Abstract
A group I intron interrupts the tRNACCUArg gene of the α-purple bacterium Agrobacterium tumefaciens (B. Reinhold-Hurek and D. A. Shub, Nature [London] 357:173–176, 1992). In this study, we assess the distribution of the corresponding intron among 12 additional species of α-purple bacteria. Of 10 newly identified tRNACCUArg genes, we found only two that contained an intron homologous to that of the Agrobacterium tRNACCUArg intron. This restricted and scattered distribution of the tRNACCUArg intron among α-purple bacteria is consistent with a recent origin and horizontal transmission. Primary and secondary structural similarities between tRNAUAALeu introns found in strains of the cyanobacterium Microcystis aeruginosa (K. Rudi and K. S. Jacobsen, FEMS Microbiol. Lett. 156:293–298, 1997) and α-purple tRNACCUArg introns suggest that these introns share a more recent common ancestor than either does with other known cyanobacterial tRNAUAALeu introns.
Group I introns are present in cellular and viral genes in eukaryotes and eubacteria (6, 15, 16). Although these introns interrupt a number of different protein-coding and RNA genes of eukaryotes and bacteriophages, the only insertion site observed so far in eubacteria is the sequence of the anticodon loop of tRNA genes. Eubacterial group I introns were first identified in the tRNAUAALeu genes of five cyanobacterial species (13, 22), prompting speculation that these introns would be widely distributed among cyanobacteria. In conjunction with the observation that most plastid genomes also possessed a homologous intron, an ancient origin predating the endosymbiotic event that gave rise to plastids was proposed for the tRNAUAALeu intron (13, 22). The phylogenetic distribution of tRNAUAALeu introns among cyanobacteria was later determined, and it was shown that the distribution was not universal but, nevertheless, was consistent with an ancient origin (17). This conclusion was challenged by the discovery of tRNAUAALeu introns in some strains of the cyanobacterium Microcystis aeruginosa which, however, seem to have originated independently of the previously characterized cyanobacterial tRNAUAALeu introns through horizontal transfer (19). Conversely, tRNA fMet group I introns are sporadically distributed among cyanobacteria (17), a finding that corroborated the initial suggestion of a relatively recent origin of these introns during cyanobacterial evolution (3).
It has previously been demonstrated that two other eubacterial tRNA genes are interrupted by group I introns, the tRNACAUIle gene of Azoarcus sp. BH72 (a β-purple bacterium) and the tRNACCUArg gene of Agrobacterium tumefaciens A136 (an α-purple bacterium) (18). Initial data suggested that similar introns are likely to be widespread among proteobacteria, as the Azoarcus and Agrobacterium intron probes cross-hybridized to genomic DNA from a number of purple bacteria (11, 18). Because determining their phylogenetic distribution is a valuable tool to assess the evolutionary history of group I introns (2, 17), we decided to survey the distribution of the tRNACCUArg intron among α-purple bacteria.
A tRNACCUArg intron in Azospirillum halopraeferens.
It has been shown that a restriction fragment from the genomic DNA of the α-purple bacterium A. halopraeferens Au5 hybridized to an Azoarcus intron probe (18). The signal was also observed when the Southern hybridization was repeated with an Agrobacterium tRNACCUArg intron probe (11). The hybridizing region was cloned in pBSM13 as a 4.3-kb PstI-SstI restriction fragment (pAGAU1.1) and subsequently subcloned as a 400-bp AvaII fragment (pAGAU1.2). The latter insert was completely sequenced on both strands, revealing a tRNACCUArg gene interrupted by a potential group I intron inserted after the U of the CCU anticodon (the same position as the Agrobacterium intron). The 217-bp Azospirillum intron sequence folds into a bona fide group I secondary structure (Fig. 1) and shares 69% identity with the Agrobacterium intron (Table 1). In addition, like the Agrobacterium intron, it self-splices in vitro (11, 18).
FIG. 1.
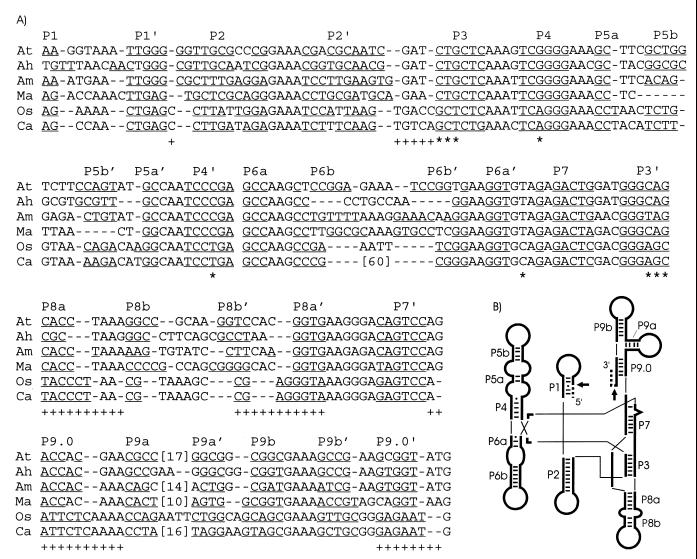
Intron sequence alignment and secondary structures. (A) DNA sequences of tRNACCUArg introns of the α-purple bacteria A. tumefaciens (At) (18), A. halopraeferens (Ah), and A. marginale (Am) are aligned with tRNAUAALeu introns from three distantly related cyanobacteria, M. aeruginosa (Ma) (19), Oscillatoria PCC 6304 (Os) (17), and Calothrix PCC 7101 (Ca) (17). Phylogenetically conserved base-paired regions (P1 to P9) (15) are indicated, with putative base-pairings underlined. Proposed base-pairing patterns of the Microcystis intron have been altered from the original report (19) to better conform to the conserved secondary structure of group I introns (15). Numbers in brackets indicate omitted sequence, and dashes represent gaps introduced to improve the alignment. Primary sequence (*) or secondary structural (+) similarities pointing to a closer relationship of the Microcystis tRNAUAALeu introns to the tRNACCUArg introns, rather than to the other known cyanobacterial tRNAUAALeu introns, are shown below the alignment. Note that these emphasized primary and secondary structural motifs are conserved among the cyanobacterial tRNAUAALeu introns (17), excluding the Microcystis introns. (B) Schematic representation of the consensus secondary structure of eubacterial tRNA group I introns, drawn according to Cech et al. (5). Phylogenetically conserved stems (P1 to P9), Watson-Crick base pairs (bars), and G-U pairs (dots) are shown according to Michel and Westhoff (15). Exons (dashed lines), the intron (thick lines), and splice sites (arrows) are indicated. Thin lines are used to join helical domains.
TABLE 1.
Identity of intron and small subunit rRNA sequencesa
| % Identity
|
||||||
|---|---|---|---|---|---|---|
| At | Ah | Am | Ma | Os | Ca | |
| At | 69 | 66 | 66 | 47 | 47 | |
| Ah | 85 | 62 | 56 | 45 | 45 | |
| Am | 83 | 80 | 63 | 53 | 51 | |
| Ma | 77 | 77 | 76 | 50 | 54 | |
| Os | 76 | 74 | 73 | 85 | 85 | |
| Ca | 74 | 73 | 72 | 83 | 83 | |
Values above the diagonal are from intron sequences, those below are from rRNA sequences. At, A. tumefaciens; Ah, A. halopraeferens; Am, A. marginale; Ma, M. aeruginosa; Os, Oscillatoria PCC 6304; Ca, Calothrix PCC 7101.
PCR amplification of tRNACCUArg genes from various α-purple bacteria.
Based upon the exonic tRNA sequences of Agrobacterium and Azospirillum, we designed degenerate primers for amplifying tRNACCUArg genes from various α-purple bacteria. We performed PCRs (17) on either the extracted DNAs or cell pellets from 11 species, with the primers ARG-5′ (5′-GTCC[G/A/T]CGATGCTCA[G/A][C/T]A[A/G]GATA-3′) and ARG-3′ (5′-TGGTGTCC[G/A/C]C[G/T][G/A][G/C][G/A/T]GGA[A/T]TCGAACC-3′). These primers were expected to amplify a sequence that includes the entire anticodon stem and loop of the tRNA. Although PCR products of ∼75 bp, the expected size of an uninterrupted tRNA gene, were amplified from all the samples, a PCR product of approximately 300 bp, the expected size of an intron-containing gene was also amplified from the DNA of Anaplasma marginale (Fig. 2). Cloning and sequencing of three independent clones confirmed the identity of this product as a tRNACCUArg gene interrupted by a group I intron inserted after the U of the CCU anticodon. The Anaplasma intron is similar both in primary sequence and secondary structure to the other known tRNACCUArg group I introns (Fig. 1; Table 1).
FIG. 2.
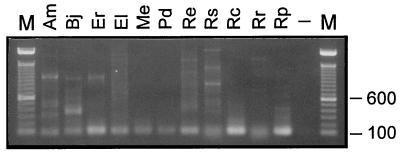
PCR amplification of tRNAArg genes. The migration in a 1% agarose gel of the PCR amplification products from DNA or cell pellets of various α-purple bacteria is shown. Am, A. marginale; Bj, B. japonicum; Er, E. risticii; El, Erythrobacter longus; Me, M. extorquens; Pd, Paracoccus denitrificans; Re, R. etli; Rs, Rhodobacter sphaeroides; Rc, R. centenaria; Rr, Rhodospirillum rubrum; Rp, R. prowazekii; −, no DNA (negative control); M, 100-bp DNA marker (Gibco/BRL) (sizes are indicated on the right side of the gel). Some of the samples shown here are the results of a reamplification (Am, Bj, Me, and Pd). R. etli and R. prowazekii are the two species from which the tRNACCUArg genes could not be detected.
To substantiate the apparent absence of introns in the tRNACCUArg genes of the other 10 species (Fig. 2), we sequenced up to 10 different tRNA-sized clones for each of them. Interestingly, the primers amplified various tRNAArg genes, not exclusively tRNACCUArg genes as intended (Table 2). For instance, the tRNA-sized PCR product from Anaplasma resulted from the amplification of a cognate tRNACCGArg gene, not from a second, intronless copy of the tRNACCUArg gene. In 4 of the 10 species we surveyed, the tRNACCUArg gene was not amplified. Alignment of tRNA gene sequences revealed that the first four nucleotides of tRNAYCUArg as amplified by our initial set of primers were invariably 5′-GAGC-3′ (Fig. 3), whereas most of the other tRNAArg sequences started with 5′-GAGT-3′ (data not shown). We exploited this discrepancy at the fourth position by designing a primer, ARG-5′EXT (5′-CC[G/A/T]CGATGCTCA[G/A][C/T]A[A/G]GATAGAGC-3′), to specifically amplify tRNAYCUArg from those four species for which we failed to detect the tRNACCUArg gene. Using the ARG-5′EXT and ARG-3′ primers, we were able to amplify the tRNACCUArg genes from two additional species (Table 2). All PCR mixtures were subjected to Southern hybridization with a radiolabeled Agrobacterium intron-specific probe, and as expected, only the intron-containing product of Anaplasma hybridized to the probe (data not shown). In summary, we identified a tRNACCUArg gene from 11 of 13 species, three of which contained an intron (Table 2). We could not amplify the corresponding gene from two species, Rhizobium etli and Rickettsia prowazekii. The genome of R. prowazekii has been completely sequenced and, as in some other bacterial genomes (e.g., Mycoplasma genitalium [10] and Haemophilus influenzae [9]), the tRNACCUArg gene is lacking (1). Assuming a G-U wobble, tRNAUCUArg can substitute for tRNACCUArg in decoding AGG triplets. Therefore, it is possible that the tRNACCUArg gene is also missing from the genome of R. etli.
TABLE 2.
PCR results and distribution of the tRNACCUArg intron
| Species | tRNAArg genes amplifieda
|
Intron in tRNACCUArg | ||||
|---|---|---|---|---|---|---|
| CCU | UCU | ACG | CCG | Total | ||
| Agrobacterium tumefaciens A136b | +d | |||||
| Anaplasma marginale | 3 | 0 | 0 | 3 | 6 | + |
| Azospirillum halopraeferens Au5b | + | |||||
| Bradyrhizobium japonicum | 1 | 2 | 4 | 0 | 7 | − |
| Ehrlichia risticii | 1 | 2 | 1 | 0 | 4 | − |
| Erythrobacter longus OCh 119 | 0 (1) | 0 (1) | 6 (1) | 4 | 13 | − |
| Methylobacterium extorquens | 1 | 2 | 7 | 0 | 10 | − |
| Paracoccus denitrificans | 1 | 1 | 0 | 2 | 4 | − |
| Rhizobium etli CE3 | 0 | 3 (4) | 0 | 7 (2) | 17c | ? |
| Rhodobacter sphaeroides ATH 2.4.1 | 0 (2) | 10 | 0 | 0 | 12 | − |
| Rhodocista centenaria | 1 | 0 | 0 | 0 | 1 | − |
| Rhodospirillum rubrum S.1 | 2 | 0 | 0 | 0 | 2 | − |
| Rickettsia prowazekii | 0 | 3 (7) | 7 | 0 | 18c | −e |
The identities of the amplified genes were determined by the anticodon sequence of their encoded tRNAs. PCR was performed with the pair of primers ARG-5′ and ARG-3′ or ARG-5′EXT and ARG-3′ (numbers in parentheses). After these data were obtained we discovered that the nucleotides complementary to positions 8 and 9 of the Agrobacterium tRNA of both 5′ PCR primers were transposed, resulting in potential mismatches. Both primers were resynthesized to be complementary at those positions, and PCR revealed no new products indicating the presence of an intron. Sequencing of 12 clones from these PCRs of R. etli DNA revealed the same anticodon species that had been determined previously. Two clones were obtained from PCR of R. prowazekii DNA; neither had an anticodon for Arg.
These species were not screened by PCR. The data were deduced from genomic clones.
A tRNAUUULys gene (R. etli) and a tRNAGCUAsn gene (R. prowazekii) were also amplified by PCR with the ARG-5′EXT and ARG-3′ primers.
Data from reference 18.
The tRNACCUArg gene is lacking from the R. prowazekii genome (1).
FIG. 3.
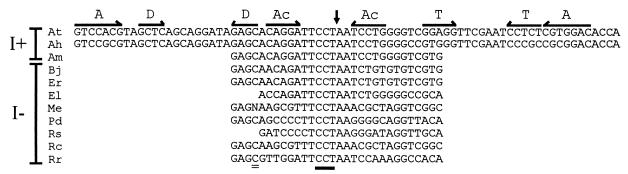
Alignment of tRNACCUArg gene sequences. Base-pairing in the acceptor stem (A), anticodon stem (Ac), D stem (D), and T stem (T), the intron insertion site (arrow), the anticodon sequence (underline), and the position used to restrict amplification to tRNAYCUArg genes (double underline) are indicated. N, any of the four possible nucleotides. I+ and I−, intron plus and minus, respectively. At, A. tumefaciens; Ah, A. halopraeferens; Am, A. marginale; Bj, B. japonicum; Er, E. risticii; El, E. longus; Me, M. extorquens; Pd, P. denitrificans; Rs, R. sphaeroides; Rc, R. centenaria; Rr, R. rubrum. Except for At and Ah, the tRNACCUArg genes were amplified by PCR. Sequences of the introns (shown in Fig. 1) and primers are omitted. The shorter sequences for E. longus and R. sphaeroides denote the use of the longer primer, ARG-5′EXT.
Distribution and evolution of the tRNACCUArg intron among α-purple bacteria.
The 13 species we have selected for this study represent only a sampling of the diversity of α-purple bacteria. Nevertheless, the distribution of the tRNACCUArg intron is informative because of the phylogenetic relationship among the species we surveyed. We believe the distribution of tRNACCUArg introns is best explained by a recent origin and horizontal transmission, rather than by an ancient origin and differential loss in various α-purple lineages. Our reasoning is as follows. First, the three species possessing a tRNACCUArg intron, A. tumefaciens, A. halopraeferens, and A. marginale, are widely divergent α-purple bacteria (Fig. 4; see also the phylogenetic tree compiled by the Ribosomal Database Project [RDP] in reference 14). Second, A. marginale and Ehrlichia risticii branch within a monophyletic clade of closely related species, but only the Anaplasma tRNACCUArg gene contains an intron. The same is also true for A. halopraeferens (intron present) and Rhodocista centenaria (also known as Rhodospirillum centenum) (no intron).
FIG. 4.
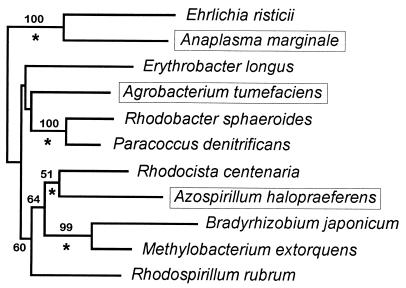
rRNA phylogenetic tree. Prealigned small subunit rRNA sequences were extracted from the RDP web site (14) for the 11 species whose tRNACCUArg genes were detected. The tree was inferred with DNADIST (Kimura two-parameter model [12], with a transition/transversion ratio of 2) and NEIGHBOR (20) as implemented in PHYLIP (8). Bootstrap (7) values were deduced from 100 replicates (only those values higher than 50 are shown). Branches conserved between this tree and that published on the RDP web site (14) are indicated (*), and species harboring a tRNACCUArg intron are boxed. Note the scattered distribution of the three tRNACCUArg intron-containing species; a monophyletic clade grouping these three species is supported by a bootstrap value of <1%. The topology shown has been suggested by the branching position of an outgroup (γ-purple bacterium Ectothiorhodospira shaposhnikovii).
Group I intron mobility is mediated by intron-encoded homing endonucleases or, alternatively, occurs via reverse splicing (for a review of group I intron mobility, see reference 21). Unlike reverse splicing, in which only the intron sequence is transferred, endonuclease-dependent mobility is accompanied by coconversion of flanking sequences. Interestingly, pairwise comparisons of the amplified regions of the tRNA genes shown in Fig. 3 reveal a striking similarity among the three intron-containing species (1, 2, or 3 differences, respectively, in 30 bp). On the other hand, with the exception of two identical pairs (E. risticii-Bradyrhizobium japonicum and Methylobacterium extorquens-R. centenaria) all of the other pairwise comparisons display 6 to 17 differences with a mean of 11 differences (note that, as expected, the 7-bp sequence defining the anticodon loop is virtually identical among all 11 species). Considering the fact that the three intron-containing species are not particularly closely related (Fig. 4), this observation is reminiscent of a recent endonuclease-dependent invasion. Although none of the introns described in this study contains an open reading frame (ORF), it would not be surprising if an ORF-containing intron is present in some other, unsurveyed species. For example, in a recent survey of cyanobacteria, tRNA-fMet genes of seven distantly related species were interrupted by highly similar introns. However, only one of these introns contained an endonuclease-encoding ORF (4, 17).
Recently, Rudi and Jacobsen showed that group I introns in tRNAUAALeu genes were sporadically distributed in strains of the cyanobacterium M. aeruginosa; six introns were found in 16 strains (19). Three of these introns were sequenced and shown to be almost identical (>99.5% identity) but only <61.5% identical to the previously described cyanobacterial tRNAUAALeu introns (19), whereas the latter are highly similar among themselves (Table 1; see also reference 17). Therefore, it was suggested that the tRNAUAALeu introns in cyanobacteria are polyphyletic and that the Microcystis introns originated independently through horizontal transfer (19). The Microcystis tRNAUAALeu introns most likely share a more recent common ancestor with the α-purple tRNACCUArg introns than either does with the other cyanobacterial tRNAUAALeu introns. This is supported by comparisons of primary sequences (Table 1; see also reference 19) and secondary structures (Fig. 1), as well as phylogenetic analyses (19 and data not shown). However, the origin of the tRNAUAALeu introns, excluding the Microcystis introns, appears to be monophyletic (17). Taken together, these results suggest that the Microcystis introns originated from a tRNACCUArg-like intron by horizontal transfer. This represents an interesting case of intron transposition and horizontal transmission between widely divergent species (cyanobacteria and α-purple bacteria).
Nucleotide sequence accession numbers.
The intron sequences reported here have been deposited in GenBank under accession numbers AF081791 (A. marginale) and AF081792 (A. halopraeferens).
Acknowledgments
We thank David Edgell and an anonymous reviewer for helpful comments and critical readings of the manuscript, David Mulbauher and Markus Landthaler for assistance in cloning and sequencing some of the clones, and Siv Andersson for sharing unpublished data.
This work was supported by NIH grant GM37746. B.P. was a postdoctoral fellow of NSERC (Canada).
REFERENCES
- 1.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Pontén T, Alsmark U C M, Podowski R M, Näslund A K, Eriksson A-S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya D, Surek B, Rüsing M, Damberger S, Melkonian M. Group I introns are inherited through common ancestry in the nuclear-encoded rRNA of Zygnematales (Charophyceae) Proc Natl Acad Sci USA. 1994;91:9916–9920. doi: 10.1073/pnas.91.21.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biniszkiewicz D, Cesnaviciene E, Shub D A. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonocora, R. P., and D. A. Shub. Unpublished results.
- 5.Cech T R, Damberger S H, Gutell R R. Representation of the secondary structure of group I introns. Nat Struct Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- 6.Damberger S H, Gutell R R. A comparative database of group I intron structures. Nucleic Acids Res. 1994;22:3508–3510. doi: 10.1093/nar/22.17.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 9.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirely R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchinson III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Heinfling, A., and D. A. Shub. Unpublished results.
- 12.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 13.Kuhsel M G, Strickland R, Palmer J D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 14.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel F, Westhoff E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 16.Nishida K, Suziki S, Kimura Y, Nomura N, Fujie M, Yamada T. Group I introns found in Chlorella viruses: biological implications. Virology. 1998;242:319–326. doi: 10.1006/viro.1998.9030. [DOI] [PubMed] [Google Scholar]
- 17.Paquin B, Kathe S D, Nierzwicki-Bauer S A, Shub D A. Origin and evolution of group I introns in cyanobacterial tRNA genes. J Bacteriol. 1997;179:6798–6806. doi: 10.1128/jb.179.21.6798-6806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhold-Hurek B, Shub D A. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature (London) 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 19.Rudi K, Jacobsen K S. Cyanobacterial tRNAUAALeu group I introns have polyphyletic origin. FEMS Microbiol Lett. 1997;156:293–298. doi: 10.1016/s0378-1097(97)00446-1. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Saldanha R, Mohr G, Belfort M, Lambowitz A M. Group I and group II introns. FASEB J. 1993;7:15–24. doi: 10.1096/fasebj.7.1.8422962. [DOI] [PubMed] [Google Scholar]
- 22.Xu M-Q, Kathe S D, Goodrich-Blair H, Nierzwicki-Bauer S A, Shub D A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990;250:1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]