Abstract
Since the outbreak of coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection, global public health and the economy have suffered unprecedented damage. Based on the increasing related literature, the characteristics and pathogenic mechanisms of the virus, and epidemiological and clinical features of the disease are being rapidly discovered. The spike glycoprotein (S protein), as a key antigen of SARS‐CoV‐2 for developing vaccines, antibodies, and drug targets, has been shown to play an important role in viral entry, tissue tropism, and pathogenesis. In this review, we summarize the molecular mechanisms of interaction between S protein and host factors, especially receptor‐mediated viral modulation of host signaling pathways, and highlight the progression of potential therapeutic targets, prophylactic and therapeutic agents for prevention and treatment of SARS‐CoV‐2 infection.
Keywords: ACE2 receptor, S protein, SARS‐CoV‐2, signaling pathway
1. INTRODUCTION
Acute respiratory diseases (ARD) are considered as the major cause of all acute morbidities and mortalities worldwide. 1 Among these, acute viral respiratory tract infection, including the influenza virus, respiratory syncytial virus, coronavirus, adenovirus, and rhinovirus, is the leading cause of ARD. In the case of adenovirus and rhinovirus, they are associated with lower mortality but significant morbidity, and cause vast health and economic burdens. 2 The observation of the mode of virus transmission from a natural vertebrate animal host to the human host, leading to cause of “zoonotic infection” spread in the human being, could be the case with COVID‐19 (The Coronavirus Disease 2019) (Figure 1A). 4 The zoonotic virus spillover or transfer to human beings can be attributed to various factors, including global warming, climate changing, and loss of biodiversity due to deforestation, poaching, decreased habitat quality, effects of expanded global food production on natural environments. 5 Therefore, emerging infectious diseases have an intense impact in terms of the global burden, threat, and evolutionary origin. For example, in recent years, global populations are facing the threat of relatively unusual but concerning new diseases with severe pandemic impacts such as SARS, COVID‐19, or MERS‐CoV. 6 Research studies have demonstrated that these viruses could be categorized as the simple variants of prediscovered pathogens, new detections of old pathogens in different circumstances, or the case of re‐emergence of old pathogens in new geographical areas (Figure 1B). 7
Figure 1.
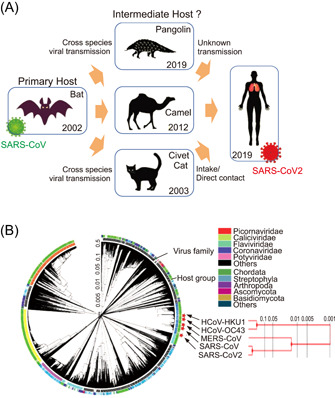
Evolutionary origin and transmission routes of SARS‐CoV‐2. (A) The evolutionary origin and journey of SARS‐CoV‐2 through different primary and intermediate hosts. Different paths of potential viral transmissions are indicated with yellow arrows. (B) The circular phylogenetic tree illustrates the positions of human coronavirus HKU1 (HCoV‐HKU1), human coronavirus OC43 (HCoV‐OC43), Middle East respiratory syndrome coronavirus (MERS‐CoV), Severe acute respiratory syndrome (SARS)‐associated coronavirus (SARS‐CoV and SARS‐CoV‐2) in different host groups. The complete genome sequences were collected from National Center for Biotechnology Information (NCBI) database and analyzed in The Interactive Tree Of Life (https://itol.embl.de) online software tool. 3 A separate phylogenetic tree of HCoV‐HKU1, HCoV‐OC43, MERS‐CoV, SARS‐CoV, and SARS‐CoV‐2 is shown on the right panel.
The COVID‐19 disease caused by severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) infection is one of the most widespread and devastating pandemics in human history. 8 The emergence and spread of such deadly viral diseases become the major cause of destroying global health and the economy. 9 According to a statistical report from World Health Organization (WHO, till March 17, 2022), SARS‐CoV‐2 infection has spread over 472 million people and caused more than 6 million deaths (https://covid19.who.int/). In this situation, remarkable efforts are being carried out by the worldwide scientific community to develop effective and safe vaccine candidates. To this end, it is extremely important to comprehend the complex patterns of host–virus interaction and structure–function analysis of different SARS‐CoV‐2 proteins. 10
It has been shown that several human pathogenic viruses including coronaviruses (CoVs) employ their envelope glycoproteins for receptor recognition and binding with the host leading to membrane fusion, and entry into the host cell to initiate viral infection. 11 Particularly, SARS‐CoV‐2 spike glycoprotein (S protein) plays an important role in initiating the host–pathogen interactions through the host cell surface angiotensin‐converting enzyme‐2 receptor (ACE2), enabling the entry of the virus into the host cell by membrane fusion. 12 Meanwhile, the spike homotrimers can protrude from the viral surface and act as a direct target for host immune responses, eliciting protective B‐cell and T‐cell mediated immune responses in the hosts during SARS‐CoV‐2 infection. 13 The consequence of viral S protein‐ACE2 interaction has a great impact on eliciting cell signaling cascades leading to diverse cellular responses. 14 Several studies have demonstrated that SARS‐CoV‐2 S protein alone is enough to elicit cell signaling events, which signifying a unique biological mechanism of SARS‐CoV‐2 spike protein. It has been proved that the recombinant spike protein S1 subunit promotes cell signaling independent of other viral components in smooth muscle cells or endothelial cells of the human pulmonary artery, leading to the pulmonary vascular remodeling and pulmonary arterial hypertension during SARS‐CoV‐2 infection. 15 , 16 Of note, S protein has gained huge attention in antiviral therapeutic strategies, and the revelation of the sophisticated structure of S protein has provided the much‐needed blueprint for COVID‐19 vaccination strategies. 17
1.1. SARS‐CoV‐2: A glimpse of evolutionary perspectives
Coronavirus is enveloped single‐stranded positive‐sense RNA virus, and belongs to the family of Coronaviridae, subfamily Coronavirinae. As shown in Figure 1B, based on genotype and serological tests, CoVs are classified into four genera assigned as alpha, beta, gamma, and delta coronavirus. 18 Seven viruses are currently identified to infect human beings within the Coronaviridae family, including NL63 and 229E from the genus alpha, OC43, HKU1, SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2 from the genus beta. 18 The SARS‐CoV‐2 is clustered with SARS‐CoVs in the species tree of severe acute respiratory syndrome‐related coronavirus in the Sarbecovirus of genus beta‐coronavirus. 19
In the past few decades, the appearance of MERS‐CoV, SARS‐CoV, and SARS‐CoV‐2 with epidemic capabilities has become the third coronavirus. Of these, SARS‐CoV‐2 causes a lower fatality rate than the other two, but with a higher transmission rate that results in a pandemic shape of COVID‐19. 18 So far, analysis by various bioinformatics tools has provided a deep insight into the genome of COVID‐19 causative pathogen. The SARS‐CoV‐2 genome analysis has revealed high similarity with bat coronavirus genome, and a highly similar receptor‐binding domain (RBD) of the spike protein as Malayan pangolin coronavirus. 20 The genetic proximity indicates that SARS‐CoV‐2 is likely to have originated from bat‐derived CoV, as a consequence of the accumulation of mutations and the acquisition of new genomic regions by recombination events between bat and pangolin coronaviruses (Figure 1A). 21 , 22 However, the exact mechanisms involved with the evolution of this coronavirus as a human pathogen are not fully understood yet. The missing links in the evolution of SARS‐CoV‐2 include the exact immediate reservoir and all other possible hosts. In addition, the spillover at the animal–human interface leading to new human infection is influenced by several factors, such as pathogen pressure human and reservoir host behavior, and the factors linked with human infection susceptibility. 23 In these human‐dominated landscapes, primates, and bats are reservoir hosts of more viruses than other mammal species, which increases the possibility of new infections in humans. 24 For instance, the potential risk of SARS‐CoV spillover from horseshoe bat population to humans was demonstrated by Menachery et al., using a reverse genetics system entailing a chimeric virus expressing the bat SHCO14 in a mouse‐adapted backbone. This study illustrates scenarios for the emergence of bat SARS‐CoV in humans, probably infection of an intermediate nonhuman host might be followed by human infection. Direct bat–human transmission could be followed by selection in the human population. 25 In another scenario, the circulation of quasi‐species pools in the animal reservoirs (camel or civet cat) might maintain multiple virus strains, some of which capable of causing infection in humans without the need for additional mutations. Both alpha‐coronaviruses and beta‐coronaviruses are identified in free‐ranging bats from Myanmar, indicating that the potential for zoonotic virus emergence in humans in close contact with sylvatic animals in forest areas are disturbed by an ongoing process of changes in land use. 26
1.2. Viral entry in cells
SARS‐CoV‐2 infects human cells via S protein binding to the receptor ACE2 on the host cell that allows the virus entry into the cell. 10 As shown in Figure 2A, the entry of coronaviruses into target cells depends on the binding of the S1 subunit of S protein to specific cellular receptor ACE2. 28 The viral protein‐receptor binding helps viral attachment to the cell surface. In addition, viral entry requires an important step known as “protein priming” by cellular proteases (TMPRSS2), which involves the cleavage of S protein at the S1/S2 and the S2′ sites enabling the fusion of viral and cellular membranes. 29 Although the binding of viral S protein and ACE2 receptors has been explained in detail, and ACE2 is considered as the prime determinant of SARS‐CoV transmissibility, 30 it is still not clear whether SARS‐CoV‐2‐S follows the same mechanisms as SARS‐CoV‐S which employs ACE2 and TMPRSS2 for cellular entry. In addition to depending on ACE2 for host cell entry, both SARS‐CoV and SARS‐CoV‐2 depend on entry activation by host cell proteases at the S1/S2 and S2′ sites, regardless of whether entry occurs by fusion or endocytosis. 31 In contrast to SARS‐CoV, SARS‐CoV‐2 also has a furin cleavage site that can be recognized by neuropilin‐1 for infectivity. 32 Like other trimeric class I fusion proteins, S proteins from SARS‐CoV, SARS‐CoV‐2, and MERS‐CoV undergo dramatic structural changes to fuse membranes, which occurs after proteolytic activation at the S1/S2 boundary. 31 S1 dissociates, and conformational changes allow the fusion peptide (FP), which is hydrophobic residues‐rich and inserted into the host cell membrane. 33 S2 forms an elongated structure, and the two heptad repeats, HR1 and HR2, eventually form a six‐helix bundle to complete the fusion process and deliver the viral genome into the cytoplasm. 34
Figure 2.
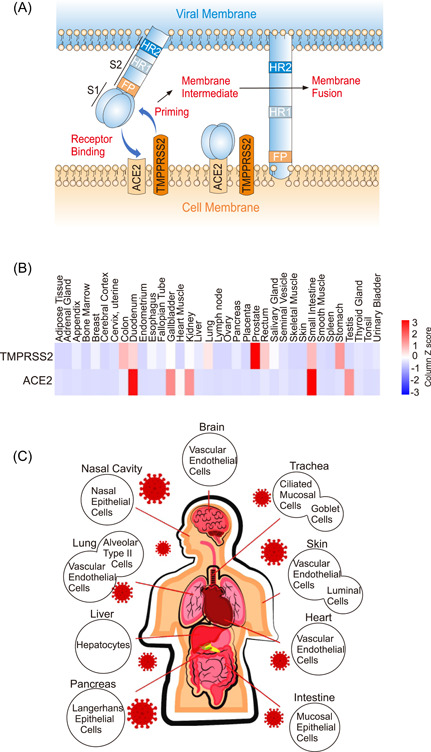
Regulation of host cell receptors by SARS‐CoV‐2‐Spike protein. (A) Sequential events of viral‐host cell membrane fusion were diagrammatically represented involving SARS‐CoV‐2 spike protein, ACE2, and TMPRSS2 receptors. (B) The expression patterns of human ACE2 and TMPRSS2 receptors in normal tissues were shown by heatmap. The expression patterns were analyzed by the Open Access FunRich functional enrichment analysis tool (http://www.funrich.org/index.html). 27 (C) Cell and tissue tropism of SARS‐CoV‐2. After successful infection, the virus disseminates in different organ tissues of the human body including the lung, pancreas, liver, brain, intestine, heart, and skin.
1.3. Cell and tissue tropism of SARS‐CoV‐2
In general, viral tropism is considered the ability of a given virus to effectively infect a particular cell, tissue, or host species. 35 The degree and establishment of viral tropism mainly depend on the susceptibility and permissiveness of a specific host cell. 36 Host cellular receptors play key roles in the determination of virus tropism and pathogenesis. The widespread distribution and expression of ACE2 across multiple organs are critical to helping understand the varied clinical outcomes of COVID‐19. 37 COVID‐19 patients often showed respiratory‐like illnesses leading to severe pneumonia, suggesting that the SARS‐CoV‐2 mainly targets the lung as the primary site of tropism. 38 Many studies have also demonstrated that important mutations within the RBD of SARS‐CoV‐2 spike protein make additional adjacent contact points with ACE2, establishing the viral characteristics of higher binding affinity and increased infectivity. 39 In the context of cellular tropism of SARS‐CoV‐2, researchers identified that airway and alveolar epithelial cells, vascular endothelial cells, and alveolar macrophages are mainly targeted by the virus during the respiratory tract infection. 40 The cells in the respiratory tract are suitable for establishing SARS‐CoV‐2 early infection and subsequent replication due to their expression of ACE2. 41 Moreover, it has been assumed that the respiratory cells in the lung may be contingent on the regulation of ACE2 expressions at the transcriptional and protein levels, favoring SARS‐CoV‐2 entry, as several studies showed that SARS‐CoV‐2 can proficiently infect the nasopharyngeal and oropharyngeal tissues, possibly due to its higher affinity for ACE2 which is also expressed in those tissues (Figure 2B,C). 42 , 43 SARS‐CoV‐2 internalizes inside the cells via ACE2, the downregulation of ACE2 expression can lead to altered tissue function and exacerbate chronic diseases. 44 ACE2 has also been identified to be critical for maintaining tissue homeostasis, as it negatively regulates the renin–angiotensin–aldosterone system (RAAS), which is extremely important for regulating normal functions in different organs including, the lungs, heart, kidney, and vasculatures. 45 In addition, several studies have shown the involvement of ACE2 in ARDS in the animal model, which may be triggered by multiple diseases including the infection of SARS‐CoV and SARS‐CoV‐2. 46 In particular, ACE2 knockout mice showed severe onset of ARDS and could be rescued from ARDS condition if ACE2 knockout mice are treated with AR1R blocker, 47 implicating the vital role of ACE2 and the critical balance between protective and proinflammatory immune reaction influenced by RAAS. 47
Despite the respiratory route is dominant for SARS‐CoV‐2 infection, the highest levels of ACE2 expression are found in the small intestine, testis, kidney, heart muscle, colon, and thyroid gland (Figure 2B). 48 The phenomenon of ACE2 and TMPRSS2 abundant expression in the gastrointestinal tract is consistent with the observation that many other coronaviruses are transmitted via the fecal–oral route and respiratory route, and may be maintained for a long time in the intestinal tissue (Figure 2C). 49 , 50 Supporting this speculation, gastrointestinal disorders are frequently found in patients with COVID‐19, and nearly 20% of patients are identified with detectable SARS‐CoV‐2 mRNA in fecal samples. 51 Cardiac infection by SARS‐CoV‐2 is frequently found in autopsy cases, and the presence of ACE2 in colon and kidney cells has been suggested as an explanation for gastrointestinal and renal complications of SARS‐CoV‐2 infection. 52 Results from organoid models showed that the SARS‐CoV‐2 can directly attack the liver tissue and cause liver damage, and can also destroy cells that control blood sugar in pancreatic organoids, which adds to mounting evidence that the virus can trigger diabetes in some people. In addition, like other coronaviruses, SARS‐CoV‐2 can enter the nervous system by crossing the neural–mucosal interface in olfactory mucosa, exploiting the close vicinity of olfactory mucosal, endothelial, and nervous tissue, including delicate olfactory and sensory nerve endings. 53 The tissue expression pattern of coronavirus receptors (ACE2, BSG, NRP1) and proteases (TMPRSS2, TMPRSS11A, TMPRSS11B, and Furin) in excitatory and inhibitory neurons, astrocytes, oligodendrocytes, and microglia, as well as in cranial vascular endothelial cells, is an indicator for SARS‐CoV‐2's vulnerability for these cells. 54 RNA‐seq profiling of the expression pattern of ACE2 in adult human testis indicated that ACE2 is predominantly enriched in spermatogonia and Leydig and Sertoli cells. 55 SARS‐CoV‐2 infection might damage the male reproductive system via ACE2 receptor, which is required to identify potential short‐term and long‐term effects of SARS‐CoV‐2 on male fertility. 56
Interestingly, low expression of ACE2 mRNA was detected in human and other mammalian lung tissues (bat, ferret, cat, dog, etc.) compared with extrapulmonary tissues. 57 ACE2 alone cannot explain the multiorgan tropism of SARS‐CoV‐2 nor the clinical differences between SARS‐CoV‐2 and SARS‐CoV, suggesting the involvement of other receptors(s) or additional cell‐intrinsic factors. 58 Several studies have demonstrated that TMPRSS2 (transmembrane serine protease‐2) plays an important role in viral entry, as the nearly untraceable level of ACE2 in lung tissue still provides support for SARS‐CoV entry as long as TMPRSS2 is present. 59 Gu et al. identified that ASGR1 and KREMEN1, as alternative functional receptors, play essential roles in ACE2‐independent virus entry, providing other insight into SARS‐CoV‐2 tropism and pathogenesis. 60 In addition, higher mRNA expression of several cellular genes, including gene members of endosomal sorting complex required for transport machinery (i.e., CHMP3, CHMP5, CHMP1A, and VPS37B which are important for the early SARS‐CoV‐2 lifecycle), are identified in human type II alveolar cells with abundant ACE2, compared to ACE2‐deficient cells. 61 Such experimental evidence indicates the way by which SARS‐CoV‐2 employs to takeover a small population of type II alveolar cells with high expression of ACE2, as well as regulates other pro‐viral genes for its successful replication inside the host cells.
1.4. SARS‐CoV‐2 spike protein: Structure–function variability
SARS‐CoV‐2 S protein has two functional subunits (S1 and S2). As shown in Figure 3, the S1 subunit contains the N‐terminal domain (NTD) and RBD, which can bind with the cellular receptor. Compared to S1, the S2 subunit contains a bit enriched structure, including FP, heptad repeat‐1 (HR1), central helix (CH), connector domain (CD), heptad repeat‐2 (HR2), transmembrane domain (TM), and cytoplasmic tail (CT). 62 The furin cleavage site at the border between the S1 and S2 subunits is called the “S1/S2 protease cleavage site.” Importantly, for all the CoVs, host proteases cut the spike glycoprotein at the S2′ cleavage site to activate proteins for membrane fusion through the irreversible conformational changes. 63 The mutation rate analysis of the S protein domain shows that S1/S2 protease cleavage site is associated with the maximum mutation density, which is much higher than that of the RBD region directly binding to the host receptor. 64 This suggests that mutations at this site in the S protein may be of advantage for the virus to undergo proteolytic cleavage by a large number of host enzymes during evolution. 64 S protein undergoes significant structural rearrangement important for ACE2 receptor binding and subsequent entry of virus by membrane fusion. 39 At the prefusion conformational stage, S1 and S2 subunits bind non‐covalently. However, some reports have shown that coronaviruses use “special domains” in the S1 subunit to identify different cellular receptors for viral entry. In particular, SARS‐CoV and SARS‐CoV‐2 recognize the ACE2 via the RBD. 65 Structural data reveals that the S protein has two forms of structural conformations such as the “closed” and “open” state. In the open state conformation, the RBD remains in the “up” position. While in the closed state conformation, the three recognition motifs do not project from the interface. The open state conformation is extremely important for the fusion of the virus with the host cell membranes, leading to SARS‐CoV‐2 entry in the host cells. 66 Importantly, both SARS‐CoV‐2 S and SARS‐CoV S proteins converge on an ACE2 epitope (17‐residue long) with the receptor‐binding motif (RBM). 67 Studies have also revealed the finer molecular architecture of RBM itself is made up of a concave surface stabilized by two β‐hairpins. The RBD of S protein is responsible for further stabilization of the concave structure. 33 On the other hand, the ACE2‐long N‐terminal helix embraced on top of the RBM, and the ACE2 residues K31 and K353 are critical for interacting with SARS‐CoV S protein. 68 In contrast, SARS‐CoV‐2 S protein residues including Y449, Q493, Q498, and N501 have been identified to be of immense importance for ACE2 binding. 69 Upon mutations of these residues, SARS‐CoV‐2 exhibits altered affinity towards S‐ACE2 complex formation. 70 Therefore, SARS‐CoV‐2 Spike protein shows conformational dynamicity and its RBD has been observed as very important for virus–host membrane fusion and cellular entry. Interestingly, S protein can only bind with ACE2 when the RBD remains in the upstate conformation, 71 while heparan sulfate interaction with the RBD maintains the open state conformation for ACE2 binding, 72 and the closed state conformation is ideally observed at endosomal pH 5.5. 73 Although different structures of the SARS‐CoV‐2 S‐ACE2 complex have been identified, both SARS‐CoV‐2 and SARS‐CoV bound with the ACE2 receptor are similar. 37
Figure 3.
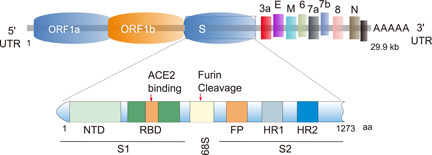
Structural overview of SARS‐CoV‐2‐Spike protein. Diagrammatic representation of SARS‐CoV‐2 genome, and spike glycoprotein (S) structures. Illustration of full‐length SARS‐CoV‐2 S‐protein domains (N‐terminal, receptor binding domain, C‐terminal), furin cleavage site, fusion peptide, and Heptad repeat regions (HR1 and HR2) are highlighted.
SARS‐CoV‐2 S protein is highly glycosylated, each protomer in the trimeric spike has 22 N‐linked glycosylation sites. 74 , 75 N‐linked glycans are crucial for proper protein folding, making neutralizing antibodies, and arranging the spike protein trimers extensively. 76 Glycosylation modification of S protein will create a glycan shield which complicates antibody neutralization. However, some of the spike glycans may directly regulate the interaction between virus and receptor. For example, elegant molecular dynamics simulations show that N‐glycans at sites N165 and N234 of S protein in modulating the conformational dynamics of the spike's RBD, which is responsible for ACE2 recognition, 77 while glycans at N090, N322, and N546 of ACE2 directly mediate the interaction with S trimer. 75 Aptamer studies also show that glycosylation of the RBD protein has significant effects on values of dissociation constants and the relative efficacy of the aptamer binding. 78 Thus, variations in glycan occupancy or processing at these sites could alter the affinity of the SARS‐CoV‐2–ACE2 interaction and modulate infectivity. Furthermore, S protein can interact with SARS‐CoV‐2 attachment receptor type C lectin through a high‐mannose N‐glycan structure, and this interaction can be blocked by glycosylation inhibitors. 79 , 80 , 81 In addition, an in silico study shows that the S1 subunit of S protein has a high affinity with toll‐like receptor TLR‐4, which may also be caused by the complex glycoglycan structure of S protein. 82 Therefore, glycosylation modification is diverse, which is varied by tissue cell type, host race, 83 and age, 84 and each glycosylation site can be modified with one of several glycan structures to produce site‐specific glycosylation combinations. 75 Glycosylation of S protein will increase the heterogeneity of its structure, affect the binding of SARS‐CoV‐2 to its receptor, and the individual susceptibility. Interestingly, studies have shown that D614G mutation may increase glycosylation of neighboring asparagine 616, 80 , 85 and glycosylation mutant N331 and N343 will significantly reduce SARS‐CoV‐2 infectivity, 86 indicating that the mutation of the S protein will also affect its glycosylation level.
1.5. SARS‐CoV‐2 spike protein‐mediated signaling: A molecular‐level analysis
To be more efficient for spreading infections in the respiratory tract, it has been reported that SARS‐CoV‐2 often uses different attachment factors presented on the host cell surface (Figure 4). Emerging evidence from in vitro and in silico experiments indicates that there are a series of molecular interactions between SARS‐CoV‐2 S protein and receptors associated with immune functions, including neuropilin‐1 (NRP1), C‐lectin type receptors (CLR)‐mannose receptor (MR), dendritic cell‐specific intracellular adhesion molecule‐3‐grabbing non‐integrin receptor (DC‐SIGN), homolog dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing nonintegrin related receptor (L‐SIGN), and macrophage galactose‐type lectin (MGL) and toll‐like receptors including TLR1, TLR4, and TLR6. 87 Researchers have also identified the association of nonimmune receptor glucose‐regulated protein 78 (GRP78) with SARS‐CoV‐2 S protein. 88 Likewise, TIM1 and AXL as members of phosphatidylserine receptor families were also suggested to be alternative SARS‐CoV‐2 receptors, respectively. 89 , 90 Such interactions greatly improve their adhesion as well as the virus's concentration to the cell surface. 91 S protein‐other attachment factors association was found very crucial for potentiating access and final engagement with their prime entry receptors. Such molecular interactions not only augment SARS‐CoV‐2 infection of target cells but also potentiate the viral entry in nonpermissive cells by the process of “trans infection.” 92 Importantly, these host factors have their unique functions in cells. The interaction between S protein and host factors often triggers the expression changes of downstream signal pathways, resulting in a series of pathological reactions.
Figure 4.
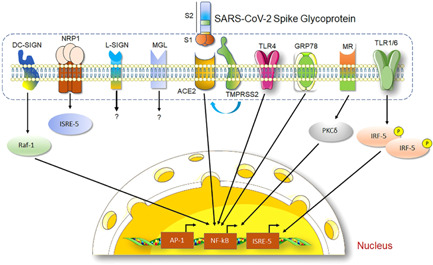
The SARS‐CoV‐2‐Spike protein mediates signaling transduction pathways. The diagram shows SARS‐CoV‐2‐Spike glycoprotein (S) interacts with several host cell receptor proteins mainly with ACE2 by its RBD region. TMPRSS2 initiates the activation of S protein for viral–host cell membrane fusion and subsequent internalization of viral particles inside the cell. SARS‐CoV‐2‐S and ACE2 interaction elicit NF‐κB signaling to regulate different cellular functions. S protein also shows the interaction with cytokine receptors (TLR‐1, 4, 6), glucose‐regulated protein (GRP78), mineralocorticoid receptor (MR), and important growth factor receptor‐neuropilin‐1 (NRP1). Apart from these, SARS‐CoV‐2‐Spike glycoprotein was shown to interact with dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing non‐integrin (DC‐SIGN) for regulating different immunological functions in the host cell.
1.6. Spike protein mediates hypertension and inflammatory factor storm via interaction with ACE2 receptor
ACE2, as the most important receptor for SARS‐CoV‐2 to enter cells, is also the master regulator of the local and central renin–angiotensin system (RAS). 93 ACE2 is widely expressed in lung, kidney, testis, intestine, adipose tissue, and brain, and catalyzes the conversion of angiotensin I to angiotensin‐(1–9) and angiotensin II (Ang II) to angiotensin‐(1–7). 94 , 95 To serve its regulatory role in the RAS, membrane‐bound ACE2 (mACE2) is the first required to be transported to the cell surface, where it is cleaved by host proteases, to release an enzymatically active soluble form of ACE2 (sACE2) into the plasma. 96 As sACE2 preserves the binding site for SARS‐CoV‐2, sequestration of SARS‐CoV‐2 by sACE2 may enable cell entry of tissues where cACE2 is poorly expressed. 97 Research shows that SARS‐CoV‐2 infection may lead to ACE2 shedding and the concentrations of sACE2 may correlate with the level of systemic inflammation that occurs. 98 The binding of SARS‐CoV‐2 S protein to ACE2 will reduce the number of membrane‐bound ACE2 and affect its normal physiological function, resulting in the imbalance of the ACE2/ACE ratio. 99 Consequently, elevated ACE/Ang II/AT1R axis activity or decreased ACE2/Ang‐(1–7)/Masr axis activity can lead to the pulmonary artery and neurogenic hypertension, which may lead to thrombosis, cardiovascular complications, and ultimately lung injury, heart failure, multiple organ failure, and bleeding, 100 , 101 which supporting clinical evidence that RAS imbalance is associated with the development of multiple organ damage during SARS‐CoV‐2 infection. 102
The interaction between SARS‐CoV‐2 spike protein and ACE2 receptor is also related to the excessive inflammatory response. On the one hand, the spike protein promoted an angiotensin II type 1 receptor (AT1) mediated signaling cascade, induced the transcriptional regulatory molecules NF‐κB and AP‐1/c‐Fos via MAPK activation, and increased IL‐6 release. 103 On the other hand, the interaction of the ACE2 receptor with the SARS‐CoV‐2 spike protein directly activates the Nlrp3 inflammasome in human VSELs and HSCs, which, if hyperactivated, may lead to cell death by pyroptosis. 104 Meanwhile, once SARS‐CoV‐2 spike protein interacts with ACE2, macrophages/microglia cells can potentiate the immune response through the cleavage of fragments complement component 3a and 5a (C3a and C5a, respectively) and non‐lytic C5b‐C9 membrane attack complex by ComC, thus activating the NLRP3 inflammasome. 105 Barreda et al. found the expression of ACE2 during the differentiation of human monocytes to mature DCs and interdonor is different. Both SARS‐CoV‐2 S protein and its RBD fragments could promote an inflammatory response in dendritic cells, and increase the expression of maturation markers (including MHC molecules and costimulatory receptors, and active signaling molecules of MAPK, AKT, STAT1, and NF‐κB), which correlates with the expression and secretion of distinctive proinflammatory cytokines. 106 Therefore, upon S protein binding, DC maturation is expected to be a key step to induce long‐lasting immunity against SARS‐CoV‐2. In addition, the study of SARS‐CoV‐2 infection in the nervous system shows that virus infection causes the release of cellular ATP, and extracellular ATP can robustly trigger activation of the NLRP3 inflammasome through P2X7 receptor activation. 107 With NLRP3 inflammasome activation, ATP is also released into the extracellular milieu, which promotes a strong positive feedback loop. 108 In this scenario, proinflammatory factors are intensely released, the so‐called “cytokine storm,” and enter the bloodstream and reach other organs, causing extensive complications. 109
1.7. Spike protein mediates inflammatory response via interaction with immune function related receptors
TLRs can identify a repertoire of pathogen‐associated molecular patterns (PAMP), 110 and induce a robust inflammatory response through the myeloid differentiation factor‐88 (MyD88)‐dependent pathway 111 or the Toll/IL‐1‐domain‐containing adapter‐inducing interferon‐beta (TRIF)‐dependent pathway. 112 Molecular docking studies have demonstrated that SARS‐CoV‐2 S protein can directly bind to TLR1, TLR4, and TLR6. 82 Biochemical studies revealed that SARS‐CoV‐2 S protein triggers inflammation via activation of the NF‐κB pathway in a MyD88‐dependent manner. Khan et al. demonstrated that SARS‐CoV‐2 S protein is a potent viral PAMP that sense dimers of TLR2/TLR1 or TLR2/TLR6 to activate the NF‐κB pathway, leading to the expression of inflammatory mediators in innate immune and epithelial cells. 113 Recent studies have also shown that the S1 subunit of SARS‐CoV‐2 S protein may play a role as a pathogen‐related molecular model (PAMP) to induce a neuroinflammatory process independent of virus infection. For example, S1 treatment could increase gene expression of microglia/brain macrophage activation markers (Iba1, Cd11b, MHC‐IIα), astrocyte activation markers (Gfap), TLRs (Tlr4), inflammasomes (Nlrp3), and proinflammatory cytokines (IL‐1β); and S1 could activate expression of TLR2 and TLR4 in HEK293 cells and TLR2 and TLR4 in microglia. 114 Due to microglia do not express ACE2, this indicates that S1 may signal through these TLRs to produce a neuroinflammatory response in microglia. 114 Following this hypothesis, Shirato et al. demonstrated that the SARS‐CoV‐2 spike protein S1 subunit activates TLR4 signaling to induce proinflammatory responses in murine and human macrophages. 115 Utilizing the microglia cell line BV‐2, Olajide et al. reported that the S1 subunit of the S protein activates NF‐κB and p38 MAPK signaling pathways and in turn induces TNF‐α, IL‐6, IL‐1β, and iNOS/NO production by targeting uses TLR4. 116 Likewise, the S protein binds to TLR4 with high affinity and induces a proinflammatory response in THP‐1 and RAW 264.7 cells, which is blocked by a TLR4 inhibitor. 117
In addition, several studies have also shown that DC‐SIGN and L‐SIGN promote SARS‐CoV‐2 transfer to permissive Vero E6 and Calu‐3 cells with ACE2 expression. 80 , 118 Apart from the sole contribution of Spike protein for inducing DC/L‐SIGN expression, it has been demonstrated that other cellular factors are equally important for their functional activation. For example, researchers observed that proinflammatory cytokines, including IL‐4, IL‐6, IL‐10, and IL‐13 (which are known to be overexpressed in severe COVID‐19 cases), may activate DC/L‐SIGN. 119 It is, therefore, speculated that DC‐SIGN binding to SARS‐CoV‐2 and increased IL‐10 levels in severe COVID‐19 patients may serve as an immune‐inhibitory mechanism stimulated by the rapid accumulation of inflammatory cytokines, via a negative feedback loop. While on the other hand, the significant increase of IL‐10 will stimulate the production of other cytokine storm mediators and potentially lead to hyperactivation of the immune system exacerbating COVID‐19 severity. 120 , 121 In addition, mannose‐binding lectin (MBL) has been reported to associate with SARS‐CoV‐2 infection severity. 122 The interaction of MBL with SARS‐CoV‐2 spike protein requires a trimeric conformation of the viral protein, which does not involve direct recognition of the RBD but is glycan dependent. 122 Unlike other receptors, it has been shown that the interaction between MBL and S protein may prevent SARS‐CoV‐2 from entering cells and activate the complement pathway. 122
1.8. Spike protein mediates the signaling pathway via other cofactors
Despite host cellular receptors playing key roles in the determination of virus tropism and pathogenesis, however, ACE2 alone cannot explain the multiorgan tropism of SARS‐CoV‐2 nor the clinical differences between SARS‐CoV‐2 and SARS‐CoV, suggesting the involvement of other receptors(s) (Figure 4). In addition to the above receptors, SARS‐CoV‐2 spike protein has been shown to interact with glucose‐regulated protein 78 (GRP78), cluster of differentiation 147 (CD147), Neuropilin‐1 (NRP1), and other molecules. 121 , 123 These molecules are not yet clear in SARS's entry into cells and pathogenesis, so they are classified as cofactors. These molecules are not yet clear in the role of SARS‐CoV‐2 entry into cells and pathogenesis, so they are classified as cofactors. GRP78 is a nonimmune receptor and essential endoplasmic reticulum chaperone protein, 124 which has been identified as a DAMP for specific TLRs. 125 In silico study has demonstrated that it is preferred potential binding of RBD regions III and IV of S glycoprotein to substrate‐binding domain‐beta of GRP78. 126 Therefore, GRP78 can cooperate with TLRs to enhance the inflammatory reaction caused by SARS infection. In another case, NRP1 is a transmembrane polypeptide, which acts as a coreceptor for different growth factors, 127 and regulating angiogenesis, gangliogenesis, and vascular permeability. 128 Recent reports have demonstrated that the SARS‐CoV‐2 spike protein can bind to the b1b2 domain of the NRP1, 129 , 130 and thereby blocking VEGF‐A/NRP‐1 signal transduction and affecting pain behavior. 131 Given the facts that KREMEN1 as an alternative functional receptor of SARS‐CoV‐2, 60 and KREMEN1/2 plus FUT8 are all negative regulators of the Wnt/β‐catenin signaling pathway which is critical in taste bud cell renewal and behavioral taste perception, 132 as well as loss of smell and taste has frequently been observed in COVID‐19 patients, 133 suggesting that SARS‐CoV‐2 may act through these factors to affect Wnt/β‐catenin signaling and in turn, promote taste loss.
In addition, some studies have also shown that SARS‐CoV‐2 spike protein may mediate cellular signals with an unknown receptor. For example, Suzuki et al. found that the S protein region without the RBD structure of SARS‐CoV‐2 alone can induce the cellular signals of human pulmonary vascular smooth muscle and endothelial cells, especially the activation of the MEK/ERK pathway. 15 Given the MEK/ERK pathway is a well‐known signaling pathway to facilitate viral replication, 134 , 135 SARS‐CoV‐2 spike protein‐mediated signaling could promote the hyperplasia and/or hypertrophy of vascular smooth muscle and endothelial cells, which contributes to the complex cardiovascular outcomes in COVID‐19. 15
1.9. Therapeutic strategies to target SARS‐CoV‐2 spike protein
During the COVID‐19 pandemic, the main approach employed by physicians to treat COVID‐19 patients is to control the COVID‐pneumonia and inhibit the deadly cytokine storm. Therefore, the therapeutic approach for completely curing and eradicating this viral disease is a major challenge to scientists worldwide. 136 Fortunately, researchers have learned from other RNA virus, including human immunodeficiency virus or Ebola virus, SARS‐CoV‐2 infection may be controlled by using the same strategies which previously applied for curing other RNA viruses‐associated infections. 137 Several clinical trials have been recently undergoing combination of antiretroviral drugs along with remdesivir. 138 Other repurposed drugs are also applied in COVID‐19 treatment including chloroquine and Tocilizumab, although there are controversies for their significant side effects on COVID‐19 patients. 139 , 140 Researchers also realized that besides newly synthesized or discovered antiviral drugs, alternative approaches, including monoclonal antibodies and repurposing drugs, should be taken to cope with the rapid COVID‐19 spread in the global population. 141 , 142 Regardless of tremendous efforts by scientific communities, no specific antiviral drugs or vaccines have been discovered to combat this deadly virus, albeit intensive research continues to seek efficient therapeutic interventions for fighting against the COVID‐19 pandemic. We will summarize some important therapeutic strategies from the perspective of S protein and its related receptors against SARS‐CoV‐2 infection below.
1.10. Vaccines and antibodies targeting SARS‐CoV‐2 spike protein
Many researchers have identified the importance of SARS‐CoV‐2 spike protein as the primary antigenic target for developing potential antiviral therapeutics and vaccine development. So far, there are three major types of SARS‐CoV‐2 vaccines based on the S protein: (1) recombinant protein vaccines that express S proteins in vitro and are used for immunization, including full‐length S protein (FLSP) recombinant vaccines, RBD recombinant vaccines, and virus‐like particle vaccines that carry S proteins on their surfaces; (2) recombinant vector vaccines that use viral vectors to carry S protein genes, including replication‐incompetent and replication‐competent vector vaccines; (3) nucleic acid vaccines that introduce the S protein gene into vaccinated individual, including DNA and RNA vaccines. Recombinant protein vaccines are currently the most developed, accounting for 34% of vaccines entering the clinical phase (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines), and studies have shown that the FLSP, RBD domain, S1 subunit, S2 subunit, NTD and FP of S proteins can all be potential components of antigens. 143 Recombinant vector vaccine technology is relatively mature and a wide range of vectors are available, including modified adenovirus, Ankara poxvirus, human parainfluenza virus, influenza virus, and Sendai virus. Most advanced candidate is ChAdOx1 nCoV‐19 (AZD1222), developed by AstraZeneca and the University of Oxford, which induces a balanced cellular and humoral immune response, and has been approved by EUA in England. 144 , 145 , 146 RNA vaccines also present greater promising future, with features of prompt modification of the encoded immunogen and quick manufacturing process. 147 Moderna (mRNA‐1273) or Pfizer‐BioNTech (BNT162b2) vaccines in Phase III clinical trials, elicited monoclonal antibodies (mAbs) potently neutralize SARS‐CoV‐2. 148 However, the development of S protein‐based vaccines is still facing many dilemmas. First, full‐length recombinant S proteins are difficult to express and have poor stability, while recombinant RBD vaccines recognize fewer neutralizing epitopes compared to FLSP vaccines. 149 Second, the safety of the vaccine remains controversial, 150 as clinical studies show that mRNA vaccines may induce immune thrombotic thrombocytopenia. 151 In addition, the continuous emergence of SARS‐CoV‐2 mutant strains requires researchers to detect S protein mutations and update the vaccine in a timely manner.
A wide range of neutralizing antibodies have been found for targeting RBD and the NTD of the SARS‐CoV‐2 S1 subunit to prevent viral infections. 10 As the RBD of SARS‐CoV‐2 and other CoVs structurally resemble each other, the possible cross‐neutralizing effects of current SARS‐CoV neutralizing antibodies against SARS‐CoV‐2 infection have been thoroughly examined. 152 For example, scientists have discovered the SARS‐CoV RBD‐specific monoclonal antibody (CR3022) is capable of binding with a conserved epitope of the SARS‐CoV‐2 RBD region, 153 and human 47D11 antibodies are able to neutralize both SARS‐CoV‐2 and SARS‐CoV in the experimental settings with Vero‐E6 cell lines. 154 Convalescent plasma sera from the SARS‐CoV infected patients are also observed to cross‐neutralize SARS‐CoV‐2 particles. 155 Therefore, using antibody cocktails containing the antibodies for targeting the different epitopes of the spike protein could be a promising therapeutic intervention. 156 In terms of the S2 subunit that contains an N‐terminal FP with heptad repeat 1 and 2 (HR1, HR2) domains, a transmembrane domain, and a cytoplasmic domain, is required for the virus‐cell membrane fusion, 157 in vitro and in vivo studies have indicated that the EK1C4 (an inhibitor of coronavirus‐host cell membrane fusion) inhibits SARS‐CoV‐2 infection by potentially targeting the HR1 domain. 158
1.11. Specific drugs targeting SARS‐CoV‐2 spike protein
The effective strategy for preventing SARS‐CoV‐2 infection is to stop the viral entry into the host cell. Therefore, extensive studies have been undergoing on specific proteins, peptides, or small molecule compounds that can bind to the S protein, thereby preventing the interaction between virus and host cell membrane (Table 1). For example, ivermectin 159 and arbidol 160 , 161 can block viral membrane fusion by competitively inhibiting the interaction between S protein RBD and ACE2, while lipopeptide EK1 (and its derivative ek1c4), 158 as a pan coronavirus fusion inhibitor, inhibits membrane fusion by targeting HR1 of S2 domain. Different from the above strategies, some studies have demonstrated that human recombinant soluble ACE2 (hrsAC32) may act as a decoy to bind SARS‐CoV‐2‐S protein for preventing virus entry into the host cell. 162 It has been also proven that clinical‐grade hrsACE2 can inhibit SARS‐CoV‐2 infection in the human blood vessel and kidney organoid models. 163
Table 1.
Therapeutic drugs targeting SARS‐CoV‐2 Spike protein.
| Target | Compound | Approval for clinical diseases | Clinical phase | Related mechanisms |
|---|---|---|---|---|
| RBD | Ivermectin | Broad spectrum antiparasitic agent | 4 | Inhibit the replication of SARS‐CoV‐2 in vitro 164 |
| Arbidol | Treatment for respiratory viral infections | 4 | Block both viral entry and postentry stages in vitro 165 | |
| hrsACE2 | – | 2 | Inhibit the infection of SARS‐CoV‐2 in human vascular and renal organs in the early stage of infection 166 | |
| ACE2‐like molecules (CTC‐445.2d) | – | – | Protect Syrian hamsters from a subsequent lethal SARS‐CoV‐2 challenge 167 | |
| LCB1/LCB1v1.3 | – | – | Inhibit SARS‐CoV‐2 infection in vitro 168 | |
| Inhibit SARS‐CoV‐2 and B.1.1.7 variant infections in hACE2 mice 169 | ||||
| HR1 | EK1/EK1C4 | – | – | Inhibit SARS‐CoV‐2 S protein‐mediated membrane fusion 170 |
| Protect hACE2 mice from SARS‐CoV‐2 infection 158 | ||||
| IPB02 | – | – | Inhibit SARS‐CoV‐2 S protein‐mediated cell–cell fusion and pseudovirus transduction 171 | |
| HR2 | [SARS HRC‐PEG4] 2 ‐chol | – | – | Prevent SARS‐CoV‐2 transmission in a ferret model 172 |
| Posaconazole | Triazole antifungal drug 173 | – | Inhibit SARS‐CoV‐2 infection in Caco‐2 cells 174 | |
| 5‐Helix | ‐ | – | Inhibit infection by pseudotyped SARS‐CoV‐2 and its variants 175 |
Recently, some scientists have also made efforts to look for effective natural compounds that may target and modulate specific novel sites within the SARS‐CoV‐2 spike protein. 176 For example, extensive in silico analysis using molecular docking experiments has shown that natural compounds (flavonoids) can potentially bind to the functional domains of the SARS‐CoV‐2 spike protein, 76 and several natural compounds can effectively bind to the C‐terminal domains of SARS‐CoV‐2 S protein and may prevent the association with the hACE2 receptor. 177
1.12. Inhibitory drugs targeting SARS‐CoV‐2 spike protein‐related receptors
The inhibitors used to downregulate the activity of the SARS receptor and cofactor and block the binding of S protein to the receptor are also one of the strategies to combat SARS‐CoV‐2 infection (Table 2). As the main receptor for SARS‐CoV‐2 to enter host cells, ACE2 is an important target for many COVID‐19 treatment strategies. Importantly, the critical residues for ACE2 receptors have been identified to binding with the C‐terminal SD‐1 domain (CTD) of the SARS‐CoV‐2 S protein, which could be a promising target for antiviral therapeutics. 178 Some studies have shown the use of ACE2 antibodies, which antagonize the binding of SARS‐CoV‐2 to ACE2 without interfering with the normal function of ACE2, may crosslink and downregulate ACE2, and thereby destroy the RAS. 179 Similarly, ACE2 mimetics are also ideal therapeutic drugs, such as ACE2 ectodomain, its Fc‐fusion form, and their variants selected for higher affinity can prevent the initial step of virus binding to the receptor without interfering with the RAS. 180 In addition, some drugs can affect the binding of SARS‐CoV‐2 to ACE2 through different mechanisms. For instance, amitriptyline and other FIASMAs inhibit ASM and the formation of ceramide‐enriched membrane domains, which serve viral entry and infection by clustering ACE2, thereby preventing infection with SARS‐CoV‐2. 181 , 182 Diltiazem, a blocker of L‐type calcium channel calcium, can interact and colocalizes with SARS‐CoV‐2 spike protein and ACE2, thereby affecting cell attachment and internalization of SARS‐CoV‐2. 183 Recent studies have also shown that peptide‐based drugs could be used to compete for binding of ACE2 receptor with the SARS‐CoV‐2 spike protein by computational prediction of drug repurposing strategy. 184 For example, Gu et al. discovered two small compounds protoporphyrin IX and verteporfin, which had been approved by the FDA for the treatment of cancers and age‐related macular degeneration respectively, sharing a porphyrin ring structure, are able to bind viral receptor ACE2 and interfere with the interaction between ACE2 and the RBD of viral S protein. 185 The compounds present an effective antiviral concentration and a broad margin of safety in the cell and mouse model, but it is required for clinical evaluation in vivo.
Table 2.
Therapeutic drugs targeting SARS‐CoV‐2 Spike protein‐related receptors.
| Target | Compound | Approval for clinical diseases | Clinical phase | Related mechanisms |
|---|---|---|---|---|
| ACE2 | Dalbavancin | Anti‐cooperative ligand | 4 | Reduce SARS‐CoV‐2 load and pneumonia in both mouse and rhesus macaque models 186 |
| Diltiazem | Acute heart rate control | 3 | Inhibit SARS‐CoV‐2 cell attachment and internalization and decrease the viral infection in mouse lung 183 | |
| Protoporphyrin IX | Cancer screening biomarkers | 1 | Prevent SARS‐CoV‐2 infection in mice adenovirally transduced with ACE2 185 | |
| Verteporfin | Treatment for choroidal neovascularization | – | Prevent SARS‐CoV‐2 infection in mice adenovirally transduced with ACE2 185 | |
| FIASMA amitriptyline | Antidepressant | – | Inhibit of the infection of nasal epithelial cells from volunteers with pp‐VSV‐SARS‐CoV‐2 spike particle 181 | |
| TMPRSS2 | Bromhexine | Mucolytic agent | 4 | Early oral therapy reduces the ICU transfer, intubation, and the mortality rate in patients with COVID‐19 187 |
| Camostat mesylate | Acute pancreatitis | 3 | Block SARS‐CoV‐2 infection of lung cells 59 , 188 | |
| Nafamostat | Anticoagulant | 3 | Reduce SARS‐CoV‐2 pulmonary infection in mouse models 188 | |
| α1‐antitrypsin (α1AT) | – | – | Inhibit SARS‐CoV‐2 entry and suppress viral replication in cell lines and primary cells 189 | |
| N‐0385 | – | – | Afford a high level of prophylactic and therapeutic benefit in the K18‐human ACE2 transgenic mouse model 190 | |
| CatB/L | Amantadine | Anti‐influenza | 3 | Inhibit SARS‐CoV‐2 cell entry with little cytotoxicity 191 |
| E‐64d | Treatment for Alzheimer's disease | 2 | Block SARS‐CoV‐2 infection of lung cells with Camostat mesylate 59 | |
| K777 | Antiparasite drug | – | Block SARS‐CoV‐2 infection of human and monkey cells 192 | |
| Furin | Decanoyl‐RVKR‐chloromethylketone (CMK) | – | – | Inhibit SARS‐CoV‐2 infection in Vero‐E6 cell 193 |
Given the activation of SARS‐CoV‐2 S protein depends on the host protease cleavage of S1/S2 and S2 sites, 29 it is another group of potential targets for developing inhibitors against SARS‐CoV‐2. In this aspect, the relevant drugs supported by experimental data include furan inhibitors, 194 TMPRSS2 inhibitors (camostat mesylate, 59 Nafamostat 195 ), and cathepsin L inhibitors (E‐64d 59 ). It is worthy to mention that inhibitors of S protein cofactors also have antiviral effects. For example, PM26, a glycomimetic antagonist of DC‐SIGN, is able to inhibit the interaction of the S protein with the lectin receptor and blocks DC‐SIGN‐mediated SARS‐CoV‐2 infection. 80 However, if these drugs are used for the prevention or treatment of SARS‐CoV‐2 infection, they still need to determine the time point and dosage of the best curative effect, clinical therapeutic effect, and side effects.
2. CONCLUSION
In summary, even though several COVID‐19 vaccines are currently in distribution worldwide, new threats are posed by the emerging circulating variants of SARS‐CoV‐2. The high mortality rate of critical illness is still a serious threat, due to the lack of safe and effective drugs for clinical treatments. We review and highlight the signal pathway modulation by SARS‐CoV‐2 S protein and its receptors, particularly current therapies and vaccines targeting the viral entry machinery. In‐depth research is still required to comprehend more details of the SARS‐CoV‐2 spike protein mutations and mode of interactions with receptors and cofactors by innovative structural biology techniques and will provide valuable information for successful antiviral therapeutics and vaccination against COVID‐19.
AUTHOR CONTRIBUTIONS
Shuvomoy Banerjee drafted review parts of introduction, evolution, tropism, and structure, as well as performing analysis. Xinyu Wang, Yuping Jia, and Yuyan Wang drafted review parts of host factors, signaling, therapeutic, and conclusion. Shujuan Du and Caixia Zhu modified figures and critical reading. Yuyan Wang and Qiliang Cai supervised and editing paper.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program (2021YFA1300803), National Natural Science Foundation of China (81971930, 32120103001, 82102386), Shanghai Municipal Science and Technology Major Project (ZD2021CY001), and Jinan University and Institute Innovation Program (2020GXRC043). Qiliang Cai is a scholar of New Century Excellent Talents at the University of China.
Banerjee S, Wang X, Du S, et al. Comprehensive role of SARS‐CoV‐2 spike glycoprotein in regulating host signaling pathway. J Med Virol. 2022;94:4071‐4087. 10.1002/jmv.27820
Shuvomoy Banerjee and Xinyu Wang contributed equally to this study.
Contributor Information
Shuvomoy Banerjee, Email: shuvomoy.banerjee@iar.ac.in.
Yuping Jia, Email: jiayupingygs@163.com.
Yuyan Wang, Email: yuyanss@fudan.edu.cn.
Qiliang Cai, Email: qiliang@fudan.edu.cn.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study. The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) database and analyzed in The Interactive Tree Of Life online software tool at https://itol.embl.de; the Open Access FunRich functional enrichment analysis tool at http://www.funrich.org/index.html.
REFERENCES
- 1. Madjid M, Miller CC, Zarubaev VV, et al. Influenza epidemics and acute respiratory disease activity are associated with a surge in autopsy‐confirmed coronary heart disease death: results from 8 years of autopsies in 34,892 subjects. Eur Heart J. 2007;28(10):1205‐1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ljubin‐Sternak S, Mestrovic T, Luksic I, Mijac M, Vranes J. Seasonal coronaviruses and other neglected respiratory viruses: a global perspective and a local snapshot. Front Public Health. 2021;9:691163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293‐W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haider N, Rothman‐Ostrow P, Osman AY, et al. COVID‐19—zoonosis or emerging infectious disease? Front Public Health. 2020;8:596944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El‐Sayed A, Kamel M. Climatic changes and their role in emergence and re‐emergence of diseases. Environ Sci Pollut Res Int. 2020;27(18):22336‐22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J, Xie W, Wang Y, et al. A comparative overview of COVID‐19, MERS and SARS: review article. Int J Surg. 2020;81:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mourya DT, Yadav PD, Ullas PT, et al. Emerging/re‐emerging viral diseases & new viruses on the Indian horizon. Indian J Med Res. 2019;149(4):447‐467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma A, Tiwari S, Deb MK, Marty JL. Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2): a global pandemic and treatment strategies. Int J Antimicrob Agents. 2020;56(2):106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakraborty I, Maity P. COVID‐19 outbreak: migration, effects on society, global environment and prevention. Sci Total Environ. 2020;728:138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol Sin. 2020;41(9):1141‐1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A. 2020;117(21):11727‐11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu B, Yan Y, Dong L, et al. Integrated characterization of SARS‐CoV‐2 genome, microbiome, antibiotic resistance and host response from single throat swabs. Cell Discov. 2021;7(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuhenn J, Meister TL, Todt D, et al. Differential interferon‐alpha subtype induced immune signatures are associated with suppression of SARS‐CoV‐2 infection. Proc Natl Acad Sci U S A. 2022;119(8):e2111600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suzuki YJ, Nikolaienko SI, Dibrova VA, et al. SARS‐CoV‐2 spike protein‐mediated cell signaling in lung vascular cells. Vascul Pharmacol. 2021;137:106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suzuki YJ. The viral protein fragment theory of COVID‐19 pathogenesis. Med Hypotheses. 2020;144:110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sternberg A, Naujokat C. Structural features of coronavirus SARS‐CoV‐2 spike protein: targets for vaccination. Life Sci. 2020;257:118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abdelghany TM, Ganash M, Bakri MM, Qanash H, Al‐Rajhi AMH, Elhussieny NI. SARS‐CoV‐2, the other face to SARS‐CoV and MERS‐CoV: future predictions. Biomed J. 2021;44(1):86‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . The species Severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5(4):536‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang S, Qiao S, Yu J, et al. Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS‐CoV‐2 evolution. Nat Commun. 2021;12:1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Touati R, Haddad‐Boubaker S, Ferchichi I, et al. Comparative genomic signature representations of the emerging COVID‐19 coronavirus and other coronaviruses: high identity and possible recombination between Bat and Pangolin coronaviruses. Genomics. 2020;112(6):4189‐4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sironi M, Hasnain SE, Rosenthal B, et al. SARS‐CoV‐2 and COVID‐19: a genetic, epidemiological, and evolutionary perspective. Infect Genet Evol. 2020;84:104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson CK, Hitchens PL, Pandit PS, et al. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc Biol Sci. 2020;287(1924):20192736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valitutto MT, Aung O, Tun KYN, et al. Detection of novel coronaviruses in bats in Myanmar. PLoS One. 2020;15(4):e0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fonseka P, Pathan M, Chitti SV, Kang T, Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. 2021;433(11):166747. [DOI] [PubMed] [Google Scholar]
- 28. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS‐CoV‐2 entry into cells. Nat Rev Mol Cell Biol. 2021;23:3‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zipeto D, da Fonseca Palmeira JD, Arganaraz GA, Arganaraz ER. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID‐19. Front Immunol. 2020;11:576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2021;19(3):155‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR. Proteolytic activation of SARS‐CoV‐2 spike at the S1/S2 boundary: potential role of proteases beyond furin. ACS Infect Dis. 2021;7(2):264‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cantuti‐Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science. 2020;370(6518):856‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finkelstein MT, Mermelstein AG, Miller EP, Seth PC, Stancofski ED, Fera D. Structural analysis of neutralizing epitopes of the SARS‐CoV‐2 spike to guide therapy and vaccine design strategies. Viruses. 2021;13(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia X. Domains and functions of spike protein in SARS‐CoV‐2 in the context of vaccine design. Viruses. 2021;13(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McFadden G, Mohamed MR, Rahman MM, Bartee E. Cytokine determinants of viral tropism. Nat Rev Immunol. 2009;9(9):645‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harrison AG, Lin T, Wang P. Mechanisms of SARS‐CoV‐2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100‐1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ni W, Yang X, Yang D, et al. Role of angiotensin‐converting enzyme 2 (ACE2) in COVID‐19. Crit Care. 2020;24(1):422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu J, Li Y, Liu Q, et al. SARS‐CoV‐2 cell tropism and multiorgan infection. Cell Discov. 2021;7(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barton MI, MacGowan SA, Kutuzov MA, Dushek O, Barton GJ, van der Merwe PA. Effects of common mutations in the SARS‐CoV‐2 spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife. 2021;10:e70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuki K, Fujiogi M, Koutsogiannaki S. COVID‐19 pathophysiology: a review. Clin Immunol. 2020;215:108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jia HP, Look DC, Shi L, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79(23):14614‐14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith JC, Sausville EL, Girish V, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS‐CoV‐2 receptor ACE2 in the respiratory tract. Dev Cell. 2020;53(5):514‐529.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ashraf UM, Abokor AA, Edwards JM, et al. SARS‐CoV‐2, ACE2 expression, and systemic organ invasion. Physiol Genomics. 2021;53(2):51‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muñoz‐Durango N, Fuentes CA, Castillo AE, et al. Role of the renin‐angiotensin‐aldosterone system beyond blood pressure regulation: molecular and cellular mechanisms involved in end‐organ damage during arterial hypertension. Int J Mol Sci. 2016;17(7):797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hulsmann S, Khabbazzadeh S, Meissner K, Quintel M. A potential role of the renin‐angiotensin‐system for disturbances of respiratory chemosensitivity in acute respiratory distress syndrome and severe acute respiratory syndrome. Front Physiol. 2021;11:588248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus‐induced lung injury. Nat Med. 2005;11(8):875‐879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Y, Wang Y, Luo W, et al. A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS‐CoV‐2, in human tissues and blood cells. Int J Med Sci. 2020;17(11):1522‐1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang H, Kang Z, Gong H, Xu D, Xu H. The digestive system is a potential route of 2019‐nCov infection: a bioinformatics analysis based on single‐cell transcriptomes. Plast Reconstr Surg. 2020;146:1275‐1284.33234957 [Google Scholar]
- 51. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS‐CoV‐2. Gastroenterology. 2020;158(6):1831‐1833.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lindner D, Fitzek A, Bräuninger H, et al. Association of cardiac infection with SARS‐CoV‐2 in confirmed COVID‐19 autopsy cases. JAMA Cardiol. 2020;5(11):1281‐1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meinhardt J, Radke J, Dittmayer C, et al. Olfactory transmucosal SARS‐CoV‐2 invasion as a port of central nervous system entry in individuals with COVID‐19. Nat Neurosci. 2021;24(2):168‐175. [DOI] [PubMed] [Google Scholar]
- 54. Veleri S. Neurotropism of SARS‐CoV‐2 and neurological diseases of the central nervous system in COVID‐19 patients. Exp Brain Res. 2022;240(1):9‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Z, Xu X. scRNA‐seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS‐CoV‐2 infection in spermatogonia, Leydig and Sertoli cells. Cells. 2020;9(4):920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clayton Edenfield R, Easley CA 4th. Implications of testicular ACE2 and the renin‐angiotensin system for SARS‐CoV‐2 on testis function. Nat Rev Urol. 2022;19(2):116‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16(7):e9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trougakos IP, Stamatelopoulos K, Terpos E, et al. Insights to SARS‐CoV‐2 life cycle, pathophysiology, and rationalized treatments that target COVID‐19 clinical complications. J Biomed Sci. 2021;28(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gu Y, Cao J, Zhang X, et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS‐CoV‐2. Cell Res. 2022;32(1):24‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single‐cell RNA expression profiling of ACE2, the receptor of SARS‐CoV‐2. Am J Respir Crit Care Med. 2020;202(5):756‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell. 2020;181(4):894‐904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peacock TP, Goldhill DH, Zhou J, et al. The furin cleavage site in the SARS‐CoV‐2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6(7):899‐909. [DOI] [PubMed] [Google Scholar]
- 64. Guruprasad L. Human SARS CoV‐2 spike protein mutations. Proteins. 2021;89(5):569‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ke Z, Oton J, Qu K, et al. Structures and distributions of SARS‐CoV‐2 spike proteins on intact virions. Nature. 2020;588(7838):498‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yi C, Sun X, Ye J, et al. Key residues of the receptor binding motif in the spike protein of SARS‐CoV‐2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol. 2020;17(6):621‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lim H, Baek A, Kim J, et al. Hot spot profiles of SARS‐CoV‐2 and human ACE2 receptor protein protein interaction obtained by density functional tight binding fragment molecular orbital method. Sci Rep. 2020;10:16862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Laurini E, Marson D, Aulic S, Fermeglia A, Pricl S. Computational mutagenesis at the SARS‐CoV‐2 spike protein/angiotensin‐converting enzyme 2 binding interface: comparison with experimental evidence. ACS Nano. 2021;15(4):6929‐6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ghorbani M, Brooks BR, Klauda JB. Critical sequence hotspots for binding of novel coronavirus to angiotensin converter enzyme as evaluated by molecular simulations. J Phys Chem B. 2020;124(45):10034‐10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gur M, Taka E, Yilmaz SZ, Kilinc C, Aktas U, Golcuk M. Conformational transition of SARS‐CoV‐2 spike glycoprotein between its closed and open states. J Chem Phys. 2020;153(7):075101. [DOI] [PubMed] [Google Scholar]
- 72. Zhang Q, Chen CZ, Swaroop M, et al. Heparan sulfate assists SARS‐CoV‐2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020;6(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Finkelstein MT, Mermelstein AG, Miller EP, Seth PC, Stancofski E‐SD, Fera D. Structural analysis of neutralizing epitopes of the SARS‐CoV‐2 spike to guide therapy and vaccine design strategies. Viruses. 2021;13(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science. 2020;369(6501):330‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhao P, Praissman JL, Grant OC, et al. Virus‐receptor interactions of glycosylated SARS‐CoV‐2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28(4):586‐601.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS‐CoV2 spike protein and human ACE2. Sci Rep. 2020;10:17699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Casalino L, Gaieb Z, Goldsmith JA, et al. Beyond shielding: the roles of glycans in the SARS‐CoV‐2 spike protein. ACS Cent Sci. 2020;6(10):1722‐1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Grabovenko F, Nikiforova L, Yanenko B, et al. Glycosylation of receptor binding domain of SARS‐CoV‐2 S‐protein influences on binding to immobilized DNA aptamers. Int J Mol Sci. 2022;23(1):557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lempp FA, Soriaga LB, Montiel‐Ruiz M, et al. Lectins enhance SARS‐CoV‐2 infection and influence neutralizing antibodies. Nature. 2021;598(7880):342‐347. [DOI] [PubMed] [Google Scholar]
- 80. Thépaut M, Luczkowiak J, Vivès C, et al. DC/L‐SIGN recognition of spike glycoprotein promotes SARS‐CoV‐2 trans‐infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021;17(5):e1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Guo L, Liang Y, Li H, et al. Epigenetic glycosylation of SARS‐CoV‐2 impact viral infection through DC&L‐SIGN receptors. iScience. 2021;24(12):103426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS‐CoV‐2 spike glycoprotein with ACE‐2 receptor homologs and human TLRs. J Med Virol. 2020;92(10):2105‐2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gebrehiwot AG, Melka DS, Kassaye YM, et al. Healthy human serum N‐glycan profiling reveals the influence of ethnic variation on the identified cancer‐relevant glycan biomarkers. PLoS One. 2018;13(12):e0209515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Krištić J, Vučković F, Menni C, et al. Glycans are a novel biomarker of chronological and biological ages. J Gerontol A Biol Sci Med Sci. 2014;69(7):779‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182(4):812‐827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284‐1294.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gadanec LK, Mcsweeney KR, Qaradakhi T, Ali B, Zulli A, Apostolopoulos V. Can SARS‐CoV‐2 virus use multiple receptors to enter host cells? Int J Mol Sci. 2021;22(3):992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sabirli R, Koseler A, Goren T, Turkcuer I, Kurt O. High GRP78 levels in Covid‐19 infection: a case‐control study. Life Sci. 2021;265:118781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mori Y, Fink C, Ichimura T, et al. KIM‐1/TIM‐1 is a receptor for SARS‐CoV‐2 in lung and kidney. medRxiv . 2022.
- 90. Wang S, Qiu Z, Hou Y, et al. AXL is a candidate receptor for SARS‐CoV‐2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang ZY, Huang Y, Ganesh L, et al. pH‐dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC‐SIGN. J Virol. 2004;78(11):5642‐5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Navarro‐Sanchez E, Altmeyer R, Amara A, et al. Dendritic‐cell‐specific ICAM3‐grabbing non‐integrin is essential for the productive infection of human dendritic cells by mosquito‐cell‐derived dengue viruses. EMBO Rep. 2003;4(7):723‐728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gheblawi M, Wang K, Viveiros A, et al. Angiotensin‐converting enzyme 2: SARS‐CoV‐2 receptor and regulator of the renin‐angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126(10):1456‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Donoghue M, Hsieh F, Baronas E, et al. A novel angiotensin‐converting enzyme‐related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1‐9. Circ Res. 2000;87(5):E1‐E9. [DOI] [PubMed] [Google Scholar]
- 95. Turner AJ, Hooper NM. The angiotensin‐converting enzyme gene family: genomics and pharmacology. Trends Pharmacol Sci. 2002;23(4):177‐183. [DOI] [PubMed] [Google Scholar]
- 96. Lambert DW, Yarski M, Warner FJ, et al. Tumor necrosis factor‐alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe‐acute respiratory syndrome‐coronavirus (SARS‐CoV) receptor, angiotensin‐converting enzyme‐2 (ACE2). J Biol Chem. 2005;280(34):30113‐30119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yeung ML, Teng JLL, Jia L, et al. Soluble ACE2‐mediated cell entry of SARS‐CoV‐2 via interaction with proteins related to the renin‐angiotensin system. Cell. 2021;184(8):2212‐2228.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kornilov SA, Lucas I, Jade K, Dai CL, Lovejoy JC, Magis AT. Plasma levels of soluble ACE2are associated with sex, metabolic syndrome, and its biomarkers in a large cohort, pointing to a possible mechanism for increased severity in COVID‐19. Crit Care. 2020;24(1):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kulkarni PG, Sakharkar A, Banerjee T. Understanding the role of nACE2 in neurogenic hypertension among COVID‐19 patients. Hypertens Res. 2022;45(2):254‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sriramula S, Xia H, Xu P, Lazartigues E. Brain‐targeted angiotensin‐converting enzyme 2 overexpression attenuates neurogenic hypertension by inhibiting cyclooxygenase‐mediated inflammation. Hypertension. 2015;65(3):577‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Moriguchi T, Harii N, Goto J, et al. A first case of meningitis/encephalitis associated with SARS‐Coronavirus‐2. Int J Infect Dis. 2020;94:55‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019‐nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Patra T, Meyer K, Geerling L, et al. SARS‐CoV‐2 spike protein promotes IL‐6 trans‐signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16(12):e1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ratajczak MZ, Bujko K, Ciechanowicz A, et al. SARS‐CoV‐2 entry receptor ACE2 is expressed on very small CD45− precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev Rep. 2021;17(1):266‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19(8):477‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Barreda D, Santiago C, Rodríguez JR, et al. SARS‐CoV‐2 spike protein and its receptor binding domain promote a proinflammatory activation profile on human dendritic cells. Cells. 2021;10(12):3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ribeiro DE, Oliveira‐Giacomelli Á, Glaser T, et al. Hyperactivation of P2X7 receptors as a culprit of COVID‐19 neuropathology. Mol Psychiatry. 2021;26(4):1044‐1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Albalawi F, Lu W, Beckel JM, Lim JC, McCaughey SA, Mitchell CH. The P2X7 receptor primes IL‐1β and the NLRP3 inflammasome in astrocytes exposed to mechanical strain. Front Cell Neurosci. 2017;11:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sun X, Wang T, Cai D, et al. Cytokine storm intervention in the early stages of COVID‐19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Takeda K, Akira S. Toll‐like receptors in innate immunity. Int Immunol. 2005;17(1):1‐14. [DOI] [PubMed] [Google Scholar]
- 111. Bagchi A, Herrup EA, Warren HS, et al. MyD88‐dependent and MyD88‐independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol. 2007;178(2):1164‐1171. [DOI] [PubMed] [Google Scholar]
- 112. Yamamoto M, Sato S, Hemmi H, et al. Role of adaptor TRIF in the MyD88‐independent toll‐like receptor signaling pathway. Science. 2003;301(5633):640‐643. [DOI] [PubMed] [Google Scholar]
- 113. Khan S, Shafiei MS, Longoria C, Schoggins JW, Savani RC, Zaki H. SARS‐CoV‐2 spike protein induces inflammation via TLR2‐dependent activation of the NF‐kappaB pathway. eLife. 2021;10:e68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Frank MG, Nguyen KH, Ball JB, et al. SARS‐CoV‐2 spike S1 subunit induces neuroinflammatory, microglial and behavioral sickness responses: evidence of PAMP‐like properties. Brain Behav Immun. 2022;100:267‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shirato K, Kizaki T. SARS‐CoV‐2 spike protein S1 subunit induces pro‐inflammatory responses via toll‐like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7(2):e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Olajide OA, Iwuanyanwu VU, Adegbola OD, Al‐Hindawi AA. SARS‐CoV‐2 spike glycoprotein S1 induces neuroinflammation in BV‐2 microglia. Mol Neurobiol. 2022;59(1):445‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhao Y, Kuang M, Li J, et al. SARS‐CoV‐2 spike protein interacts with and activates TLR41. Cell Res. 2021;31(7):818‐820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Katz DH, Tahir UA, Ngo D, et al. Proteomic profiling in biracial cohorts implicates DC‐SIGN as a mediator of genetic risk in COVID‐19. medRxiv . 2020.
- 119. Relloso M, Puig‐Kroger A, Pello OM, et al. DC‐SIGN (CD209) expression Is IL‐4 dependent and is negatively regulated by IFN, TGF‐β, and anti‐inflammatory agents. J Immunol. 2002;168(6):2634‐2643. [DOI] [PubMed] [Google Scholar]
- 120. Lu L, Zhang H, Dauphars DJ, He YW. A potential role of interleukin 10 in COVID‐19 pathogenesis. Trends Immunol. 2021;42(1):3‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Trbojević‐Akmačić I, Petrović T, Lauc G. SARS‐CoV‐2 S glycoprotein binding to multiple host receptors enables cell entry and infection. Glycoconj J. 2021;38(5):611‐623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Stravalaci M, Pagani I, Paraboschi EM, et al. Recognition and inhibition of SARS‐CoV‐2 by humoral innate immunity pattern recognition molecules. Nature Immunol. 2022;23(2):275‐286. [DOI] [PubMed] [Google Scholar]
- 123. Evans JP, Liu S‐L. Role of host factors in SARS‐CoV‐2 entry. J Biol Chem. 2021;297(1):100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35(4):373‐381. [DOI] [PubMed] [Google Scholar]
- 125. Shimasaki S, Koga T, Shuto T, et al. Endoplasmic reticulum stress increases the expression and function of toll‐like receptor‐2 in epithelial cells. Biochem Biophys Res Commun. 2010;402(2):235‐240. [DOI] [PubMed] [Google Scholar]
- 126. Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID‐19 spike‐host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Huang X, Ye Q, Chen M, et al. N‐glycosylation‐defective splice variants of neuropilin‐1 promote metastasis by activating endosomal signals. Nat Commun. 2019;10(1):3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Maden CH, Gomes J, Schwarz Q, Davidson K, Tinker A, Ruhrberg C. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev Biol. 2012;369(2):277‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cantuti‐Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science. 2020;370(6518):856‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Daly JL, Simonetti B, Klein K, et al. Neuropilin‐1 is a host factor for SARS‐CoV‐2 infection. Science. 2020;370(6518):861‐865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Moutal A, Martin LF, Boinon L, et al. SARS‐CoV‐2 spike protein co‐opts VEGF‐A/neuropilin‐1 receptor signaling to induce analgesia. Pain. 2021;162(1):243‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mao B, Wu W, Davidson G, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta‐catenin signalling. Nature. 2002;417(6889):664‐667. [DOI] [PubMed] [Google Scholar]
- 133. Xydakis MS, Dehgani‐Mobaraki P, Holbrook EH, et al. Smell and taste dysfunction in patients with COVID‐19. Lancet Infect Dis. 2020;20(9):1015‐1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12(1):9‐18. [DOI] [PubMed] [Google Scholar]
- 135. Bonjardim CA. Viral exploitation of the MEK/ERK pathway—a tale of vaccinia virus and other viruses. Virology. 2017;507:267‐275. [DOI] [PubMed] [Google Scholar]
- 136. Jean SS, Lee PI, Hsueh PR. Treatment options for COVID‐19: the reality and challenges. J Microbiol Immunol. 2020;53(3):436‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Castel AD, Wilbourn B, Magnus M, Greenberg AE. SARS‐CoV‐2 and HIV: epidemiology, treatment, and lessons learned from HIV. AIDS Rev. 2020;22(3):133‐142. [DOI] [PubMed] [Google Scholar]
- 138. Sun DX. Remdesivir for treatment of COVID‐19: combination of pulmonary and IV administration may offer aditional benefit. AAPS J. 2020;22(4):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Khuroo MS. Chloroquine and hydroxychloroquine in coronavirus disease 2019 (COVID‐19). Facts, fiction and the hype: a critical appraisal. Int J Antimicrob Agents. 2020;56(3):106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Samaee H, Mohsenzadegan M, Ala S, Maroufi SS, Moradimajd P. Tocilizumab for treatment patients with COVID‐19: recommended medication for novel disease. Int Immunopharmacol. 2020;89:107018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Saha RP, Sharma AR, Singh MK, et al. Repurposing drugs, ongoing vaccine, and new therapeutic development initiatives against COVID‐19. Front Pharmacol. 2020;11:1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Taylor PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL. Neutralizing monoclonal antibodies for treatment of COVID‐19. Nat Rev Immunol. 2021;21(6):382‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Malik JA, Mulla AH, Farooqi T, Pottoo FH, Anwar S, Rengasamy KRR. Targets and strategies for vaccine development against SARS‐CoV‐2. Biomed Pharmacother. 2021;137:111254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV‐19 vaccine prevents SARS‐CoV‐2 pneumonia in rhesus macaques. Nature. 2020;586(7830):578‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Voysey M, Costa Clemens SA, Madhi SA, et al. Single‐dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV‐19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397(10277):881‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Muhammed Y, Yusuf Nadabo A, Pius M, et al. SARS‐CoV‐2 spike protein and RNA dependent RNA polymerase as targets for drug and vaccine development: a review. Biosaf Health. 2021;3(5):249‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature. 2021;592(7855):616‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Krammer F. SARS‐CoV‐2 vaccines in development. Nature. 2020;586(7830):516‐527. [DOI] [PubMed] [Google Scholar]
- 150. Gallo K, Goede A, Mura C, et al. A comparative analysis of COVID‐19 vaccines based on over 580,000 cases from the vaccination adverse event reporting system. Vaccines. 2022;10(3):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Cines DB, Bussel JB. SARS‐CoV‐2 vaccine‐induced immune thrombotic thrombocytopenia. N Engl J Med. 2021;384(23):2254‐2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Fu Y, Pan Y, Li Z, Li Y. The utility of specific antibodies against SARS‐CoV‐2 in laboratory diagnosis. Front Microbiol. 2020;11:603058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rattanapisit K, Shanmugaraj B, Manopwisedjaroen S, et al. Rapid production of SARS‐CoV‐2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana . Sci Rep. 2020;10(1):17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS‐CoV‐2 infection. Nat Commun. 2020;11(1):2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Chen RE, Zhang X, Case JB, et al. Resistance of SARS‐CoV‐2 variants to neutralization by monoclonal and serum‐derived polyclonal antibodies. Nat Med. 2021;27(4):717‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS‐CoV‐2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014‐1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Xia S, Liu M, Wang C, et al. Inhibition of SARS‐CoV‐2 (previously 2019‐nCoV) infection by a highly potent pan‐coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. de Oliveira OV, Rocha GB, Paluch AS, Costa LT. Repurposing approved drugs as inhibitors of SARS‐CoV‐2 S‐protein from molecular modeling and virtual screening. J Biomol Struct Dyn. 2021;39(11):3924‐3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS‐CoV‐2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents. 2020;56(2):105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Padhi AK, Seal A, Khan JM, Ahamed M, Tripathi T. Unraveling the mechanism of arbidol binding and inhibition of SARS‐CoV‐2: insights from atomistic simulations. Eur J Pharmacol. 2021;894:173836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chan KK, Tan TJC, Narayanan KK, Procko E. An engineered decoy receptor for SARS‐CoV‐2 broadly binds protein S sequence variants. Sci Adv. 2021;7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181(4):905‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA‐approved drug ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. Antiviral Res. 2020;178:104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wang X, Cao R, Zhang H, et al. The anti‐influenza virus drug, arbidol is an efficient inhibitor of SARS‐CoV‐2 in vitro. Cell Discov. 2020;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181(4):905‐913.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Linsky TW, Vergara R, Codina N, et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS‐CoV‐2. Science. 2020;370(6521):1208‐1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Cao L, Goreshnik I, Coventry B, et al. De novo design of picomolar SARS‐CoV‐2 miniprotein inhibitors. Science. 2020;370(6515):426‐431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Case JB, Chen RE, Cao L, et al. Ultrapotent miniproteins targeting the SARS‐CoV‐2 receptor‐binding domain protect against infection and disease. Cell Host Microbe. 2021;29(7):1151‐1161.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Xia S, Zhu Y, Liu M, et al. Fusion mechanism of 2019‐nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17(7):765‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zhu Y, Yu D, Yan H, Chong H, He Y. Design of potent membrane fusion inhibitors against SARS‐CoV‐2, an emerging coronavirus with high fusogenic activity. J Virol. 2020;94(14):e00635‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. de Vries RD, Schmitz KS, Bovier FT, et al. Intranasal fusion inhibitory lipopeptide prevents direct‐contact SARS‐CoV‐2 transmission in ferrets. Science. 2021;371(6536):1379‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Clark NM, Grim SA, Lynch JP, 3rd . Posaconazole: use in the prophylaxis and treatment of fungal infections. Semin Respir Crit Care Med. 2015;36(5):767‐785. [DOI] [PubMed] [Google Scholar]
- 174. Jana ID, Bhttacharya P, Mayilsamy K, et al. Targeting an evolutionarily conserved "E‐L‐L" motif in the spike protein to develop a small molecule fusion inhibitor against SARS‐CoV‐2. bioRxiv . 2022. [DOI] [PMC free article] [PubMed]
- 175. Xing L, Xu X, Xu W, et al. A five‐helix‐based SARS‐CoV‐2 fusion inhibitor targeting heptad repeat 2 domain against SARS‐CoV‐2 and its variants of concern. Viruses. 2022;14(3):597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Bhuiyan FR, Howlader S, Raihan T, Hasan M. Plants metabolites: possibility of natural therapeutics against the COVID‐19 pandemic. Front Med. 2020;7:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Pandey P, Rane JS, Chatterjee A, et al. Targeting SARS‐CoV‐2 spike protein of COVID‐19 with naturally occurring phytochemicals: an in silico study for drug development. J Biomol Struct Dyn. 2021;39(16):6306‐6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Jamwal S, Gautam A, Elsworth J, Kumar M, Chawla R, Kumar P. An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID‐19 pandemic. Life Sci. 2020;257:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS‐CoV‐2 entry into cells. Nat Rev Mol Cell Bio. 2022;23(1):3‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Tada T, Fan C, Chen JS, et al. An ACE2 microbody containing a single immunoglobulin Fc domain is a potent inhibitor of SARS‐CoV‐2. Cell Rep. 2020;33(12):108528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Carpinteiro A, Edwards MJ, Hoffmann M, et al. Pharmacological Inhibition of acid sphingomyelinase prevents uptake of SARS‐CoV‐2 by epithelial cells. Cell Rep Med. 2020;1(8):100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Carpinteiro A, Gripp B, Hoffmann M, et al. Inhibition of acid sphingomyelinase by ambroxol prevents SARS‐CoV‐2 entry into epithelial cells. J Biol Chem. 2021;296:100701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Wang X, Luo J, Wen Z, et al. Diltiazem inhibits SARS‐CoV‐2 cell attachment and internalization and decreases the viral infection in mouse lung. PLoS Pathog. 2022;18(2):e1010343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Egieyeh S, Egieyeh E, Malan S, Christofells A, Fielding B. Computational drug repurposing strategy predicted peptide‐based drugs that can potentially inhibit the interaction of SARS‐CoV‐2 spike protein with its target (humanACE2). PLoS One. 2021;16(1):e0245258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Gu C, Wu Y, Guo H, et al. Protoporphyrin IX and verteporfin potently inhibit SARS‐CoV‐2 infection in vitro and in a mouse model expressing human ACE2. Sci Bull. 2021;66(9):925‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wang G, Yang M‐L, Duan Z‐L, et al. Dalbavancin binds ACE2 to block its interaction with SARS‐CoV‐2 spike protein and is effective in inhibiting SARS‐CoV‐2 infection in animal models. Cell Res. 2021;31(1):17‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ansarin K, Tolouian R, Ardalan M, et al. Effect of bromhexine on clinical outcomes and mortality in COVID‐19 patients: A randomized clinical trial. Bioimpacts. 2020;10(4):209‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Li K, Meyerholz DK, Bartlett JA, McCray PB. The TMPRSS2 Inhibitor Nafamostat Reduces SARS‐CoV‐2 Pulmonary Infection in Mouse Models of COVID‐19. mBio. 2021;12(4):e0097021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Wettstein L, Weil T, Conzelmann C, et al. Alpha‐1 antitrypsin inhibits TMPRSS2 protease activity and SARS‐CoV‐2 infection. Nat Commun. 2021;12(1):1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Shapira T, Monreal IA, Dion SP, et al. A TMPRSS2 inhibitor acts as a pan‐SARS‐CoV‐2 prophylactic and therapeutic. Nature . 2022. [DOI] [PMC free article] [PubMed]
- 191. Zhao M‐M, Yang W‐L, Yang F‐Y, et al. Cathepsin L plays a key role in SARS‐CoV‐2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther. 2021;6(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Mellott DM, Tseng C‐T, Drelich A, et al. A clinical‐stage cysteine protease inhibitor blocks SARS‐CoV‐2 infection of human and monkey cells. ACS Chem Biol. 2021;16(4):642‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Cheng Y‐W, Chao T‐L, Li C‐L, et al. Furin inhibitors block SARS‐CoV‐2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2):108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Bollavaram K, Leeman TH, Lee MW, et al. Multiple sites on SARS‐CoV‐2 spike protein are susceptible to proteolysis by cathepsins B, K, L, S, and V. Protein Sci. 2021;30(6):1131‐1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Hoffmann M, Schroeder S, Kleine‐Weber H, Müller MA, Drosten C, Pöhlmann S. Nafamostat mesylate blocks activation of SARS‐CoV‐2: new treatment option for COVID‐19. Antimicrob Agents Chemother. 2020;64(6):e00754‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study. The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) database and analyzed in The Interactive Tree Of Life online software tool at https://itol.embl.de; the Open Access FunRich functional enrichment analysis tool at http://www.funrich.org/index.html.


