Abstract
We investigated whether peripheral blood levels of SARS‐CoV‐2 Spike (S) receptor binding domain antibodies (anti‐RBD), neutralizing antibodies (NtAb) targeting Omicron S, and S‐reactive‐interferon (IFN)‐γ‐producing CD4+ and CD8+ T cells measured after a homologous booster dose (3D) with the Comirnaty® vaccine was associated with the likelihood of subsequent breakthrough infections due to the Omicron variant. An observational study including 146 nursing home residents (median age, 80 years; range, 66–99; 109 female) evaluated for an immunological response after 3D (at a median of 16 days). Anti‐RBD total antibodies were measured by chemiluminescent immunoassay. NtAb were quantified by an Omicron S pseudotyped virus neutralization assay. SARS‐CoV‐2‐S specific‐IFNγ‐producing CD4+ and CD8+ T cells were enumerated by whole‐blood flow cytometry for intracellular cytokine staining. In total, 33/146 participants contracted breakthrough Omicron infection (symptomatic in 30/33) within 4 months after 3D. Anti‐RBD antibody levels were comparable in infected and uninfected participants (21 123 vs. 24 723 BAU/ml; p = 0.34). Likewise, NtAb titers (reciprocal IC50 titer, 157 vs. 95; p = 0.32) and frequency of virus‐reactive CD4+ (p = 0.82) and CD8+ (p = 0.91) T cells were similar across participants in both groups. anti‐RBD antibody levels and NtAb titers estimated at around the time of infection were also comparable (3445 vs. 4345 BAU/ml; p = 0.59 and 188.5 vs. 88.9; p = 0.70, respectively). Having detectable NtAb against Omicron or SARS‐CoV‐2‐S‐reactive‐IFNγ‐producing CD4+ or CD8+ T cells after 3D was not correlated with increased protection from breakthrough infection (OR, 1.50; p = 0.54; OR, 0.0; p = 0.99 and OR 3.70; p = 0.23, respectively). None of the immune parameters evaluated herein, including NtAb titers against the Omicron variant, may reliably predict at the individual level the risk of contracting COVID‐19 due to the Omicron variant in nursing home residents.
Keywords: anti‐spike antibodies, breakthrough infection, Comirnaty® COVID‐19 vaccine, neutralizing antibodies, nursing home residents, SARS‐CoV‐2 Omicron variant, spike‐reactive T cells
1. INTRODUCTION
The SARS‐CoV‐2 Omicron variant has become dominant in many countries 1 due to its increased transmissibility compared to the Delta variant, owing at least partly to its remarkable ability to escape from SARS‐CoV‐2 Spike (S) neutralizing antibodies (NtAb) elicited during natural infection or after vaccination with regular or booster schedules. 2 , 3 , 4 , 5 Adaptive immunity is critically involved in preventing SARS‐CoV‐2 infection 6 ; nevertheless, protective thresholds remain elusive for both S‐binding NtAb and T cells. 7 Herein, taking advantage of SARS‐CoV‐2 Omicron variant outbreaks in several nursing homes, whose congregate nature facilitates wide exposure to the virus, we investigated whether levels of antibodies targeting the receptor‐binding domain (RBD) of S (anti‐RBD), NtAb targeting Omicron S, and S‐reactive functional T cells measured after a homologous booster dose (3D) with Comirnaty® were associated with the likelihood of subsequently contracting breakthrough infections in elderly nursing home residents.
2. MATERIALS AND METHODS
2.1. Participants
The current prospective and observational study included 146 participants (median age, 80 years; range, 66–99; 109 female; median Charlson comorbidity index of 7, range 1–14) institutionalized in four nursing homes in the Valencian Community (Spain). Participants vaccinated with the Comirnaty® vaccine (two doses) were evaluated for their immunological response after a homologous booster dose (a median to 16 days; range, 15–18), 8 which was given to all participants irrespective of their SARS‐CoV‐2 infection status, and were followed for up 4 months. SARS‐CoV‐2 infection status at the time of 3D (naïve vs. experienced) was defined according to historical records in the electronic Valencia Health System Integrated Databases and/or the presence or absence of anti‐SARS‐CoV‐2 nucleocapsid (N) IgG antibodies. None of the residents included in the study had a documented immunosuppression condition or were under immunosuppressive therapy within the follow‐up period. Anti‐RBD and anti‐N antibody detection and T‐cell assays were performed at the Microbiology Service of the Hospital Clínico Universitario of Valencia. NtAb was measured at the Institute for Integrative Systems Biology, Universitat de Valencia‐CSIC. Whole‐genome sequencing was performed at the Foundation for the promotion of Health and Biomedical Research of the Valencian Community (FISABIO) (Valencia, Spain).
2.2. Virological diagnosis of SARS‐CoV‐2 breakthrough infections
Residents suspected of having developed COVID‐19 were tested within 24 h after symptoms onset by reverse transcription polymerase chain reaction (RT‐PCR) for detection of SARS‐CoV‐2 RNA in nasopharyngeal specimens. Asymptomatic residents were tested by RT‐PCR within 48 h of diagnosis of the index case and two to three times afterward throughout the outbreak. Involvement of the SARS‐CoV‐2 Omicron BA.1 variant was documented by whole‐genome sequencing performed at the FISABIO (Valencia, Spain), as previously described. 8
2.3. Immunological testing
Anti‐RBD total antibodies and N‐reactive IgGs were detected by the Roche Elecsys® Anti‐SARS‐CoV‐2 S and the Elecsys® Anti‐SARS‐CoV‐2 N assays (Roche Diagnostics), respectively, with values ≥ 0.4 BAU/ml and cut‐off index ≥1.0 considered positive results in the respective tests, according to the manufacturer.
NtAb targeting the S protein was measured using a Green fluorescent protein (GFP)‐expressing vesicular stomatitis virus pseudotyped with the Omicron variant, as previously described. 8 , 9 In brief, we introduced mutations in a mammalian expression vector encoding a codon‐optimized SARS‐CoV‐2 S sequence from the Wuhan‐Hu‐1 (ancestral) reference variant. First, the D614G mutation was introduced by site‐directed mutagenesis. Subsequently, additional site‐directed mutagenesis and/or the cloning of synthetic fragments harboring the mutations (Gblocks, IDT) was performed to introduce all mutations using the NEBuilder HiFi DNA assembly mix (NEB). Pseudotype vesicular stomatitis virus (VSV) carrying the S protein of Omicron was generated using a codon‐optimized Omicron S expression plasmid (Genescript MC_0101274) that was modified to delete the C‐terminal 19 amino acids to improve pseudotyping efficiency. The neutralization capacity of circulating antibodies (NtAb) against the SARS‐CoV‐2 ancestral (Wuhan‐Hu‐1) and Omicron S was then assessed in plasma samples using this GFP‐expressing VSV pseudotyped virus on A549‐ACE2‐TMPRSS2 cells (InvivoGen catalog code: a549‐hace2tps). All tests were done in duplicate using fivefold serum dilutions ranging from 1:20 to 1:62 500, with ~500 focus forming units per well. Following 16 h of infection, the GFP signal in each well was quantified using a live‐cell microscope (Incucyte S3; Sartorius). Background fluorescence from uninfected wells was subtracted from all infected wells, and the GFP fluorescence in each antibody‐treated dilution was standardized to the average fluorescence observed in mock‐treated wells. Any value resulting in a relative GFP signal of <0.001 versus was assigned a value of 0.001 to eliminate negative values. Finally, the reciprocal antibody dilution resulting in 50% virus neutralization was calculated using the drc package (version 3.0‐1) in R via a three‐parameter log‐logistic regression model (LL.3 model). Sera testing negative (undetectable) were arbitrarily ascribed a titer of 1/20 (limit of quantitation of the assay for all variants).
SARS‐CoV‐2‐S specific‐IFNγ‐producing CD4+ and CD8+ T‐cell immunity were measured by whole‐blood flow cytometry for intracellular cytokine staining (BD Fastimmune, Becton Dickinson, and Company Biosciences) as previously described. 10 Heparinized whole blood (0.5 ml) was simultaneously stimulated for 6 h with two sets of 15‐mer overlapping peptides (11‐mer overlap) encompassing the SARS‐CoV‐2 Spike (S) glycoprotein (S1, 158 peptides and S2, 157 peptides) at a concentration of 1 μg/ml per peptide, in the presence of 1 μg/ml of costimulatory monoclonal antibodies (mAbs) to CD28 and CD49d. Peptide mixes were obtained from JPT Peptide Technologies GmbH. Samples mock‐stimulated with phosphate‐buffered saline (PBS)/dimethyl sulfoxide and costimulatory antibodies were run in parallel. Brefeldin A (10 μg/ml) was added for the last 4 h of incubation. Blood was then lysed (BD FACS lysing solution) and frozen at −80°C until tested. On the day of testing, stimulated blood was thawed at 37°C, washed, permeabilized (BD permeabilizing solution), and stained with a combination of labeled mAbs (anti‐IFNγ‐FITC, anti‐CD4‐APC‐H7, anti‐CD8‐PerCP‐Cy5.5, and anti‐CD3‐APC) for 1 h at room temperature. Appropriate positive (phytohemagglutinin) and isotype controls were used. Cells were then washed, resuspended in 200 μl of 1% paraformaldehyde in PBS, and analyzed within 2 h on a FACSCanto flow cytometer using DIVA v8 software (BD Biosciences Immunocytometry Systems). CD3+/CD8+or CD3+/CD4+ events were gated then analyzed for IFN‐γ production. All data were corrected for background IFN‐γ production (FITC‐labeled isotype control antibody). Data are expressed as the number of SARS‐CoV‐2‐reactive IFN‐γ‐producing CD4+or CD8+ T cells relative to the absolute number of CD4+ and CD8+ T cells, respectively, x100 (%). Any frequency value of SARS‐CoV‐2‐reactive IFN‐γ‐producing CD4+ or CD8+ T cells after background subtraction was considered a positive (detectable) result and used for analysis purposes.
2.4. Statistical methods
Frequency comparisons for categorical variables were carried out using the Fisher exact test. Differences between medians were compared using the Mann–Whitney U‐test. Regression logistic models were built to assess the association between immune parameters and the risk of breakthrough infection. The odds ratio for each of these parameters is reported). The analyses were performed using SPSS version 20.0 (SPSS). Statistical significance was set at p < 0.05. Data on anti‐SARS‐CoV‐2 RBD total antibody levels in sequential specimens collected from 33 residents at a median of 20 days and3 months after 2D, and at the time of 3D, were used to build an exponential decay model 11 to estimate antibody half‐life on an individual basis.
3. RESULTS
3.1. SARS‐CoV‐2 Omicron breakthrough infections in nursing home residents
Out of the 146 participants, 33 (22.6%) developed a breakthrough infection due to the Omicron BA.1 variant, at a median of 105 days after 3D (range, 76–123). Case distribution across participating nursing homes is shown in Supporting Information: Table 1. SARS‐CoV‐2 infection was symptomatic in 30/33 participants (two requiring hospitalization). Incidence of breakthrough infection was comparable (p = 0.12) across SARS‐CoV‐2‐naïve (22/80) and experienced (11/66) residents at 3D.
3.2. Anti‐RBD antibody response following the third vaccine dose and subsequent occurrence of SARS‐CoV‐2 Omicron infection
In total, 145/146 participants had detectable anti‐RBD total antibodies after 3D (median, 16 days). Residents developing breakthrough infection displayed comparable median plasma levels of anti‐RBD antibodies to those who did not (21 123 vs. 24 723 BAU/ml; p = 0.34) (Figure 1A).
Figure 1.
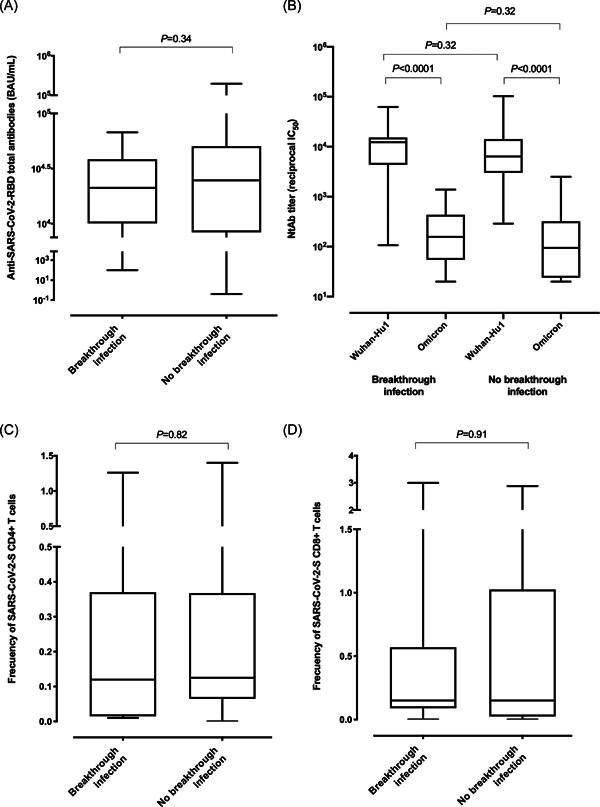
SARS‐CoV‐2‐S‐reactive antibody and T‐cell responses following a booster dose of the Comirnaty® vaccine in nursing home residents either with or without subsequent Omicron variant breakthrough infection. The reverse transcription polymerase chain reaction assays used for diagnosis were the Roche Cobas 6800 SARS‐CoV‐2 test (Roche Diagnostics) and the TaqPath COVID‐19 Combo Kit (Thermo Fisher Scientific). Box‐and‐whisker plots depicting total anti‐RBD antibody levels (A), neutralizing antibodies against the Spike protein of the Omicron variant and Wuhan‐Hu1 ‐ancestral‐variant (B), frequency of peripheral blood SARS‐CoV‐2‐S specific‐IFNγ‐producing CD4+ (C) and CD8+ (D) T cells in comparison groups. p Values (Mann–Whitney test) for comparisons across infected and uninfected residents are shown. Statistical significance was set at p < 0.05.
3.3. NtAb responses following the third vaccine dose and subsequent occurrence of SARS‐CoV‐2 Omicron infection
Data on S‐reactive NtAb against the ancestral Wuhan Hu‐1 and Omicron variants were available from 68 residents. No differences between those who contracted the infection or not were observed for the fraction of residents with detectable NtAb responses targeting the Omicron variant (24/28 vs. 32/40, respectively; p = 0.75), nor median NtAb titers (reciprocal IC50 titer of 157 vs. 95, respectively; p = 0.32; Figure 1B). As expected, 2 , 3 , 4 , 5 overall, NtAb titers against the Omicron variant were significantly lower than those against the Wuhan Hu‐1 variant and were comparable in magnitude across participants either contracting or not breakthrough infection (p = 0.32). Having detectable NtAb against Omicron was not associated with a decreased risk of developing breakthrough infection (OR, 1.50; 95% CI, 0.40–5.57; p = 0.54).
3.4. SARS‐CoV‐2‐S‐reactive‐IFNγ‐producing T‐cell responses following the third vaccine dose and subsequent occurrence of SARS‐CoV‐2 Omicron infection
Data on SARS‐CoV‐2‐S‐reactive‐IFNγ‐producing T cells were available from 59 participants. The frequency of detectable CD4+ and CD8+ T‐cell responses after 3D was comparable in infected and uninfected participants (13/13 vs. 40/46 for CD4+, respectively; p = 0.32; 12/13 vs. 36/46 for CD8+, respectively; p = 0.42). Moreover, median frequencies of both S‐reactive T‐cell subsets were similar across participants in both groups (p = 0.82 and p = 0.91, shown in Figure 1C,D respectively). Having detectable SARS‐CoV‐2‐S‐reactive‐IFNγ‐producing CD4+ or CD8+ T cells after 3D was not associated with a decreased risk of developing breakthrough infection (OR, 0.0; 95% CI, 0.0 to >50; p = 0.99; OR, 3.70; 95% CI, 0.42–32.76; p = 0.23, respectively).
3.5. Estimated anti‐RBD and NtAb levels at the time of breakthrough infections
Sequential data on anti‐RBD total antibodies before receipt of 3D were available from 33 residents. As shown in Figure 2A, the data fit well to an exponential decay model in 22 participants (5 SARS‐CoV‐2‐experienced and 17 naïve), yielding an antibody half‐life of 53.6 days (range, 14.1–164). As a secondary analysis, we used this derived half‐life measurement and the antibody levels quantified after 3D to estimate anti‐RBD and NtAb within 2 days before diagnosis of breakthrough infection as well as at a comparable time in uninfected residents. Median predicted anti‐RBD antibody levels for 2 days pre‐infection were comparable across groups (3445 BAU/ml; IQR, 80.2–21,713 vs. 4345 BAU/ml; IQR, 89.7–33,618, respectively; p = 0.59; Figure 2B). Similarly, predicted NtAb against Omicron 2 days before diagnosis also did not differ between groups (188.5 reciprocal IC50; IQR, 0–267 vs. 88.9; IQR, 0–903.6; p = 0.70; Figure 2C).
Figure 2.
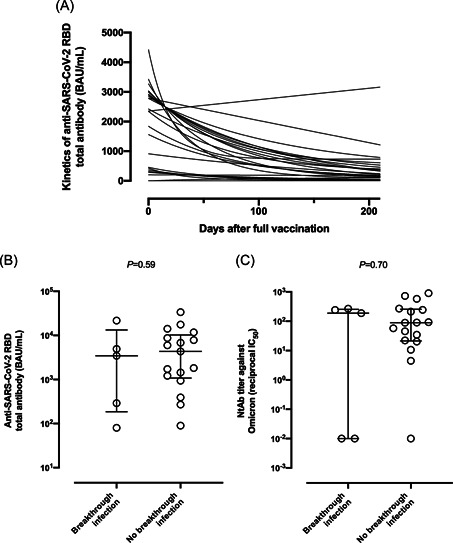
(A) Kinetics of anti‐SARS‐CoV‐2‐RBD total antibody level waning before receipt of the Comirnaty® vaccine booster dose in nursing home residents. Estimated levels of anti‐SARS‐CoV‐2‐RBD total antibodies (B) and neutralizing antibodies against the Spike protein of the Omicron variant (C) at 48 h before diagnosis of breakthrough infection and at comparable times in uninfected participants. p Values (Mann–Whitney test) for comparisons across infected and uninfected residents are shown. Statistical significance was set at p < 0.05.
4. DISCUSSION
In this study, we have been unable to identify a threshold level that would protect against Omicron breakthrough infection within 4 months following 3D in elderly nursing home residents using anti‐RBD antibody levels, Omicron‐binding NtAb titer, and frequencies of SARS‐CoV‐2‐S specific‐IFNγ‐producing CD4+ and CD8+ T cells in whole blood as predictive parameters; indeed, not even the presence or absence of detectable NtAb responses after 3D were predictive of breakthrough success. In the two antibody specificities evaluated (anti‐RBD and NtAb), antibody levels as estimated at around the time of molecular diagnosis of infection were also unpredictive. Omicron BA.1 infection occurred despite strong anti‐RBD antibody levels, as previously reported, 12 and moderate‐to‐high frequencies of peripheral blood S‐reactive T cells. Omicron‐S‐binding NtAb levels were overall rather low after 3D, in line with previously reported data, 2 , 3 , 4 , 5 but this was the case whether or not residents contracted Omicron infection.
The current study has several limitations, including the low number of breakthrough infections in the cohort and the lack of available specimens collected near the time of Omicron infection. In addition, we cannot rule out that the enumeration of monofunctional T cells other than those producing IFNγ or polyfunctional T cells may behave as a reliable marker of protection. Likewise, the assumption that NtAb decay kinetics are similar to those of anti‐RBD antibodies may prove wrong; in fact, a previous study showed NtAb response dynamics in patients who have recovered from COVID‐19 to vary greatly at the individual level. 13 Moreover, antibody decay models were not built separately for SARS‐CoV‐2 naïve and SARS‐CoV‐2 experienced individuals, due to the scarce number of participants in the latter category (n = 5). Finally, the policy of RT‐PCR testing of asymptomatic individuals in nursing homes could have not been optimal for detecting cases, so the possibility of misclassification regarding SARS‐CoV‐2 infection status cannot be ruled out.
Population‐based studies concur that NtAb levels are strongly associated with vaccine efficacy, 14 , 15 yet our data suggested that none of the immune parameters evaluated herein, including NtAb titers against the Omicron variant, may reliably predict the risk of contracting Omicron breakthrough COVID‐19 on an individual basis.
AUTHOR CONTRIBUTIONS
Ignacio Torres, Estela Giménez, Eliseo Albert, Joao Zulaica, Beatriz Álvarez‐Rodríguez, and Ron Geller performed SARS‐CoV‐2 antibody and T‐cell assays and analyzed the data. Javier S. Burgos, Salvador Peiró, Ramón Limón, Hermelinda Vanaclocha, Javier Díez‐Domingo, José Sánchez‐Payá: Conceptualization and data analysis. Celia Rodado, Pilar Botija, Amelia Sifre, Borja Tur, Rosa A. Lozano, Iria Orosa, MªÁngeles Vicente‐Ruiz, Ramón J. Carrión were in charge of managing SARS‐CoV‐2 outbreaks at Nursing Home residences. Iñaki Comas and Fernando González‐Candelas carried out whole‐genome sequencing analyses. David Navarro: Conceptualization, data analysis and writing the original draft. All authors reviewed the original draft.
MEMBERS OF THE VALENCIAN VACCINE RESEARCH PROGRAM (PROVAVAC) STUDY GROUP
J. S. Burgos (General Directorate of Research and Healthcare Supervision, Department of Health, Valencia Government, Valencia, Spain); R. Meneu de Guillerna (Vice‐President Foundation Research Institute in Public Services, Valencia, Spain); H. Vanaclocha Luna (General Directorate of Public Health, Department of Health, Valencia Government, Valencia, Spain); D. J. Burks (The Prince Felipe Research Center‐CIPF, Valencia, Spain; A. Cervantes (INCLIVA Health Research Institute, Valencia, Spain); I. Comas (Biomedicine Institute of Valencia, Spanish Research Council (CSIC); J. Díez‐Domingo (Foundation for the promotion of health and biomedical research of the Valencian Community‐FISABIO, Valencia, Spain); S. Peiro (Foundation for the promotion of health and biomedical research of the Valencian Community‐FISABIO, Valencia, Spain); F. González‐Candelas (CIBER in Epidemiology and Public Health, Spain; Joint Research Unit “Infection and Public Health” FISABIO‐University of Valencia, Valencia, Spain; Institute for Integrative Systems Biology [I2SysBio], CSIC‐University of Valencia, Valencia, Spain); C. Ferrer Albiach (Fundación Hospital Provincial de Castelló); I. Hernández‐Aguado (University Miguel Hernández, Alicante, Spain); N. Oliver Ramírez (DataPop Alliance); J. Sánchez‐Payá (Preventive Medicine Service, Alicante General and University Hospital, Alicante, Spain; Alicante Institute of Health and Biomedical Research [ISABIAL], Alicante, Spain; M. Vento Torres (Instituto de Investigación Sanitaria La Fe); E. Zapater Latorre (Fundación Hospital General Universitario de València); D. Navarro (Microbiology Service, Clinic University Hospital, INCLIVA Health Research Institute, Valencia, Spain; Department of Microbiology, School of Medicine, University of Valencia, Valencia, Spain).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was approved by the institutional ethical review board of the Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana (FISABIO) in Valencia, Spain. This study was undertaken under the epidemiological surveillance competencies of the Valencia Government Health Department (Law 16/2003/May 28 on Cohesion and Quality of the National Health System, and Law 10/2014/December 29 on Public Health of the Valencian Community).
Supporting information
Supplementary information.
ACKNOWLEDGEMENT
We are grateful to the Vice‐presidency and Ministry of Equality and Inclusive Policies of the Valencia Community, the Corporate Association of Residences and Services for People with Dependency of the Valencian Community (AERTE), the Valencia Health System nursing home departmental committees, and the staff and residents of the participant nursing homes for their collaboration in developing the ProVaVac program. We would also like to thank Ana Berenguer, General Director of Analysis and Public Policies of the Presidency of the Generalitat. Ignacio Torres (Río Hortega Contract; CM20/00090), Estela Giménez (Juan Rodés Contract, JR18/00053), and Eliseo Albert (Juan Rodés Contract; JR20/00011) hold contracts funded by the Carlos III Health Institute (cofinanced by the European Regional Development Fund, ERDF/FEDER). Ron Geller holds a Ramon y Cajal fellowship from the Spanish Ministerio de Economía y Competitividad (RYC‐2015‐17517). This study work was supported by Instituto de Salud Carlos III, Madrid, Spain (FIS, PI21/00563) to David Navarro, and by the European Commission NextGenerationEU fund (EU 2020/2094), through CSIC's Global Health Platform (PTI Salud Global) to Ron Geller.
Torres I, Giménez E, Albert E, et al. SARS‐CoV‐2 Omicron BA.1 variant breakthrough infections in nursing home residents after an homologous third dose of the Comirnaty® COVID‐19 vaccine: looking for correlates of protection. J Med Virol. 2022;94:4216‐4223. 10.1002/jmv.27867
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Lino A, Cardoso MA, Martins‐Lopes P, Gonçalves HMR. Omicron—the new SARS‐CoV‐2 challenge? Rev Med Virol. 2022: e2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffmann M, Krüger N, Schulz S, et al. The Omicron variant is highly resistant against antibody‐mediated neutralization: implications for control of the COVID‐19 pandemic. Cell. 2022;185:447‐456.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654‐656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu L, Mok BWY, Chen LL, et al. Neutralization of SARS‐CoV‐2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021: ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature. 2022;602:671‐675. [DOI] [PubMed] [Google Scholar]
- 6. Moss P. The T cell immune response against SARS‐CoV‐2. Nat Immunol. 2022;23:186‐193. [DOI] [PubMed] [Google Scholar]
- 7. Feng S, Phillips DJ, White T, et al. Correlates of protection against symptomatic and asymptomatic SARS‐CoV‐2 infection. Nat Med. 2021;27:2032‐2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giménez E, Albert E, Zulaica J, et al. SARS‐CoV‐2 adaptive immunity in nursing home residents following a third dose of the Comirnaty® COVID‐19 vaccine. Clin Infect Dis. 2022: ciac223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sánchez‐Sendra B, Albert E, Zulaica J, et al. Neutralizing antibodies against SARS‐CoV‐2 variants of concern elicited by the comirnaty COVID‐19 vaccine in nursing home residents. Sci Rep. 2022;12:3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Albert E, Burgos JS, Peiró S, et al. Immunological response against SARS‐CoV‐2 following full‐dose administration of Comirnaty® COVID‐19 vaccine in nursing home residents. Clin Microbiol Infect. 2022;28:279‐284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dimeglio C, Herin F, Da‐Silva I, et al. Post‐vaccination SARS‐CoV‐2 antibody kinetics and protection duration. Clin Infect Dis. 2021: ciab984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dimeglio C, Migueres M, Mansuy JM, et al. Antibody titers and breakthrough infections with Omicron SARS‐CoV‐2. J Infect. 2022;84(4):e13‐e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chia WN, Zhu F, Ong SWX, et al. Dynamics of SARS‐CoV‐2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microb. 2021;2:e240‐e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cromer D, Steain M, Reynaldi A, et al. Neutralising antibody titres as predictors of protection against SARS‐CoV‐2 variants and the impact of boosting: a meta‐analysis. Lancet Microbe. 2022;3:e52‐e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng SMS, Mok CKP, Leung YWY, et al. Neutralizing antibodies against the SARS‐CoV‐2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022;28:486‐489. 10.1038/s41591-022-01704-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


