Abstract
The constantly emerging severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2) variants of concerns (VOCs) with mutations in the receptor‐binding domain (RBD) spread rapidly and has become a severe public health problem worldwide. Effective vaccines and optimized booster vaccination strategies are thus highly required. Here, the gene encoding six different RBD (Alpha, Beta, Gamma, Kappa, Delta, and Epsilon variants) along with the Fc fragment of human IgG1 (RBD‐Fc) was cloned into plant expression vector and produced in Nicotiana benthamiana by transient expression. Further, the immunogenicity of plant‐produced variant RBD‐Fc fusion proteins were tested in cynomolgus monkeys. Each group of cynomolgus monkeys was immunized three times intramuscularly with variant RBD‐Fc vaccines at Day 0, 21, 42, and neutralizing antibody responses were evaluated against ancestral (Wuhan), Alpha, Beta, Gamma, and Delta variants. The results showed that three doses of the RBD‐Fc vaccine significantly enhanced the immune response against all tested SARS‐CoV‐2 variants. In particular, the vaccines based on Delta and Epsilon mutant RBD elicit broadly neutralizing antibodies against ancestral (Wuhan), Alpha, and Delta SARS‐CoV‐2 variants whereas Beta and Gamma RBD‐Fc vaccines elicit neutralizing antibodies against their respective SARS‐CoV‐2 strains. The Delta and Epsilon RBD‐Fc based vaccines displayed cross‐reactive immunogenicity and might be applied as a booster vaccine to induce broadly neutralizing antibodies. These proof‐of‐concept results will be helpful for the development of plant‐derived RBD‐Fc‐based vaccines against SARS‐CoV‐2 and its variants.
Keywords: COVID‐19, immunogenicity, plant‐produced recombinant protein, receptor‐binding domain, SARS‐CoV‐2, subunit vaccine, variant vaccine
Abbreviations
- A450
absorbance at 450 nm
- Alum
aluminum hydroxide
- Al content
aluminum content
- BSL
biosafety level
- CI
confidence interval
- COVID‐19
coronavirus disease 2019
- DMEM
Dulbecco's Modified Eagle Medium
- Fc region
fragment crystallizable region
- GMT
geometric mean titer
- hACE2
human angiotensin‐converting enzyme 2
- HRP
horseradish peroxidase
- IgG
immunoglobulin G
- LOD
limit of detection
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- MN50 titer
50% microneutralizing titer
- N
nucleocapsid glycoprotein
- NHP
nonhuman primate
- NPRCT‐CU
National Primate Research Center of Thailand‐Chulalongkorn University
- PBS
phosphate buffered saline
- PBST
1×phosphate buffered saline with 0.05% Tween 20
- PS
Positive serum
- PVNT50
50% pseudovirus neutralizing titer
- RBD
receptor‐binding domain
- RT
room temperature
- S
spike or surface glycoprotein
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TCID50
50% tissue culture infective dose
- TMB
tetramethylbenzidine
- VOC
variant of concern
- WHO
World Health Organization
1. INTRODUCTION
The world is currently dealing with pandemic coronavirus disease 2019 (COVID‐19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 1 , 2 As of May 20, 2022, more than 521 million confirmed cases, and 6.2 million deaths were reported by World Health Organization (WHO). 3 As soon as the virus outbreak, several research groups, and biopharmaceutical companies attempt to develop effective and promising SARS‐CoV‐2 vaccines utilizing different technologies. 4 Subunit vaccines based on synthetic peptides or recombinant proteins have been established for SARS‐CoV and Middle East respiratory syndrome coronavirus (MERS‐CoV), which are shown to be effective in animal models. 5 Earlier reports showed that the receptor‐binding domain (RBD) located on the Spike (S) protein of SARS‐CoV‐2 is considered the main target for neutralizing antibodies and therapeutic vaccine development. 6 The subunit‐based vaccines developed against SARS‐CoV‐2 such as NVX‐CoV2373 (Novavax), ZF2001 (Anhui Zhifei Longcom), and KBP‐COVID‐19 (Kentucky Bioprocessing Inc.) showed encouraging clinical trial results. 7 , 8 , 9
Recently, few COVID‐19 vaccines have been approved by WHO including those produced by Pfizer/BioNTech, Oxford/AstraZeneca‐SK Bio, Janssen, Moderna, Sinopharm/BIBP, Sinovac, and Bharat Biotech. 10 Several countries introduced massive vaccination campaigns to combat the virus infection. However, the virus evolves into new variants that evade host immunity with increasing infectivity, virulence, and replication fitness. 11 , 12 Five variants of concerns (VOCs) were categorized by WHO including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529), as of April, 2022. 13 These VOCs contain multiple mutations on the spike and RBD region that plays a vital role in viral entry by interaction with the host cell receptor Angiotensin‐Converting Enzyme 2 (ACE2). Some of the key mutations identified in variants are reported to reduce the efficacy of vaccines. 14 , 15 BNT162b2 (Pfizer/BioNTech) elicited antibodies showed a 2.6‐fold and 8.8‐fold reduction in neutralizing Alpha and Beta variants compared with the ancestral (Wuhan) variant. 16 However, 3‐fold and 16‐fold decrease in the neutralization titers against the Delta and the Beta variants compared to the Alpha variant were reported. Furthermore, 5‐fold and 9‐fold reduction in neutralization titers against the Delta and the Beta variants relative to the Alpha variant were reported for the ChAdOx1_nCoV‐19 vaccine (Oxford/AstraZeneca). Both Pfizer and AstraZeneca vaccines showed notable neutralizing antibody responses against the Delta variant after the second dose. 17 In addition, BNT162b2 recipient sera showed neutralizing antibody response against the Omicron 40‐fold lower than the ancestral (Wuhan) variant and none of the CoronaVac (Sinovac) recipients sera showed neutralizing response against the Omicron variant. 18 Therefore, urgent research is required to fill the knowledge gaps on SARS‐CoV‐2 and its variants for effective vaccine design and treatment to combat its infection. In addition, the efficacy of available vaccines and other potential candidates against VOCs should be investigated.
The cost‐effective platform for producing recombinant vaccines could reduce the overall vaccine cost, which, in turn, reduce the financial burden and improve vaccine accessibility. Recently, the plant‐based platform has been gaining popularity for the production of recombinant proteins, enzymes, vaccine antigens, antimicrobial peptides, diagnostic/research reagents, and monoclonal antibodies. 19 , 20 , 21 , 22 , 23 Rapid scale‐up of recombinant proteins, low risk of human pathogen contamination, ability to perform posttranslational modifications, and low cost are some of the major advantages of plant expression system. 24 , 25 , 26 , 27 , 28 Previously, our group has successfully demonstrated the production of SARS‐CoV‐2 antigens and monoclonal antibodies in Nicotiana benthamiana in response to emergency demands. 29 , 30 We have also shown the potential of plant‐produced recombinant SARS‐CoV‐2 RBD‐ Fc fusion protein adjuvanted with alum in inducing the neutralizing antibodies in both mice and cynomolgus monkeys. 31 In addition, we have demonstrated the role of adjuvants in enhancing the immune response of plant‐produced subunit vaccine candidates against SARS‐CoV‐2. 32 In this study, SARS‐CoV‐2 variant RBD proteins, Alpha (N501Y, A570D, and D614G), Beta (K417N, E484K, N501Y, and D614G), Gamma (K417T, E484K, N501Y, and D614G), Kappa (L452R, E484Q, and D614G), Delta (L452R, T478K, and D614G), and Epsilon (L452R and D614G) were fused with Fc region of human IgG1 for subunit vaccine development. Fc‐fused protein vaccines against several diseases have been evaluated including SARS‐CoV and influenza. 33 , 34 Here, six recombinant variant RBD proteins were fused with the Fc region of human IgG1 and produced in N. benthamiana by transient expression. The yield of purified plant‐produced variant RBD‐Fc proteins were found to be in the range of 20–28 µg/g fresh weight. Further, the in vivo immunogenicity of the plant‐produced variant RBD‐Fc proteins were evaluated in cynomolgus macaques, and the ability to neutralize antibodies elicited by these vaccines against ancestral (Wuhan) and mutant viruses (variants) were also investigated.
2. MATERIALS AND METHODS
2.1. Preparation of recombinant RBD proteins
The ancestral RBD sequence (Wuhan) of SARS‐CoV‐2 RBD (F318‐C617) containing 3XGGGGS linker fused with Fc region of human IgG1 (P35‐K255) in the recombinant plasmid pBYR2eK2Md (Figure 1A) 31 was used as a template to generate a series of RBD point mutation (Alpha, Beta, Gamma, Kappa, Delta, and Epsilon RBD‐Fc) (Figures 1B and S1). The construct of Alpha (N501Y, A570D, and D614G) and Beta RBD‐Fc (K417N, E484K, N501Y, and D614G) were produced as previously described. 35 Gamma (K417T, E484K, N501Y, and D614G), Kappa (L452R, E484Q, and D614G), Delta (L452R, T478K, and D614G), and Epsilon (L452R and D614G) RBD were constructed using the set of primers by polymerase chain reaction (PCR) (Table S1). Briefly, Gamma RBD was constructed by using Beta RBD‐Fc as a template with SP‐F/K417T‐R primers and K417T‐F/D614G‐R primers to introduce K417T mutation. The ancestral (Wuhan) RBD‐Fc construct was used as the template for generating Epsilon RBD by using SP‐F/L452R‐R and L452R‐F/D614G‐R primers to introduce L452R and D614G mutations. Kappa and Delta RBD were developed by using Epsilon RBD as the template with SP‐F/E484Q‐R and E484Q‐F/D614G‐R primers for Kappa (E484Q), and SP‐F/T478K‐R and T478K‐F/D614G‐R primers for Delta (T478K). Then, all variant RBD were ligated with linker and Fc region via., BamHI site by T4 DNA ligase (New England Biolabs). Then, each variant RBD‐Fc was ligated into geminiviral vector pBYR2eK2Md (pBY2eK) 36 via., XbaI, and SacI sites. The recombinant plasmids were transformed to Agrobacterium tumefaciens GV3101 by electroporation. Wild‐type N. benthamiana plants were agroinfiltrated with Agrobacterium harboring each variant RBD‐Fc constructs. Infiltrated plants were harvested 3 days post infiltration and the recombinant protein was purified by protein A affinity chromatography (GE Healthcare) as previously described. 35
Figure 1.
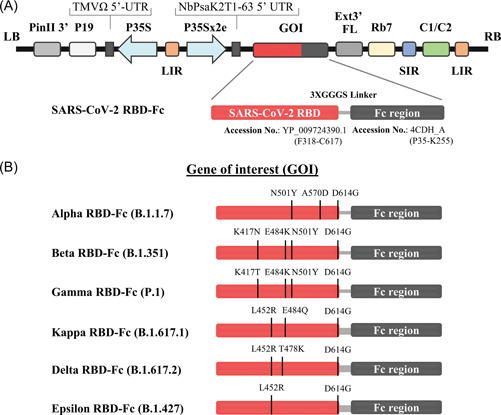
Schematic representation of geminiviral vector (pBY2eK) map of SARS‐CoV‐2 variant RBD‐Fc fusion proteins produced in N. benthamiana. T‐DNA region between LB (left border) and RB (right border) of pBY2eK consists of PinII 3′ (terminator from potato proteinase inhibitor II gene), P19 (P19 gene from tomato bushy stunt virus (TBSV)), TMVΩ 5′‐UTR (5′ untranslated region of tobacco mosaic virus Ω), P35S (cauliflower mosaic virus (CaMV) 35S promoter), LIR (long intergenic region of BeYDV genome), NbPsaK2T 5′UTR (5′ untranslated region), GOI (gene of interest), Ext3′FL (3′ full length of the tobacco (Nicotiana tabacum) extension gene), Rb7 (tobacco RB7 promoter), SIR (short intergenic region of BeYDV genome), and C2/C1 (bean yellow dwarf virus (BeYDV) open reading frames C1 and C2 encoding for replication initiation protein (Rep) and RepA) (A). Diagrammatic representation of the SARS‐CoV‐2 variant RBD‐Fc fusion proteins. The RBD protein with the predicted mutation sites in the SARS‐CoV‐2 variants compared to the RBD of original strain Wuhan was highlighted (B). RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
2.2. Vaccine formulation
The purified plant‐produced SARS‐CoV‐2 variant RBD‐Fc fusion proteins at a dose of 10 μg protein were formulated with 0.5 mg alum, Alhydrogel® 2% (Croda), as an adjuvant. The phosphate‐buffered saline (PBS) containing alum without the antigen was used as a control. A total volume of 0.5 ml of the vaccine was used for immunization.
2.3. Immunization in cynomolgus monkeys
The study was performed at the National Primate Research Center of Thailand‐Chulalongkorn University (NPRCT‐CU; AAALAC International Accredited facility). The animal use and the experimental procedures were approved by the NPRCT‐CU Animal Care and Use Committee (Protocol review no. 2175005 and 2175007).
Thirty‐five male and female cynomolgus monkeys (Macaca fascicularis) aged 2.5–9 years and body weight between 2.5 and 6.4 kg, supplied by the NPRCT‐CU breeding facility were used for the study. Animals were randomly divided into seven groups; plant‐produced SARS‐CoV‐2 Alpha, Beta, Gamma, Kappa, Delta, Epsilon RBD‐Fc vaccines (n = 5) and control group (n = 3). Monkeys were intramuscularly injected in the quadriceps femoris muscle with 0.5 mL of vaccines or alum alone on Days 0, 21, and 42 (3‐week interval). The blood samples were collected on Day 0 (before the first injection) and 14 days after each immunization on Day 14, 35, and 56, to assess the antigen‐specific antibody titer, live virus‐neutralizing antibody, and pseudovirus neutralization antibody titers. The immunization schedule was shown in Figure 3A.
Figure 3.
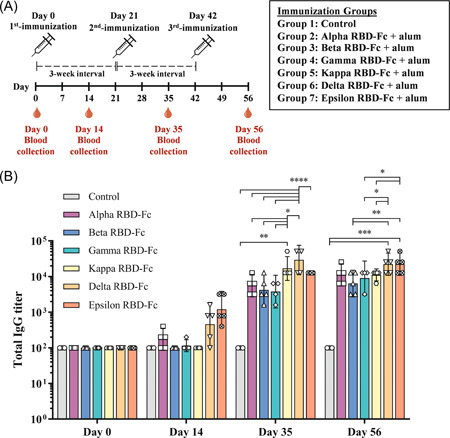
Immunization and blood collection schedule in cynomolgus monkeys (A). Monkeys were divided into seven groups, ie., plant‐produced SARS‐CoV‐2 Alpha, Beta, Gamma, Kappa, Delta, Epsilon RBD‐Fc vaccine group (n = 5), and control group (n = 3). Monkeys were immunized with a 3‐dose regimen (Day 0, 21, and 42) and were bled on Days 14, 35, and 56. Response of SARS‐CoV‐2 RBD‐specific IgG titers in immunized animals was presented (B). Data presented as GMT ± 95% CI of the endpoint titer in each group, n = 5 (control, n = 3). Two‐way ANOVA, Tukey test was used. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). ANOVA, analysis of variance; CI, confidence interval; GMT, geometric mean titer; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
2.4. Evaluation of RBD‐specific antibody titer by enzyme‐linked immunosorbent assay (ELISA)
SARS‐CoV‐2 spike protein RBD (Cat. No. Z03479; GenScript) at 100 ng/well was used to coat a high binding 96‐well plate (Greiner bio‐one) and incubated overnight at 4°C. Next, the wells were blocked with 3% w/v bovine serum albumin (BSA; HiMedia Laboratories) in 1×PBS pH 7.4 for 1 h at 37°C. Then, monkey sera were diluted with 1% w/v BSA in 1×PBS with 2‐fold serial dilution by starting at 1:100. The diluted sera were loaded in each well as duplicates and incubated for 1 h at room temperature (RT). After washing, goat anti‐monkey IgG HRP conjugate (Abcam) at the dilution of 1:20,000 in 1×PBS was added and incubated for 1 h at RT. Subsequently, TMB substrate (Promega) was added to the plates as substrate, and then the reactions were stopped by adding 1 M H2SO4. The absorbance at 450 nm (A450) was measured by SpectraMax® M3 Microplate Reader (Molecular Devices). The plates were washed with 1×PBS with 0.05% Tween 20 (PBST) three times between each step. The endpoint titers were determined as the highest dilution of immunized sera, which had A450 more than the cut‐off value calculated from A450 of pre‐immunized sera at 1:100 dilution. 37
2.5. Microneutralization assay
Microneutralization assay was performed in 96‐well microplates containing confluent Vero E6 cells and live SARS‐CoV‐2 virus, ancestral (SARS‐CoV‐2/human/THA/LJ07_P3/2020), Alpha (B.1.1.7, SARS‐CoV‐2/human/THA/NH657_P3/2021), Beta (B.1.351, SARS‐CoV‐2/human/THA/NH088_P3/2021), and Delta (B.1.617.2, SARS‐CoV2/human/THA/OTV007_P3/2021), isolated from COVID‐19 patients in Thailand. The experiment was conducted in a certified biosafety level (BSL) 3 facility of Microbiology Department, Faculty of Science, Mahidol University, Thailand, as previously described 31 , 38 with some modifications.
Briefly, immunized monkey sera and the convalescent serum from COVID‐19 patients (positive control) were heat‐inactivated at 56°C for 30 min. Two‐fold serially diluted sera were mixed with 100 of 50% tissue culture infective dose (TCID50) of SARS‐CoV‐2 and its variants in Dulbecco's Modified Eagle Medium (DMEM) at 37°C for 1 h. Virus control and cell control wells were included in all plates. Then, the mixture was applied to a Vero E6 cell monolayer and incubated at 37°C for 2 days. Subsequently, the cells were washed once with 1×PBS, fixed, and permeabilized with chilled 1:1 methanol/acetone fixative solution at 4°C for 20 min. After washing three times with 1×PBST and the plates were blocked with 2% BSA in 1×PBS containing 0.1% Tween 20 at room temperature (RT) for 1 h. Viral infection was then assessed using 1:5,000 of SARS‐CoV/SARS‐CoV‐2 nucleocapsid (N) monoclonal antibody (Sino Biological) in 1×PBS containing 0.5% BSA and 0.1% Tween 20 as a primary antibody and incubated at 37°C for 1 h followed by adding 1:2000 of HRP‐conjugated goat anti‐rabbit polyclonal antibodies (Dako) in 1×PBS as a secondary antibody and incubated at 37°C for 1 h. The KPL Sureblue™ TMB substrate (SeraCare) was added, and the reaction was stopped by 1 N HCl. The absorbance was measured using a Sunrise™ microplate reader (Tecan). The differences of A450 of samples were compared with the 50% of the cut point, which was calculated as previously described. 38 The assay was performed in duplicates.
2.6. Pseudovirus neutralization assay
Lentiviral pseudoviruses bearing CoV spike were constructed as previously described 39 with minor modifications. Briefly, the combination of plasmids including the lentivirus backbone expressing a firefly luciferase reporter gene (pCSFLW, kindly provided by Dr. Nigel James Temperton, University of Kent), the expression plasmid expressing HIV‐1 structural/regulatory proteins (pCMV∆R8.91), and pCAGGS expressing the codon‐optimized spike gene (ancestral (Wuhan), Alpha, Beta, Gamma, and Delta) was used to generate pseudoviruses. Unless otherwise indicated, HEK293T/17 producer cells were seeded in 6‐well plates at 7.5 × 105/well 24 h before being transfected with the following plasmids: 600 ng pCMV∆R8.91, 600 ng pCSFLW, and 500 ng of pCAGGS‐Spike, in Opti‐MEM (Gibco) with 10 μl polyethyleneimine (PEI). Transfected cells were incubated at 37°C, 5% CO2. At 12 h after transfection, cells were washed and cultured in DMEM‐10%. Pooled harvests of supernatants containing pseudoviruses were taken at 72 h posttransfection, centrifuged at 1500×g for 10 min at 4°C to remove cellular debris, aliquoted, and stored at −80°C.
To titrate pseudoviruses, HEK 293T/17‐ACE2 cells were first transfected with the expression plasmid encoding for human TMPRSS2 using Fugene HD (Promega) according to the manufacturer's instructions. At 24 h after transfection, the supernatant was replaced by DMEM containing 10% FBS and subsequently used as pseudovirus target cells. Supernatants containing pseudoviruses were serially two‐fold diluted in a DMEM medium in 96‐well, flat‐bottomed culture plates, and TMPRSS2‐expressing HEK 293 T/17‐ACE2 target cells (1 × 104 cells/well) were added to each well. After 72 h, the luminescence of cell cultures (in Relative Luminescence Units or RLUs) was evaluated by luminometry (Synergy Plate Reader) using the Bright‐Glo assay system (Promega).
To measure the neutralizing activity of the serum samples, a two‐fold serial dilution of heat‐inactivated sera was prepared, starting from 1:40, in a culture medium (DMEM high glucose without FBS). The sera were mixed with pseudoviruses displaying the CoV spike of interest in a 1:1 vol/vol ratio in a 96‐well culture plate. The pseudovirus input used was normalized to 1 × 105 RLU/well. The serum‐pseudovirus mixture was then incubated for 1 h at 37°C. Subsequently, cell suspensions of HEK293T‐ACE‐2 pre‐transfected with pCAGGS expressing human TMPRSS2 (2 × 104 cells/ml) were mixed with the serum‐pseudovirus mixture seeded into each well of CulturPlate™ Microplates (PerkinElmer). The plates were incubated at 37°C for 48 h, and the neutralizing antibodies were determined based on luciferase activity as previously described. 40
2.7. Statistical analysis
Statistical significance was calculated across groups by Two‐way analysis of variance (ANOVA) and multiple comparisons using GraphPad Prism 9 (GraphPad Software, Inc.). The data were plotted as a geometric mean titer (GMT) with ± 95% confidence interval (CI). The p‐value <0.05 was considered statistically significant.
3. RESULTS
3.1. Expression of variant RBD‐Fc fusion proteins in N. benthamiana
SARS‐CoV‐2 variant RBD‐Fc fusion proteins were successfully expressed in N. benthamiana and purified from the plant crude extracts using Protein A affinity chromatography. The purified proteins were confirmed by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and western blot analysis. The SDS‐PAGE followed by western blot analysis of plant‐produced variant RBD‐Fc proteins performed under reducing and nonreducing conditions and the result was presented in Figure 2. All samples showed the major band at approximately 150 kDa and 75 kDa in nonreducing and reducing conditions as expected. The yield of purified plant‐produced variant RBD‐Fc were found to be in the range of 20–28 µg/g fresh weight.
Figure 2.
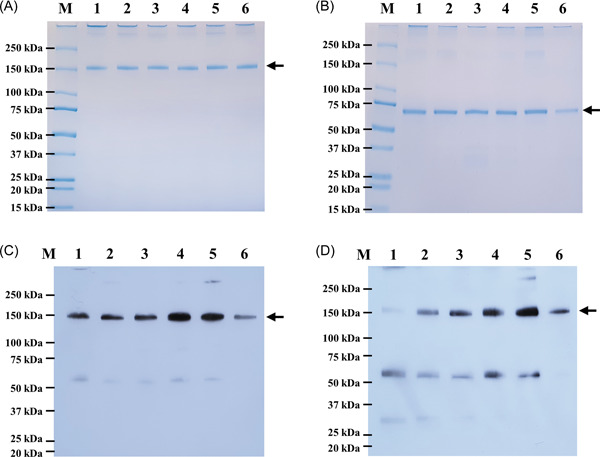
Purified plant‐produced SARS‐CoV‐2 variant RBD‐Fc proteins. Proteins were analyzed by SDS‐PAGE under nonreducing condition (A) and reducing condition (B), and western blot analysis with antihuman gamma chain conjugated HRP (C) and anti‐SARS‐CoV‐2 RBD conjugated HRP (D) under nonreducing condition. Lane M: marker; Lane 1: Alpha RBD‐Fc; Lane 2: Beta RBD‐Fc; Lane 3: Gamma RBD‐Fc; Lane 4: Kappa RBD‐Fc; Lane 5: Delta RBD‐Fc; Lane 6: Epsilon RBD‐Fc. The arrowhead indicates the major band. HRP, horseradish peroxidase; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.2. Plant‐produced variant RBD‐Fc vaccines induced antibody responses in monkeys
To investigate the in vivo immunogenicity of variant RBD‐Fc proteins produced in N. benthamiana, cynomolgus macaques were immunized with 10 μg of variant RBD‐Fc vaccines on Days 0, 21, and 42 and the blood was collected for the detection of RBD‐specific IgG, antibodies by ELISA. As shown in Figure 3B, Alpha, Gamma, Delta, and Epsilon RBD‐Fc vaccines induced considerably higher IgG titers 14 days after the first immunization with geometric mean titer (GMT) of 174, 115, 459, and 1213, respectively. After second immunization (Day 35), Kappa vaccine group (GMT = 16 890) showed significantly higher IgG titer than control group (GMT = 100) with p < 0.01, and Alpha (GMT = 5572), Beta (GMT = 4222), and Gamma (GMT = 3805) RBD‐Fc vaccine groups with p < 0.05, but not with Epsilon RBD‐Fc vaccinated group (GMT = 12 800). Delta RBD‐Fc vaccinated group (GMT = 29 407) exhibited significantly higher IgG antibody titer than control, Alpha, Beta, Gamma, and Epsilon RBD‐Fc groups with p < 0.0001, and Kappa group with p < 0.05. IgG titers were elicited after the third immunization on day 56 with those of Delta (GMT = 22 286) and Epsilon vaccine groups (GMT = 22 286) exhibiting significantly higher titer than the control group (GMT = 100) with p < 0.001. Notably, the IgG titers of both Delta and Epsilon groups were significantly higher than those of Beta group (GMT = 6400) with p < 0.01, and the Gamma (GMT = 9051) and Kappa (GMT = 11 143) groups with p < 0.05, but not with Alpha (GMT = 11 143) RBD‐Fc immunized group.
3.3. Plant‐produced variant RBD‐Fc vaccines induced neutralizing antibodies against SARS‐CoV‐2 variants
3.3.1. Microneutralization assay
Plant‐produced variant Alpha, Beta, Gamma, Kappa, Delta, and Epsilon, RBD‐Fc vaccines induced detectable neutralizing antibodies against live SARS‐CoV‐2 ancestral (Wuhan), Alpha, Beta, and Delta strains at 14 days after first immunization (Day 14) in monkeys as shown in Figure 4.
Figure 4.
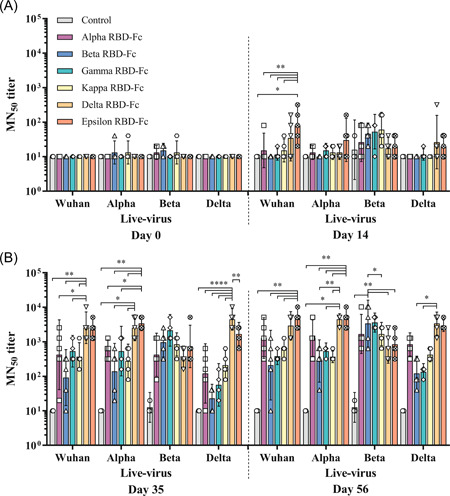
50% microneutralizing (MN50) titers against SARS‐CoV‐2 Wuhan, Alpha, Beta, and Delta viruses in immunized animals. The animals were immunized with plant‐produced Alpha, Beta, Gamma, Kappa, Delta, or Epsilon vaccines, or alum only (control), on Days 0, 21, and 42. The monkey sera were collected on Days 0, 14, (A) 35, and 56 (B), and the neutralizing titer was estimated. Values smaller than the limit of detection (LOD) are plotted as 0.5*LOD. Data presented as GMT ± 95% CI of the endpoint titer in each group, n = 5 (control, n = 3). Two‐way ANOVA, Tukey test was used. (*p < 0.05, **p < 0.01, ****p < 0.0001). ANOVA, analysis of variance; CI, confidence interval; GMT, geometric mean titer; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
We have measured the neutralizing antibody titers against ancestral (Wuhan) strain; Epsilon RBD‐Fc vaccinated group (GMT = 80) induced significantly higher neutralizing titer than Alpha (GMT = 15), Beta (GMT = 10), Gamma (GMT = 11), and Kappa (GMT = 15) RBD‐Fc immunized groups with p < 0.01 and control (GMT = 10) with p < 0.05 on Day 14, but not in Delta group (GMT = 35). After second immunization (Day 35), the neutralizing titer of Delta RBD‐Fc immunized group (GMT = 2560) was significantly increased compared with Beta (GMT = 92), Kappa (GMT = 368), and control (GMT = 10) groups with p < 0.01, and Alpha (GMT = 422) and Gamma (GMT = 538) RBD‐Fc immunized groups with p < 0.05, with no significant differences in Epsilon group (GMT = 2229). The neutralizing titer after third immunization (Day 56) showed that the Epsilon RBD‐Fc (GMT = 4457) significantly induced the neutralizing antibodies when compared to Beta (GMT = 211), Gamma (GMT = 381), and control (GMT = 10) groups with p < 0.01, with no significant differences in Alpha (GMT = 1280) and Delta (GMT = 2941) RBD‐Fc immunized groups.
The neutralizing titer against Alpha strain showed that Delta RBD‐Fc vaccinated group (GMT = 2560) was significantly higher than Beta (GMT = 139), Kappa (GMT = 279) and control (GMT = 10) groups with p < 0.05 on Day 35. Further, Epsilon RBD‐Fc vaccine group (GMT = 3378) showed significantly higher titer than Beta, Kappa and control groups with p < 0.01, and Alpha (GMT = 557) and Gamma (GMT = 538) RBD‐Fc groups with p < 0.05. After 14 days of third immunization (Day 56), neutralizing titer induced by Delta RBD‐Fc vaccine group (GMT = 4457) against Alpha strain was significantly higher than Beta (GMT = 279) and Kappa (GMT = 368) RBD‐Fc groups with p < 0.01; Gamma (GMT = 538) and control (GMT = 10) groups with p < 0.05. Further, Epsilon RBD‐Fc vaccine group (GMT = 4457) induced significantly higher neutralizing antibodies than Beta, Gamma, Kappa RBD‐Fc and control groups with p < 0.01, whereas no significant differences was observed when compared to Alpha (GMT = 1280) and Delta groups.
The potency of the neutralizing response against Beta strain was also evaluated. Alpha (GMT = 422), Beta (GMT = 970), Gamma (GMT = 2153), Kappa (GMT = 844), Delta (GMT = 368), Epsilon (GMT = 735) vaccine groups, and control group (GMT = 13) were not significant different after 14‐day of second immunization (Day 35). After third immunization (Day 56), Beta RBD‐Fc vaccine group (GMT = 3378) induced significantly higher titer than Delta (GMT = 735), Epsilon (GMT = 844), and control (GMT = 13) groups with p < 0.01, and Kappa group (GMT = 1689) with p < 0.05, with no significant differences compared to Alpha (GMT = 1689) and Gamma RBD‐Fc (GMT = 3620) immunized groups.
For the neutralizing titer against Delta strain, Delta vaccine group (GMT = 4457) induced significantly higher titer than Alpha (GMT = 121), Beta (GMT = 23), Gamma (GMT = 57), Kappa RBD‐Fc (GMT = 211), and control (GMT = 10) groups with p < 0.0001, and Epsilon (GMT = 1689) with p < 0.01 on Day 35. The neutralizing titer of Delta vaccine (GMT = 3378) was significantly higher than Beta (GMT = 121) and Gamma (GMT = 135) groups with p < 0.05, with no significant differences compared with Alpha (GMT = 844), Kappa (GMT = 422), Epsilon (GMT = 2,941), and control (GMT = 10) groups on Day 56.
The results showed that the levels of neutralizing antibodies found in the sera on Day 56 were slightly higher than Day 35 whereas Delta RBD‐Fc vaccinated group showed a slightly lower titer on the sera collected on Day 56 compared to Day 35. Overall, Delta and Epsilon vaccines elicited broadly neutralizing antibodies against ancestral (Wuhan), Alpha, and Delta except for the Beta variant.
3.3.2. Pseudovirus neutralization assay
We investigated the breadth of inhibition of virus entry by plant‐produced variant RBD‐Fc against Alpha, Beta, Gamma, and Delta variants using spike pseudovirus neutralization assay. The detectable neutralizing antibodies against all SARS‐CoV‐2 variants were observed 14 days after first immunization (Day 14) as shown in Figure 5.
Figure 5.
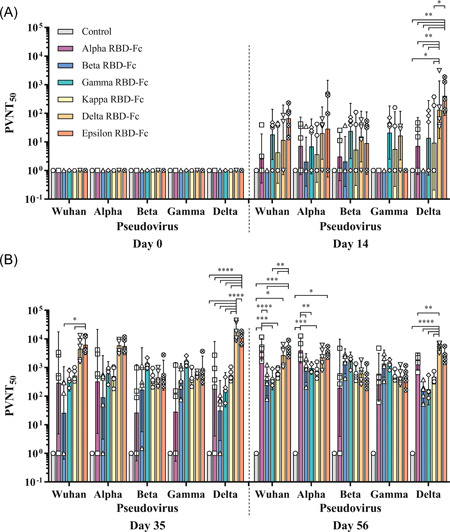
50% pseudovirus neutralization titers (PVNT50) of the immunized animals against SARS‐CoV‐2 and its variants, Wuhan, Alpha, Beta, Gamma, and Delta. Animals were immunized with plant‐produced Alpha, Beta, Gamma, Kappa, Delta, or Epsilon vaccines, or alum only (control), on Days 0, 21, and 42. The monkey sera were collected on Days 0, 14, (A) 35, and 56 (B). Data presented as GMT ± 95% CI of the endpoint titer in each group, n = 5 (control, n = 3). Two‐way ANOVA, Tukey test was used. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). ANOVA, analysis of variance; GMT, geometric mean titer; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Sera Collected from Epsilon RBD‐Fc immunized group on day 35 (GMT = 6228) neutralized pseudovirus bearing the spike protein of the ancestral (Wuhan) strain significantly higher than those of Beta (GMT = 26) and Kappa (GMT = 520) RBD‐Fc groups with p < 0.05, whereas the difference is not significant compared to Alpha (GMT = 291), Gamma (GMT = 377), Delta (GMT = 4512) RBD‐Fc and control (GMT = 1). After the third immunization (Day 56), the neutralizing titer of Alpha RBD‐Fc vaccine group (GMT = 4157) was significantly higher than Gamma (GMT = 411), Kappa (GMT = 670), and control (GMT = 1) with p < 0.001, and Beta group (GMT = 366) with p < 0.0001. The titer of Delta RBD‐Fc vaccine group (GMT = 2674) against the ancestral (Wuhan) strain was substantially higher than the control with p < 0.05. Furthermore, the Epsilon RBD‐Fc vaccinated group (GMT = 4222) was significantly higher than the Beta and control groups with p < 0.001, and Gamma and Kappa RBD‐Fc groups with p < 0.01.
For PVNT against the Alpha variant on Day 35, Alpha (GMT = 327), Beta (GMT = 91), Gamma (GMT = 617), Kappa (GMT = 502), Delta (GMT = 5969), Epsilon (GMT = 5626), and control (GMT = 1) groups were not significantly different. On Day 56, the neutralizing titer of Alpha vaccine (GMT = 3951) was significantly higher than Kappa (GMT = 748) and control (GMT = 1) groups with p < 0.001, and Beta (GMT = 968) and Gamma (GMT = 959) with p < 0.01 whereas, Epsilon vaccine group (GMT = 3243) induced significantly higher titer than control group with p < 0.05. No significant difference was observed in Delta vaccine (GMT = 2312).
Neutralizing titers against Beta variant on Day 35 showed that there is no significant difference was observed in Alpha (GMT = 27), Beta (GMT = 168), Gamma (GMT = 1357), Kappa (GMT = 413), Delta (GMT = 445), Epsilon (GMT = 540), and control (GMT = 1) groups. After the third immunization (Day 56), neutralizing titers of Alpha (GMT = 196), Beta (GMT = 1653), Gamma (GMT = 1632), Kappa (GMT = 587), Delta (GMT = 619), Epsilon (GMT = 444), and control (GMT = 1) groups were also not significantly different.
Similarly, the neutralizing titer of Alpha (GMT = 28, 455), Beta (GMT = 354, 1368), Gamma (GMT = 1639, 1278), Kappa (GMT = 424, 487), Delta (GMT = 587, 484), Epsilon (GMT = 780, 472), and control (GMT = 1, 1) groups against Gamma variant did not show any significant difference on both Day 35 and 56, respectively.
Neutralization of the Delta variant pseudovirus tested using sera collected on Day 14 showed that the Delta vaccine (GMT = 133) induced significantly higher than Kappa (GMT = 9) and control (GMT = 1) groups with p < 0.05, and Alpha (GMT = 7), Beta (GMT = 1), and Gamma (GMT = 14) groups with p < 0.01. Further, Epsilon group (GMT = 407) induced significantly higher neutralizing antibodies than Alpha, Beta, Gamma, and control groups with p < 0.01, and Kappa group with p < 0.05, but not in Delta group. On Day 35, neutralizing titer of the Delta vaccine group (GMT = 19 684) was significantly higher than in Alpha (GMT = 181), Beta (GMT = 31), Gamma (GMT = 169), Kappa (GMT = 536), Epsilon (GMT = 10,889), and control (GMT = 1) groups with p < 0.0001. The Epsilon vaccine was also significantly higher than in Alpha, Beta, Gamma, Kappa, and control groups with p < 0.0001. After the third immunization (Day 56), the neutralizing titer of the Delta vaccine group (GMT = 5241) was significantly higher than Beta (GMT = 148), Gamma (GMT = 174), Kappa (GMT = 469), and control (GMT = 1) groups with p < 0.0001, and Alpha group (GMT = 1603) with p < 0.01, but not significantly higher compared with the Epsilon group (GMT = 2584).
Taken together, these results suggested that Delta and Epsilon RBD‐Fc vaccines elicit broadly neutralizing antibodies against ancestral (Wuhan), Alpha, and Delta, except Beta and Gamma variants, while Beta and Gamma RBD‐Fc based vaccines significantly neutralize their respective strains.
4. DISCUSSION
Currently, SARS‐CoV‐2 variants with multiple mutations are predominately emerging globally. SARS‐CoV‐2 variants with mutations in their RBD, increase the affinity of the virus binding to the host receptor ACE2 and also result in immune escape. The mutations E484K, N501Y, and D614G were reported as the enhancer for virus binding with human ACE2. 41 , 42 , 43 The mutations K417N/T, E484K, and L452R were reported to be associated with the ability to evade the immunity induced by convalescent plasma and vaccinated sera. 41 , 44 , 45 , 46 , 47 Recently, the neutralizing activity of the vaccine recipients against VOCs has been reviewed and compared with the ancestral (Wuhan) strain of SARS‐CoV‐2. 14 The neutralizing titer is reduced against the Alpha strain among vaccinated individuals, and varying effects were observed against the Beta strain for mRNA, viral vector, inactivated, and subunit vaccines. For Gamma and Delta variants, minimal to moderate neutralizing activity was also reported. Furthermore, a substantial reduction of neutralizing response against the Omicron variant was reported among BNT162b2 recipients, whereas no detectable neutralizing antibody titer was observed among Coronavac recipients. 18 The variant‐specific SARS‐CoV‐2 vaccines are likely required to cope with the emerging new variants, especially those that are less susceptible to immunity elicited by currently available vaccines. In this study, we have evaluated different variant‐specific subunit vaccines which are developed based on the variant SARS‐CoV‐2 RBD protein produced in the plant expression system. Previously, plant‐produced SARS‐CoV‐2 RBD‐Fc subunit vaccine (the ancestral (Wuhan) strain) adjuvanted with alum has been shown to elicit robust immune responses in both mice and monkeys against the original SARS‐CoV‐2. 31 Besides, our group has also reported that the plant‐produced Alpha and Beta RBD‐Fc can be successfully expressed from N. benthamiana and it was found that these RBDs have shown reduced affinity against plant‐produced anti‐SARS‐CoV‐2 mAbs. 35
To gain more insights into the effect of key mutations commonly found in each VOC, respective mutations were introduced in the RBD sequence of SARS‐CoV‐2 to generate Alpha (N501Y, A570D, and D614G), Beta (K417N, E484K, N501Y, and D614G), Gamma (K417T, E484K, N501Y, and D614G), Kappa (L452R, E484Q, and D614G), Delta (L452R, T478K, and D614G), and Epsilon (L452R and D614G) RBD and fused with Fc region of human IgG1. The fusion proteins were transiently expressed in N. benthamiana and purified. The major bands of variant RBD‐Fc proteins showed a similar molecular weight of approximately 150 kDa and 75 kDa in nonreducing and reducing conditions, respectively. However, in the western blot analysis, the affinity of anti‐RBD HRP‐conjugated antibody was found to be different for each variant RBD‐Fc protein. The most intense major band was observed in Delta RBD‐Fc protein, whereas the band intensity was found to be lower in Kappa, Epsilon, Gamma, Beta, and Alpha RBD‐Fc proteins.
Immunogenicity of plant‐produced variant vaccines were assessed in monkeys and the results revealed that both Delta and Epsilon RBD‐Fc vaccines induced 2–3.5‐fold higher RBD‐specific total IgG titer than the other tested variant vaccines after third immunization (Figure 3B). Subsequently, neutralizing antibodies response against SARS‐CoV‐2 variants in vitro and pseudoviruses was tested using the vaccinated monkey sera. The results showed that both Delta and Epsilon RBD‐Fc (sharing L452R and D614G mutations) vaccinated groups showed neutralization activities against ancestral (Wuhan), Alpha and Delta strains higher than other vaccinated groups. On the other hand, neutralizing titer of Beta and Gamma RBD‐Fc (sharing E484K, N501Y, D614G sites) vaccinated groups, was found to be higher against Beta and Gamma strains respectively compared to other immunized groups. Our results are in line with Amanat et al., 48 who reported that the mice immunized with recombinant spike proteins from the wild‐type Wuhan‐1 strain, B.1.1.7, B.1.351, and P.1 displayed high neutralization titers against the homologous viruses and decline in neutralization were detected for B.1.1.7–B.1.351 (4.8‐fold), from B.1.1.7 to P.1 (4.4‐fold), and from B.1.351 to P.1 (4.2‐fold).
Based on our results, the SARS‐CoV‐2 variant‐specific vaccine might not cross‐react or neutralize all the circulating variants. Recently, the concept of cocktail vaccine has been proposed by combining the antigens for eliciting a broad immune response against SARS‐CoV‐2 variants. Wang et al. revealed that application of RBD with multiple mutations as the cocktail vaccine increased variant‐specific antibodies than the single antigen vaccine formulation or infection with wild‐type SARS‐CoV‐2. 49
In conclusion, our study demonstrated that the SARS‐CoV‐2 variant vaccine might not induce broad protection against all the SARS‐CoV‐2 variants. The variant vaccine could be considered for booster dose to increase the breadth of the immune response. However, research warrants further efficacy studies to corroborate the findings. In addition, another possible approach is that the variant antigens could be combined to develop as a cocktail vaccine. Further studies are needed to validate the cocktail vaccine strategy and its efficacy against SARS‐CoV‐2 and the circulating variants. Overall, these proof‐of‐concept results facilitate the design and development of plant‐produced variant‐specific subunit vaccines against SARS‐CoV‐2 variants.
AUTHOR CONTRIBUTIONS
Arunee Thitithanyanont, Anan Jongkaewwattana, Balamurugan Shanmugaraj, and Waranyoo Phoolcharoen designed all experiments. Narach Khorattanakulchai, Balamurugan Shanmugaraj, Kaewta Rattanapisit, and Chalisa Panapitakkul performed protein expression, protein purification, and ELISA. Taratorn Kemthong, Nutchanat Suttisan, and Suchinda Malaivijitnond performed vaccination in nonhuman primates. Suwimon Manopwisedjaroen and Arunee Thitithanyanont performed a microneutralization assay. Kanjana Srisutthisamphan performed a pseudovirus neutralization assay. Narach Khorattanakulchai and Balamurugan Shanmugaraj drafted and revised the manuscript. All authors analyzed the data, revised the manuscript, and approved it for publication.
CONFLICT OF INTEREST
WP from Chulalongkorn University is a founder/shareholder of Baiya Phytopharm Co., Ltd., Thailand.
ETHICS STATEMENT
The animal study protocol was approved by the National Primate Research Center of Thailand‐Chulalongkorn University (NPRCT‐CU) Animal Care and Use Committee, Chulalongkorn University, Thailand (Protocol No. 2175005, Approval date:27 May 2021, and 2175007, Approval date:15 Jul 2021).
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The author (NK) would like to thank The Second Century Fund (C2F), Chulalongkorn University, Thailand, for the doctoral fellowship. This study was funded by Baiya Phytopharm Co., Ltd., Thailand.
Khorattanakulchai N, Manopwisedjaroen S, Rattanapisit K, et al. Receptor binding domain proteins of SARS‐CoV‐2 variants produced in Nicotiana benthamiana elicit neutralizing antibodies against variants of concern. J Med Virol. 2022;94:4265‐4276. 10.1002/jmv.27881
Contributor Information
Balamurugan Shanmugaraj, Email: Balamurugan.S@baiyaphytopharm.com.
Waranyoo Phoolcharoen, Email: Waranyoo.P@chula.ac.th.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization . WHO Director‐General's opening remarks at the media briefing on COVID‐19 ‐ 11 March, 2020. Accessed May 23, 2022. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- 3. World Health Organization . WHO coronavirus (COVID‐19) dashboard. 2020. Accessed May 23, 2022. https://covid19.who.int/
- 4. World Health Organization . DRAFT landscape of COVID‐19 candidate vaccines. 2021. Accessed May 23, 2022. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 5. Wang N, Shang J, Jiang S, Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tai W, He L, Zhang X, et al. Characterization of the receptor‐binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pollet J, Chen WH, Strych U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv Drug Deliv Rev. 2021;170:71‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keech C, Albert G, Cho I, et al. Phase 1‐2 trial of a SARS‐CoV‐2 recombinant spike protein nanoparticle vaccine. N Engl J Med. 2020;383(24):2320‐2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang S, Li Y, Dai L, et al. Safety and immunogenicity of a recombinant tandem‐repeat dimeric RBD‐based protein subunit vaccine (ZF2001) against COVID‐19 in adults: two randomised, double‐blind, placebo‐controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1107‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. World Health Organization . Status of COVID‐19 vaccines within WHO EUL/PQ evaluation process. Accessed May 23, 2022. https://extranet.who.int/pqweb/sites/default/files/documents/Status_COVID_VAX_02April2022.pdf
- 11. Abdool Karim SS, de Oliveira T. New SARS‐CoV‐2 variants ‐ clinical, public health, and vaccine implications. N Engl J Med. 2021;384(19):1866‐1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS‐CoV‐2 variants of concern are emerging in India. Nat Med. 2021;27:1131‐1133. [DOI] [PubMed] [Google Scholar]
- 13. World Health Organization . Tracking SARS‐CoV‐2 variants. 2021. Accessed May 23, 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ [PubMed]
- 14. Fiolet T, Kherabi Y, MacDonald C‐J, Ghosn J, Peiffer‐Smadja N. Comparing COVID‐19 vaccines for their characteristics, efficacy and effectiveness against SARS‐CoV‐2 and variants of concern: a narrative review. Clin Microbiol Infect. 2022;28(2):202‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shanmugaraj B, Malla A, Khorattanakulchai N, Phoolcharoen W. SARS‐CoV‐2 omicron variant: could it be another threat? J Med Virol. 2022;94(4):1284‐1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bates TA, Leier HC, Lyski ZL, et al. Neutralization of SARS‐CoV‐2 variants by convalescent and BNT162b2 vaccinated serum. Nat Commun. 2021;12(1):5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS‐CoV‐2 variant delta to antibody neutralization. Nature. 2021;596(7871):276‐280. [DOI] [PubMed] [Google Scholar]
- 18. Lu L, Mok BW, Chen LL, et al. Neutralization of SARS‐CoV‐2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021: ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shanmugaraj B, Phoolcharoen W. Addressing demand for recombinant biopharmaceuticals in the COVID‐19 era. Asian Pac J Trop Med. 2021;14(2):49‐51. [Google Scholar]
- 20. Capell T, Twyman RM, Armario‐Najera V, Ma JK, Schillberg S, Christou P. Potential applications of plant biotechnology against SARS‐CoV‐2. Trends Plant Sci. 2020;25(7):635‐643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar M, Kumari N, Thakur N, et al. A comprehensive overview on the production of vaccines in plant‐based expression systems and the scope of plant biotechnology to combat against SARS‐CoV‐2 virus pandemics. Plants. 2021;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lobato Gomez M, Huang X, Alvarez D, et al. Contributions of the international plant science community to the fight against human infectious diseases ‐ part 1: epidemic and pandemic diseases. Plant Biotechnol J. 2021. [DOI] [PMC free article] [PubMed]
- 23. Shanmugaraj B, Siriwattananon K, Malla A, Phoolcharoen W. Potential for developing plant‐derived candidate vaccines and biologics against emerging coronavirus infections. Pathogens. 2021;10(8):1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gleba Y, Klimyuk V, Marillonnet S. Magnifection: a new platform for expressing recombinant vaccines in plants. Vaccine. 2005;23(17‐18):2042‐2048. [DOI] [PubMed] [Google Scholar]
- 25. LeBlanc Z, Waterhouse P, Bally J. Plant‐based vaccines: the way ahead? Viruses. 2021;13(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sohrab SS, Suhail M, Kamal MA, Husen A, Azhar EI. Recent development and future prospects of plant‐based vaccines. Curr Drug Metab. 2017;18(9):831‐841. [DOI] [PubMed] [Google Scholar]
- 27. Shanmugaraj B, Bulaon CJI, Phoolcharoen W. Plant molecular farming: a viable platform for recombinant biopharmaceutical production. Plants. 2020;9(7):842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shanmugaraj B, Khorattanakulchai N, Phoolcharoen W. SARS‐CoV‐2 vaccines: current trends and prospects of developing plant‐derived vaccines. Biomedical Innovations to Combat COVID‐19. Elsevier; 2022:213‐229. [Google Scholar]
- 29. Shanmugaraj B, Rattanapisit K, Manopwisedjaroen S, Thitithanyanont A, Phoolcharoen W. Monoclonal antibodies B38 and H4 produced in Nicotiana benthamiana neutralize SARS‐CoV‐2 in vitro. Front Plant Sci. 2020;11:(‐) 1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rattanapisit K, Shanmugaraj B, Manopwisedjaroen S, et al. Rapid production of SARS‐CoV‐2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana . Sci Rep. 2020;10(1):17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siriwattananon K, Manopwisedjaroen S, Shanmugaraj B, et al. Plant‐produced receptor‐binding domain of SARS‐CoV‐2 elicits potent neutralizing responses in mice and non‐human primates. Front Plant Sci. 2021;12:682953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siriwattananon K, Manopwisedjaroen S, Shanmugaraj B, et al. Immunogenicity studies of plant‐produced SARS‐CoV‐2 receptor binding domain‐based subunit vaccine candidate with different adjuvant formulations. Vaccines. 2021;9(7):744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du L, Zhao G, He Y, et al. Receptor‐binding domain of SARS‐CoV spike protein induces long‐term protective immunity in an animal model. Vaccine. 2007;25(15):2832‐2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y, Du L, Qiu H, et al. A recombinant protein containing highly conserved hemagglutinin residues 81‐122 of influenza H5N1 induces strong humoral and mucosal immune responses. Biosci Trends. 2013;7(3):129‐137. [PubMed] [Google Scholar]
- 35. Rattanapisit K, Bulaon CJI, Khorattanakulchai N, Shanmugaraj B, Wangkanont K, Phoolcharoen W. Plant‐produced SARS‐CoV‐2 receptor binding domain (RBD) variants showed differential binding efficiency with anti‐spike specific monoclonal antibodies. PLoS One. 2021;16(8):e0253574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Q, He J, Phoolcharoen W, Mason HS. Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum Vacc. 2011;7(3):331‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frey A, Di Canzio J, Zurakowski D. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods. 1998;221(1‐2):35‐41. [DOI] [PubMed] [Google Scholar]
- 38. Vacharathit V, Aiewsakun P, Manopwisedjaroen S, et al. CoronaVac induces lower neutralising activity against variants of concern than natural infection. Lancet Infect Dis. 2021;21(10):1352‐1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hyseni I, Molesti E, Benincasa L, et al. Characterisation of SARS‐CoV‐2 lentiviral pseudotypes and correlation between pseudotype‐based neutralisation assays and live virus‐based micro neutralisation assays. Viruses. 2020;12(9):1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferrara F, Temperton N. Pseudotype neutralization assays: from laboratory bench to data analysis. Methods Protoc. 2018;1(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barton MI, MacGowan SA, Kutuzov MA, Dushek O, Barton GJ, van der Merwe PA. Effects of common mutations in the SARS‐CoV‐2 spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife. 2021;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Groves DC, Rowland‐Jones SL, Angyal A. The D614G mutations in the SARS‐CoV‐2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2021;538:104‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y, Liu J, Plante KS, et al. The N501Y spike substitution enhances SARS‐CoV‐2 infection and transmission. Nature. 2022;602:294‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feder KA, Patel A, Vepachedu VR, et al. Association of E484K spike protein mutation with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection in vaccinated persons: Maryland, January–May 2021. Clin Infect Dis. 2021: ciab762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jensen B, Luebke N, Feldt T, et al. Emergence of the E484K mutation in SARS‐COV‐2‐infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Regional Health‐Europe. 2021;8:100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS‐CoV‐2 variant P. 1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747‐751.e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS‐CoV‐2 variants B. 1.351 and B. 1.1. 7. Nature. 2021;593(7857):130‐135. [DOI] [PubMed] [Google Scholar]
- 48. Amanat F, Strohmeier S, Meade PS, et al. Vaccination with SARS‐CoV‐2 variants of concern protects mice from challenge with wild‐type virus. PLoS Biol. 2021;19(12):e3001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang E, Chakraborty AK Design of immunogens for eliciting antibody responses that may protect against SARS‐CoV‐2 variants. bioRxiv. 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


