The average neutralization percentage was shown for each data point (n = 3).
Total RNA was then extracted from shark blood cells and converted into cDNA to construct a phage library. The immune phage library contained over 2 × 108 clones with diversity exceeding 95%. Using RBD protein of SARS‐CoV‐2 Wuhan as the binding antigen, 26 RBD‐specific vnarbodies were obtained after three rounds of bio‐panning. Nine of them presented significant RBD binding activities. Vnarbodies 20G6 and 17F6 showed the highest EC50 at 0.003 and 0.007 µg mL−1, respectively (Figure 1B). We chose a prokaryotic system with low production cost to express 20G6 and 17F6 monomers. The production yield was up to 100 mg L−1 under laboratory conditions, with good antibody homogeneity and no production of dimers and multimers (Figure S2A,B, Supporting Information). The vnarbodies have good thermal stability and retain strong binding activity even after treatment at 90° for 1 h (Figure S2C, Supporting Information).
We constructed 20G6 and 17F6 dimer fusion proteins using the human IgG1 Fc domain. The divalent 20G6‐Fc and 17F6‐Fc were evaluated for binding to RBDs of SARS‐CoV‐2 variants, including Alpha (N501Y), Beta (K417N, E484K, N501Y), Kappa (L452R, E484Q), Delta (L452R, T478K), Delta plus (K417N, L452R, T478K) and Lambda (L452Q, F490S). Both 20G6‐Fc and 17F6‐Fc could bind to all these RBDs that 20G6‐Fc and 17F6‐Fc had similar affinities to RBDs of SARS‐CoV‐2 Wuhan variants Alpha, Beta, Kappa, Delta, Delta plus, and Lamda with KD below 10 × 10−12 m (Figure 1C and Figure S3A,B, Supporting Information). These results suggest that 20G6 and 17F6 are broadly reactive against RBDs of SARS‐CoV‐2 variants of concerns (VOCs).
2.2. Vnarbodies 20G6 and 17F6 Exhibit Potent and Broadly Neutralizing Activities Against SARS‐CoV‐2 Variants
A surrogate virus‐neutralizing test (sVNT) showed that 20G6 and 17F6 could compete with hACE2 to bind to RBDs of Wuhan, Alpha, Beta, and Delta (Figure S4A, Supporting Information). The biolayer interferometry (BLI) competition assay revealed that the pre‐bound 20G6 to RBD was highly effective in blocking the further binding of hACE2 to RBD protein with no competition tolerance, while the pre‐bound 17F6 to RBD was less effective in blocking the binding of hACE2 to RBD protein with 40.94% competition tolerance (Figure S4B‐D, Supporting Information). We also used the pseudotyped virus neutralization assay to access the neutralizing activities of 20G6 and 17F6 to SARS‐CoV‐2 Wuhan, Beta, Delta, Delta plus, Kappa, and Lambda variants (Figure 1D). The 20G6 monomer, 20G6‐Fc dimer, 17F6 monomer, and 17F6‐Fc dimer can neutralize SARS‐CoV‐2 variants with the half‐maximal inhibitory concentration (IC50) at the nanomolar range (Table S1, Supporting Information). We then tested the neutralizing activities of monovalent and divalent 20G6 and 17F6 to authentic SARS‐CoV‐2 Wuhan strain, Beta, and Delta variants (200 focus forming units, FFU) using focus reduction neutralization test 50 (FRNT50) assay (Figure S4E, Supporting Information). All monovalent and divalent 20G6 and 17F6 vnarbodies could neutralize the authentic viruses in a dose‐dependent manner. 20G6‐Fc dimer showed neutralizing IC50 at 11.79 × 10−9 m to Wuhan strain, 9.36 × 10−9 m to Beta variant, and 10.26 × 10−9 m to Delta variant. 17F6‐Fc dimer showed neutralization with IC50 at 34.36 × 10−9 m to Wuhan strain, 19.87 × 10−9 m to Beta variant, and 33.85 × 10−9 m to Delta variant, respectively (Figure 1E, Table S1, Supporting Information). The divalent 20G6‐Fc and 17F6‐Fc were 2–10 times more efficient than their monovalent counterparts in neutralization. These results showed that 20G6 and 17F6, either in monovalent or divalent form, can potentially neutralize SARS‐CoV‐2 variants.
2.3. Intranasal Administration of vnarbody 20G6 Conferred Effective Prophylactic and Therapeutic Protection in Mice Challenged with SARS‐CoV‐2
We evaluated the in vivo protection efficacy in mice challenged with SARS‐CoV‐2 Wuhan or Beta variant. 20G6 was chosen for further evaluation as it exhibits the highest potency against SARS‐CoV‐2 Wuhan and other variants in vitro (Figure 2A). We first evaluated the prophylactic efficacy of vnarbody 20G6 against SARS‐CoV‐2 Wuhan in hACE2‐transgenic C57BL/6 mice. 20G6‐Fc was administered intranasally 3 h before the challenge. At 3 days after intranasal challenge with SARS‐CoV‐2 Wuhan (5 × 105 FFU), mice were sacrificed, and the lungs were collected for determination of viral genomic RNA (gRNA) and subgenomic RNA (sgRNA) and for histopathological analysis. The viral gRNA copies average was 1.5 logs (0.44–2.48 logs) lower and sgRNA average was 1.5 logs (1.15–1.82 logs) lower as compared to the control group (Figure 2B,C). Since the SARS‐CoV‐2 Beta variant can infect wild‐type mice, 20G6‐Fc was tested in BALB/c mice by intranasal administration 3 h before the challenge. At 3 days after intranasal challenge with SARS‐CoV‐2 Beta variant (1 × 104 FFU), mice were sacrificed, and the lungs were collected to determine viral gRNA and sgRNA. The viral gRNA copies decreased 4.5 logs (3.98–4.99 logs) (Figure 2D), and sgRNA decreased 2.8 logs (2.61–3.16) (Figure 2E), as compared to an unrelated vnarbody control group.
Figure 2.
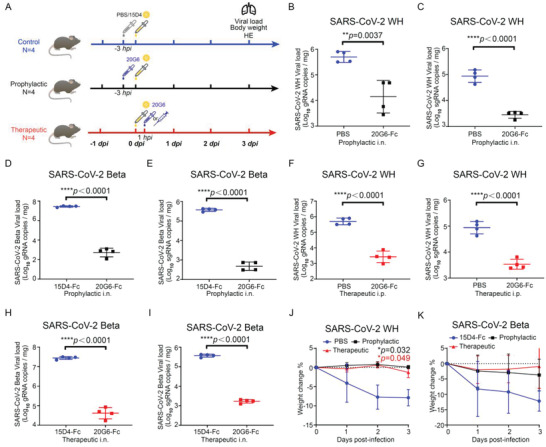
Prophylactic and therapeutic efficacy of neutralizing vnarbody 20G6‐Fc against SARS‐CoV‐2 infection in mice. A) Experimental design for protection of 20G6 in mouse model. For prophylactic group, mice were administered with 20G6‐Fc dimer or 15D4 (a control VNAR antibody) via intranasal instillation. After 3 h, mice were challenged with Wuhan (WH), Beta, or Delta SARS‐CoV‐2 virus via intranasal instillation. For therapeutic group, mice were administered with 20G6‐Fc or 15D4 via intranasal or intraperitoneal 1 h after challenged with SARS‐CoV‐2 virus. Lungs were harvested at day 3 postinfection and fixed or homogenized for downstream analysis (n = 4). B,C) Viral burden in the lungs of SARS‐CoV‐2 WH prophylactic model was measured with gRNA (B) and sgRNA (C) by RT‐qPCR. D,E) Viral burden in the lungs of SARS‐CoV‐2 Beta prophylactic model was measured with gRNA (D) and sgRNA (E) by RT‐qPCR. F,G) Viral burden in the lungs of SARS‐CoV‐2 WH therapeutic model was measured with gRNA (F) and sgRNA (G) by RT‐qPCR. H,I) Viral burden in the lungs of SARS‐CoV‐2 Beta therapeutic model was measured with gRNA (H) and sgRNA (I) by RT‐qPCR. J,K) Body weight was recorded daily and the mean percentage weight change from baseline was plotted. Data are represented as mean ± SD, two‐tailed t‐test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Blue, red, and green dots denote control group (15D4 or PBS), therapeutic group, and prophylactic group. Each dot represents an individual mouse (B–I).
We also evaluated the therapeutic potential of 20G6‐Fc in mice after infection with SARS‐CoV‐2. hACE2‐transgenic C57BL/6 mice were challenged intranasally with SARS‐CoV‐2 Wuhan (5 × 105 FFU). 20G6‐Fc was then administrated via intraperitoneal injection at 3 h postchallenge. At 3 days after intranasal challenge, mice were sacrificed, and the lungs were collected for determination of viral gRNA and sgRNA. The viral gRNA copies average reduced 2.2 logs (1.65–2.77 logs) (Figure 2F), and the viral sgRNA copies average reduced 1.4 logs (1.02–1.77 logs) (Figure 2G) in 20G6‐Fc treated mice. In BALB/c mice infected with SARS‐CoV‐2 Beta strain (1 × 104 FFU), 20G6‐Fc was administrated via intranasal instillation. The viral gRNA copies average reduced 2.7 logs (2.43–3.03 logs) (Figure 2H), and the viral sgRNA copies average reduced 2.4 logs (2.20–2.48 logs) (Figure 2I) in 20G6‐Fc treated mice. Further, mice in the untreated group showed a significant weight loss (10%–20% by day 3), whereas all mice treated prophylactically and therapeutically showed no significant weight loss (Figure 2J,K). We evaluated the protective effects of 20G6‐Fc on lung histopathology using lung tissue sections with hematoxylin‐eosin (H&E) staining. There were severe bronchopneumonia and interstitial pneumonia in the control group with alveolar collapse, edema, bronchial epithelia cell desquamation, and infiltration of lymphocytes within alveolar spaces. In contrast, there was significantly reduced lung pathology in 20G6 prophylactic and therapeutic groups (Figure S5, Supporting Information). These results demonstrated that 20G6 could confer effective protection against SARS‐CoV‐2 Wuhan and Beta variant in both prophylactic and therapeutic models.
2.4. Vnarbodies 20G6 and 17F6 Bind to a Conserved Region on RBD via β‐Sheet Interaction to Block its Interaction with ACE2
To explore the molecular mechanisms that underlie the potent neutralizing activities of these two vnarbodies, we first determined the crystal structures of 20G6 complexed with RBD (N501Y) at 1.90 Å and 17F6 with RBD (WT) at 2.85 Å (Table S2, Supporting Information). Interestingly, both 20G6 and 17F6 bind to a conserved epitope region (365–380) on the RBD (Figure 3A,B, Supporting Information), which belongs to class 4 binding antibodies targeting outside of ACE2 binding site[ 29 ] (Figure 3C and Figure S6,Supporting Information). These epitopes are only exposed when RBD adapts the up conformation. Although 20G6 binds to a surface region not overlapping with ACE2 binding site, 20G6 can compete with ACE2 for binding to RBD, specifically with the glycan at Asn322 of ACE2 (Figure 3D).
Figure 3.
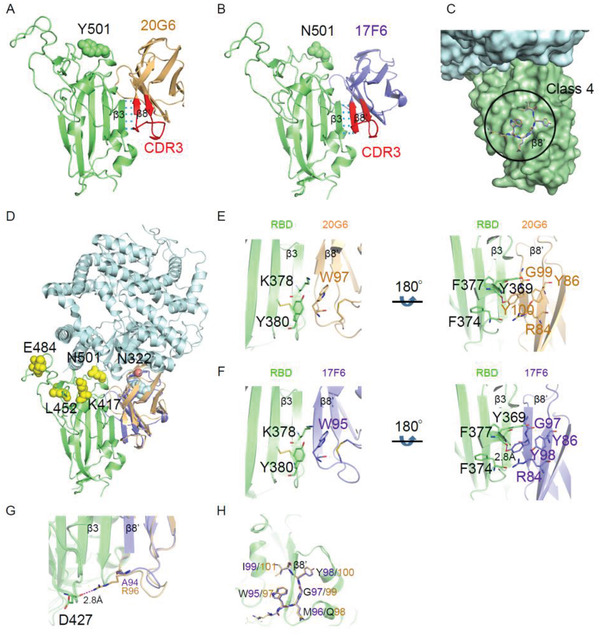
Crystal structures of 17F6 or 20G6 vnarbodies in complex with the SARS‐CoV‐2 receptor binding domain (RBD). A,B) A cartoon representation showing the localization of 20G6 (A) or 17F6 (B) bound to RBDN501Y. RBDN501Y, 20G6, and 17F6 are colored in lime, light orange, and slate, respectively. The CDR3 region of 20G6 or 17F6 is colored in red. C) The binding epitope of 17F6/20G6 belongs to Class 4. D) Clashing effect of 17F6 (slate) or 20G6 (light orange) on hACE2‐RBD (PDB ID: 7mjn) interactions. N322 glycosylation site (ACE2) is presented as red dot. The sugar chain is shown as cyan sphere. Emerging mutations found in RBD are marked in yellow. E,F) π‐interaction and hydrophobic interaction clusters around W97 and Y100 of 20G6 (E) and W95 and Y98 of 17F6 (F) with RBD. RBD is colored in lime, 20G6 is colored in light orange, and 17F6 is colored in slate. G) The R96 may be responsible for the higher affinity of 20G6 than 17F6 to RBD. H) Zoomed‐in view of the binding interface. The conserved “WXGY” motif of 17F6/20G6 using for binding to RBD was shown as sticks.
In the complex structure, only the second β‐strand of CDR3 (β8′) of 20G6 interacts with RBD. Through this interaction, the five‐stranded β‐sheet of RBD and the three‐stranded β‐sheet of 20G6 form a continuous antiparallel intermolecular β‐sheet (Figure 3A), resulting in high structural stability of the complex and high binding affinity between RBD and 20G6. On β8′ strand of 20G6, the side chains of W97 and Y100 are involved in two hydrophobic interactions. W97 of 20G6 forms π–π interactions with K378 and Y380 of RBD. The Y100 side chain forms a hydrogen bond with the main chain of Y369 on the α2 helix of RBD, and is sandwiched between R84, Y86 from 20G6 and F374, F377 from the hydrophobic core of RBD, which potentially increase the stability of the complex (Figure 3E). Similar to 20G6, Y98 and W95 in 17F6 also form strong interactions with RBD (Figure 3F). However, the equivalent residue of R96 in 20G6 at the N‐terminus of β8′ strand changes to A94 in 17F6 (Figure 3G). The side chain of Arginine can form an additional hydrogen bond interaction with RBD, which enhances the affinity of 20G6 with RBD and potentially contributes to the stronger competitive ability against ACE2. The amino acid sequences of 20G6 and 17F6 were presented in Figure S7 (Supporting Information). Interestingly, both 17F6 and 20G6 contain a “WXGY motif” in the CDR3, which forms the center of interaction between these vnarbodies and RBD (Figure 3H). Among them, the side chain of W and Y form the main hydrophobic interactions with RBD. X is a variable amino acid, and G is the smallest amino acid with no side chain, which provides a space for insertion of the side chain of Y369 from RBD.
2.5. S375F Substitution in the RBD of Omicron Variant Contributes to Escape to 20G6‐Class Antibodies
There are 15 mutations in the RBD of Omicron (G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H). Crystal structure revealed that the 365–380 region of RBD was the binding epitope of 20G6 and 17F6, and this epitope was highly conserved in SARS‐CoV‐2 variants and other sarbecoviruses (Figure 4A,B), suggesting that 20G6 and 17F6 have broad neutralizing activities. We tested the binding and neutralizing activity of 20G6‐Fc and 17F6‐Fc to MERS‐CoV and other sarbecoviruses, including SARS‐CoV‐1, Pangolin CoV, and Bat CoV. 20G6 and 17F6 have only weak binding ability to SARS‐CoV‐1 RBD, whereas the binding to Pangolin CoV and Bat CoV RBDs was comparable with SARS‐CoV‐2 RBD. In contrast, 20G6‐Fc and 17F6‐Fc showed no binding to MERS‐CoV RBD (Figure 4C). Furthermore, 20G6 and 17F6 vnarbodies also could not neutralize the Omicron and SARS‐CoV‐1 pseudovirus but effectively neutralize the Pangolin CoV and Bat CoV pseudovirus (Figure 4D).
Figure 4.
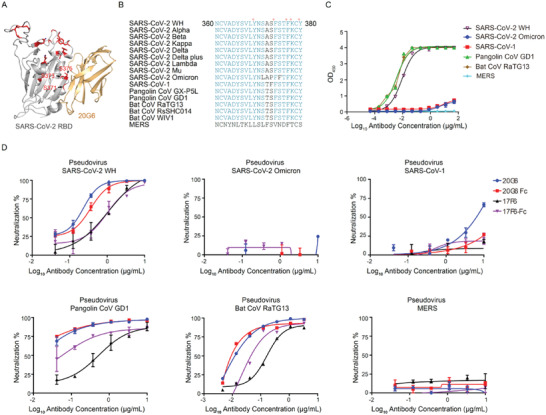
Binding and neutralizing ability of 20G6 to betacoronavirus. A) Mapping the mutation sites of Omicron on receptor binding domain (RBD). B) Sequence alignment of conserved regions of SARS‐CoV‐2 and other beta coronavirus. C) Binding capacity of 20G6‐Fc to RBDs of betacoronavirus. D) The neutralization potency of 20G6 and 17F6 were calculated based on the pseudotyped betacoronavirus neutralization assay (luciferase).
Of note, three mutations S371L, S373P, and S375F on Omicron RBD are close to the interaction interface between RBD and 20G6 (Figure 4A). The side chains of these three serine residues point inward RBD and are not involved in direct interaction with 20G6. These mutations may induce some conformational changes that interfere with 20G6 interaction. To further explore the mechanism of Omicron resistance to 20G6, we constructed single amino acid mutations on S371L/S373P/S375F residues on Omicron RBD. The binding results showed that the single S375F RBD led to a poor association with 20G6, while the single S371L or S373P RBD had little effect on the binding with 20G6 (Figure 5A). In SARS‐CoV‐1, when Serine is replaced by Phenylalanine at position 373 (S373F), the binding of 20G6 is also significantly weakened. Structural analysis showed that the benzene ring structure of 375F in Omicron and 373F in SARS‐CoV‐1 could disrupt the β‐strand structure of RBD and thus impair the antibody–antigen interaction, allowing the Omicron variant and SARS‐CoV‐1 to evade 20G6‐class antibodies (Figure 5B). These results indicated that the S375F mutation is the critical residue contributing to Omicron resistance to 20G6‐class antibodies.
Figure 5.
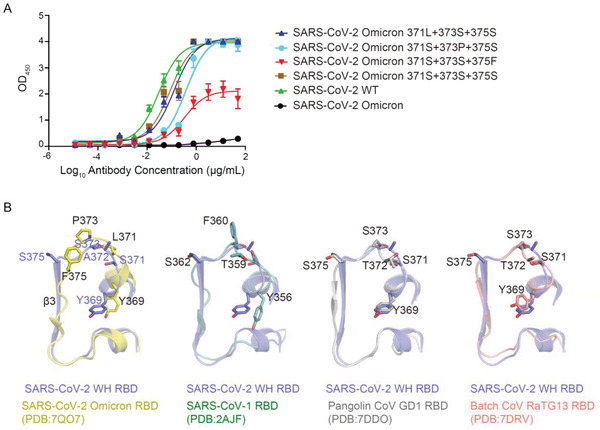
Mechanism of 20G6‐like antibody escaped by Omicron. A) Binding capacity of 20G6 to receptor binding domains (RBDs) with different mutations. B) Structure comparison of sarbecovirus RBD (amino acid residues 369–385).
3. Discussion
In this study, we constructed a phage library from immunized Bamboo shark (C. plagiosum) and isolated SARS‐CoV‐2 RBD‐specific single domain antibodies we termed vnarbodies. The immunization and maintenance of Bamboo sharks are relatively simple and cost‐effective, as it does not require a farm to house camelids and so is more environmentally friendly. Two lead vnarbodies, 20G6 and 17F6, possess a picomolar binding affinity to RBDs of seven SARS‐CoV‐2 variants, including Wuhan, Alpha, Beta, Kappa, Delta, Delta plus, and Lamda. Virus neutralization assay using pseudoviruses and authentic SARS‐CoV‐2 viruses, together with in vivo evaluation in two different mouse infection models (SARS‐CoV‐2 Wuhan and Beta variant), demonstrated their prophylactic and therapeutic potential. Intriguingly, 20G6 and 17F6 represent a new class of vnarbodies that can bind to a conserved region on RBDs of SARS‐related coronaviruses through a new mechanism. These vnarbodies contain a “WXGY” motif in the CDR3 region that forms the β‐strand to hinder the interaction between RBD and the viral receptor ACE2.
A variety of SARS‐CoV‐2 variants have appeared worldwide, including Alpha, Beta, Delta, Delta plus, Kappa, Lambda, and most recently, Omicron. The Delta variant is highly transmissible and dominantly spread around the world. RBDs of these variants possess a variety of amino acid mutations, such as K417, T478, E484, and N501, which are the key sites for t he binding of RBD to ACE2. The Alpha RBD carries the N501Y mutation, which increases the binding affinity of S protein and ACE2 by sevenfold.[ 13 , 30 ] The Beta RBD carries K417N, E484K, and N501Y mutations, which poses a huge challenge to current vaccines and monoclonal antibodies. The neutralizing activity of COVID‐19 survivor's serum against the Beta variant could be reduced by 6.9‐fold.[ 31 ] Many monoclonal antibodies isolated earlier failed to effectively inhibit Beta and Delta variants.[ 31 ] Only a few human monoclonal antibodies showed broad neutralizing activities against multiple variants. Nanobodies derived from camelids or vnarbodies derived from sharks, given their smaller size, may offer unique binding capabilities to spike protein, especially in the regions which are not readily susceptible to human antibodies. A few nanobodies isolated from camelids showed potent protection against SARS‐CoV‐2.[ 32 , 33 , 34 , 35 ] Several shark vnarbodies screened from a synthetic library were reported to have neutralizing activities against the SARS‐CoV‐2 Wuhan strain in vitro[ 36 , 37 ] with no studies on neutralizing variants and animal models. So far, limited studies have been carried out to evaluate the protective ability of single‐domain antibodies in animal models. 20G6 showed the best neutralizing activities against 7 SARS‐CoV‐2 VOCs that we tested. Unlike the traditional systemic antibody administration, we used the intranasal administration to demonstrate that the 20G6‐Fc can confer protection against SARS‐CoV‐2 Wuhan and Beta variants in both prophylactic and therapeutic models. For respiratory viruses, the nasal cavity and respiratory tract are the main routes of infection. Therefore, building a first defense barrier in the nasopharynx can offer an effective prevention advantage. Vnarbodies have the advantages of good thermal stability and low cost of production. 20G6 and 17F6 may be worthwhile for further development into a prophylactic nasal spray to lower the risk of SARS‐CoV‐2 infection for healthcare workers, travelers in high incidence areas, and people in need.
We solved the crystal structures to illustrate the neutralization mechanism of vnarbodies 20G6 and 17F6. Previously, camelid‐derived nanobodies have been reported to recognize RBD through the extended CDR loops.[ 38 ] Interestingly, we found that both 20G6 and 17F6 used the same mechanism that relies on the β‐sheet (β8′) of the CDR3 to form a continuous antiparallel intermolecular β‐sheet with the five‐strand β‐sheet of RBD. Therefore, the β‐sheet interaction mechanism appears to be a common mechanism in shark vnarbodies, as substantiated by another group recently.[ 36 ] The interaction between β‐sheets is often stronger than the interaction between α‐helices. Notably, in almost all SARS‐CoV‐2 RBD specific vnarbodies that we screened, there is a “WXGY” motif in the CDR3, which was not found in the vnarbodies against influenza that we screened in another project. “W (Tryptophan)” and “Y (Tyrosine)” form a strong interaction between vnarbody and RBD. “G (Glycine)” is the smallest amino acid without a side chain, which provides enough space for the interaction between 20G6 and RBD or 17F6 and RBD. The “WXGY” motif may be a unique motif from shark‐derived vnarbodies that binds to RBD. In addition to this motif, the interaction of other amino acids in the CDR3 region can further enhance the binding of vnarbodies with RBD. For example, 20G6 has better binding and neutralizing activity than 17F6, because an additional Arg96 on 20G6 forms a salt bridge with the Asp427 on RBD. Therefore, targeted amino acid mutations on the vnarbodies may achieve stronger RBD binding vnarbodies. One reason that 20G6 and 17F6 can bind to a variety of SARS‐CoV‐2 mutant RBD proteins is that these two vnarbodies bind to a highly conserved region in sarbecoviruses (corresponding to amino acid 365–380 on RBD of SARS‐CoV‐2 Wuhan), suggesting that 20G6 and 17F6 are likely the pan‐sarbecovirus neutralizing antibodies. This epitope is hidden and is only exposed in the “up” conformation of RBD, which belongs to Class 4 in RBD epitope classification, and does not overlap with the ACE2‐RBD binding interface.[ 29 ] Many class 4 binding antibodies, such as CR3022[ 39 ] and EY6A,[ 40 ] have no or weak neutralizing activity because they do not compete with ACE2. The spatial orientation of 17F6 and 20G6 binding with RBD is opposite to CR3022, enabling them to form a steric hindrance with the glycan at Asn322 of ACE2 and play a neutralizing role by clashing with ACE2. During the preparation of this manuscript, the emergence of the SARS‐CoV‐2 Omicron variant raised the concern of immune evasion from established immunity.[ 41 ] The Omicron variant has three major sublineages: BA.1, BA.2, and BA.3.[ 42 ] Omicron shows unprecedented resistance to most of all six types of human‐derived neutralizing antibodies reported[ 43 , 44 ] and shark‐derived vnarbodies 20G6 and 17F6 in this study. Recently, a study revealed that the S375F mutation is closely associated with the explosive spread of Omicron.[ 45 ] Our study found that three mutations S371L, S373P, and S375F on Omicron RBD are close to the interaction interface between RBD and 20G6. Further structural analysis and binding assays illustrated that the S375F mutation is the critical residue that confers resistance to 20G6‐like antibodies. The phenylalanine mutation disrupts the β‐sheet structure of RBD and thus impairs the binding to 20G6‐like antibodies. We are now performing affinity maturation for 20G6 using yeast display and screening phage display library to obtain new vnarbodies that may have broader neutralizing activities.
We intend to develop lead vnarbodies as an intranasal or topical shield for prophylactic usage in the future. The sequence of shark‐derived vnarbodies shares only 25%–30% identity to human heavy chains. The structure of a vnarbody has classic immunoglobulin folding. Superimposition of human variable heavy and light chains onto vnarbodies revealed a structural relatedness within the core framework.[ 46 ] For example, E06, a shark‐derived anti‐human serum albumin (HSA) single domain antibody, showed very low immunogenic responses comparable with its humanized versions. Since vnarbodies have a similar framework, they are likely to have low immunogenicity. Nevertheless, humanization may render vnarbodies more suitable for in vivo applications.
This study obtained two shark‐derived vnarbodies that can broadly bind to a conserved region on RBD of SARS‐related coronaviruses and neutralize most SARS‐CoV‐2 variants, pangolin CoV and Bat CoV except Omicron. Intranasal administration of these vnarbodies may offer a topical application for preventing respiratory infection of most SARS‐CoV‐2 variants. The study on the neutralizing mechanism of vnarbodies and resistance by the Omicron provides new insights for searching broadly neutralizing antibodies. Our future work will identify vnarbodies with a broad neutralizing spectrum against Omicron and other variants.
4. Experimental Section
Proteins
The SARS‐COV‐2 extracellular spike protein (S1+S2 ECD), the S1 domain of S, the hACE2 protein were purchased from Sino Biological (Beijing, China).
Cell Lines and Animals
Cell lines used in this study were obtained from the ATCC (293T, Vero E6) or Themo Fisher Scientific (Expi293). hACE2‐transgenic C57BL/6 mice were presented by Cyagen Biosciences Inc. (C001191). Balb/c mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd.
Cloning, Expression, and Purification of SARS‐CoV‐2 RBD Variants
The RBD of S genes (NC_045512.2) was synthesized and cloned into pSecTag2A vector. RBD mutants were constructed using site‐directed mutagenesis kit (TIANGEN). The constructs were transfected into the Expi293 cells using PEI (PolyScience). Four days after transfection, the supernatant was purified by Ni sepharose, then by ion exchange chromatography Source 15Q, Source 15S, and size‐exclusion chromatography Superdex 75 Increase (Cytiva).
Immunization
One male bamboo shark (C. plagiosum) was immunized with 100 µg S1 protein of SARS‐CoV‐2 Wuhan in 100 µL PBS with Complete Freund′s Adjuvant (Thermo Fisher Scientific). Two weeks later, the shark received a booster immunization using 100 µg S1 protein of SARS‐CoV‐2 Wuhan in 100 µL PBS with Incomplete Freund′s Adjuvant. Ten days after the booster immunization, 5 mL blood was collected from the tail vain. Serum was tested for binding antibody titer and pseudovirus neutralization. Blood cells were collected for construction of phage display library.
Construction of Phage Display Library and Isolation of Vnarbodies
Shark blood cells were lysised by trizol, and total RNA were extracted by chloroform, then precipitation with isopropanol. iScript cDNA Synthesis Kit (Bio‐rad,170889) was used for cDNA synthesis. Vnarbody sequences were amplified by the specific primers which performed similarly as described before[ 47 ] and cloned into pcantab5e phagemid vector using SfiI and NotI restriction enzymes (New England Biolabs). Ligation product was electro‐transformed into TG1 competent cells (Lucigen). Phage library was rescued by M13KO7 helper phage (New England Biolabs) and precipitated with PEG/NaCl. After three round panning by RBD immobilized on immune tube, monoclonal phages were detected by phage enzyme‐linked immunosorbent assay (ELISA).
Vnarbody Expression in E. coli and Purification
Vnarbodies were amplified from ELISA positive phage clones and cloned into a prokaryotic expression vector with his tag and transformed into Arctic express competent cells (Miaoling Bio). Monoclonal clones were grown in the LB medium until OD = 0.6 and induced by 0.1 × 10−3 m IPTG at 12 °C for 20 h. Bacteria were crushed by high pressure and the supernatant was purified by Ni sepharose, then by ion exchange chromatography Source 15Q and size‐exclusion chromatography Superdex 75 Increase (Cytiva).
SDS‐PAGE and Western Blot
Purified 20G6 was boiled in SDS‐PAGE loading buffer for denaturation, and the other 20G6 was directly resuspended in the nondenaturing loading buffer. The samples were separated on a 15% SDS‐PAGE. One gel was stained with Coomassie brilliant blue for 30 min at room temperature and destained with ethanol and the other gel was transferred to PVDF membranes (Millipore). Membranes were blocked in 5% non‐fat milk‐PBS with 0.05% Tween 20 for 1 h at room temperature. Blots were then incubated with secondary anti‐His HRP labeled antibodies (Sino Biological) for 1 h at room temperature. After five washes with PBS 0.05% Tween 20, detection was performed using ECL Western Blotting Substrate (Bio‐rad).
Fc Tag Vnarbody Expression and Purification
Vnarbody sequences were amplified and cloned into pCMV3‐IgG1 vector with human IgG1 Fc fusion. The constructs were transfected into the Expi293 cells using PEI (PolyScience). Five days after transfection, the supernatant was purified using MabSelect antibody purification chromatography resin (Cytiva).
Enzyme‐Linked Immunosorbent Assay (ELISA)
SARS‐COV‐2 variants RBD were respectively coated onto 96‐well ELISA plate at 50 ng well−1 in 0.1 m carbonate buffer (pH 9.6) overnight at 4 °C and blocked with blocking buffer (PBS, 0.05% Tween‐20, 5% milk) at 37 °C for 2 h. Serial fourfold dilutions of vnarbodies in blocking buffer were incubated with antigen‐coated plates for 2 h. After wash five times with PBST (PBS with 0.05% tween 20), HRP‐conjugated secondary antibodies were incubated for 1 h, and washed with PBST. The plates were incubated with TMB (Millipore) solution in dark for 15 min. Reactions were stopped with 1 m H2SO4 and measured absorption at 450 nm. The data was analyzed by GraphPad Prism 8 using nonlinear regression.
Affinity Assay and Competitive Assay by Biolayer Interferometry (BLI)
Affinity assays were performed on a GatorPrime (Gator Bio) biolayer interferometry instrument at room temperature with shaking at 1000 rpm. Anti‐his or anti‐hFc probes were used for vnarbody affinity assays. Probes were soaked in the kinetic buffer (0.01 m PBS, 0.02% Tween‐20, 0.2% BSA) for 5 min before assay. Vnarbodies were diluted to 10 µg mL−1 in the kinetic buffer, and loaded to the probes to reach the level of 1 nm. After baseline equilibration, association of vnarbody in a twofold dilution series from 200 × 10−9 to 3.125 × 10−9 m was performed prior to dissociation for 300 s. One probe was used as reference and only dipped into kinetic buffer. Probes were regeneration with 0.1 m glycine‐HCl (pH 2.0) for three times between each assay. Data for which association responses were >0.15 nm were aligned, reference‐subtracted, and analyzed using Gator data analysis software. Ka, Kd, and KD values were evaluated with a global fit applied to all data.
Competitive assay was performed following the “in‐tandem assay” protocol. RBD was loaded on the probes to reach the level of 0.5‐1 nm. The first vnarbody was loaded on the probes until saturation. The ACE2 was added for 180 s to measure the binding in the presence of the first saturating ACE2 or vnarbody. Competition tolerance was calculated by the percentage of the binding rate of the second antibody by the presence of the first antibody. Competition tolerance larger than 50% indicates a high possibility of no overlapping epitope.
Surrogate Virus Neutralizing Test (sVNT)
A sVNT based on ACE2 competition was performed with cPass SARS‐CoV‐2 neutralization antibody detection kit (GenScript) following the manual.[ 48 ] In brief, SARS‐COV‐2 RBD variants were incubated with the 96‐well ELISA plate for 15 min at 37 °C, then a series of fourfold diluted vnarbodies were added to the plate. After incubated for 15 min at 37 °C, HRP‐conjugated ACE2 were added into the plate for 15 min at 37 °C. The plates were washed with washing buffer for five times and incubated with TMB solution in dark for 15 min. Reactions were stopped with 1 m stopping buffer and measured absorption at 450 nm. The data was analyzed by GraphPad Prism 8 using nonlinear regression.
Crystallization of Vnarbodies 20G6, 17F6 with RBD
Vnarbodies were incubated with RBD (WT or N501Y) at a molar ratio of 1.5:1 for 1 h. The complex was subjected to size‐exclusion chromatography using a Superdex 75 column (10/300 GL; GE Healthcare, New York, USA) in 20 × 10−3 m Tris pH 7.5, 150 × 10−3 m NaCl. The purified complex was then concentrated to 15.0 mg mL−1 and used to prepare hanging‐drop crystallization trays. 17F6‐RBDWH crystals were grown in 0.18 m Sodium citrate tribasic dihydrate, 20% PEG3350, and 20G6‐RBDN501Y crystals were grown in 70 × 10−3 m Citric acid, 30 × 10−3 m Bis‐Tris propane pH 3.4, 20% PEG3350. All crystals were frozen in liquid nitrogen.
Data Collection and Structural Determination
Diffraction data were collected at the Shanghai Synchrotron Radiation Facility at 100 K, and processed using DIALS[ 49 ] or XDS.[ 50 ] Then, data sets were merged and scaled using Aimless from the CCP4 program suite.[ 51 ] Five percent of the data was randomly selected for the R‐free calculation. The initial structure solution of RBD in complex with 17F6 was obtained by Phaser‐MR using shark VNAR structure (PDB ID: 2i26) and RBD structure (from PDB ID:7eam) with truncations on the variable or flexible regions as the search models. Then the structure was refined and rebuilt by REFMAC5[ 52 ] and Coot.[ 53 ] TLS refinement was used in the final stages. The final 17F6 VNAR structure was served as the template to solve 20G6‐RBDN501Y structure. Structure figures were prepared using the program PyMOL (The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA).
Neutralization Assay Using Pseudovirus
Fourfold serially diluted vnarbodies were incubated with the SARS‐COV‐2 luciferase pseudovirus (500 TCID50 per well) for 1 h 37 °C. An amount of 100 µL of the mixtures was added to 100 µL of 293T‐hACE2 cells (2 × 105 mL−1) in the 96‐well plate and cultured at 37 °C with 5% CO2 for 72 h. Cells were lysed, and the luciferase signal was measured by the Bio‐Lite Luciferase Assay System (Vazyme). The data was analyzed by GraphPad Prism 8 using nonlinear regression and to calculate the IC50.
Focus Reduction Neutralization Test (FRNT) Using Authentic SARS‐CoV‐2 Wuhan, Beta, and Delta Variants
Focus reduction neutralization test (FRNT) was performed in a Biosafety Level 3 laboratory. The clinical isolate of authentic SARS‐CoV‐2 Wuhan, Beta, and Delta were used for the neutralizing assays. Briefly, four‐fold diluted vnarbodies were incubated with authentic SARS‐COV‐2 containing 200 focus forming units (FFU) for 1 h at 37 °C. An amount of 100 µL of the mixtures was added to a monolayer of Vero E6 cells in 96‐well plate. After 1 h incubation at 37 °C, the mixture was removed, and cells was added with 100 µL DMEM medium supplied with 1.6% carboxymenthylcellulose and 4% FBS and incubated at 37 °C with 5% CO2 for 24 h, then fixed with 4% paraformaldehyde. PFA was removed, and cells were blocked with 1% BSA contained 0.2% Triton for 30 min. Then the cells were incubated with anti‐SARS‐N primary antibody (SinoBiological) 37 °C for 1 h. After wash three times with PBST (PBS with 0.05% tween 20), HRP‐conjugated goat anti‐rabbit secondary antibodies (Jackson) were incubated for 1 h, and washed with PBST. Focuses were visualized by staining with TrueBlue. Percentage of focus reduction was calculated as: percent of focus reduction = 100‐focus number with NAb/focus number without NAb × 100. The IC50 values were calculated by GraphPad Prism 8 using nonlinear regression.
Evaluation in Mouse Challenge Models Using SARS‐CoV‐2 Wuhan and Beta Variant
All protocols for animal experiments were approved by the Institutional Animal Care and Use Committees of the Guangzhou Institutes of Biomedicine and Health, China (IACUC: 2020025). All work with live SARS‐CoV‐2 was conducted in the Biosafety Level 3 (BLS3) Laboratories. For SARS‐CoV‐2 Wuhan challenge model, 20‐weeks old hACE2‐transgenic C57BL/6 mice were divided into three groups (4 mice per group). Mice were intranasally instilled with 5 × 105 FFU of SARS‐CoV‐2 Wuhan clinical isolate and were monitored for a 3‐day time course. 20G6‐Fc was administered intranasally at a dose of 10 mg kg−1 3 h before challenge for prophylactic evaluation or by intraperitoneal 1 h post challenge for therapeutic evaluation. PBS was used as an untreated control. For SARS‐CoV‐2 Beta challenge model, 20‐weeks old Balb/c mice were divided into three groups (4 mice per group). Mice were intranasally instilled with 1 × 104 FFU of a clinical isolate of SARS‐CoV‐2 Beta and were monitored for a 3‐day time course. 20G6‐Fc was administered intranasally at a dose of 10 mg kg−1 3 h before challenge for prophylactic evaluation or by intraperitoneal 1 h post challenge for therapeutic evaluation. 15D4, an unrelated vnarbody, was injected as control. Three days after infection, mice were sacrificed and the lungs were collected for viral load analysis and pathological analysis. For the viral load assay, RNA was extracted from the supernatants of lung homogenate. For the pathological analysis, the lungs were fixed in 4% paraformaldehyde solution for 2 weeks, and cut into 3 µm sections after embedded with paraffin. H&E stain was used to observe histopathological changes.
Quantitative PCR (RT‐qPCR)
Three days after infection, lung lobes were harvested and homogenized in TRIzol (Invitrogen). Total RNA was extracted according to the manufacturer's instructions. The SARS‐CoV‐2 genomic and subgenomic RT‐qPCR was performed by using one‐step QuantiTect SYBR Green RT‐PCR Kit (Qiagen). The S gene was used to evaluate the genomic RNA of SARS‐CoV‐2 and the sequences of the primers were: RBD‐qF1: 5′ CAATGGTTTAACAGGCACAGG 3′, RBD‐qR1: 5′ CTCAAGTGTCTGTGGATCACG 3′. The amplification was performed as followed: 50 °C for 30 min, 95 °C for 15 min followed by 45 cycles of 95 °C for 15 s, 50 °C for 30 s, and 72 °C for 30 s. The sgE gene was used to evaluate the subgenomic RNA of SARS‐CoV‐2, and the sequences of the primers were: sgLeader F: 5′ CGATCTCTTGTAGATCTGTTCTC 3′, sgRNA‐E R: 5′ ATATTGCAGCAGTACGCACACA 3′. The amplification was performed as followed: 50 °C for 30 min, 95 °C for 15 min followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Quantification was carried out using a standard curve based on 10‐fold serial dilutions of a plasmid DNA comprising the target gene ranging from 101 to 107 copies.
Statistical Analysis
In vitro neutralization activity was estimated using microneutralization assay with fourfold diluted antibody concentration. Four‐parameter nonlinear regression model fit was used for IC50 calculation. Continuous variables with normal distribution were presented as mean ± SD. For statistical analyses in which two groups with normally distributed date were compared, an unpaired two‐tailed Student's t test was used, and p values were indicated by *p < 0.05, **p < 0.01, ***p<0.001, ****p<0.0001. All statistical analysis was performed using GraphPad Prism 8.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
B.F., Z.C., J.S., and T.X. contributed equally to this work. L.C. conceived the study. B.F. and Z.C. performed most of the experiments with assistance from H.Y., M.F., R.H., Y.S., S.H., C.L., J.L., P.H., P.Z., P.H., and Q.Y. Q.W. performed pseudovirus neutralization assay. J.Z. and J.S. directed and performed live virus assays and animal studies. J.L., T.X., and J.Z. collected and analyzed the crystal structure data. L.C., B.F., and Z.C. integrated the data and wrote the original draft. J.L., J.Z., X.X., and X.N. participated in manuscript writing and editing. All authors reviewed and approved the final version of the manuscript.
Supporting information
Supporting Information
Acknowledgements
The authors thank the staff members of beamline 18U1, 19U1 at the National Center for Protein Science Shanghai and the Shanghai Synchrotron Radiation Facility, People's Republic of China, for assistance during the collection of diffraction data. This work was partly supported by the Chinese Academy of Sciences Pilot Strategic Science and Technology Projects (XDB29050701), Guangzhou Science and Technology Program Key Projects (2021904020037), the grant from the State Key Lab of Respiratory Disease, Guangzhou Medical University (SKLRD‐QN‐201901), the Youth Innovation Promotion Association of CAS (2021357), Guangzhou Institute of Respiratory Health Open Project (Funds provided by China Evergrande Group, 2020GIRHHMS08), and China Postdoctoral Science Foundation (2020T130023ZX).
Feng B., Chen Z., Sun J., Xu T., Wang Q., Yi H., Niu X., Zhu J., Fan M., Hou R., Shao Y., Huang S., Li C., Hu P., Zheng P., He P., Luo J., Yan Q., Xiong X., Liu J., Zhao J., Chen L., A Class of Shark‐Derived Single‐Domain Antibodies can Broadly Neutralize SARS‐Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape. Small Methods 2022, 6, 2200387. 10.1002/smtd.202200387
Contributor Information
Jinsong Liu, Email: liu_jinsong@gibh.ac.cn.
Jincun Zhao, Email: zhaojincun@gird.cn.
Ling Chen, Email: chen_ling@gibh.ac.cn.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- 1. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., China Novel Coronavirus I., Research T., N. Engl. J. Med. 2020, 382, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kirby T., Lancet Respir. Med. 2021, 9, e20.33417829 [Google Scholar]
- 4. Pan D., Mudalige N. L., Sze S., Koeckerling D., Oyefeso O., Barker J., Williams C. M., Tang J. W., Pareek M., Clin. Med. 2021, 21, e295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang J. W., Toovey O. T. R., Harvey K. N., Hui D. D. S., J. Infect. 2021, 82, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dougherty K., Mannell M., Naqvi O., Matson D., Stone J., Morb. Mortal. Wkly. Rep. 2021, 70, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Silva M. S. D., Demoliner M., Hansen A. W., Gularte J. S., Silveira F., Heldt F. H., Filippi M., Pereira V., Silva F. P. D., Mallmann L., Fink P., Silva L. L. D., Weber M. N., Almeida P. R., Fleck J. D., Spilki F. R., Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaughan A., New Sci. 2021, 252, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bloch E. M., Shoham S., Casadevall A., Sachais B. S., Shaz B., Winters J. L., van Buskirk C., Grossman B. J., Joyner M., Henderson J. P., Pekosz A., Lau B., Wesolowski A., Katz L., Shan H., Auwaerter P. G., Thomas D., Sullivan D. J., Paneth N., Gehrie E., Spitalnik S., Hod E. A., Pollack L., Nicholson W. T., Pirofski L. A., Bailey J. A., Tobian A. A., J. Clin. Invest. 2020, 130, 2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoepler W. P., Weidner L., Traugott M. T., Neuhold S., Meyer E. L., Zoufaly A., Seitz T., Kitzberger R., Baumgartner S., Pawelka E., Karolyi M., Grieb A., Hind J., Laferl H., Friese E., Wenisch C., Aberle S. W., Aberle J. H., Weseslindtner L., Jungbauer C., Infect. Dis. 2021, 53, 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walls A. C., Park Y. J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Cell 2020, 181, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K. Y., Wang Q., Zhou H., Yan J., Qi J., Cell 2020, 181, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Nature 2020, 581, 215. [DOI] [PubMed] [Google Scholar]
- 14. Huang Y., Yang C., Xu X. F., Xu W., Liu S. W., Acta Pharmacol. Sin. 2020, 41, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ho M., Antibiot. Ther. 2020, 3, 109. [Google Scholar]
- 16. Hamers‐Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R., Nature 1993, 363, 446. [DOI] [PubMed] [Google Scholar]
- 17. Nguyen V. K., Hamers R., Wyns L., Muyldermans S., EMBO J. 2000, 19, 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bathula N. V., Bommadevara H., Hayes J. M., Cancer Biother. Radiopharm. 2021, 36, 109. [DOI] [PubMed] [Google Scholar]
- 19. Cheong W. S., Leow C. Y., Abdul Majeed A. B., Leow C. H., Int. J. Biol. Macromol. 2020, 147, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steeland S., Vandenbroucke R. E., Libert C., Drug Discovery Today 2016, 21, 1076. [DOI] [PubMed] [Google Scholar]
- 21. Wu Y., Jiang S., Ying T., Front. Immunol. 2017, 8, 1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duggan S., Drugs 2018, 78, 1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greenberg A. S., Avila D., Hughes M., Hughes A., McKinney E. C., Flajnik M. F., Nature 1995, 374, 168. [DOI] [PubMed] [Google Scholar]
- 24. Chen F., Liu Z., Jiang F., Front. Immunol. 2021, 12, 690742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zebardast A., Hosseini P., Hasanzadeh A., Latifi T., Mol. Biol. Rep. 2022, 49, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W. P., Schneidman‐Duhovny D., Zhang C., Shi Y., Science 2020, 370, 1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bessalah S., Jebahi S., Mejri N., Salhi I., Khorchani T., Hammadi M., 3 Biotech 2021, 11, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kovaleva M., Ferguson L., Steven J., Porter A., Barelle C., Expert Opin. Biol. Ther. 2014, 14, 1527. [DOI] [PubMed] [Google Scholar]
- 29. Barnes C. O., Jette C. A., Abernathy M. E., Dam K. A., Esswein S. R., Gristick H. B., Malyutin A. G., Sharaf N. G., Huey‐Tubman K. E., Lee Y. E., Robbiani D. F., Nussenzweig M. C., West A. P. Jr., Bjorkman P. J., Nature 2020, 588, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laffeber C., de Koning K., Kanaar R., Lebbink J. H. G., J. Mol. Biol. 2021, 433, 167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., Liao H., Feng Y., Li X., Shi X., Zhang Z., Zhang F., Ding Q., Zhang T., Wang X., Zhang L., Immunity 2021, 54, 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schepens B., Schie L. v., Nerinckx W., Roose K., Breedam W. V., Fijalkowska D., Devos S., Weyts W., Cae S. D., Vanmarcke S., Lonigro C., Eeckhaut H., Herpe D. V., Borloo J., Oliveira A. F., Catani J. P. P., Creytens S., Vlieger D. D., Michielsen G., Marchan J. C. Z., Moschonas G. D., Rossey I., Sedeyn K., Hecke A. V., Zhang X., Langendries L., Jacobs S., Horst S. t., Seldeslachts L., Liesenborghs L., et al, Sci. Transl. Med. 2021, 13, eabi7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li T., Cai H., Yao H., Zhou B., Zhang N., van Vlissingen M. F., Kuiken T., Han W., GeurtsvanKessel C. H., Gong Y., Zhao Y., Shen Q., Qin W., Tian X. X., Peng C., Lai Y., Wang Y., Hutter C. A. J., Kuo S. M., Bao J., Liu C., Wang Y., Richard A. S., Raoul H., Lan J., Seeger M. A., Cong Y., Rockx B., Wong G., Bi Y., et al, Nat. Commun. 2021, 12, 4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu X., Cheng L., Fu M., Huang B., Zhu L., Xu S., Shi H., Zhang D., Yuan H., Nawaz W., Yang P., Hu Q., Liu Y., Wu Z., Cell Rep. 2021, 37, 109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nambulli S., Xiang Y., Tilston‐Lunel N. L., Rennick L. J., Sang Z., Klimstra W. B., Reed D. S., Crossland N. A., Shi Y., Duprex W. P., Sci. Adv. 2021, 7, eabh0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ubah O. C., Lake E. W., Gunaratne G. S., Gallant J. P., Fernie M., Robertson A. J., Marchant J. S., Bold T. D., Langlois R. A., Matchett W. E., Thiede J. M., Shi K., Yin L., Moeller N. H., Banerjee S., Ferguson L., Kovaleva M., Porter A. J., Aihara H., LeBeau A. M., Barelle C. J., Nat. Commun. 2021, 12, 7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gauhar A., Privezentzev C. V., Demydchuk M., Gerlza T., Rieger J., Kungl A. J., Walsh F. S., Rutkowski J. L., Stocki P., FASEB J. 2021, 35, e21970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun D., Sang Z., Kim Y. J., Xiang Y., Cohen T., Belford A. K., Huet A., Conway J. F., Sun J., Taylor D. J., Schneidman‐Duhovny D., Zhang C., Huang W., Shi Y., Nat. Commun. 2021, 12, 4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T., Emerging Microbes Infect. 2020, 9,382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou D., Duyvesteyn H. M. E., Chen C. P., Huang C. G., Chen T. H., Shih S. R., Lin Y. C., Cheng C. Y., Cheng S. H., Huang Y. C., Lin T. Y., Ma C., Huo J., Carrique L., Malinauskas T., Ruza R. R., Shah P. N. M., Tan T. K., Rijal P., Donat R. F., Godwin K., Buttigieg K. R., Tree J. A., Radecke J., Paterson N. G., Supasa P., Mongkolsapaya J., Screaton G. R., Carroll M. W., Gilbert‐Jaramillo J., et al, Nat. Struct. Mol. Biol. 2020, 27, 950. [DOI] [PubMed] [Google Scholar]
- 41. Thakur V., Ratho R. K., J. Med. Virol. 2022, 94, 1821. [DOI] [PubMed] [Google Scholar]
- 42. Viana R., Moyo S., Amoako D. G., Tegally H., Scheepers C., Althaus C. L., Anyaneji U. J., Bester P. A., Boni M. F., Chand M., Choga W. T., Colquhoun R., Davids M., Deforche K., Doolabh D., du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N. Y., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S. L., Kraemer M. U. G., et al, Nature 2022, 603, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X. S., Nature 2022, 602, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dejnirattisai W., Huo J., Zhou D., Zahradnik J., Supasa P., Liu C., Duyvesteyn H. M. E., Ginn H. M., Mentzer A. J., Tuekprakhon A., Nutalai R., Wang B., Dijokaite A., Khan S., Avinoam O., Bahar M., Skelly D., Adele S., Johnson S. A., Amini A., Ritter T. G., Mason C., Dold C., Pan D., Assadi S., Bellass A., Omo‐Dare N., Koeckerling D., Flaxman A., Jenkin D., et al, Cell 2022, 185, 467.35081335 [Google Scholar]
- 45. Kimura I., Yamasoba D., Nasser H., Zahradnik J., Kosugi Y., Wu J., Nagata K., Uriu K., Tanaka Y. L., Ito J., Shimizu R., Tan T. S., Butlertanaka E. P., Asakura H., Sadamasu K., Yoshimura K., Ueno T., Takaori‐Kondo A., Schreiber G., Toyoda M., Shirakawa K., Irie T., Saito A., Nakagawa S., Ikeda T., Sato K., bioRxiv 2022, 10.1101/2022.04.03.486864. [DOI] [Google Scholar]
- 46. Kovalenko O. V., Olland A., Piche‐Nicholas N., Godbole A., King D., Svenson K., Calabro V., Muller M. R., Barelle C. J., Somers W., Gill D. S., Mosyak L., Tchistiakova L., J. Biol. Chem. 2013, 288, 17408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu J. L., Anderson G. P., Delehanty J. B., Baumann R., Hayhurst A., Goldman E. R., Mol. Immunol. 2007, 44, 1775. [DOI] [PubMed] [Google Scholar]
- 48. Tan C. W., Chia W. N., Qin X., Liu P., Chen M. I., Tiu C., Hu Z., Chen V. C., Young B. E., Sia W. R., Tan Y. J., Foo R., Yi Y., Lye D. C., Anderson D. E., Wang L. F., Nat. Biotechnol. 2020, 38, 1073. [DOI] [PubMed] [Google Scholar]
- 49. Winter G., Waterman D. G., Parkhurst J. M., Brewster A. S., Gildea R. J., Gerstel M., Fuentes‐Montero L., Vollmar M., Michels‐Clark T., Young I. D., Sauter N. K., Evans G., Acta Crystallogr. Sect. D: Struct. Biol. 2018, 74, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kabsch W., Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Potterton E., Briggs P., Turkenburg M., Dodson E., Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003, 59, 1131. [DOI] [PubMed] [Google Scholar]
- 52. Murshudov G. N., Skubak P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., Lohkamp B., Scott W. G., Cowtan K., Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


