Dear Editor,
1.
At the time of writing, the world continues to witness the extraordinarily rapid evolution and selection of SARS‐CoV‐2, with the Omicron variants comprising five lineages known as BA.1, BA.2, BA.3, BA.4, and BA.5. 1 The Omicron variant shows a large number of mutations leading to amino‐acid changes, which are widely distributed on many structural and nonstructural regions of SARS‐CoV‐2. 2 The spike protein of the Omicron variant is characterized by at least 32 amino acid substitutions, some small deletions, and insertions. 2 From the beginning of the COVID‐19 pandemics, a large number of SARS‐CoV‐2 whole‐genome sequences have been generated from all around the world and shared, mostly through GISAID (https://www.gisaid.org/). 3 Pyrosequencing has played an important role to discover and track the spread of SARS‐CoV‐2 variants having a large number of mutations nearly in real‐time by worldwide genome sequencing efforts. 3 , 4 Unsurprisingly, given the vaccine‐ and prior infection‐driven selection pressures, many of the changes are located in the receptor‐binding domain of the spike protein that are important for virus–host cell interaction mediated by ACE‐2 receptor. 5 In the virus–host cell interaction, the nucleocapsid protein binds directly to host mRNAs in cells and suppresses the physiological stress response of host cells. 6 Of note, these mutations are associated with more efficient cell entry, immune escape, and increased infectivity of the Omicron variant. 7 BA.1 was first found in southern Africa in November 2021 during the investigation of an unexplained burst in positive case numbers. 8 BA.1 was reported to be associated with an S‐gene target failure or SGTF (also called S‐gene dropout) on the Thermo Fisher TaqPath COVID‐19 RT‐PCR assay because of the small deletion at the positions 69 and 70 in the spike gene. 7 After the initial discovery, BA.1 had spread rapidly and WHO estimated that this variant was already in most countries. 9 The United Kingdom reported the first death associated with BA.1 infection. 10 In the United States, BA.1 displaced Delta to become the predominant variant, accounting for more than 98% by the end of January 2022 (https://covid.cdc.gov/covid-data-tracker/#variant-proportions). Chen et al. 11 reported low neutralizing antibodies against the Omicron variant, even at 3‐ to 5‐week postvaccination. 11
Since Omicron is not effectively neutralized by monoclonal antibody treatments and, unfortunately, cannot be distinguished from other SARS‐COV2 variants using the clinically qualified NAAT tests, we recently developed a one‐step qualitative RT‐PCR assay to determine if the SARS‐CoV‐2 is of the BA.1 lineage. 12 The RT‐PCR BA.1 assay was designed based on the deletion del69‐70 and the insertion ins214EPE seen in the BA.1 spike gene. A specimen is BA.1 detected only if two targets (del69‐70 and ins214EPE) are positive, or as designated as “del69‐70 positive/ins214EPE positive.” The RT‐PCR BA.1 assay can detect two copies/μl and does not have any cross‐reactivity with other SARS‐CoV‐2 variants, showing high sensitivity and specificity. 12 To assess the prevalence of SARS‐CoV‐2 Omicron variants, some randomly selected nasopharyngeal swabs, which were determined to harbor SARS‐CoV‐2 per the Cepheid Xpert Xpress SARS‐CoV‐2/Flu/RSV test, were tested using the RT‐PCR BA.1 assay. We performed one run of the RT‐PCR BA.1 assay every 2 weeks, and the number of nasopharyngeal specimens tested in each run varied depending on their availability as shown in Figure 1. Briefly, the specimens were extracted using the bioMérieux EasyMag, and the RT‐PCR reactions were run on the Applied Biosystems 7500 Fast Dx real‐time PCR system using the cycling condition, which was previously described. 12 In addition, BA.1 negative specimens undergo whole‐genome sequencing to determine their genetic lineages.
Figure 1.
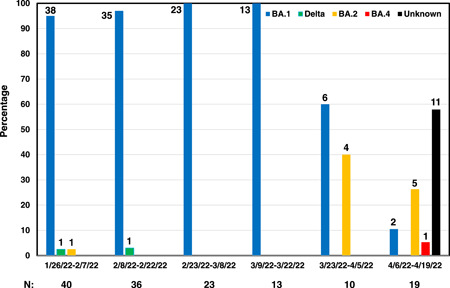
The graph presented the changing prevalence of SARS‐CoV‐2 variants in the UPMC health care system in Allegheny County, Pennsylvania. The numbers below the graph are the number of SARS‐CoV‐2 positive specimens tested in each time window.
In total, there were 141 nasopharyngeal specimens from patients (with ages ranging from 15 to 92 years, and 44% being male) tested using the RT‐PCR BA.1 assay from January to April 2022. Of these, 83.0% (117/141) specimens were BA.1 and this variant was found to be predominant until mid‐March 2022 with the prevalence rate ranging from 95% (35/40) up to 100% (23/23 and 13/13). However, the prevalence rate of BA.1 dropped to 60% (6/10) during March 23 to May 4, 2022 and continued to decrease to 10.5% (2/19) during April 6–19, 2022, as shown in Figure 1.
There were 24 BA.1 negative specimens. Of seven BA.1 negative specimens being “del69‐70 negative/ins214EPE negative” during January 26 to May 4, 22, two specimens were Delta and five specimens were BA.2 as determined by sequencing. It is interesting that BA.2 emerged during March 23 to May 4, 2022, accounting for 40% (4/10). Of 17 BA.1 negative specimens from April 6 to 19, 2022, 16 specimens were “del69‐70 negative/ins214EPE negative” and one specimen was “del69‐70 positive/ins214EPE negative.” The genomic analysis revealed that five specimens having the “del69‐70 negative/ins214EPE negative” result were BA.2, and one specimen having the “del69‐70 positive/ins214EPE negative” result was BA.4.
While the genetic lineage of 11 specimens having the “del69‐70 negative/ins214EPE negative” result is not known as they were not sequenced, we predicted that they were all BA.2, based upon the initial results of local circulation confirmed by sequencing. Since BA.2 does not have both del69‐70 and ins214EPE in the spike gene that is seen in BA.1, the RT‐PCR BA.1 assay on BA.2 is negative for both targets del69‐70 and ins214EPE. 12 While lacking the ins214EPE, BA.3 through BA.5 have the del69‐70 in the spike gene (https://cov-lineages.org/lineage_list.html). Therefore, BA.3 through BA.5 are positive for del69‐70 and negative for ins214EPE tested by the RT‐PCR BA.1 assay. It is obvious that the “del69‐70 negative/ins214EPE negative” result or the “del69‐70 positive/ins214EPE negative” result can be used as a rapid proxy to flag potential BA.2 or BA.3 through BA.5 cases, respectively.
Omicron BA.4 and BA.5 are new variants, which were first discovered in Botswana and South Africa (https://www.afro.who.int/news/botswana-south-africa-deepen-probe-new-omicron-sub-variants). According to the World Health Organization, BA.4 and BA.5 are circulating at low levels in several countries, and currently, there is no significant epidemiological difference observed between BA.4 and BA.5 and other known lineages of the Omicron variant. The data on BA.4 is scarce, and our search in PubMed did not reveal any trials or experiments of BA.4 in animal models at the time of this writing. Currently, Qiagen offers LNA‐based qPCR assays for the genotyping of SARS‐CoV‐2 variants, including Omicron, based on the detection of lineage‐specific mutations (https://www.qiagen.com/us/applications/infectious-disease/coronavirus/research-solutions/pcr-based-epidemiology). However, this assay cannot differentiate among BA.1 through BA.5. While developing a multiplexed RT‐PCR test for the simultaneous qualitative detection of Omicron and other respiratory viruses is potentially interesting, it would be challenging due to the rapid evolution of SARS‐CoV‐2 and the possible appearance of new variants. While the unexpected detection of BA.4 in our study is interesting, and even a single case, our finding underscores the importance of genomic surveillance as a critical tool for tracking emerging variants of SARS‐CoV‐2.
AUTHOR CONTRIBUTIONS
Tung Phan and Alan Wells designed the study and wrote the manuscript; Stephanie Boes, Jamie Gribschaw, and Melissa McCullough performed and managed the PCR‐specific testing; Jane W. Marsh and Lee H. Harrison performed and managed WGS. All read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
All testing was performed as a part of routine clinical care and performed according to CLIA '88 regulations by the appropriate personnel. The entire study was deemed to be a Quality Improvement initiative by the UPMC IRB and approved by the UPMC QI Review Board.
ACKNOWLEDGMENTS
The authors thank the UPMC Clinical Microbiology Laboratory for testing the specimens and performing the evaluation. We thank the Microbial Genome Sequencing Center (MiGS) for expert SARS‐CoV‐2 whole‐genome sequencing. The study was internally funded by the UPMC Clinical Laboratories as part of a Quality Improvement initiative.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no data sets were generated or analyzed during the current study.
REFERENCES
- 1. Tegally H, Moir M, Everatt J, et al. Continued emergence and evolution of Omicron in South Africa: new BA.4 and BA.5 lineages. medRxiv . 2022. 10.1101/2022.05.01.22274406 [DOI]
- 2. Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS‐CoV‐2, a global urgent public health alert! J Med Virol. 2022;94:1255‐1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oude Munnink BB, Worp N, Nieuwenhuijse DF, et al. The next phase of SARS‐CoV‐2 surveillance: real‐time molecular epidemiology. Nat Med. 2021;27:1518‐1524. [DOI] [PubMed] [Google Scholar]
- 4. Goswami C, Sheldon M, Bixby C, et al. Identification of SARS‐CoV‐2 variants using viral sequencing for the centers for disease control and prevention genomic surveillance program. BMC Infect Dis. 2022;22:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim S, Nguyen TT, Taitt AS, et al. SARS‐CoV‐2 Omicron mutation is faster than the chase: multiple mutations on spike/ACE2 interaction residues. Immune Netw. 2021;21:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nabeel‐Shah S, Lee H, Ahmed N, et al. SARS‐CoV‐2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience. 2022;25:103562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Araf Y, Akter F, Tang YD, et al. Omicron variant of SARS‐CoV‐2: genomics, transmissibility, and responses to current COVID‐19 vaccines. J Med Virol. 2022;94:1825‐1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaughan A. Omicron emerges. New Sci. 2021;252:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omicron May Already Be in Most Countries, WHO Warns. Accessed December 15, 2021. https://www.downtoearth.org.in/news/health/omicron-may-already-be-in-most-countries-who-warns-80704.
- 10. Thakur V, Ratho RK. Omicron (B.1.1.529): a new SARS‐CoV‐2 variant of concern mounting worldwide fear. J Med Virol. 2022;94:1821‐1824. [DOI] [PubMed] [Google Scholar]
- 11. Cheng SMS, Mok CKP, Leung YWY, et al. Neutralizing antibodies against the SARS‐CoV‐2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022;28:486‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phan T, Boes S, McCullough M, et al. Development of a one‐step qualitative RT‐PCR assay to detect the SARS‐CoV‐2 Omicron (B.1.1.529) variant in respiratory specimens. J Clin Microbiol. 2022;60:e0002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no data sets were generated or analyzed during the current study.


