Abstract
Background:
Pathologic complete response (pCR) to neoadjuvant chemotherapy (NAC) is strongly associated with favorable outcome. We examined the utility of serial circulating tumor DNA (ctDNA) testing for predicting pCR and risk of metastatic recurrence.
Patients and methods:
Cell-free DNA (cfDNA) was isolated from 291 plasma samples of 84 high-risk early breast cancer patients treated in the neoadjuvant I-SPY 2 TRIAL. Blood was collected at pretreatment (T0), 3 weeks after initiation of paclitaxel (T1), between paclitaxel and anthracycline regimens (T2), or prior to surgery (T3). A personalized ctDNA test was designed to detect up to 16 patient-specific mutations (from whole exome sequencing of pretreatment tumor) in cfDNA by ultra-deep sequencing. The median follow-up time for survival analysis was 4.8 years.
Results:
At T0, 61 of 84 (73%) patients were ctDNA-positive, which decreased over time (T1–35%; T2–14%; T3–9%). Patients who remained ctDNA-positive at T1 were significantly more likely to have residual disease after NAC (83% non-pCR) compared to those who cleared ctDNA (52% non-pCR; OR 4.33, P=0.012). After NAC, all patients who achieved pCR were ctDNA-negative (n=17, 100%). For those who did not achieve pCR (n=43), ctDNA-positive patients (14%) had significantly increased risk of metastatic recurrence (HR 10.4; 95% CI, 2.3–46.6); interestingly, patients who did not achieve pCR but were ctDNA-negative (86%) had excellent outcome, similar to those who achieved pCR (HR 1.4; 95% CI, 0.15–13.5).
Conclusions:
Lack of ctDNA clearance was a significant predictor of poor response and metastatic recurrence, while clearance was associated with improved survival even in patients who did not achieve pCR. Personalized monitoring of ctDNA during NAC of high-risk early breast cancer may aid in real-time assessment of treatment response and help fine-tune pCR as a surrogate endpoint of survival.
Keywords: breast cancer, circulating tumor DNA, neoadjuvant chemotherapy, pathologic complete response
Introduction
Circulating tumor ctDNA (ctDNA) in blood offers a minimally invasive approach for disease monitoring and evaluation of response to therapy [1–3]. Findings from recent clinical studies have shown that ctDNA may play a role in detecting minimal residual disease (MRD) and emerging therapy resistance, i.e., molecular relapse (MR), in early stage breast cancers [4–7], as well as in monitoring of disease progression in patients with advanced breast cancer [8–10]. However, it is not yet known if failure to clear ctDNA during therapy could provide guidance for escalation of treatment to prevent early disease recurrence [11].
Neoadjuvant chemotherapy (NAC) has become a standard-of-care for patients with locally-advanced breast cancer [12]. First, NAC provides a unique opportunity for real-time monitoring of tumor response and evaluation of drug efficacy [13–15]. Second, NAC may downstage tumors and thus improve chances of breast-conserving surgery [12, 16, 17]. Third, response to NAC provides prognostic information which can supplement those derived from standard clinico-pathologic characteristics of the primary tumor, such as subtype, nodal status, and grade [12, 16–20].
Pooled analysis by Cortazar and colleagues has shown that patients who achieved a pathologic complete response (pCR, or the absence of residual cancer in the breast and lymph nodes after NAC) have significant survival advantage over those who did not [21]. Standard NAC alone or in combination with other agents has resulted in pCR for 10–50% of patients depending on subtype [21]. Data from the I-SPY 2 TRIAL, a multicenter phase 2 trial that evaluates investigational drugs in combination with standard NAC (paclitaxel followed by anthracycline treatment) [22], have shown that pCR in women with molecularly high-risk stage II or III tumors, whether from standard or targeted therapies, unequivocally conferred a survival advantage (hazard ratio of 0.2) [23].
While pCR accurately identifies patients with low risk of relapse, studies have shown that predicting early metastatic recurrence in those with residual disease (non-pCR) is less robust [21, 23]. For example, survival analysis in the I-SPY 2 TRIAL (median follow-up of 3.8 years) showed that the 3-year distant disease-free survival (DRFS) of patients who achieved pCR was 95% [23]. Among non-pCR patients, only 22% of experienced metastatic recurrence. In this study, we evaluated the potential role of ctDNA as a biomarker for monitoring of response to NAC and assessed the additive value of ctDNA to further stratify patients with residual disease to predict early metastatic recurrence. We hypothesized that early changes in ctDNA are predictive of response to NAC and that ctDNA dynamics during NAC as well as ctDNA status (positive vs. negative) at each time point are associated with patient outcomes. To address these hypotheses, we performed a correlative study in the I-SPY 2 TRIAL to detect ctDNA in serial plasma samples collected before, during and after NAC [24]. We used a previously analytically validated personalized ctDNA test composed of a panel of up to 16 most clonal somatic variants present in the pretreatment tumor [10, 25–27]. The test involves multiplex polymerase chain reaction amplification followed by ultra-deep sequencing to detect tumor-specific mutations (i.e., ctDNA) in cell-free DNA (cfDNA). This approach enables more accurate monitoring of disease burden than pre-fixed driver mutation panels, as each test reflects tumor heterogeneity at the individual patient level [5, 8, 28].
Patients and Methods
Patients
We performed a retrospective ancillary ctDNA study on prospectively collected samples from high-risk early breast cancer patients enrolled in the multicenter neoadjuvant I-SPY 2 TRIAL (NCT01042379). Women with ≥2.5 cm stage II/III breast cancer were eligible. Patients were screened for metastatic disease by imaging (CT or PET) prior to enrollment, and those with de novo metastatic disease were excluded. Restaging scans were not performed after NAC prior to surgery. Eligibility was limited to patients with a MammaPrint high score, and thus the trial was enriched for those with increased risk of metastatic recurrence within 5 years after diagnosis. Patients received standard NAC combined with MK-2206 (AKT inhibitor) or standard NAC alone. Detailed descriptions of the design, eligibility, and study assessments in the I-SPY 2 TRIAL have been reported previously [22, 29]. Institutional Review Boards at all participating institutions approved the protocol. All patients signed informed consent to allow research on their biospecimen samples.
ctDNA analysis
Detailed description of the clinical samples and the methods for ctDNA analysis [26, 27, 30] (Supplementary Fig S1–S3) are found in the Supplementary Methods.
Statistical analysis
To determine the cutoff for ctDNA-positivity, a large set of negative control samples (~1000) were pre-processed to build a background error model. For each target variant identified in the plasma, a confidence score was calculated based on the depth of read for mutant and reference alleles [25]. In addition, simulation studies were performed as previously described [10, 26, 27] to determine limits of detection and quality control thresholds for stringent assessment of ctDNA results (Supplementary Methods, Supplementary Fig S4 and Supplementary Fig S5). A plasma sample with at least 2 variants with a confidence score above a predefined threshold (0.97) was defined as ctDNA-positive.
Logistic regression was used to assess association between pCR and ctDNA clearance. Survival curves were generated by Kaplan-Meier analysis and compared using log rank test. Cox regression analysis was used to estimate hazard ratio and 95% CI. Survival data were available for 75 of the 84 patients. Detailed description of the study design and the statistical methods can be found in the Supplementary Methods.
Results
ctDNA analysis in I-SPY 2 TRIAL patients
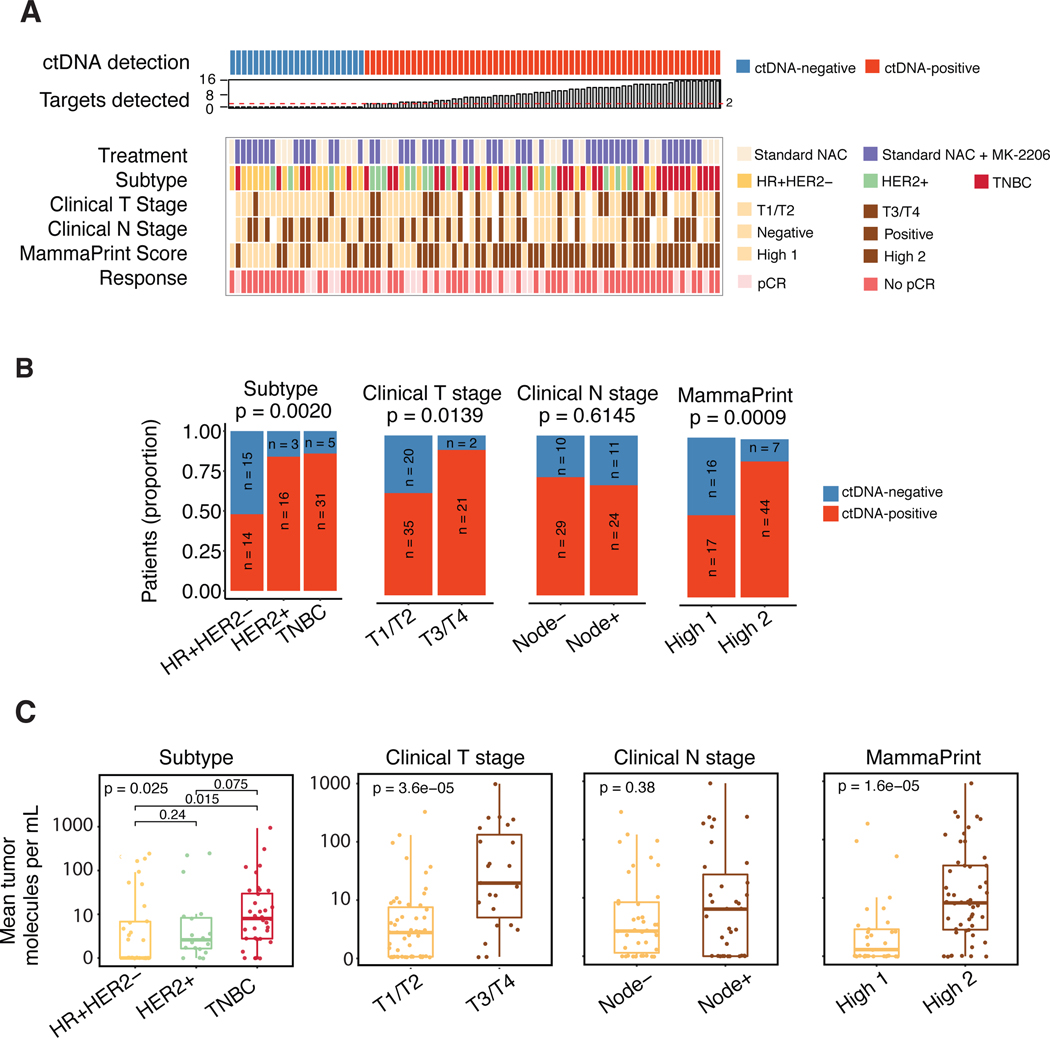
This ctDNA study was performed retrospectively on samples collected from I-SPY 2 TRIAL patients who received standard NAC alone or combined with MK-2206 (AKT inhibitor) treatment (Fig 1A). Primary tumor and matched normal samples for whole exome sequencing was available for 90 patients (Fig 1B, 1C, Supplementary Fig S1). Of these, 6 were excluded due to poor quality sequencing data, resulting in an analytic cohort of 84 patients (Supplementary Table S1). Whole exome sequencing detected a mean of 181 mutations in the 84 untreated primary tumor tissue analyzed (median 159; range: 32–772; Supplementary Table S2). CfDNA was isolated from plasma samples collected from pretreatment (T0), 3 weeks after initiation of treatment (T1), between paclitaxel and anthracycline regimens (T2), and after NAC prior to surgery (T3) (Fig 1A, Supplementary Fig S2). From the list of variants derived from whole exome sequencing, a unique personalized panel consisting of up to 16 highly ranked somatic mutations were selected (median: 16; range: 12–16). Multiplex polymerase chain reaction assays were designed and used to interrogate cfDNA for the presence of these mutations (Fig 1C, Supplementary Table S2, Supplementary Methods). Amplicons were subjected to ultra-deep sequencing to detect ctDNA (Supplementary Fig S3). ctDNA analysis was successfully performed on 291 (87%) of the potential 336 total plasma samples (84 patients x 4 time points). Samples with at least two detectable somatic variants were considered ctDNA-positive (Supplementary Fig S4, Supplementary Fig S5) [10, 25–27].
Fig 1. Study schema, methods for ctDNA analysis, patients and samples.
A) Diagram showing the study schema of the I-SPY 2 TRIAL. Prior to study entry, tumor biopsy from each patient is analyzed to assess hormone-receptor (HR) and human epidermal growth factor receptor 2 (HER2) status and MammaPrint scores. Blood samples are collected at the following time points: T0- baseline/pretreatment, T1– 3 weeks after initiation of therapy, T2- between two treatment regimens (paclitaxel +/− MK-2206 and anthracycline (AC)), T3- after neoadjuvant chemotherapy (NAC) prior to surgery. B) Flow chart showing patients and samples evaluated in the study and sample performance at different quality control (QC) points. C) Schema of the methods for ctDNA analysis. PCR-polymerase chain reaction.
Of the 84 patients, 35% were hormone receptor-positive (HR+)/human epidermal growth factor receptor 2-negative (HER2-), 23% HER2+, and 43% triple negative breast cancers (TNBC); 30% had T3 or T4 tumors; 53% were node-negative and 61% were considered to be MammaPrint High 2 (ultra-high risk) (Supplementary Table S1). There were no significant differences in the clinicopathologic characteristics between patients who were excluded (n=67) and those who were included in the study (n=84).
Baseline ctDNA is associated with tumor burden and aggressive phenotype
At pretreatment (T0), 73% of the patients had detectable ctDNA (Fig 2A). ctDNA detection rates in patients who received standard NAC (n=27, 73%) was similar to those who received additional MK-2206 (n=57, 72%) (Fig 2A, Supplementary Table S1). The proportion of ctDNA-positive samples was significantly higher among HER2+ (84%) and TNBC (86%) subtypes as compared to HR+/HER2- (48%) subtype (p<0.01, Supplementary Table S1, Fig 2A–B). ctDNA positivity was also associated with larger tumors (T3/T4, 91%, P=0.014) but not with nodal status at the time of diagnosis. A significantly higher proportion of MammaPrint High 2 patients were ctDNA-positive (86%) compared to 52% in MammaPrint High 1 (p< 0.01).
Fig 2. Association between ctDNA and clinicopathologic characteristics.
A) Overview of patient and tumor characteristics according to ctDNA status at baseline (T0). HR-hormone receptor, TNBC-triple negative breast cancer, pCR- pathological complete response. B) Proportion of ctDNA-positive and negative patients at baseline (T0) according to clinical characteristics. P values were calculated using Fisher’s exact test. C) Mean tumor molecules per mL of plasma according to clinical characteristics. Distributions were compared using Wilcoxon rank sum (binary variable) or Kruskal Wallis (ternary variable) tests.
We also evaluated the absolute ctDNA levels (i.e., mean tumor molecules per mL of plasma) in the different groups stratified according to these same clinical variables and observed the same trend. The mean tumor molecules per mL in TNBC patients was significantly higher compared to that of HR+/HER2- patients (Fig 2C). Significantly higher levels of ctDNA were also observed for clinical T-stage T3/T4 vs. T1/T2 and MammaPrint high 2 vs. high 1
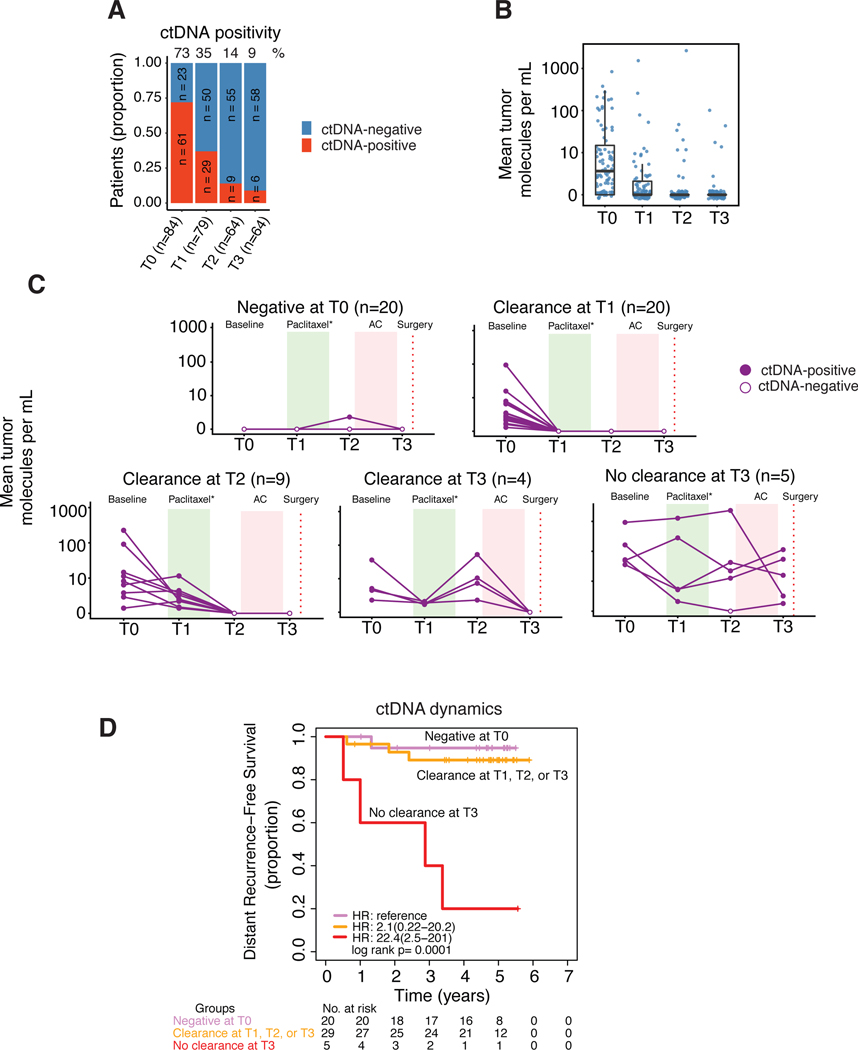
ctDNA positivity decreases with distinct dynamics during NAC
In the population as a whole, ctDNA positivity decreased during the course of NAC, from 73% before treatment (T0), to 35% at 3 weeks (T1), to 14% at the inter-regimen time point (T2), and down to 9% after NAC (T3) (Fig 3A). Similarly, the absolute ctDNA levels decreased over time (Fig 3B). Although, on average the ctDNA positivity decreased with time, at the individual patient level, five main patterns were observed. Fig 3C shows ctDNA positivity as a function of time during treatment for 58 of the 84 patients who had complete serial data available at all four time points: Patients with undetectable ctDNA at T0 who remained undetectable at T3 (n=20, 34.5%), patients who respectively, cleared at T1 (n=20, 34.5%), at T2 (n=9, 15.5%), or at T3 (n=4, 6.9%), or patients who remained ctDNA-positive after NAC (T3) (n=5, 8.6%).
Fig 3. ctDNA dynamics over the course of neoadjuvant chemotherapy.
A) Proportion of patients according to ctDNA positivity based on number of samples available per time point. B) Mean tumor molecules per mL of plasma across time points. C) Patients with complete ctDNA data for four time points (n=58) grouped according to observed patterns of ctDNA clearance or non-clearance. D) Survival in patients grouped according to ctDNA clearance. Of the 58 patients, 54 had survival data. Patients who cleared ctDNA at T1, T2 or T3 were combined into one group and their survival was compared with those of patients who did not clear ctDNA at T3 and those who were ctDNA-negative at T0 (reference group).
Clearance dynamics of ctDNA is associated with NAC response
We evaluated ctDNA clearance as a predictor of response to NAC. The rates for pCR across subtypes were 13.8%, 47.4% and 27.8% for HR+/HER2-, HER2+ and TNBC, respectively. 56 patients who were ctDNA-positive at T0 had a corresponding T1 plasma measurement (Fig 4A); and of these, 29 (52%) remained ctDNA-positive at T1, 3 weeks after the initiation of treatment. 83% of patients who did not clear their ctDNA at T1 had residual disease at surgery (24/29 non-pCR) compared to 52% in patients who cleared ctDNA at T1 (14/27 non-pCR). This association was significant (OR 4.33, P=0.012, adjusted for subtype and treatment received). Among the 39 non-pCR patients who had undetectable ctDNA after NAC, 17 (43%) were ctDNA-negative at baseline, 10 (26%) cleared ctDNA by T1, 12 (31%) cleared ctDNA by T2 or T3. The positive predictive value (PPV) of the test (for predicting non-pCR) increased with time (Fig 4B).
Fig 4. Association of ctDNA with response to neoadjuvant chemotherapy and its positive predictive value.
A) Sankey plot showing ctDNA dynamics (clearance or non-clearance) early during treatment vs. response (pathologic complete response, pCR, or no pCR). Analysis was focused on patients who were ctDNA-positive at baseline (T0) and had corresponding ctDNA testing results at T1, 3 weeks after initiation of therapy. B) Positive predictive value (PPV) of ctDNA-positivity in predicting failure to achieve pCR. PPV is the proportion of patients with a positive ctDNA test (at a specific time point) with residual cancer after NAC.
Clinical events are frequent in patients with detectable ctDNA
Survival data was available for 75 of the 84 patients, with a median follow-up of 4.8 years (range: 0.5 to 6.3 years). In this period, 8 had local recurrences, 10 experienced distant metastases, of whom 8 died (Fig 5A). Detectable ctDNA in at least one time point was observed in 6 of the 8 patients (75%) with local recurrence, 9 of the 10 patients (90%) who had distant recurrence and in all 8 patients who died (100%). Of note, a patient who experienced brain metastasis did not have detectable ctDNA at all time points.
Fig 5. ctDNA and clinical outcomes.
A) Overview of the ctDNA detection across different time points [T0: baseline/pretreatment, T1: 3 weeks after initiation of therapy, T2: between two treatment regimens (paclitaxel and AC), T3: after NAC prior to surgery]. The right panel shows a swimmer plot depicting the length of follow-up and events in 75 patients with survival data. The primary endpoint of the study was distant recurrence-free survival. B) Proportion of subtypes according to groups based on pCR and ctDNA status at T3. C) Patient survival stratified based on ctDNA status after NAC (T3) and response to treatment (pathological complete response, pCR). Inset table shows the numbers and percentages of patients according to subtype and response/ctDNA status.
ctDNA dynamics is significantly associated with metastatic recurrence
We examined whether ctDNA dynamic patterns (see Fig 3C) were associated with DRFS, the secondary endpoint of the I-SPY 2 TRIAL. Of 58 patients with ctDNA data at all time points, 54 had follow-up information. Patients who had cleared ctDNA at T1, T2 or T3 (n=29) had similar risk of metastatic recurrence compared to those who were ctDNA-negative at T0 (n=20) (hazard ratio (HR), 2.1; 95% confidence interval (CI), 0.22–20.2) (Fig 3D). Patients who did not clear ctDNA at T3 (n=5) had significantly higher risk of metastatic recurrence (HR, 22.4; 95%, CI, 2.5–201, p<0.001)
ctDNA at T1, T2, and T3 but not T0 is associated with increased risk of metastatic recurrence
Next, we examined whether ctDNA status (positive or negative) at different time points was associated with DRFS (Supplementary Fig S6). At baseline (T0), ctDNA-positive patients had increased risk of metastatic recurrence, but this association did not reach statistical significance (HR, 4.11; 95% CI, 0.52–32.4). In contrast, ctDNA positivity at 3 weeks after initiation of therapy (T1; HR, 4.5; 95% CI, 1.2–17.4), between regimens (T2; HR, 5.4; 95% CI, 1.3–22.5), and after NAC (T3; HR, 11.5; 95% CI, 2.9–46.1) was significantly associated with increased risk of metastatic recurrence.
Clearance of ctDNA after NAC (T3) is associated with improved survival
Patients were stratified according to pCR and ctDNA status after NAC (n=60). The proportion of subtypes varied across groups based on pCR and ctDNA-positivity (Fisher exact p=0.0257, Fig 5B). 17 patients who achieved pCR (100%), all of whom were ctDNA-negative, showed favorable DRFS (Fig 5C). In patients who did not achieve pCR (n=43), ctDNA-positivity (n=6; Supplementary Fig S7) was a significantly associated with worse DRFS (n=37; HR 10.4, 95% CI, 2.3–46.6). Interestingly, risk of metastatic recurrence in patients who failed to achieve pCR but were ctDNA-negative was similar to those who achieved pCR (HR 1.4, 95% CI, 0.15–13.5). PPV and NPV were 67% (4/6) and 93% (50/54), respectively. A landmark analysis (using T3 as the starting point) was performed and revealed similar results (Supplementary Fig S8). In an exploratory multivariable Cox regression analysis, ctDNA-positivity after NAC was a significant predictor of poor DRFS (Table 1).
Table 1.
Multivariate Cox regression analysis to determine association between ctDNA-positivity after NAC (T3) and distant disease-free survival (DRFS) while controlling for pCR and subtype. HR-hormone receptor; TN-triple negative, pCR-pathologic complete response, CI-confidence interval.
| DRFS | ||||
|---|---|---|---|---|
|
|
||||
| Variable | Hazard Ratio | lower 0.95 CI | upper 0.95 CI | p-value |
|
| ||||
| ctDNA+ vs. ctDNA- at T3 | 14.9 | 2.66 | 83.11 | 0.0021 |
| pCR vs. no pCR | 0.4 | 0.04 | 4.45 | 0.4579 |
| HR+HER2- vs. TN | 0.8 | 0.16 | 3.98 | 0.7833 |
| HER2+ vs. TN | 3.6 | 0.39 | 33.80 | 0.2559 |
Discussion
In this study, we examined the role of personalized ctDNA as a predictive biomarker for response and outcome in the neoadjuvant setting. The cohort included early stage breast cancer patients with high-risk of recurrence and who were treated in the I-SPY 2 TRIAL.
ctDNA studies in neoadjuvant setting in breast cancer have recently been reported [28, 31–33] (Supplementary Table S3). Two of the four studies were limited to a particular breast cancer subtype (i.e., only TNBC [28] or only HER2+ [31]). Rothé and colleagues observed that ctDNA detection before NAC was associated with decreased likelihood of achieving a pCR [31]. McDonald and colleagues showed that non-responding patients have higher ctDNA levels after NAC compared to those achieved a pCR [32]. Two of the studies examined association between ctDNA and survival [28, 31], but none were able to demonstrate the prognostic impact of residual ctDNA after NAC. To our knowledge, our work represents the most comprehensive study on ctDNA detection in all subtypes, before, during, and after NAC and examined for the first time its association with response and survival in early breast cancer.
Here, we report on the use of a personalized ctDNA test informed by each patient’s tumor genotype. We found that ctDNA is frequently detected in untreated high-risk early stage population (~70% of patients). The patterns of change in ctDNA during NAC were significantly correlated with risk of metastatic recurrence. We also found that ctDNA testing early during NAC (at 3 weeks) provided potentially actionable information as persistent ctDNA identified patients who were unlikely to achieve a pCR, whereas clearance was associated with improved response.
ctDNA positivity rate at baseline was significantly different among breast cancer subtypes (HR+/HER2-: 52%, HER2+: 82%, TNBC: 89%). We speculate that the lower rate of ctDNA positivity in HR+ breast cancer compared to HER2+ and TNBC is due in part to the lower proliferation rates (lower expression of Ki67 [34]) in this subtype, as was observed by Abbosh and colleagues in lung cancer [34]. Elucidating molecular and genomic factors predictive of ctDNA presence in the blood may shed on light on the biology of ctDNA release and clearance during treatment.
We examined whether ctDNA status at different time points was associated with risk of metastatic recurrence. We found that ctDNA-positive patients at T1, T2, and T3 (but not at T0) had significantly inferior DRFS compared to those who were ctDNA-negative. We observed that PPV and hazard ratio increased with time, indicating that the last time point (i.e., after NAC before surgery) may be most informative for risk-stratification of patients, and thus potentially help guide treatment in the adjuvant setting.
Our study showed that ctDNA status after NAC can potentially stratify patients who did not achieve pCR into low- and high-risk groups (Fig 5C). We found that clearance of ctDNA after NAC was associated with improved survival even in patients who did not achieve pCR. If validated, these findings could have a profound impact on treatment management in the neoadjuvant and adjuvant settings.
Recent clinical studies in breast cancer have shown that additional adjuvant therapy for non-responders to NAC can lead to improvements in patient outcomes [35, 36]. Future studies should take into account the potential confounding effects of adjuvant treatment on the prognostic performance of ctDNA and other biomarkers analyzed in the neoadjuvant setting.
Differences in prognostic value of pCR by subtype have been reported, including its poor association with prognosis in HR+ breast cancer [21, 37]. Survival analysis in I-SPY 2 involving 950 patients have shown that pCR and subtype (including HR+) were strongly associated with DRFS [23]. In this subset, the individual prognostic impact of pCR and subtype were not observed, perhaps due to the modest sample size. Our exploratory survival analysis did show that ctDNA after NAC was a strong prognostic factor for DRFS. Further studies in larger cohorts are warranted to examine the contributions of ctDNA, pCR and subtype in predicting outcomes of patients who received NAC.
The I-SPY 2 schema includes the collection of serial magnetic resonance imaging (MRI) data during NAC to assess tumor response [38, 39]. We have previously analyzed paired ctDNA and MRI data collected at the same time points in the same cohort as this present study [40]. We found that MRI-based functional tumor volume (FTV)—a clinically established measure of residual disease in the breast [38, 41]—was significantly correlated with ctDNA levels at all time points [40]. Furthermore, we found that ctDNA status after NAC improved the performance of FTV as predictor of metastatic recurrence and death. ctDNA testing could therefore serve as complementary tool to MRI for risk stratification of patients post-NAC.
A number of technologies for detection of ctDNA have been developed and are described in detail in a recently published review [42]. Our approach provides several advantages over other methods of ctDNA analysis. The upfront whole exome sequencing of primary tumors enables personalized selection of ctDNA targets that is independent of driver status. Our assay simultaneously tracks up to 16 patient-specific somatic variants and thus offers a more robust representation of the heterogeneity of a patient’s tumor [26, 27, 43]. In contrast, other methods like droplet digital polymerase chain reaction (ddPCR) [8] or BEAMing [44] track only one to a few somatic variants. Our ctDNA test does have certain limitations including the inability to detect new second primary cancers which are often genetically unrelated to the original cancer [45]; also, it will miss novel somatic variants that arise during tumor evolution in response to therapy-mediated selection pressures [46].
Clonal hematopoiesis of indeterminate potential (CHIP) mutations are potential sources of false positives in sequencing analyses of cfDNA [47, 48]. The ctDNA detection approach used in this study filters out CHIP mutations by focusing only on tumor-specific mutations that were initially detected by whole exome sequencing of paired pretreatment tumor and germline DNA.
In the light of our findings, novel paradigms for ctDNA-directed treatment can be envisioned in future clinical trials. The current I-SPY 2 schema provides patients a single therapeutic opportunity to achieve a pCR [49] (Fig 1A). In the next iteration of the trial, patients will be given options to receive additional treatment to improve their chances of achieving a pCR, i.e., if the initial agent does not result in a predicted complete response. For example, the decision to switch therapy for patient without an early clinical or imaging response to a novel therapeutic agent would be supported if the patient fails to clear ctDNA. On the other hand, patients who clear their ctDNA could continue treatment. Furthermore, information from ctDNA testing after NAC may help guide clinical decisions on whether to escalate or de-escalate treatment in the adjuvant setting. For example, if clearance of ctDNA is confirmed as a predictor of low risk of metastatic recurrence, such information can support treatment de-escalation.
Analysis of pooled serial circulating tumor cell (CTC) data obtained during neoadjuvant treatment of early breast cancer revealed that the prognostic impact of CTCs was the strongest at pretreatment (prior to NAC) compared to other time points [50]. In contrast, our study showed that ctDNA status after NAC appeared to be the most important time point for prognostication. While both blood-based biomarkers display prognostic impact, their clinical value may not be redundant [51]. Further studies that contemporaneously assess ctDNA and CTCs in the neoadjuvant setting are needed to elucidate the relative contributions of each biomarker in predicting response and outcome.
The focus of this study was to examine the clinical significance of ctDNA monitoring in the neoadjuvant setting. The detection of minimal residual disease after surgery is of great clinical importance and can provide a unique opportunity for treatment redirection to delay metastatic recurrence and improve patient outcomes. We have now expanded our studies to include post-surgical monitoring of ctDNA in the adjuvant setting with the focus on residual disease detection after surgery and recurrence prediction.
In summary, our study shows promise that early response prediction by highly sensitive ctDNA analysis in high-risk early breast cancer patients may facilitate a timely and judicious change in treatment to improve patients’ chances of achieving favorable long-term outcomes. The I-SPY 2 TRIAL provides an excellent platform to investigate how personalized ctDNA testing can complement imaging [52] and pathologic evaluation [53] of tumor response to fine-tune pCR as a surrogate endpoint for improved survival. Dynamic monitoring of ctDNA during NAC could facilitate evaluation of new agents by providing an early endpoint of treatment efficacy. Response over time as measured by imaging and ctDNA in the setting of early (pCR) and late (DRFS) outcomes will provide a robust framework for elucidating the potential clinical utility of ctDNA in the neoadjuvant setting.
Supplementary Material
Acknowledgments:
The authors thank the patients and their families and the I-SPY 2 TRIAL Investigators for their participation in this study.
Funding:
This study was funded in part by the Breast Cancer Research Foundation (BCRF) (L.J.V. Veer), and the Breast Cancer Research Fund at UCSF (L.J.V. Veer) and NIH/NCI P01CA210961 (L.J. Esserman). We are grateful to Dr. Matthew Rabinowitz for his support and insightful discussions of the project. ctDNA analysis was provided by Natera without cost.
Footnotes
Conflict of interest disclosures: The following authors are employees of Natera, Inc. (Himanshu Sethi, Hsin-Ta Wu, Raheleh Salari, Antony Tin, Svetlana Shchegrova, Hemant Pawar, Paul Billings, Alexey Aleshin, Maggie Louie, Bernhard Zimmermann). Laura van ‘t Veer is co-founder, stockholder and part-time employee of Agendia NV. The rest of the authors declare no potential conflicts of interest.
References:
- 1.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nature reviews Clinical oncology 2017; 14: 531–548. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson J, Park BH. Circulating Tumor DNA: Measurement and Clinical Utility. Annu Rev Med 2018; 69: 223–234. [DOI] [PubMed] [Google Scholar]
- 3.Rossi G, Ignatiadis M. Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine. Cancer Res 2019; 79: 2798–2804. [DOI] [PubMed] [Google Scholar]
- 4.Chen YH, Hancock BA, Solzak JP et al. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 2017; 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Murillas I, Schiavon G, Weigelt B et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015; 7: 302ra133. [DOI] [PubMed] [Google Scholar]
- 6.Olsson E, Winter C, George A et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 2015; 7: 1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Murillas I, Chopra N, Comino-Mendez I et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson SJ, Tsui DW, Murtaza M et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 9.Murtaza M, Dawson SJ, Pogrebniak K et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015; 6: 8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coombes RC, Page K, Salari R et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res 2019. [DOI] [PubMed] [Google Scholar]
- 11.Merker JD, Oxnard GR, Compton C et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018; 36: 1631–1641. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Hou L, Chen M et al. Neoadjuvant Chemotherapy Creates Surgery Opportunities For Inoperable Locally Advanced Breast Cancer. Sci Rep 2017; 7: 44673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hylton NM, Gatsonis CA, Rosen MA et al. Neoadjuvant Chemotherapy for Breast Cancer: Functional Tumor Volume by MR Imaging Predicts Recurrence-free Survival-Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology 2016; 279: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMichele A, Yee D, Berry DA et al. The Neoadjuvant Model Is Still the Future for Drug Development in Breast Cancer. Clin Cancer Res 2015; 21: 2911–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry DA, Hudis CA. Neoadjuvant Therapy in Breast Cancer as a Basis for Drug Approval. JAMA Oncol 2015; 1: 875–876. [DOI] [PubMed] [Google Scholar]
- 16.Derks MGM, van de Velde CJH. Neoadjuvant chemotherapy in breast cancer: more than just downsizing. Lancet Oncol 2018; 19: 2–3. [DOI] [PubMed] [Google Scholar]
- 17.Early Breast Cancer Trialists’ Collaborative G. Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol 2018; 19: 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Minckwitz G, Untch M, Blohmer JU et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 2012; 30: 1796–1804. [DOI] [PubMed] [Google Scholar]
- 19.Symmans WF, Wei C, Gould R et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J Clin Oncol 2017; 35: 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Symmans WF, Peintinger F, Hatzis C et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 2007; 25: 4414–4422. [DOI] [PubMed] [Google Scholar]
- 21.Cortazar P, Zhang L, Untch M et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014; 384: 164–172. [DOI] [PubMed] [Google Scholar]
- 22.Rugo HS, Olopade OI, DeMichele A et al. Adaptive Randomization of Veliparib-Carboplatin Treatment in Breast Cancer. N Engl J Med 2016; 375: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium IST, Yee D, DeMichele AM et al. Association of Event-Free and Distant Recurrence-Free Survival With Individual-Level Pathologic Complete Response in Neoadjuvant Treatment of Stages 2 and 3 Breast Cancer: Three-Year Follow-up Analysis for the I-SPY2 Adaptively Randomized Clinical Trial. JAMA Oncol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathy D, Chien AJ, Hylton N et al. Adaptively randomized trial of neoadjuvant chemotherapy with or without the Akt inhibitor MK-2206: Graduation results from the I-SPY 2 Trial. Journal of Clinical Oncology 2015; 33: 524-524.25584001 [Google Scholar]
- 25.Abbosh C, Birkbak NJ, Wilson GA et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017; 545: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen E, Birkenkamp-Demtroder K, Sethi H et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol 2019; JCO1802052. [DOI] [PubMed] [Google Scholar]
- 27.Reinert T, Henriksen TV, Christensen E et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riva F, Bidard FC, Houy A et al. Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin Chem 2017; 63: 691–699. [DOI] [PubMed] [Google Scholar]
- 29.Park JW, Liu MC, Yee D et al. Adaptive Randomization of Neratinib in Early Breast Cancer. N Engl J Med 2016; 375: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bratman SV, Yang SYC, Iafolla MAJ et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nature Cancer 2020; 1: 873–881. [DOI] [PubMed] [Google Scholar]
- 31.Rothe F, Silva MJ, Venet D et al. Circulating tumor DNA in HER2 amplified breast cancer: a translational research substudy of the NeoALTTO phase 3 trial. Clin Cancer Res 2019. [DOI] [PubMed] [Google Scholar]
- 32.McDonald BR, Contente-Cuomo T, Sammut SJ et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler TM, Boniface CT, Johnson-Camacho KM et al. Circulating Tumor DNA Dynamics using Patient-Customized Assays are Associated with Outcome in Neoadjuvantly-Treated Breast Cancer. Cold Spring Harb Mol Case Stud 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang Q, Ma D, Gao RF, Yu KD. Effect of Ki-67 Expression Levels and Histological Grade on Breast Cancer Early Relapse in Patients with Different Immunohistochemical-based Subtypes. Sci Rep 2020; 10: 7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Minckwitz G, Huang CS, Mano MS et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N Engl J Med 2019; 380: 617–628. [DOI] [PubMed] [Google Scholar]
- 36.Masuda N, Lee SJ, Ohtani S et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N Engl J Med 2017; 376: 2147–2159. [DOI] [PubMed] [Google Scholar]
- 37.Gustavo Werutsky G, Untch M, Hanusch C et al. Locoregional recurrence risk after neoadjuvant chemotherapy: A pooled analysis of nine prospective neoadjuvant breast cancer trials. Eur J Cancer 2020; 130: 92–101. [DOI] [PubMed] [Google Scholar]
- 38.Hylton NM, Blume JD, Bernreuter WK et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy--results from ACRIN 6657/I-SPY TRIAL. Radiology 2012; 263: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Newitt DC, Gibbs J et al. Predicting breast cancer response to neoadjuvant treatment using multi-feature MRI: results from the I-SPY 2 TRIAL. npj Breast Cancer 2020; 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magbanua MJ, Li W, Wolf DM et al. Abstract: Circulating tumor DNA (ctDNA) and magnetic resonance imaging (MRI) for monitoring and predicting response to neoadjuvant therapy (NAT) in high-risk early breast cancer patients in the I-SPY 2 TRIAL. AACR Advances in Liquid Biopsies Conference 2020. [Google Scholar]
- 41.Tromberg BJ, Zhang Z, Leproux A et al. Predicting Responses to Neoadjuvant Chemotherapy in Breast Cancer: ACRIN 6691 Trial of Diffuse Optical Spectroscopic Imaging. Cancer Res 2016; 76: 5933–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buono G, Gerratana L, Bulfoni M et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat Rev 2019; 73: 73–83. [DOI] [PubMed] [Google Scholar]
- 43.Coombes RC, Page K, Salari R et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res 2019; 25: 4255–4263. [DOI] [PubMed] [Google Scholar]
- 44.Diehl F, Schmidt K, Choti MA et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Begg CB, Ostrovnaya I, Geyer FC et al. Contralateral breast cancers: Independent cancers or metastases? Int J Cancer 2018; 142: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiavon G, Hrebien S, Garcia-Murillas I et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 2015; 7: 313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Chen X, Wang J et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann Oncol 2019; 30: 464–470. [DOI] [PubMed] [Google Scholar]
- 48.Razavi P, Li BT, Brown DN et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019; 25: 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esserman L, Hylton N, Asare S et al. I-SPY2: Unlocking the Potential of the Platform Trial. In Antonijevic Z, Beckman RA (eds): Platform Trial Designs in Drug Development: Umbrella Trials and Basket Trials Chapman and Hall/CRC) 2018; 3–22. [Google Scholar]
- 50.Bidard FC, Michiels S, Riethdorf S et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J Natl Cancer Inst 2018; 110: 560–567. [DOI] [PubMed] [Google Scholar]
- 51.Pierga J-Y, Silveira A, Girard E et al. Abstract 3390: Predictive and prognostic value of circulating tumor DNA (ctDNA) compared to circulating tumor cells (CTC) in a prospective cohort of metastatic breast cancer patients: The UCBG COMET trial. Cancer Research 2020; 80: 3390-3390. [Google Scholar]
- 52.Li W, Newitt DC, Wilmes LJ et al. Additive value of diffusion-weighted MRI in the I-SPY 2 TRIAL. J Magn Reson Imaging 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell JI, Yau C, Krass P et al. Comparison of residual cancer burden, American Joint Committee on Cancer staging and pathologic complete response in breast cancer after neoadjuvant chemotherapy: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res Treat 2017; 165: 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.