Abstract
The Ambr15 system is an automated, high‐throughput bioreactor platform which comprises 24 individually controlled, single‐use stirred‐tank reactors. This system plays a critical role in process development by reducing reagent requirements and facilitating high‐throughput screening of process parameters. However, until now, the system was used to simulate processes involving cells in suspension or growing on microcarriers and has never been tested for simulating cells growing on macrocarriers. Moreover, to our knowledge, a complete production process including cell growth and virus production has never been simulated. Here, we demonstrate, for the first time, the amenability of the automated Ambr15 cell culture reactor system to simulate the entire SARS‐CoV‐2 vaccine production process using macrocarriers. To simulate the production process, accessories were first developed to enable insertion of tens of Fibra‐Cel macrocarries into the reactors. Vero cell adsorption to Fibra‐Cels was then monitored and its adsorption curve was studied. After incorporating of all optimized factors, Vero cells were adsorbed to and grown on Fibra‐Cels for several days. During the process, culture medium was exchanged, and the quantity and viability of the cells were followed, resulting in a typical growth curve. After successfully growing cells for 6 days, they were infected with the rVSV‐ΔG‐Spike vaccine virus. The present results indicate that the Ambr15 system is not only suitable for simulating a process using macrocarriers, but also to simulate an entire vaccine production process, from cell adsorption, cell growth, infection and vaccine virus production.
Keywords: Ambr15 system, culture‐based vaccine, Fibra‐Cel, Vero cells, virus vaccine production
1. INTRODUCTION
As a response to the SARS‐CoV‐2 pandemic, different vaccine candidates have been approved for emergency use and a few hundred are in clinical trials. One of the vaccines being tested in clinical trials and developed by the Israel Institute for Biological Research (IIBR) is based on the vesicular stomatitis virus (VSV), in which its native surface glycoprotein has been replaced with the surface glycoprotein of SARS‐CoV‐2 (rVSV‐ΔG‐Spike). 1 The vaccine generated by our institute is produced in Vero cells grown on macrocarriers (Fibra‐Cels). Cell‐based vaccines are often touted as a solution to the vaccine demand spike in a pandemic scenario. 2 The advantages of cell‐based vaccine production include the independence from supply of chicken eggs along with minimization of cross contamination or allergic reactions. In addition, the use of defined and serum free cell culture allows a more consistent process. 3 The rising use of cell culture‐based vaccine production has led to the carrier technology development. Micro and macrocarriers serve as small, compact surface support matrices for growing adherent cells. The carriers are ideally used for high density cell culture in batch‐ or perfusion modes with minimized time and resources. The macrocarrier Fibra‐Cels was found to be suitable for many anchorage‐dependent cell types, including Vero cells. 4 , 5
Maximizing production yield in a chemically defined cell culture process format has been the common goal for cell culture process scientists. In the recent years, the biopharmaceutical industry has faced ever increasing demands to deliver quality therapeutic products to the market, with shrinking timelines and limited resources. As a consequence, there is a need to conduct large number of optimization experiments under different conditions. This need led to the development of miniaturized high‐throughput technologies for process development, including the Ambr15 system (Sartorius AP). 6 This technology is very cost‐effective, high throughput, and relies on disposable reactors vessels with maximum working volume of 15 ml controlled by an automated workstation. The system provides parallel processing with the use of 24–48 disposable reactors with online monitoring of pH, temperature, and dissolved oxygen (DO) for each reactor. The Ambr15 system provides a new way forward, with independent control of dissolved O2 and dissolved CO2 per reactor by automatic gassing through a sparge tube or into the headspace, and automatic liquid handling for reactor set‐up, feeding, base addition, and sampling. Full control of impeller speed and culture temperature is also achievable on this system.
The Ambr15 system is increasingly used to accelerate development of newer approaches, such as cell and gene‐based therapies, next generation vaccines and antibody‐drug conjugates. The system has proven to be a very effective scaled‐down model. In its initial years, applications of the Ambr15 system have focused exclusively on suspension culture. Specifically, most of the studies involved Chinese Hamster Ovary (CHO) cells. A decade ago, Lewis et al. 7 used the Ambr15 system to determine if the system could replicate the characteristics of classical bioreactors at microscale in fed‐batch mode. Experiments were performed to compare CHO cell viability and antibody titer. Subsequently, Hsu et al. 8 evaluated the Ambr15 system to determine its ability to support cell culture process development in four representative CHO cell lines and compare its performance to that of 2 L bioreactors. In the same year, Moses et al. 9 reported a study evaluating and implementing the Ambr15 system for CHO cell culture. Production processes of two established monoclonal antibodies were compared. Two years later, Rameez et al. 10 evaluated CHO culture performance in the Ambr15 compared to larger bioreactors across various scales. Other cells growing in suspension were also examined in the Ambr15 system, including human hematopoietic stem/progenitor cells (hHSPC) which were shown to expand in the Ambr15 system. 11
Yet, many cell culture applications involve anchorage‐dependent cells and/or incorporation of larger particles such as carriers or Dynabeads. Therefore, in the last several years, an additional application for the Ambr15 system was tested, with the involvement of adherent cells or large particles. Rafiq et al. 12 demonstrated the amenability of Ambr15 system for the development of a scalable adherent human mesenchymal multipotent stroma/stem cell microcarrier process. Costariol et al. 13 demonstrated that T‐cells could be cultivated and their growth consistently and significantly improved compared to T‐flask static cultures, with equivalent cell quality. The T‐cell activation was achieved by using magnetic Dynabeads covalently coupled to anti‐CD3 and anti‐CD28 antibodies to mimic the role of antigen‐presenting cells. These studies demonstrated the ability to use this system with adherent cells and large particles. However, the system proved to be sub‐optimal for cell therapy candidates that require suspension of microcarriers or other particles such as Dynabeads. As a result, new reactors were later designed and developed for improved cell and gene therapy applications. 14
Thus far, all applications of the ambr15 system have focused on suspension cells or micro carriers cultured cells, but never macrocarriers. The aim of the present work was to demonstrate that the Ambr15 system is a suitable scale‐down model for production processes involving macrocarriers. Several adaptations and optimization were necessary since the system was not tailored to macrocarriers. Once established, the Ambr15 system was used to simulate the entire SARS‐CoV‐2 vaccine production process, from cell adsorption, cell growth, infection and vaccine virus production. The resulting titer vaccine virus confirmed the relevance of the Ambr15 system for process optimization studies. This relevance was proved by subsequent experiments with the adjusted Ambr15 system that were able to optimize the vaccine production process. 15
2. MATERIALS AND METHODS
2.1. Materials
Vero cell were purchased from WHO (RCB 10–87). NutriVero FLEX‐20, L‐alanine L‐glutamine, pen‐strep antibiotics, recombinant trypsin–EDTA solution, Trypan blue solution, and phosphate buffered saline (PBS) were purchased from Biological Industries (05‐069‐1A, 03‐022‐1B, 03‐031‐1C, 03‐079‐1A, 03‐102‐1B, and 02‐020‐1A, respectively). Fibra‐Cel disks were purchased from Eppendorf (New Brunswick Scientific, M1292‐9984). Alamar blue was purchased from Promega (Cell titer blue, G808). Calcein AM was purchased from Sigma (C1359).
2.2. Vero cells cultivation
Vero cells were maintained at 37°C under 5% CO2 in T‐flasks in FLEX 20 medium supplemented with 2 mM L‐alanine L‐glutamine and 0.1% pen‐strep antibiotics. Cells were harvested by washing the flask with PBS and incubating with trypsin–EDTA. The cells were centrifuged for 5 min at 1200 rpm. The supernatant was removed, and the pellet resuspended in fresh medium. Cell counts were performed with cell countessTM (Invitrogen) after 1:1 mixing with trypan blue.
2.3. Preparing reactors with Fibra‐Cels
Fibra‐Cels were weighed and 0.28 g (equal to 60 Fibra‐Cels) sterilized in 50 ml Costar centrifuge tubes containing 15 ml PBS and autoclaved at 121°C for 20 min. The autoclaved Fibra‐Cels in PBS were inserted to reactors using a specific funnel (described in 3.1.1). After insertion of Fibra‐Cels in PBS, all reactors were drained with a “rapid vessel drain” step. The default time of this step was 10 s, but was changed to 25 s to ensure complete draining (as the Fibra‐Cels might temporary block the draining tips). The reactors were then filled with 14 ml fresh medium with “add to reactor” step. This volume covers the Fibra‐Cels and allows their free movement. The agitation speed was set to 300 rpm (upstir) with the “stirring speed” step. The reactors were stabilized for 24 h in 37°C and 50% DO.
2.4. Adsorption of cells to Fibra‐Cels
The next day, the reactors were inoculated either controlled by software or manually with a dispenser. Since Vero cells adhere very rapidly, manual inoculation was preferred. Three ml of medium were first drained from each reactor with the “drain from reactor” step (the rectors were drained to 11 ml volume) after which 3 ml containing different numbers of cells (equivalent to the desired cell quantity) were manually added to each reactor. The new volume of the reactors (14 ml) was updated manually in the “vessels data tabs”.
To follow cell adsorption, medium was sampled from each reactor every 30 min. At the end of the adsorption time, the reactors were drained using the “rapid vessel drain” step (as before, for 25 s) and 14 ml of fresh medium was added. At this point, the system was fixed to 50% DO, 7.05–7.2 pH, 37°C and an agitation speed of 300 rpm (upstir). The DO 50% was controlled with “control DO pH” step. The pH was controlled by two different steps: 1. “control DO pH”—that was used to control pH above 7.2 limit by CO2 flow, and 2. “background pH addition”—that was used to control pH below 7.05 limit by additions of bicarbonate buffer. The temperature was controlled by “temperature control” step.
2.5. Growing cells on Fibra‐Cels in reactors
Cells were allowed to grow for 6 days. Replacement of medium with fresh medium was done routinely based on glucose consumption, with a limit of 1 g/L glucose in the medium. To follow glucose, samples were taken regularly every few hours with the “sample from reactor” step. Medium exchange (different percentages) was performed with the “media exchange” step. Glucose concentration was monitored with Cobas Integra 400® Plus (Roche Diagnostics GmbH, Mannheim, German). This analyzer is an automated wet chemistry analyzer with comprehensive testing capabilities. One hundred micro liters of each sample were inserted into capless Eppendorf tubes and tested for glucose (GLC2B) using a commercially available kit from Roche.
During cell growth, DO and pH were monitored by spots (two spots, white and red) located at the bottom of the reactors. Since the pH spots need calibration, samples were taken every 12 h for the pH analysis module (AM). Results from the module were automatically updated in the software, and the offset was calculated. Additions of anti‐foam were performed when required.
2.6. Virus production
The rVSV‐ΔG‐spike virus was constructed as described in. 1 Virus stock was loaded into the system (the stock can be stored in 1/4/24 well plates or in Eppendorf tubes using a Beckman adapter). Infection was carried out by “add to reactor” step to the desired multiplicity of infection (MOI). The amount to be added was calculated in advance, by multiplying the total number of cells in reactor by 0.1 (for a 0.1 MOI). During the virus production process, a sample was taken every 24 h and analyzed using a PFU assay.
2.7. Quantification of cells with Alamar assay
Quantification was done as described in Rosen et al. 16 Briefly, a calibration curve of 50,000–800,000 cells/well was seeded in a first row of 24 well plate (COSTAR). To assess the number of cells grown on Fibra‐Cel, one randomly taken representative Fibra‐Cel was added to a well containing 1 ml medium. All wells were supplemented with 100 μl Alamar blue. The plate was covered with aluminum foil to protect it from light and incubated for 1 hour at 37°C without CO2 after which the plate was read in a SPARK plate reader, TECAN, NEOTEC BIO, with Ex 550 nm, Em 580 nm filters. The number of cells grown on Fibra‐Cel was calculated based on FORCAST of the calibration curve. All data were analyzed by Excel and Prism.
2.8. Quantification of cells by glucose consumption rate
Cell quantity on each Fibra‐Cel carrier was estimated by a correlation between glucose consumption rate and number of cells. During the first experiment, the glucose concentration was measure at different time intervals in each reactor. Glucose consumption rate was then calculated based on: (residual glucose concentration at time 2—residual glucose concentration at time 1) / (time 2—time 1). In parallel, for the same time points, the numbers of cells on Fibra‐Cel carriers was quantified by Alamar blue as described in previous section. 16 , 17 A strong correlation between glucose consumption rate and number of cells on Fibra‐Cel carrier was found and was used in our next experiments to estimate the number of cells on Fibra‐Cel carrier in every time point
2.9. Calcein Staining
Cells on Fibra‐Cel were stained with Calcein AM. Calcein solution (1 μl of 4 mM Calcein in 1 ml of PBS) was added to each well containing Fibra‐Cel and incubated at 37°C for 10 min. The plate was analyzed with fluorescence microscopy.
2.10. Plaque forming unit (PFU)
The virus titer was determined with a plaque forming unit (PFU) assay (Dulbecco et al., 1952). The rVSV‐ΔG‐Spike creates a center of destruction on a monolayer cell culture. Samples from reactors were spread, in different dilutions, on E6 Vero cells in wells of a six well plate (the PFU assay was performed with E6 Vero cells. The virus production was performed in Vero cells mentioned in 2.1). After 1 hour incubation, a layer of 0.8% tragacanth was added to each well. After 3 days of incubation (37°C, 5% CO2) the cells were fixed, stained with crystal violet, and the number of plaques was counted. The virus titer (PFU/ml) was calculated by multiplying the number of plaques formed in a well by the dilution factor and the sow factor volume.
3. RESULTS AND DISCUSSION
Our VSV‐based vaccine is produced in Vero cells. The cells are first grown for 6 days on Fibra‐Cels and then infected. After infection, the virus is produced and released into the culture medium. At the end of the production process, 2 days post infection, the medium containing the vaccine virus is collected. As part of our VSV‐based vaccine production project, we first planned to optimize the main parameters with the Ambr15 system. To that end, the process must be first simulated in the system. However, the Ambr15 system is designed to simulate processes involving suspension cells or cells growing on microcarriers and had never been tested for simulating cells growing on macrocarriers, such as Fibra‐Cel. As a result, some creative adaptive optimizations were needed and are described herein. After system adjustment, absorbance of cells to Fibra‐Cel was tracked, cell growth was monitored, and the virus production step was performed thereby simulating the entire vaccine production process.
3.1. Optimizing process parameters
3.1.1. Design and preparation of a funnel for Fibra‐Cel insertion
The VSV‐based vaccine production process is performed in the BioBLU® 5p Eppendorf single‐use bioreactors (Cino et al. 2011), pre‐packed with tens of thousands of Fibra‐Cels. Therefore, to simulate the vaccine production process in the Ambr15 system, it was necessary to insert Fibra‐Cels into the reactors. The bioreactors' working volume is >200 times more than that of Ambr15 reactors (3.5 L vs. 15 ml). Based on this ratio, a few tens of Fibra‐Cel is need to be inserted into each single use reactor.
The reactor sample port diameter is about 7 mm while Fibra‐Cel diameter is 6 mm (Figure 1 (a) and (b)), making their insertion challenging. Moreover, when the Fibra‐Cel is wet (as part of sterilization and preparation for production) it adsorbs fluid and swells to an even a larger volume. As a solution, a funnel that would enable fast and easy insertion of the Fibra‐Cel to the reactor was suggested. A few funnels were tested, some made of glass and others of plastic. A plastic funnel was selected with a hose that narrows toward the bottom of the funnel. As a consequence, a cut in the funnels hose was needed to make its diameter equal to that of the reactor port. A cut, 0.5 cm above the funnel's nose end allowed a smooth connection between the funnel and the reactor port. A 2 cm long rubber hose was wrapped over the funnel's end, giving 0.5 cm to be wrapped around the entry of the reactor port (Figure 1 [c]). The rubber hose was used to avoid leaking and to permit direct connection of the funnel to the reactor. The funnel can be autoclaved and is therefore reusable. Insertion of tens of Fibra‐Cel to 12 reactors with the above funnel was easy and took less than 10 min (Figure 1 [d]).
FIGURE 1.

Design of a funnel for Fibra‐Cel insertion. (a) Ambr15 reactor, indicating its sample port, through which Fibra‐Cels are inserted. (b) Illustration of Fibra‐Cels. (c) The funnel designed for Fibra‐Cel insertion, indicating the rubber hose wrapped around the funnel and the reactor port. (d) 12 reactors filled each with 60 Fibra‐Cels
3.1.2. Optimizing agitation and Fibra‐Cels macrocarriers quantity
Cells growing adherent to a solid matrix, such as macrocarriers, are exposed to shear forces from the moving fluid which may have critical impact on these cells. Shear forces that are too high may cause the cells to detach from the carriers. Therefore, the agitation speed should be optimized to ensure homogenous mixing on one hand and minimal shear‐force damage to cells on the other. To visualize the influence of different agitation speeds, three reactors were each filled with 10 ml FLEX‐20 medium and differing numbers of Fibra‐Cels (30, 60 or 120). The reactors were loaded on the Ambr15 deck and were agitated at 300 rpm, the lowest speed possible. The medium in all reactors was mixed efficiently, as indicated by same cell growth in different Fibra‐Cels, while the Fibra‐Cels stayed in the same place with no influence of agitation. Elevating the agitation speed to 500 rpm resulted in significant movement of Fibra‐Cels and their exit from the medium, especially with the higher number of Fibra‐Cels (60 and 120). In light of these results, an agitation speed of 300 rpm was selected.
The BioBLU® 5p Eppendorf single‐use bioreactors (Cino et al. 2011) have a working volume of 3.5 L and approximately 35,000 Fibra‐Cels; thus, there are ~10 Fibra‐Cels per ml medium. Ambr15 reactors working volume is 12–15 ml. Consequently, to keep the same ratio, it is necessary to use 120–150 Fibra‐Cels in each reactor. However, the Fibra‐Cels within the bioreactors are circumscribed to a specific area, while no such area exists in Ambr15 reactors. Therefore, it was necessary to test the suitability of different numbers of Fibra‐Cels in a reactor.
Between 30 and 120 Fibra‐Cels were inserted into reactors filled with 15 ml (max volume) FLEX‐20. Reactors were agitated at 300 rpm for 30 min. No fluid exit from the reactors was seen as a result of mixing in any reactor. However, after a few minutes of mixing, the higher Fibra‐Cels number interrupted the mixing process, resulting in exit of Fibra‐Cels from the medium and non‐homogeneous mixing. Consequently, to avoid these problems, 60 Fibra‐Cels were chosen as an optimal number. In ~12 ml medium in the reactor, this represents 5 Fibra‐Cels per ml, which is half of that in a bioreactor. Although not ideal, this quantity allows proper mixing while best simulating the process in bioreactors.
3.2. Bioprocess development using the Ambr15 system
To determine whether the Ambr15 system can be used to optimize our vaccine production process, it was necessary to demonstrate that all production steps can be simulated in the system. Production comprises three major steps, adsorption of cells to Fibra‐Cels; growth of cells for 6 days; and virus production by these cells.
3.2.1. Adsorption of cells to Fibra‐Cels in Ambr15 reactors
To test adsorption of cells to Fibra‐Cels in the Ambr15 system, experiments that mimic the procedure executed in bioreactors were performed. Ten reactors were loaded with 60 Fibra‐Cels each in PBS as described above. A “rapid vessel drain” step was carried out, to drain the PBS, leaving reactors with only 60 Fibra‐Cels. At this point, reactors were filled with fresh FLEX‐20 medium, and were left to stabilize for 24 h. The temperature was set to 37°C with a fixed flow of 0.02 ml/minute of nitrogen. After 24 h, 3 ml of medium were taken from the reactors and replaced manually with 3 ml of Vero cells at different concentrations. The maximum average number of Vero cells per Fibra‐Cel was calculated from these initial concentrations, by dividing the total amount by 60 Fibra‐Cels. The agitation speed was set to 300 rpm and the dissolved oxygen (DO) was set to 50%.
Every 30 min (and up to 3 h) a 100 μl medium sample was taken from each reactor. The concentration of cells in these samples was quantified with a cytometer. The percentage of cells adsorbed to Fibra‐Cels at each time point was calculated as follows:
First, the total number of un‐adsorbed cells was calculated for each reactor by multiplying the reactor volume with the concentration of cells in the samples.
The result of step 1 was subtracted from the total number of cells that had been inoculated into the reactor.
The result of step 2 was divided by 60 (number of Fibra‐Cels per reactor) to find the average number of cells adsorbed to Fibra‐Cels.
The results of step 3 were divided by the maximum theoretical number of cells adsorbed to Fibra‐Cels based on the inoculation concentration (maximum adsorption). The results are the percentage of adsorption.
The adsorption percentages found in all experiments were consistent, achieving 70–80% adsorption after 2 h, regardless inoculation concentration. Results of 4 representative reactors, inoculated with different initial concentration, are shown in Figure 2. As can be seen, the adsorption percentages increased over time until reaching a maximum after 2 h. Longer adsorption times (up to 3 h) did not increase the percentages (data not shown).
FIGURE 2.
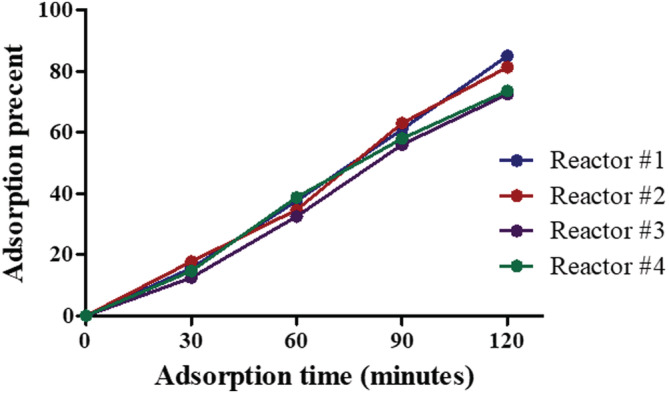
Adsorption percentages over time in representative reactors. Each reactor was inoculated with a different number of cells, and adsorption was monitored every 30 min up to 3 h. Percentages of adsorption were calculated for every reactor and each time point
To examine adsorbent visually, a few Fibra‐Cels were taken from different reactors 2 h after inoculation with 12,500–100,000 cells/Fibra‐Cel. The Fibra‐Cels were stained with Calcein AM, a cell permeant dye that indicates cell viability. In live cells, the non‐fluorescent Calcein AM is converted to a green fluorescent Calcein after acetoxy‐methyl ester hydrolysis by intracellular esterases. Figure 3 demonstrate cells adsorbed to an individual representative Fibra‐Cel taken from 3 different reactors, inoculated with 12,500 (a), 42,000 (b) or 100,000 (c) cells/Fibra‐Cel. The variance between cells adsorption is clearly seen as is a clear correlation between the number of cells adsorbed to Fibra‐Cels and the initial cell inoculum.
FIGURE 3.
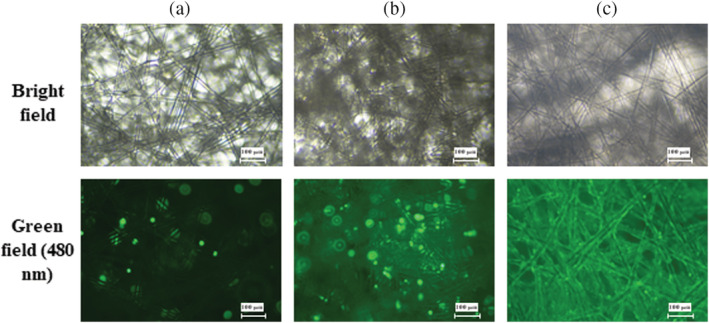
Calcein AM staining of cells adsorbed to Fibra‐Cels. Different numbers of cells/Fibra‐Cel—12,500 (a), 42,000 (b), and 100,000 (c)—were inoculated and allowed to adsorb for 2 h. Sample Fibra‐Cels were taken from reactors and stained with Calcein AM. The polypropylene fibers inside the Fibra‐Cel material is visualized using Bright filed microscopy (upper row). Live cells adsorbed to Fibra‐Cel are visualized as bright green spots (lower row)
Since the cells produce the virus, high cell biomass is desired. In addition, adsorption of 70–80% was seen regardless of inoculation concentrations and better homogenous coverage of cells on Fibra‐Cel was seen at 100,000 cells/Fibra‐Cel. Therefore, at this point the results indicated a need for ~100,000 cells/Fibra‐Cel at the beginning of the cell growth phase. Based on ~70–80% adsorption percentages, to reach ~100,000 cells/Fibra‐Cel at the beginning of the growth phase, inoculations contained >120,000 cells/Fibra‐Cel with the adsorption time set to 2 h.
3.2.2. Cell growth on Fibra‐Cel in Ambr15 reactors
After demonstrating cells' adsorption to macro carriers, obtaining adsorption curves and deciding upon adequate adsorption time (2 h), the cells' ability to grow on Fibra‐Cels in the reactors was tested. At the end of the adsorption time, reactors were drained, and 14 ml of fresh medium was added. The system was fixed to 50% DO, 7.05–7.2 pH, 37°C and agitation speed of 300 rpm (upstir). The growth phase in the bioreactors continued for 6 days at the end of which cells biomass was expected to be sufficient for virus production. During growth, the cells' condition was examined continuously to ensure both their viability and that all the nutrients needed for their growth would be provided. As done in bioreactors, the culture medium needed to be replaced in the Ambr15 reactors to supply new nutrients on one hand and remove waste products on the other, allowing for a desirable growth environment.
To monitor the cells' condition, a sample of 100 μl was taken from each reactor every 8–12 h and was analyzed by the automatic chemistry analyzer Cobas Integra 400 Plus. Depending on the measured glucose concentration, medium exchange was performed to maintain a glucose concentration above 1 g/L. During the first days, only 10–20% exchange was performed every 12–24 h. As the number of cells grew, a higher volume (up to 70%) of medium was exchanged in shorter intervals (8 h).
To estimate cell quantity on Fibra‐Cel, a correlation between glucose consumption rate and number of cells was obtained and used (as described in Section 2.8). It was found that an average of 40 pg glucose/hour per cell was consumed (Figure 4). An example of typical cell growth curves in the Ambr15 system is shown in Figure 5. In this experiment, different inoculation cell numbers were used, including 12,500, 42,000 and 100,000 cells/Fibra‐Cel. Cell growth was monitored for 6 days (144 h). Estimated cell numbers on Fibra‐Cel at each time point was based on glucose consumption rate (the estimated cell numbers were divided by 60 Fibra‐Cel/reactor to get cells/Fibra‐Cel). In reactors that were inoculated with fewer cells (12,500 and 42,000 cells/Fibra‐Cel), a lag phase was seen, and growth began after 2 days. With more cells inoculated (100,000 cells/Fibra‐Cel) the cells adapted and thrived with a much shorter lag phase, until a few hours. The jump in cell numbers in specific time points (for example 90–100 hours for 100,000 cells/Fibra‐Cel) is probably as an artificial result caused by media exchange and the fact that cell numbers were estimated by glucose consumption rate. After 6 days of growth, cells numbers on Fibra‐Cel reached 0.6 × 106, 1 × 106, and 2 × 106 (for the inoculation of 12,500, 42,000, and 100,000 cells/Fibra‐Cel, respectively). These results, together with the adsorption studies described above, set the ground for vaccine production process and demonstrate that the Ambr15 system is competent to mimic the adsorption and cell growth steps. The next and critical step was to simulate virus production.in the Ambr15 system.
FIGURE 4.
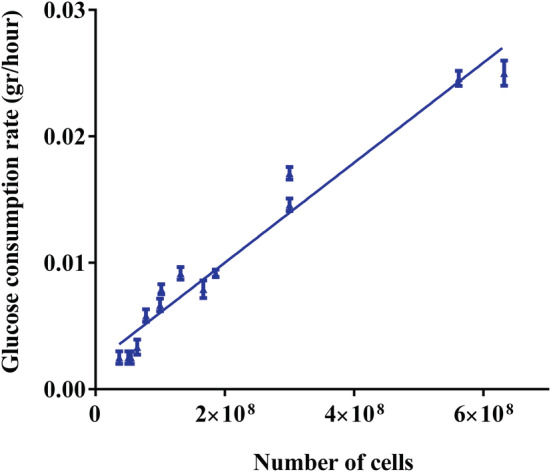
Correlation between glucose consumption rate and number of cells. Glucose concentration was determined in several time points and its consumption rate was calculated by the difference in glucose concentration divided by the time interval. In parallel, the numbers of cells on Fibra‐Cel carriers was quantified by Alamar blue. A strong correlation between glucose consumption rate and number of cells was found to be of 40 pg/h/cell with R 2 = 0.959
FIGURE 5.
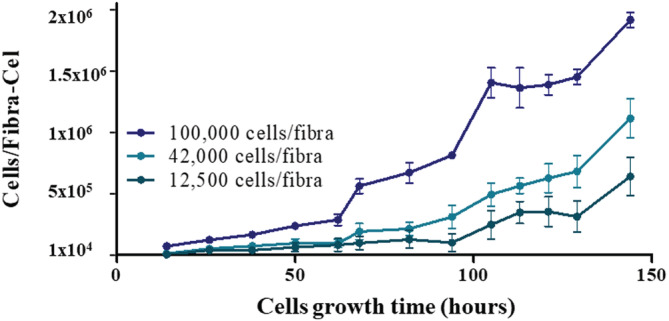
Cell numbers on Fibra‐Cel estimated by glucose consumption rate. Reactors were inoculated with 12,500, 42,000, and 100,000 cells/Fibra‐Cel. Growth was followed for 6 days and cell numbers attained were estimated by glucose consumption rate. Bars represent the mean of the results from three different reactors ± standard deviation
3.2.3. Vaccine virus production
After demonstrating that all the previous steps were well simulated in the Ambr15 system, simulation of the last step—virus production—had to be demonstrated. For that purpose, cells adsorbed to Fibra‐Cels were grown for 6 days in the same conditions described above. At day 6, the number of cells was estimated by glucose consumption rate to be 750,000 cells/Fibra‐Cels. For infection, MOI of 0.1 was used (The MOI refers to the number of virions that are added per cell during infection; an MOI of 0.1 means that 1 virion is added per 10 cells). The total number of cells within a reactor was estimated as 45 × 106 (750,000 cells/Fibra‐Cels multiply by 60 Fibra‐Cels per reactor). Therefore, for an MOI of 0.1, 4.5x106 virions were needed. Because the reactor contains 14 ml, therefore, 14 ml with of 3.2 × 105 virions/ml was added.
The number of virions per ml was tested by the PFU assay every day for 4 consecutive days. The results are shown in Figure 6 (red). After infection with 3.2 × 105 virions/ml (day 0), there was a small increase in virion concentration at day 1, indicating that the process started. More than an order of magnitude increase was seen at day 2, demonstrating virus production. The concentration remained the same at days 3 and 4, probably as a result of limited nutrient resources for production and/or byproduct accumulation. These results clearly demonstrate the suitability of the Ambr15 system for simulating the virus production step. Establishment of the process simulation was a basic requirement for later experiments conducted for vaccine production process optimization with the system. 15 In addition, virus production kinetics in the Ambr15 system, using the same basic process parameters, was compared in Bioreactors. The similar results that were achieved (Figure 6, blue) further demonstrate the Ambr15 ability to model processes in larger reactors.
FIGURE 6.
Infection of Vero cells grown on Fibra‐Cels with rVSV‐ΔG‐Spike. Infectious viral titers expressed as PFU/ml are plotted against the time post infection. Bars represent the mean of the samples from three different biological replicates ± standard deviation
4. CONCLUSIONS
While some studies have evaluated the Ambr15 system as a model for process development, none have shown a process involving cells grown on macrocarriers. In addition, there have been no reports of using the system for a complex process comprising both cell growth and virus production. In this paper we have shown, for the first time, a simulation of an entire vaccine production process. The process includes inoculation and adsorption of cells to Fibra‐Cels, cell growth on these carriers, infection with vaccine virus and vaccine virus production. This simulation was later served as a basis for vaccine production process optimization with the system. 15 Moreover, these capabilities of the Ambr15 system can be incorporated to other processes and can promote cell‐based vaccine production. Integration of macrocarriers expands the Ambr15 spectrum, which had been limited to suspension cells or microcarriers only. The introduction of macrocarriers into this system enables sustained periods of high‐density cell growth in perfusion, without the risk of clogging and eliminates the need for cell filtration steps to separate cells from end products.
AUTHOR CONTRIBUTIONS
Avital Jayson: Data curation (lead); formal analysis (equal); methodology (supporting); writing – original draft (supporting); writing – review and editing (equal). Michael Goldvaser: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Eyal Dor: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Arik Monash: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Lilach Levin: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Lilach Cherry: Formal analysis (supporting); funding acquisition (supporting); writing – review and editing (supporting). Edith Lupu: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Niva Natan: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Meni Girshengorn: Data curation (supporting); formal analysis (supporting); writing – review and editing (supporting). Eyal Epstein: Conceptualization (supporting); investigation (equal); methodology (supporting); project administration (lead); writing – review and editing (supporting). Osnat Rosen: Conceptualization (lead); data curation (equal); formal analysis (equal); methodology (equal); writing – original draft (lead); writing – review and editing (equal).
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare that are relevant to the content of this article.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/btpr.3277.
ACKNOWLEDGMENTS
We would like to thank Dr. Sandy Livnat for editorial assistance.
Jayson A, Goldvaser M, Dor E, et al. Application of Ambr15 system for simulation of entire SARS‐CoV‐2 vaccine production process involving macrocarriers. Biotechnol. Prog. 2022;e3277. doi: 10.1002/btpr.3277
Contributor Information
Eyal Epstein, Email: eyale@iibr.gov.il.
Osnat Rosen, Email: osnatr@iibr.gov.il.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Yahalom‐Ronen Y, Tamir H, Melamed S, et al. A single dose of recombinant VSV‐∆G‐spike vaccine provides protection against SARS‐CoV‐2 challenge. Nat Commun. 2020;11(1):6402. doi: 10.1038/s41467-020-20228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ng SK. Current cell‐based influenza vaccine production technology as pandemic contingency. Hum Vaccin Immunother. 2012;8(2):267‐271. doi: 10.4161/hv.18336 [DOI] [PubMed] [Google Scholar]
- 3. Aubrit F, Perugi F, Léon A, Guéhenneux F, Champion‐Arnaud P, Lahmar M, Schwamborn K. Cell substrates for the production of viral vaccines. Vaccine 2015;33(44):5905–5912. doi: 10.1016/j.vaccine.2015.06.110 [DOI] [PubMed] [Google Scholar]
- 4. Cino J, Mirro R, Kedzierski S. An update on the advantages of Fibra‐Cel® disks for cell. Culture. 2011. https://www.eppendorf.com/product‐media/doc/en/70429/New‐Brunswick_Fermentors‐Bioreactors_Application‐Note_313_Fibra‐Cel_An‐Update‐Advantages‐Fibra‐Cel‐Disks‐Cell‐Culture.pdf [Google Scholar]
- 5. Han X(K), Becken U, Sha M. Vero perfusion, packed‐bed vessels intensify vaccine production. Genet Eng Biotechnol News. 2018;38(15):S16‐S18. doi: 10.1089/gen.38.15.13 [DOI] [Google Scholar]
- 6. Fletcher T. Using a microscale bioreactor to develop cell culture media explicitly for steady state processes.; 2014.
- 7. Lewis G, Lugg R, Lee K, Wales R. Novel automated micro‐scale bioreactor technology: a qualitative and quantitative mimic for early process development. BioProcess J. 2010;9(1):22‐25. [Google Scholar]
- 8. Hsu WT, Aulakh RPS, Traul DL, Yuk IH. Advanced microscale bioreactor system: a representative scale‐down model for bench‐top bioreactors. Cytotechnology. 2012;64(6):667‐678. doi: 10.1007/s10616-012-9446-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moses S, Manahan M, Alexandre A, Ling WLW. Assessment of AMBRTM as a model for high‐throughput cell culture process development strategy. Adv Biosci Biotechnol. 2012;3:918‐927. [Google Scholar]
- 10. Rameez S, Mostafa SS, Miller C, Shukla AA. High‐throughput miniaturized bioreactors for cell culture process development: reproducibility, scalability, and control. Biotechnol Prog. 2014;30(3):718‐727. doi: 10.1002/btpr.1874 [DOI] [PubMed] [Google Scholar]
- 11. Ratcliffe E, Glen KE, Workman VL, Stacey AJ, Thomas RJ. A novel automated bioreactor for scalable process optimisation of haematopoietic stem cell culture. J Biotechnol. 2012;161(3):387‐390. [DOI] [PubMed] [Google Scholar]
- 12. Rafiq QA, Hanga MP, Heathman TRJ, et al. Process development of human multipotent stromal cell microcarrier culture using an automated high‐throughput microbioreactor. Biotechnol Bioeng. 2017;114(10):2253‐2266. doi: 10.1002/bit.26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costariol E, Rotondi M, Amini A, et al. Establishing the scalable manufacture of primary human T‐cells in an automated stirred‐tank bioreactor. Biotechnol Bioeng. 2019;116(10):2488‐2502. doi: 10.1002/bit.27088 [DOI] [PubMed] [Google Scholar]
- 14. Rotondi M, Grace N, Betts J, et al. Design and development of a new ambr250® bioreactor vessel for improved cell and gene therapy applications. Biotechnol Lett. 2021;43(5):1103‐1116. doi: 10.1007/s10529-021-03076-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosen O, Jayson A, Goldvaser M, et al. Optimization of VSV‐ΔG‐spike production process with the Ambr15 system for a SARS‐COV‐2 vaccine. Biotechnol Bioeng. 2022. 10.1002/bit.28088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosen O, Jayson A, Natan N, et al. Novel method for quantifying cells on carriers and its demonstration during SARS‐2 vaccine development. Biotechnol Bioeng. 2021;118(10):3811‐3820. doi: 10.1002/bit.27856 [DOI] [PubMed] [Google Scholar]
- 17. Rosen O, Jayson A, Natan N, Epstein E. A simple method for in situ quantification of cells on carriers. Biol Protoc. 2021;11(23):e4254. doi: 10.21769/BioProtoc.4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


