Abstract
The current COVID‐19 pandemic is severely threatening public healthcare systems around the globe. Some supporting therapies such as remdesivir, favipiravir, and ivermectin are still under the process of a clinical trial, it is thus urgent to find alternative treatment and prevention options for SARS‐CoV‐2. In this regard, although many natural products have been tested and/or suggested for the treatment and prophylaxis of COVID‐19, carotenoids as an important class of natural products were underexplored. The dietary supplementation of some carotenoids was already suggested to be potentially effective in the treatment of COVID‐19 due to their strong antioxidant properties. In this study, we performed an in silico screening of common food‐derived carotenoids against druggable target proteins of SARS‐CoV‐2 including main protease, helicase, replication complex, spike protein and its mutants for the recent variants of concern, and ADP‐ribose phosphatase. Molecular docking results revealed that some of the carotenoids had low binding energies toward multiple receptors. Particularly, crocin had the strongest binding affinity (−10.5 kcal/mol) toward the replication complex of SARS‐CoV‐2 and indeed possessed quite low binding energy scores for other targets as well. The stability of crocin in the corresponding receptors was confirmed by molecular dynamics simulations. Our study, therefore, suggests that carotenoids, especially crocin, can be considered an effective alternative therapeutics and a dietary supplement candidate for the prophylaxis and treatment of SARS‐CoV‐2.
Practical applications
In this study, food‐derived carotenoids as dietary supplements have the potential to be used for the prophylaxis and/or treatment of SARS‐CoV‐2. Using in silico techniques, we aimed at discovering food‐derived carotenoids with inhibitory effects against multiple druggable sites of SARS‐CoV‐2. Molecular docking experiments against main protease, helicase, replication complex, spike protein and its mutants for the recent variants of concern, and ADP‐ribose phosphatase resulted in a few carotenoids with multitarget inhibitory effects. Particularly, crocin as one of the main components of saffron exhibited strong binding affinities to the multiple drug targets including main protease, helicase, replication complex, mutant spike protein of lineage B.1.351, and ADP‐ribose phosphatase. The stability of the crocin complexed with these drug targets was further confirmed through molecular dynamics simulations. Overall, our study provides the preliminary data for the potential use of food‐derived carotenoids, particularly crocin, as dietary supplements in the prevention and treatment of COVID‐19.
Keywords: Crocin, dietary supplement candidates, food‐derived carotenoids, molecular docking, molecular dynamics simulation, multitarget inhibitors, SARS‐CoV‐2
The molecular docking and molecular dynamics simulations of carotenoids against multiple SARS‐CoV‐2 druggable targets revealed that carotenoids are promising therapeutic candidates for COVID‐19. Particularly, crocin showed strong binding affinity against multiple drug targets and thus can be considered as a dietary supplement option in the prevention and treatment of COVID‐19.
1. INTRODUCTION
The current COVID‐19 pandemic caused by the emerging severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) is significantly impacting public healthcare systems around the globe and has paralyzed societal and economical norms. As of March 19, 2022, the number of cases has reached around 500 million, while more than 6 million people died due to the life‐threatening disease. The novel CoV belongs to the Coronaviridae family and is an enveloped, positive polarity, single‐stranded RNA betacoronavirus. Its genome with a 79.6% sequence identity to SARS‐CoV encodes nonstructural proteins (Nsps), structural proteins, and several accessory proteins (Wu et al., 2020; Zhou et al., 2020).
To develop therapeutics against SARS‐CoV‐2, potentially druggable virus proteins have been identified. These proteins are mostly involved in viral replication and play roles in the control of host cellular machineries (Gil et al., 2020; Saxena, 2020). First, spike (S) protein is one of the main drug targets due to its key role in the viral entry to the host cells. S protein has two subunits S1 and S2. S1 binds the receptor‐binding domain (RBD) of angiotensin‐converting enzyme 2 (ACE2), whereas the S2 subunit helps the virus in membrane fusion to complete the viral entry and infection process (Zhang & Kutateladze, 2020). Nucleocapsid (N) protein is highly immunogenic, making it a candidate for vaccine development (Dutta et al., 2020). Interestingly, the crystal structure of the N protein of SARS‐CoV‐2 revealed that it performs multiple critical functions during the viral life cycle. Particularly, its unique RNA‐binding pocket renders it a perfect target for the drug discovery process (Kang et al., 2020). Moreover, the nonstructural proteins are important targets for the treatment of COVID‐19. For instance, one of the most important and druggable target proteins is the main protease (Mpro) (also known as chymotrypsin‐like protease (3CLpro)) due to its essential role in the viral life cycle (de Vries et al., 2021). It is responsible for the cleavage of 13 (Nsp4‐Nsp16) of the 16 nonstructural proteins, including the polymerase subunits, and enables the proper folding and assembly of polymerase subunits into the active polymerase complex (Gadlage & Denison, 2010). Similar to Mpro, papain‐like protease (PLpro) cleaves Nsp1‐Nsp3 and removes ubiquitin‐like ISG15 protein modifications, dampening inflammation and antiviral signaling (Klemm et al., 2020). Furthermore, viral RNA‐dependent RNA polymerase (RdRp) has a crucial function in the replication and transcription of the viral genome. The architecture of mini replication and transcription complex of SARS‐CoV‐2 was elucidated by cryo‐electron microscopy, which indicated that it includes the assembly of the catalytic subunit of RdRp (Nsp12) with accessory subunits (Nsp7 and Nsp8) and two helicases (Nsp13‐1 and Nsp13‐2) (Yan et al., 2020). Some of the currently used drugs for COVID‐19, such as favipiravir and remdesivir target RdRp, indicate that it is an important target protein for the development of new therapeutics (Kokic et al., 2021; Naydenova et al., 2021). Nsp16, an S‐adenosylmethionine (SAM)‐dependent methyltransferase, is also considered to be essential for the viral life cycle. Thus, Nsp16 with its cofactor Nsp10 is a very promising druggable protein as it is indispensable for the replication of coronavirus in cell cultures (Krafcikova et al., 2020). In the replication of SARS‐CoV‐2, replicase (Nsp9) is an important protein as well, which binds and stabilizes the viral RNA as it emerges from the replication and transcription complex and protects it from nucleases (Farias et al., 2021; Littler et al., 2020). Moreover, NTPase/helicase (Nsp13) as a potential druggable target catalyzes the separation of double‐stranded oligonucleotides into a single strand (Gurung, 2020; Habtemariam et al., 2020). ADP‐ribose phosphatase (Nsp3, ADRP) of SARS‐CoV was known to remove the terminal phosphate group of ADP‐ribose phosphate in SARS‐CoV. The ADRP inhibitors were reported to potentially block the functional nsP‐complex formation. Therefore, they can be potential therapeutic agents in combatting COVID‐19 with the reduction in the replication of the virus (Debnath et al., 2020; Saikatendu et al., 2005).
For the treatment of COVID‐19, there is still an urgent need for an effective drug and/or alternative prevention options such as dietary supplementation although several drugs have been used as supporting therapies to reduce the severity of the pandemic such as remdesivir, favipiravir, and ivermectin. Several drugs have been in the clinical trials as part of drug repurposing studies (Tarighi et al., 2021). For instance, nafamostat was found to prevent the entry of viruses via the inhibition of transmembrane protease serine 2 (TMPRSS2), leading to the blockage of S protein‐mediated membrane fusion (Hoffmann et al., 2020). A case study on three patients (above 60 years) with COVID‐19 pneumonia revealed that nafomostat can promisingly alleviate the clinical symptoms (Jang & Rhee, 2020). For the urgent need to develop new specific drugs and alternative preventive measures for the treatment of SARS‐CoV‐2, many natural products and synthetic compounds have been tested and/or suggested for the treatment and prophylaxis of COVID‐19 (Boozari & Hosseinzadeh, 2021; Gasmi et al., 2022; Huang et al., 2020). However, one important class of natural products (carotenoids) was not yet investigated for the development of drugs and/or dietary supplement candidates for SARS‐CoV‐2. The dietary supplementation of some antioxidants including carotenoids was suggested to be potentially effective in the treatment of COVID‐19 since oxidative stress and inflammation are key aspects of the pathogenicity of SARS‐CoV‐2 (Barciszewska, 2021; Lammi & Arnoldi, 2021).
Previous studies showed the potential of food‐derived carotenoids in the treatment of SARS‐CoV‐2 due to their antioxidant and anti‐inflammatory activities (Barciszewska, 2021; Lammi & Arnoldi, 2021). Carotenoids have also been tested against some viruses. For example, bacterioruberin as a carotenoid was found in an in vitro study to be a strong inhibitor of RNA‐dependent RNA polymerases of Hepatitis B and C viruses (Hegazy et al., 2020). Similarly, crocin exhibited promising anti‐HSV‐1 and anti‐HIV‐1 activities (Soleymani et al., 2018). However, to our knowledge, there is no report on the potential antiviral activities of common food‐derived carotenoids against SARS‐CoV‐2. In this study, therefore, the food‐derived carotenoids were investigated using in silico approaches, molecular docking, and molecular dynamics simulation, to discover their potential as dietary supplements against various druggable targets of SARS‐CoV‐2. More than 750 different natural carotenoids have been reported in the literature (Fidan & Zhan, 2019). However, 20 widely used carotenoids were chosen for in silico analysis against druggable proteins of SARS‐CoV‐2 as they exist in various daily consumed food products. Some of them have been used as a food‐coloring agent and dietary supplement, whereas some are available in various fruits and vegetables such as tomatoes, pumpkin, saffron, pepper, carrot, etc. (Eggersdorfer & Wyss, 2018; Melendez‐Martinez, 2019; Zakynthinos & Varzakas, 2016). Additionally, some of the chosen carotenoids have already been used as a dietary supplement (Böhm et al., 2021; Kiokias & Gordon, 2003), whereas others have the potential to be candidate dietary supplements, particularly during the COVID‐19 pandemic (Michele et al., 2020; Singh et al., 2020). Molecular docking revealed that some of the chosen carotenoids had strong binding affinities to the tested drug targets. Particularly, crocin as the primary pigment for the color of saffron possessed strong binding affinities to most of the target proteins including Mpro, ADRP, replication complex, helicase, and spike protein. The mutant spike proteins for the concerning variants of SARS‐CoV‐2 were also docked with the chosen carotenoids. Crocin had an even stronger binding affinity to the mutant spike protein for the Beta variant compared with wild type, indicating its promising potential in the treatment and prophylaxis of concerning SARS‐CoV‐2 variants. The molecular docking results were validated with the molecular dynamics simulations, in which crocin was stably bound to the target receptors during the simulation process. This study provides the therapeutic potential of food‐derived carotenoids and supports the idea of carotenoid use as a dietary supplement to prevent and treat COVID‐19.
2. MATERIALS AND METHODS
2.1. Ligand and receptor preparation
The crystal structures of all the receptor proteins (6LU7: main protease, 6W02: ADP‐ribose phosphatase, 6ZSL: helicase, 7BV2: Nsp12‐Nsp7‐Nsp8 complex, 6M0J: spike protein) were downloaded from the protein data bank (PDB) of Research Collaboratory for Structural Bioinformatics (RCSB). The crystal structures of mutant spike proteins (6M0JR: spike protein of lineage B.1.617.1, Delta/Indian variant; 6M0JSA: spike protein of lineage B.1.351, Beta/South African variant; 6M0JBr: the spike of protein of lineage P.1, Gamma/Brazilian variant; 6M0JUK: spike protein of lineage B.1.1.7, Alpha/United Kingdom variant) were prepared through mutating the specific amino acid of the macromolecular structure followed by the energy minimization of the whole structure using Desmond software. The United Kingdom variant of the viral spike protein was characterized by N501Y mutation, whereas the Brazil and the South Africa variant was characterized by E484K, N501Y, and K417N mutations (Mujwar, 2021). The ligand files were downloaded from PubChem Database with their 3D structures in sdf format. For crocin, lycopene, phytoene, and phytofluene, 3D structures were not available on the PubChem webpage. Thus, they were downloaded with their 2D structures in sdf format and then converted to 3D structures with Open Babel GUI software. Subsequently, all SDF files were converted to PDB files for each ligand using the same software. Both proteins and the ligands were prepared for the molecular docking experiments using AutoDock Tools (ADT) 1.5.6 [17]. In the protein preparation process, water molecules were deleted, and polar hydrogen atoms and Kollmans charges were added. For ligands, Gasteiger charges were added, aromatic carbon and rotatable bonds were detected, torsion number was automatically set, and nonpolar hydrogens were merged. Then, each receptor and ligand file were saved as PDBQT files.
2.2. Molecular docking
The grid boxes for each receptor were determined based on the position of the endogenous ligand interacting with the specific target. The imaginary grid box for each antiviral target has been prepared by casing the complexed endogenous ligand as well as the interacting macromolecular residues (Mujwar et al., 2019). The grid box for 6LU7 was at x = −13.58, y = 12.60, and z = 68.93 with sizes of 52, 56, and 52 Å. The grid box for 6W02 was at x = 3.70, y = −5.93, and z = −22.58 with sizes of 40, 40, and 40 Å. The grid box for 6ZSL was at x = −13.60, y = 25.93, and z = −70.22 with sizes of 58, 58, and 58 Å. The grid box for 7BV2 was at x = 91.78, y = 91.56, and z = 104.86 with sizes of 58, 58, and 58 Å. The grid box for 6M0J and mutants were at x = −33.36, y = 28.55, and z = 9.19 with sizes of 50, 50, and 50 Å. This utilized docking protocol was validated for all the macromolecular targets used in the current study by redocking the reference ligand complexed with the macromolecular target. The redocked reference ligand was evaluated by overlay methods as well as the chemical resemblance of the docked conformation with respect to its crystallized bioactive conformation for validation purposes (Jain & Mujwar, 2020). After determining the grid boxes, the prepared flexible ligands were docked against the fixed rigid receptor protein using AutoDock Vina in the Windows command line window after preparing config files with necessary information (receptor, grid box coordinates, number of modes: 10, energy range: 8 and exhaustiveness: 16) [18]. Pymol version 2.4.1 and Discovery Studio 2020 Client software were used for the visualization and protein–ligand interactions, respectively.
2.3. Molecular dynamics simulation
The molecular docking screening of the food‐derived carotenoids against various drug targets of SARS‐CoV‐2 such as Mpro, helicase, ADRP, RdRp, and spike protein revealed that the crocin molecule consistently had strong binding affinities toward most of the antiviral targets of SARS‐CoV‐2 (Kwarteng et al., 2020; Mujwar, 2021). With the intention to strengthen the molecular docking results and validating the stability of the macromolecular complexes of crocin with specific antiviral drug targets, molecular dynamics (MD) simulation was performed for each macromolecular complex. The NPT ensemble MD simulation for each of the macromolecular complex was conducted using similar parameters at 300 K temperature for a timeframe of 100 ns (Duan et al., 2019). The NPT ensemble condition was selected so as to keep the temperature and pressure constant while allowing the volume of the system to change during the simulation process. The drug molecule has to perform its action within the human body under constant temperature and pressure conditions. Thus, the NPT ensemble condition was enabled during the simulation process.
2.4. Postdynamics complex stability analysis
The main objective of performing the MD simulation of a drug receptor complex was to observe the stability of the ligand and the macromolecular target with respect to time. Thus, the individual behavior of the ligand as well as the macromolecular antiviral targets was observed during the whole 100 ns timeframe through the root mean square deviation (RMSD) separately for both ligand and the target receptor. The initial conformational changes both in the macromolecular target as well as the complexed ligand were within the permissible limits of 1–3 Å, leading to the stabilized conformation of the complex. Therefore, to analyze the stability of the various drug receptor complexes of the crocin against various antiviral drug targets, the fluctuating behaviors of both the ligand and the macromolecular target were observed by considering the change in their RMSD values during MD simulation of each of the macromolecular complex with crocin.
3. RESULTS AND DISCUSSION
3.1. Molecular docking results of food‐derived carotenoids
The utilized docking protocols were successfully validated as the docked conformation of the reference ligand was found to be flawlessly overlapping over its crystallized bioactive conformation. Also, the reference ligands were found to have similar binding interactions as present in the crystallized macromolecular complex of all the target proteins used in the current study. The resemblance in the binding pattern with a similar type of chemical interaction shown by the docked confirmation of the reference ligand with respect to their bioactive crystallized conformation was clearly an indication that the utilized docking tool was exactly simulating the physiological process during a drug–receptor interaction (Mujwar, 2021). For postsuccessful validation, Auto Dock Vina was used in molecular docking of the selected carotenoids against various structural and nonstructural proteins of SARS‐CoV‐2. The receptors are Mpro, ADRP, replication complex, helicase, and spike protein including the mutant S proteins for the variants of concern of SARS‐CoV‐2. Carotenoids were, respectively, docked against each receptor and their binding energies are listed in Table 1. For the main protease (6LU7), crocin had the strongest binding affinity (−8.0 kcal/mol), it interacted with His41, Tyr54, Asn142, Ser144, His164, Glu166, Asp187, and Gln189 through hydrogen bonds as well as with Tyr118 and Leu141 via alkyl and Pi‐alkyl interactions (Figure S1). Additionally, alpha‐carotene and fucoxanthin also bound relatively strongly with the binding energies of −7.6 kcal/mol (Table 1).
TABLE 1.
Binding energies of ligands docked against receptors
| PubChem ID | Compounds | 6LU7 | 6W02 | 6ZSL | 7BV2 | 6M0J | 6M0JR | 6M0JSA | 6M0JBr | 6M0JUK |
|---|---|---|---|---|---|---|---|---|---|---|
| 11433225 | Cucurbitaxanthin A | −7.5 | −9.3 | −9.7 | −9.2 | −7.9 | −8.0 | −7.4 | −7.7 | −7.1 |
| 16061212 | Cucurbitaxanthin B | −7.3 | −9.1 | −9.9 | −9.1 | −7.6 | −7.7 | −7.2 | −7.0 | −8.1 |
| 4369188 | Alpha‐carotene | −7.6 | −9.6 | −8.3 | −8.7 | −7.8 | −7.7 | −7.3 | −8.1 | −7.1 |
| 446925 | Lycopene | −6.0 | −7.3 | −7.5 | −7.0 | −6.4 | −6.8 | −6.6 | −6.6 | −6.4 |
| 448438 | Violaxanthin | −7.4 | −9.0 | −9.5 | −8.7 | −7.2 | −6.9 | −6.7 | −7.4 | −7.1 |
| 5280489 | Beta‐carotene | −7.3 | −9.0 | −8.9 | −8.4 | −7.1 | −7.4 | −8.0 | −7.7 | −8.0 |
| 5280784 | Phytoene | −5.2 | −7.5 | −6.0 | −5.9 | −5.6 | −5.7 | −5.6 | −6.0 | −4.9 |
| 5280899 | Zeaxanthin | −7.2 | −7.9 | −9.3 | −8.6 | −7.3 | −7.7 | −7.0 | −7.0 | −6.8 |
| 5281224 | Astaxanthin | −7.3 | −7.3 | −9.7 | −8.9 | −7.4 | −7.3 | −7.0 | −7.9 | −7.5 |
| 5281226 | Bixin | −5.8 | −7.1 | −6.6 | −7.5 | −6.3 | −5.6 | −6.1 | −5.9 | −6.2 |
| 5281227 | Canthaxanthin | −6.9 | −8.1 | −9.2 | −8.7 | −7.4 | −7.5 | −7.4 | −7.4 | −7.2 |
| 5281228 | Capsanthin | −7.0 | −9.4 | −9.4 | −9.1 | −6.7 | −6.9 | −7.0 | −6.7 | −6.7 |
| 5281229 | Capsarubin | −7.3 | −9.1 | −9.0 | −9.1 | −6.5 | −7.1 | −6.7 | −6.4 | −6.7 |
| 5281232 | Crocetin | −5.2 | −7.5 | −6.5 | −6.8 | −6.0 | −5.8 | −5.7 | −6.0 | −5.5 |
| 5281233 | Crocin | −8.0 | −8.2 | −9.5 | −10.5 | −6.7 | −6.8 | −7.6 | −6.3 | −6.4 |
| 5281235 | Beta‐cryptoxanthin | −7.0 | −9.2 | −8.6 | −8.7 | −6.6 | −7.5 | −7.2 | −7.1 | −7.7 |
| 5281239 | Fucoxanthin | −7.6 | −8.5 | −8.5 | −9.0 | −7.7 | −7.1 | −7.0 | −7.1 | −7.3 |
| 5281243 | Lutein | −7.3 | −8.0 | −9.3 | −8.5 | −6.9 | −7.1 | −7.1 | −6.8 | −6.7 |
| 5281247 | Neoxanthin | −7.3 | −9.4 | −9.5 | −8.9 | −7.6 | −7.3 | −6.8 | −7.2 | −7.0 |
| 6436722 | Phytofluene | −4.8 | −7.5 | −6.2 | −6.5 | −4.1 | −5.5 | −5.3 | −4.4 | −5.1 |
| 492405 | Favipiravir | −5.7 | −5.9 | −5.8 | −6.7 | −5.3 | −5.4 | −5.3 | −5.5 | −5.3 |
| 6321424 | Ivermectin | −7.8 | −7.3 | −9.9 | −10.6 | −7.7 | −7.5 | −8.2 | −7.2 | −8.2 |
| 121304016 | Remdesivir | −7.8 | −8.0 | −7.9 | −8.5 | −6.8 | −6.9 | −6.9 | −7.1 | −6.9 |
| 155903259 | Paxlovid | −7.5 | −9.0 | −7.8 | −9.1 | −6.4 | −6.7 | −6.4 | −5.8 | −6.1 |
| 145996610 | Molnupiravir | −6.7 | −7.7 | −7.6 | −9.9 | −6.0 | −6.0 | −6.0 | −6.0 | −5.8 |
| Native ligands | −7.0 | −8.4 | −7.0 | −8.5 | NA | NA | NA | NA | NA |
Note: Native ligand for 6LU7: N3; Native ligand for 6W02: ADP ribose; Native ligand for 6ZSL: Z2327226104; Native ligand for 7BV2: Remdesivir. NA: Not applicable for S protein because its native ligand is human ACE2.
With the molecular docking scores of less than −7 kcal/mol, ADRP and its ligands revealed that all the carotenoids had strong binding affinities toward ADRP (W. Wu et al., 2021). Particularly, alpha‐carotene, capsanthin, and neoxanthin showed the lowest binding energies of −9.6, 9.4, and − 9.4 kcal/mol, respectively (Table 1). Crocin also binds to ADRP with a good binding affinity (−8.2 kcal/mol). Alpha‐carotene interacts with the certain amino acid residues (Asn40, Gly46, Gly47, Gly48, Pro125, Ser128, Gly130, Phe156, Asp157, and Leu160) through van der Waals interactions and with the residues (Ala38, Tyr42, Lys44, Val49, Ala50, Leu126, Ala129, Ile131, Phe132, and Val155) through the alkyl and Pi‐alkyl interactions (Figure S2). The majority of van der Waals interactions also exist on the ADRP interacting with its original substrate (ADP‐ribose), indicating that these interacting residues may play important role in the inhibition of ADRP (Michalska et al., 2020). Helicase (6ZSL) is another target protein and its docking results showed strong binding energy scores for some carotenoids including cucurbitaxanthin B and crocin. The interacting residues of helicase with cucurbitaxanthin B are TRP167 and ARG560 through conventional hydrogen bonds; Arg173, Pro174, Asn177, Asn179, Asp207, Leu405, Ala407, Arg409, Leu417, Asp483, Ser485, Ser486, Hiss554, and Asn557 through van der Waals interactions; Leu165, Pro175, Leu176, Tyr180, Val209, Pro406, Pro408, Leu412, and Val484 through alkyl and Pi‐alkyl interactions (Figure S3). Moreover, crocin binds strongly with RdRp (7BV2) including replication complex, which had the lowest binding affinity of −10.5 kcal/mol. Crocin interacts with replication complex through van der Waals interaction, conventional hydrogen bond, carbon‐hydrogen bond, and Pi‐alkyl interactions (Figure S4). Crocin interacts with the RNA‐binding site of the replication complex, which implies its potential antiviral activity.
For the S protein, we conducted molecular docking experiments with the S protein of original SARS‐CoV‐2 as well as Brazilian, South African, UK, and Indian variants. Crocin had relatively strong binding affinities toward S protein and its mutants, particularly the mutant S protein for the South African variant. Additionally, some other carotenoids bind to S protein, and its mutants are stronger than crocin. For instance, cucurbitaxanthin A showed the lowest binding scores for S protein and its mutant for the Indian variant, whereas cucurbitaxanthin B had the strongest binding energy with mutant S protein for the UK variant. Also, alpha‐carotene and beta‐carotene possessed the lowest binding affinities to the mutant S proteins for Brazilian and South African variants, respectively (Table 1). The interacting residues of S protein and its mutants with the strongly bound ligands are shown in Figures S5–S9.
The molecular docking of current therapeutic agents such as remdesivir, favipiravir, molnupiravir, paxlovid, and ivermectin was also conducted against all the receptors (Table 1). It is obvious that the docking scores of carotenoids were quite comparable to the ones of current therapeutics. In some cases, carotenoids outperformed the current drug molecules based on molecular docking experiments. For example, alpha‐carotene had the strongest binding affinity (−9.6 kcal/mol) toward ADRP, whereas remdesivir and molnupiravir had the scores of −8.0 and − 7.7 kcal/mol, respectively. Similarly, ivermectin is bound to the replication complex with the lowest binding energy of −10.6 kcal/mol. One of the chosen carotenoids, crocin also had the strongest binding affinity with a score of −10.5 kcal/mol, indicating the potential of carotenoids in the treatment and prevention of SARS‐CoV‐2.
3.2. Molecular dynamic simulations
To validate the molecular docking results, we performed MD simulations with the best docking models. We conducted MD simulations for crocin since it had strong binding energy scores for the majority of target proteins. For S protein and its mutants, MD simulations were only performed for the mutant of the South African variant because its binding affinity was the lowest among all the S proteins.
MD simulation for Mpro of SARS‐CoV‐2 with crocin revealed that the RMSD value was well within the acceptable limits during the simulation process with some initial conformational changes up to 2.7 Å during initial 20 ns (Figure 1a). The complexed ligand adjusted itself in the binding cavity of the viral Mpro and fluctuated a bit for initial 20 ns and then made a major conformational change to stabilize itself at 40 ns. The RMSD value of crocin was also lied within the acceptable range of 3 Å during the whole simulation of 100 ns. Except some terminal residues, the root mean square fluctuation (RMSF) value of the macromolecular residues was within the acceptable limit of 3 Å with an average value of 1 Å during the whole simulation of 100 ns. The macromolecular structure of viral Mpro had 40.79% of secondary structure elements (SSE) in total, including 16.81% α‐helix and 23.98% β‐strands, remained conserved throughout the simulation process. The protein–ligand contacts between viral Mpro and crocin (Figure 2a) remained stable during the simulation process and the number of contacts between the ligand and the macromolecule fluctuated between 8 and 24 contacts with an average value of 16 contacts during the 100 ns simulation. The interacting residues, type of interaction, and their fractions for the viral Mpro with crocin are given in Table 2 and Figure 3a. In particular, Glu166, His172, and Gln189 residues were previously reported to be among the important interacting residues in Mpro with N3 inhibitor (Jin et al., 2020), indicating the potential of crocin for the inhibition of Mpro.
FIGURE 1.
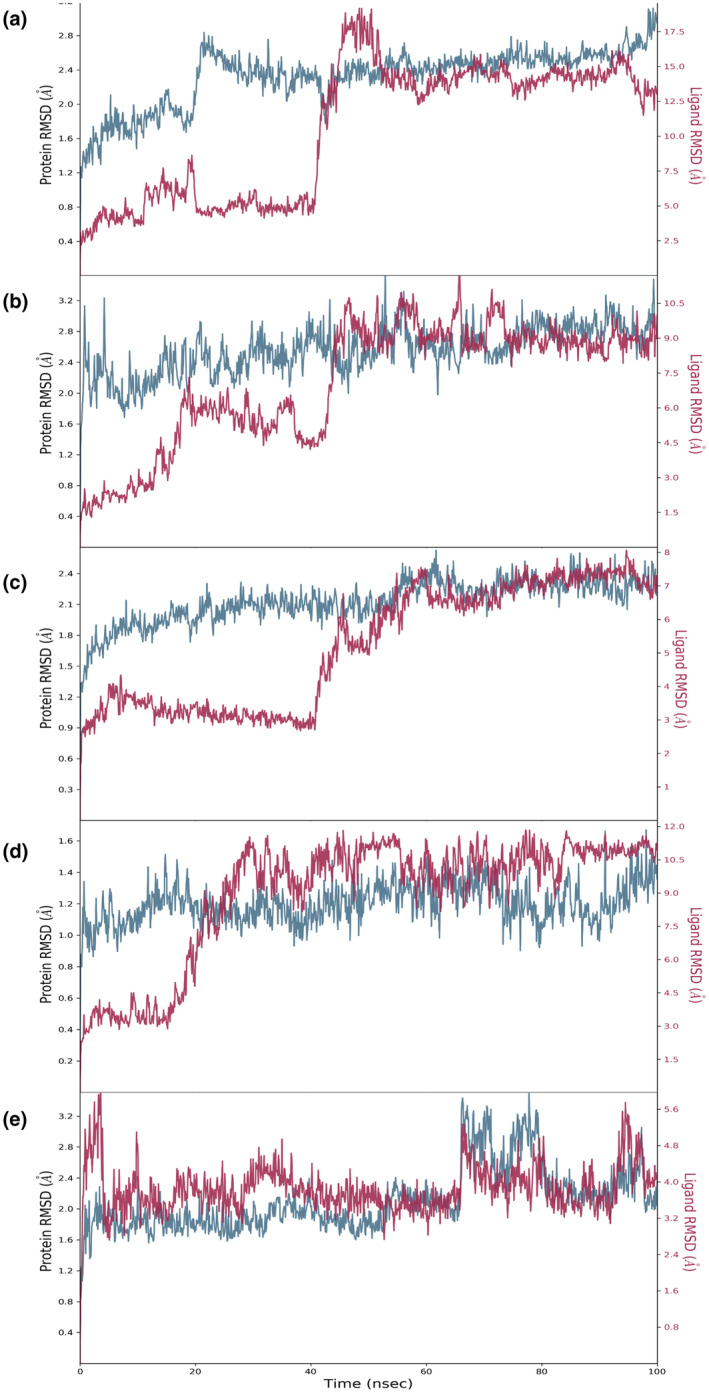
The protein–ligand RMSD value observed after performing 100 ns MD simulation for all the macromolecular complexes of crocin with (a) Mpro, (b) Helicase, (c) Replication complex, (d) ADRP, and (e) B.1.351 variant of S protein. Lines with blue color indicate the RMSD profiles for the receptors complexed with crocin, whereas lines with red color show the RMSD profiles for crocin complexed with receptors
FIGURE 2.
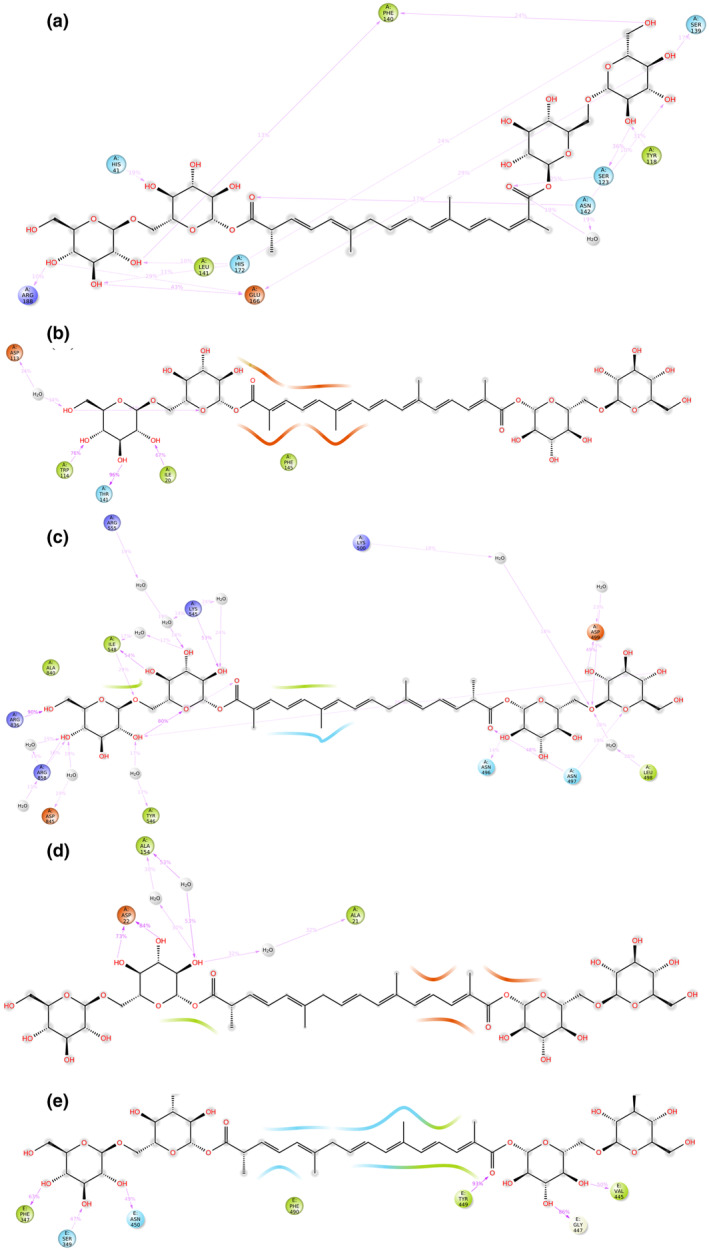
The protein–ligand contacts observed after performing 100 ns MD simulation for all the macromolecular complexes of crocin with (a) Mpro, (b) Helicase, (c) Replication complex, (d) ADRP, and (e) B.1.351 variant of S protein. The macromolecular residues demonstrated in green color have hydrophobic interaction, whereas sky blue‐colored residues have polar interactions with the complexed ligand molecule. The orange‐colored residues are negatively charged, whereas the dark blue residues are positively charged
TABLE 2.
The protein–ligand contacts observed after performing 100 ns MD simulation for all the macromolecular complexes of crocin with the drug target receptors
| Viral drug target | PDB ID | Interacting residues |
|---|---|---|
| Main Protease | 6LU7 | Ala116, Ser123, Ser139, Phe140, Leu141, Asn142, Glu166, His172, Gln189 |
| Helicase | 6ZSL | Ala18, Ile20, Cys112, Asp113, Trp114, Thr141, Phe145, Gly415, His482, Asp483, Val484, Ser485, Tyr515, Thr552, His554 |
| Replication complex | 7BV2 | Asn497, Asp499, Lys500, Lys545, Ile548, Arg836, Asp845, Arg858 |
| ADRP | 6W02 | Asp22, Lys44, Gly48, Ala154, Phe156 |
| Spike Protein (B.1.351) | 6M0JSA | Arg346, Phe347, Ser349, Tyr351, Leu441, Lys444, Val445, Gly447, Asn448, Tyr449, Asn450, Phe490 |
FIGURE 3.
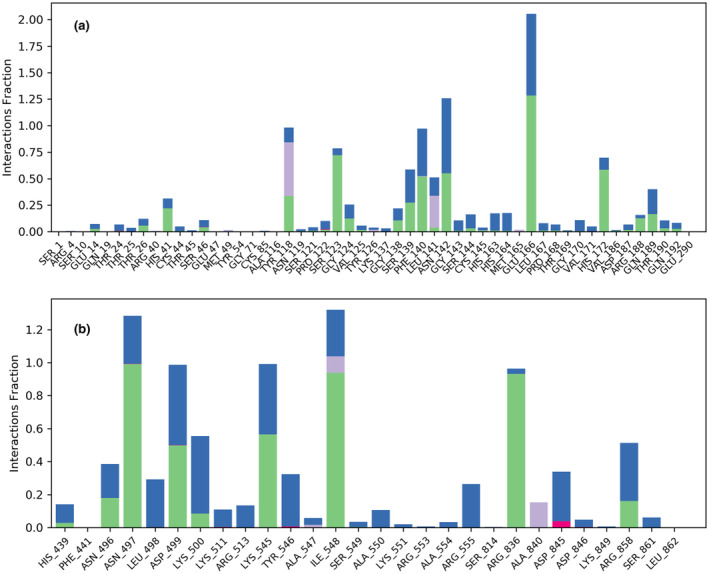
Type of interaction of crocin with the key interacting amino acid residues of (a) Mpro and (b) replication complex of SARS‐CoV‐2. The green‐colored and blue‐colored bars refer to the presence of H‐bonding and the presence of water bridges, respectively. Purple‐colored and dark pink‐colored bars demonstrate the presence of hydrophobic interactions and the presence of ionic interactions between the macromolecular residues and the complexed ligand, respectively
The RMSD value for helicase was also well within the acceptable limits during the simulation process with some initial conformational changes up to 3.2 Å, followed by its fluctuation between 2.0 and 3.2 Å throughout the simulation process (Figure 1b). The complexed ligand adjusted itself in the binding cavity of the viral helicase and fluctuated a bit for initial 40 ns, and then made a major conformational change to reach the final stabilization within the macromolecular binding cavity. The RMSD value of crocin was also well within the acceptable range of 3.2 Å during the whole simulation of 100 ns. Except for few macromolecular residues, the RMSF value of the target macromolecule fluctuated from 1.2 to 2.4 Å. The macromolecular structure of helicase remained conserved during the whole simulation time. The protein–ligand contacts between helicase and crocin (Figure 2b) also remained stable during the simulation process and the number of contacts between the ligand and the macromolecule fluctuated between 6 and 22 contacts with an average value of 12 contacts. The interacting residues, type of interaction, and their fractions for helicase interacted with crocin are given in Table 2 and Figure 4a. Among these residues, Tyr515 and Thr552 were listed to form polar contacts with tested ligands (Newman et al., 2021). These contacts including Ser486 and Asn516 are similar to the contacts formed by two successive RNA phosphates in the UPF1 helicase–RNA complex (Chakrabarti et al., 2011; Newman et al., 2021). Therefore, crocin could be an attractive molecule as a potential RNA‐competitive inhibitor.
FIGURE 4.
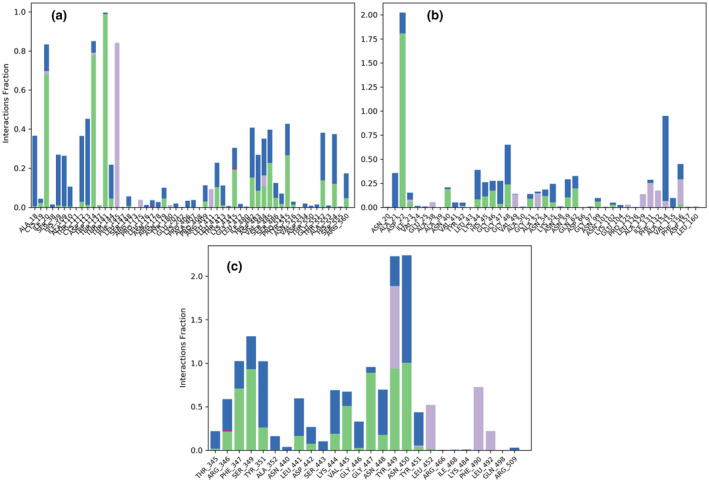
Type of interaction of crocin with the key interacting amino acid residues of (a) Helicase, (b) ADRP, and (c) RBD of S protein of SARS‐CoV‐2 for Beta variant. The green‐colored and blue‐colored bars refer to the presence of H‐bonding and the presence of water bridges, respectively. Purple‐colored and dark pink‐colored bars demonstrate the presence of hydrophobic interactions and the presence of ionic interactions between the macromolecular residues and the complexed ligand, respectively
The replication complex is an important target in the drug development for COVID‐19 due to its crucial role in the replication of the virus (Gil et al., 2020; Zhu et al., 2020). The MD simulation for the replication complex docked with crocin showed that the RMSD value for the receptor was acceptable during the simulation process with some initial conformational changes up to 2.4 Å, followed by its fluctuation between 1.5 and 2.4 Å for the rest of the simulation process. The complexed ligand fluctuated a bit for initial 40 ns, followed by adjusting itself in the binding cavity of the replication complex, and then a major conformational change led to the final stabilization within the macromolecular binding cavity. The RMSD value for crocin was within the range of 2.4 Å during the whole simulation of 100 ns (Figure 1c). Except a very few receptor residues, the RMSF value of the receptor fluctuated from 0.5 to 2.4 Å during the whole simulation of 100 ns. The macromolecular structure had 46.44% of SSE in total including 11.99% α‐helix and 34.85% β‐strands, which remained conserved during the whole simulation time. The protein–ligand contacts between receptor and ligand (Figure 2c) also remained stable during the simulation process. The number of contacts between the ligand and the macromolecule fluctuated between 6 and 24 contacts with an average value of 12 contacts during the 100 ns simulation. The interacting residues, type of interaction, and their fractions for the viral replication with crocin are given in Table 2 and Figure 3b. The list of interacting residues of the replication complex with crocin includes Asn497, Asp499, Lys500, Lys545, Ile548, Arg836, Asp845, Arg858, some of which were included in the interacting residues of RdRP of SARS‐CoV‐2 complexed with remdesivir. Particularly, Lys545 and Arg555 are the residues interacting with the primer strand RNA and thus stabilize incoming nucleotide in the correct position for replication (Yin et al., 2020). Besides remdesivir, EIDD‐2801 as a nucleotide analog was reported to efficiently inhibit SARS‐CoV‐2 replication in cell‐based assays and its 3–10 times higher potency was reported to be due to the extra hydrogen bond of the N4 hydroxyl group of EIDD‐2801 with Lys545 residue (Sheahan et al., 2020). Therefore, crocin interacting with this key residue holds significant potential in the treatment and prophylaxis of SARS‐CoV‐2.
ADRP of SARS‐CoV‐2 has 165 amino acids, and a total of 2506 atoms including 1248 heavy atoms. MD simulation of ADRP with crocin showed that its RMSD was highly stable with very little fluctuation, which was lying within the acceptable range of 0.8–1.4 Å during the whole simulation process of 100 ns. The complexed ligand adjusted itself in the binding cavity of the viral ADRP by changing its conformation at 20 Å, leading to the final stabilization within the macromolecular binding cavity till the end of the simulation process (Figure 1d). Except a very few macromolecular residues, the RMSF value of the target receptor fluctuated from 0.4 to 1.4 Å during the whole simulation process of 100 ns. The macromolecular structure remained conserved during the whole simulation time. The protein–ligand contacts between viral ADRP and crocin (Figure 2d) remained stable and the number of contacts between the ligand and the macromolecule fluctuated between 5 and 15 contacts with an average value of 10 contacts during the 100 ns simulation. The interacting residues, type of interaction, and their fractions for the viral ADRP with crocin are given in Table 2 and Figure 4b. The interacting residues of the viral ADRP with crocin in the MD simulation were Asp22, Lys44, Gly48, Ala154, and Phe156. ADP‐ribose is the natural substrate of ADRP and was found to be sandwiched between α2 and β7 of ADRP, in which Asp22, Ala154, and Phe156 were involved in the interaction of ADP‐ribose with ADRP through hydrogen bonds and hydrophobic interactions (Michalska et al., 2020). This indicated that crocin might be a good candidate molecule in the inhibition of ADRP due to its interactions with the important residues of ADRP similar to the original ligand.
S protein of the B.1.351 variant of SARS‐CoV‐2 emerged in South Africa was found to be highly contagious and was characterized by triple mutations, that is, E484K, N501Y, and K417N. The MD simulation of mutant S protein complexed with crocin was conducted, and the RMSD value was found to be well within the acceptable limits during the simulation process with some initial conformational changes up to 3.6 Å, followed by its fluctuation between 1.6 and 3.2 Å. The complexed ligand was constantly stabilized in the binding cavity of the RBD of mutant S protein without any fluctuation or major conformational change. The RMSD value of crocin was also within the acceptable range of 3.2–5.4 Å throughout the simulation process (Figure 1e). Except some of the receptor residues, the RMSF value of the receptor fluctuated from 0.6 to 1.8 Å during the whole simulation process of 100 ns. The macromolecular structure of RBD remained conserved during the whole simulation time. The protein–ligand contacts between RBD of S protein and crocin (Figure 2e) remained stable during the simulation process and the number of contacts between the ligand and the macromolecule fluctuated between 10 and 20 contacts with an average value of 16 contacts during the 100 ns simulation process. The interacting residues, type of interaction, and their fractions for the viral RBD of S protein for Beta variant with crocin are given in Table 2 and Figure 4c.
3.3. Postdynamics complex stability analysis
The MD simulation of the macromolecular complex of crocin complexed with viral Mpro revealed that both crocin and its macromolecular target had major conformational changes as the RMSD value fluctuates up to 2.8 Å till the initial 40 ns of the simulation. But after taking certain moves, the ligand stabilized within the macromolecular cavity, resulting in the stabilization of the whole complex in the remaining part of the simulation process. The MD simulation of the macromolecular complex of crocin complexed with viral ADRP suggested that only the macromolecular target underwent slight conformational changes as the RMSD value fluctuated to 1.4 Å in the initial 20 ns of the simulation. But afterward, the complex remained stable during the rest of the simulation process. The MD simulation of the macromolecular complex of crocin complexed with helicase revealed that only the macromolecular target underwent conformational changes as the RMSD value fluctuated up to 2.4 Å at the initial 40 ns of the simulation by gradually achieving stabilized conformation. The MD simulation of the macromolecular complex of crocin complexed with viral RdRp revealed that both the macromolecular target underwent initial conformational changes within the acceptable RMSD fluctuation between 0.9 and 2.1 Å till the initial 45 ns of the simulation. But afterward, both the ligand and the macromolecular target receptor perfectly stabilized in the remaining part of the simulation process. MD simulation of the macromolecular complex of crocin complexed with the South Africa variant of the mutated S protein showed that despite slight conformational change by the macromolecular target till 0–4 ns, both the crocin and the mutant S protein remained stable without the presence of any major conformational changes during the 100 ns simulation process.
3.4. Carotenoids and their biological activities against SARS‐CoV‐2
There are more than 750 carotenoids, which are widely found in fruits, vegetables, and algae. They exhibit various biological activities, such as antioxidant, anticancer, anti‐inflammatory, and immunity booster properties (Fidan & Zhan, 2019). Some carotenoids serve as precursors for the synthesis of vitamin A. Nevertheless, human beings cannot biosynthesize carotenoids, which makes them a group necessary for diet supplementation (Olson & Krinsky, 1995). Carotenoids have also been reported to hold significant potential in the development of effective therapeutics for the treatment and prophylaxis of COVID‐19 (Fakhri et al., 2021). They attracted special attention due to their involvement in various steps of the viral life cycle and host proteins (Santoyo et al., 2011). For instance, as strong antioxidant agents, carotenoids play important roles in viral infections due to cytokine storms and thus oxidative stress. To alleviate the oxidative damage, vitamin C as an antioxidant molecule was tested in COVID‐19 patients and found to improve pulmonary function and decreased the risk of acute respiratory distress syndrome (Liu et al., 2020). Vitamin E was also a candidate for the alleviation of oxidative damage and inflammation induced by SARS‐COV‐2 (Soto et al., 2020). On the other hand, astaxanthin is an important dietary supplement with a strong antioxidant activity than vitamins C and E and might be a promising carotenoid candidate for the treatment of COVID‐19 complications (Fakhri et al., 2020). Additionally, carotenoids such as β‐Carotene and lycopene were shown to have good anti‐inflammatory properties with their reactive oxygen species (ROS)‐scavenging activities. This indicated their potential use in the prophylaxis and treatment of COVID‐19 (Kawata et al., 2018). Overall, due to the aforementioned biological activities of carotenoids, they have been attractive dietary supplement molecules as an alternative strategy for the prevention and/or treatment of SARS‐CoV‐2 (Lammi & Arnoldi, 2021).
A carotenoid, capsanthin, was reported to be potentially an effective inhibitor of Mpro of SARS‐CoV‐2 through an in silico study (Ibrahim et al., 2020). Our in silico experiments revealed that a few carotenoids are particularly attractive due to targeting multiple druggable targets of SARS‐CoV‐2. Cucurbitaxanthin A and B were isolated from pumpkin Cucurbita maxima and paprika Capsicum annuum (Hornero‐Méndez & Mínguez‐Mosquera, 1998; Matsuno et al., 1986). To our knowledge, there is few studies on their potential antiviral activities. In molecular docking studies, cucurbitaxanthin A and B showed quite strong affinities toward some target proteins, such as ADRP, helicase, replication complex, and spike protein including mutant spike protein for Delta variant (Table 1). Furthermore, alpha‐ and beta‐carotenes indicated strong binding energies against multiple target proteins, which were reported to be potential inhibitors of the main peptidase of SARS‐CoV (Costa et al., 2021). These compounds are commonly found in various foods such as carrot, sweet potato, spinach, pumpkin, etc. (Costa et al., 2021). Particularly, beta‐carotene as pro‐vitamin A was shown to reduce the overall severity and consequent fatalities associated with HIV, malaria, and measles. Thus, it was considered to be an important micronutrient to support the host immunocompetence and reduce the oxidative stress in COVID‐19 (Stipp, 2020; Trujillo‐Mayol et al., 2021). In addition to these biological benefits for COVID‐19 patients, our study proves to some extent that it might also have an inhibitory effect on the multiple drug targets of SARS‐CoV‐2. Another attractive carotenoid was beta‐cryptoxanthin, which is commonly found in egg yolk, butter, apples, and papaya. It was found to have good binding affinities toward multiple SARS‐CoV‐2 receptors and reported antiviral potential due to its tight affinity toward Mpro and PLpro (Karpiński et al., 2021; Wang et al., 2020). Beta‐cryptoxanthin has also shown to be a potential antiviral agent because it is bound to S protein in the in silico studies, leading to the prevention of viral entry to human cells (Umashankar et al., 2021). These preliminary in silico results are required to be confirmed through in vitro and in vivo experiments.
Crocin as one of the main components of saffron had the strongest binding affinity toward the replication complex of SARS‐CoV‐2. It also bound strongly toward multiple drug targets including Mpro, ADRP, helicase, and mutant S protein of Beta variant (Table 1). Its stability in these receptors was also confirmed with MD simulations. Interestingly, saffron has been suggested to be used as a drug supplement for the treatment of SARS‐CoV‐2 in the recent reviews and a commentary (Ghasemnejad‐Berenji, 2021; Husaini et al., 2021; Mentis et al., 2021). Furthermore, saffron was reported to exhibit antitumor and antiviral properties. For example, the anti‐HSV‐1 and anti‐HIV‐1 properties of saffron and its main components, including crocin and picrocrocin, were investigated and found that the aqueous saffron extract possessed a mild antiviral activity; whereas crocin and picrocrocin exhibited effective antiviral activity (Soleymani et al., 2018). Through in silico approaches, saffron crocetin, for instance, had a high affinity for both Mpro and S protein (Aanouz et al., 2021). Crocin was also tested in silico against S protein and showed strong hydrogen bonding with RBD of SARS‐CoV‐2 (Stalin et al., 2021). In this study, we simulated crocin against multiple drug targets of SARS‐CoV‐2 and found that it binds to multiple receptors with strong binding affinities. Molecular docking indicated that the binding affinities of crocin toward replication complex, helicase, ADRP, and Mpro were much stronger than toward RDB of S protein. This clearly renders crocin a promising dietary supplement in the treatment and prophylaxis of SARS‐CoV‐2. The ADME analysis for crocin was also conducted using SWISSADME online server. Crocin has a molecular weight of 976.96 g/mol, the lipophilicity of 3.06, 24 hydrogen acceptors, and 14 hydrogen donors, indicating that it violates three criteria of Lupinski's rule of five. Yet, it was predicted to be water‐soluble, which is an important aspect of the bioavailability of drug molecules (Savjani et al., 2012). Also, crocin was not an inhibitor for pharmacokinetically important cytochrome P450 oxidases. Even though ADME analysis might not appear to be promising in terms of druggability criteria, crocin has indeed been tested in animal and human studies without any medical issues. For example, crocin showed to provide genoprotective properties for DNA damage and to improve glycemic control and insulin resistance in patients with type 2 diabetes (Behrouz et al., 2020; Hosseinzadeh et al., 2008). Overall, our in silico findings needs to be in fact supported by in vitro and in vivo studies to draw a solid conclusion. Since some structurally similar apocarotenoids, including bixin and β‐apo‐8′carotenoic acid, were tested against SARS‐CoV‐2 in Vero E6 cells and had remarkable antiviral activities (Bereczki et al., 2021), crocin is potential to be used in the treatment of anti‐SARS‐CoV‐2.
4. CONCLUSION
The molecular docking studies of the 20 most widely used food‐derived carotenoids against multiple drug targets of SARS‐CoV‐2 revealed that carotenoids have significant inhibitory effects on different target proteins. Some of them showed strong binding affinities toward multiple targets, including replication complex, helicase, ADRP, Mpro, and S protein. Crocin, particularly, exhibited quite low binding energies to multiple receptors and had the most stable binding with the replication complex of SARS‐CoV‐2. The molecular docking experiments were further validated with MD simulations, which confirmed the stability and affinity of crocin against the target proteins. The MD simulations also indicated that crocin interacts with some of the key amino acid residues of the receptors. Therefore, crocin can be considered an effective supplementary therapeutic, which targets multiple proteins for the treatment and prophylaxis of COVID‐19. Carotenoids have already been acknowledged for their health‐promoting properties in the treatment of various diseases including COVID‐19. Our study reveals that crocin, a water‐soluble carotenoid, can provide further benefits with its promising antiviral activity. The in silico study will be further investigated through wet‐lab experiments to draw a more solid conclusion.
CONFLICT OF INTEREST
No potential conflict of interest was reported by the authors.
AUTHOR CONTRIBUTIONS
Somdutt Mujwar: Data curation; methodology; software; visualization; writing – original draft; writing – review and editing. Lei Sun: Conceptualization; writing – original draft; writing – review and editing. Ozkan Fidan: Conceptualization; data curation; methodology; visualization; writing – original draft; writing – review and editing.
Supporting information
Figure S1‐S9
Mujwar, S. , Sun, L. & Fidan, O. (2022). In silico evaluation of food‐derived carotenoids against SARS‐CoV‐2 drug targets: Crocin is a promising dietary supplement candidate for COVID‐19. Journal of Food Biochemistry, 00, e14219. 10.1111/jfbc.14219
Funding information
The current study was not granted by any funding agencies.
Contributor Information
Somdutt Mujwar, Email: somduttmujwar@gmail.com.
Lei Sun, Email: nj.sunleith@outlook.com.
Ozkan Fidan, Email: ozkan.fidan@agu.edu.tr.
DATA AVAILABILITY STATEMENT
All data were included in the original manuscript and the supplementary data.
REFERENCES
- Aanouz, I. , Belhassan, A. , El‐Khatabi, K. , Lakhlifi, T. , El‐Ldrissi, M. , & Bouachrine, M. (2021). Moroccan Medicinal plants as inhibitors against SARS‐CoV‐2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 39(8), 2971–2979. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barciszewska, A. M. (2021). Elucidating of oxidative distress in COVID‐19 and methods of its prevention. Chemico‐Biological Interactions, 344, 109501. 10.1016/j.cbi.2021.109501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrouz, V. , Dastkhosh, A. , Hedayati, M. , Sedaghat, M. , Sharafkhah, M. , & Sohrab, G. (2020). The effect of crocin supplementation on glycemic control, insulin resistance and active AMPK levels in patients with type 2 diabetes: A pilot study. Diabetology & Metabolic Syndrome, 12(1), 59. 10.1186/s13098-020-00568-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereczki, I. , Papp, H. , Kuczmog, A. , Madai, M. , Nagy, V. , Agócs, A. , Batta, G. , Milánkovits, M. , Ostorházi, E. , Mitrović, A. , Kos, J. , Zsigmond, Á. , Hajdú, I. , Lőrincz, Z. , Bajusz, D. , Keserű, G. M. , Hodek, J. , Weber, J. , Jakab, F. , … Borbás, A. (2021). Natural apocarotenoids and their synthetic glycopeptide conjugates inhibit SARS‐CoV‐2 replication. Pharmaceuticals (Basel), 14(11), 1111. 10.3390/ph14111111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm, V. , Lietz, G. , Olmedilla‐Alonso, B. , Phelan, D. , Reboul, E. , Bánati, D. , Borel, P. , Corte‐Real, J. , de Lera, A. R. , Desmarchelier, C. , Dulinska‐Litewka, J. , Landrier, J. F. , Milisav, I. , Nolan, J. , Porrini, M. , Riso, P. , Roob, J. M. , Valanou, E. , Wawrzyniak, A. , … Bohn, T. (2021). From carotenoid intake to carotenoid blood and tissue concentrations—Implications for dietary intake recommendations. Nutrition Reviews, 79(5), 544–573. 10.1093/nutrit/nuaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boozari, M. , & Hosseinzadeh, H. (2021). Natural products for COVID‐19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phytotherapy Research, 35(2), 864–876. 10.1002/ptr.6873 [DOI] [PubMed] [Google Scholar]
- Chakrabarti, S. , Jayachandran, U. , Bonneau, F. , Fiorini, F. , Basquin, C. , Domcke, S. , Le Hir, H. , & Conti, E. (2011). Molecular mechanisms for the RNA‐dependent ATPase activity of Upf1 and its regulation by Upf2. Molecular Cell, 41(6), 693–703. 10.1016/j.molcel.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Costa, A. N. , de Sa, E. R. A. , Bezerra, R. D. S. , Souza, J. L. , & Lima, F. (2021). Constituents of buriti oil (Mauritia flexuosa L.) like inhibitors of the SARS‐Coronavirus main peptidase: An investigation by docking and molecular dynamics. Journal of Biomolecular Structure and Dynamics, 39(13), 4610–4617. 10.1080/07391102.2020.1778538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, M. , Mohamed, A. S. , Prescott, R. A. , Valero‐Jimenez, A. M. , Desvignes, L. , O'Connor, R. , Steppan, C. , Devlin, J. C. , Ivanova, E. , Herrera, A. , Schinlever, A. , Loose, P. , Ruggles, K. , Koralov, S. B. , Anderson, A. S. , Binder, J. , & Dittmann, M. (2021). A comparative analysis of SARS‐CoV‐2 antivirals characterizes 3CL(pro) inhibitor PF‐00835231 as a potential new treatment for COVID‐19. Journal of Virology, 95(10), e01819‐20. 10.1128/JVI.01819-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath, P. , Debnath, B. , Bhaumik, S. , & Debnath, S. (2020). In silico identification of potential inhibitors of ADP‐ribose phosphatase of SARS‐CoV‐2 nsP3 by combining E‐pharmacophore‐ and receptor‐based virtual screening of database. ChemistrySelect, 5(30), 9388–9398. 10.1002/slct.202001419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, L. , Guo, X. , Cong, Y. , Feng, G. , Li, Y. , & Zhang, J. Z. H. (2019). Accelerated molecular dynamics simulation for helical proteins folding in explicit water. Frontiers in Chemistry, 7, 540. 10.3389/fchem.2019.00540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, N. K. , Mazumdar, K. , & Gordy, J. T. (2020). The nucleocapsid protein of SARS‐CoV‐2: A target for vaccine development. Journal of Virology, 94(13), e00647‐20. 10.1128/JVI.00647-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggersdorfer, M. , & Wyss, A. (2018). Carotenoids in human nutrition and health. Archives of Biochemistry and Biophysics, 652, 18–26. 10.1016/j.abb.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Fakhri, S. , Nouri, Z. , Moradi, S. Z. , Akkol, E. K. , Piri, S. , Sobarzo‐Sanchez, E. , Farzaei, M. H. , & Echeverria, J. (2021). Targeting multiple signal transduction pathways of SARS‐CoV‐2: Approaches to COVID‐19 therapeutic candidates. Molecules, 26(10), 2917. 10.3390/molecules26102917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhri, S. , Nouri, Z. , Moradi, S. Z. , & Farzaei, M. H. (2020). Astaxanthin, COVID‐19 and immune response: Focus on oxidative stress, apoptosis and autophagy. Phytotherapy Research, 34(11), 2790–2792. 10.1002/ptr.6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias, A. B. , Candiotto, G. , Siragusa, L. , Goracci, L. , Cruciani, G. , Oliveira, E. R. , & Horta, B. A. (2021). Targeting Nsp9 as an anti‐SARS‐CoV‐2 strategy. New Journal of Chemistry, 45(2), 522–525. 10.1039/D0NJ04909C [DOI] [Google Scholar]
- Fidan, O. , & Zhan, J. (2019). Discovery and engineering of an endophytic Pseudomonas strain from Taxus chinensis for efficient production of zeaxanthin diglucoside. Journal of Biological Engineering, 13(1), 66. 10.1186/s13036-019-0196-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadlage, M. J. , & Denison, M. R. (2010). Exchange of the coronavirus replicase polyprotein cleavage sites alters protease specificity and processing. Journal of Virology, 84(13), 6894–6898. 10.1128/JVI.00752-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmi, A. , Chirumbolo, S. , Peana, M. , Noor, S. , Menzel, A. , Dadar, M. , & Bjorklund, G. (2022). The role of diet and supplementation of natural products in COVID‐19 prevention. Biological Trace Element Research, 200(1), 27–30. 10.1007/s12011-021-02623-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemnejad‐Berenji, M. (2021). Immunomodulatory and anti‐inflammatory potential of ccrocin in COVID‐19 treatment. Journal of Food Biochemistry, 45(5), e13718. 10.1111/jfbc.13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, C. , Ginex, T. , Maestro, I. , Nozal, V. , Barrado‐Gil, L. , Cuesta‐Geijo, M. Á. , Urquiza, J. , Ramírez, D. , Alonso, C. , Campillo, N. E. , & Martinez, A. (2020). COVID‐19: Drug targets and potential treatments. Journal of Medicinal Chemistry, 63(21), 12359–12386. 10.1021/acs.jmedchem.0c00606 [DOI] [PubMed] [Google Scholar]
- Gurung, A. B. (2020). In silico structure modelling of SARS‐CoV‐2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Reports, 21, 100860. 10.1016/j.genrep.2020.100860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtemariam, S. , Nabavi, S. F. , Banach, M. , Berindan‐Neagoe, I. , Sarkar, K. , Sil, P. C. , & Nabavi, S. M. (2020). Should we try SARS‐CoV‐2 helicase inhibitors for COVID‐19 therapy? Archives of Medical Research, 51(7), 733–735. 10.1016/j.arcmed.2020.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy, G. E. , Abu‐Serie, M. M. , Abo‐Elela, G. M. , Ghozlan, H. , Sabry, S. A. , Soliman, N. A., & Abdel‐Fattah, Y. R. (2020). In vitro dual (anticancer and antiviral) activity of the carotenoids produced by haloalkaliphilic archaeon Natrialba sp. M6. Sci Rep, 10(1), 5986. 10.1038/s41598-020-62663-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Schroeder, S. , Kleine‐Weber, H. , Muller, M. A. , Drosten, C. , & Pohlmann, S. (2020). Nafamostat mesylate blocks activation of SARS‐CoV‐2: New treatment option for COVID‐19. Antimicrobial Agents and Chemotherapy, 64(6), e00754‐20. 10.1128/AAC.00754-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornero‐Méndez, D. , & Mínguez‐Mosquera, M. I. (1998). Isolation and identification of the carotenoid capsolutein from Capsicum annuum as cucurbitaxanthin A. Journal of Agricultural and Food Chemistry, 46(10), 4087–4090. 10.1021/jf980401m [DOI] [Google Scholar]
- Hosseinzadeh, H. , Abootorabi, A. , & Sadeghnia, H. R. (2008). Protective effect of Crocus sativus stigma extract and crocin (trans‐crocin 4) on methyl methanesulfonate–induced DNA damage in mice organs. DNA and Cell Biology, 27(12), 657–664. 10.1089/dna.2008.0767 [DOI] [PubMed] [Google Scholar]
- Huang, J. , Tao, G. , Liu, J. , Cai, J. , Huang, Z. , & Chen, J. X. (2020). Current prevention of COVID‐19: Natural products and herbal medicine. Frontiers in Pharmacology, 11, 588508. 10.3389/fphar.2020.588508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husaini, A. M. , Jan, K. N. , & Wani, G. A. (2021). Saffron: A potential drug‐supplement for severe acute respiratory syndrome coronavirus (COVID) management. Heliyon, 7(5), e07068. 10.1016/j.heliyon.2021.e07068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, M. A. A. , Abdelrahman, A. H. M. , Hussien, T. A. , Badr, E. A. A. , Mohamed, T. A. , El‐Seedi, H. R. , Pare, P. W. , Efferth, T. , & Hegazy, M. F. (2020). In silico drug discovery of major metabolites from spices as SARS‐CoV‐2 main protease inhibitors. Computers in Biology and Medicine, 126, 104046. 10.1016/j.compbiomed.2020.104046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R. , & Mujwar, S. (2020). Repurposing metocurine as main protease inhibitor to develop novel antiviral therapy for COVID‐19. Structural Chemistry, 31, 1–13. 10.1007/s11224-020-01605-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S. , & Rhee, J. Y. (2020). Three cases of treatment with nafamostat in elderly patients with COVID‐19 pneumonia who need oxygen therapy. International Journal of Infectious Diseases, 96, 500–502. 10.1016/j.ijid.2020.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z. , Du, X. , Xu, Y. , Deng, Y. , Liu, M. , Zhao, Y. , Zhang, B. , Li, X. , Zhang, L. , Peng, C. , Duan, Y. , Yu, J. , Wang, L. , Yang, K. , Liu, F. , Jiang, R. , Yang, X. , You, T. , Liu, X. , … Yang, H. (2020). Structure of M(pro) from SARS‐CoV‐2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kang, S. , Yang, M. , Hong, Z. , Zhang, L. , Huang, Z. , Chen, X. , He, S. , Zhou, Z. , Zhou, Z. , Chen, Q. , Yan, Y. , Zhang, C. , Shan, H. , & Chen, S. (2020). Crystal structure of SARS‐CoV‐2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B, 10(7), 1228–1238. 10.1016/j.apsb.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpiński, T. M. , Kwaśniewski, M. , Ożarowski, M. , & Alam, R. (2021). In silico studies of selected xanthophylls as potential candidates against SARS‐CoV‐2 targeting main protease (Mpro) and papain‐like protease (PLpro). Herba Polonica, 67(2), 1–8. 10.2478/hepo-2021-0009 [DOI] [Google Scholar]
- Kawata, A. , Murakami, Y. , Suzuki, S. , & Fujisawa, S. (2018). Anti‐inflammatory activity of beta‐carotene, lycopene and tri‐n‐butylborane, a scavenger of reactive oxygen species. In Vivo, 32(2), 255–264. 10.21873/invivo.11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiokias, S. , & Gordon, M. H. (2003). Dietary supplementation with a natural carotenoid mixture decreases oxidative stress. European Journal of Clinical Nutrition, 57(9), 1135–1140. 10.1038/sj.ejcn.1601655 [DOI] [PubMed] [Google Scholar]
- Klemm, T. , Ebert, G. , Calleja, D. J. , Allison, C. C. , Richardson, L. W. , Bernardini, J. P. , Lu, B. G. , Kuchel, N. W. , Grohmann, C. , Shibata, Y. , Gan, Z. Y. , Cooney, J. P. , Doerflinger, M. , Au, A. E. , Blackmore, T. R. , van der Heden van Noort, G. J. , Geurink, P. P. , Ovaa, H. , … Komander, D. (2020). Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2. EMBO Journal, 39(18), e106275. 10.15252/embj.2020106275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokic, G. , Hillen, H. S. , Tegunov, D. , Dienemann, C. , Seitz, F. , Schmitzova, J. , Farnung, L. , Siewert, A. , Höbartner, C. , & Cramer, P. (2021). Mechanism of SARS‐CoV‐2 polymerase stalling by remdesivir. Nature Communications, 12(1), 279. 10.1038/s41467-020-20542-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova, P. , Silhan, J. , Nencka, R. , & Boura, E. (2020). Structural analysis of the SARS‐CoV‐2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nature Communications, 11(1), 3717. 10.1038/s41467-020-17495-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwarteng, A. , Asiedu, E. , Sakyi, S. A. , & Asiedu, S. O. (2020). Targeting the SARS‐CoV2 nucleocapsid protein for potential therapeutics using immuno‐informatics and structure‐based drug discovery techniques. Biomedicine and Pharmacotherapy, 132, 110914. 10.1016/j.biopha.2020.110914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi, C. , & Arnoldi, A. (2021). Food‐derived antioxidants and COVID‐19. Journal of Food Biochemistry, 45(1), e13557. 10.1111/jfbc.13557 [DOI] [PubMed] [Google Scholar]
- Littler, D. R. , Gully, B. S. , Colson, R. N. , & Rossjohn, J. (2020). Crystal structure of the SARS‐CoV‐2 non‐structural protein 9, Nsp9. IScience, 23(7), 101258. 10.1016/j.isci.2020.101258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Zhu, Y. , Zhang, J. , Li, Y. , & Peng, Z. (2020). Intravenous high‐dose vitamin C for the treatment of severe COVID‐19: study protocol for a multicentre randomised controlled trial. BMJ Open, 10(7), e039519. 10.1136/bmjopen-2020-039519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno, T. , Tani, Y. , Maoka, T. , Matsuo, K. , & Komori, T. (1986). Isolation and structural elucidation of cucurbitaxanthin a and b from pumpkin Cucurbita maxima . Phytochemistry, 25(12), 2837–2840. 10.1016/s0031-9422(00)83753-x [DOI] [Google Scholar]
- Melendez‐Martinez, A. J. (2019). An overview of carotenoids, apocarotenoids, and vitamin A in agro‐food, nutrition, health, and disease. Molecular Nutrition and Food Research, 63(15), e1801045. 10.1002/mnfr.201801045 [DOI] [PubMed] [Google Scholar]
- Mentis, A. A. , Dalamaga, M. , Lu, C. , & Polissiou, M. G. (2021). Saffron for “toning down” COVID‐19‐related cytokine storm: Hype or hope? A mini‐review of current evidence. Metabolism Open, 11, 100111. 10.1016/j.metop.2021.100111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska, K. , Kim, Y. , Jedrzejczak, R. , Maltseva, N. I. , Stols, L. , Endres, M. , & Joachimiak, A. (2020). Crystal structures of SARS‐CoV‐2 ADP‐ribose phosphatase: From the apo form to ligand complexes. IUCrJ, 7(Pt 5), 814–824. 10.1107/S2052252520009653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michele, C. A. , Angel, B. , Valeria, L. , Teresa, M. , Giuseppe, C. , Giovanni, M. , Michele, D. P. , Ernestina, P. , & Mario, B. (2020). Vitamin supplements in the Era of SARS‐Cov2 pandemic. GSC Biological and Pharmaceutical Sciences, 11(2), 007‐019. 10.30574/gscbps.2020.11.2.0114 [DOI] [Google Scholar]
- Mujwar, S. (2021). Computational repurposing of tamibarotene against triple mutant variant of SARS‐CoV‐2. Computers in Biology and Medicine, 136, 104748. 10.1016/j.compbiomed.2021.104748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujwar, S. , Deshmukh, R. , Harwansh, R. K. , Gupta, J. K. , & Gour, A. (2019). Drug repurposing approach for developing novel therapy against mupirocin‐resistant Staphylococcus aureus . Assay and Drug Development Technologies, 17(7), 298–309. 10.1089/adt.2019.944 [DOI] [PubMed] [Google Scholar]
- Naydenova, K. , Muir, K. W. , Wu, L. F. , Zhang, Z. , Coscia, F. , Peet, M. J. , Castro‐Hartmann, P. , Qian, P. , Sader, K. , Dent, K. , Kimanius, D. , Sutherland, J. D. , Löwe, J. , Barford, D. , & Russo, C. J. (2021). Structure of the SARS‐CoV‐2 RNA‐dependent RNA polymerase in the presence of favipiravir‐RTP. Proceedings of the National Academy of Sciences of the United States of America, 118(7), e2021946118. 10.1073/pnas.2021946118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, J. A. , Douangamath, A. , Yadzani, S. , Yosaatmadja, Y. , Aimon, A. , Brandao‐Neto, J. , Dunnett, L. , Gorrie‐Stone, T. , Skyner, R. , Fearon, D. , Schapira, M. , von Delft, F. , & Gileadi, O. (2021). Structure, mechanism and crystallographic fragment screening of the SARS‐CoV‐2 NSP13 helicase. Nature Communications, 12(1), 4848. 10.1038/s41467-021-25166-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, J. A. , & Krinsky, N. I. (1995). Introduction: the colorful, fascinating world of the carotenoids: important physiologic modulators. FASEB Journal, 9(15), 1547–1550. 10.1096/fasebj.9.15.8529833 [DOI] [PubMed] [Google Scholar]
- Saikatendu, K. S. , Joseph, J. S. , Subramanian, V. , Clayton, T. , Griffith, M. , Moy, K. , Velasquez, J. , Neuman, B. W. , Buchmeier, M. J. , Stevens, R. C. , & Kuhn, P. (2005). Structural basis of severe acute respiratory syndrome coronavirus ADP‐ribose‐1″‐phosphate dephosphorylation by a conserved domain of nsP3. Structure, 13(11), 1665–1675. 10.1016/j.str.2005.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo, S. , Jaime, L. , Plaza, M. , Herrero, M. , Rodriguez‐Meizoso, I. , Ibañez, E. , & Reglero, G. (2011). Antiviral compounds obtained from microalgae commonly used as carotenoid sources. Journal of Applied Phycology, 24(4), 731–741. 10.1007/s10811-011-9692-1 [DOI] [Google Scholar]
- Savjani, K. T. , Gajjar, A. K. , & Savjani, J. K. (2012). Drug solubility: Importance and enhancement techniques. ISRN Pharmaceutics, 2012, 195727. 10.5402/2012/195727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, A. (2020). Drug targets for COVID‐19 therapeutics: Ongoing global efforts. Journal of Biosciences, 45(1), 1–24. 10.1007/s12038-020-00067-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan, T. P. , Sims, A. C. , Zhou, S. , Graham, R. L. , Pruijssers, A. J. , Agostini, M. L. , Leist, S. R. , Schäfer, A. , Dinnon, K. H., 3rd , Stevens, L. J. , Chappell, J. D. , Lu, X. , Hughes, T. M. , George, A. S. , Hill, C. S. , Montgomery, S. A. , Brown, A. J. , Bluemling, G. R. , Natchus, M. G. , … Baric, R. S. (2020). An orally bioavailable broad‐spectrum antiviral inhibits SARS‐CoV‐2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Science Translational Medicine, 12(541), eabb5883. 10.1126/scitranslmed.abb5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. , Tripathi, M. K. , Yasir, M. , Khare, R. , Tripathi, M. K. , & Shrivastava, R. (2020). Potential inhibitors for SARS‐CoV‐2 and functional food components as nutritional supplement for COVID‐19: A review. Plant Foods for Human Nutrition, 75(4), 458–466. 10.1007/s11130-020-00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleymani, S. , Zabihollahi, R. , Shahbazi, S. , & Bolhassani, A. (2018). Antiviral effects of saffron and its major ingredients. Current Drug Delivery, 15(5), 698–704. 10.2174/1567201814666171129210654 [DOI] [PubMed] [Google Scholar]
- Soto, M. E. , Guarner‐Lans, V. , Soria‐Castro, E. , Manzano Pech, L. , & Perez‐Torres, I. (2020). Is antioxidant therapy a useful complementary measure for Covid‐19 treatment? An algorithm for its application. Medicina (Kaunas), 56(8), 386. 10.3390/medicina56080386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalin, A. , Lin, D. , Senthamarai Kannan, B. , Feng, Y. , Wang, Y. , Zhao, W. , Ignacimuthu, S. , Wei, D. Q. , & Chen, Y. (2021). An in‐silico approach to identify the potential hot spots in SARS‐CoV‐2 spike RBD to block the interaction with ACE2 receptor. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2021.1897682. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Stipp, M. (2020). SARS‐CoV‐2: Micronutrient optimization in supporting host immunocompetence. International Journal of Clinical Case Reports and Reviews, 2(2), 1–10. 10.31579/2690-4861/024 [DOI] [Google Scholar]
- Tarighi, P. , Eftekhari, S. , Chizari, M. , Sabernavaei, M. , Jafari, D. , & Mirzabeigi, P. (2021). A review of potential suggested drugs for coronavirus disease (COVID‐19) treatment. European Journal of Pharmacology, 895, 173890. 10.1016/j.ejphar.2021.173890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo‐Mayol, I. , Guerra‐Valle, M. , Casas‐Forero, N. , Sobral, M. M. C. , Viegas, O. , Alarcon‐Enos, J. , Ferreira, I. M. , & Pinho, O. (2021). Western dietary pattern antioxidant intakes and oxidative stress: Importance during the SARS‐CoV‐2/COVID‐19 pandemic. Advances in Nutrition, 12(3), 670–681. 10.1093/advances/nmaa171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umashankar, V. , Deshpande, S. H. , Hegde, H. V. , Singh, I. , & Chattopadhyay, D. (2021). Phytochemical moieties from Indian traditional medicine for targeting dual hotspots on SARS‐CoV‐2 Spike protein: An integrative in‐silico approach. Frontiers in Medicine (Lausanne), 8, 672629. 10.3389/fmed.2021.672629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Zhang, X. , Omarini, A. B. , & Li, B. (2020). Virtual screening for functional foods against the main protease of SARS‐CoV‐2. Journal of Food Biochemistry, 44(11), e13481. 10.1111/jfbc.13481 [DOI] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , Hu, Y. , Tao, Z. W. , Tian, J. H. , Pei, Y. Y. , Yuan, M. L. , Zhang, Y. L. , Dai, F. H. , Liu, Y. , Wang, Q. M. , Zheng, J. J. , Xu, L. , Holmes, E. C. , & Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W. , Wang, S. , Zhang, H. , Guo, W. , Lu, H. , Xu, H. , Zhan, R. , Fidan, O. , & Sun, L. (2021). Biosynthesis of novel naphthoquinone derivatives in the commonly‐used chassis cells Saccharomyces cerevisiae and Escherichia coli . Applied Biochemistry and Microbiology, 57(1), S11–S26. 10.1134/S0003683821100124 [DOI] [Google Scholar]
- Yan, L. , Zhang, Y. , Ge, J. , Zheng, L. , Gao, Y. , Wang, T. , Jia, Z. , Wang, H. , Huang, Y. , Li, M. , Wang, Q. , Rao, Z. , & Lou, Z. (2020). Architecture of a SARS‐CoV‐2 mini replication and transcription complex. Nature Communications, 11(1), 5874. 10.1038/s41467-020-19770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W. , Mao, C. , Luan, X. , Shen, D. D. , Shen, Q. , Su, H. , Wang, X. , Zhou, F. , Zhao, W. , Gao, M. , Chang, S. , Xie, Y. C. , Tian, G. , Jiang, H. W. , Tao, S. C. , Shen, J. , Jiang, Y. , Jiang, H. , Xu, Y. , … Xu, H. E. (2020). Structural basis for inhibition of the RNA‐dependent RNA polymerase from SARS‐CoV‐2 by remdesivir. Science, 368(6498), 1499–1504. 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakynthinos, G. , & Varzakas, T. (2016). Carotenoids: From plants to food industry. Current Research in Nutrition and Food Science Journal, 4(1), 38–51. 10.12944/CRNFSJ.4.Special-Issue1.04 [DOI] [Google Scholar]
- Zhang, Y. , & Kutateladze, T. G. (2020). Molecular structure analyses suggest strategies to therapeutically target SARS‐CoV‐2. Nature Communications, 11(1), 2920. 10.1038/s41467-020-16779-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C. L. , Chen, H. D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R. D. , Liu, M. Q. , Chen, Y. , Shen, X. R. , Wang, X. , … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Chen, C. Z. , Gorshkov, K. , Xu, M. , Lo, D. C. , & Zheng, W. (2020). RNA‐dependent RNA polymerase as a target for COVID‐19 drug discovery. SLAS Discovery, 25(10), 1141–1151. 10.1177/2472555220942123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S9
Data Availability Statement
All data were included in the original manuscript and the supplementary data.


