Abstract
Background
Antibody‐based tests are available for measuring SARS‐CoV‐2‐specific immune responses but fast T‐cell assays remain scarce. Robust T cell‐based tests are needed to differentiate specific cellular immune responses after infection from those after vaccination.
Methods
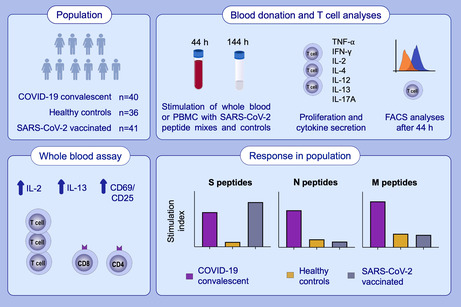
One hundred seventeen individuals (COVID‐19 convalescent patients: n = 40; SARS‐CoV‐2 vaccinees: n = 41; healthy controls: n = 36) were evaluated for SARS‐CoV‐2‐specific cellular immune responses (proliferation, Th1, Th2, Th17, and inflammatory cytokines, activation‐induced marker [AIM] expression) by incubating purified peripheral blood mononuclear cells (PBMC) or whole blood (WB) with SARS‐CoV‐2 peptides (S, N, or M), vaccine antigens (tetanus toxoid, tick borne encephalitis virus) or polyclonal stimuli (Staphylococcal enterotoxin, phytohemagglutinin).
Results
N‐peptide mix stimulation of WB identified the combination of IL‐2 and IL‐13 secretion as superior to IFN‐γ secretion to discriminate between COVID‐19‐convalescent patients and healthy controls (p < .0001). Comparable results were obtained with M‐ or S‐peptides, the latter almost comparably recalled IL‐2, IFN‐γ, and IL‐13 responses in WB of vaccinees. Analysis 10 months as opposed to 10 weeks after COVID‐19, but not allergic disease status, positively correlated with IL‐13 recall responses. WB cytokine responses correlated with cytokine and proliferation responses of PBMC. Antigen‐induced neo‐expression of the C‐type lectin CD69 on CD4+ (p < .0001) and CD8+ (p = .0002) T cells informed best about the SARS‐CoV‐2 exposure status with additional benefit coming from CD25 upregulation.
Conclusion
Along with N‐ and S‐peptide‐induced IL‐2 and CD69 neo‐expression, we suggest to include the type 2 cytokine IL‐13 as T‐cellular recall marker for SARS‐CoV‐2 specific T‐cellular immune responses after infection and vaccination.
Keywords: COVID, flow cytometry, lymphocytes, SARS‐CoV‐2, T cells
S‐ versus N‐ and M‐derived SARS‐CoV‐2 peptides allow discrimination between SARS‐CoV‐2 post‐vaccination and post‐infection cellular immune responses. T‐cell responses to SARS‐CoV‐2 peptides are accurately determined by a short‐term whole blood assay by analysis of secreted IL‐2 and IL‐13 along with CD69 and CD25 expression on T cells. The fast whole blood assay has a better performance than proliferation and cytokine secretion assays performed with gradient‐isolated peripheral blood mononuclear cells.
Abbreviations
- ACE2
angiotensin converting enzyme 2
- AIM
activation‐induced marker
- AUC
Area under the curve
- CD
cluster of differentiation
- CoV‐2
coronavirus 2
- COVID‐19
coronavirus disease 2019
- ELISA
enzyme‐linked immunosorbent assay
- HC
healthy control
- HLA
human leukocyte antigen
- HRP
horseradish peroxidase
- Ig
immunoglobulin
- M
matrix protein
- N
nucleocapsid protein
- NPV
negative predictive value
- OD
optical density
- PB
peripheral blood
- PBMC
peripheral blood mononuclear cells
- PHA
phytohemagglutinin
- PPV
positive predictive value
- RBD
receptor‐binding domain
- S
spike protein
- SARS
severe acute respiratory syndrome
- SEB
Staphylococcal enterotoxin B
- TBE
tick‐borne encephalitis antigens
- TT
tetanus toxoid
- WB
whole blood
1. INTRODUCTION
An outbreak of an epidemic respiratory infection causing severe pneumonia in a subset of patients was reported in Wuhan City, Hubei province, China, in December 2019. 1 , 2 , 3 Shortly thereafter, the disease was shown to be caused by a novel beta‐coronavirus (CoV) termed SARS‐CoV‐2 and the respiratory disease associated with it was named COVID‐19. 4 , 5 Since December 2019 and as of May 20, 2022, SARS‐CoV‐2 has infected 526 million individuals and caused 6.2 million fatalities world‐wide, 6 corresponding to an average fatality rate of 1.2%.
Typically, COVID‐19 induces both humoral and cellular immune responses by inducing antiviral antibodies that may block or even enhance the binding of the virus to its cellular receptor ACE2 to a varying degree 7 and activation of specific CD4+ and CD8+ T cells. 8 , 9 , 10 SARS‐CoV‐2 vaccines, which elicit various degrees of immunity, have been rapidly developed and licensed 11 , 12 , 13 resulting in the vaccination of 11.44 billion doses till May 2022. The vaccines currently authorized for use in Europe by the European Medical Association (EMA) are primarily mRNA‐ or vector‐based (Comirnaty, Spikevax, Vaxzevria, COVID‐19 Vaccine Janssen) and enable in vivo transfected or infected body cells, respectively, to exclusively express the SARS‐CoV‐2 S protein, but no other viral proteins. While in most cases, infection and vaccination induce humoral and cellular memory, 14 , 15 , 16 , 17 certain groups of individuals may have a considerable failure rate in doing so, 18 for example, those who suffer from (i) solid or hematologic malignancies and/or are in complete remission from such diseases, 19 , 20 or from (ii) primary 21 and secondary 22 , 23 immunodeficiency, have undergone (iii) solid 24 or bone marrow 25 transplantation, or are (iv) vaccine non‐responders. Apart from that, we have observed a considerable number (40%) of RBD non‐responders among COVID‐19 convalescent patients. 26 Accordingly, the mere fact that a particular patient had survived COVID‐19 or had been vaccinated with an mRNA‐ or vector‐based SARS‐CoV‐2 vaccine does not guarantee with certainty that the respective individual has also developed a protective humoral and cellular immunity. Moreover, recent data have shown that post‐vaccination side‐effects may not be differentiable with high accuracy from COVID‐19 by solely applying symptom profiles or machine‐derived algorithms. 27 This may be especially a problem if PCR tests were not performed in the time period directly after vaccination, since vaccination itself (especially during the first 3 days after vaccination) may cause fever and fatigue, symptoms also associated with mild COVID‐19 infection. 9 Therefore, in vitro assays are needed that can distinguish specific T‐cellular immune responses following infection from those following vaccination, especially in situations where humoral immune responses are absent (e.g., vaccine non‐responders) or have already diminished because the infection occurred some time ago 28 , or are masked due to immunoglobulin (Ig) substitution or Ig‐based immunomodulation therapy.
In fact, after cognate interaction with foreign peptide presented by self‐MHC molecules 29 , 30 within the immunological synapse, 31 T lymphocytes react in at least three typical ways. Firstly, they neo‐express activation‐induced molecules (AIM) on their surface, 32 secondly, they start to produce and secrete soluble effector molecules, such as cytokines, 33 and thirdly, they initiate proliferation if the interaction between them and the antigen‐presenting cell exceeds a critical period of time. 34 All three parameters should be helpful when it comes to the determination of T‐cellular immune responses of a patient either after COVID‐19 or upon SARS‐CoV‐2 specific vaccination. 35 , 36 , 37
Here, we established the basis for distinguishing T‐cellular SARS‐CoV‐2‐specific immune responses following infection from those following vaccination by identifying robust biomarkers. For that purpose, we tested a panel of S‐, N‐, and M‐protein specific peptides in a newly established 2‐day whole blood (WB) assay, which was bench‐marked to a standard antigen‐specific proliferation and cytokine secretion assay based on gradient‐isolated peripheral blood mononuclear cells (PBMC) which takes approximately 8–9 days for completion. 35 In the WB assay, we determined the SARS‐CoV‐2‐specific cellular immune response and compared it with the one induced by classic vaccine antigens (tetanus toxoid, tick borne encephalitis virus) and polyclonal T‐cell stimuli. The assay allows to measure the cumulative secretion of Th1, Th2, Th17, and inflammatory cytokines into the supernatant and to monitor the antigen‐specific T‐cell activation status by virtue of the expression of AIMs by CD4+ and CD8+ T cells alike. The WB assay is easy to perform, provides robust results in a short period of time (within 2 days), allows to differentiate COVID‐19 convalescent patients from vaccinated individuals and healthy controls, and identifies the type 2 cytokine IL‐13 along with the type 1 cytokines IL‐2 and IFN‐γ as well as the activation‐induced C‐type lectin CD69 and CD25 as biomarkers of high significance for determination of T‐cellular immune responses against SARS‐CoV‐2 after both COVID‐19 infection or SARS‐CoV‐2 vaccination.
2. MATERIALS AND METHODS
2.1. Patients, vaccinees, and control subjects
All subjects gave their written informed consent in accordance with the Declaration of Helsinki. This retrospective, observational, monocentric investigator‐driven study, which was performed at the Center of Pathophysiology, Infectiology, and Immunology at the Medical University of Vienna was approved by the Ethics Committee of the Medical University of Vienna registration number EK No.: 1302/2020. This study was supported by grants from the Austrian Science Fund, grant number: DK W1248; the Medizinisch‐Wissenschaftlicher Fonds des Bürgermeisters der Bundeshauptstadt Wien (Stiftungsfonds zur Förderung der Bekämpfung der Tuberkulose und anderer Lungenkrankheiten), grant numbers: COVID001 and COVID006, in part by a research grant from Viravaxx AG, Vienna, Austria and a grant from the Federal State of Lower Austria, Grant: Danube Allergy Research Cluster (Danube ARC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Convalescent patients had rtPCR‐ and/or SARS‐CoV‐2 antibody‐confirmed 7 COVID‐19 disease 10 weeks (n = 28) or 10 months (n = 12) previously. In parallel, healthy control subjects, who were reportedly asymptomatic for the last 10 weeks and who were SARS‐CoV‐2 negative by SARS‐CoV‐2 antibody test 7 and had a negative SARS‐CoV‐2 test (antigen or rtPCR) at the time of venipuncture (n = 36) or were vaccinated once (n = 8), twice (n = 11), or three times (n = 22) were enrolled into the study between November 1, 2020, and March 23, 2022. Table 1 shows the baseline demographic, serological and clinical characteristics for each group. Of the eight vaccinees, who were vaccinated once, five received only one of two required doses of Vaxzevria, one received only one of two required doses of Comirnaty and two received the licensed single dose of COVID‐19 Vaccine Janssen. Of the 11 vaccinees, who were vaccinated twice, seven received Comirnaty, three Vaxzevria, and one the combination of the latter (Tables 1 and S1). From the three times vaccinated individuals, 11 received Comirnaty, 9 two times Vaxzevria and once Comirnaty, and two individuals two times Vaxzevria and once Spikevax.
TABLE 1.
Demographic, serological and clinical characteristics of study participants
| Characteristics (%) | COVID‐19 convalescent patients | Healthy control subjects | COVID‐19 vaccinees |
|---|---|---|---|
| N | 40 | 36 | 41 |
| Age Median (Range) | 42 (24–82) | 37 (20–71) | 32 (21–69) |
| (Mean ± SD) | 43.4 ± 14.6 | 41.1 ± 16.0 | 36.8 ± 13.3 |
| Female (%) | 28 (70) | 21 (58.3) | 23 (56.1) |
| PCR positive (%) a | 39 (97.5) | 0 (0.0) | 0 (0.0) |
| Anti‐S IgG Ab positive (%) | 40 (100) | 0 (0) | 41 (100) |
| Anti‐S IgG4 Ab positive (%) | 0 (0) | 0 (0) | 15 (36.6) |
| Anti‐RBD IgG Ab positive (%) | 20 (50.0) | 0 (0) | 35 (85.4) |
| Anti‐RBD IgG4 Ab positive (%) | 0 (0) | 0 (0) | 10 (24.4) |
| Clinical symptoms (%) d | 39 (97.5) | 0 (0.0) | 0 (0.0) |
| Fever (days) | 3.3 ± 4.6 | 0 (0.0) | 0 (0.0) |
| Fever (%) | 24 (60.0) | 0 (0.0) | 0 (0.0) |
| <37.5°C | 2 (5.0) | ||
| 37.5–38.0°C | 9 (22.5) | ||
| 38.1–39.0°C | 4 (10.0) | ||
| >39.0°C | 9 (22.5) | ||
| Hospitalization | 9 (22.5) | ||
| 10 weeks after 1° infection, n (%) | 28 (70.0) | ||
| Days after 1° infection (Mean ± SD) | 101 ± 30 | ||
| 10 months after 1° infection, n (%) | 12 (30.0) | ||
| Days after 1° infection (Mean ± SD) | 296 ± 62 | ||
| Post‐vaccination (days after last injection, Mean ± SD) | 80.2 ± 46.6 | ||
| Median (Range) | 87 (12–209) | ||
| Vaccinated subjects | 41 | ||
| Vaccinated once (%) | 8 (19.5) | ||
| Vaccinated twice (%) | 11 (26.8) | ||
| Vaccinated three times (%) | 22 (53.7) | ||
| Preexisting medical conditions b (%) | 25 (62.5) | 12 (33.3) | 15 (36.6) |
| Allergy/asthma c (%) | 12 (30.0) | 8 (22.2) | 12 (29.3) |
One patient was PCR negative but S‐antibody positive and was thus included into the group of COVID‐19 convalescent individuals.
The following preexisting medical conditions were inquired for: Cardiovascular diseases, chronic lung diseases, allergy/asthma, diabetes mellitus, hematopoietic diseases, immunosuppressive conditions, liver diseases, metabolic diseases, neurological disorders, or renal diseases.
Allergy/asthma patients have been singled out from patients with preexisting medical conditions to control for/exclude a putative recruitment bias.
Short term symptoms (1‐3 days) directly after vaccination were considered vaccine‐induced and thus were not taken into consideration as COVID‐19‐related clinical symptoms.
Venous blood was drawn from all subjects and was Li‐heparin‐anticoagulated (whole blood assays and preparation of PBMC), or silicon dioxide coagulated (to obtain serum for determining specific antibodies).
2.2. T‐cell proliferation assays
2.2.1. Plate assays with isolated peripheral blood mononuclear cells
Peripheral blood mononuclear cells of heparinized blood were isolated from COVID‐19 convalescent individuals, healthy control subjects, or SARS‐CoV‐2 vaccinees according to standard protocols. 11 , 12 , 14 Briefly, heparinized blood was diluted 1:2 with IMDM medium containing 20 U/ml heparin, 10% FCS and antibiotics (15 μg/ml Gentamicin; 0.5 μg/ml Amphotericin) and was overlaid onto Ficoll–Hypaque gradients in 50 ml tubes followed by centrifugation at 500 g for 15 min. The PBMC rich interphase was collected, washed with fresh medium and adjusted to 1 x 106 cells/ml with RPMI 1640 medium (Hyclone, Cytiva, Pasching, Austria) containing 2% human serum (Sigma‐Aldrich, St. Louis, MO, USA). One‐hundred microliters of freshly isolated PBMC (1 x 105/well) were incubated in 96‐well round bottom plates (Sarstedt, Nümbrecht, Germany) with the indicated stimuli (Table S2) in a total volume of 200 μl. After 144 h (6 days), 100 μl of supernatants were collected from each well for subsequent cytokine determinations and stored frozen at −80°C until further use (see online repository Appendix S1). The cells were replenished with 100 μl of fresh medium and pulsed with methyl‐[3H]‐thymidine (1 μCi/well) for 18 h and T‐cell proliferation was quantified on a Betaplate Counter (Perkin Elmer, Waltham, MA, USA).
2.2.2. Whole blood assays in tubes
Three hundred microliters of whole blood anticoagulated with Li‐heparin were incubated with the indicated stimuli (Table S2) or medium alone in 300 μl culture medium (RPMI 1640, Hyclone, Cytiva, Pasching, Austria) in sterile 5 ml polystyrene round‐bottom tubes (12 x 75 mm tubes with caps, BD Biosciences, San Diego, CA, USA) in a final volume of 600 μl. In order to maximally increase IFN‐γ release from activated memory cells, tubes were spiked with 25 μl of IL‐18 (2.25 μg/ml), so that a final concentration of IL‐18 of 90 ng/ml was reached. 38 , 39 After addition of 300 μl of whole blood, the cell/medium suspension was mixed by vortexing 3‐times for 1 s. Tubes with semi‐closed lids were then incubated at 37°C, in a 5% CO2, and 95% humidity atmosphere for 44 h. Subsequently, cultures were mixed by vortexing 3‐times for 1 s, then cells were spun down at 600 g for 10 min, and 300 μl of the supernatant was collected in 1.5 ml microfuge tubes and stored frozen at −80°C until further use. The cellular part of stimulations was used to determine activation induced markers (AIM) on CD3+CD4+ and CD3+CD8+ and T cells. Cells from 31 out of the 40 COVID‐19 convalescent patients, 32 out of the 36 healthy control individuals and 40 out of the 41 vaccinees could be tested with the WB‐ and PBMC‐based assays in parallel.
2.3. Statistical analyses
Distribution analysis was performed for each dependent variable. Best fit was obtained in any case for a log‐normal distribution. Therefore, variables were analyzed by a General Linear Model (GLM) with a log link. All stimuli and the different groups were analyzed simultaneously for each cytokine, and groups (within stimuli) were compared by Tukey's honestly significant difference (HSD) test holding the family‐wise error rate constant at the chosen level of significance of 5% to account for multiple testing. For illustrative purposes, p‐values below .05 were indicated as: * p < .05, ** p < .01, *** p < .001, respectively. However, since seven cytokines were measured, in addition to the univariate approach, a discriminant analysis was performed with stimulation indices for all cytokines and the S‐peptide, N‐peptide, and M‐peptide mixtures as stimulants of WB. Exploratory analyses were done comparing stimulation indices in vaccinees by number of vaccinations received and in COVID‐19 patients according to the severity of the disease (hospitalized vs. not hospitalized) and the time since recovery (10 weeks vs. 10 months). In these latter analyses, healthy controls were included as contrast group. In all these exploratory analyses, orthogonal Helmert contrast was computed allowing independent interpretation of the differences obtained. For these exploratory analyses, power was still sufficient (80%) to detect a relevant effect corresponding to a Cohen's d = 1. While within endpoint, analyses were corrected for multiple testing and family‐wise error rate was held constant, no correction for multiple endpoints was applied. Therefore, results have to be interpreted with caution. Receiver operating characteristics (ROC) curves were calculated based on 95% confidence intervals by the method of Wilson/Brown. Stata 17.0 (StataCorp), Statistica 10.0 (Statsoft), and GraphPad 9.2.0 (GraphPad Software) were used for analyses and graphs.
Further Materials and methods can be found in the online repository (Appendix S1).
3. RESULTS
3.1. Description of the study population
We enrolled a total of 117 individuals into this study consisting of 40 COVID‐19 convalescent patients, 36 healthy control, and 41 vaccinated individuals (Table 1). COVID‐19‐specific T‐cell responses toward S‐, N‐, and M‐protein‐derived peptides (Table S3) were compared in a newly developed 2‐day WB test evaluating COVID‐19 specific effector cytokine release and CD4+ and CD8+ T‐cellular activation with that of classical lymphocyte proliferation and cytokine secretion assays using gradient‐isolated PBMC. The WB‐ and PBMC‐based tests were performed in 25 independent experiments (Table S4) and in addition contained the recall antigens tetanus toxoid (TT) and tick born encephalitis virus antigen (TBE) as well as the polyclonal stimuli Staphylococcal enterotoxin B (SEB) and PHA as positive and medium alone as negative controls. Sera from COVID‐19 convalescent patients and healthy control individuals were screened for the presence of SARS‐CoV‐2 specific antibodies against SARS‐CoV‐2 S‐ and RBD‐protein, to confirm or exclude their SARS‐CoV‐2 exposure status independent of the self‐reported medical history. Each out of the enrolled 40 COVID‐19 convalescent patients had a SARS‐CoV‐2 S‐protein specific antibody response (Table 1), with only 20 out of the 40 patients also revealing RBD‐specific ELISA reactivity above OD 0.3 (Table 1), confirming their SARS‐CoV‐2 specific seroconversion as established previously. 7 , 26 In contrast, none of the 36 healthy control individuals presented with SARS‐CoV‐2 S‐ or RBD‐protein specific antibody reactivity (Table 1).
3.2. SARS‐CoV‐2 peptide‐specific proliferation of gradient‐isolated PBMC discriminates post‐infection from post‐vaccination subjects
We here aimed to establish the basis for a robust, easy‐to‐perform bioassay able to distinguish SARS‐CoV‐2 specific T‐cell immune responses of COVID‐19 convalescent patients, vaccinated individuals, and SARS‐CoV‐2 non‐exposed healthy controls. Such distinction may not be trivial if solely based on antibody testing since antibody non‐responders to important constituents of the SARS‐CoV‐2 virus (e.g., S‐ or RBD protein) have been described. 26 For instance, only 50% of the COVID‐19 convalescent individuals enrolled into this study presented with RBD antibodies (Table 1) and also S‐antibodies are known to drop below background in a fraction of COVID‐19 convalescent patients 10 months after infection (manuscript in preparation). By simultaneous monitoring of the elaboration of Th1, Th2, and Th17 and inflammatory cytokines, we also wanted to understand which cytokines are primarily associated with the T‐cellular immune response against major SARS‐CoV‐2 antigens, such as surface (S), nucleocapsid (N), and matrix (M) antigen.
Accordingly, and to benchmark the test system, we first evaluated whether stimulation of gradient‐isolated PBMC with peptide‐pools of SARS‐CoV‐2 S‐, N‐, or M‐proteins would allow for such differentiation. Therefore, we selected a well‐defined study population consisting of individuals without SARS‐CoV‐2 exposure (healthy controls) and individuals with a clear history of either SARS‐CoV‐2 infection or vaccination (Table 1). Notably, we found that PBMC of COVID‐19 convalescent patients strongly proliferated upon incubation with the S‐, N‐, and M‐peptide mixes, which was in significant contrast to PBMC of healthy control subjects. The proliferative response of PBMC of COVID‐19 convalescent patients upon stimulation with S‐peptide was 1.7‐fold and 2.6‐fold higher when compared with N‐ or M‐peptide mix, respectively (Figures 1, S1 and S2; Tables S5 and S6), which identified S‐protein peptides as major targets for the human T‐cell response. Apart from incubation with SARS‐CoV‐2 peptide mixes, virus‐like particles 40 pseudotyped with S‐, RBD‐, or N‐proteins led to robust and specific T‐cell stimulation (not shown). S‐ but not N‐ or M‐peptide mixes also well stimulated PBMC of vaccinees (Figures 1, S1 and S2). In contrast to the differential SARS‐CoV‐2 peptide‐induced proliferation in the three study populations, proliferation of PBMC of COVID‐19 convalescent patients or vaccinees was not superior to those of healthy control subjects when incubated with the vaccine recall antigens from TT or TBE or upon incubation with the polyclonal stimuli Staphylococcal enterotoxin B or PHA (Tables S5 and S6).
FIGURE 1.
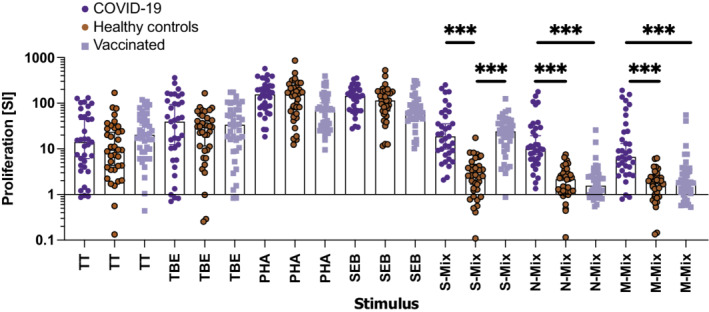
Results of cellular proliferation assays performed with gradient‐isolated peripheral blood mononuclear cells (PBMC) stimulated with the indicated antigen‐specific and polyclonal stimuli in classical plate assays for 7 days. Shown is the summary of stimulation indices (SI, y‐axis) of PBMC which were incubated with the indicated stimuli (x‐axis). The bars represent the median, whiskers the Hodges‐Lehmann 95% confidence intervals, dark blue circles show proliferation of PBMC of COVID‐19 convalescent patients, red circles those of non‐exposed healthy controls and light blue squares those of vaccinees. M‐mix, SARS‐CoV‐2 matrix protein peptide mix; N‐mix, SARS‐CoV‐2 nucleocapsid protein peptide mix; PHA, phytohemagglutinin; S‐mix, SARS‐CoV‐2 spike protein peptide mix; SEB, Staphylococcal enterotoxin B; TBE, tick borne encephalitis antigen; TT, tetanus toxoid. Data show the summary of 35 COVID‐19 convalescent patients, except 31 for SEB, 36 healthy controls, except 32 for SEB, and 40 vaccinees. p values were calculated by Tuckey's test. Only significant differences are shown. ***p < .001
3.3. Prominent cellular IL‐13 recall responses in gradient‐isolated PBMC post‐infection and post‐vaccination
To better understand the role and differential contribution of cytokines during SARS‐CoV‐2‐specific T‐cell recall responses, we studied in an unbiased approach the secretion of signature Th1, Th2, Th17, and inflammatory cytokines upon incubation of gradient‐isolated PBMC with the three peptide mixes, classic vaccine antigens, polyclonal T‐cell stimuli, and medium alone. Notably the highest discriminative accuracy between COVID‐19 convalescent patients and HC subjects was obtained upon determination of IL‐2 in supernatants of PBMC stimulated with N‐ (AUC: 0.8710 [CI: 0.7711–0.9708]; p < .0001; Figures 2 and S3) or M‐ (AUC: 0.8356 [CI: 0.7250–0.9461] p < .0001; Figure S3) peptide mixes immediately followed by IFN‐γ (AUC: 0.8439 (CI: 0.7470–0.9408) p < .0001 and AUC: 0.7924 (CI: 0.678–0.9080) p < .0001, respectively), and IL‐13 (AUC: 0.8033 (CI: 0.6884–0.9183) p < .0001 and AUC: 0.7388 (CI: 0.6147–0.8629) p = .0012, respectively). A similarly high discriminatory power was obtained with IFN‐γ (AUC: 0.8944, [CI: 0.8022–0.9866] p < .0001) and IL‐13 (AUC: 0.8927 [CI: 0.8211–0.9643] p < .0001) upon S‐, but not N‐ or M‐peptide mix‐specific stimulation of PBMC of vaccinees compared with healthy control subjects while IL‐2 was somewhat less discriminatory (AUC: 0.7383, [CI: 0.6138–0.8628] p = .0006; Figures 2 and S4). Absolute levels of secreted cytokines upon incubation with the different stimuli are shown in Figure S5 and corroborated the SI results, although they revealed a modestly inferior diagnostic accuracy (Tables S7–S9). IL‐4, IL‐12, and IL‐17A secretion levels were not informative in assays performed with gradient‐isolated PBMC.
FIGURE 2.
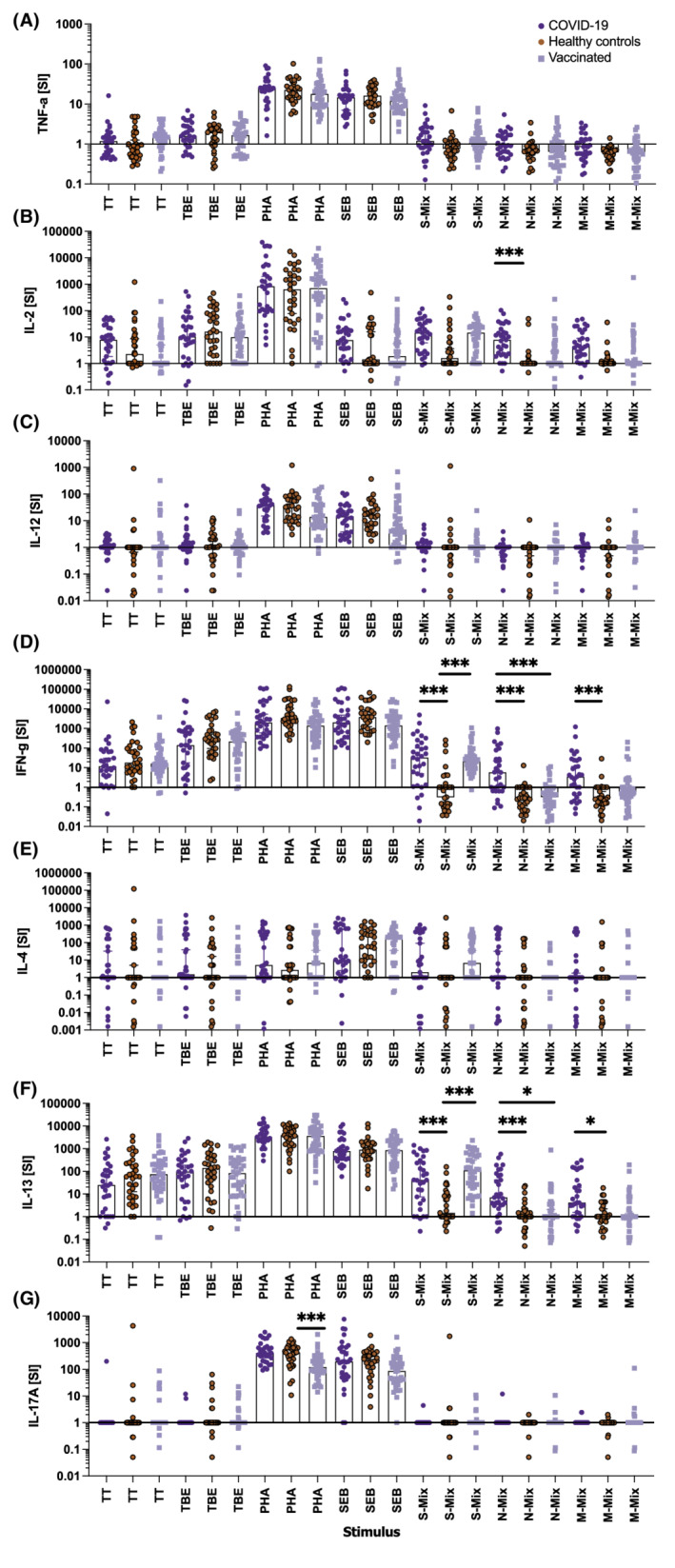
Results of cytokine secretion obtained with gradient‐isolated peripheral blood mononuclear cells (PBMC) stimulated with the indicated antigen‐specific and polyclonal stimuli in classical plate assays. Shown are the stimulation indices (y‐axes) for (A) TNF‐α, (B) IL‐2, (C) IL‐12p70, (D) IFN‐γ, (E) IL‐4, (F) IL‐13, and (G) IL‐17A produced after incubation of 1 x 105 PBMC per well co‐incubated with the indicated stimuli (x‐axes) for 144 h. Bars represent the median, whiskers the Hodges‐Lehmann 95% confidence intervals, dark blue circles show COVID‐19 convalescent patients, red circles those of non‐exposed healthy controls, and light blue squares vaccinees. M‐mix, SARS‐CoV‐2 matrix protein peptide mix; N‐mix, SARS‐CoV‐2 nucleocapsid protein peptide mix; PHA, phytohemagglutinin; S‐mix, SARS‐CoV‐2 spike protein peptide mix; SEB, Staphylococcal enterotoxin B; TBE, tick borne encephalitis antigen; TT, tetanus toxoid. Data show the summary of 31 COVID‐19 convalescent patients, 32 healthy controls, and 40 vaccinees. p values were calculated by Tuckey's test. Only significant differences are shown. *p < .05; ***p < .001
3.4. SARS‐CoV‐2 specific IL‐13 recall responses are significantly elevated independent of allergy status
The prominent IL‐13 recall responses upon incubation of gradient‐isolated PBMC of COVID‐19 convalescent patients and vaccinees with SARS‐CoV‐2 peptide mixes were intriguing and could have been due to different representation of allergic/asthmatic patients in the respective groups, representing a putative recruiting bias. To investigate this possibility, each study group was divided into non‐allergic and allergic subjects (Figure 3 and Table 1). Of note, SARS‐CoV‐2 peptide‐induced IL‐13 secretion levels did neither positively correlate with the allergy status of COVID‐19 convalescent patients, nor with those of vaccinees or healthy control subjects (Figure 3). The present study was not powered to detect differences in IL‐13 levels between the approximately 30% of allergic/asthmatic individuals in the COVID‐19 convalescent and vaccinated groups versus the 20% allergic/asthmatic individuals in the healthy control group. Irrespective of this limitation, we assume that a relevant effect size could have been detected in this unplanned comparison but the actually observed one is so small that we conclude that the allergy/asthmatic status most likely does not account for the differences in release of IL‐13.
FIGURE 3.
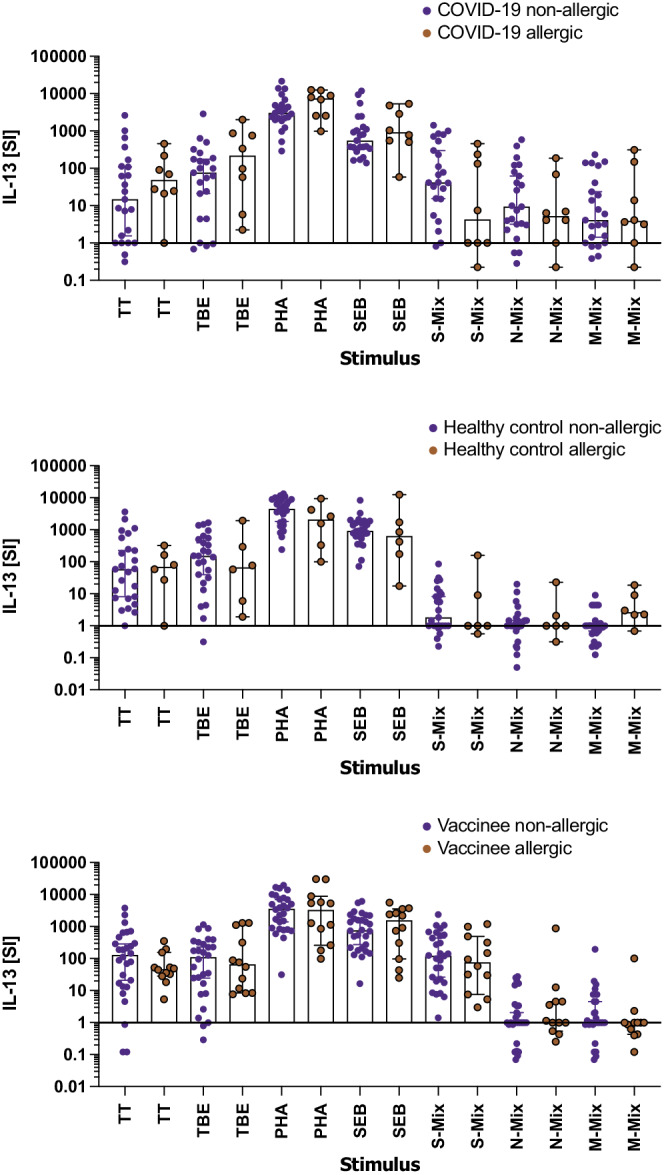
Comparison of IL‐13 production by gradient‐isolated peripheral blood mononuclear cells (PBMC) of non‐allergic versus allergic patients in groups of COVID‐19 convalescent patients, healthy control subjects and vaccinated subjects. Shown are the summaries of the stimulation indices (SI, y‐axes) for cytokine production of PBMC incubated with the indicated stimuli (x‐axes). The bars represent the median, whiskers the Hodges‐Lehmann 95% confidence interval, blue circles show non‐allergic, red circles allergic COVID‐19 convalescent patients (upper panel), healthy controls (middle panel) and vaccines (lower panel). M‐mix, SARS‐CoV‐2 matrix protein peptide mix; N‐mix, SARS‐CoV‐2 nucleocapsid protein peptide mix; PHA, phytohemagglutinin, S‐mix, SARS‐CoV‐2 spike protein peptide mix; SEB, Staphylococcal enterotoxin B; TBE, tick borne encephalitis antigen; TT, tetanus toxoid. Data show the summary of 31 COVID‐19 convalescent patients, 31 healthy controls and 40 vaccinees. p values were calculated by Helmert contrasts test. Only significant differences are shown
3.5. Antigen‐specific, cellular Th1 and Th2 recall responses characterize COVID‐19 convalescent patients and vaccinees in a two‐day WB assay
Virus‐specific recall responses determined with gradient‐isolated PBMC well‐discriminated the three study groups and have the power to discriminate between the post‐infection and post‐vaccination status of subjects. Next, we investigated if such results can be obtained with a rapid whole blood (WB) assay using cytokine secretion and AIM expression as read‐outs. Short term assays for T‐cellular recall responses typically rely on the early release of IFN‐γ (interferon gamma release assay, IGRA, https://www.quantiferon.com/products/quantiferon‐tb‐gold‐plus‐qft‐plus/) from antigen‐activated memory T cells. 41 , 42
To show how individuals with similar history of SARS‐CoV‐2 exposure (infection vs. vaccination) and healthy control individuals group, discriminant analyses were performed with all data obtained upon WB stimulation with the three SARS‐CoV‐2 peptide mixes and using the seven cytokines as read‐out. These analyses showed that both roots highly significantly (p < .001) contributed to the separation of our three study populations. The first root discriminated between vaccinated individuals on one side and controls and COVID‐19 patients on the other (Figure 4). Large values of IL‐13, TNF‐α, IL‐2, and IFN‐γ upon S‐mix stimulation speak in favor of vaccination‐induced responses. The second root discriminates between healthy controls and COVID‐19 patients with high stimulation of IL‐2 by the S‐ or M‐peptide mix, of IL‐17A and IFN‐γ by the N‐peptide mix, and IFN‐γ and IL‐13 by the S‐peptide being in favor of previous infection (Figure 4). Accordingly, COVID‐19 convalescent patients are primarily positioned in the upper right quadrant, healthy controls are found in the lower right quadrant, while vaccinated individuals distribute to the two left quadrants. Detailed factor and stimulus dependent disentanglement of data showed that incubation with all three individual peptide mixes induced significant elaboration of IL‐2 and IL‐13 by WB of COVID‐19 convalescent patients compared with that of healthy control subjects (Figure 5). In fact, the best discrimination between COVID‐19 convalescent patients and healthy control subjects based on a single cytokine was obtained upon stimulation of their WB with the SARS‐CoV‐2 M‐peptide mix and subsequent determination of secreted levels compared to medium levels (stimulation indices, SI) of IL‐2 (AUC: 0.9418) followed by IL‐13 (AUC: 0.8129) and IFN‐γ (AUC: 0.6944; Figure S6C; Tables S10–S12). Similar, albeit somewhat weaker results were obtained when WB was stimulated with S‐ (Figure S6A) or N‐ (Figure S6B) peptide mixes. Notably, IL‐4, TNF‐α, IL‐12, and IL‐17A (except IL‐17A upon stimulation of WB with S‐peptide mix) were not discriminatory in single factor analyses. While upon stimulation of WB with all three peptide mixes, IL‐2 and IL‐13 secretion levels were highly discriminatory according to ROC analyses, with significance levels exceeding p < .001, the discriminatory power of IFN‐γ after similar stimulation was less pronounced, with corresponding p‐values in ROC analyses ranging from p = .0024 (S‐peptide mix) to only p = .0493 (N‐peptide mix; Figures S6A‐C). When comparing WB of vaccinees with that of healthy control individuals, it turned out that S‐ but not N‐ or M‐peptide stimulation highly significantly (p < .0001) discriminated between groups (Figure S7). Discrimination followed the order IL‐2 > IL‐13 > IFN‐γ with, however, only subtle differences in the respective highly significant AUC (0.9310, 0.8994, 0.8316) values (all p < .0001; Figure S7A), which was similar to the results obtained with gradient‐purified PBMC with regard to IL‐13 and IFN‐γ while IL‐2 levels were clearly lower and less discriminatory after 6 days (Figure S4A). In contrast to the discriminant analyses, TNF‐α had no weight in single factor analysis. For both SARS‐CoV‐2 antigen‐experienced study groups, convalescents and vaccinees alike, the SI results were corroborated by absolute cytokine levels (Figure S8).
FIGURE 4.
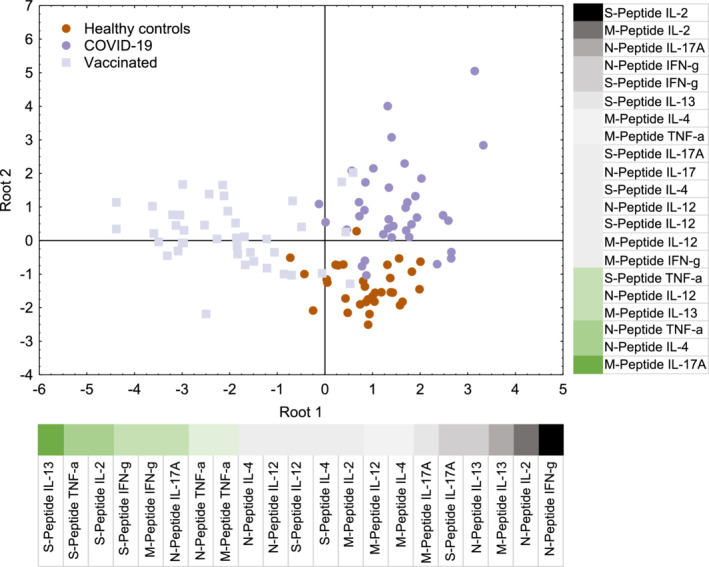
Discriminant analysis of whole blood (WB) cytokine responses after stimulation with SARS‐CoV‐2 peptide mixes and using Th1, Th2, Th17, and inflammatory cytokines as readout. Shown is the impact on discrimination of the release of the 7 cytokines upon stimulation of the WB of the three study groups with the S‐, N‐, or M‐peptide mixes. High positive weights are indicated by black and large negative ones by dark green color. Both roots are highly significant (p < .001)
FIGURE 5.
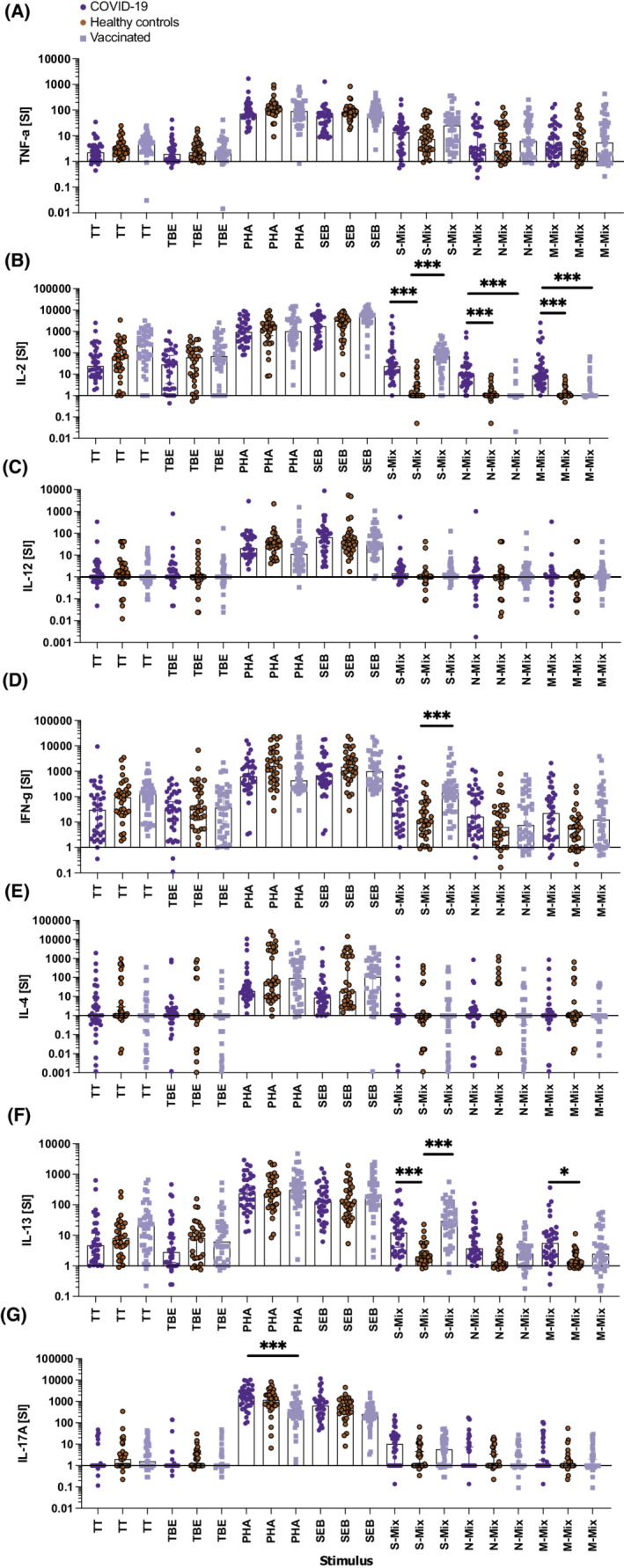
Cytokine responses of whole blood (WB) cultures clearly separate COVID‐19 convalescent patients and SARS‐CoV‐2 vaccinated subjects from non‐exposed healthy control individuals. Shown are the stimulation indices (SI, y‐axes) for (A) TNF‐α, (B) IL‐2, (C) IL‐12p70, (D) IFN‐γ, (E) IL‐4, (F) IL‐13, and (G) IL‐17A produced after incubation of whole blood samples with the indicated stimuli (x‐axes) for 44 h. Bars represent the median, whiskers the Hodges‐Lehmann 95% confidence intervals dark blue circles show COVID‐19 convalescent patients, red circles those of non‐exposed healthy controls and light blue squares vaccinees. M‐mix, SARS‐CoV‐2 matrix protein peptide mix; N‐mix, SARS‐CoV‐2 nucleocapsid protein peptide mix; PHA, phytohemagglutinin; S‐mix, SARS‐CoV‐2 spike protein peptide mix; SEB, Staphylococcal enterotoxin B; TBE, tick borne encephalitis antigen; TT, tetanus toxoid. Data show the summary of 36 COVID‐19 convalescent patients, 32 healthy controls, and 41 vaccinees. p values were calculated by Tuckey's test Only significant differences are shown. *p < .05; ***p < .001
No significant differences in IL‐2, IL‐13, and IFN‐γ cytokine secretion levels from WB cells of convalescent COVID‐19 subjects or vaccinees compared with those of healthy control subjects were detected after incubation with the vaccine‐recall antigens TT and TBE or the polyclonal stimuli PHA and SEB (Figure 5 ).
3.6. New cellular biomarkers for the discrimination of COVID‐19 convalescent patients and vaccinees from healthy control subjects
Next, we studied whether the discriminatory accuracy of the SARS‐CoV‐2 peptide‐induced IL‐2 secretion of WB could be further improved by creating biomarkers representing the products of the S‐, N‐, or M‐peptide mix‐induced SI of IL‐2 and IL‐13 or IFN‐γ, respectively. The product of the SI for IL‐2 and IL‐13 proved to be superior (Figure S9A) to the individual SI of IL‐2 or IL‐13 alone when the N‐peptide mix was used to stimulate WB of COVID‐19 convalescent patients (AUC of IL‐2 x IL13: 0.9444 (CI: 0.8944–0.9945) p < .0001 vs. AUC of IL‐2: 0.9314 (CI: 0.8714–0.9914) p < .0001 and AUC of IL‐13: 0.8008 (CI: 0.6981–0.9035) p < .0001; respectively; Figures S6C,D). A cut‐off value of 10.6 for the product of the IL‐2 SI with the IL‐13 SI resulted in a sensitivity of 83.3% and a specificity of 90.6%, resulting in a PPV of 90.9% and a NPV of 82.9% for accurately discriminating COVID‐19 convalescent patients from healthy control subjects. A comparably albeit somewhat weaker discriminative power was obtained with the SI products of IL‐2 and IL‐13 upon stimulation of WB of COVID‐19 convalescent individuals with S‐ (AUC: 0.9210 [CI: 0.853–0.989] p < .0001) or M‐ (AUC: 0.9271 [CI: 0.8575–0.9967] p < .0001) peptide mix (Figure S6D). With regard to the discrimination of vaccinated individuals from HC, a cut‐off value of 20.0 for the product of the IL‐2 SI with the IL‐13 SI upon stimulation of WB with S‐peptide mix resulted in a sensitivity of 92.7% and a specificity of 84.4%, with a corresponding PPV of 88.4% and a NPV of 90.0%. The novel cytokine‐based WB biomarker generated by combining the SI of IL‐2 and IL‐13 upon S‐peptide mix stimulation of WB from COVID‐19 convalescent and vaccinated individuals significantly correlated with the same biomarker and the proliferation results of gradient‐purified PBMC (Figure S9B,C).
3.7. Subgroup analyses by severity of disease and time period of WB analyses after disease onset
Stratification of COVID‐19 patients according to the severity of disease (hospitalization vs. no hospitalization) showed that hospitalized patients mounted somewhat stronger WB IL‐2 memory responses upon stimulation with S‐ (SI: 211.0 (CI: −474.9–24.88) vs. SI: 18.0 (CI: −46.2–283.8, p = .0027) as compared with N‐ (SI: 28.4 (CI: −122.5–398.3) vs. SI: 8.6 (CI: −6.4–67.7), p = .0339), or M‐ (SI: 65.6 (CI: −47.7–494.0) vs. SI: 8.0 (CI: −67.9–306.5) p = .0164) peptide mix when compared with non‐hospitalized individuals (Figure S10). Moreover, stratification of patients according to venipuncture 10 weeks versus 10 months after COVID‐19 revealed that WB IL‐2 recall responses for all three peptide mixes were significantly stronger when blood was collected 10 months after disease onset (S‐peptide mix SI 55.2 (CI: −184.6–1798.0) vs. SI: 15.5 (CI: −1.578–143.4) p = .0033; N‐peptide mix SI: 25.0 (CI: −41.7–314.7) vs. SI: 6.5 (CI: 4.7–314.7) p = .0006; M‐peptide mix SI: 27.0 (CI: −90.7–847.6) vs. SI: 7.3 (CI: −2.6–51.5) p < .0001 (Figure S11), and significantly correlated with the amounts of CD3+CD4+CD45RO+CCR7+ central memory T cells (R = .71; p = .0118; Figure S12). This was compatible with our recent finding that central memory T cells in COVID‐19 convalescent patients are significantly increased at the 10 months as compared with the 10 weeks time point (manuscript in preparation). Interestingly, this was not observed for IFN‐γ or IL‐13 production (Figure S11), which may indicate that different cell types contribute to the different responses.
3.8. Subgroup analyses of vaccination groups
Of note, stratification of vaccinees according to the number of vaccines received did not reveal significant differences between the groups of 1×‐, 2×‐, and 3×‐vaccinated individuals, which could be due to the limited number of analyzed subjects and/or the varying time points of analyses and, for example, the increasing intervals between the last vaccine and the number of vaccines received (Table S1). However, subdivision of the 3×‐vaccinated subjects into those who were vaccinated with 3× Comirnaty versus those vaccinated with 2× Vaxzevria followed by 1× Comirnaty versus those vaccinated with 2× Vaxzevria followed by 1× Spikevax and who, on average, had all received the last vaccination at a comparable amount of time before blood sampling (96.5 ± 20.6 vs. 107.7 ± 15.7 vs. 113.5 ± 3.5 days) indicated that the cellular reactivity of the 3× Comirnaty‐vaccinated subjects was better with respect to both S‐peptide stimulated IL‐2 and IL‐13 production (Table S1). Moreover, correlating the days after the 3rd vaccination to venipuncture with the secreted cytokine levels indicated that IL‐13 constantly increased beyond months four, which seems to be at opposite to IL‐2 and IFN‐γ secretion levels, which tended to stay constantly high (not shown).
3.9. AIM expression of CD4 + and CD8 + T lymphocytes upon incubation with SARS‐CoV‐2 peptides in WB of COVID‐19 convalescent patients, vaccinees and healthy control individuals are associated with T‐cell cytokine memory responses
Finally, we analyzed the expression of a number of activation‐induced cell surface markers (AIM) on CD3+CD4+ and CD3+CD8+ T cells in response to 2‐day SARS‐CoV‐2 peptide stimulation of WB. Figure 6 shows that the C‐type lectin CD69 expressed on CD3+CD4+ T cells upon S‐peptide mix stimulation best separated COVID‐19 convalescent patients from healthy control individuals (SI: 5.0 (CI; −2.5–60.2) vs. SI: 1.3 (CI: 0.7–4.9) p < .001). The discriminative power was high, with an AUC of 0.8720 ([CI: 0.872–0.988], p < .001), which was immediately followed by CD69 neo‐expression on CD3+CD8+ T cells (SI: 15.4 (CI: 12.6–38.7) vs. SI: 3.5 (CI: 2.9–10.8) p < .001), resulting in an AUC of 0.8251 ([CI: 0.700–0.950] p = .0002; Figure S13). Representative flow cytometry diagrams depicting S‐peptide mix induced CD69 neo‐expression on CD4+ and CD8+ T cells are shown in Figure 6A. Other AIMs, such as CD25 (Figure 6B) and CD38 (Figure 6C) also pointed toward significant differences between COVID‐19 convalescent patients and healthy control subjects, however, compared with CD69 (Figure 6D) their discriminatory power was less pronounced, while HLA‐DR, CD137, and CD154 appeared to be less powerful or even futile (Figures 6E,G). Overall similar, albeit less significant results were obtained when changes of percent values of AIM positive cells instead of SI were analyzed between groups (Figure S14), indicating the importance to take differences in the pre‐activation status of WB samples of different individuals into account. N‐ and M‐peptide mix stimulation (Figures S13B,C) revealed generally similar albeit weaker results with regard to CD69 expression, however, only on CD4+ but not on CD8+ T cells, which did, however, not reach significance upon correction for multiple comparisons.
FIGURE 6.
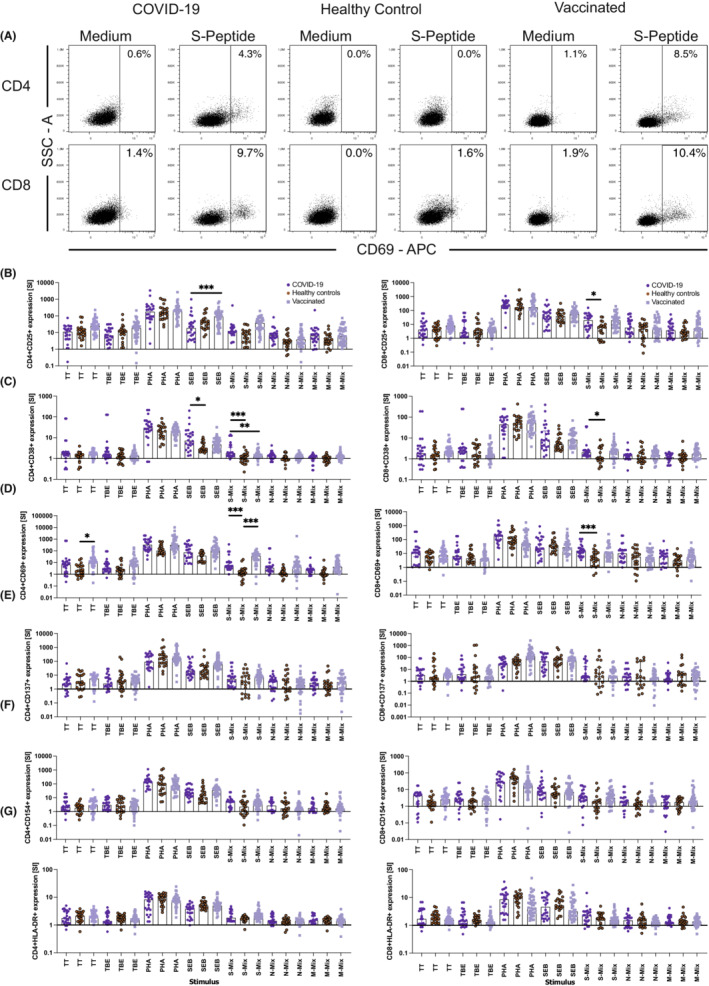
Activation‐induced marker (AIM) expression on CD4+ and CD8+ T lymphocytes help to distinguish COVID‐19 convalescent patients and SARS‐CoV‐2 vaccinated subjects from non‐exposed healthy control individuals. Shown are representative flow cytometry dot plots depicting side scatter (y‐axes) and CD69 expression (x‐axes) (A) and summary graphs (B to G) quantifying the changes (SI, y‐axes) in (B) CD25, (C) CD38, (D) CD69, (E) CD137, (F) CD154, and (G) HLA‐DR cell surface expression on CD4+ (left panels) and CD8+ (right panels) T cells of COVID‐19 convalescent individuals (dark blue circles), unexposed healthy control individuals (red circles), and vaccinated subjects (light blue squares). Data show the summary of 24 COVID‐19 convalescent patients, 23 non‐exposed healthy controls and 39 vaccinees. p values were calculated by Tuckey's test. Only significant differences are shown. *p < .05; **p < .01; ***p < .001
Similar encouraging results were obtained upon AIM determination using WB of vaccinated subjects stimulated with S‐peptide mix, however, in this case, the best discriminatory power followed the order CD4+CD69+ > CD4+CD25+ > CD8+CD25+ > CD8+CD69+ with AUC of 0.9048, 0.8669, 0.7789, and 0.7268, respectively (Figure S15) when compared with healthy control subjects. Of note, 6 instead of 44 h of stimulation did not lead to antigen‐specific CD69 and/or CD25 up‐regulation, neither upon incubation with the SARS‐CoV‐2 peptide mixes nor with the classical vaccine antigens TT and TBE, while polyclonal stimuli such as PHA were able to do so. Both findings are perfectly in line with previous reports. 43
Together, these results suggested that staining for AIM after short‐term activation with SARS‐CoV‐2 peptides in WB is practicable and reliable and almost reaches the discriminative power of secreted IL‐2 and IL‐13.
4. DISCUSSION
We describe a novel, simple and robust biomarker approach to discriminate between post‐infection and post‐vaccination T‐cellular SARS‐CoV‐2‐specific immune responses by combined assessment of S‐ and N‐specific T‐cellular IL‐2 and IL‐13 secretion and CD69 neo‐expression. The WB assay developed here monitors cytokine secretion and the expression of AIM after a 2‐day cultivation period in the presence of SARS‐CoV‐2 peptide mixes and had a superior discriminative power over classical proliferation and cytokine secretion experiments performed with gradient‐isolated PBMC requiring 8–9 days for completion. In principle, virus‐specific T‐cellular proliferation could also be achieved with recombinant proteins (e.g., receptor binding domain [RBD] of S‐protein) or virus‐like nanoparticles 40 pseudotyped with SARS‐CoV‐2 S‐, RBD‐, or N‐proteins (not shown). Notably, we found that both type 1 and intriguingly also type 2 immune responses 44 significantly contributed to the T‐cellular recall response to SARS‐CoV‐2 peptide antigens in COVID‐19 convalescent patients and subjects vaccinated with currently registered genetic vaccines based on S antigen. In fact, IL‐2 and IL‐13 cytokine secretion and the stimulation with the N‐peptide mix had the most significant discriminatory power to distinguish COVID‐19 convalescent patients from healthy control subjects, with an AUC of 0.9444 (CI: 0.8944–0.9945), p < .0001, a sensitivity of 83.3% and a specificity of 90.6% (cut‐off of 10.6 for the product of the two SIs), resulting in a PPV of 90.9% and a NPV of 82.9% for accurately discriminating COVID‐19 convalescent patients from healthy control subjects.
Similarly strong discriminatory power was found upon stimulation of WB with S‐ or M‐peptide mixes. Moreover, the combined assessment of IL‐2 and IL‐13 production in the WB test is able to discriminate with high accuracy between COVID‐19 convalescent patients, reacting with S‐, N‐, and M‐peptide mixes alike and vaccinees, who lack cytokine production upon incubation with N‐ and M‐peptide mixes (Figures S7A‐C). Such discrimination on the T‐cellular level is of interest because it informs on the nature of antigenic exposure of patients (S antigen‐based vaccination only or SARS‐CoV‐2 infection ± vaccination) and will help guide clinicians when patients present with (long‐) COVID‐19‐like or post‐vaccination symptoms. Moreover, the determination of SARS‐CoV‐2‐specific T‐cell immune responses will contribute to the monitoring of SARS‐CoV‐2 vaccination in individuals with impaired humoral immunity, for example, due to primary or secondary antibody‐production deficiencies. 45 Furthermore, our assay will contribute to the better understanding of the overall T‐cellular immune response to SARS‐CoV‐2, as differences within groups of convalescent patients upon stratification according to disease severity and time post‐infection have been observed herein. Of note, the secretion of IL‐2 in response to the S‐peptide mix was stronger in the subgroup of patients with severe COVID‐19, which may suggest that patients direct their T‐cell immune response with prolongation of disease duration increasingly toward the S‐protein, which possibly also explains the higher levels of anti‐S‐protein antibodies in this subgroup of individuals. 26 Moreover, longer‐term presence of virus‐specific peptide‐antigen(s) in the body may drive T cells more effieciently than B cells and contribute to this phenomenon. Routine application of the test with higher sample sizes will help to understand, how T‐cell immunity against SARS‐CoV‐2 is building up and persists. Since our test relies on the measurement of IL‐2, IL‐13 and IFN‐γ directly from whole blood after 44 h, it can easily be performed in outpatient clinics and hospitals without the need for specialized equipment other than those required for performing cytokine ELISAs.
Fast and accurate diagnosis of the SARS‐CoV‐2 memory status of peripheral blood lymphocytes (COVID‐19 convalescent, SARS‐CoV‐2 vaccinated, non‐exposed) represents an important parameter for the global control of the COVID‐19 pandemic since it informs, apart from the determination of specific antibody levels, about the population at risk for severe COVID‐19 infection. At the same time, it helps to identify individuals with already existing T‐cell immunity, who may have a lower risk for severe disease. 46 The T‐cell‐based test may also provide important information about the individual status of immunity of single individuals. The gold standard for the detection of acute SARS‐CoV‐2 infection is polymerase chain reaction‐based virus detection in oropharyngeal specimen sampled by different means (oropharyngeal swabs, saline gargles). 47 While the determination of acquired humoral memory by evaluation of serum for reactivity against the SARS‐CoV‐2 S‐ or N‐proteins or subdomains thereof, reliably informs about the exposure status of otherwise healthy individuals, these tests become futile, for example, in case of primary or secondary B‐cell deficiency. 22 , 23 , 45 , 48 The distinction between SARS‐CoV‐2 infection or vaccination by testing N‐ versus S‐ and RBD‐specific T‐cellular responses could help in the diagnosis of clinical symptoms that can occur both after vaccination and in the context of COVID‐19, as the symptoms may be quite similar. This testing modality would allow patients suffering from the symptoms of “long‐COVID‐19” to detect or exclude their viral exposure even after vaccination and independent of serological tests (https://www.cdc.gov/coronavirus/2019‐ncov/long‐term‐effects/index.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019‐ncov%2Flong‐term‐effects.html). 49 We know today that so‐called “vaccine breakthroughs” are quite frequent and are often asymptomatic. Still, even clinically silent COVID‐19 may eventually cause the sequelae of “long‐COVID‐19”, which gives an additional high relevance to the question whether somebody is only vaccinated or vaccinated and previously infected or only previously infected. Therefore, T‐cellular assays could prove particularly useful for patients currently undergoing immunoglobulin substitution therapy (e.g., for primary and/or secondary antibody formation disorders or as immunomodulators in the setting of autoimmune diseases), in whom the presence of SARS‐CoV‐2 IgG transferred along with the hyperimmunoglobulin preparations then prevents unequivocal serologic diagnosis of SARS‐CoV‐2 exposure. Since our test allows to detect distinct cellular immune responses even 10 months after SARS‐CoV‐2 infection, with an even stronger IL‐2 release compared with the steadily decreasing anti‐S and anti‐N antibody levels, the WB test will be useful for the long‐term follow‐up of the COVID‐19 status. Moreover, this test may also be used for various farm animals such as ferrets or also ordinary hamsters, which were in the focus of recent attention, 50 , 51 as reservoir for infections and mutations and some countries even ordered the culling of those animals (https://www.bbc.com/news/world‐europe‐54818615, https://www.nature.com/articles/d41586‐021‐00531‐z).
Currently, the distinction between vaccinated and vaccinated and exposed individuals could be very useful also in SARS‐CoV‐2 research to (i) make predictions about further measures to determine the role of vaccinated individuals in virus spread, (ii) accurately determine the efficacy of vaccination, (iii) determine, whether T‐cell immunity alone is sufficient to prevent severe forms of COVID‐19, (iv) examine how mild COVID‐19 infections affect cellular immune responses in those already vaccinated, (v) observe whether vaccine non‐responders more likely fail to respond in terms of antibody production alone or also fail to mount T‐cellular immune responses in parallel.
That the WB assay and its elaborated cytokines provides reliable information is evidenced by the high‐degree of correlation with results from experiments performed in parallel with gradient‐isolated PBMC stimulated with the same sets of SARS‐CoV‐2 peptides and reading out classical parameters such as proliferation and cytokine secretion in 6–7‐day cell culture‐based plate assays (Figure S9B,C). The observation that stimulation with M‐peptide mix leads to higher production of IL‐2 in the WB compared with the plate assay performed with gradient‐isolated PBMC may, in fact, be the result of differences in the production and consumption of cytokines at 2 versus 6 days. The highly significant M‐peptide mix‐induced proliferation is also indicative for this (Figure 1).
Whole blood stimulation with SARS‐CoV‐2‐specific peptides has also shown that apart from determination of secreted T‐cell effector cytokines, the measurement of distinct T‐cell activation molecules (CD69 and CD25) is practical and has high diagnostic power. The flow cytometry‐based determination of CD69 and CD25 puts another dimension to the whole blood test, because it allows for the discrimination between CD4+ and CD8+ T‐cell memory responses. In fact, the expression levels of the C‐type lectin CD69 reliably informed about the SARS‐CoV‐2 exposure status with an AUC of 0.8720 (CI: 0.7564–0.9877) and when taken a cut‐off SI of 2.9 with a sensitivity of 79.2% and a specificity of 81.0% with a respective PPV of 82.7% and a NPV of 77.2% for CD4+ T cells and an AUC of 0.8251 (CI: 0.7004–0.9497) and at a respective cut‐off SI of 9.3 with a sensitivity of 73.9% and a specificity of 76.2% and a resulting PPV of 77.2% and a NPV of 69.6% for CD8+ T cells. Of note, SARS‐CoV‐2‐induced neo‐expression of CD69 was equally high in samples obtained from patients 10 weeks or else 10 months after COVID‐19 infection, suggesting that CD69 neo‐expression can be used for both short‐ and long‐term monitoring of the COVID‐19‐specific T helper memory status (data not shown). Interestingly, T cells of vaccinated individuals showed a somewhat higher induction of distinct AIM also in response to superantigen SEB which might be related to their elevated basic levels of HLA‐DR‐expressing T cells (Figure S14F). This elevated expression, together with putatively elevated HLA‐DR expression on antigen presenting cells may explain the increased responsiveness to SEB. Alternatively, the increased SEB‐stimulation may be the result of a TCRβ bias possibly induced by vaccine‐induced expansion of clonotypic T cells expressing SEB reactive T‐cell receptors, e.g., Vβ3, Vβ8.1, Vβ8.2, and Vβ8.3 or others. 52
The fact that IL‐13 is so prominently produced during the SARS‐CoV‐2 specific T‐cell recall response was highly intriguing. IL‐13 is widely known as a cytokine involved in allergic immune responses, promoting (i) B‐cell immunoglobulin class switch recombination toward IgE, 53 (ii) mucus hyperproduction in the lung epithelium, 54 and (iii) bronchial smooth muscle hyperplasia, 54 the latter two phenomena in particular further aggravating allergic asthma due to sustained remodeling of the organ. 55
But what could be the role of IL‐13 in antiviral T‐cell immunity? In this regard, research on HIV infection has recently shown that apart from IFN‐γ, higher IL‐13 secretion levels as part of T‐cell adaptive responses were associated with lower virus load along with a lack of disease progression. 56 Moreover, previous reports have revealed that in addition to the Th1 response, the Th2 response, that is, among others, the release of IL‐13, which is decreased in HIV T helper cells with increasing duration of infection, can be efficiently restored by inhibiting the PD‐1 checkpoint. 57 In addition, the importance of IL‐13 for anti‐viral immune responses was also shown in hepatitis B virus (HBV) vaccination trials indicating that in study subjects the serum IL‐13 levels, apart from the serum IFN‐γ levels, significantly correlated with anti‐HBs antigen‐induced antibody responses after HBV vaccination. 58 The mechanisms, how IL‐13 may contribute to improved anti‐viral immune responses are several‐fold, by augmenting expression of integrins and HLA class II molecules on monocytes/macrophages 54 or by directly enhancing the antigen‐presenting function of CD14+ monocytic cells, as has been shown by supplementing PBMC of chronically infected HIV patients with IL‐13 which increased subsequent HIV p24‐specific T‐cell responses. 59 Furthermore, IL‐13 is a well‐known B‐cell stimulating factor, 60 inducing proliferation and upregulation of HLA Class II, and apart of IgE production, contributing to the production of neutralizing IgG4 antibodies. 53 While COVID‐19 convalescent individuals generally showed negligible levels of IgG4 antibodies against SARS‐CoV‐2 S‐ or RBD‐proteins, 26 low levels of IgG4 anti S‐ and anti RBD‐antibodies could be detected in a subset of individuals vaccinated with mRNA SARS‐CoV‐2 vaccines. 61 , 62 The slow but sustained build‐up of IgG4 levels is typical for this immunoglobulin isotype, 63 as also observed in allergen‐specific immunotherapy trials 64 in which IgG4 increased only after a series of 5–7 injections with antigen over a prolonged period of time (1–2 years). This may explain why IgG4 build‐up is rather slow, even if T cells elaborate its switch factor, IL‐13. Interestingly, IL‐4, although produced in extenso upon polyclonal stimulation, was not found to be produced in significant amounts by SARS‐CoV‐2 specific T cells. These results were also confirmed by our data, which showed that RBD‐specific IgG4 were detectable only in a fraction of triple‐vaccinated individuals but not in the serum of COVID‐19 convalescent patients. (Table 1). Moreover, the levels of SARS‐CoV‐2‐specific IgG4 in those vaccinated individuals did not correlate with S‐protein mix induced IL‐13 levels (Figure S16).
This study also has a number of potential limitations. For instance, the SARS‐CoV‐2 peptides used herein were optimized for HLA class II presentation and thus, may have preferentially visualized CD4+ T‐cell responses. This could be improved in future studies by adjusting and optimizing the collection of stimulatory peptides for HLA class I presentation and subsequent CD8+ T cell stimulation. However, highly significant co‐expression of AIM on CD8+ T cells from COVID‐19 convalescent patients (CD69, p < .001, Figure S13A), or vaccinees (CD25, p < .001, Figure S1A) shows that this does not seem to be the case. Another potential limitation of the study might be the selection of a restricted set of seven prototypic signature cytokines, while a much larger panel could have been studied. Moreover, the time‐point at which cytokines and AIMs were analyzed in the WB assay may bias results. However, in pilot experiments, we compared cytokine analysis after 1 and 2 days of culture and learned that the 2‐day culture supernatants of the WB assay gave more robust results along with comparable levels of CD69 expression, except for CD154, which was expressed better after the 1‐day incubation period. Would it not suffice to incubate WB with specific antigens/peptides for 6 h to detect specific CD69 upregulation? We also checked this possibility and analyzed antigen‐specific surface expression of CD69 after 6 h of incubation. However, as has been shown previously, 43 antigen‐specific CD69 neo‐expression was very weak to non‐existent after 6 as compared with 44 h of incubation, irrespective whether SARS‐CoV‐2 peptide mixes or bona fide vaccine antigens such as TT or TBE were applied, which was in clear contrast to the stimulation with polyclonal stimulation such as PHA.
Another potential limitation was the fact that our novel test, which reads out the combination of IL‐2 and IL13 after SARS‐CoV‐2 peptide‐specific stimulation, does not achieve 100% discriminatory power in separating COVID‐19 patients, vaccinees and healthy controls. However, compared with other commonly used diagnostic tests, we found it to be in a similar range in terms of its predictive power as, for example, the commonly used troponin 1 0/1 h test for predicting myocardial infarction, which has a PPV of 76.8%. 65
In summary, the herein developed test provides a simple and accurate solution for the differentiation between post‐vaccination and post‐infection T‐cellular SARS‐CoV‐2‐specific immune responses from those of naïve (non‐exposed), healthy control individuals. Such differentiation will complement results obtained by evaluation of humoral responses and thus help to correctly assign the different forms of antigen contact in different populations. Determination of the presence and degree of T‐cellular memory in patients with impaired or masked humoral immunity, for example, due to antibody substitution therapy, will help identifying patients at risk of becoming reservoirs for the development of variants of concern (VOC) due to their inability to efficiently clear virus, as has been shown previously. 46 The here developed test is practicable, needs little hands‐on‐time, can be performed within 2 days with only two cytokines required for analysis. This makes the test also attractive to physicians in outpatient clinics while promoting a better understanding of the development of T cell‐mediated SARS‐CoV‐2 immunity.
AUTHOR CONTRIBUTIONS
B. K., R. V., and W. F. P. designed research. B. K., L. C. S., P. W‐S, D. T., P. G., P. A. T., A. N. A. S., U. K., A. R., M. F., T. O., K. G‐P., K. B., Y. D., I. T., M. W., B. M., R. V., W. F. P., A. K., B. N, and M.K. performed research and analyzed data. B. K., R. V., and W. F. P. wrote the paper. All authors critically read the paper and approved the manuscript.
CONFLICT OF INTEREST
With regard to the authors disclosure of potential conflicts of interest, we would like to indicate that Winfried F. Pickl has received honoraria from Novartis, Astra Zeneca, and Roche. Rudolf Valenta has received research grants from HVD Life‐Sciences, Vienna, Austria, WORG Pharmaceuticals, Hangzhou, China, and from Viravaxx, Vienna, Austria. He serves as consultant for Viravaxx and WORG Pharmaceuticals. The other authors have no conflict of interest to declare.
Supporting information
Appendix S1
Figures S1‐S16
ACKNOWLEDGMENTS
We thank Doris Werjant‐Locmele and Anna Guentcheva for their help regarding recruitment and administration of study subjects. We thank Dr. Ferdinand Fast and Dr. Armin Rieger for expert assistance in blood sampling. We are indebted to all individuals who participated in our study.
Kratzer B, Schlax LC, Gattinger P, et al. Combined assessment of S‐ and N‐specific IL‐2 and IL‐13 secretion and CD69 neo‐expression for discrimination of post–infection and post‐vaccination cellular SARS‐CoV‐2‐specific immune response. Allergy. 2022;00:1‐18. doi: 10.1111/all.15406
Funding information
This study was supported by grants from the Austrian Science Fund, grant number: DKW1248; the Medizinisch‐Wissenschaftlicher Fonds des Bürgermeisters der Bundeshauptstadt Wien (Stiftungsfonds zur Förderung der Bekämpfung der Tuberkulose und anderer Lungenkrankheiten), grant numbers: COVID001 and COVID006, funded in part by a research grant from Viravaxx AG, Vienna, Austria to RV and a grant from the Federal State of Lower Austria, Grant: Danube Allergy Research Cluster (Danube ARC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Rudolf Valenta, Email: rudolf.valenta@meduniwien.ac.at.
Winfried F. Pickl, Email: winfried.pickl@meduniwien.ac.at.
REFERENCES
- 1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang J‐J, Dong X, Cao Y‐Y, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020;75(7):1730‐1741. [DOI] [PubMed] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park M, Thwaites RS, Openshaw PJM. COVID‐19: lessons from SARS and MERS. Eur J Immunol. 2020;50(3):308‐311. [Google Scholar]
- 5. World Health Organization (WHO) . Coronavirus disease (COVID‐19) outbreak; 2020. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019
- 6. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20(5):533‐534. doi: 10.1016/S1473-3099(20)30120-1. Epub 2020 Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gattinger P, Borochova K, Dorofeeva Y, et al. Antibodies in serum of convalescent patients following mild COVID‐19 do not always prevent virus‐receptor binding. Allergy. 2021;76(3):878‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Agerer B, Koblischke M, Gudipati V, et al. SARS‐CoV‐2 mutations in MHC‐I‐restricted epitopes evade CD8(+) T cell responses. Sci Immunol. 2021;6(57):eabg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kratzer B, Trapin D, Ettel P, et al. Immunological imprint of COVID‐19 on human peripheral blood leukocyte populations. Allergy. 2021;76(3):751‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nielsen SS, Vibholm LK, Monrad I, et al. SARS‐CoV‐2 elicits robust adaptive immune responses regardless of disease severity. EBioMedicine. 2021;68:103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383(27):2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrett JR, Belij‐Rammerstorfer S, Dold C, et al. Phase 1/2 trial of SARS‐CoV‐2 vaccine ChAdOx1 nCoV‐19 with a booster dose induces multifunctional antibody responses. Nat Med. 2021;27(2):279‐288. [DOI] [PubMed] [Google Scholar]
- 15. Sahin U, Muik A, Derhovanessian E, et al. COVID‐19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594‐599. [DOI] [PubMed] [Google Scholar]
- 16. Widge AT, Rouphael NG, Jackson LA, et al. Durability of responses after SARS‐CoV‐2 mRNA‐1273 vaccination. N Engl J Med. 2021;384(1):80‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalimuddin S, Tham CY, Qui M, et al. Early T cell and binding antibody responses are associated with Covid‐19 RNA vaccine efficacy onset. Med (N Y). 2021;2(6):682‐688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bird S, Panopoulou A, Shea RL, et al. Response to first vaccination against SARS‐CoV‐2 in patients with multiple myeloma. Lancet Haematol. 2021;8:e389‐e392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monin L, Laing AG, Muñoz‐Ruiz M, et al. Safety and immunogenicity of one versus two doses of the COVID‐19 vaccine BNT162b2 for patients with cancer: interim analysis of a prospective observational study. Lancet Oncol. 2021;22(6):765‐778. doi: 10.1016/S1470-2045(21)00213-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kratzer B, Trapin D, Gattinger P, et al. Lack of induction of RBD‐specific neutralizing antibodies despite repeated heterologous SARS‐CoV‐2 vaccination leading to seroconversion and establishment of T cell‐specific memory in a patient in remission of multiple myeloma. Vaccines (Basel). 2022;10(3):374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta S, Su H, Narsai T, Agrawal S. SARS‐CoV‐2‐associated T‐cell responses in the presence of humoral immunodeficiency. Int Arch Allergy Immunol. 2021;182(3):195‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Achiron A, Dolev M, Menascu S, et al. COVID‐19 vaccination in patients with multiple sclerosis: what we have learnt by February 2021. Mult Scler. 2021;27(6):864‐870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonelli MM, Mrak D, Perkmann T, Haslacher H, Aletaha D. SARS‐CoV‐2 vaccination in rituximab‐treated patients: evidence for impaired humoral but inducible cellular immune response. Ann Rheum Dis. 2021;80:1355‐1356. [DOI] [PubMed] [Google Scholar]
- 24. Boyarsky BJ, Werbel WA, Avery RK, et al. Immunogenicity of a single dose of SARS‐CoV‐2 messenger RNA vaccine in solid organ transplant recipients. JAMA. 2021;325(17):1784‐1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thakkar A, Pradhan K, Jindal S, et al. Patterns of seroconversion for SARS‐CoV‐2 IgG in patients with malignant disease and association with anticancer therapy. Nat Cancer. 2021;2:392‐399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gattinger P, Niespodziana K, Stiasny K, et al. Neutralization of SARS‐CoV‐2 requires antibodies against conformational receptor‐binding domain epitopes. Allergy. 2021;77:230‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Canas LS, Österdahl MF, Deng J, et al. Disentangling post‐vaccination symptoms from early COVID‐19. EClinicalMedicine. 2021;42:101212. doi: 10.1016/j.eclinm.2021.101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaebler C, Wang Z, Lorenzi JCC, et al. Evolution of antibody immunity to SARS‐CoV‐2. Nature. 2021;591(7851):639‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419‐466. [DOI] [PubMed] [Google Scholar]
- 30. Wang J‐H, Reinherz EL. Structural basis of T cell recognition of peptides bound to MHC molecules. Mol Immunol. 2002;38(14):1039‐1049. [DOI] [PubMed] [Google Scholar]
- 31. Dustin ML. T‐cell activation through immunological synapses and kinapses. Immunol Rev. 2008;221:77‐89. [DOI] [PubMed] [Google Scholar]
- 32. Montoya MC, Sancho D, Vicente‐Manzanares M, Sanchez‐Madrid F. Cell adhesion and polarity during immune interactions. Immunol Rev. 2002;186:68‐82. [DOI] [PubMed] [Google Scholar]
- 33. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity. 1998;8(1):89‐95. [DOI] [PubMed] [Google Scholar]
- 35. Bach FH, Segall M, Zier KS, Sondel PM, Alter BJ, Bach ML. Cell mediated immunity: separation of cells involved in recognitive and destructive phases. Science. 1973;180(4084):403‐406. [DOI] [PubMed] [Google Scholar]
- 36. Doroszczak N, Yoshida T, Cohen S. Subpopulations of lymphocytes to produce various lymphokines. I. Function of subpopulations separated by velocity sedimentation. J Immunol. 1977;119(5):1617‐1620. [PubMed] [Google Scholar]
- 37. Cotner T, Williams JM, Christenson L, Shapiro HM, Strom TB, Strominger J. Simultaneous flow cytometric analysis of human T cell activation antigen expression and DNA content. J Exp Med. 1983;157(2):461‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riber U, Boesen HT, Jakobsen JT, Nguyen LTM, Jungersen G. Co‐incubation with IL‐18 potentiates antigen‐specific IFN‐gamma response in a whole‐blood stimulation assay for measurement of cell‐mediated immune responses in pigs experimentally infected with Lawsonia intracellularis. Vet Immunol Immunopathol. 2011;139(2–4):257‐263. [DOI] [PubMed] [Google Scholar]
- 39. Chang JT, Segal BM, Nakanishi K, Okamura H, Shevach EM. The costimulatory effect of IL‐18 on the induction of antigen‐specific IFN‐gamma production by resting T cells is IL‐12 dependent and is mediated by up‐regulation of the IL‐12 receptor beta2 subunit. Eur J Immunol. 2000;30(4):1113‐1119. [DOI] [PubMed] [Google Scholar]
- 40. Derdak SV, Kueng HJ, Leb VM, et al. Direct stimulation of T lymphocytes by immunosomes: virus‐like particles decorated with T cell receptor/CD3 ligands plus costimulatory molecules. Proc Natl Acad Sci USA. 2006;103(35):13144‐13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andersen P, Munk ME, Pollock JM, Doherty TM. Specific immune‐based diagnosis of tuberculosis. Lancet. 2000;356(9235):1099‐1104. [DOI] [PubMed] [Google Scholar]
- 42. Mazurek GH, Villarino ME, CDC . Guidelines for using the QuantiFERON‐TB test for diagnosing latent mycobacterium tuberculosis infection. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2003;52(RR‐2):15‐18. [PubMed] [Google Scholar]
- 43. Caruso A, Licenziati S, Corulli M, et al. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry. 1997;27(1):71‐76. [DOI] [PubMed] [Google Scholar]
- 44. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136(7):2348‐2357. [PubMed] [Google Scholar]
- 45. Apostolidis SA, Kakara M, Painter MM, et al. Cellular and humoral immune responses following SARS‐CoV‐2 mRNA vaccination in patients with multiple sclerosis on anti‐CD20 therapy. Nat Med. 2021;27:1990‐2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weigang S, Fuchs J, Zimmer G, et al. Within‐host evolution of SARS‐CoV‐2 in an immunosuppressed COVID‐19 patient as a source of immune escape variants. Nat Commun. 2021;12(1):6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. LeBlanc JJ, Pettipas J, Di Quinzio M, Hatchette TF, Patriquin G. Reliable detection of SARS‐CoV‐2 with patient‐collected swabs and saline gargles: a three‐headed comparison on multiple molecular platforms. J Virol Methods. 2021;295:114184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mira E, Yarce OA, Ortega C, et al. Rapid recovery of a SARS‐CoV‐2‐infected X‐linked agammaglobulinemia patient after infusion of COVID‐19 convalescent plasma. J Allergy Clin Immunol Pract. 2020;8(8):2793‐2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rubin R. As their numbers grow, COVID‐19 "long haulers" stump experts. JAMA. 2020;324(14):1381‐1383. [DOI] [PubMed] [Google Scholar]
- 50. Au GG, Marsh GA, McAuley AJ, et al. Characterisation and natural progression of SARS‐CoV‐2 infection in ferrets. Sci Rep. 2022;12(1):5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thakur N, Gallo G, Newman J, et al. SARS‐CoV‐2 variants of concern alpha, beta, gamma and delta have extended ACE2 receptor host ranges. J Gen Virol. 2022;103(4): doi: 10.1099/jgv.0.001735 [DOI] [PubMed] [Google Scholar]
- 52. White J, Herman A, Pullen AM, Kubo R, Kappler JW, Marrack P. The V beta‐specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell. 1989;56(1):27‐35. [DOI] [PubMed] [Google Scholar]
- 53. Punnonen J, Aversa G, Cocks BG, et al. Interleukin 13 induces interleukin 4‐independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA. 1993;90(8):3730‐3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hershey GK. IL‐13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol. 2003;111(4):677‐690. quiz 691. [DOI] [PubMed] [Google Scholar]
- 55. Zhu Z, Homer RJ, Wang Z, et al. Pulmonary expression of interleukin‐13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103(6):779‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bailer RT, Holloway A, Sun J, et al. IL‐13 and IFN‐gamma secretion by activated T cells in HIV‐1 infection associated with viral suppression and a lack of disease progression. J Immunol. 1999;162(12):7534‐7542. [PubMed] [Google Scholar]
- 57. Porichis F, Hart MG, Zupkosky J, et al. Differential impact of PD‐1 and/or interleukin‐10 blockade on HIV‐1‐specific CD4 T cell and antigen‐presenting cell functions. J Virol. 2014;88(5):2508‐2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sabry R, Mohamed ZAZ, Abdallah AM. Relationship between Th1 and Th2 cytokine serum levels and immune response to hepatitis B vaccination among Egyptian health care workers. J Immunoassay Immunochem. 2018;39(5):496‐508. [DOI] [PubMed] [Google Scholar]
- 59. Papasavvas E, Sun J, Luo Q, et al. IL‐13 acutely augments HIV‐specific and recall responses from HIV‐1‐infected subjects in vitro by modulating monocytes. J Immunol. 2005;175(8):5532‐5540. [DOI] [PubMed] [Google Scholar]
- 60. Defrance T, Carayon P, Billian G, et al. Interleukin 13 is a B cell stimulating factor. J Exp Med. 1994;179(1):135‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klingler J, Lambert GS, Itri V, et al. Detection of antibody responses against SARS‐CoV‐2 in plasma and saliva from vaccinated and infected individuals. Front Immunol. 2021;20(12):759688. doi: 10.3389/fimmu.2021.759688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gattinger P, Kratzer B, Tulaeva I, et al. Vaccine based on folded RBD‐PreS fusion protein with potential to induce sterilizing immunity to SARS‐CoV‐2 variants. Allergy. 2022. doi: 10.1111/all.15305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aalberse RC, Stapel SO, Schuurman J, Rispens T. Immunoglobulin G4: an odd antibody. Clin Exp Allergy. 2009;39(4):469‐477. [DOI] [PubMed] [Google Scholar]
- 64. Eckl‐Dorna J, Weber M, Stanek V, et al. Two years of treatment with the recombinant grass pollen allergy vaccine BM32 induces a continuously increasing allergen‐specific IgG4 response. EBioMedicine. 2019;50:421‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boeddinghaus J, Nestelberger T, Koechlin L, et al. Early diagnosis of myocardial infarction with point‐of‐care high‐sensitivity cardiac troponin I. J Am Coll Cardiol. 2020;75(10):1111‐1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Figures S1‐S16


