Abstract
A novel outer membrane lipoprotein in Pseudomonas aeruginosa is encoded by the omlA gene, which was identified immediately upstream of the fur (ferric uptake regulator) gene. The omlA and fur genes were divergently transcribed and had overlapping promoter regions. The proximal fur P2 promoter and the omlA promoter shared a 5-bp DNA motif for their −10 promoter elements. The distal fur P1 promoter was located within the omlA coding sequence, and the omlA and fur T1 mRNAs overlapped by 154 nucleotides. Optimal expression of both fur and omlA required roughly 200 bp of DNA upstream of the promoter regions, suggesting the presence of cis-acting transcriptional activation elements located within the omlA and fur genes, respectively. The levels of Fur and OmlA proteins had no influence on omlA or fur expression, excluding any trans-acting cross-regulation between fur and omlA. Expression of omlA was constitutive regardless of growth phase, oxygen tension, iron concentration, pH, and temperature. OmlA contained a signal sequence typical of bacterial lipoproteins, with a cysteine as a putative cleavage and lipid attachment site. Inhibition of signal peptidase II by globomycin resulted in failure to process OmlA, thus giving strong evidence that OmlA is a lipoprotein. Cell fractionation followed by Western blot analysis indicated that all OmlA protein is localized in the outer membrane. Mature OmlA was an acidic (pI = 4.5) protein of 17.3 kDa and had close to 40% amino acid sequence identity to SmpA (small protein A) of Escherichia coli, Vibrio cholerae, and Haemophilus influenzae, a protein of unknown function. All P. aeruginosa strains tested as well as Pseudomonas fluorescens were found to produce OmlA. A mutant strain with impaired production of OmlA but no change in the expression of the overlapping fur gene was constructed. The omlA mutant was hypersusceptible to anionic detergents such as sodium dodecyl sulfate and deoxycholate, and it showed increased susceptibility to various antibiotics, including nalidixic acid, rifampin, novobiocin, and chloramphenicol. A structural role of OmlA in maintaining the cell envelope integrity is proposed.
The opportunistic pathogen Pseudomonas aeruginosa has the capacity to produce a large variety of virulence factors that play a role in the infection of injured or immunocompromised hosts (28, 56). The production of virulence factors is regulated in response to the environmental conditions, such as iron and oxygen availability (23, 33). Iron is frequently limiting for P. aeruginosa, which prefers an aerobic metabolism that requires iron-containing respiratory enzymes. P. aeruginosa has thus evolved powerful iron acquisition systems which can be activated upon iron starvation. The ferric uptake regulator (Fur) plays the central role in the control of the iron-regulated genes. Fur is an iron-responsive, DNA binding repressor which employs Fe(II) as a corepressor and binds as a dimer to a so-called Fur box in the promoter regions of iron-regulated genes (31). Roughly 30 targets of the P. aeruginosa Fur protein have been identified and were shown to be expressed in an iron-dependent manner in vivo (32). The P. aeruginosa fur gene is transcribed from two separate promoters which are 170 bp apart. While examining the region upstream of fur, we identified a novel gene which we designated omlA. Although fur and omlA possessed overlapping promoter regions, their functions appeared to be unrelated. OmlA represented a novel outer membrane lipoprotein which seemed to play a role in maintaining the cell envelope integrity. A large number of outer membrane proteins in P. aeruginosa have been characterized, and they appear to be highly conserved among the Pseudomonadaceae (for a review see reference 16). General functions include pore formation, transport of specific substrates, cell structure determination, and membrane stabilization. The porin class includes the major outer membrane protein OprF, which is a homolog of Escherichia coli OmpA (55); the highly homologous OprO and OprP, which are induced under phosphate limitation (45); OprC, a copper-binding channel protein (59); OprE, an anaerobically induced channel-forming protein (58); and components of multidrug-resistance efflux pumps such as OprM (formerly OprK) (35), OprJ (34), OprD (19), and OprN (21). Involved in the maintenance of the cell envelope are the very small OprI, which is 30% identical to Braun’s lipoprotein of E. coli (9); OprH, which is associated with lipopolysaccharide and replaces outer membrane-stabilizing divalent cations (2); and OprL, a peptidoglycan-associated lipoprotein (22). Only three of the above proteins, OprI, OprM, and OprL, are lipoproteins, and the novel OmlA described here falls also in this category. The posttranslational modification and processing of prolipoproteins had been studied in vitro (50) and in vivo (42). They involve a few enzymatic modification steps, which include the transfer of a diacylglyceryl moiety to the sulfhydryl group of the prospective N-terminal cysteine, cleavage of the signal sequence by signal peptidase II, and acylation of the new amino terminus.
Outer membrane proteins are of great importance as vaccine candidates, since they are typically very immunogenic and have adjuvant activity. OprF and OprI have been successfully demonstrated as potential vaccines in mice and humans, either alone, or as OprF-OprI fusion proteins, or as carriers for foreign epitopes (14, 17, 52). Also, due to their conserved occurrence, outer membrane proteins are of considerable importance in clinical diagnostics. In fact, the oprI lipoprotein gene has been used as a very small but specific DNA probe and as a reliable PCR target within RNA group I of the Pseudomonadaceae (39). Similarly, a PCR assay based on the simultaneous amplification of both oprI and oprL lipoprotein genes has been used to detect P. aeruginosa in clinical material with very high sensitivity and absolute specificity (8). The lipoprotein OmlA, as characterized in this report, constitutes another candidate for testing as a potential vaccine or drug target.
MATERIALS AND METHODS
Strains, plasmids, primers, and media.
The relevant strains, plasmids, and primers used in this study are shown in Table 1. Luria broth (LB) was used for strain maintenance, and M9 medium containing 0.2% glucose was used for growth and susceptibility assays (40). Chelex-treated and dialyzed tryptic soy broth (D-TSB) containing 1% glycerol and 50 mM glutamate was used as low-iron medium and was supplemented with 50 μg of FeCl3 per ml in high-iron medium (36). P. aeruginosa was grown at 32°C aerobically in shake flasks or microaerobically (5% oxygen) in static CampyPak jars. Antibiotics were used as follows: for E. coli, ampicillin (100 μg/ml), gentamicin (15 μg/ml), kanamycin (100 μg/ml), tetracycline (15 μg/ml), and globomycin (100 and 250 μg/ml); for P. aeruginosa, carbenicillin (750 μg/ml), gentamicin (75 μg/ml), tetracycline (150 μg/ml), and streptomycin (500 μg/ml); and for Pseudomonas fluorescens, streptomycin (500 μg/ml).
TABLE 1.
Strains, plasmids, and primers used in this studya
| Strain, plasmid, or primer | Description | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PAO1 (ATCC 15692) | Prototroph | 18 |
| PA103 | Prototroph | 24 |
| PAK | Prototroph | 30 |
| PG201 | Prototroph | 15 |
| PA1277 | Prototroph | 51 |
| Ps388 | Prototroph, hypotoxigenic | 51 |
| WR5 | Prototroph, exotoxin A deficient | 51 |
| 6B omlA::Tc | omlA mutant producing small amounts of OmlA | This study |
| 3A omlA::Tc | omlA 3′-truncated mutant | This study |
| CS fur | Cold-sensitive fur mutant of PAO1 | 20 |
| CSR fur | Spontaneous revertants of CS | This study |
| P. fluorescens | ||
| ATCC 15453 | Prototroph | ATCC |
| SBW25 | Prototroph | 37 |
| E. coli | ||
| DH5α | hsdR recA lacZYA φ80 lacZΔM15 | BRL |
| HB101 | hsdS recA proA lacY | 5 |
| QC1732 | fur null mutant | M. McIntosh |
| BL21(DE3)/pLysE | High-stringency T7 expression host, hsdS DE3, Cmr | Novagen |
| Plasmids | ||
| pBluescript SK(+) | AmprlacZ′, cloning vector | Stratagene |
| pCRII 2.1 | Ampr Kmr, PCR cloning vector | Invitrogen |
| pGEX-2T | Ampr, lacIq, Ptac-gst expression vector for protein fusions | Pharmacia |
| pSUP203 | Ampr Tcr Cmr mob, suicide vector | 46 |
| pVLT35 | Smr mob, lacIq, broad-host-range Ptrp-lac expression vector | 7 |
| pRK2013 | Kmr conjugation helper plasmid | 11 |
| pUCP19 | Ampr PlacZ, broad-host-range cloning and expression vector | 43 |
| pUCP24 | Gmr PlacZ, broad-host-range cloning and expression vector | 43 |
| pPZ20, pPZ30 | Ampr, ′lacZ-based promoter probe vector | 44 |
| pGEX-omlA | pGEX-2T containing a gst-omlA fusion under tac promoter control | This work |
| pET-omlA | pET23a containing omlA under T7 promoter control | This work |
| pOML15 | pUCP19 containing omlA as a 0.8-kb PstI-BglII fragment under lac promoter control | This work |
| pOML24 | pUCP24 containing omlA as a 0.8-kb PstI-BglII fragment | This work |
| pOML36 | pUCP19 containing a 3.6-kb PstI fragment of the omlA region | This work |
| pCRII-omlA-444a and pCRII-omlA-444b | pCRII containing the 444-bp omlA-fur promoters in omlA sense (a) or fur sense (b) orientation relative to the T7 promoter | This work |
| pPZ-PomlA | Series of pPZ20 containing various omlA promoter fragments | This work |
| pPZ-Pfur | Series of pPZ30 containing various fur promoter fragments | This work |
| pPZ-PF-PomlA | pPZ20 containing a 399-bp fragment of the P. fluorescens omlA promoter fused to lacZ | This work |
| pSUP-omlA 6B | pPSUP203 containing a 370-bp 5′ omlA fragment | This work |
| pSUP-omlA 3A | pPSUP203 containing a 291-bp internal fragment of omlA | This work |
| Primersb | ||
| omlA-(−101) | CCTCGCCTGCTTCCATC | |
| omlA-(−35) | (PstI)-CGAGCATCTGCAGGATCTTG | |
| omlA-139 | (NdeI)-catATGCAAAACGCCAAGCTCATGC | |
| omlA-156 | (HindIII)-aAGCTTGGCGTTTTGCATCG | |
| omlA-205 | (NdeI)-catATGTCGTTTCCTGGCGTCTATAAAATCG | |
| omlA-343 | GGAAGGTATCGACGATGA | |
| omlA-495 | (Stop)-tcaGAGGATCGCTTCGTCGCGGCT | |
| omlA-508 | TGCCTTCCTTGCCGAGGATC | |
| omlA-676 | (BamHI)-ggatCCAGCCGTCATTGCGGGCT |
Abbreviations: Ampr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Smr, streptomycin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; ATCC, American Type Culture Collection; mob, mobilizable.
The primers are numbered with respect to the DNA sequence as shown in Fig. 1. Lowercase letters at the 5′ end are nonmatching nucleotides to form the indicated motif as shown underlined.
General genetic methods.
PCRs were performed with Taq DNA polymerase (Bethesda Research Laboratories [BRL]) and appropriate custom-made primers (BRL) in a Perkin-Elmer Cetus thermal cycler, with 30 cycles of denaturing (1 min, 94°C), annealing (1 min, 54°C), and extending (1 min, 72°C), and the amplified DNA fragments were purified in a preparative agarose gel and subsequently cloned into pCRII-2.1 (Invitrogen). All cloned PCR fragments and the omlA gene were sequenced by the dideoxy chain termination method (41) with Sequenase 2.0 (United States Biochemical) and M13 primers or custom-made 18-mer oligonucleotides. Published procedures were followed for Southern blotting (47) and colony hybridization (12).
RNase protection analyses were performed with the Riboprobe system (Promega), and radiolabeled riboprobes from suitable cloned PCR fragments were generated by runoff transcription from the T7 promoter of linearized pCRII-2.1 as described in detail elsewhere (1). Complementary antisense RNA probes for omlA and fur were synthesized from a PCR fragment generated with the primers omlA-343 and omlA-(−101), which was cloned separately in both orientations behind the T7 promoter of pCRII-2.1 (pCRII-omlA-444a and pCRII-omlA-444b).
Translational fusions of omlA to the lacZ reporter gene on plasmid pPZ20 were constructed by directional cloning of appropriate PCR products as EcoRI-HindIII fragments into pPZ20. To generate the PomlA PCR fragments, the primer omlA-156, which contains the HindIII restriction site, was used in combination with six primers located further upstream to yield the DNA fragments as mentioned in the text. Similarly, translational fusions of fur to lacZ were constructed by transferring previously cloned PCR products as EcoRI-PstI fragments into plasmid pPZ30. To produce the Pfur PCR fragments, the primer omlA-(−35), which contains the PstI restriction site, was used together with six different primers upstream of fur.
Overexpression and labeling of OmlA.
The omlA coding sequence from the ATG start codon to 7 bp downstream of the TGA stop codon was amplified from chromosomal PAO1 DNA with the primers omlA-139 and omlA-676 (Table 1). The resulting 538-bp fragment was gel purified, cloned into pCRII-2.1, sequenced, and transferred as an NdeI-BamHI fragment into the T7 expression vector pET23a, yielding pET-omlA. For induction and labeling experiments, E. coli BL21(DE3)/pLysE containing pET-omlA or the pET23a control vector was grown in M9 minimal medium at 37°C and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density at 600 nm (OD600) of 0.4. Rifampin (200 μg/ml) was added to 1-ml culture aliquots 30 min postinduction, and incubation was continued for 20 min. A mixture of 14C-labeled amino acids (5 μCi) was added, and the cultures were shaken for 1 h. The cells were harvested, lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (40), and analyzed by SDS-PAGE followed by autoradiography to detect radiolabeled proteins. The above protocol was also used to monitor the processing of OmlA, with the addition of globomycin in methanol (100 and 250 μg/ml) together with the rifampin.
Generation of polyclonal anti-OmlA antibodies and Western blot analysis.
A 472-bp NdeI/BamHI fragment containing the omlA gene minus the first 66 bp of the coding sequence was generated by PCR with the primers omlA-205 and omlA-676 (Table 1). The DNA fragment was cloned into pCRII-2.1 and then transferred as an EcoRI fragment into pGEX-2T, yielding pGEX-omlA. The correct DNA sequence, orientation, and in-frame fusion of gst to omlA in pGEX-omlA was verified by sequencing. The E. coli fur null mutant QC1732 was transformed with pGEX-omlA, and 1 liter of culture was grown in LB at 37°C. The production of glutathione S-transferase (GST)–OmlA fusion protein was induced with 1 mM IPTG during log phase (OD600 = 0.4). The cells were harvested 4 h later (OD600 = 2.5) and resuspended in 30 ml of 50 mM Tris · HCl (pH 8.5)–150 mM NaCl. Lysis of the cells was achieved by freezing and thawing followed by sonication (six bursts of 30 s each on ice-NaCl). Cell debris were removed by centrifugation (10,000 × g, 10 min), and 20 ml of the soluble fraction was mixed with 2 ml of glutathione-Sepharose 4B (Pharmacia), gently agitated for 30 min at 25°C, and poured into a 5-ml spin column. After three washes of the matrix with 10 ml of phosphate-buffered saline, the bound GST-OmlA protein was specifically eluted with 4 ml of 10 mM glutathione–50 mM Tris · HCl (pH 8.0). A portion of this affinity-purified GST-OmlA fraction (2 ml, 6 mg) was mixed with 2 ml of 2× SDS sample buffer and loaded onto a 10-cm preparative 11% polyacrylamide–SDS tube gel (Bio-Rad) topped with 1.5 cm of 4% stacking gel. The gel was run at 23 ml/h at 4°C, and fractions of 2 ml were collected and subsequently analyzed for proteins on 15% minigels. Fractions containing purified GST-OmlA were pooled, and 100-μg aliquots were used without further processing for the immunization of two female New Zealand White rabbits. The antigen was administered in complete Freund’s adjuvant in three weekly intervals and in incomplete Freund’s adjuvant for two monthly boosters. Serum samples prepared 7 days after the last booster were used for the subsequent immunologic detection of OmlA, and preimmunization serum served as a control. Whole-cell extracts or overproduced soluble proteins were separated by SDS-PAGE on 15% acrylamide gels and blotted onto BA S-85 nitrocellulose (Schleicher & Schuell). The membranes were probed with 2,000-fold-diluted anti-OmlA serum and developed by using the Western-Light chemiluminescence detection system (Tropix Inc.). The anti-OmlA serum reacted specifically to P. aeruginosa OmlA that had been overproduced in E. coli, and preimmunization serum did not react with P. aeruginosa whole-cell extracts (data not shown).
Cell fractionation procedure.
P. aeruginosa cells were fractionated following a published protocol (27) with modifications. In brief, P. aeruginosa was grown for 6 h in 30 ml of LB and the cells were collected by centrifugation (10,000 × g, 10 min), washed with 5 ml of ice-cold 20% sucrose and resuspended in 4.5 ml ice-cold 20% sucrose. The following ice-chilled solutions were slowly added: 2.25 ml of 2 M sucrose, 2.5 ml of 0.1 M Tris · HCl (pH 7.8), 0.2 ml of 25 mM Na3EDTA (pH 8), and 0.45 ml of 0.5% lysozyme. The mixture was then incubated for 1 h at 30°C without agitation. Spheroplasts were removed by centrifugation (17,000 × g, 15 min), and the outer membranes were separated from the periplasmic fraction by ultracentrifugation (SW41, 30,000 rpm, 1 h). The spheroplasts were lysed osmotically in 4 volumes of 5 mM MgCl2 and fractionated into intracellular proteins and inner membranes by centrifugation (20,000 × g, 20 min). Crude inner and outer membrane fractions were resuspended in water, while the periplasmic and intracellular proteins as well as culture supernatants were concentrated by 80% saturated ammonium sulfate and dissolved in 50 mM Tris · HCl (pH 7.5) before subsequent SDS-PAGE and Western blot analysis.
Disruption of the omlA gene.
The PAO1 omlA::Tc mutant 6B, which produces extremely small amounts of OmlA protein, was constructed as follows. A 370-bp fragment comprising the 5′ portion of the omlA coding sequence from the ATG start codon was generated with the primers omlA-139 and omlA-508 (Table 1), cloned into pCRII-2.1, sequenced, and transferred as an EcoRI fragment into pSUP203 linearized with EcoRI. The resulting plasmid, pSUP-omlA-6B, was mobilized into P. aeruginosa PAO1 in a triparental mating by using E. coli HB101/pRK2013 as the helper strain (11, 46), and tetracycline-resistant transconjugants were isolated.
The PAO1 omlA::Tc mutant 3A, which lacks a functional omlA gene, was generated similarly, using a 291-bp PCR fragment amplified with the primers omlA-205 and omlA-495 (Table 1). A stop codon was introduced into primer omlA-495, so that the subsequent PAO1 omlA::Tc 3A transconjugants harbor a mutated version of the omlA gene with a stop codon 171 nucleotides earlier than wild-type omlA and thus express an OmlA protein lacking the 57 carboxy-terminal amino acids. The mutations were confirmed by Southern blotting and PCR analysis.
Nucleotide sequence accession numbers.
The DNA sequence of the 2.3-kb fur-omlA-ORF1-ORF2 region has been deposited in GenBank under accession no. AF050676; the omlA gene from P. fluorescens has been deposited under accession no. AF050677.
RESULTS
fur-omlA locus and DNA sequence of the omlA gene.
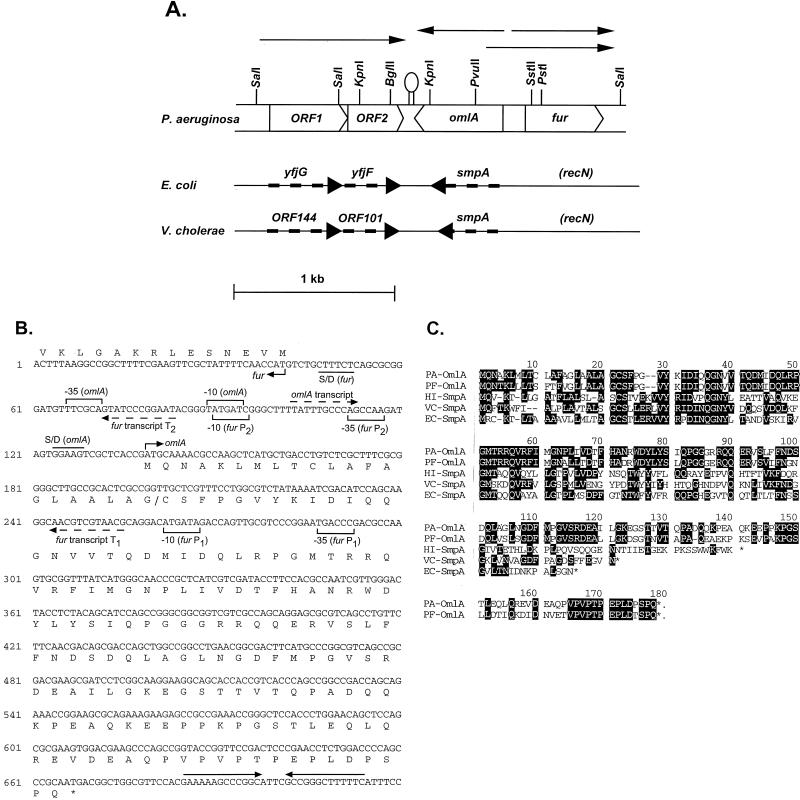
The fur promoter region (36) was used in Southern blots to probe chromosomal PAO1 DNA cut with various enzymes, and a 3.6-kb PstI fragment containing the region upstream of fur was subsequently cloned (pOML36). DNA sequence analysis of pOML36 revealed the presence of three putative open reading frames upstream of the fur gene (Fig. 1A). Immediately upstream and in the opposite orientation to fur was a 531-bp open reading frame (omlA), and located further upstream were two open reading frames (ORF1 and ORF2). A strong stem-loop structure between the ends of the omlA gene and ORF2 suggested the presence of a terminator. A portion of the DNA sequence of the 2.3-kb fur-omlA-ORF1-ORF2 region with the omlA transcriptional and translational elements and the omlA translation is shown in Fig. 1B. The hypothetical protein sequences deduced from omlA, ORF1, and ORF2 were found to be homologous to corresponding E. coli proteins encoded in a region at 59 min of the chromosome (YfjG, YfiF, and SmpA) and to Vibrio cholerae proteins encoded near a pathogenicity island (ORF144 protein, ORF101 protein, and SmpA). However, these homologous genes in E. coli and V. cholerae did not map adjacent to fur as in P. aeruginosa or P. fluorescens but were located downstream of recN, which is located downstream of fur in P. aeruginosa (Fig. 1A). The putative 16-kDa protein encoded by ORF1 was 44% identical to E. coli YfjG and 49% identical to the V. cholerae ORF144 protein. ORF2 encoded a putative 11-kDa protein which was 43% identical to E. coli YfjF and 41% identical to the V. cholerae ORF101 protein. However, the functions of the ORF1 and ORF2 proteins and of their homologs in E. coli and V. cholerae are unknown. The deduced amino acid sequence for OmlA was homologous to a group of proteins named SmpA, for “small protein A” (Fig. 1C), although OmlA was larger and extended beyond the carboxy termini of all the SmpA proteins. P. aeruginosa OmlA was 73% identical to P. fluorescens OmlA and had also substantial identity to SmpA from V. cholerae (40%), E. coli (39%), and Haemophilus influenzae (29%). An additional homolog of OmlA could be found in Alcaligenes eutrophus, and interestingly, this A. eutrophus gene was located directly upstream of fur, while none of the smpA genes map close to fur. However, the A. eutrophus homolog, which had not been annotated as a gene, may be a pseudogene, or, in the case of a sequencing error, at least needed one frameshift introduced in the reported DNA sequence to translate into a protein similar to OmlA.
FIG. 1.
(A) P. aeruginosa ORF1-ORF2-omlA-fur region. The transcripts are indicated by arrows above the corresponding genes, and an experimentally confirmed transcriptional terminator is shown between ORF2 and omlA. Also shown are the homologous regions of E. coli and V. cholerae, with the corresponding genes indicated by thick dashed arrows. (B) DNA sequence of the omlA gene. The corresponding amino acid sequence of OmlA is given below the DNA coding region. Also shown is the start of the divergent fur gene on the opposite DNA strand, with the amino-terminal protein sequence given above the 5′ fur coding sequence. Promoter elements for omlA and fur are indicated by brackets, dashed arrows represent transcriptional start sites for omlA, fur T1, and fur T2, and a transcriptional terminator is shown by head-to-head arrows. Also included are translational signals such as Shine-Dalgarno sequences (S/D) and the translational start sites for both omlA and fur. (C) Amino acid sequence alignment of OmlA from P. aeruginosa and P. fluorescens with SmpA from H. influenzae (GenBank no. 1175311), V. cholerae (GenBank no. U39068), and E. coli (GenBank no. D90888).
Expression of omlA and fur.
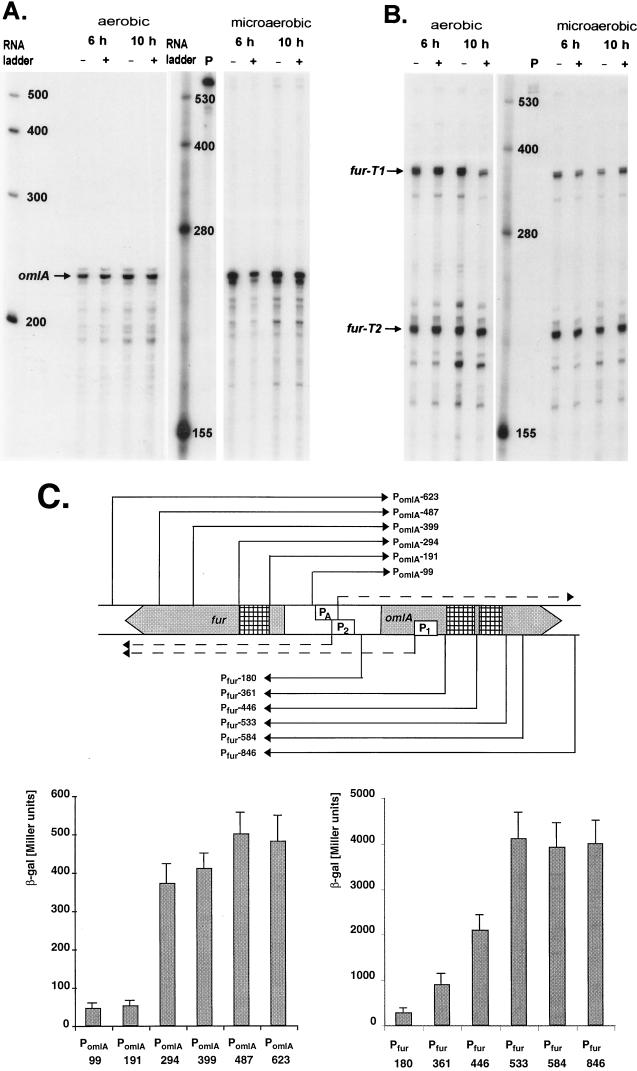
omlA gene expression was monitored at the transcriptional level by RNase protection using a riboprobe covering the omlA promoter. A single protected omlA mRNA fragment of 242 ± 3 nucleotides was detected (Fig. 2A), corresponding to a transcriptional start site at 36 ± 3 nucleotides upstream of the omlA start codon. Typical ς70-like −35 and −10 promoter elements were located within the expected distance relative to the mapped mRNA start, as indicated in Fig. 1B. OmlA expression appeared to be constitutive regarding growth phase, iron concentration, and oxygen tension (Fig. 2A), as well as pH and temperature (data not shown). The fur and omlA genes were divergently transcribed and had overlapping promoters. In P. aeruginosa, transcription of fur was driven by two separate promoters, P1 and P2, about 170 bp apart, resulting in two fur transcripts, T1 and T2 (Fig. 2B). Interestingly, the distal fur P1 promoter and thus the start site of the T1 transcript were located well within the omlA coding sequence (Fig. 1B). In fact, the divergent fur T1 and the omlA transcripts overlapped by 154 bases and had the potential to form antiparallel RNA-RNA hybrids that may affect fur or omlA translation. The proximal fur promoter P2 driving expression of the shorter fur transcript T2 overlapped the omlA promoter so that the −10 elements of fur P2 and of omlA shared the same base pair motif on the complementary DNA strands (Fig. 1B), suggesting a potential competition for RNA polymerase binding in this region.
FIG. 2.
(A) RNase protection analysis of omlA expression. A 444-nucleotide riboprobe spanning the region from 343 to −101 of the omlA sequence was hybridized to total RNA isolated from P. aeruginosa PAO1 grown for 6 or 10 h aerobically or microaerobically under low-iron (−) or high-iron (+) iron conditions. The protected omlA mRNA is indicated with an arrow. (B) RNase protection analysis of fur expression. The 444-nucleotide riboprobe covered from −101 to 343 of the omlA sequence, and the protected transcripts T1 and T2 are indicated by arrows. (C) Expression of omlA and fur. The map of the fur-omlA locus shows the omlA promoter PA, the fur promoters P1 and P2, the relevant transcripts (dashed arrows), and regions for cis activation (hatched boxes). Promoter fragments (e.g., PomlA, Pfur) of increasing size as indicated by arrows were translationally fused to the lacZ reporter gene. The β-galactosidase activities were measured in triplicate cultures grown for 4 h in D-TSB medium.
Expression of omlA and fur was also studied with translational fusions to the lacZ gene, using a series of omlA and fur promoter fragments containing increasing upstream DNA sequence as outlined in Fig. 2C. Optimal expression of omlA required the presence in cis of roughly 250 bp upstream of the −35 omlA promoter element. The expression of fur-lacZ was threefold higher when both P1 and P2 promoters were present than with P2 alone. Furthermore, fur-lacZ expression increased twofold and fourfold when roughly 100 and 200 bp of additional upstream DNA were included in the promoter fragment (Fig. 2C). In either case, the maximal gene expression did not require the presence of the complete upstream gene, excluding any trans-acting regulatory mechanism due to high levels of Fur or OmlA protein caused by multiple gene copies. Thus, it appeared rather that cis activation domains for optimal fur and omlA expression which were located within the coding sequence of the respective upstream gene existed (Fig. 2C).
Mutants affected in the omlA-fur intergenic region.
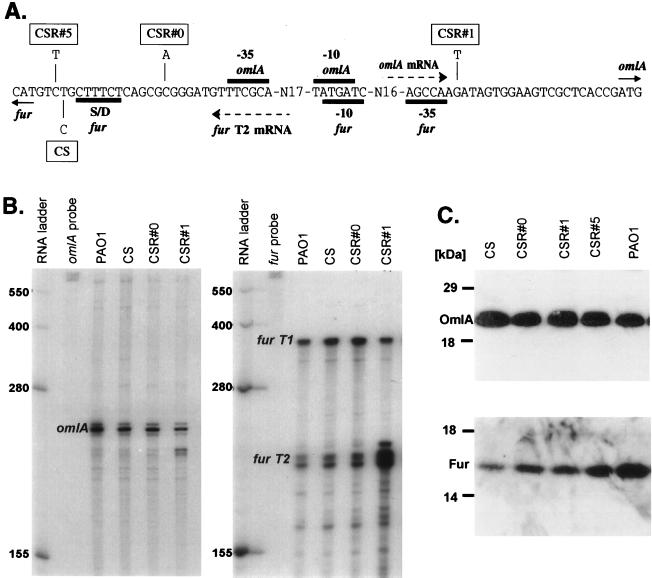
Since the omlA and fur genes were highly linked to each other, we further examined the possibility whether the expression of the two genes was interdependent or whether the OmlA protein was somehow involved in Fur function and/or modification. Although fur was found to be essential in P. aeruginosa, mutant strains producing an altered Fur were obtained by screening for manganese resistance (1). Also, a conditional, cold-sensitive Fur mutant (CS) which had a Fur− phenotype at 25°C, but not at 37°C (20), was isolated. CS was unstable and reverted spontaneously, allowing the isolation of several different revertants (CSR#0, CSR#1, and CSR#5) which appeared to be normally iron regulated and were no longer resistant to manganese. Genetic analysis of CS revealed a single base pair transition (T→C) 4 bp upstream of the fur start codon. The revertants had individual single base pair changes close to the original CS point mutation: CSR#0 had a C→A mutation 13 bp upstream, CSR#1 had a G→T point mutation 73 bp upstream, and CSR#5 had a C→T change 1 bp downstream of the original CS mutation site with respect to fur gene orientation (Fig. 3A). All these mutants were affected in the omlA-fur intergenic region but not in the coding regions, and the mutations had the potential to influence either transcription, mRNA stability, or translation of both genes. Analysis of omlA and fur mRNA revealed virtually identical omlA transcript levels in PAO1, CS, and all CSR strains (Fig. 3B, left panel). Fur transcripts T1 and T2 were detected at very similar levels in PAO1 wild-type, CS, and CSR#0; however, in CSR#1 fur T2 was up-regulated at least fivefold (Fig. 3B, right panel). Translation of omlA and fur was measured by translational fusions of CS and CSR omlA and fur to the lacZ gene (data not shown). The results were in good agreement with the direct determination of OmlA and Fur protein levels by Western blot analyses, indicating that the OmlA levels were identical (Fig. 3C, upper panel) and that the Fur levels were roughly 10-fold lower in CS, 4-fold lower in CSR#1, and 2-fold lower in CSR#1 and CSR#5 than in the PAO1 wild type (Fig. 3C, lower panel). Taken together, it appeared that the CS fur phenotype was caused by a translational rather than a transcriptional effect. The CS mutation between the fur start codon and the Shine-Dalgarno motif seemed to negatively affect translation of fur, resulting in a low level of Fur, which was insufficient to maintain proper iron-dependent control of Fur-regulated genes. In the revertants CSR#0 and CSR#5 the additional point mutations described above partially suppressed the translational deficiency, and the Fur levels, although still lower than in the wild type, were above the threshold concentration required for Fur-dependent gene regulation. In CSR#1, the suppressor mutation was located in the −35 region of fur P2 and had an up-regulating effect on fur T2, ultimately increasing the Fur levels. OmlA did not seem to be involved in any way in fur transcription or translation, and this finding was supported by the fact that the omlA gene on a plasmid (pOML15) did not complement the CS fur phenotype (data not shown).
FIG. 3.
(A) Locations of single base pair mutations in CS and in its revertants, CSR#0, CSR#1, and CSR#5. The relevant promoter elements, transcripts, and translational motifs within the omlA-fur intergenic region are indicated. S/D, Shine-Dalgarno site. (B) RNase protection of omlA (left) and fur (right) transcripts in PAO1, CS, CSR#0, and CSR#1. The RNA was isolated after 8 h of growth in high-iron D-TSB at 25°C. (C) Western blot analyses of OmlA (top) and Fur (bottom) proteins in CS, CSR#0, CSR#1, CSR#5, and wild-type PAO1. The cells were grown for 10 h in high-iron D-TSB at 25°C, and whole-cell extract samples were prepared and normalized for cell densities.
Characterization of OmlA as an outer membrane lipoprotein.
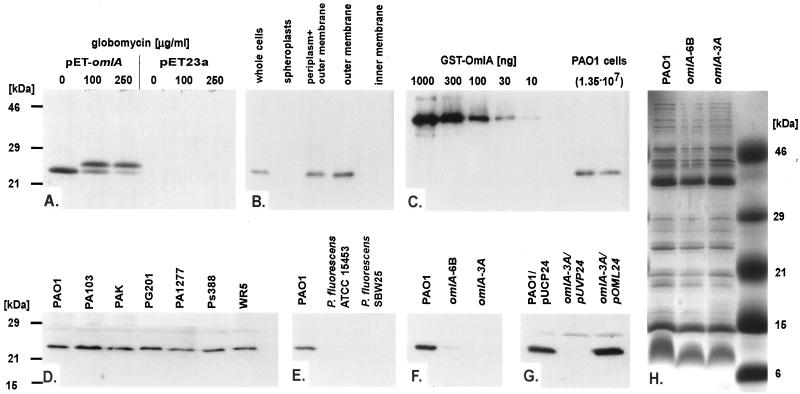
The omlA gene encoded an acidic protein (isoelectric point = 4.5) of 19.3 kDa which contained a 21-amino-acid-long hydrophobic signal sequence typical for bacterial lipoproteins, followed by a characteristic Cys residue at position 22, which could serve as the lipid attachment site. According to the common processing of prolipoproteins, the mature OmlA protein had an expected mass of roughly 18 kDa, which was the 17.3 kDa of the protein after cleavage of the signal sequence plus the masses of the modifying elements such as diacylglyceryl and the amino-terminal acyl group. To test whether OmlA was indeed a lipoprotein, inhibition of the lipoprotein maturation-specific signal peptidase II by globomycin was performed. Overproduction and selective labeling of OmlA in a T7 expression system in E. coli yielded a single radiolabeled protein migrating at roughly 24 kDa, and the addition of globomycin resulted in a slower-migrating form, presumably unmodified OmlA (Fig. 4A). Moreover, Western blot analysis of P. aeruginosa cell fractions clearly demonstrated that the OmlA protein was localized in the outer membrane (Fig. 4B).
FIG. 4.
(A) Autoradiography of overexpressed and radiolabeled OmlA. The omlA gene was expressed in a T7 system in E. coli/pET-omlA in the absence or presence of globomycin. Also shown is the pET23a vector control. (B through G) Western blot analysis of P. aeruginosa cell fractions, standardized GST-OmlA, and total protein from 1.35 × 107 PAO1 cells and of whole-cell extracts prepared from different P. aeruginosa wild-type strains, from P. fluorescens, from omlA mutants, and from a complemented omlA strain carrying pOML24. (H) Outer membrane protein profiles of wild-type PAO1 and omlA mutants on a Coomassie-stained 15% acrylamide SDS gel.
A rough estimate of the number of OmlA molecules per cell was obtained by comparing Western blot signal obtained from whole-cell extracts of a known number of bacteria to the signals of serially diluted purified GST-OmlA fusion protein as a standard (Fig. 4C). Similar signal intensities were obtained with 30 ng of the 43-kDa GST-OmlA fusion protein and with total proteins from 1.35 × 107 P. aeruginosa PAO1 cells, resulting in a calculated number of 31,000 OmlA molecules per cell.
The OmlA protein was found to be well conserved among clinical and environmental isolates of P. aeruginosa and was readily detectable in all strains tested, as shown for a few representative strains in Fig. 4D. Interestingly, the anti-OmlA serum did not cross-react with whole-cell extracts prepared from P. fluorescens (Fig. 4E). Southern blot analysis clearly demonstrated the presence of the omlA gene in P. fluorescens ATCC 15453 (data not shown), which we subsequently isolated (PF-omlA) to construct a lacZ fusion (pPZ-PF-PomlA). Expression of PF-omlA was detectable in P. aeruginosa PAO1, in P. fluorescens ATCC 15453, and in E. coli DH5α and was even somewhat higher than expression of P. aeruginosa omlA (data not shown).
OmlA is involved in maintaining the integrity of the cell envelope.
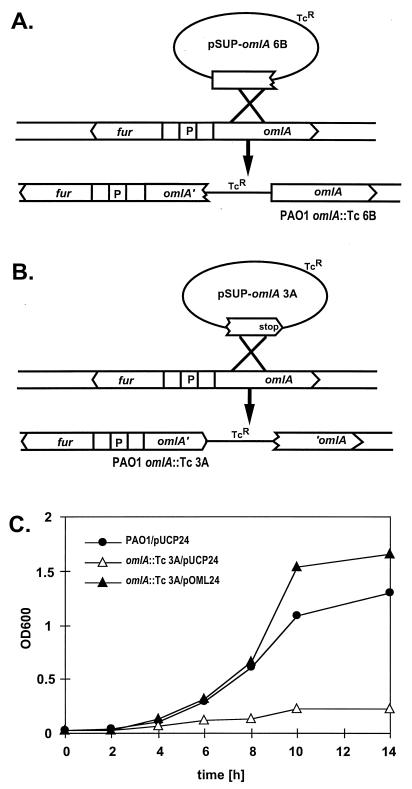
Mutants affected in the omlA gene were constructed in order to determine the function of the novel OmlA lipoprotein. Care had to be taken not to simultaneously alter the overlapping fur gene, i.e., not to disconnect the upstream fur P1 promoter and the activation elements which were located within the omlA coding sequence as demonstrated above. This was achieved by duplicating parts of the omlA gene and cointegration of a plasmid into the fur-omlA locus of PAO1 through a single crossover. The PAO1 omlA::Tc mutant strain 6B harbored the complete omlA coding sequence; however, the promoter and the ribosomal binding site were lacking (Fig. 5A). The PAO1 omlA::Tc mutant strain 3A possessed a truncated version of the omlA gene due to the introduction of an early stop codon (Fig. 5B), resulting in a hypothetical short OmlA protein that lacked 57 amino acids at the carboxy terminus. Western blot analysis of whole-cell extracts revealed extremely small amounts of OmlA in 6B, and 3A completely lacked any immunoreactive protein (Fig. 4F), suggesting that the truncated OmlA protein eventually produced in 3A was readily degraded. Complementation of 3A with the multicopy plasmid pOML24, harboring a functional omlA gene, restored the production of OmlA, as demonstrated by Western blot analysis (Fig. 4G). Outer membrane protein profiles from PAO1, 6B, and 3A looked virtually identical and did not discriminate the protein band corresponding to OmlA at 24 kDa on Coomassie-stained gels (Fig. 4H), although the identical samples indicated the complete loss of OmlA in 3A by Western blot analysis. Obviously, an unrelated outer membrane protein in that size range masked the OmlA protein band, or, alternatively, OmlA had disadvantageous staining properties.
FIG. 5.
(A) Construction of PAO1 omlA::Tc mutant 6B. The integration of the plasmid creates a promoterless omlA gene without affecting the divergent fur gene. (B) Construction of PAO1 omlA::Tc mutant 3A. The single crossover at omlA generates a truncated omlA gene due to an early stop codon. (C) Growth inhibition of 3A in M9 medium containing 0.1% SDS. Growth curves are shown for PAO1/pUCP24 (wild type with a control plasmid), 3A/pUCP24 (omlA mutant with a control plasmid), and 3A/pOML24 (omlA mutant genetically complemented with the omlA gene on a multicopy plasmid).
The omlA mutant strains were tested for their susceptibility to various detergents, antibiotics, and organic solvents. Mutant 6B was more susceptible and mutant 3A was hypersusceptible to anionic detergents, such as SDS and deoxycholate, but not to cationic or nonionic surface-active compounds, such as cetyltrimethylammonium bromide (CTAB), polymyxin B, Triton X-100, and Tween-20 (Table 2). Also, mutant 3A showed increased susceptibility to some antibiotics, including nalidixic acid, rifampin, novobiocin, and chloramphenicol, but not to gentamicin, cycloserine, or polymyxin B (Table 2). P. aeruginosa PAO1 and 3A had virtually identical growth rates and cell yields in either LB or M9 medium. Extremes in osmolarity or temperature and the presence of organic solvents such as xylene or toluene did not affect the growth characteristics of the omlA mutants in liquid M9 medium compared to the PAO1 wild type (data not shown). The most pronounced growth inhibition through cell lysis of 3A was found in liquid M9 medium containing SDS at concentrations of 0.05 to 0.2%. Growth and sensitivity to SDS were complemented with plasmid pOML24 supplying the omlA gene in trans; in fact, the complemented omlA::Tc strain 3A/pOML24 exhibited a higher resistance to SDS than wild-type PAO1 during stationary phase of growth, presumably due to a high-copy-number effect of omlA on pOML24 (Fig. 5C). The susceptibility of the omlA mutant to ionic detergents suggested a role of the OmlA protein in building the cell wall structure and, being an outer membrane protein, maintaining the integrity of the cell envelope. By using the pPZ-PomlA-399 omlA-lacZ fusion in PAO1, the various compounds and conditions mentioned above were also tested for their capacity to induce up-regulation of omlA expression; however, such a specific stress response could not be detected (data not shown).
TABLE 2.
Susceptibilities of wild-type PAO1 and of omlA mutants to various detergents and antibiotics
| Compounda | Zone of growth inhibition (mm)b
|
||
|---|---|---|---|
| PAO1 | 6B omlA::Tc | 3A omlA::Tc | |
| 20% SDS | 0 | —c | 13 |
| 10% Deoxycholate | 0 | 10 | 13 |
| 5% CTAB | 11 | 11 | 11 |
| 10% Triton X-100 | 14 | 14 | 14 |
| 10% Tween-20 | 0 | 0 | 0 |
| 0.5 M EDTA | 15 | 15 | 15 |
| 2% Nalidixic acid | 27 | 29 | 30 |
| 1% Rifampin | 18 | 23 | 26 |
| 2% Novobiocin | 19 | 21 | 24 |
| 2% Chloramphenicol | 28 | 30 | 33 |
| 2% Cycloserine | 10 | 10 | 10 |
| 1% Polymyxin B | 22 | 22 | 22 |
| 1% Gentamicin | 30 | 31 | 30 |
The compounds were spotted (15 μl) on a sterile filter disk placed on M9 top agarose containing 106 cells.
The zones were measured after 16 h at 37°C and are the averages for triplicate plates.
Partial growth inhibition occurred in a zone of 12 mm.
DISCUSSION
The omlA gene encoding a novel outer membrane lipoprotein has been identified, isolated, and partially characterized. It was located directly upstream of fur and was divergently transcribed from fur. The spacing of the two genes was extremely tight; in fact, the omlA and fur P2 promoters had overlapping −10 RNA polymerase binding sites on the opposite DNA strands, and the corresponding mRNA start sites were only 20 bp apart. Moreover, the upstream fur P1 promoter and thus the start site of the longer fur T1 transcript were located within the omlA coding sequence. As a consequence of this astonishing finding, the omlA transcript and the fur T1 transcript had an antisense overlap of at least 150 ribonucleotides. The tight spacing of two divergent promoters is a common feature in many bacteria and often occurs when two divergently transcribed genes are coregulated. Well-studied examples include the divergent xylR and xylS genes on the TOL plasmid of Pseudomonas putida, with a 300-bp intergenic region carrying two tandem promoters which are coregulated by XylR and IHF (25), and the divergent tetR and tetA genes of Tn10, which have transcriptional start sites separated by only 36 bp and are subject to coregulation by the Tet repressor (4). A highly unusual feature in bacteria, and, to our best knowledge, the first such example in P. aeruginosa, was our finding of truly overlapping mRNAs as in the case of omlA and fur T1. The transcriptional start site of fur T1 had been mapped clearly within the omlA coding region. Further strong evidence to support the existence of the fur P1 promoter was obtained by engineering a fur mutant strain by cointegration of a plasmid carrying the fur P2 promoter through single crossover into the omlA-fur intergenic region. In the resulting mutant the distal fur P1 promoter was disconnected and left the fur gene under control of P2 alone, without affecting the omlA gene at all. Interestingly, this mutant produced very small amounts of Fur and exhibited a Fur− phenotype (20), which was most likely caused by the loss of the major fur T1 mRNA. The fact that both omlA and fur, although strongly intertwined, were functional and simultaneously expressed genes raised several interesting questions regarding the bacterial transcription and translation machinery and the control thereof. The mechanism of regulation and competition for RNA polymerase binding of two overlapping divergent promoters is poorly understood. A recent study of the E. coli fepA-fes ferrienterochelin uptake genes which have overlapping promoters similar to omlA and fur P2 suggested that simultaneous binding of the RNA polymerase to both promoters can occur (10). Furthermore, it had to be considered that the 5′ regions of the overlapping omlA and fur T1 mRNAs had the potential to form RNA-RNA hybrids. Generally, such duplex formation may have an impact on message stability; specifically, the translational signals, including the ribosome binding site and the start codon on the omlA mRNA, may be masked by the overlapping fur T1 mRNA, which could result in a down-regulation of omlA translation. Such a mechanism involving RNA-RNA hybrids had been postulated to control plasmid replication (54). The production of RepA, a protein required for plasmid replication, is regulated at the posttranscriptional level by the short RNA I encoded by the cop gene (29). RNA I and the 5′ end of repA mRNA are complementary and can form a stable complex in vitro, thereby controlling RepA translation (48, 49). Alternatively, transcription and translation of omlA and fur may be strongly coupled so that any newly synthesized mRNA is quickly and repeatedly bound by ribosomes, thereby impairing the hybridization of the complementary RNAs. Following the latter scenario, the tight overlapping spacing of omlA and fur can conserve some space, in analogy to the organization of some viral genomes. It is intriguing to argue that the burying of an advantageous housekeeping gene such as omlA into an essential locus such as fur would help to conserve the omlA gene in the long term.
The cellular concentrations of both Fur and OmlA protein remained virtually constant over the entire growth phase and did not respond to changes in temperature, iron concentration, or oxygen tension. However, our fur and omlA expression data obtained with series of lacZ fusions strongly suggested the presence of upstream activation sites for both genes. The locations of these potential binding sites for a transcriptional activator were mapped within roughly 100 bp; however, further experiments are required to refine these upstream activation sites more accurately and to identify the transcriptional activators.
OmlA was demonstrated to be a lipoprotein by the inhibition of processing by globomycin. It was localized exclusively in the outer membrane according to Western blot analysis of different cell fractions. The cellular distribution of OmlA was in agreement with the nature of the OmlA signal sequence, since the amino acid residue following the prospective amino-terminal cysteine of the mature OmlA protein was a serine. It has been shown in E. coli that the second amino acid residue of a lipoprotein plays a crucial role in determining its final location in the cell envelope and is therefore called the sorting signal. An aspartate residue in that position results in a cytoplasmic membrane localization, whereas other residues result in an outer membrane localization (57).
OmlA exhibited a high degree of identity to so-called SmpA (small protein A) of a few other gram-negative bacteria; however, the biological function of the SmpA homologs is unknown. Amino acid sequence analysis and motif searches revealed that SmpA proteins also harbor a lipoprotein-like signal sequence. The characterization of the highly homologous OmlA suggested that these proteins may comprise a novel family of outer membrane lipoproteins. However, the SmpA homologs were considerably smaller than OmlA because they lacked the carboxy-terminal domain of OmlA. This domain contained a helix-turn-helix motif (amino acid residues 113 to 139 of the mature OmlA protein) and the proline-rich motif PVPVPTPEPLDPSPQ (amino acid residues 141 to 155). Proline-rich proteins have been associated with aberrant migration in denaturing gels. This may explain why OmlA expressed in either E. coli or P. aeruginosa migrated at roughly 24 kDa, although its predicted size was only 18 kDa. A high proportion of prolines typically occurs immediately adjacent to α-helices, and this applied also to the OmlA protein, where the PVPVPTPEPLDPSPQ motif was located directly carboxy terminal of the helix-turn-helix. Repetitive short proline-rich sequences have been shown to play key roles in the function of E. coli TonB and OmpA (for a review, see reference 53). In the major outer membrane protein OmpA, which mediates F-dependent conjugation and is required for the structural integrity of the outer membrane in E. coli, the motif (AP)4 has been proposed to act as a hinge region. In TonB, which is a key protein involved in the transport of small molecules such as iron siderophores through the cell membranes, the motif (EP)5X13(KP)5 was shown to span the periplasmic space, with the (XP)n sequences acting as molecular triggers required for signal transduction across the membranes. The omlA::Tc mutant 3A, which produced a truncated OmlA protein lacking both the helix-turn-helix motif and the proline-rich motif, was clearly affected in its cell wall stability, suggesting that these motifs may be essential for OmlA function. Further investigations will have to focus on the topology of OmlA relative to the outer membrane. Like most outer membrane proteins, OmlA is polar overall, and hydrophobic domains are absent. Clearly, candidate motifs for the interaction of OmlA with the outer membrane are its amino terminal lipid and diacylglyceryl lipoprotein modifications and the carboxy-terminal proline-rich motif.
The precise function of OmlA remains to be elucidated. OmlA could be excluded as a porin, since the omlA mutants were hypersusceptible to some antibiotics whereas porin mutations are a frequent cause of high-level resistance to certain antibiotics (6, 13, 26). Similarly, OmlA was not part of a drug efflux pump because the omlA gene was not in a cluster with genes encoding the other efflux components typically found in all P. aeruginosa systems known so far, including the mexAB-oprM or mexCD-oprJ multidrug resistance operons (34). Furthermore, the antibiotics to which the omlA mutants were more susceptible were structurally and functionally unrelated. They belonged to different substance classes, such as quinolones, rifampins, and nitrophenyl- and dichloroacetylated compounds, and had different modes of action, such as inhibition of gyrase, transcription, and protein synthesis. Therefore, it was unlikely that the observed hypersusceptibility to these different antibiotics was due to a specific role of OmlA in binding or transport of these compounds. More plausible was a role of OmlA in maintaining the cell wall architecture, and the increased susceptibility to certain antibiotics was an indirect effect due to leakage of the cell envelope. In good agreement with this was the finding that omlA mutants were hypersusceptible to anionic detergents. A similar phenotype has been associated with P. putida mutants affected in oprL, which encodes the peptidoglycan-associated lipoprotein (38). Further experiments will investigate whether OmlA is associated with cell wall components such as peptidoglycan or lipopolysaccharide and whether omlA is somehow involved in the formation of outer membrane vesicles and/or release of periplasmic proteins into the extracellular milieu, similar to the E. coli tol-pal system (3). A location of OmlA at the exposed outer membrane would make it an ideal target for novel antimicrobial compounds.
ACKNOWLEDGMENTS
We thank M. Inouye for the generous gift of globomycin. The E. coli fur null mutant QC1732 was kindly provided by M. McIntosh (University of Missouri).
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI15940) to Michael L. Vasil.
REFERENCES
- 1.Barton H A, Johnson Z, Cox C D, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 2.Bell A, Hancock R E. Outer membrane protein H1 of Pseudomonas aeruginosa: purification of the protein and cloning and nucleotide sequence of the gene. J Bacteriol. 1989;171:3211–3217. doi: 10.1128/jb.171.6.3211-3217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernadac A, Gavioli M, Lazzaroni J C, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand K P, Postle K, Wray L V, Jr, Reznikoff W S. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene. 1983;23:149–156. doi: 10.1016/0378-1119(83)90046-x. [DOI] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Daikos G L, Lolans V T, Jackson G G. Alterations in outer membrane proteins of Pseudomonas aeruginosa associated with selective resistance to quinolones. Antimicrob Agents Chemother. 1988;32:785–787. doi: 10.1128/aac.32.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 8.De Vos D, Lim A, Jr, Pirnay J P, Struelens M, Vandenvelde C, Duinslaeger L, Vanderkelen A, Cornelis P. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol. 1997;35:1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duchene M, Schweizer A, Lottspeich F, Krauss G, Marget M, Vogel K, von Specht B U, Domdey H. Sequence and transcriptional start site of the Pseudomonas aeruginosa outer membrane porin protein F gene. J Bacteriol. 1988;170:155–162. doi: 10.1128/jb.170.1.155-162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escolar L, Perez-Martin J, de Lorenzo V. Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J Bacteriol. 1998;180:2579–2582. doi: 10.1128/jb.180.9.2579-2582.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forster A, Buluwela L, Rabbitts T H. Turbo-screening of bacterial colonies using microwave denaturation on paper filters. Trends Genet. 1990;6:141. doi: 10.1016/0168-9525(90)90135-s. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1990;34:1757–1761. doi: 10.1128/aac.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabelsberger J, Knapp B, Bauersachs S, Enz U I, von Specht B U, Domdey H. A hybrid outer membrane protein antigen for vaccination against Pseudomonas aeruginosa. Behring Inst Mitt. 1997;98:302–314. [PubMed] [Google Scholar]
- 15.Guerra-Santos L H, Kapelli O, Fiechter A. Dependence of Pseudomonas aeruginosa continuous culture biosurfactant production on nutritional and environmental factors. Appl Microbiol Biotechnol. 1986;24:443–448. [Google Scholar]
- 16.Hancock R E, Siehnel R, Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol. 1990;4:1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 17.Hancock R E, Wong R. Potential of protein OprF of Pseudomonas in bivalent vaccines. Behring Inst Mitt. 1997;98:283–290. [PubMed] [Google Scholar]
- 18.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Siehnel R J, Bellido F, Rawling E, Hancock R E. Analysis of two gene regions involved in the expression of the imipenem-specific, outer membrane porin protein OprD of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1992;76:267–273. doi: 10.1016/0378-1097(92)90347-q. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, Z., and M. L. Vasil. Unpublished data.
- 21.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 22.Lim A, Jr, De Vos D, Brauns M, Mossialos D, Gaballa A, Qing D, Cornelis P. Molecular and immunological characterization of OprL, the 18 kDa outer-membrane peptidoglycan-associated lipoprotein (PAL) of Pseudomonas aeruginosa. Microbiology. 1997;143:1709–1716. doi: 10.1099/00221287-143-5-1709. [DOI] [PubMed] [Google Scholar]
- 23.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P V. The roles of various fractions of Pseudomonas aeruginosa on its pathogenesis. II. Effects of lecithinase and protease. J Infect Dis. 1966;116:112–116. doi: 10.1093/infdis/116.1.112. [DOI] [PubMed] [Google Scholar]
- 25.Marques S, Gallegos M T, Manzanera M, Holtel A, Timmis K N, Ramos J L. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol. 1998;180:2889–2894. doi: 10.1128/jb.180.11.2889-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuno T, Kageyama M. Separation and characterization of the outer membrane of Pseudomonas aeruginosa. J Biochem (Tokyo) 1978;84:179–191. doi: 10.1093/oxfordjournals.jbchem.a132106. [DOI] [PubMed] [Google Scholar]
- 28.Nicas T I, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 29.Nordstrom K, Wagner E G, Persson C, Blomberg P, Ohman M. Translational control by antisense RNA in control of plasmid replication. Gene. 1988;72:237–240. doi: 10.1016/0378-1119(88)90148-5. [DOI] [PubMed] [Google Scholar]
- 30.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochsner U A, Vasil A I, Vasil M L. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol. 1995;177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Payne S M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993;1:66–69. doi: 10.1016/0966-842x(93)90036-q. [DOI] [PubMed] [Google Scholar]
- 34.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 35.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prince R W, Cox C D, Vasil M L. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol. 1993;175:2589–2598. doi: 10.1128/jb.175.9.2589-2598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rainey P B, Bailey M J. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol Microbiol. 1996;19:521–533. doi: 10.1046/j.1365-2958.1996.391926.x. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Herva J J, Ramos-Gonzalez M I, Ramos J L. The Pseudomonas putida peptidoglycan-associated outer membrane lipoprotein is involved in maintenance of the integrity of the cell cell envelope. J Bacteriol. 1996;178:1699–1706. doi: 10.1128/jb.178.6.1699-1706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saint-Onge A, Romeyer F, Lebel P, Masson L, Brousseau R. Specificity of the Pseudomonas aeruginosa PAO1 lipoprotein I gene as a DNA probe and PCR target region within the Pseudomonadaceae. J Gen Microbiol. 1992;138:733–741. doi: 10.1099/00221287-138-4-733. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sankaran K, Wu H C. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 43.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 44.Schweizer H P. Improved broad-host-range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene. 1991;103:87–92. doi: 10.1016/0378-1119(91)90396-s. [DOI] [PubMed] [Google Scholar]
- 45.Siehnel R J, Egli C, Hancock R E. Polyphosphate-selective porin OprO of Pseudomonas aeruginosa: expression, purification and sequence. Mol Microbiol. 1992;6:2319–2326. doi: 10.1111/j.1365-2958.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 46.Simon R, Priefer U, Puhler A. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 47.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–507. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama T, Itoh T. Control of ColE2 DNA replication: in vitro binding of the antisense RNA to the Rep mRNA. Nucleic Acids Res. 1993;21:5972–5977. doi: 10.1093/nar/21.25.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takechi S, Yasueda H, Itoh T. Control of ColE2 plasmid replication: regulation of Rep expression by a plasmid-coded antisense RNA. Mol Gen Genet. 1994;244:49–56. doi: 10.1007/BF00280186. [DOI] [PubMed] [Google Scholar]
- 50.Tokunaga M, Tokunaga H, Wu H C. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci USA. 1982;79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vasil M L, Chamberlain C, Grant C C. Molecular studies of Pseudomonas exotoxin A gene. Infect Immun. 1986;52:538–548. doi: 10.1128/iai.52.2.538-548.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Specht B U, Domdey H, Schodel F, Blum B, Lucking C, Knapp B, Muth G, Hungerer K D, Broker M. Outer membrane proteins of Pseudomonas aeruginosa as vaccine candidates. Behring Inst Mitt. 1994;95:85–96. [PubMed] [Google Scholar]
- 53.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Womble D D, Dong X, Wu R P, Luckow V A, Martinez A F, Rownd R H. IncFII plasmid incompatibility product and its target are both RNA transcripts. J Bacteriol. 1984;160:28–35. doi: 10.1128/jb.160.1.28-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodruff W A, Hancock R E. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J Bacteriol. 1989;171:3304–3309. doi: 10.1128/jb.171.6.3304-3309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods D E, Vasil M L. Pathogenesis of Pseudomonas aeruginosa infections. In: Baltch A L, Smith R P, editors. Pseudomonas aeruginosa infections and treatment. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 21–50. [Google Scholar]
- 57.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 58.Yamano Y, Nishikawa T, Komatsu Y. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1. Mol Microbiol. 1993;8:993–1004. doi: 10.1111/j.1365-2958.1993.tb01643.x. [DOI] [PubMed] [Google Scholar]
- 59.Yoneyama H, Nakae T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology. 1996;142:2137–2144. doi: 10.1099/13500872-142-8-2137. [DOI] [PubMed] [Google Scholar]