Abstract
Alveolar macrophages (AMs) are functionally important innate cells involved in lung homeostasis and immunity and whose diversity in health and disease is a subject of intense investigations. Yet, it remains unclear to what extent conditions like smoking or chronic obstructive pulmonary disease (COPD) trigger changes in the AM compartment. Here, we aimed to explore heterogeneity of human AMs isolated from healthy nonsmokers, smokers without COPD, and smokers with COPD by analyzing BAL fluid cells by flow cytometry and bulk and single-cell RNA sequencing. We found that subpopulations of BAL fluid CD206+ macrophages could be distinguished based on their degree of autofluorescence in each subject analyzed. CD206+ autofluorescenthigh AMs were identified as classical, self-proliferative AM, whereas autofluorescentlow AMs were expressing both monocyte and classical AM-related genes, supportive of a monocytic origin. Of note, monocyte-derived autofluorescentlow AMs exhibited a functionally distinct immunoregulatory profile, including the ability to secrete the immunosuppressive cytokine IL-10. Interestingly, single-cell RNA-sequencing analyses showed that transcriptionally distinct clusters of classical and monocyte-derived AM were uniquely enriched in smokers with and without COPD as compared with healthy nonsmokers. Of note, such smoking-associated clusters exhibited gene signatures enriched in detoxification, oxidative stress, and proinflammatory responses. Our study independently confirms previous reports supporting that monocyte-derived macrophages coexist with classical AM in the airways of healthy subjects and patients with COPD and identifies smoking-associated changes in the AM compartment that may favor COPD initiation or progression.
Keywords: lung, airway macrophages, smoking, single-cell and bulk RNA-seq, COPD
Mammalian lungs are at the interface between the host and the environment and are continuously exposed to noxious substances, microorganisms, and foreign antigens, and a tightly regulated cellular network of immune cells is required to sustain respiratory function while coping with insults the lung is subjected to. Among lung immune cells, alveolar macrophages (AMs) populate the airways and are therefore particularly exposed (1, 2). Like for other tissue macrophages, the functional identity of AMs is thought to be determined by the niche of residence, their origin, and the nature and extent of inflammation-related signals (3). Functionally, AMs act as a first line of defense against external threats and exert tissue-supportive functions such as surfactant homeostasis (1). Moreover, they can contribute to the regulation of various infectious or noninfectious disorders in an insulted lung (1, 4). The extent to which such versatility is driven by ontogeny, the local niche or stress signals remains a subject of intense investigations (5). Indications about the origin of AMs are mainly derived from laboratory mouse studies supporting that AMs are seeded early in life from embryonic progenitors and can self-maintain with a minimal contribution of monocytes under homeostatic conditions (6–9), whereas monocytes can contribute to the AM pool in conditions in which homeostasis is broken (10–12). Several studies indicate that human monocytes can indeed give rise to functionally distinct AMs in various disease contexts (10, 13–15). Recently, evidence emerged that monocyte-derived cells also populate airways of healthy adults (16), thereby providing further insights into the diversity of human BAL fluid (BALF) cells.
Chronic obstructive pulmonary disease (COPD) is a progressive inflammatory lung disease mainly triggered by inhalation of toxic substances such as cigarette smoke or pollution and characterized by irreversible airway obstruction and persistent inflammatory responses in the lung (17). COPD is the third leading cause of death worldwide and represents a huge socioeconomic burden (18, 19), emphasizing the need to better understand disease pathogenesis and elaborate novel therapies.
Here, we aimed to evaluate human AM diversity in healthy nonsmokers, smokers without COPD, and smokers with COPD by analyzing BALF cells by flow cytometry, bulk and single-cell (sc) RNA-sequencing (RNA-seq). Our data provide experimental evidence that monocytes can contribute to the pool of AMs over the lifespan in humans, and that smoking is associated with yet-unidentified distinct clusters of AMs, both in healthy subjects and patients with COPD, which exhibit profiles consistent with a potential contribution to inflammatory events associated with COPD.
Methods
Study Design
We recruited three smokers without COPD, three healthy nonsmokers, and three patients with COPD more than 40 years old between March 2017 and October 2019 (see Table 1 and the data supplement). BALFs and blood samples were collected at the pneumology department of the University Hospital (CHU Liège, Belgium) for bulk RNA-seq and scRNA-seq experiments. Our study received approval from the local ethical committee, and all patients signed an informed consent.
Table 1.
Demographic and Clinical Characteristics of Human Patients from Whom Originate the BALF Cells Analyzed by Bulk and Single-Cell RNA Sequencing
| HD1 | HD2 | HD3 | HD4 | HD5 | HD6 | HD7 | HD8 | HD9 | |
|---|---|---|---|---|---|---|---|---|---|
| Status | Healthy nonsmoker |
Smoker without COPD |
Smoker with COPD |
||||||
| Age | 60 | 56 | 56 | 56 | 56 | 40 | 63 | 60 | 65 |
| BMI | 28.95 | 26.2 | 25.7 | 20.83 | 24.1 | 30.1 | 20.34 | 23.5 | 25.95 |
| Males/Female | F | M | M | F | M | M | M | M | M |
| Ethnicity | Caucasian |
||||||||
| Smoker status | Nonsmoker |
Current smoker |
|||||||
| Pack-years | 0 | 0 | 0 | 13 | 41 | 24 | 40 | 35 | 55 |
| FEV, % predicted | 123 | 98 | 108 | 106 | 94 | 96 | 45 | 63 | 81 |
| FEV/FVC, % | 89.9 | 74 | 78 | 88.7 | 80 | 84 | 47 | 59 | 64 |
| Leucocytes counts, 106 c/L | 53.1 | 46.22 | 41.7 | 91.4 | 564 | 46.2 | 61.7 | 100 | 206 |
| GOLD COPD stage | NA | NA | NA | NA | NA | NA | III | II | II |
| Corticoids, yes/no | NA | NA | NA | NA | NA | NA | No | Yes | Yes |
Definition of abbreviations: BMI = body mass index; COPD = chronic obstructive pulmonary disease; FEV = forced expiratory volume; FVC = forced vital capacity; GOLD = Global Initiative for COPD; HD = human donor; NA = not applicable.
BALF Processing
BALFs were processed directly after collection. The first collection tube, typically containing large amounts of mucus and epithelial cells, was discarded. The following fractions were pooled together. The sample was centrifuged at 1,400 rpm for 10 minutes. The cell pellet was suspended and filtered in a PBS solution with 10 mM EDTA.
Blood Monocyte Isolation
Monocytes were isolated directly from blood with the EasySep Direct Human Monocyte Isolation Kit (Stemcell). To eliminate debris and increase purity, the cells were then FACS sorted directly in Trizol reagent on the basis of their CD14 expression.
Flow Cytometry
For phenotyping and sorting, cells were stained during 30 minutes with antibodies and Fc block (BD Biosciences) to avoid nonspecific binding. Subpopulations of macrophages were FACS-sorted on the basis of their expression of CD45 and CD206 and their autofluorescence (visualized in phycoerythrin [PE] channel). The macrophages were FACS-sorted directly in Trizol reagent (Life Technologies) for bulk RNA-seq or in PBS for cytospin and cell culture. The majority of the experiments have been performed on a FACSAria III (BD Biosciences), except for the CCR2 staining that has been performed on a FACS Sony MA900. References for all antibodies can be found in Table E1 in the data supplement.
Bulk and Single-Cell RNA-Seq and Analyses
Methods related to bulk and single-cell RNA-seq and analyses can be found in the data supplement.
Data and Codes Availability
Resource data are available in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE183982, and raw data were deposed in the Sequence Read Archive database under accession numbers SRP336731 and SRP336735. Additional details can be found in the data supplement.
Cytologic Examination and Culture of FACS-sorted Macrophages
Detailed methods can be found in the data supplement.
Statistical Analysis
Statistical analyses were performed using Prism 7 (GraphPad Software), except for bulk and scRNA-seq data, for which the R packages “DESeq2” or “Seurat” were used, respectively. For transcriptional data, the adjusted P values are shown. We performed two-tailed paired Student’s t tests and two-way ANOVA, as mentioned in the respective figure legends. We considered a P value lower than 0.05 as significant. *P < 0.05, **P < 0.01, and ***P < 0.001; ns, not significant.
Results
A Population of Autofluorescentlow Small Macrophages Is Present in Human BALF
First, we sought to analyze human BALF cells by flow cytometry. BALFs were obtained from routine complementary exams performed on patients suffering from diverse lung diseases at the pneumology department of the University Hospital (ULiège, Belgium) (Table E2). We used a fluorescent-conjugated antibody directed against the CD206 (C-type lectin mannose receptor) to identify AMs by flow cytometry, defined as singlet CD45+CD206+ cells (Figure 1A) (20, 21). Although we tested additional antibodies directed against CD11B, CD11C, HLA-DR, CD169, or CD163, macrophage-specific expression of those markers was homogeneous (Figure E1), supporting no evidence for phenotypical heterogeneity. Nevertheless, by looking at auto-fluorescence, a hallmark of resident AMs (20, 21), we consistently found distinct amounts of autofluorescence on CD206+ macrophages (Figure 1A). Autofluorescenthigh (AFhi) macrophages represented the predominant population of AMs and were forward scatter (FSC)/side scatter (SSC)hi, as opposed to autofluorescentlow (AFlo) AMs, which were FSC/SSClo (Figure 1B). Cytologic examination of FACS-sorted AFlo and AFhi macrophages showed that both AM subpopulations contained vacuoles in their cytoplasm, but AFhi AMs displayed a larger size than AFlo AMs (Figures 1C and 1D), consistent with the idea that AFhi AMs corresponded to resident “classical” AM, whereas AFlo AM represented a less characterized population.
Figure 1.
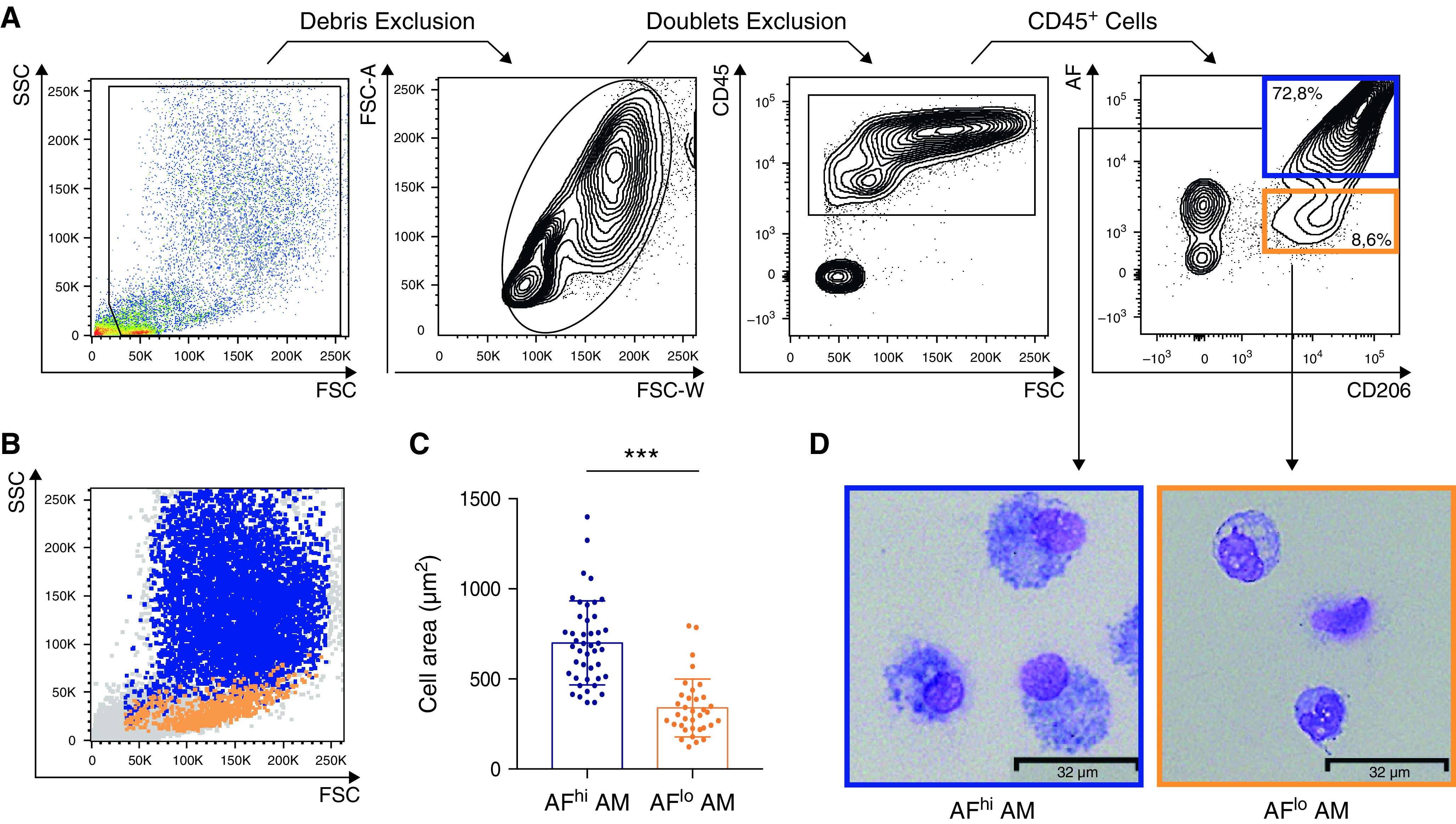
Alveolar macrophages (AMs) collected by BAL encompass small autofluorescentlow (AFlo) and large autofluorescenthigh (AFhi) CD206+ macrophages. (A) Representative flow cytometry gating strategy to delineate BAL fluid (BALF) CD206+ AFlo (orange box) and AFhi (blue box) macrophages. (B) Representative side scatter (SSC) versus forward scatter (FSC) plot depicting AFlo (orange) and AFhi (blue) macrophages. (C) Quantification of the size of AFlo and AFhi AMs. (D) Representative photographs of FACS-sorted AFlo and AFhi AMs. (A, B, and D) Data are representative of 1 of more than 10 experiments, each showing similar results. (C) Data show mean + SEM, as well as individual cells (n = 10 donors, 2–8 cells per donor). P values were calculated using a two-tailed paired Student’s t test. Scale bars = 32 μm.
BALF AFlo Macrophages Share Expression of Both Monocyte- and AM-associated Genes
We were further interested in investigating the identity of AFlo and AFhi AMs in distinct categories of patients using high-throughput technologies. To this end, we recruited three healthy nonsmokers, three smokers without COPD, and three smokers with COPD (i.e., stages II or III according to the Global Initiative for Chronic Obstructive Lung Disease COPD stage [22]), as presented in Table 1. BALF cells were collected and divided into two fractions that were used for bulk RNA-seq and scRNA-seq (Figure 2A).
Figure 2.
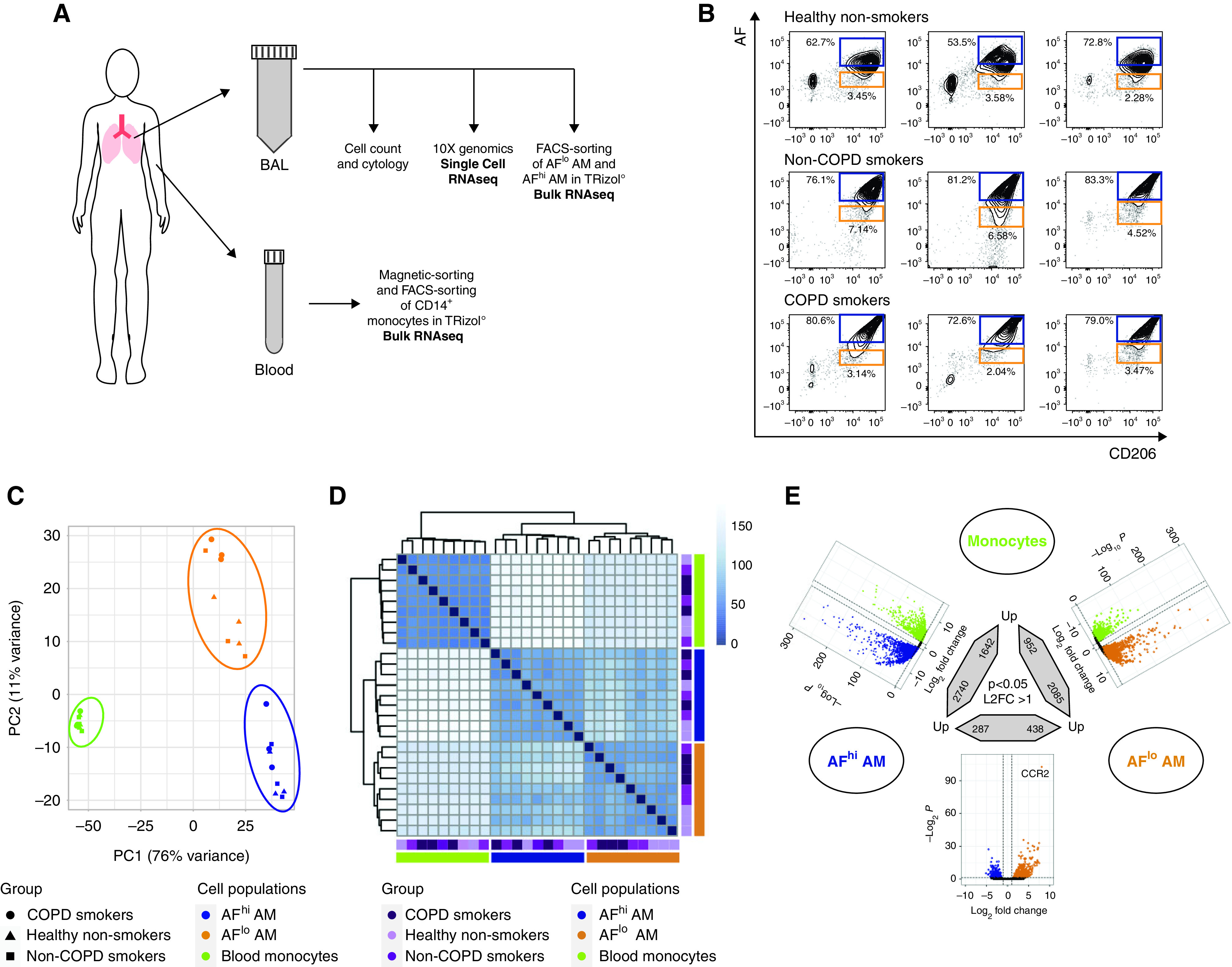
Bulk RNA-sequencing (RNAseq) of blood monocytes and AFlo and AFhi AMs reveals conserved cell type–specific signatures that are independent of the health status. (A) Experimental pipeline for bulk and single-cell RNAseq. (B) Flow cytometry plots depicting AF versus CD206 expression of BALF singlet CD45+ cells collected from the healthy nonsmokers (n = 3), smokers without chronic obstructive pulmonary disease (COPD) (n = 3) and smokers with COPD (n = 3) included in this study. The orange and blue boxes indicate the gating used for FACS sorting of AFlo and AFhi AMs before bulk RNAseq. (C) Two-dimensional principal component (PC) analysis comparing blood monocytes, BALF AFlo and AFhi AMs in the indicated groups of individuals. Percentages indicate the variability explained by each compartment. (D) Unsupervised hierarchical clustering of the biological samples analyzed. The color scale reflects the clustering distance (0 = highest correlation; 150 = the lowest correlation) between the transcriptomic profiles of each biological sample. (E) Summary of differentially expressed (DE) genes (P < 0.05; fold change > 2) in the indicated pairwise comparisons showing significant DE genes in color in the volcano plots and the total numbers of upregulated genes in the bidirectional arrows.
For bulk RNA-seq, CD14+ blood monocytes and FACS-sorted AFlo and AFhi CD45+CD206+ BALF cells were included (Figures 2A and 2B). A total of 27 samples were collected and analyzed. Principal component (PC) analysis revealed a cell population-based clustering, with AFlo AMs being more closely related to AFhi AMs than blood monocyte on PC1 (Figure 2C). Of note, the three isolated cell populations analyzed were largely similar among healthy nonsmokers, smokers without COPD, and smokers with COPD (Figure 2C).
Unsupervised hierarchical clustering further confirmed a cell type–based clustering that was independent of the health status (Figure 2D). Using a log2 fold-change (FC) of 1 and an adjusted P value (Padj) of less than 0.05, a total of 3,037, 4,382, and 725 genes were differentially expressed between monocytes and AFlo AMs, monocytes and AFhi AMs, and AFlo and AFhi AMs, respectively (Figure 2E). We defined a common “macrophage” signature as the list of differentially expressed genes that were jointly upregulated in AFlo and AFhi AMs as compared with monocytes (Figure 3A). Such signature encompassed 1,785 genes, including MARCO, FABP4, PPARG, and MRC1 (Figure 3B), confirming that both AM subsets expressed core AM-associated genes (13, 14, 23). Because AFlo AMs were located between monocytes and AFhi AM on PC1 (Figure 2B), we also defined a signature of commonly upregulated genes between AFlo AMs and monocytes as compared with AFhi AMs, which encompassed 262 genes (Figure 3C), among which were core monocyte-associated genes, such as CCR2, CX3CR1, or ADAM19 (Figure 3D) (24, 25). Of note, only 27 genes were jointly upregulated between monocytes and AFhi AMs as compared with AFlo AMs (Figure 3E).
Figure 3.
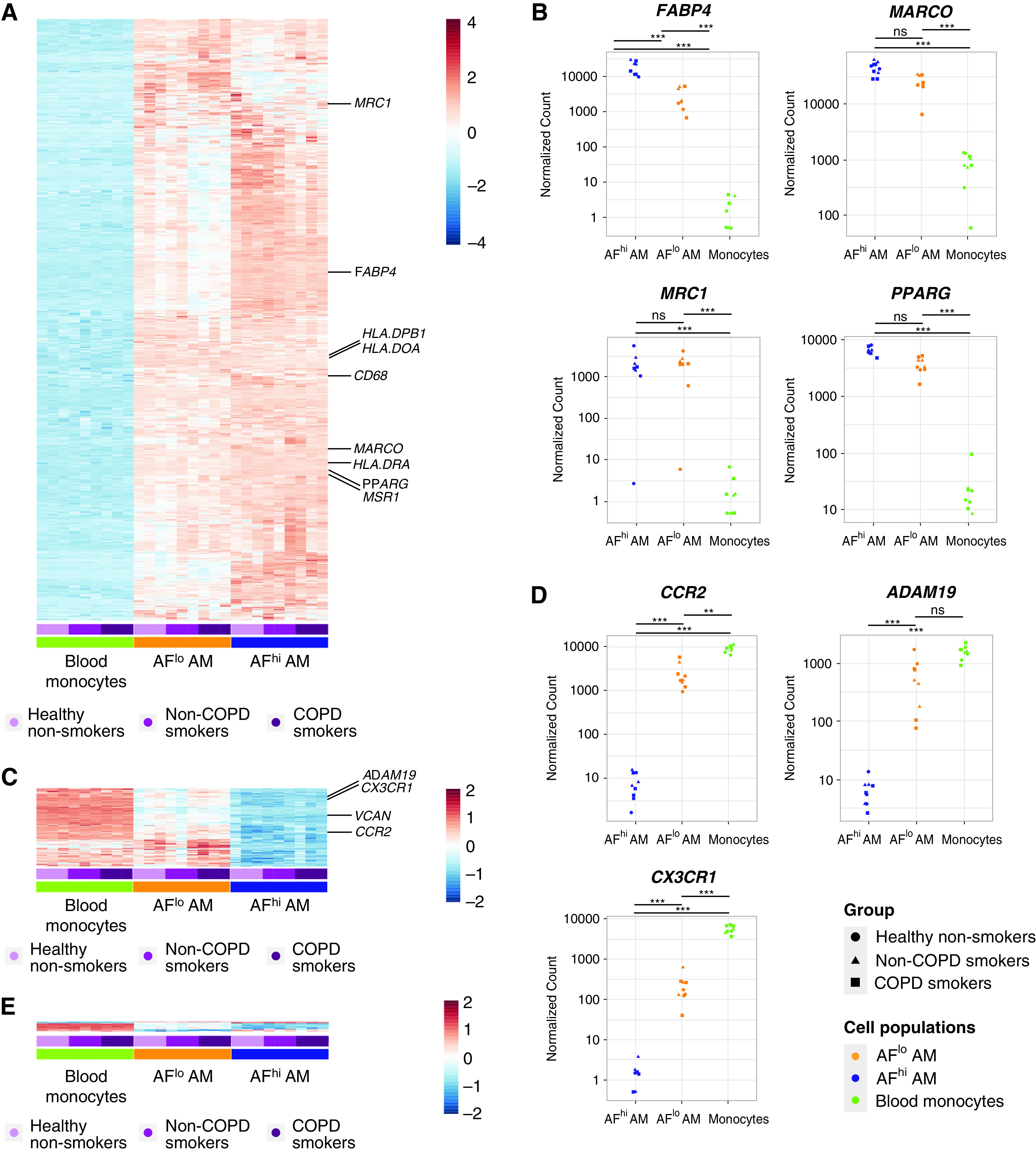
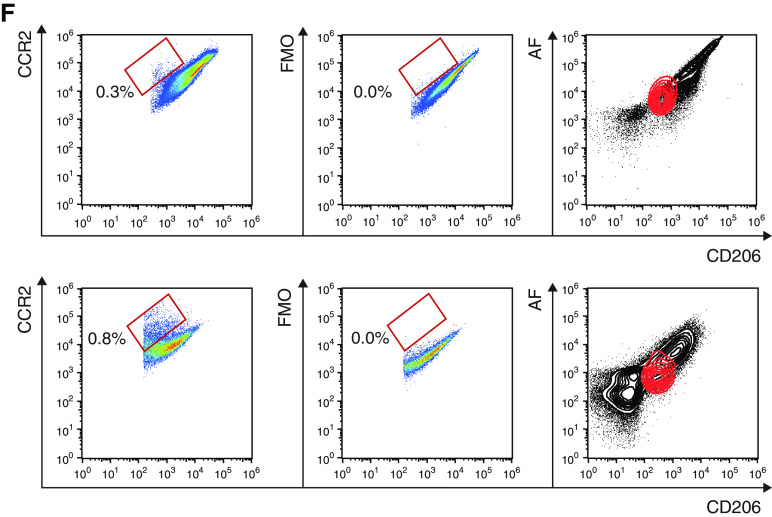
AFlo AMs share both AM- and monocyte-associated transcriptional signatures. (A) Heatmap depicting the list of significant DE genes jointly upregulated in AFlo or AFhi AMs as compared with blood monocytes. (B) Individual expression, shown as normalized counts, of the indicated genes within the indicated cell populations. (C) Heatmap depicting the list of significant DE genes commonly upregulated in blood monocytes or AFlo AMs as compared with AFhi AMs. (D) Individual expression, shown as normalized counts, of the indicated genes within the indicated cell populations. (E) Heatmap depicting the list of significant DE genes commonly upregulated in blood monocytes or AFhi AMs as compared with AFlo AMs. (F) CCR2 protein expression on BALF CD45+CD206+ macrophages (HD20 and HD21) (see Table E1). Representative flow cytometry plots showing CCR2 versus CD206 expression on BALF macrophages stained with (left) an anti-CCR2 or (center) FMO. Inserts indicated % of CCR2+ cells in the parent gate; (right) flow cytometry plots showing AF versus CD206 expression on total BALF CD45+ cells (gray) or CCR2+ macrophages (red). (B and D) P adjusted values are shown and were estimated thanks to the DESeq2 package. AF, auto-fluorescent; DE, differentially expressed; FMO = fluorescence minus one; HD = human donor; ns = nonsignificant. **P < 0.01 and ***P < 0.001.
Strikingly, CCR2, a marker of CD14+ monocytes (24), was the most significantly upregulated gene within AFlo AMs as compared with AFhi AMs (Figure 3D) (log2 FC, 8.13; Padj = 2.01 × 10−103). Of note, assessment of CCR2 protein expression revealed a small population of CCR2+ macrophages that were all AFlo (Figure 3F), consistent with the hypothesis that AFlo AM are derived from CCR2+ monocytes that have differentiated into AM to acquire expression of core AM genes.
AFlo AMs Are Functionally Distinct from Classical AFhi AMs and Contain IL-10–Producing Cells
Even though bulk RNA-seq analysis indicates that AFlo AMs and AFhi AMs jointly express AM-associated genes, their distinct transcriptional profiles suggested that they may have specific functionalities. We used gene set enrichment analyses to compare the transcriptome of AFhi and AFlo AMs with defined gene sets (26). AFhi AMs expressed genes involved in lipid or fatty acid metabolism and in PPAR signaling pathways (Figure 4A and Table E3), further supporting that AFhi AMs represent “bona fide” AMs involved in surfactant recycling and removal. Conversely, AFlo AMs expressed genes enriched in cytokine receptor activity, chemokine binding, and the negative regulation of T-cell activation (Figure 4A and Table E3), highlighting their potential roles in the regulation of immune and inflammatory responses. To assess this possibility, we cultured FACS-sorted AFlo AMs and AFhi AMs ex vivo and measured the concentrations of IL-10, a hallmark of regulatory macrophages (27, 28). We found that AFlo AMs were uniquely able to secrete IL-10, as opposed to AFhi AMs (Figure 4B and Table E1). IL-10 transcript concentrations were also higher in AFlo AMs than AFhi AMs or blood monocytes, even though it did not reach statistical significance (Figure 4C).
Figure 4.
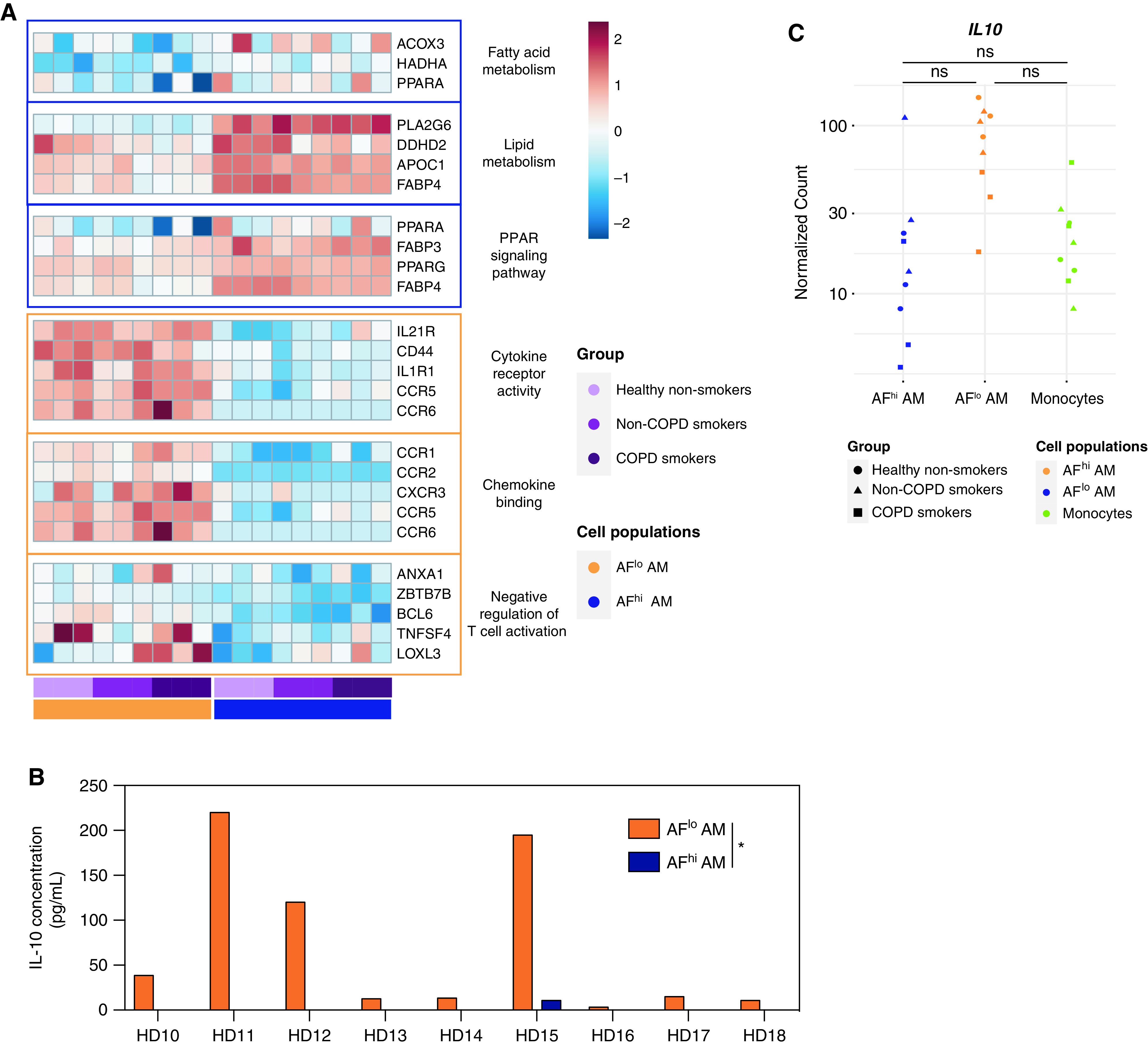
Monocyte-derived AFlo AMs are functionally distinct from classical AFhi AMs. (A) Heatmaps showing expression of the indicated genes involved in the biological responses that were found to be enriched in AFhi and AFlo AMs analyzed by gene set enrichment analysis (also see Table E3). (B) IL-10 concentrations measured by ELISA in culture supernatant of FACS-sorted AFlo and AFhi AMs from nine patients (HD1 to HD9) (Table E1). (C) Individual expression, shown as normalized counts, of IL-10 gene within the indicated cell populations. Data show individual mean values from technical duplicates. P values were calculated using a two-way ANOVA. *P < 0.05.
In line with others (16), our data suggest that, in adult lungs, AMs encompass resident homeostatic AMs as well as monocyte-derived AFlo AMs endowed with distinct functional properties. Importantly, such AM heterogeneity is found in the BALF of healthy nonsmokers, smokers without COPD, and smokers with COPD, suggesting that monocyte-derived AMs contribute to the pool of AMs over the lifespan.
ScRNA-Seq of Human BALF Cells Consistently Identifies Classical and Monocyte-derived AMs
Bulk RNA-seq analyses did not disclose any difference related to the health status but might have highlighted two extreme ends of a more complex spectrum enclosing monocyte-derived AFlo AMs and classical AFhi AMs and, hence, have missed more subtle differences. Thus, we sought to perform scRNA-seq on BALF cells obtained from the same subjects (Table 1) to further assess the functional diversity of AMs. We subjected all BALF cells to scRNA-seq using the 10X Genomics Platform (29) (Figure 2A and Figure E2). A total of 31,708 cells from nine samples passed quality control (QC) filtering, from which 29,827 cells belonged to the mononuclear phagocyte system (Figure E3 and Table E4). Graph-based clustering of merged single cells identified four transcriptionally distinct clusters of cells, as visualized on a global uniform manifold approximation and projection plot (Figure 5A). Uniform manifold approximation and projection plots of cells from each condition and from each sample are shown in Figure 5B and Figure E4, respectively. The relative contribution of each cluster within each sample is shown in Figure 5C.
Figure 5.
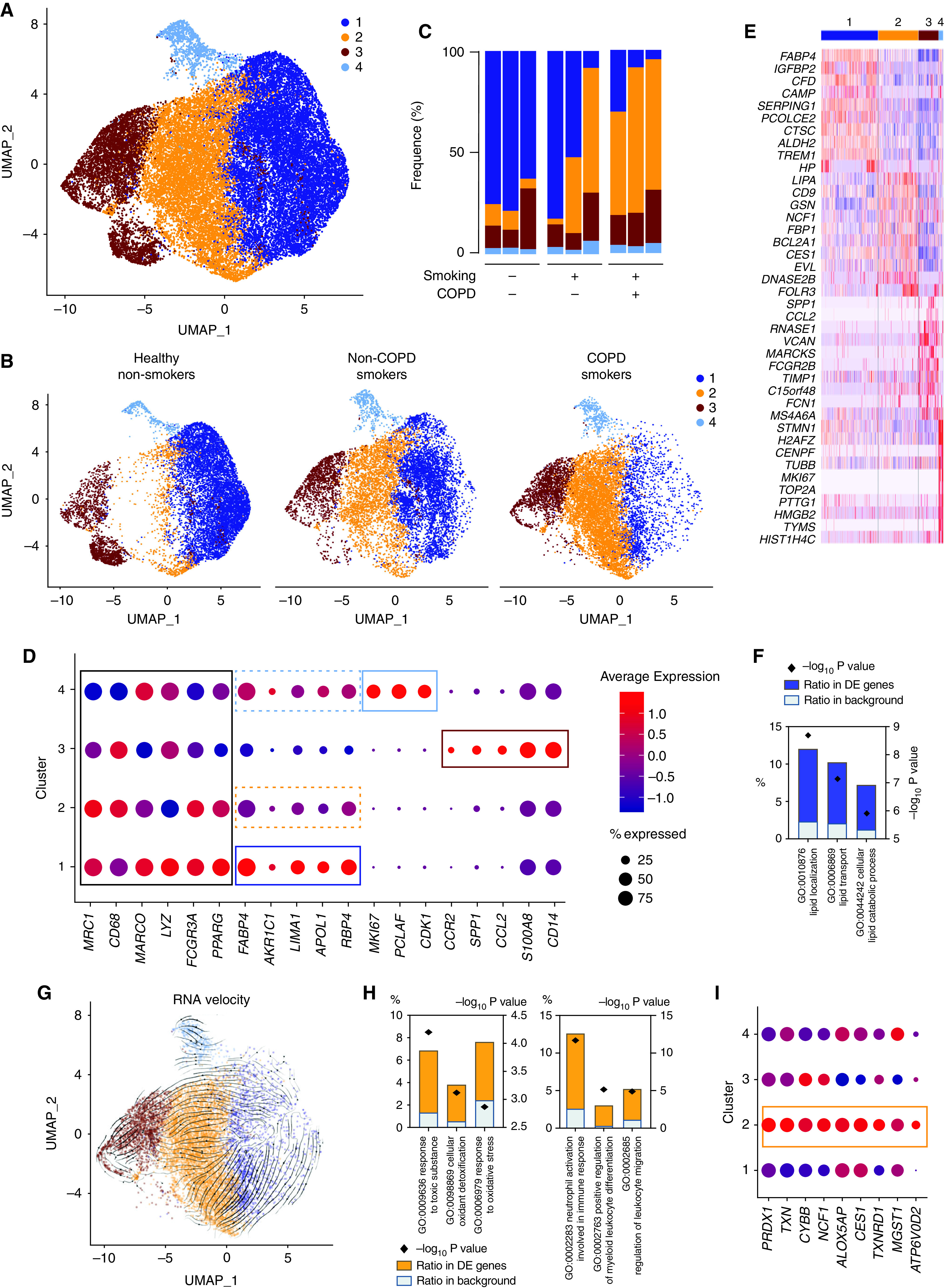
BALF myeloid cell heterogeneity revealed by single-cell (sc) RNAseq analyses. (A) Uniform manifold approximation and projection (UMAP) plot depicting BALF myeloid cells from the merged biological samples. (B) UMAP plots depicting BALF myeloid cells isolated from healthy nonsmokers (left), smokers without COPD (middle), and smokers with COPD (right). (C) Histogram showing percentage of each cluster within individual samples. (D) Dot plots showing average expression and percentage of cells expressing the indicated genes within cell clusters. (E) Heatmap depicting the 10 most upregulated genes in each cluster. (F) Histograms showing results of gene ontology (GO) enrichment tests for the upregulated genes in cluster 1 as compared with the other clusters. (G) Patterns of RNA velocities of single cells from smokers with COPD substantiated by arrows visualized on the UMAP plot. (H) Histograms showing results of GO enrichment tests for the upregulated genes in cluster 2 as compared with cluster 1. (I) Dot plots showing average expression and percentage of cells expressing the indicated genes within cell clusters.
All clusters exhibited high expression of core macrophage genes and the AM-associated transcription factor PPARG (Figure 5D). As compared with the other clusters, cluster 1 significantly upregulated transcripts encoding FAPB4, AKR1C1, LIMA1, APOL1, and RBP4 (Figures 5D and 5E), all of which are involved in lipid metabolism (Figure 5F). Of note, we also found that cells from cluster 1 expressed the highest signature score corresponding to AFhi AMs analyzed by bulk RNA-seq (Figures E5A–E5C).
Although cluster 4 also expressed high amounts of genes associated with classical AMs (dashed clear blue box, Figure 5D), it also upregulated cycling-related genes, including MKI67, PCLAF, and CDK1 (Figures 5D and 5E), supporting that it encompassed resident AMs undergoing cell proliferation. Accordingly, cluster 4 comprised cells found in phases S or G2M of their cell cycle, whereas cells from the other clusters were mainly in the gap phase G1 (Figure E5D).
Cluster 3 encompassed monocyte-derived cells, as cluster 3 overexpressed transcripts encoding monocyte lineage-associated molecules (e.g., CCR2, SPP1, CCL2, S100A8, and CD14) (Figures 5D and 5E). Moreover, cells from cluster 3 expressed the highest signature score corresponding to AFlo AMs analyzed by bulk RNA-seq (Figures E5A–E5C).
A Distinct Cluster of Classical AM Is Associated with Smoking
As opposed to the other clusters, cluster 2 was almost uniquely present in smokers without COPD and smokers with COPD as compared with healthy nonsmokers (Figure 5C). The gene expression profile of cluster 2 was relatively similar to that of cluster 1 (dashed orange box, Figure 5D), and RNA velocity analysis (30) of single cells supported the idea that cluster 2 represented an altered state of classical AMs but did not derive from monocyte-derived AMs (Figure 5G). As compared with cluster 1, the transcriptomic profile of cluster 2 was significantly enriched in genes involved in the response to toxic substances (NCF1, ALOX5AP, and CES1), cellular oxidant detoxification (ALOX5AP, TXNRD1, and MGST1), and oxidative stress (PRDX1, TXN, and CYBB) (Figures 5H and 5I). Moreover, cluster 2 upregulated genes involved in proinflammatory responses such as leukocyte migration, myeloid cell differentiation, and neutrophil activation (Figure 5H). Altogether, these data support the hypothesis that cluster 2 represents classical AMs that are stressed or damaged in response to toxic substances present in cigarette smoke and that exhibit proinflammatory capabilities.
BALF Cells Contain Dendritic Cells and Distinct Subsets of Monocyte-derived AMs
Given the heterogeneity present in cluster 3, we sought to subset and recluster it, which revealed four distinct clusters (Figure 6A), whose distribution by sample and by condition is shown in Figures 6B and 6C and in Table E4. Cluster 4 was homogeneously distributed, whereas clusters 2 and 3 were enriched in healthy nonsmokers and cluster 1 was enriched in smokers. Reclustering allowed identification of a small population of dendritic cells (cluster 4) on the basis of elevated expression of dendritic cell–associated genes as compared with the other clusters (Figures 6D and 6E). Cluster 3 significantly upregulated genes associated with matrix components, such as SPP1, SELENOP, MMP9, MARCKS, LGMN, or FOLR2, and involved in the regulation of response to wounding, regulation of wound healing, and inflammatory response to wounding (Figures 6D–6F), whereas cluster 2 exhibited a chemokine-secreting profile as exemplified by elevated expression of CCL18, CCL20, CCL4, CCL3, CXCL9, and CXCL10 (Figures 6D and 6E). Finally, cluster 1, more prominent in smokers, expressed high amounts of the monocyte lineage genes, such as CCR2, CSF1R, or FCN1 (Figure 6E), and its profile was enriched in proinflammatory genes involved in myeloid cell activation (Figure 6F). These data suggest that cluster 1 encompasses recently recruited monocyte-derived AMs with an activated phenotype, a process that is linked with smoking. Last, but not least, cluster 1 also upregulated expression of CLEC5A and VCAN (Figures 6D and 6E), two genes that have been shown to be associated with monocyte recruitment (31) and COPD pathology (32, 33).
Figure 6.
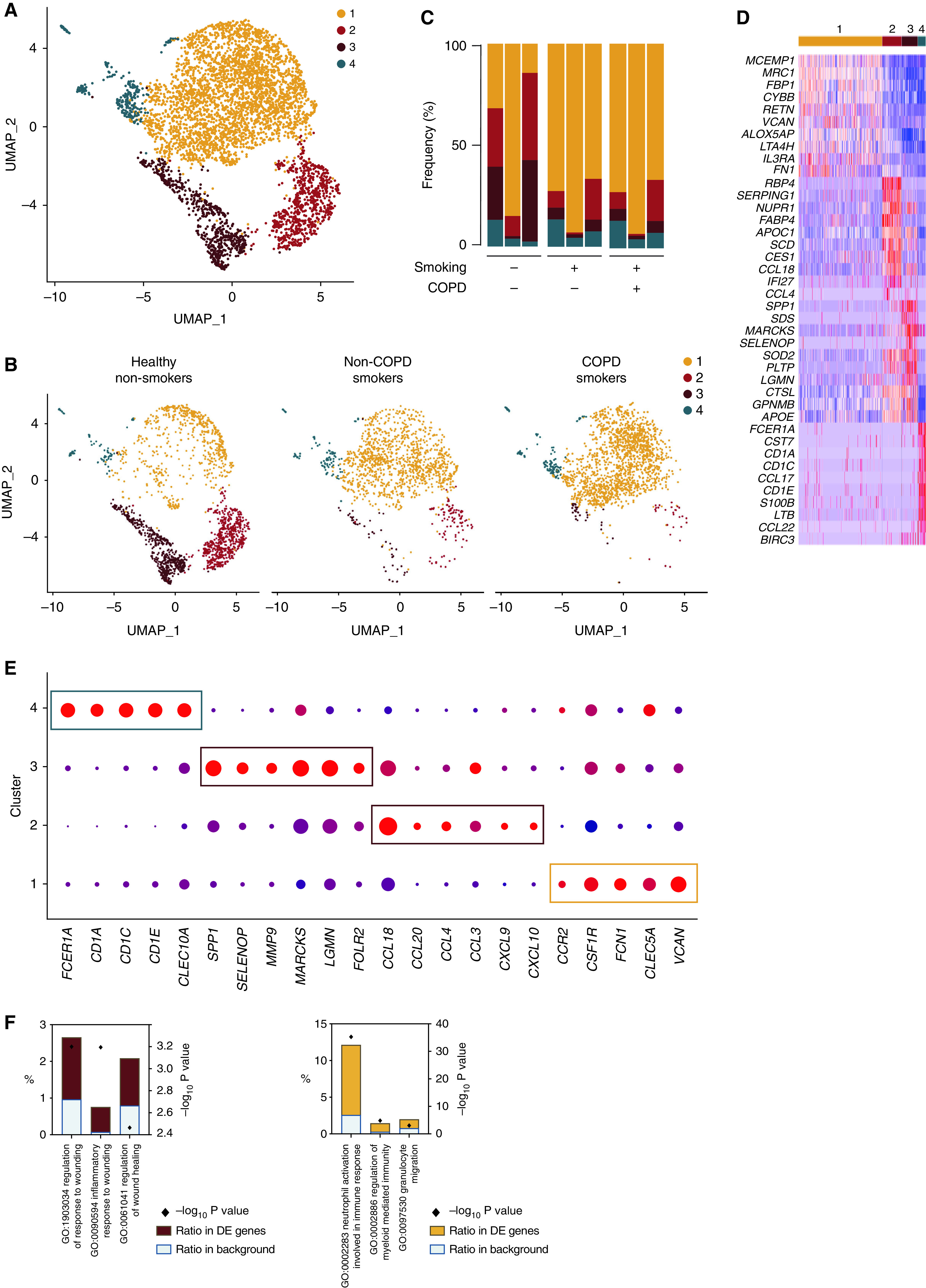
Heterogeneity of BALF monocyte-derived macrophage analyzed by scRNAseq. (A) UMAP plot depicting BALF monocyte-derived macrophage (i.e., cluster 3 of Figure 5A) from the merged biological samples. (B) UMAP plots depicting BALF monocyte-derived macrophage isolated from healthy nonsmokers (left), smokers without COPD (middle), and smokers with COPD (right). (C) Histogram showing percentage of each cluster within individual samples. (D) Heatmap depicting the 10 most upregulated genes in each cluster. (E) Dot plots showing average expression and percentage of cells expressing the indicated genes within cell clusters. (F) Histograms showing results of GO enrichment tests for the upregulated genes in clusters 3 (left) and 1 (right) as compared with the other clusters.
Discussion
Here, we show that the pool of AMs in adult healthy nonsmokers, smokers without COPD, and smokers with COPD is heterogeneous and encompasses transcriptionally distinct subpopulations. Our data also support that smoking alters the AM landscape and triggers changes that may have implications for COPD physiopathology.
In nondiseased human lungs, AMs are classically defined by flow cytometry as autofluorescent SSChi CD206+ cells (20, 21, 34). Here, we report that a population of small AFlo AMs is consistently present and equally represented between healthy subjects and patients with COPD. Of note, our results are concordant with those from Jambo and colleagues, who previously identified small and large AMs in the BALF of human adults based on their concentrations of SSC (35). Although AFlo AMs express monocyte-associated genes, supportive of a monocytic origin, they also express high amounts of AM-associated genes, suggesting that they have been imprinted by the local niche to express features of classical AMs.
Our results are consistent with the idea that monocytes contribute to the pool of homeostatic AMs in healthy lungs, as shown recently (16). In line with this, it was demonstrated that human CD14+ monocytes were able to give rise to fully differentiated CD206+ AMs in a humanized mouse model (36). Of note, as opposed to previous evidence obtained from laboratory mice supporting that AMs self-maintain with a minimal contribution from the monocyte compartment (6, 8, 9), a recent mouse study challenged this notion and also found that AMs were slowly replaced by monocyte-derived cells in old mice (37).
Functionally, monocyte-derived AFlo AMs express high amounts of the gene coding for fatty acid binding protein (i.e., FABP4) as compared with blood monocytes, consistent with the hypothesis that some AFlo AMs, like classical AMs, may also contribute to fatty acid metabolism. Nevertheless, AFlo AMs also expressed a unique transcriptional signature associated with specific immunoregulatory functions. Of note, such signature, along with their monocytic origin and their small size, is reminiscent of what is described for immunoregulatory CD206+ IL-10–producing interstitial macrophages in the murine steady-state lung (38, 39). Importantly, mouse interstitial macrophages can also be recruited to the airways under particular experimental conditions, such as after exposure to microbial products (28) or after a respiratory infection with murine herpesvirus-4 infection (11) or influenza virus (12). Thus, the abundance and functionality of AFlo AM might give a picture of the immunoregulatory potential of human lungs and might translate their ability to control excessive inflammatory and immune responses, which can have functional consequences for lung diseases.
ScRNA-seq of human BALF cells represents a highly valuable tool to decipher AM diversity in health and disease. We detected, in addition to classical AMs, a population of monocyte-derived AMs that was present in each sample analyzed, including those from healthy subjects, as previously described (16). Even though the total number of cells present in monocyte-derived AMs is relatively small, we found after reclustering a cluster of cells expressing many genes coding for chemokines, which is concordant with the discrete cluster of proinflammatory AMs identified by Mould and colleagues (16). Similarly to what was shown in the same report, we describe the existence of monocyte-derived AMs expressing high amounts of matrix-associated genes (16). Whether these cells contribute to the homeostatic remodeling of the lung tissue or to pathological profibrotic events remains unclear, but the fact that they tend to be underrepresented in smokers is consistent with the hypothesis that they may exert beneficial rather than detrimental functions.
An original edge of our study is the comparison BALF cells from healthy nonsmokers, smokers without COPD, and smokers with COPD at the single-cell level. Although performed on a limited number of samples, we were able to identify a distinct cluster of AMs that was uniquely enriched in subjects exposed to cigarette smoke. Interestingly, this cluster does not express monocyte lineage–associated genes and represents a continuum with classical AMs, supporting that it represents an altered state of classical AMs in response to exposure to cigarette smoke. Functionally, these macrophages exhibit a proinflammatory profile and may therefore be implicated in persistent inflammation associated with COPD. In addition, a cluster of proinflammatory monocyte-derived macrophages was significantly enriched in smokers with and without COPD as compared with healthy nonsmokers. Of note, these cells expressed elevated amounts of CLEC5A and VCAN, which are thought to contribute to COPD pathology. Indeed, CLEC5A has been shown to be upregulated in AMs from smoking mice or humans and can mediate features of COPD pathology in mice (33). Similarly, VCAN is coding for a proteoglycan involved in fibroblast differentiation and is also thought to contribute to tissue remodeling in COPD (32). Interestingly, VCAN and CLEC5A have also been associated with monocyte recruitment (31), and their overexpression in smokers could contribute to the regulation of monocyte influx in the airways. Altogether, our data support the hypothesis that cigarette smoke alters both classical and monocyte-derived AM to endow them with functional features that may favor either subclinical proinflammatory events in smokers without COPD or COPD-related persistent inflammation in smokers with COPD. Future prospective investigations will be needed using samples from larger cohorts of patients with the inclusion of nonsmokers with COPD or ex-smokers to better understand the fate, plasticity, functions, and persistence of such macrophage subpopulations during COPD progression.
Acknowledgments
Acknowledgment
The authors thank the GIGA Flow Cytometry and Cell Imaging Platform (GIGA Institute, Liege University, Belgium) and Cédric François, Raja Fares, and Ilham Sbai (Faculty of Veterinary Medicine, Liege University, Belgium) for their excellent technical and administrative support.
Footnotes
Supported by the Fonds de la Recherche scientifique (FRS)-FNRS, the FRFS-Walloon Excellence in Life Sciences and Biotechnology (FRFS-WELBIO) (grants 20263584 and 35049229), the Acteria Foundation, and an European Research Council starting grant (T.M.); and by the FRFS-WELBIO and an Excellence of Science grant (F.B.).
Author Contributions: M.L., L.F., T.M., and F.B. designed the research study. M.L., L.F., D.P., and C.L. conducted experiments. Q.B., M.L., and D.P. analyzed the data. J.G., F.S., and J.-L.C. collected human samples. F.B. and R.L. supervised the study. M.L., Q.B., T.M., and F.B. wrote the manuscript. All authors gave feedback on the manuscript. Co–first authorship order was determined according to the contribution to the study.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2021-0563OC on May 6, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol . 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 2. Kulikauskaite J, Wack A. Teaching old dogs new tricks? The plasticity of lung alveolar macrophage subsets. Trends Immunol . 2020;41:864–877. doi: 10.1016/j.it.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blériot C, Chakarov S, Ginhoux F. Determinants of resident tissue macrophage identity and function. Immunity . 2020;52:957–970. doi: 10.1016/j.immuni.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 4. Trapnell BC, Carey BC, Uchida K, Suzuki T. Pulmonary alveolar proteinosis, a primary immunodeficiency of impaired GM-CSF stimulation of macrophages. Curr Opin Immunol . 2009;21:514–521. doi: 10.1016/j.coi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morales-Nebreda L, Misharin AV, Perlman H, Budinger GRS. The heterogeneity of lung macrophages in the susceptibility to disease. Eur Respir Rev . 2015;24:505–509. doi: 10.1183/16000617.0031-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med . 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature . 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity . 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity . 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med . 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Machiels B, Dourcy M, Xiao X, Javaux J, Mesnil C, Sabatel C, et al. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat Immunol . 2017;18:1310–1320. doi: 10.1038/ni.3857. [DOI] [PubMed] [Google Scholar]
- 12. Aegerter H, Kulikauskaite J, Crotta S, Patel H, Kelly G, Hessel EM, et al. Influenza-induced monocyte-derived alveolar macrophages confer prolonged antibacterial protection. Nat Immunol . 2020;21:145–157. doi: 10.1038/s41590-019-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med . 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med . 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 15. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv . 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mould KJ, Moore CM, McManus SA, McCubbrey AL, McClendon JD, Griesmer CL, et al. Airspace macrophages and monocytes exist in transcriptionally distinct subsets in healthy adults. Am J Respir Crit Care Med . 2021;203:946–956. doi: 10.1164/rccm.202005-1989OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barnes PJ. Chronic obstructive pulmonary disease. N Engl J Med . 2000;343:269–280. doi: 10.1056/NEJM200007273430407. [DOI] [PubMed] [Google Scholar]
- 18. Gutiérrez Villegas C, Paz-Zulueta M, Herrero-Montes M, Parás-Bravo P, Madrazo Pérez M. Cost analysis of chronic obstructive pulmonary disease (COPD): a systematic review. Health Econ Rev . 2021;11:31. doi: 10.1186/s13561-021-00329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet . 2018;392:1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu Y-RA, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol . 2016;54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol . 2016;54:147–149. doi: 10.1165/rcmb.2015-0147LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med . 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 23. Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J . 2019;54:1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Al-Rashoudi R, Moir G, Al-Hajjaj MS, Al-Alwan MM, Wilson HM, Crane IJ. Differential expression of CCR2 and CX3CR1 on CD16+ monocyte subsets is associated with asthma severity. Allergy Asthma Clin Immunol . 2019;15:64. doi: 10.1186/s13223-019-0379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med . 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 26. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA . 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoppstädter J, Diesel B, Zarbock R, Breinig T, Monz D, Koch M, et al. Differential cell reaction upon Toll-like receptor 4 and 9 activation in human alveolar and lung interstitial macrophages. Respir Res . 2010;11:124. doi: 10.1186/1465-9921-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabatel C, Radermecker C, Fievez L, Paulissen G, Chakarov S, Fernandes C, et al. Exposure to bacterial CpG dna protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity . 2017;46:457–473. doi: 10.1016/j.immuni.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 29. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun . 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature . 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Toeda K, Nakamura K, Hirohata S, Hatipoglu OF, Demircan K, Yamawaki H, et al. Versican is induced in infiltrating monocytes in myocardial infarction. Mol Cell Biochem . 2005;280:47–56. doi: 10.1007/s11010-005-8051-4. [DOI] [PubMed] [Google Scholar]
- 32. Andersson-Sjöland A, Hallgren O, Rolandsson S, Weitoft M, Tykesson E, Larsson-Callerfelt A-K, et al. Versican in inflammation and tissue remodeling: the impact on lung disorders. Glycobiology . 2015;25:243–251. doi: 10.1093/glycob/cwu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wortham BW, Eppert BL, Flury JL, Garcia SM, Donica WR, Osterburg A, et al. Cutting edge: CLEC5A mediates macrophage function and chronic obstructive pulmonary disease pathologies. J Immunol . 2016;196:3227–3231. doi: 10.4049/jimmunol.1500978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, et al. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med . 2016;193:614–626. doi: 10.1164/rccm.201507-1376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jambo KC, Banda DH, Kankwatira AM, Sukumar N, Allain TJ, Heyderman RS, et al. Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol . 2014;7:1116–1126. doi: 10.1038/mi.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evren E, Ringqvist E, Tripathi KP, Sleiers N, Rives IC, Alisjahbana A, et al. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity . 2021;54:259–275.e7. doi: 10.1016/j.immuni.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 37. Liu Z, Gu Y, Chakarov S, Bleriot C, Kwok I, Chen X, et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell . 2019;178:1509–1525.e19. doi: 10.1016/j.cell.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 38. Schyns J, Bai Q, Ruscitti C, Radermecker C, De Schepper S, Chakarov S, et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat Commun . 2019;10:3964. doi: 10.1038/s41467-019-11843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science . 2019;363:eaau0964. doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]