Abstract
Remdesivir is the first US Food and Drug Administration (FDA)‐approved drug for the treatment of coronavirus disease 2019 (COVID‐19). We conducted a retrospective pharmacogenetic study to examine remdesivir‐associated liver enzyme elevation among Million Veteran Program participants hospitalized with COVID‐19 between March 15, 2020, and June 30, 2021. Pharmacogene phenotypes were assigned using Stargazer. Linear regression was performed on peak log‐transformed enzyme values, stratified by population, adjusted for age, sex, baseline liver enzymes, comorbidities, and 10 population‐specific principal components. Patients on remdesivir had higher peak alanine aminotransferase (ALT) values following treatment initiation compared with patients not receiving remdesivir. Remdesivir administration was associated with a 33% and 24% higher peak ALT in non‐Hispanic White (NHW) and non‐Hispanic Black (NHB) participants (p < 0.001), respectively. In a multivariable model, NHW CYP2C19 intermediate/poor metabolizers had a 9% increased peak ALT compared with NHW normal/rapid/ultrarapid metabolizers (p = 0.015); this association was not observed in NHB participants. In summary, remdesivir‐associated ALT elevations appear to be multifactorial, and further studies are needed.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Remdesivir is associated with liver injury in patients with coronavirus disease 2019 (COVID‐19), yet the mechanism of this injury is unknown.
WHAT QUESTION DID THIS STUDY ADDRESS?
We utilized a genetically guided approach to investigate whether polymorphisms in drug metabolizing genes or transporters were associated with alanine aminotransferase (ALT) elevations following remdesivir treatment.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE
Remdesivir was associated with a 30% increase in peak ALT in patients hospitalized with COVID‐19 which differs by population. Non‐Hispanic White (NHW) individuals with the CYP2C19 intermediate or poor metabolizer phenotype experienced a higher peak ALT than NHW individuals with normal, rapid, or ultrarapid metabolizer phenotype.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Pharmacogenetic approaches to investigation of severe adverse events may be useful in elucidating the mechanisms of drug metabolism and toxicity.
INTRODUCTION
Remdesivir is a nucleoside analog prodrug originally developed against Ebola virus and the first US Food and Drug Administration (FDA) approved medication for the treatment of patients hospitalized with novel coronavirus disease 2019 (COVID‐19). 1 Remdesivir is metabolized intracellularly to the pharmacologically active remdesivir triphosphate, which competes with endogenous adenosine triphosphate to prevent replication of viral RNA. 2 Due to its emergency approval by the FDA for COVID‐19, there is limited information on the safety, pharmacokinetic properties, and drug–drug interactions with remdesivir. Liver chemistry elevations were observed in early safety data in healthy volunteers and the compassionate use program. The FDA reported an overall 11.7% incidence of liver enzyme elevations among the 163 patients enrolled in the compassionate use program. 3
Remdesivir is primarily metabolized by plasma hydrolases. However, in vitro studies demonstrate that remdesivir is a substrate of several cytochrome P450 enzymes, including CYP2C8, CYP2D6, CYP3A4, and the drug transporters OATP1B and P‐glycoprotein. 4 Remdesivir is also a weak inhibitor of CYP1A2, CYP2C9, CYP2C19, and CYP2D6 in vitro. There are no published in vivo drug interaction studies of remdesivir. 4
Safety concerns with newly approved drugs may emerge in the post‐approval phase. To identify patients that may be susceptible to remdesivir‐associated liver injury, we performed a pharmacogenetic analysis utilizing a national cohort of US Veterans hospitalized with COVID‐19 with genetic information available through the VA Million Veteran Program (MVP).
METHODS
Data sources
MVP is a large multi‐ethnic genetic biobank within the Veterans Health Administration (VHA). 5 Biospecimens are linked to electronic health record data, including diagnosis codes (International Classification of Diseases ninth revision [ICD‐9] and tenth revision [ICD‐10]); current procedural terminology codes (CPT); clinical laboratory measures, and demographics. A COVID‐19 Shared Data Resource (SDR) was generated by the VA Informatics and Computing Infrastructure (VINCI) and hosted by the VA Phenomics Library, Centralized Interactive Phenomics Resource (CIPHER), documenting information regarding conditions, laboratory measures, medications, and procedures pertaining to the COVID‐19 pandemic. 6
Participants were genotyped using a custom axiom genotyping platform and imputed using the 1000 Genomes reference panel, as previously reported. 7 A composite variable derived from a combination of self‐reported survey information and genetically derived ancestry (harmonized ancestry, race, and ethnicity [HARE]) 8 was used in the models as a proxy for population stratification and global ancestry. Participants of non‐Hispanic White (NHW) and non‐Hispanic Black (NHB) populations were included for the current analysis. These populations reflect concordance among genetic and self‐reported race, ethnicity, and ancestry, and therefore, in this study, we use the term “population” to refer to these groups of participants. 8
The MVP received ethical and study protocol approval by the Veterans Affairs Central Institutional Review Board. Informed consent has been obtained from all participants. Each additional study was also approved by the local institutional review board. This project was also approved by the MVP COVID‐19 Scientific Steering Committee and the MVP Publication and Presentation Committee.
Study population
We identified 6910 MVP Veterans who were hospitalized with COVID‐19 within the VHA between March 15, 2020, and June 30, 2021. Patients were included in the cohort if they had positive severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2) polymerase chain reaction testing from a nasopharyngeal specimen and were hospitalized 14 days prior or 14 days after the positive test. Participants with pre‐existing liver disease, acute liver injury, cirrhosis, chronic hepatitis codes, and end‐stage renal disease based on ICD‐9/10 (Table S1) were excluded from the analysis (n = 1255). Further restricting our study within the HARE = NHW/NHB subgroup yielded the final study cohort of 4125 patients.
Remdesivir was the primary exposure and was determined based on review of the inpatient Bar Code Medication Administration (BCMA) files. Baseline aspartate aminotransferase (AST) and alanine aminotransferase (ALT) values were those closest to admission (including the day of admission) identified in the prior 730 days. The primary outcomes, peak AST and ALT, were identified in remdesivir users following the initiation of remdesivir. Peak AST and ALT in the control group was identified as the highest value during the hospitalization for COVID‐19.
Statistical analyses
Candidate genes selected for this analysis were based on in vitro data submitted by the manufacturer (Gilead) to the European Medicines Agency. 4 Pharmacogene phenotypes were assigned for the candidate genes CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, SLCO1B1, and SLCO1B3 using Stargazer software. 9 Phenotypes were manually assigned for CES1 (rs2244613), ABCB1 (rs2032582), CYP1A2 (rs762551), CYP2B6 (rs3745274, rs2279343, rs28399499, and rs34223104) based on variant information and functional assignment from PharmGKB 10 or PharmVar. 11 CYP2D6 ultrarapid metabolizers could not be assigned due to lack of copy number variation on the genotyping platform. Phenotype distributions by population are provided in Table S2. Analysis of covariance (ANCOVA) was used to assess the effect of remdesivir on peak liver chemistries while adjusting baseline liver chemistries. Univariate linear regression was performed on log of peak AST/ALT values during the hospitalization, in participants stratified by population (NHW and NHB separately), adjusted for age, sex, body mass index (BMI) at admission, glomerular filtration rate (GFR), baseline liver chemistries, history of pre‐existing type 2 diabetes (T2D) and hypertension, first 10 population‐specific principal components (PCs), and pharmacogene phenotypes together. For pharmacogenes with p < 0.05, multivariable models were created with both populations combined, adjusted for HARE and treatment. The models were then confirmed in each treatment group separately to evaluate the effect of remdesivir treatment. All statistical tests were two‐sided, where a p < 0.05 or a 95% confidence interval (CI) that did not contain unity was considered statistically significant. All analyses were conducted using R version 3.6.1. 12
RESULTS
Among the 4125 MVP participants hospitalized for COVID‐19, 1697 received remdesivir and 2428 did not (Table 1). Remdesivir was administered for a mean duration (SD) of 4 (2) days. Remdesivir‐treated participants were older, with a significantly greater BMI, Charlson comorbidity index, baseline ALT and AST, length of stay, and more often NHW population affiliation compared with patients not receiving remdesivir. Compared with untreated participants, higher peak ALT values were observed in the remdesivir‐treated and NHB participants (Figure 1). Among those remdesivir treated, 21.5% exhibited a peak ALT exceeding three times the upper limit of normal (×ULN) compared with 10.2% in the untreated participants (p < 0.0001). In those receiving remdesivir, the odds of an ALT greater than three times the ULN was significantly increased in NHW participants (odds ratio [OR] 1.89, 95% CI 1.33–2.71, p < 0.001) yet not in NHB participants (OR 1.23, 95% CI 0.84–1.81, p = 0.28). In an ANCOVA model adjusting for baseline ALT, remdesivir treatment versus nontreatment was associated with 33% and 24% higher peak ALT in NHW and NHB participants, respectively (p < 0.001). After adjusting for baseline ALT and population, the association between remdesivir treatment and higher peak ALT remained significant and associated with ~30% higher elevation versus the no treatment group (p < 0.001).
TABLE 1.
Characteristics of veterans hospitalized for COVID‐19, stratified by remdesivir treatment
| N = 4125, mean (SD), N (%) | No remdesivir, N = 2428 | Remdesivir, N = 1697 | p value |
|---|---|---|---|
| Age, years | 70.4 (12.2) | 71.3 (10.2) | 0.008 |
| Sex, female | 146 (6.0) | 95 (5.6) | 0.62 |
| Population a | |||
| White | 1537 (63.3) | 1192 (70.2) | <0.001 |
| Black | 891 (36.7) | 505 (29.8) | |
| BMI, kg/m2 | 29.7 (6.8) | 31.0 (7.1) | <0.001 |
| Charlson comorbidity index | 4.8 (3.8) | 5.1 (3.4) | 0.005 |
| Glomerular filtration rate, ml/min | 67.2 (24.0) | 67.8 (20.4) | 0.43 |
| Serum creatine, mg/dl | 1.3 (0.6) | 1.2 (0.4) | <0.001 |
| Diabetes | 1286 (53.0) | 938 (55.3) | 0.15 |
| Hypertension | 2022 (83.3) | 1397 (82.3) | 0.45 |
|
Length of stay, days Median (IQR) |
4 (2, 9) | 7 (5, 14) | <0.001 |
|
ALT baseline, U/L Median (IQR) |
24 (16, 36) | 27 (19, 41) | <0.001 |
|
AST baseline, U/L Median (IQR) |
27 (19, 40) | 36 (25, 52) | <0.001 |
|
Total bilirubin baseline, mg/dl Median (IQR) |
0.60 (0.40, 0.80) | 0.64 (0.50, 0.90) | <0.001 |
| Alkaline phosphatase baseline, U/L Median (IQR) | 77 (62, 98) | 71 (57, 91) | <0.001 |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; COVID‐19, coronavirus disease 2019; HARE, harmonized ancestry, race, and ethnicity; IQR, interquartile range.
Population determined by HARE ancestry.
FIGURE 1.
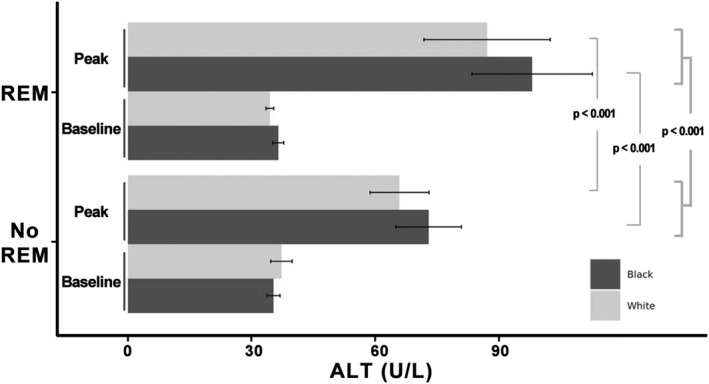
Peak ALT values by treatment group and population assignments of harmonized genetic ancestry and self‐reported race/ethnicity. Remdesivir (REM) leads to 30% increase of peak ALT in the entire study cohort with a slightly larger effect in non‐Hispanic White compared with non‐Hispanic Black participants (33% vs. 24% increase). p‐values were from ANCOVA model adjusting for baseline ALT in each population, or ANCOVA model adjusting for both baseline ALT and population. ALT, alanine aminotransferase; ANCOVA, analysis of covariance.
Higher peak AST values were also observed in the remdesivir‐treated participants versus untreated (median interquartile range [IQR] 44.5, IQR 30–71 vs. median 34, IQR 23–59 U/L, p < 0.001). After adjusting for baseline AST and population, the association between remdesivir treatment and higher peak AST was significant and associated with an ~16% increase versus the no treatment group (p < 0.001).
In a population stratified multivariable model (Table S3), with remdesivir included as a covariate, and adjusted for age, sex, baseline ALT, GFR, BMI, history of T2D, hypertension, Charlson comorbidity index, and 10 within‐population PCs, NHW participants who were CYP2C19 intermediate/poor metabolizers (IM/PM) had a 9% increased peak ALT compared with normal/rapid/ultrarapid metabolizers (NM/RM/UM; beta = 0.084, 95% CI 0.015–0.153, p = 0.017). This association was not observed in NHB participants (beta = −0.017, 95% CI −0.122–0.088, p = 0.76). In a sensitivity analysis in remdesivir‐treated participants only, CYP2C19 IM/PM had a 13% increased peak ALT compared with NM/RM/UM (beta = 0.123, 95% CI 0.024–0.221, p = 0.015) in NHW. However, this association was not observed in NHB participants (beta = −0.021, 95% CI −0.198–0.155, p = 0.81). There was no association between CYP2C19 phenotype and ALT elevation in the group not receiving remdesivir.
DISCUSSION
In over 4000 Veterans hospitalized with COVID‐19, we completed a genetically guided analysis to determine if liver enzyme elevations were more frequent in those treated with remdesivir or in individuals with pharmacogenetic variants using the MVP biobank. Among hospitalized patients without pre‐existing liver disease, ALT elevations (>3× ULN) affected twice as many remdesivir‐treated participants as those not treated with remdesivir and were higher in NHB participants. In a multivariable model adjusting for age, sex, BMI, renal function, baseline liver function tests, and comorbid conditions, NHW participants with CYP2C19 IM/PM had greater peak ALT compared to CYP2C19 NM/RM/UM; this effect was not seen in NHB participants. Although this latter observation may relate to insufficient statistical power, racial/ethnic differences in COVID‐19 outcomes and manifestations are described, 13 , 14 and so this finding still merits further exploration. Whereas our results will require validation in external cohorts, these novel findings suggest a pharmacogenetic approach using existing clinical genotyping methodologies that may help identify patients at greater risk for medication adverse events, especially for remdesivir with its limited information on metabolic and toxicity pathways.
In our hospitalized population, we observed significant ALT elevations (>3× ULN) in 21.5% of participants following remdesivir treatment as compared with 10.2% of untreated patients. In the VHA, these ALT elevations generally triggered treatment discontinuation and are described in the remdesivir package label. 1 In our MVP cohort, the rates of ALT elevation were several‐fold greater than those reported in the Adaptive COVID‐19 Treatment Trial (ACTT‐1). In the double‐blind placebo‐controlled ACTT‐1, non‐serious ALT elevations affected 2.3% and 4.7% of the remdesivir and placebo groups, respectively. 15 Compared with the ACTT‐1 cohort, the MVP cohort was older (71 vs. 59 years) with higher baseline rates of T2D (54% vs. 31%) and hypertension (83% vs. 51%), and included more NHB patients (34% vs. 21%). Advanced age, polypharmacy, and T2D may be risk factors for drug‐induced liver injury (DILI). 16 It is also possible that this real‐world Veteran cohort had greater comorbidity than those enrolled in clinical trials. Remdesivir‐associated liver injury has been observed in other real‐world cohorts of critically ill patients with COVID‐19 17 and pharmacovigilance studies 18 , 19 ; it has been associated with liver failure 20 and increased mortality. 21 Our study was not designed to analyze clinical outcomes of those observed to exhibit ALT elevations in the remdesivir‐treated group.
Pharmacogenetic variants in genes encoding for drug metabolizing enzymes and membrane transporters can substantively affect drug pharmacokinetic and pharmacodynamic properties, as well as adverse events. 22 Remdesivir is a prodrug extensively metabolized by esterases and CYP2C8, CYP2D6, and CYP3A4, as reported in preliminary in vitro data from healthy human donor livers. 4 , 23 Although specific metabolic pathways for the primary metabolite, a nucleoside core (GS‐441524), have not been reported, higher GS‐441524 plasma concentrations were observed in a critically ill patient with COVID‐19 and renal impairment, suggesting renal elimination may play an important role for the efficacy or toxicity of the metabolite. 24 Remdesivir was also found to be a weak inhibitor of CYP1A2, CYP2C9, CYP2C19, and CYP2D6 in vitro. 4 , 23 In our study, NHW participants with CYP2C19 IM/PM were observed to have greater ALT elevations compared with NM/RM/UM phenotypes. Based on our findings, in vivo drug–drug interaction studies with a CYP2C19 substrate can be performed to validate this potential pharmacokinetic mechanism. Moreover, in vitro studies also show remdesivir to inhibit the drug transporters OATP1B1/1B3, MATE1, and OCT1, which may increase intracellular remdesivir/metabolite concentrations 25 ; genetic polymorphisms encoding these transporters can also be investigated in future studies as potential mechanisms of DILI. Remdesivir is a good candidate for several drug–drug interactions, as described by Deb and colleagues, because it is a substrate for multiple CYP enzymes and transporters, and it is a CYP inhibitor and inducer. 23 Our study focused on the association of pharmacogenes with liver enzyme elevations and not drug pharmacokinetics. It is possible that pharmacogenetic variants are associated with remdesivir pharmacokinetics.
Our study included more than 4000 NHW and NHB participants with genetic data linked to well‐curated phenotype data derived from the nationally linked VHA electronic medical record data. Our study strengths include our broad analyses of pharmacogenes likely to affect remdesivir metabolism, its real‐world and clinically actionable focus, and evaluation of liver chemistry data throughout hospitalization for COVID‐19. Our relatively small study size and number of investigated genes limited a thorough genetic analysis and likely resulted in our study being underpowered to show a genetic effect in NHB participants. Additionally, we did not examine any long‐term outcomes of those with liver chemistry elevations. Yet, our findings build on and inform emerging remdesivir metabolic and safety data.
In our MVP study, remdesivir was associated with a 30% increase of peak ALT in patients hospitalized with COVID‐19, with a greater increase observed in NHW versus NHB participants. Remdesivir‐associated ALT elevations appear to be multifactorial, including genetic and non‐genetic contributions, and further studies are needed. A pharmacogenetic approach to investigation of severe adverse events may be useful assist in building the pharmacological profile of newly approved medicines while metabolic and toxicity data are emerging.
AUTHOR CONTRIBUTIONS
Wrote the manuscript: S.T., Z.Y., O.W., H.‐C.C., F.W., C.P.C., S.C.S., C.M.H., A.S., C.C., B.R.G., J.J., S.‐W.L., V.N., C.R.‐C., R.T., J.Z., K.‐M.C., A.M.H. Designed research: S.T., Z.Y., O.W., H.‐C.C., F.W., C.P.C., S.C.S., C.M.H., A.S., K.‐M.C., A.M.H. Performed research: S.T., Z.Y., O.W., H.‐C.C., F.W., C.P.C., S.C.S., C.M.H., A.S., C.C., B.R.G., J.J., S.‐W.L., V.N., C.R.‐C., R.T., J.Z., K.‐M.C., A.M.H. Analyzed data: S.T., Z.Y., O.W., H.‐C.C., F.W., C.P.C., S.C.S., C.M.H., A.S., B.R.G., K.‐M.C., A.M.H.
CONFLICT OF INTEREST
C.M.H. does consulting for Akebia Therapeutics, Inc. All other authors declared no competing interests for this work.
Supporting information
Appendix S1
ACKNOWLEDGEMENTS
The authors are grateful to our Veterans for their contribution to MVP.
Tuteja S, Yu Z, Wilson O, et al. Pharmacogenetic variants and risk of remdesivir‐associated liver enzyme elevations in Million Veteran Program participants hospitalized with COVID‐19. Clin Transl Sci. 2022;15:1880‐1886. doi: 10.1111/cts.13313
Funding information
This research is based on data from the Million Veteran Program Office of Research and Development, Veteran Health Administration, and was supported by award #MVP035 (MVP COVID‐19 Science Program) and VA Clinical Science Research and Development‐Investigator grant CX001897 (A.M.H.). Additional funding provided by K23HL143161 and the Penn Center for Precision Medicine for ST. This publication does not represent the views of the Department of Veteran Affairs or the United States Government
REFERENCES
- 1. Veklury (remdesivir) . Package insert. 2020. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/214787Orig1s000lbl.pdf. Accessed September 28, 2021.
- 2. Eastman RT, Roth JS, Brimacombe KR, et al. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID‐19. ACS Central Science. 2020;6:672‐683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grein J, Ohmagari N, Shin D, et al. Compassionate use of Remdesivir for patients with severe Covid‐19. N Engl J Med. 2020;382:2327‐2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. European Medicines Agency . Summary of compassionat use of remdesivir. https://www.ema.europa.eu/en/documents/other/summary‐compassionate‐use‐remdesivir‐gilead_en.pdf. Accessed August 1, 2020.
- 5. Gaziano JM, Concato J, Brophy M, et al. Million veteran program: a mega‐biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214‐223. [DOI] [PubMed] [Google Scholar]
- 6. Song RJ, Ho YL, Schubert P, et al. Phenome‐wide association of 1809 phenotypes and COVID‐19 disease progression in the veterans health administration million veteran program. PLoS One. 2021;16:e0251651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunter‐Zinck H, Shi Y, Li M, et al. Genotyping Array design and data quality control in the million veteran program. Am J Hum Genet. 2020;106:535‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fang H, Hui Q, Lynch J, et al. Harmonizing genetic ancestry and self‐identified race/ethnicity in genome‐wide association studies. Am J Hum Genet. 2019;105:763‐772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee SB, Wheeler MM, Patterson K, et al. Stargazer: a software tool for calling star alleles from next‐generation sequencing data using CYP2D6 as a model. Gene Med. 2019;21:361‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whirl‐Carrillo M, Huddart R, Gong L, et al. An evidence‐based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2021;110:563‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gaedigk A, Casey ST, Whirl‐Carrillo M, Miller NA, Klein TE. Pharmacogene variation consortium: a global resource and repository for Pharmacogene variation. Clin Pharmacol Ther. 2021;110:542‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2021. https://www.R‐project.org/. [Google Scholar]
- 13. Yancy CW. COVID‐19 and African Americans. Jama. 2020;323:1891‐1892. [DOI] [PubMed] [Google Scholar]
- 14. Rentsch CT, Kidwai‐Khan F, Tate JP, et al. Patterns of COVID‐19 testing and mortality by race and ethnicity among United States veterans: a nationwide cohort study. PLoS Med. 2020;17:e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19‐ final report. N Engl J Med. 2020;383:1813‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chalasani NP, Maddur H, Russo MW, Wong RJ, Reddy KR. ACG clinical guideline: diagnosis and Management of Idiosyncratic Drug‐Induced Liver Injury. Am J Gastroenterol. 2021;116:878‐898. [DOI] [PubMed] [Google Scholar]
- 17. Zampino R, Mele F, Florio LL, et al. Liver injury in remdesivir‐treated COVID‐19 patients. Hepatol Int. 2020;14:881‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rocca E, Gauffin O, Savage R, Vidlin SH, Grundmark B. Remdesivir in the COVID‐19 pandemic: an analysis of spontaneous reports in VigiBase during 2020. Drug Saf. 2021;44:987‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montastruc F, Thuriot S, Durrieu G. Hepatic disorders with the use of Remdesivir for coronavirus 2019. Clin Gastroenterol Hepatol. 2020;18:2835‐2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carothers C, Birrer K, Vo M. Acetylcysteine for the treatment of suspected Remdesivir‐associated acute liver failure in COVID‐19: a case series. Pharmacotherapy. 2020;40:1166‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hundt MA, Deng Y, Ciarleglio MM, Nathanson MH, Lim JK. Abnormal liver tests in COVID‐19: a retrospective observational cohort study of 1,827 patients in a major U.S. Hospital Network. Hepatology. 2020;72:1169‐1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394:521‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deb S, Reeves AA, Hopefl R, Bejusca R. ADME and pharmacokinetic properties of remdesivir: its drug interaction potential. Pharmaceuticals. 2021;14:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tempestilli M, Caputi P, Avataneo V, et al. Pharmacokinetics of remdesivir and GS‐441524 in two critically ill patients who recovered from COVID‐19. J Antimicrob Chemother. 2020;75:2977‐2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ambrus C, Bakos É, Sarkadi B, Özvegy‐Laczka C, Telbisz Á. Interactions of anti‐COVID‐19 drug candidates with hepatic transporters may cause liver toxicity and affect pharmacokinetics. Sci Rep. 2021;11:17810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


