Among the earliest events in many bacterial infections are the molecular interactions that occur between the pathogen and host cells. These interactions are typically required for extracellular colonization or internalization to occur and may involve a complex cascade of molecular cross talk at the host-pathogen interface. Colonization of host tissues is usually mediated by adhesins on the surface of the microbe; the adhesins are responsible for recognizing and binding to specific receptor moieties of host cells. The receptor binding event may activate complex signal transduction cascades in the host cell that can have diverse consequences including the activation of innate host defenses or the subversion of cellular processes facilitating bacterial colonization or invasion. In addition, the binding event may also activate the expression of new genes in the microbe that are important in the pathogenic process. In many instances, adhesins are assembled into hair-like appendages called pili or fimbriae that extend out from the bacterial surface. In other cases, the adhesins are directly associated with the microbial cell surface (so-called nonpilus adhesins). Collectively, these adhesins and related structures are expressed in organisms associated with a broad range of diseases (Table 1). At least four distinct mechanisms have emerged in recent years to account for the assembly of these diverse organelles: (i) the chaperone-usher pathway, (ii) the general secretion pathway, (iii) the extracellular nucleation-precipitation pathway, and (iv) the alternate chaperone pathway. This list is by no means all-inclusive but rather represents some of the best-characterized systems to date (for a recent review of other systems that do not utilize these pathways, see reference 51). Molecular blueprints of these pathways will ultimately facilitate the understanding of host-pathogen interactions as well as provide a framework for understanding how complex hetero-oligomeric interactions are orchestrated within the bacterial cell. In this minireview, we focus on the molecular architecture of the adhesive organelles assembled by these four principal pathways and on the coordinated functions of the proteins that constitute their assembly machineries.
TABLE 1.
Adhesive structures on the bacterial cell surface and their assembly pathways
| Assembly pathway | Structure | Assembly gene products | Organism | Disease(s) associated with pilus expression | Reference(s) |
|---|---|---|---|---|---|
| Chaperone-usher pathway | Thick, rigid pili | FGS chaperone/ushera | |||
| P pili | PapD/PapC | E. coli | Pyelonephritis or cystitis | 47, 48, 50 | |
| Prs pili | PrsD/PrsC | E. coli | Cystitis | 77, 112 | |
| Type 1 pili | FimC/FimD | E. coli | Cystitis | 58, 93, 109 | |
| Salmonella sp. | |||||
| Klebsiella pneumoniae | |||||
| S pili | SfaE/SfaF | E. coli | UTIb | 84, 105 | |
| Newborn meningitis | |||||
| F1C pili | FocC/FocD | E. coli | Cystitis | 96 | |
| Haemophilus influenzae fimbriae | HifB/HifC | Haemophilus influenzae | Otitis media | 39, 118 | |
| Meningitis | |||||
| H. influenzae biogroup aegyptius fimbriae | HafB/HafE | H. influenzae | Brazilian purpuric fever | 95 | |
| Type 2 and 3 pili | FimB/FimC | Bordetella pertussis | Whooping cough | 75, 85, 121 | |
| MR/P pili | MrpD/MrpC | Proteus mirabilis | Nosocomial UTI | 5 | |
| PMF pili | PmfC/PmfD | P. mirabilis | Nosocomial UTI | 6 | |
| Long polar fimbriae | LpfB/LpfC | Salmonella typhimurium | Gastroenteritis | 9 | |
| Pef pili | PefD/PefC | S. typhimurium | Gastroenteritis | 28 | |
| Ambient-temperature fimbriae | AftB/AftC | P. mirabilis | UTI | 79, 80 | |
| 987P fimbriae | FasB/FasD | E. coli | Diarrhea in piglets | 15, 25 | |
| Thin, flexible pili | |||||
| K99 pili | FaeE/FaeD | E. coli | Neonatal diarrhea in calves, lambs, and piglets | 7, 107 | |
| K88 pili | FanE/FanD | E. coli | Neonatal diarrhea in piglets | 7, 26 | |
| F17 pili | F17D/F17papC | E. coli | Diarrhea | 74 | |
| MR/K pili | MrkB/MrkC | K. pneumoniae | Pneumonia | 3, 34 | |
| REPEC fimbriae | RalE/RalD | E. coli | Diarrhea in rabbits | 1 | |
| Atypical structures | FGL chaperone/ushera | ||||
| CS31A capsule-like protein | ClpE/ClpD | E. coli | Diarrhea | 35 | |
| Antigen CS6 | CssC/CssD | E. coli | Diarrhea | 65, 122 | |
| Myf fimbriae | MyfB/MyfC | Yersinia enterocolitica | Enterocolitis | 54 | |
| pH 6 antigen | PsaB/PsaC | Yersinia pestis | Plague | 73 | |
| CS3 pili | CS3-1/CS3-2 | E. coli | Diarrhea | 57, 72 | |
| Envelope antigen F1 | Caf1M/Caf1A | Y. pestis | Plague | 32, 64 | |
| Nonfimbrial adhesins I | NfaE/NfaC | E. coli | UTI | 2, 36 | |
| Newborn meningitis | |||||
| SEF14 fimbriae | SefB/SefC | S. enteritidis | Gastroenteritis | 16, 86 | |
| Agregative adherence fimbriae I | AggD/AggC | E. coli | Diarrhea | 104 | |
| AFA-III | AfaB/AfaC | E. coli | Pyelonephritis | 33, 70 | |
| Extracellular nucleation-precipitation pathway | Curli | CsgG/CsgE/CsgF | E. coliS. enteritidis | Sepsis | 10, 11, 17, 40, 41, 76, 94, 106 |
| General secretion pathway | Type 4 pili | General secretion apparatus | Neisseria sp. | Gonorrhea | 4, 12, 23, 24, 31, 42, 43, 44, 62, 83, 89, 91, 92, 99, 100, 113 |
| P. aeruginosa | |||||
| V. cholerae | Cholera | ||||
| M. bovis | |||||
| D. nodosus | |||||
| Eikenella corrodens | |||||
| Bundle-forming pili | General secretion apparatus | E. coli | Diarrhea | 12, 21, 22, 108 | |
| Alternate chaperone pathway | CS1 pili | CooB/CooC | E. coli | Diarrhea | 102 |
| CS2 pili | CotB/CotC | E. coli | Diarrhea | 30 | |
| CS4 pili | E. coli | Diarrhea | 115 | ||
| CS14 pili | E. coli | Diarrhea | 81 | ||
| CS17 pili | E. coli | Diarrhea | 82 | ||
| CS19 pili | E. coli | Diarrhea | 38 | ||
| CFA/I pili | CfaA/CfaC | E. coli | Diarrhea | 61 | |
| Cable type II pili | B. cepacia | Opportunistic infections in cystic fibrosis patients | 101 |
The chaperone is given before the slash, and the usher is given after the slash.
UTI, urinary tract infection.
MOLECULAR STRUCTURES OF FIMBRIAL ADHESINS
We begin by looking at the architectural features of various fimbrial organelles assembled by each of the four general assembly pathways. We focus on the best-characterized systems in each pathway as prototypes for each assembly classification: P and type 1 pili (chaperone-usher pathway), type IV pili (general secretion pathway), curli (extracellular nucleation-precipitation pathway) and CS1 pili (alternate chaperone pathway). Note that for the purposes of this minireview, the term subunit will apply to the structural proteins that make up these composite organelles, while the term adhesin will be reserved for those subunits with specific receptor binding properties.
P pili and type 1 pili.
P pili are expressed on the surfaces of uropathogenic strains of Escherichia coli associated with acute pyelonephritis (63). Eleven genes organized in the pap gene cluster are required for the expression and assembly of these organelles (46, 49, 50, 78). Studies of P pili using quick-freeze, deep-etch electron microscopy have shown that P pili are composite fibers consisting of flexible fibrillae joined end to end to pilus rods (67). The tip fibrillae are comprised predominantly of PapE subunits. The rod is composed of repeating PapA subunits packed into a right-handed helical assembly, with an external diameter of 68 Å, an axial hole of 15 Å, and a pitch distance of 24.9 Å, with 3.28 subunits per turn of the helical cylinder (14, 37). The adhesin of P pili, PapG, mediates binding to Galα(1,4)Gal moieties present in the globoseries of glycolipids on uroepithelial cells and erythrocytes (71, 111). The adhesin is located at the distal end of the tip and is joined to the PapE fibrillum via a specialized adapter protein, PapF. Another adapter protein, PapK, joins the adhesin-containing tip to the PapA rod. Another minor component, PapH, is located at the base of the PapA rod; its incorporation into the growing organelle is thought to signal the termination of assembly.
Type 1 pili are important virulence determinants expressed in E. coli as well as in most members of the Enterobacteriaceae family that mediate binding to mannose-oligosaccharides (66). The expression and assembly of type 1 pili typically require at least nine genes that are present in the type 1 gene cluster (46, 50). Like P pili, type 1 pili are also composite structures in which a short tip fibrillar structure containing FimG and the FimH adhesin (and possibly the minor component FimF as well) are joined to a rod comprised predominantly of FimA subunits (58). The overall structure of the type 1 rod is very similar to that of the PapA rod of P pili. The type 1 subunits are arranged in a helix with an external diameter of 6 to 7 nm and an axial hole of 20 to 25 Å, with a pitch distance of 23.1 Å and 3.125 subunits per turn (13).
Type IV pili.
Type IV pili have been implicated in a variety of functions, including adhesion to host cell surfaces, twitching motility, modulation of target cell specificity, and bacteriophage adsorption. They are found on such bacteria as Pseudomonas aeruginosa, pathogenic Neisseria, Moraxella bovis, Dichelobacter nodosus, Vibrio cholerae, and enteropathogenic E. coli (EPEC) (113). The role of type IV pili in the virulence of EPEC strains has recently been demonstrated by Bieber and colleagues (12). These structures have a diameter of 60 Å and are typically up to 4,000 nm long, with a pitch distance of approximately 40 Å and about five subunits per turn (89). They are composed predominantly of identical pilin subunits with a number of distinctive features, including a short (6 to 7 amino acids), positively charged leader sequence, a modified amino acid (N-methylphenylalanine) at the amino terminus of the mature pilin, and a highly conserved amino-terminal domain (4, 43, 91, 92). A few other proteins also associate with these structures, including in the case of Neisseria, the tip-localized adhesin (100).
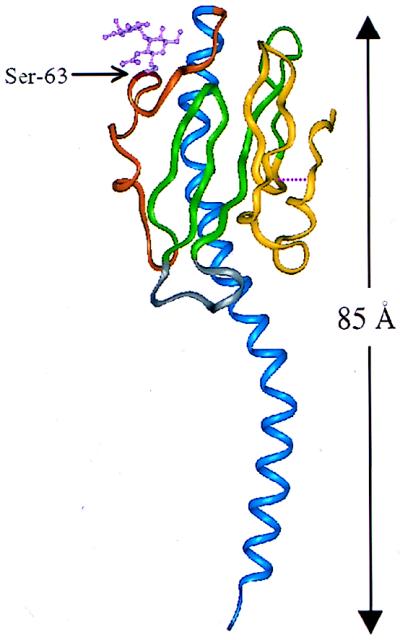
The crystal structure of a type IV pilin subunit (PilE) from Neisseria gonorrhoeae has recently been determined by Parge and coworkers at an atomic resolution of 2.6 Å (89). It reveals an α-β roll fold with a rather long hydrophobic N-terminal α1-helical spine (residues 2 to 54) that gives the molecule an overall ladle shape (Fig. 1). Other elements of the structure include the following: (i) an extended disaccharide-bound sugar loop (residues 55 to 77), with N-acetylglucosamine-α(1,3)-galactose O linked at position Ser-63, (ii) two β-hairpins forming a four-stranded antiparallel β-sheet (residues 78 to 93 and 103 to 122), (iii) a β2-β3 loop connection (residues 94 to 102), and (iv) a disulfide-containing region (residues 121 to 158), which despite its hypervariable nature, appears to be a regular β-hairpin (β5-β6) followed by a loop connection. Systematic modeling of the pilin monomer within the constraints imposed by the available biochemical and biophysical data has led to a three-layered model of the type IV pilus (27, 89). The outermost hypervariable layer in the proposed fiber model is comprised of residues 123 to 143 and 152 to 158, as well as the disaccharide at Ser-63, from each monomer. The central layer is a continuous 25-stranded β-sheet, made up of the four strands from the antiparallel β-sheet as well as the sugar loop from each of the five pilin monomers present in each turn. The innermost layer is a parallel coiled-coil made up of the highly conserved N-terminal α1-helices. A key feature of this model is that essentially only the hypervariable and sugar-binding domains of each pilin monomer are exposed in the final assembled pilus structure, which may account for the antigenic variation that these pili undergo.
FIG. 1.
Ribbon representation of pilin from N. gonorrhoeae. Colored regions indicate the secondary structural elements referred to in the text: blue, N-terminal α1-helix (residues 2 to 54); orange, extended disaccharide-bound sugar loop (residues 55 to 77); green, β-hairpins (residues 78 to 93 and 103 to 122); gray, β2-β3 loop (residues 94 to 102); yellow, disulfide bond-containing C-terminal region (residues 121 to 158). Also shown are the disulfide bridge (cysteine residues 121 and 151), signified by a broken line, and Ser-63 with covalently linked disaccharide.
Curli.
Many clinical E. coli and Salmonella enteritidis isolates produce a class of thin, irregular, and highly aggregated surface structures known as curli (17, 94). These organelles mediate binding to a variety of host proteins, including fibronectin (94), plasminogen (106), and human contact phase proteins (10). Curli are highly stable structures that require extreme chemical treatment (e.g., 90% formic acid) to depolymerize them. The major component of E. coli curli is a 15.3-kDa protein termed CsgA, which exhibits more than 86% primary sequence similarity to its counterpart in S. enteritidis, AgfA. CsgB is a minor component that may be found associated with the outer membrane (OM) or distributed along the length of the curli fiber (11). CsgE, CsgF, and CsgG are required assembly factors that do not appear to constitute part of the final curli structure and hence may serve as part of the assembly apparatus (40).
CS1 pili.
CS1 pili are found on the surface of EPEC and are thought to be involved in the colonization of the host intestine (102). Other pilus structures in this family include CS2 (30), CS4 (115), CS14 (81), CS17 (82), CS19 (38), CFA/I (61), and the cable type II pili of the cystic fibrosis-associated pathogen Burkholderia cepacia (101). CS1 pili appear to be composed predominantly of one component, CooA, with a distally located minor component, CooD. Electron microscopic examination of these structures reveals that they are morphologically similar to P and type 1 pili, although the structural proteins of CS1-like pili bear no significant sequence similarity to those of other pilus systems (102).
ASSEMBLY OF FIMBRIAL ADHESINS
The coordinated assembly of complex hetero-oligomeric organelles poses many special challenges to the bacterial cell, including the correct incorporation of individual subunits in a predefined order during biogenesis and the prevention of premature associations between intrinsically aggregative subunits. Details of how these molecular interactions are orchestrated have been worked out to various degrees in different systems and have begun to shed light on the similarities and variations employed in diverse bacterial species in the assembly of these organelles.
Chaperone-usher pathway.
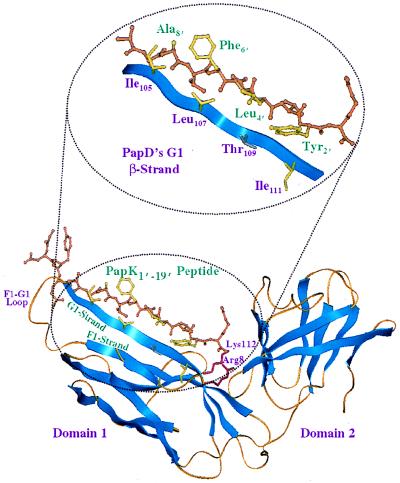
The highly conserved chaperone-usher pathway is involved in the assembly of more than 25 adhesive organelles in gram-negative bacteria. The assembly machinery is comprised of two specialized classes of proteins, a periplasmic immunoglobulin-like chaperone and an OM usher. The crystal structure of the periplasmic chaperone involved in the assembly of P pili, PapD, has been determined to a resolution of 2.0 Å, revealing two immunoglobulin-like domains oriented towards one another in such a manner so as to give the molecule an overall boomerang shape (Fig. 2) (45). Hung et al. (52) have shown that the chaperones can be organized into two structurally and functionally distinct subfamilies on the basis of conserved amino acid differences in the chaperone cleft and the length of the loop that connects the F1 and G1 β-strands of domain 1. The two subfamilies are designated FGS (for F1-G1 short) and FGL (for F1-G1 long), corresponding to loop lengths of ≤20 amino acids and ≥21 amino acids, respectively. Interestingly, these two subfamilies assemble pili with distinct architectures. FGS chaperones, of which PapD is a member, are involved in assembling pili with rod-like architecture. FGL chaperones, on the other hand, mediate the assembly of very thin or afimbrial adhesive structures on the surfaces of bacteria.
FIG. 2.
Ribbon representation of the crystal structure of PapD and PapK peptides. Inset provides a magnified view of the PapD and PapK peptide contact interface. Note how the conserved alternating hydrophobic residues of the peptide interdigitate with the residues along PapD’s G1 β-strand.
PapD is known to form periplasmic preassembly complexes with each of the pilus subunits prior to their incorporation into a pilus. The relative concentration of each subunit type in the periplasm is thought to be an important factor in regulating the length of the tip fibrillum and pilus rod. Overexpression of the PapK adapter, for example, leads to production of pili with shorter tips; similarly, overexpression of PapH leads to shortened rods (55). Although no crystal structure of any of these complexes has yet been determined, recent studies have identified some of the important determinants for chaperone-subunit recognition. Several lines of evidence indicate that chaperones recognize a highly conserved motif present in the C termini of all subunits assembled by PapD-like chaperones (68). This motif is characterized by a series of alternating hydrophobic residues flanked by a glycine located 14 residues upstream from the C terminus and by a penultimate tyrosine. Two peptides, corresponding to the C-terminal 19 amino acids of PapG and PapK, have been cocrystallized with PapD (68, 110). Despite significant sequence dissimilarities, both C-terminal fragments bound to PapD in a nearly identical manner via an extended conformation that has been termed a β-zipper motif (68, 110). The results of more recent mutagenesis experiments suggested that a highly conserved region found near the N termini of all subunits assembled by PapD-like chaperones is also recognized by the chaperone (110). This region is also characterized by an alternating pattern of hydrophobic residues, together with a cysteine residue that is involved in an intramolecular disulfide bond. This region is not present at the N terminus of the PapG adhesin. This is not unexpected, given the domain structure of the adhesin. Fimbrial adhesins can be thought of as having a receptor binding domain fused to a pilin domain. In PapG, the receptor binding domain consists of the amino-terminal half of the protein. The C-terminal half of the protein contains most of the pilin-like features, including the conserved β-zipper motif and two cysteines spaced approximately 40 amino acids apart. Interestingly, the amino-terminal region of the pilin domain of PapG has recently been shown to contain a surface that is recognized by the PapD chaperone (110, 123). This region corresponds in approximate sequence position (as measured from the COOH terminus) to the highly conserved N-terminal regions of other pilus subunits.
Initial translocation of P-pilus subunits across the cytoplasmic membrane occurs via the Sec (general secretion system) machinery, although this pathway itself is not sufficient for the efficient release of subunits into the periplasm (59). Nascent subunits are retained in the inner cytoplasmic membrane via an interaction mediated by their hydrophobic C termini. In the presence of PapD, the subunits are partitioned into the periplasmic space as chaperone-subunit preassembly complexes (Fig. 3A). Based on available crystallographic data, invariant cleft residues of the chaperone are thought to participate in the β-zippering interaction with a subunit. Mutations in these invariant cleft residues of the chaperone abolish the ability of the chaperone to import subunits and form chaperone-subunit complexes, underscoring the importance of β-zipper formation in mediating chaperone function (59, 68, 110). Release of the subunits from the inner membrane is a prerequisite for their folding into an assembly-competent conformation, and there is evidence that folding of the subunits with the chaperone serving as a template may occur concomitantly with their release from the membrane (59, 110).
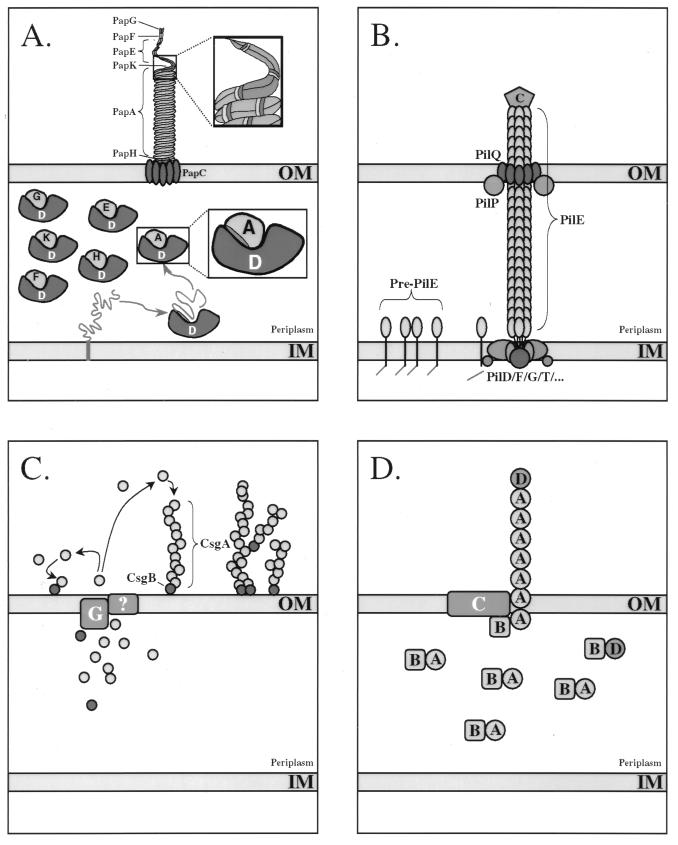
FIG. 3.
(A) Assembly of P pili from E. coli via the chaperone-usher pathway. Chaperone-mediated extraction of subunits from the inner cytoplasmic membrane (IM) is coupled with their folding into an assembly-competent state. The G1 β-strand of the immunoglobulin-like chaperones, which may serve as a template in the subunit folding pathway, protects nascently folded subunits from premature oligomerization in the periplasmic space by directly capping the newly formed assembly surfaces. These interactive surfaces remain protected by the chaperone until delivery of the preassembly complex to the OM assembly site comprised of the usher. PapG (G), PapD (D), PapE (E), PapK (K), PapA (A), and PapH (H) proteins are shown. (B) Assembly of type IV pilus from N. gonorrhoeae via the general secretion pathway. Prepilin is processed by the PilD signal peptidase, which cleaves the positively charged leader sequence from the N terminus of the pilin subunit. The mature PilE subunit is then assembled by the inner membrane (IM) assembly complex. Translocation of the pilus through the OM is mediated via PilQ, possible with the assistance of other factors such as PilP. The PilC adhesin, which is thought to ultimately be incorporated at the tip of the growing organelle, also appears to be required for translocation through the OM. C, C terminus. (C) Assembly of curli from E. coli via the extracellular nucleation-precipitation pathway. CsgA, the main component of curli, is secreted across the OM. Surface-localized CsgB serves to nucleate CsgA assembly. CsgB is also found distributed along the curli fiber, where it may serve to initiate branching of the fiber. CsgG (G) is an OM-localized lipoprotein that is required for the secretion of CsgA and CsgB, although its function is not known at this time. (D) Assembly of CS1 pili from E. coli via the alternate chaperone pathway. The CooB (B) chaperone forms periplasmic complexes with the main components of the pilus, CooA (A) and CooD (D). It also appears to bind and perhaps stabilize the OM protein CooC (C) in the absence of subunits. CooC may function as an OM channel for passage of the pilin fiber.
Once formed, the chaperone-subunit complexes are targeted to the OM PapC usher for assembly. The PapC usher was purified and shown to form a pore when reconstituted into liposomes (114). This was confirmed by high-resolution electron microscopy, which showed that PapC assembled into ring-shaped complexes containing central pores of 2 to 3 nm in diameter (114). The PapC complexes consisted of at least six subunits. PapC and other usher family members are predicted to have a largely β-sheet secondary structure, typical of bacterial OM pore-forming proteins, and likely present large regions to the periplasm for interaction with chaperone-subunit complexes. To facilitate pilus assembly, the usher must be able to translocate pilus subunits across the OM. The 2-nm-wide linear tip fibrillum would be able to pass through the 2- to 3-nm-diameter usher channel, but the 6.8-nm-wide helical pilus rod would not be able to fit through the usher. A solution to this problem was revealed in experiments that showed that P-pilus rods could be unraveled into linear fibers (114). These unraveled rods measure 2 nm in diameter and would therefore be narrow enough to pass through the usher pore. Therefore, it was proposed that the pilus rod is translocated across the OM in this linear form and adopts its final helical conformation only upon reaching the external surface. This may be part of the mechanism that drives the outward growth of the organelle.
The usher presumably has a more active role in pilus assembly than simply functioning as a diffusion pore for the translocation of pili across the OM. Dodson et al. (20) showed that PapC differentially recognized chaperone-subunit complexes depending upon their final position in the pilus. These studies have recently been extended by examining the real-time kinetics of the interaction of chaperone-subunit complexes with the usher by using surface plasmon resonance technology (103). Chaperone-adhesin complexes from both the P and type 1 pilus systems were found to bind tightest and fastest to their respective ushers, suggesting that kinetic partitioning of chaperone-adhesin complexes to the usher is a defining factor in the tip localization of the adhesin. In addition, dissociation rates for all of the chaperone-subunit complexes from the usher were found to be slow, arguing that after association of a complex with the usher, the subunit is destined for assembly into the pilus. A stable usher-chaperone-adhesin complex was purified from bacteria expressing FimD (type 1 usher), FimC (type 1 chaperone), and FimH (type 1 adhesin) (103). Expression of other combinations of chaperone-subunit complexes with the usher did not result in formation of a stable ternary complex. Formation of the FimDCH complex led to protection of the usher from degradation by trypsin in vivo, apparently due to a conformational change in the usher (103). This conformational change was maintained during pilus assembly, suggesting that interaction of FimCH with FimD stabilizes the usher in an assembly-competent conformation. Since a FimH− strain is nonpiliated, these data argue that interaction of the chaperone-adhesin complex with the usher is critical to initiate pilus biogenesis, as has been observed in several clinical strains (103).
In addition to preferential interactions of different chaperone-subunit complexes with the usher, another factor that plays a key role in dictating the relative order of subunit incorporation into the growing organelle is subunit-subunit surface complementarity. Jacob-Dubuisson et al. (55) demonstrated that the PapF and PapK adapter proteins were required for the efficient initiation of tip fibrillae and pilus rods, respectively. Deletion of both the papF and papK genes abolished piliation altogether, suggesting that other pilus subunits do not possess the structural determinants necessary to initiate the formation of tip fibrillae and pilus rods. The highly conserved N- and C-terminal regions of pilus subunits have recently been identified as serving as the primary assembly surfaces that mediate subunit-subunit interactions in the quaternary structure of the mature pilus. Subtle differences in these primary assembly regions from one subunit to another may be responsible for controlling the order of incorporation of pilus subunits (110).
Additional insight can be gained by grouping the sequences of individual subunits assembled by FGS chaperones according to their known structural roles (Fig. 4). According to this scheme, there are three basic classes of subunits. Class I subunits are the major subunits of thick rod-like assemblies. Class II subunits are minor components of pili, including those that function as adapters and those that assemble into open helical fibers. Class III subunits are similar to class I subunits in that they comprise the major subunit of the respective fiber but differ from class I subunits in that they typically assemble into structures with thin, flexible morphologies.
FIG. 4.
Alignment of subunits assembled by FGS chaperones. Amino acid sequence alignment of structural subunits assembled by members of the FGS subfamily of immunoglobulin-like chaperones. Sequences have been grouped into three classes, based upon whether they represent a major or minor subunit and on the morphology of the assembled structures (i.e., thick rods versus thin fimbrillae). Only those residues that are conserved in at least 90% of the sequences within a class or across classes are shaded and coded with the following colors: pink, invariant; yellow, conserved hydrophobic (A, L, V, I, P, M, W, F, C, Y, and G); purple, conserved polar and charged (N, Q, S, T, H, D, E, K, and R). Residues marked by an asterisk appear to be conserved within a given class but not across classes.
An alignment of subunits assembled by FGS chaperones revealed seven homology regions (HR) consisting of distinct patterns of residues conserved among all subunits. We further divided class II subunits into two subclasses (II-A and II-B) based upon distinct patterns of conservation, as described below. The most extensively conserved regions are the aforementioned N- and C-terminal regions (designated HR-1 and HR-7, respectively). The region (HR-3) near the second cysteine residue, which is linked to the cysteine in HR-1 by disulfide bonds, is also highly conserved across all subunits, as is a region near the center of all the sequences examined (HR-5). However, there are notable differences between corresponding regions among the different classes of subunits. For example, the HR-1 motif of class I subunits begins with a highly conserved glycine that is absent in most class II and class III subunits. Class II-A subunits appear to be closely related to class I subunits, with only subtle differences in the HR-1, -2, -4, and -5 motifs. In contrast, class II-B subunits have characteristic vicinal prolines immediately preceding the first cysteine in HR-1, as well as distinct HR-2, -3, -5, and -6 motifs. The PapF adapter protein of P pili falls within this class, whereas other minor components of P pili fall within the class II-A grouping. These differences may be a determinant in allowing PapF to function as an adapter between the PapG adhesin and the PapE tip fibrillum-forming subunit. It is intriguing to speculate that these conserved regions are important structural determinants that may dictate subunit function in pilus assembly and that differences in these regions may account in part or in whole for differences in function. HR-1 and HR-7 have already been shown to play a role in mediating subunit-subunit interactions in the mature pilus (110). HR-3 may also make an important contribution to the assembly surface formed by HR-1, as the position of the conserved alternating pattern of hydrophobic residues relative to the cysteines suggests these two regions could be two adjacent parallel β-strands within a sheet. It is interesting to note that there is no difference in the HR-7 motif among classes. This likely reflects the importance of this region as a common recognition motif for the periplasmic chaperones.
General secretion pathway.
The formation of type 4 pili requires the expression of several proteins that are involved in the assembly of these structures, including the following: (i) a prepilin peptidase that cleaves a short leader peptide from the subunits; (ii) an integral membrane protein located in the inner cytoplasmic membrane that may serve as a platform for fimbrial assembly; (iii) a hydrophilic nucleotide-binding protein located in the cytoplasm or associated with the cytoplasmic face of the inner membrane that may energize secretion by ATP hydrolysis; and (iv) an OM component that forms a channel allowing the translocation of assembled pili through the OM (4). Donnenberg and colleagues (22) have recently identified a total of 14 genes that are sufficient for the biogenesis of type IV pili in a heterologous E. coli host, including the periplasmic disulfide-bond oxidoreductase DsbA. This assembly system appears to function independently of any chaperone activity, which likely reflects the localization of the assembly platform within the inner membrane and the absence of a need to transport pilin monomers across the periplasm in a soluble form. It is noteworthy that a number of pilin-like proteins, possessing similar leader peptides and hydrophobic amino termini, are thought to assemble into a pilus-like secretion tube used in the secretion of a variety of proteases, toxins, and other extracellular factors across the OM (91, 92). This export machinery is known as the general secretion apparatus and has been shown to be dependent on some of the gene products involved in the assembly of type 4 pili (e.g., signal peptidase), hence, the grouping of the type 4 assembly machinery as part of the general secretion pathway. Our knowledge of type IV pilus biogenesis remains incomplete, but work from several groups has laid a solid foundation for understanding how these complex organelles are assembled. We herein present an overview of our current understanding of type IV pilus biogenesis in N. gonorrheae (Fig. 3B). For discussions describing the assembly of these organelles in P. aeruginosa, we refer the reader to the recent reviews by Alm and Mattick (4) and Hahn (42).
Following translocation of the pre-PilE precursor subunits into the periplasmic compartment by the general secretion apparatus, these molecules are retained in the inner membrane by their hydrophobic N-terminal segments, with their hydrophilic C-terminal domains oriented towards the periplasm (31, 83). The PilD signal peptidase removes the positively charged leader sequence from the cytoplasmic side of the prepilin to generate mature PilE, which can then undergo assembly as subunits associate with their hydrophobic stems. PilF, PilG, and PilT are among the factors required for this assembly, although their functions are not well understood. It has been suggested based on studies of its homologues that PilF may function as an ATPase or kinase (116, 117). PilG has been proposed to play a role in the optimal localization or stabilization of PilD and or PilF (116). PilT is a putative nucleotide-binding protein that has been postulated to play a role in twitching motility and pilus retraction (43).
The assembled pili are thought to be translocated across the OM by a gated pore formed by a multimeric form of PilQ (23). A lipoprotein, PilP, appears to function in stabilizing the expression of PilQ as a multimer (24). The PilC adhesin appears to facilitate passage of the growing organelle through this pore, although the molecular basis for the role of PilC in this process is not well understood (60, 88, 99).
As our understanding of type IV pilus biogenesis continues to expand, the importance of other players in the assembly and regulation of these organelles will no doubt become apparent. Recent work by Kaiser and colleagues with Myxococcus xanthus underscores the many subtle factors that influence type IV pilus assembly and function. M. xanthus has two genetic systems, called adventurous (A) and social (S) motility, which control its gliding motility and swarming behavior, respectively (44). S motility is dependent upon type IV pili, as mutants lacking pili do not display this type of motility (62). It has been shown that strains with mutations in a particular S motility gene called tgl (for transient gliding) lack S motility and type IV pili but that these qualities can be transiently restored by contact with tgl+ (donor) cells in a process called stimulation (44, 62). Stimulation does not involve a diffusable factor but rather depends upon physical contact between cells. Furthermore, stimulation is transient and occurs only phenotypically, as the offspring of stimulated cells remain S− and lack pili. The tgl gene product is a putative lipoprotein that appears to be localized to the periplasm, probably attached to the outer membrane (97, 98), and contains multiple tetratrico peptide repeat domains that are thought to be important in protein-protein interactions (69). At present, the exact mechanism of Tgl action is unknown. The identification of the proteins that interact with Tgl will certainly shed light on the process of stimulation and how this protein modulates the assembly of type IV pili.
Extracellular nucleation-precipitation pathway.
The formation of curli represents a departure from the chaperone-usher pathway and the general assembly pathway typified by P and type 4 pili, respectively. Whereas those structures undergo assembly from the base (i.e., the distal end containing the adhesin is assembled first), curli formation occurs from the outside of the microbe by the precipitation of secreted soluble subunits into thin fibers on the surface of the microbe (41). In E. coli, the products of two divergently transcribed operons are required for curli assembly (40). The csgBA operon encodes the principal fiber-forming subunit, CsgA, which is secreted directly into the extracellular milieu as a soluble protein. It also encodes CsgB, which is proposed to be a nucleator that induces polymerization of CsgA on the cell surface (Fig. 3C) (11). In support of this model, it has been demonstrated that a CsgA+ CsgB− donor strain can secrete CsgA subunits that can be assembled into curli on the surface of a CsgA− CsgB+ recipient strain (41). Furthermore, CsgB appears to be distributed along the length of the curli fiber, where it has been suggested to be able to initiate branching of the fibrillar structure (11). Interestingly, in the absence of CsgA, overexpressed CsgB appears to be able to form short polymers on the bacterial cell surface (11).
The csgDEFG operon encodes a transcriptional activator for curli production (CsgD) and three putative assembly factors (40). One of these factors, CsgG, has recently been shown to be a lipoprotein that is localized to the OM (76). In its absence, curli assembly does not take place and it appears that CsgA and CsgB are subjected to rapid proteolytic degradation. The precise role of CsgG is not known at this time. Loferer and colleagues (76) have proposed that CsgG might be a chaperone that works in concert with another, as yet unidentified OM translocator to export CsgA and CsgB and protect these subunits from premature degradation. Alternatively, a multimeric form of CsgG itself may function as a Csg-specific channel within the OM. The roles of CsgE and CsgF have not been established at this time; however, it has been reported that a strain deficient in these two assembly factors can export assembly-competent CsgA, suggesting that expression of CsgG is sufficient for production and assembly of CsgA (76).
Alternate chaperone pathway.
The operons for CS1 and the related CS2 and CFA/I fimbrial structures are each composed of four functional genes (29, 30, 61). The pathway for the assembly of these structures also employs a specialized set of periplasmic chaperones that appear to be distinct from those of the chaperone-usher pathway; hence we term the mode of assembly for these organelles the alternate chaperone pathway. In the case of CS1 pili, the chaperone CooB has been shown to form periplasmic complexes with the pilin components CooA and CooD, which are transported into the periplasm in a Sec-dependent manner (Fig. 3D). The former serves as the major pilin component, while the latter is a minor component that appears to be tip localized and may serve to initiate the assembly of CooA (102). Both subunits appear to share a conserved sequence motif near their C termini, which may function as a chaperone recognition motif (102, 120). Note that this motif shares no homology with the conserved C-terminal motif of alternating hydrophobic residues found in subunits assembled by the chaperone-usher pathway.
CooC is an OM protein that may function as an OM channel for passage of the pilin fiber. Interestingly, CooB also appears to stabilize CooC in the OM and is able to bind CooC in the absence of the other pilin subunits (120). Despite apparent functional similarities, CS1 and related structures do not appear to be related to those assembled by the classic chaperone-usher pathway, suggesting that these two systems arose independently through convergent evolution.
LINKS BETWEEN PILUS BIOGENESIS AND HOST PATHOGENESIS
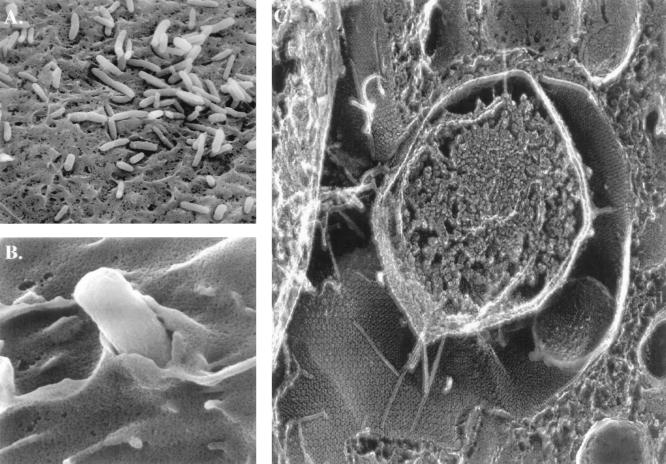
Colonization is not a single event but rather a dynamic process that involves a panoply of changes in both the bacterium and host alike as a result of attachment. Mulvey and colleagues (87) have recently used scanning and high-resolution transmission electron microscopy in a murine cystitis model to investigate the structural basis and consequences of in vivo interactions between type 1-piliated E. coli and host superficial bladder cells. These studies revealed that type 1 pilus tips interacted directly with a class of integral membrane glycoproteins known as uroplakins that are situated on the luminal surface of the bladder epithelial cells (Fig. 5). Attached pili were shortened to an average length of 0.12 ± 0.01 μm. In contrast, type 1 pili present on bacteria in broth culture are typically 1 to 2 μm long (13). The mechanism by which this apparent shortening occurs remains to be elucidated, but retraction of the pilus upon attachment has been suggested as one possible means (87). Alternatively, contact of the type 1 pilus tips with the host epithelium could impede the growth of nascent pili (87). Either pilus retraction or a hindrance of pilus growth mechanism would likely result in a buildup of unassembled pilin subunits in the periplasm.
FIG. 5.
Type 1 pilus-mediated bacterial adherence to the mouse bladder epithelium at 2 h postinfection. (A) Scanning electron micrograph (EM) of bacteria on surface of bladder epithelial cells. The bacteria often appear to be situated in grooves and niches formed by the apical membrane of the superficial cells. (B) Scanning EM of a bacterium being enveloped by the membrane of bladder epithelial cells. (C) High-resolution, freeze-fracture, deep-etch EM of infected bladder epithelia, showing a centrally located bacterium making intimate contact with the luminal surface of the epithelia. Type 1 pili can be seen radiating out from the organisms and spanning the distance between the outer membrane and host cell surface. The FimH adhesin at the tips of these pili mediates contact with the hexagonal uroplakin plaques embedded in the epithelial cell membrane.
The consequences of such a buildup can be inferred from studies showing that the expression of pilus subunits in the absence of the chaperone is toxic in E. coli strains lacking the DegP periplasmic protease (59). Toxicity presumably results from the formation of subunit aggregates in the periplasm that the DegP protease normally breaks down. By using lacZ fusions to degP and cpxP, it was demonstrated that expression of subunits in the absence of PapD activates the CpxA-CpxR two-component system in which CpxA is the membrane-bound sensor/kinase and CpxR is the DNA binding response regulator (18, 19, 59, 90). This pathway up-regulates degP transcription as well as a number of other chaperone-like proteins, such as the disulfide isomerase DsbA and cis-trans prolyl isomerases. These factors facilitate subunit folding: DsbA is required for pilus biogenesis (56). These studies suggested that Cpx monitors pilus biogenesis and responds by controlling the expression of factors that facilitate pilus biogenesis. It is intriguing to speculate that activation of the CpxA-CpxR pathway in response to pilus-mediated attachment leads to the expression of an array of virulence genes necessary for establishing an infection. Hung and colleagues (53) refer to this state as the attached phenotype.
PERSPECTIVE AND FUTURE DIRECTIONS
Bacteria have developed a number of distinct mechanisms for the assembly of a diverse range of adhesive organelles. Despite the variations, several common themes do emerge from the study of these assembly pathways. For example, the inner membrane appears to have the capacity to function as a temporary reservoir for nascently translocated subunits assembled by the chaperone-usher pathway and the general secretion pathway. Those pathways that require subunits to be transported through the periplasm prior to their assembly appear to require the function of a periplasmic chaperone to prevent premature subunit oligomerization. In the case of the chaperone-usher and alternate chaperone pathways, the corresponding chaperones appear to interact with their target proteins either immediately or shortly following subunit translocation into the periplasm. In the case of the extracellular nucleation-precipitation pathway, an OM-localized protein may function as a chaperone and/or usher to transform curlin monomers into an assembly-competent conformation just before their export, thereby perhaps minimizing the chances for premature associations in the periplasmic compartment. In contrast, molecular chaperones appear to be absent in the general secretion pathway, as the pilin subunits assembled by this route are assembled directly on the inner membrane. For all four pathways, it appears that specific OM channels are involved in the export of pilin subunits either in an assembled or nonassembled state.
Understanding the molecular events involved in the biogenesis of these organelles will be crucial for the development of novel therapeutic strategies. Elucidating common themes in these pathways will be a prerequisite for any efforts targeted towards developing a therapeutic strategy with broad-spectrum activity. The identification of those processes that occur following attachment will undoubtedly open up further avenues of therapeutic possibilities, as we come closer to understanding how host-pathogen interactions lead to the expression of bacterial genes that are important in pathogenesis.
ACKNOWLEDGMENTS
We gratefully acknowledge our ongoing collaboration with the lab of T. Silhavy. It is largely through their enthusiastic sharing of results and ideas that our labs jointly formed the current model concerning the role of Cpx in pilus biogenesis. We thank M. Mulvey for kindly providing us with electron micrographs of bacteria expressing type 1 pili interacting with mouse bladder epithelial cells.
Some of the work described was supported by National Institutes of Health grants R01AI29549 and R01DK51406.
REFERENCES
- 1.Adams L M, Simmons C P, Rezmann L, Strugnell R A, Robins-Browne R M. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect Immun. 1997;65:5222–5230. doi: 10.1128/iai.65.12.5222-5230.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrens R, Ott M, Ritter A, Hoschutzkky H, Buhler T, Lottspeich F, Boulnois G J, Jann K, Hacker J. Genetic analysis of the gene cluster encoding nonfimbrial adhesin I from an Escherichia coli uropathogen. Infect Immun. 1993;61:2505–2512. doi: 10.1128/iai.61.6.2505-2512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen B L, Gerlach G F, Clegg S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J Bacteriol. 1991;173:916–920. doi: 10.1128/jb.173.2.916-920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 5.Bahrani F K, Johnson D E, Robbins D, Mobley H L T. Proteus mirabilis flagella and MR/P fimbriae: isolation, purification, N-terminal analysis, and serum antibody response following experimental urinary tract infection. Infect Immun. 1991;59:3574–3580. doi: 10.1128/iai.59.10.3574-3580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahrani F K, Cook S, Hull R, Massad G, Mobley H L T. Proteus mirabilis fimbriae: N-terminal amino acid sequence of a major fimbrial subunit and nucleotide sequences of the genes from two strains. Infect Immun. 1993;61:884–891. doi: 10.1128/iai.61.3.884-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakker D, Vader C E, Roosendaal B, Mooi F R, Oudega B, de Graff F K. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol Microbiol. 1991;5:875–886. doi: 10.1111/j.1365-2958.1991.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 8.Balley M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 9.Baumler A J, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben Nasr A, Olsen A, Sjöbring U, Müller-Esterl W, Björk L. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol Microbiol. 1996;20:927–935. doi: 10.1111/j.1365-2958.1996.tb02534.x. [DOI] [PubMed] [Google Scholar]
- 11.Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bieber D, Ramer S W, Wu C-Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 13.Brinton C C., Jr The structure, function, synthesis, and genetic control of bacterial pili and a model for DNA and RNA transport in gram negative bacteria. Trans N Y Acad Sci. 1965;27:1003–1065. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 14.Bullitt E, Makowski L. Structural polymorphism of bacterial adhesion pili. Nature. 1995;373:164–167. doi: 10.1038/373164a0. [DOI] [PubMed] [Google Scholar]
- 15.Cao J, Chan A S, Bayer M E, Schifferli D M. Ordered translocation of 987P fimbrial subunits through the outer membrane of Escherichia coli. J Bacteriol. 1995;177:3704–3713. doi: 10.1128/jb.177.13.3704-3713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clouthier S C, Muller K H, Doran J L, Collinson S K, Kay W W. Characterization of three fimbrial genes, sefABC, of Salmonella enteriditis. J Bacteriol. 1993;175:2523–2533. doi: 10.1128/jb.175.9.2523-2533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collinson S K, Emödy L, Müller K-H, Trust T J, Kay W W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding systems in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 19.De Las Penas A, Connolly L, Gross C A. The ςE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of ςE. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 20.Dodson K W, Jacob-Dubuisson F, Striker R T, Hultgren S J. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donnenberg M, Giron J, Nataro J, Kaper J. A plasmid encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 22.Donnenberg M S, Zhang H-Z, Stone K D. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability. Gene. 1997;192:33–38. doi: 10.1016/s0378-1119(96)00826-8. [DOI] [PubMed] [Google Scholar]
- 23.Drake S L, Koomey M. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol. 1995;18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 24.Drake S L, Sandstedt S A, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular weight multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- 25.Edwards R A, Cao J, Schifferli D M. Identification of major and minor chaperone proteins involved in the export of 987P fimbriae. J Bacteriol. 1996;178:3426–3433. doi: 10.1128/jb.178.12.3426-3433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson A K, Willgohs J A, McFarland S Y, Benfield D A, Francis D H. Identification of two porcine brush border glycoproteins that bind the K88ac adhesin of Escherichia coli and correlation of these glycoproteins with the adhesive phenotype. Infect Immun. 1992;60:983–988. doi: 10.1128/iai.60.3.983-988.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forest K T, Tainer J A. Type-4 pilus-structure: outside to inside and top to bottom. Gene. 1997;192:165–169. doi: 10.1016/s0378-1119(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 28.Friedich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of a fimbrial biosynthetic gene. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 29.Froehlich B J, Karakashian A, Melsen L R, Wakefield J C, Scott J R. CooC and CooD are required for assembly of CS1 pili. Mol Microbiol. 1994;12:387–401. doi: 10.1111/j.1365-2958.1994.tb01028.x. [DOI] [PubMed] [Google Scholar]
- 30.Froehlich B J, Karakashian A, Sakellaris H, Scott J R. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect Immun. 1995;63:4849–4856. doi: 10.1128/iai.63.12.4849-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 32.Galyov E E, Karlishev A V, Chernovskaya T V, Dolgikh D A, Smirnov O Y, Volkovoy K I, Abramov V M, Zav′yalov V P. Expression of the envelope antigen F1 of Yersinia pestis is mediated by the product of cal1M gene having homology with the chaperone protein PapD of Escherichia coli. FEBS Lett. 1991;286:79–82. doi: 10.1016/0014-5793(91)80945-y. [DOI] [PubMed] [Google Scholar]
- 33.Garcia M I, Labigne A, LeBouguenec C. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J Bacteriol. 1994;176:7601–7613. doi: 10.1128/jb.176.24.7601-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerlach G F, Allen B L, Clegg S. Molecular characterization of the type 3 (MR/K) fimbriae of Klebsiella pneumoniae. J Bacteriol. 1988;170:3547–3553. doi: 10.1128/jb.170.8.3547-3553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girdeau J P, Vartanian M D, Ollier J L, Contrepois M. CS31A, a new K88-related fimbrial antigen on bovine enteropathogenic and septicemic Escherichia coli strains. Infect Immun. 1988;56:2180–2188. doi: 10.1128/iai.56.8.2180-2188.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldhar J, Perry R, Golecki J R, Hoschutzky H, Jann B, Jann K. Nonfimbrial, mannose-resistant adhesins from uropathogenic Escherichia coli O83:K1:H4 and O14:K?:H11. Infect Immun. 1987;55:1837–1842. doi: 10.1128/iai.55.8.1837-1842.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong M, Makowski L. Helical structure of P pili from Escherichia coli. J Mol Biol. 1992;228:735–742. doi: 10.1016/0022-2836(92)90860-m. [DOI] [PubMed] [Google Scholar]
- 38.Grewal H M, Valvatne H, Bhan M K, van Dijk L, Gaastra W, Sommerfelt H. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect Immun. 1997;65:507–513. doi: 10.1128/iai.65.2.507-513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerina N G, Langerman S, Clegg H W, Kessler T W, Goldman D A, Gilsdorf J R. Adherence of piliated Haemophilus influenzae to human oropharyngeal cells. J Infect Dis. 1982;146:564. doi: 10.1093/infdis/146.4.564. [DOI] [PubMed] [Google Scholar]
- 40.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 41.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 43.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;1:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 44.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 45.Holmgren A, Brändén C I. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature. 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 46.Hull R A, Gill R E, Hsu P, Minshaw B H, Falkow S. Construction and expression of recombinant plasmids encoding type 1 and d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hull R A, Hull S I, Falkow S. Frequency of gene sequences necessary for pyelonephritis-associated pili expression among isolates of Enterobacteriaceae from human extraintestinal infections. Infect Immun. 1984;43:1064–1067. doi: 10.1128/iai.43.3.1064-1067.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hull R A, Hull S I. Adherence mechanisms in urinary tract infections. In: Miller V L, Kaper J B, Portnoy D A, Isberg R R, editors. Molecular genetics of bacterial pathogenesis. Washington, D.C: ASM Press; 1994. pp. 79–90. [Google Scholar]
- 49.Hultgren S J, Normark S. Biogenesis of the bacterial pilus. Curr Opin Genet Dev. 1991;1:313–318. doi: 10.1016/s0959-437x(05)80293-x. [DOI] [PubMed] [Google Scholar]
- 50.Hultgren S J, Normark S, Abraham S N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 51.Hultgren S J, Jones C H, Normark S. Bacterial adhesins and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2730–2756. [Google Scholar]
- 52.Hung D L, Knight S D, Woods R M, Pinkner J S, Hultgren S J. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 1996;15:3792–3805. [PMC free article] [PubMed] [Google Scholar]
- 53.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. Unpublished data.
- 54.Iriarte M, Vanooteghem C, Delor I, Diaz R, Knutton S, Cornelis G R. The Myf fibrillae of Yersinia enterocolitica. Mol Microbiol. 1993;9:507–520. doi: 10.1111/j.1365-2958.1993.tb01712.x. [DOI] [PubMed] [Google Scholar]
- 55.Jacob-Dubuisson F, Heuser J, Dodson K, Normark S, Hultgren S. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 1993;12:837–847. doi: 10.1002/j.1460-2075.1993.tb05724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacob-Dubuisson F, Pinkner J, Xu Z, Striker R, Padmanhaban A, Hultgren S J. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc Natl Acad Sci USA. 1994;91:11552–11556. doi: 10.1073/pnas.91.24.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jalajakumari M B, Thomas C J, Halter R, Manning P A. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol Microbiol. 1989;3:1685–1695. doi: 10.1111/j.1365-2958.1989.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 58.Jones C H, Pinkner J S, Roth R, Heuser J, Nicholes A V, Abraham S N, Hultgren S J. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonsson A B, Nyberg G, Normark S. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 1991;10:477–488. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jordi B J A M, Willshaw G A, van der Zeist B A M, Gaastra W. The complete nucleotide sequence of region 1 of the CFA/I fimbrial operon of human enterotoxigenic Escherichia coli. DNA Sequence. 1992;2:257–263. doi: 10.3109/10425179209020811. [DOI] [PubMed] [Google Scholar]
- 62.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kallenius G, Svenson S B, Hultberg H, Molby R, Helin I, Cedergen B, Windberg J. Occurrence of P fimbriated Escherichia coli in urinary tract infection. Lancet. 1981;ii:1369–1372. doi: 10.1016/s0140-6736(81)92797-5. [DOI] [PubMed] [Google Scholar]
- 64.Karlyshev A, Galyov E, Smirnov O, Guzayev A, Abramov V, Zav′yalov V. A new gene of the f1 operon of Y. pestis involved in the capsule biogenesis. FEBS Lett. 1992;297:77–80. doi: 10.1016/0014-5793(92)80331-a. [DOI] [PubMed] [Google Scholar]
- 65.Knutton S, McConnel M M, Rowe B, McNeish A S. Adhesin and ultrastructural properties of human enterotoxigenic Escherichia coli producing colonization factor antigens III and IV. Infect Immun. 1989;57:3364–3371. doi: 10.1128/iai.57.11.3364-3371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krogfelt K A, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58:1995–1999. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuehn M J, Heuser J, Normark S, Hultgren S J. P pili in uropathogenic E. coli are composite fibers with distinct fibrillar adhesive tips. Nature. 1992;356:252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- 68.Kuehn M J, Ogg D J, Kihlberg J, Slonim L N, Flemmer K, Bergfors T, Hultgren S J. Structural basis of pilus subunit recognition by the PapD chaperone. Science. 1993;262:1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 69.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interaction: to TPR or not to TPR. Trends Biochem Sci. 1995;20:257–258. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 70.Le Bouguenec C, Garcia M I, Oulin V, Desperrier J-M, Gounon P, Labigne A. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun. 1993;61:5106–5514. doi: 10.1128/iai.61.12.5106-5114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leffler H, Svanborg-Eden C. Chemical identification of a glycosphingolipid receptor for Escherichia coli attaching to human urinary tract epithelial cells and agglutinating human erythrocytes. FEMS Microbiol Lett. 1980;8:127–134. [Google Scholar]
- 72.Levine M M, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements M L, Cheney C, Patnaik R. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect Immun. 1984;44:409–429. doi: 10.1128/iai.44.2.409-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindler L E, Tall B D. Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol Microbiol. 1993;8:311–324. doi: 10.1111/j.1365-2958.1993.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 74.Lintermans P F, Pohl P, Bertels A, Charlier G, Vandekerckhove J, Van Damme J, Shoup J, Schlicker C, Korhonen T, De Greve H, Van Montagu M. Characterization and purification of the F17 adhesin on the surface of bovine enteropathogenic and septicemic Escherichia coli. Am J Vet Res. 1988;49:1794–1799. [PubMed] [Google Scholar]
- 75.Locht C, Geoffroy M-C, Renauld G. Common accessory genes for the Bordetella pertussis filamentous hemagglutinin and fimbriae share sequence similarities with the papC and papD gene families. EMBO J. 1992;11:3175–3183. doi: 10.1002/j.1460-2075.1992.tb05394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 77.Lund B, Marklund B I, Stromberg N, Lindberg F, Karlsson K A, Normark S. Uropathogenic Escherichia coli can express serologically identical pili of different receptor binding specificities. Mol Microbiol. 1988;2:255–263. doi: 10.1111/j.1365-2958.1988.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 78.Marklund B I, Tennent J M, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 79.Massad G, Bahrani F K, Mobley H L T. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infect Immun. 1994;62:1989–1994. doi: 10.1128/iai.62.5.1989-1994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Massad G, Fulkerson J F, Jr, Watson D C, Mobley H L T. Proteus mirabilis ambient-temperature fimbriae: cloning and nucleotide sequence of the aft gene cluster. Infect Immun. 1996;64:4390–4395. doi: 10.1128/iai.64.10.4390-4395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McConnell M M, Chart H, Field A M, Hibberd M, Rowe B. Characterization of a putative colonization factor (PCFO166) of enterotoxigenic Escherichia coli of serogroup O166. J Gen Microbiol. 1989;135:1135–1144. doi: 10.1099/00221287-135-5-1135. [DOI] [PubMed] [Google Scholar]
- 82.McConnell M M, Hibberd M, Field A M, Chart H, Rowe B. Characterization of a new putative colonization factor (CS17) from a human enterotoxigenic Escherichia coli of serogroup O114:H21 which produces only heat-labile enterotoxin. J Infect Dis. 1990;161:343–347. doi: 10.1093/infdis/161.2.343. [DOI] [PubMed] [Google Scholar]
- 83.Meyer T F. Variation of pilin and opacity-associated protein in pathogenic Neisseria species. In: Iglewski B, Clark V, editors. The bacteria 43—molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press; 1990. pp. 137–159. [Google Scholar]
- 84.Moch T, Hoschutzky H, Hacker J, Kroncke K-D, Jann K. Isolation and characterization of the α-sialyl-β-2,3-galactosyl-specific adhesin from fimbriated Escherichia coli. Proc Natl Acad Sci USA. 1987;84:3462–3466. doi: 10.1073/pnas.84.10.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mooi F R, Jansen W H, Brunings H, Gielen H, van der Heide H G J, Walvoort H C, Guniee P A M. Construction and analysis of Bordetella pertussis mutants defective in the production of fimbriae. Microb Pathog. 1992;12:127–135. doi: 10.1016/0882-4010(92)90115-5. [DOI] [PubMed] [Google Scholar]
- 86.Muller K H, Collinson S K, Trust T J, Kay W W. Type 1 fimbriae of Salmonella enteriditis. J Bacteriol. 1991;173:454–457. doi: 10.1128/jb.173.15.4765-4772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 88.Nassif X, Beretti J L, Lowy J, Stenberg P, O’Gaora P, Pfeifer J, Normark S, So M. Roles of pilin and PilC in adhesin of Neisseria meningitidis to human epithelial and endothelial cells. Proc Natl Acad Sci USA. 1994;91:3769–3773. doi: 10.1073/pnas.91.9.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parge H E, Forest K T, Hickey M J, Christensen D A, Getzoff E D, Tainer J A. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 90.Pogliano J, Lynch A S, Berlin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 91.Pugsley A P. Protein traffic in bacteria. In: Lederberg J, editor. Encyclopedia of microbiology. Vol. 3. New York, N.Y: Academic Press; 1992. pp. 461–479. [Google Scholar]
- 92.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ofek I, Mirelman D, Sharon S. Adherence of Escherichia coli to human mucosal cells mediated by mannose receptors. Nature. 1977;265:623–625. doi: 10.1038/265623a0. [DOI] [PubMed] [Google Scholar]
- 94.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 95.Read T D, Dowdell M, Satola S W, Farley M M. Duplication of pilus gene complexes of Haemophilus influenzae biogroup aegyptius. J Bacteriol. 1996;178:6564–6570. doi: 10.1128/jb.178.22.6564-6570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Riegman N, Kusters R, van Veggel H, Bergmans H, Van Bergen En Henegouwen P, Hacker J, Van Die I. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J Bacteriol. 1990;172:1114–1120. doi: 10.1128/jb.172.2.1114-1120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodriguez-Soto J P, Kaiser D. The tgl gene: social motility and stimulation in Myxococcus xanthus. J Bacteriol. 1997;179:4361–4371. doi: 10.1128/jb.179.13.4361-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rodriguez-Soto J P, Kaiser D. Identification and localization of the Tgl protein, which is required for Myxococcus xanthus social motility. J Bacteriol. 1997;179:4372–4381. doi: 10.1128/jb.179.13.4372-4381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rudel T, Hans-Jürgen B, Meyer T F. Pilus biogenesis and epithelial cell adherence of Neisseria gonorrhoeae pilC double knock-out mutants. Mol Microbiol. 1995;17:1057–1071. doi: 10.1111/j.1365-2958.1995.mmi_17061057.x. [DOI] [PubMed] [Google Scholar]
- 100.Rudel T, Scheuerpflug I, Meyer T F. Neisseria PilC protein identified as type 4 pilus tip-located adhesin. Nature. 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 101.Sajjan U S, Sun L, Goldstein R, Forstner J F. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sakellaris H, Balding D P, Scott J R. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- 103.Saulino E T, Thanassi D G, Pinkner J, Hultgren S J. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Savarino S, Fox P, Yikang D, Nataro J P. Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J Bacteriol. 1994;176:4949–4957. doi: 10.1128/jb.176.16.4949-4957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmoll T, Morschhauser J, Ott M, Luwig B, van Die I, Hacker J. Complete genetic organization and functional aspects of the Escherichia coli S fimbrial adhesin determinant: nucleotide sequence of the genes sfa B, C, D, E, F. Microb Pathog. 1990;9:331–343. doi: 10.1016/0882-4010(90)90067-z. [DOI] [PubMed] [Google Scholar]
- 106.Sjöbring U, Pohl G, Olsen A. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA) Mol Microbiol. 1994;14:443–452. doi: 10.1111/j.1365-2958.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 107.Smit H, Gaastra W, Kamerling J P, Vliegenthart J F G, de Graaf F K. Isolation and structural characterization of the equine erythrocyte receptor for enterotoxigenic Escherichia coli K99 fimbrial adhesin. Infect Immun. 1984;46:578–584. doi: 10.1128/iai.46.2.578-584.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sohel I, Puente J L, Murray W J, Vuopio-Varkila J, Schoolnik G K. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 109.Sokurenko E V, Courtney H S, Abraham S N, Klemm P, Hasty D L. Functional heterogeneity of type 1 fimbriae of Escherichia coli. Infect Immun. 1992;60:4709–4719. doi: 10.1128/iai.60.11.4709-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soto G E, Dodson K W, Ogg D, Liu C, Heuser J, Knight S, Kihlberg J, Jones C H, Hultgren S J. Periplasmic chaperone recognition motif of subunits mediates quaternary interactions in the pilus. EMBO J. 1998;17:6155–6167. doi: 10.1093/emboj/17.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Striker R, Nilsson U, Stonecipher A, Magnusson G, Hultgren S J. Structural requirements for the glycolipid receptor of human uropathogenic E. coli. Mol Microbiol. 1995;16:1021–1029. doi: 10.1111/j.1365-2958.1995.tb02327.x. [DOI] [PubMed] [Google Scholar]
- 112.Stromberg N, Marklund B I, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson K A, Normark S. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galα(1-4)Gal-containing isoreceptors. EMBO J. 1990;9:2001–2010. doi: 10.1002/j.1460-2075.1990.tb08328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tennent J M, Mattick J S. Type 4 fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Ann Arbor, Mich: CRC Press; 1994. pp. 127–146. [Google Scholar]
- 114.Thanassi D G, Saulino E T, Lombardo M J, Roth R, Heuser J, Hultgren S J. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thomas L V, McConnell M M, Rowe B, Field A M. The possession of three novel coli surface antigens by enterotoxigenic Escherichia coli strains positive for the putative colonization factor PCF8775. J Gen Microbiol. 1985;131:2319–2326. doi: 10.1099/00221287-131-9-2319. [DOI] [PubMed] [Google Scholar]
- 116.Tønjum T, Koomey M. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships. Gene. 1997;192:155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 117.Turner L R, Lara J C, Nunn D N, Lory S. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:4962–4969. doi: 10.1128/jb.175.16.4962-4969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Alphen L, Geelen-van Den Broek L, Blaas L, van Ham M, Dankert J. Blocking of fimbria-mediated adherence of Haemophilus influenzae by sialyl gangliosides. Infect Immun. 1991;59:4473–4477. doi: 10.1128/iai.59.12.4473-4477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 120.Voegele K, Sakellaris H, Scott J R. CooB plays a chaperone-like role for the proteins involved in formation of CS1 pili of enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 1997;94:13257–13261. doi: 10.1073/pnas.94.24.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Willems R J L, van der Heide H G J, Mooi F R. Characterization of a Bordetella pertussis fimbrial gene cluster which is located directly downstream of the filamentous haemagglutinin gene. Mol Microbiol. 1992;6:2661–2671. doi: 10.1111/j.1365-2958.1992.tb01443.x. [DOI] [PubMed] [Google Scholar]
- 122.Wolf M K, Andrews G P, Tall B D, McConnel M M, Levine M M, Boedeker E C. Characterization of CS4 and CS6 antigenic components of PCF8775, a putative colonization factor complex from enterotoxigenic Escherichia coli E8775. Infect Immun. 1989;57:164–173. doi: 10.1128/iai.57.1.164-173.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu Z, Jones C H, Haslem D, Pinkner J S, Dodson K, Kihlberg J, Hultgren S J. Molecular dissection of PapD interaction with PapG reveals two chaperone-binding sites. Mol Microbiol. 1995;16:1011–1020. doi: 10.1111/j.1365-2958.1995.tb02326.x. [DOI] [PubMed] [Google Scholar]