Abstract
The mannose-sensitive hemagglutinin (MSHA) of the Vibrio cholerae O1 El Tor biotype is a member of the family of type 4 pili. Type 4 pili are found on the surface of a variety of gram-negative bacteria and have demonstrated importance as host colonization factors, bacteriophage receptors, and mediators of DNA transfer. The gene locus required for the assembly and secretion of the MSHA pilus has been localized to a 16.7-kb region of the V. cholerae chromosome. Sixteen genes required for hemagglutination, including five that encode prepilin or prepilin-like proteins, have been identified. Examination of MSHA-specific cDNAs has localized two promoters that drive expression of these genes. This evidence indicates that the MSHA gene locus is transcriptionally organized into two operons, one encoding the secretory components and the other encoding the structural subunits, an arrangement unique among previously characterized type 4 pilus loci. The genes flanking the MSHA locus encode proteins that show homology to YhdA and MreB of Escherichia coli. In E. coli, the yhdA and mreB genes are adjacent to each other on the chromosome. The finding that the MSHA locus lies between these two E. coli homologs and that it is flanked by a 7-bp direct repeat suggests that the MSHA locus may have been acquired as a mobile genetic element.
Type 4 pili are thin, 6- to 7-nm fibers elaborated by a wide variety of gram-negative bacterial species (50). Type 4 pili are composed of pilin subunits and are classified as either type 4a or type 4b according to amino acid sequence similarities within the amino terminus of the subunit polypeptide. The pilin monomers are synthesized as precursor proteins with a hydrophilic leader peptide of variable length that is processed at a consensus cleavage site by a type 4 prepilin peptidase during pilin secretion. The majority of type 4 pili belong to the type 4a subclass. Type 4a prepilin subunits are characterized by a short five- to six-amino acid leader sequence, which upon cleavage results in a mature pilin subunit with an N-terminal methylated phenylalanine residue. Type 4a pili tend to be distributed either peritrichously or in a polar position on the bacterial cell surface. Type 4b pili are primarily associated with enteric bacteria, namely, Vibrio cholerae, enteropathogenic Escherichia coli, and enterotoxigenic E. coli. A type 4b pilus is also encoded on the broad-host-range conjugal plasmid R64 (29). Type 4b prepilins tend to have leader peptides longer than those of the type 4a subclass and the N-terminal amino acid of the mature pilin subunit is variable, being either methionine, leucine, or tryptophan (13, 17, 53). Pili composed of type 4b pilin subunits have a tendency to form large bundles of laterally associated fibers. Type 4 pili of either subclass can promote bacterial aggregation, a property which may contribute to colonization mechanisms, biofilm formation, or stabilization of mating pairs (29, 37, 53).
Assembly and secretion of type 4 pili requires the expression of numerous gene products, including the structural prepilin subunit and its cognate prepilin peptidase (22). In addition, biogenesis of type 4 pili requires many ancillary proteins whose exact function in pilus assembly is unclear. These include predicted inner and outer membrane proteins and cytoplasmic nucleotide-binding proteins which may provide the energy necessary for translocation (22). The genetic organization of the loci required for type 4 pilus biogenesis tends to vary according to the pilus subclass. Genes required for the synthesis and secretion of type 4b pili tend to be organized as a single operon associated with a plasmid or pathogenicity island, whereas the genes encoding similar functions with respect to type 4a pili tend to be distributed to several locations throughout the chromosome (16, 56).
V. cholerae O1, the etiologic agent of the acute diarrheal disease cholera, expresses both subclasses of type 4 pili. The toxin coregulated pilus (TCP), a member of the family of type 4b pili, is a major determinant in the establishment of V. cholerae colonization of the small intestine (21, 51, 53). In addition, TCP serves as the receptor for the filamentous phage, CTXΦ, which encodes the potent cholera exotoxin that is ultimately responsible for the severe diarrhea associated with the disease (58). The genes responsible for TCP elaboration, as well as accessory colonization factor (ACF) genes, are encoded within a 12-kb operon located on the TCP-ACF pathogenicity island (28, 30). Expression of TCP and cholera toxin genes is coordinately regulated in response to specific environmental cues as part of the ToxR regulatory cascade (48).
Epidemic cholera is associated with strains of the O1 and O139 serogroups. The O1 serogroup is further divided into two biotypes, El Tor and classical, based on a variety of phenotypic characteristics such as differential antibiotic and phage sensitivities. Strains of the O139 serogroup are closely related to those of the O1 El Tor biotype (19). One feature which distinguishes the El Tor from the classical O1 biotype is the presence of the mannose-sensitive hemagglutinin (MSHA) pilus on the El Tor cell surface (14, 25). MSHA is also expressed by strains of the O139 serogroup (1). The MSHA pilus, a member of the type 4a family of pili, is not required for colonization of humans or colonization in the infant mouse cholera model (4, 51, 54). While the exact function of MSHA is unknown, recent evidence demonstrates that the MSHA pilus serves as the receptor for the filamentous bacteriophage 493, which was isolated from a V. cholerae O139 strain and has been suggested to have a role in the horizontal evolution of V. cholerae (27). The MSHA pilus may also have a role in the environmental persistence or survival of El Tor V. cholerae. Recent studies have demonstrated that mshA mutants are unable to produce biofilms on abiotic surfaces (59). In this regard, biofilm formation may play an important role in mediating V. cholerae survival outside the living host.
Initial transposon mutagenesis identified two genetic loci required for mannose-sensitive hemagglutination of El Tor V. cholerae. One locus contained a set of contiguous open reading frames that showed significant homology to proteins of the general secretory pathway of gram-negative bacteria (20, 41). In a separate study, the gene encoding the major structural subunit of the pilus, mshA, was identified and shown to lie within a locus that included three additional genes encoding type 4 prepilin-like proteins (26). The subsequent finding that the secretory and structural gene loci are physically linked on the V. cholerae chromosome suggested that the genes encoding the MSHA pilus are organized into a potential pilus biogenesis operon (33). The organization of pilus biogenesis genes into a discrete cluster is atypical of type 4a pilus-encoding loci. In the present study, this genetic arrangement has been investigated by delineating the boundaries and examining the transcriptional organization of the MSHA gene locus.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids used in this study are described in Table 1. Bacteria were grown in Luria-Bertani (LB) liquid culture or solid media. Antibiotics were used at the following concentrations unless stated otherwise: ampicillin, 100 μg/ml; polymyxin B, 50 IU/ml; streptomycin, 100 μg/ml; and kanamycin, 45 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Refer-ence(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| SM10λpir | thi thr leu tonA lacY supE recA [RP4-2-Tc::Mu] λpirR6K Kmr | 52 |
| S17-1λpir | thi pro recA hsdR [RP4-2Tc::Mu-Km::Tn7] λpir Tpr Smr | 10 |
| MC4100 | F−araD139 Δ(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi Smr | 45 |
| V. cholerae | ||
| C6706str2 | C6706, spontaneous Smr | 54 |
| JM127 | C6706str2, mshL::pGP704 Apr | This study |
| JM157 | C6706str2, yhdA::pGP704 Apr | This study |
| JM191 | C6706str2, Φ(mshA-phoA)Hyb1 | This study |
| JM222 | JM191, mshF::TT500::mshB | This study |
| JM223 | C6706str2, mshF::TT500::mshB | This study |
| JM225 | C6706str2, yhdA::TT500::mshI | This study |
| JM227 | C6706str2, mshQ::pKAS32 Apr | This study |
| JM242 | JM191, ssb::TT500::yhdA | This study |
| JM265 | JM191, yhdA::TT500::mshI | This study |
| JM278 | C6706str2, mreB::pKAS32 Apr | This study |
| JM282 | C6706str2, ssb::TT500::yhdA | This study |
| Plasmids | ||
| pGP704 | oriR6K mobRP4 Apr | 32, 35 |
| pKAS32 | pGP704, rpsL Apr | 47 |
| pKAS46 | pKAS32, Apr Kmr | 47 |
| pRS415 | lac operon fusion vector, Apr | 46 |
| pBAD24 | araC rrnBT1T2 Apr | 18 |
| pJM8 | pGP704, ′mshL′ Apr | This study |
| pJM133 | pGP704, JM127 NcoI chromosomal capture, Apr | This study |
| pJM139 | pGP704, JM127 SmaI chromosomal capture, Apr | This study |
| pJM150 | pGP704, ′yhdA′ Apr | This study |
| pJM218 | pKAS46, ′mshF::TT500::mshB′ Apr | This study |
| pJM220 | pKAS46, ′yhdA::TT500::mshI′ Apr | This study |
| pJM226 | pKAS32, ′mshQ′ Apr | This study |
| pJM245 | pKAS32, JM227 NsiI chromosomal capture, Apr | This study |
| pJM246 | pRS415, mshB::lacZ Apr | This study |
| pJM247 | pRS415, mshB(rev)::lacZ Apr | This study |
| pJM271 | pKAS32, ′mreB′ Apr | This study |
| pJM281 | pKAS46, ′ssb::TT500::yhdA′ Apr | This study |
| pJM307 | pRS415, mshI::lacZ Apr | This study |
| pJM318 | pRS415, mshI(rev)::lacZ Apr | This study |
Abbreviations: Tpr, trimethoprim resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Gmr, gentamicin resistance.
Mutant construction.
All mutants used in this study were constructed by sequence-specific suicide plasmid integration into the V. cholerae chromosome. Plasmid disruption of mshL in C6706str2 resulted in JM127. Briefly, primers JM3 and JM4 (Table 2), carrying unique restriction enzyme sites, were used to PCR amplify an internal fragment of the mshL gene from C6706str2 genomic DNA. The resulting product was digested with the appropriate restriction enzymes and ligated into the multiple cloning site of the suicide vector pGP704. Following electroporation of this reaction mixture into SM10λpir cells and selection on LB agar containing ampicillin, transformants were screened by colony PCR with primer pair JM3-JM4, which identified plasmid pJM8 carrying the mshL fragment. Plasmid pJM8 was integrated into the V. cholerae chromosome by conjugation of SM10λpir(pJM8) with C6706str2 and selection on LB agar containing ampicillin and streptomycin. Southern blot analysis utilizing a digoxygenin-labeled probe complementary to mshK sequences confirmed the mshL plasmid integrant, JM127. Plasmid disruption of mshQ, mreB, and yhdA in C6706str2 was carried out in a similar fashion. Amplification of C6706str2 genomic DNA with gene-specific primer pair MSH33-MSH36, REB1-REB2, or EC1-EC2, each carrying unique restriction enzyme sites, resulted in the production of PCR fragments homologous to internal portions of mshQ, mreB, and yhdA, respectively. Each gene-specific fragment was digested with the appropriate restriction enzymes for ligation into the suicide vector pKAS32 (mshQ and mreB) or pGP704 (yhdA). S17-1λpir cells were electroporated with the mshQ or mreB ligation reaction mixture followed by selection on LB agar containing ampicillin. The yhdA construct was electroporated into SM10λpir cells followed by selection on ampicillin. The resulting transformants were screened for the correct insert by colony PCR with the appropriate sets of gene-specific primers described above. In this manner, plasmid constructs pJM226, pJM271, and pJM150, corresponding to gene fragments mshQ, mreB, and yhdA, respectively, were identified. Integration of each construct into the V. cholerae chromosome was carried out by conjugation with C6706str2. The mshQ and mreB exconjugants were selected on LB agar containing polymyxin B and ampicillin, while the yhdA exconjugants were selected in the presence of streptomycin and ampicillin. The mreB plasmid integrant strain, JM278, was identified by colony PCR using primer R6K1, which is specific to the origin of replication on the integrated pKAS32 vector, directed toward the 5′ end of mreB on the V. cholerae chromosome and primer MSH56, which is specific to sequences upstream of mreB but outside the region of homology on the integrating plasmid and directed toward the insertion site. The mshQ plasmid insertion mutant, JM227, was identified by colony PCR in a similar fashion, using primers R6K1 and MSH30. Southern blot analysis using a digoxygenin-labeled probe specific to mshK confirmed yhdA plasmid integrant strain, JM157.
TABLE 2.
Oligonucleotides used in this study
| Oligo-nucleotide | Sequence |
|---|---|
| EC1 | 5′-CTAGAGATCTGGCGATTAAGTTGATTGAGCGC-3′ |
| EC2 | 5′-CTAGGAATTCGCGAATCCAGCGAGCATACCG-3′ |
| JM3 | 5′-CTAGTCTAGACATCGTGATCCTGTTGAAGC-3′ |
| JM4 | 5′-CTAGGAATTCGCCTTGGCTTCGACATCTC-3′ |
| KAN4 | 5′-TCGAGCAAGACGTTTCCCG-3′ |
| MSP1 | 5′-CTAGTCTAGACACTCTCGGTAAACTTGCAC-3′ |
| MSP2 | 5′-CTAGGAATTCCGACACCGCCATGCTTTGC-3′ |
| MSP3 | 5′-CTAGGAGCTCTGACACCCAGCATCTGTAAGG-3′ |
| MSP4 | 5′-CTAGGAGCTCGGCAAACGTAATCGATGGCG-3′ |
| MSP5 | 5′-CTAGTCTAGATCGTGAAGTCGACTATG-3′ |
| MSP6 | 5′-CTAGGAGCTCTCAGCGCATCAAAAAACTGGC-3′ |
| MSP7 | 5′-TTTTGCCTGCTCATGGCTAG-3′ |
| MSP8 | 5′-CTAGGAATTCTCTTTCATGTGAATACGCAGC-3′ |
| MSH30 | 5′-GGATTCCCTACACCAGTAGCG-3′ |
| MSH33 | 5′-CTAGTCTAGAGTTGGGTGTCCGCTTACATG-3′ |
| MSH36 | 5′-CTAGGAATTCAGATCTTGCTCATCACCGCT-3′ |
| MSH38 | 5′-CTAGGAGCTCAATACTAAAACGGGATGTCG-3′ |
| MSH42 | 5′-CTAGTCTAGAGTGCGTTATATGCCAAGC-3′ |
| MSH43 | 5′-CTAGGAGCTCGCACATAAGGAACGTGGC-3′ |
| MSH44 | 5′-CTGAGAATTCAATACGATCAGTAGCACCGC-3′ |
| MSH49 | 5′-ACAAATTCATGTTGGCTGCC-3′ |
| MSH50 | 5′-GATCAAGCTTTCCGCCACTACATCCGTCAC-3′ |
| MSH56 | 5′-TGTGATGGCAAGGCATTGG-3′ |
| REB1 | 5′-CATGAGATCTCGATTTAGGTACCGCCAAC-3′ |
| REB2 | 5′-CATGAATTCGCACTTCGATCTCTTGCAC-3′ |
| TT3 | 5′-TTTAGAGCTCGCTTGGCTGTTTTGGCGGATGAGAG-3′ |
| TT4 | 5′-GATGGAGCTCAAGAGTTTGTAGAAACGCAAAAAGGCCATCCG-3′ |
| LAC1 | 5′-GTCATAGCTGTTTCCTGTGTG-3′ |
| R6K1 | 5′-GGTTTAACGGTTGTGGACAAC-3′ |
Transcription terminator constructs.
A 500-bp fragment of the E. coli rrnB transcription terminator (6) (TT500) was inserted upstream of the yhdA, mshI, or mshB coding sequence in both C6706str2 and the isogenic mshA-phoA strain, JM191, by the following method. Two 500-bp fragments flanking either the yhdA, mshI, or mshB upstream region were generated by amplification of C6706str2 genomic DNA with a set of primer pairs, MSH42-MSH38 and MSH43-MSH44, MSP1-MSP3 and MSP4-MSP2, or MSP5-MSP6 and MSP7-MSP8, respectively. The primers carry restriction enzyme sites for ligation into the multiple cloning site of the suicide allelic exchange vector, pKAS46. A SacI site was specifically incorporated into the primers of each set that flank the region to be exchanged with TT500. Ligation reaction mixtures were electroporated into S17-1λpir cells, and transformants were selected on LB agar containing ampicillin. Isolated colonies were screened by PCR with the relevant flanking 5′ and 3′ primer pair described above to identify correct yhdA, mshI, and mshB plasmid constructs. TT500, generated by PCR amplification of pBAD24 DNA with primers TT3 and TT4, each of which carries a SacI site, was introduced into the SacI site of each plasmid construct. The correct orientation of the TT500 insert was determined by PCR using TT4 and the corresponding 5′ primer for each TT500 plasmid construct—MSH42, MSP1, or MSP5. S17-1λpir cells carrying the yhdA, mshI, and mshB TT500 plasmid derivatives pJM281, pJM220, and pJM218, respectively, were mated with either C6706str2 or the isogenic mshA-phoA gene fusion strain, JM191, and exconjugants were selected on LB agar in the presence of polymyxin B and kanamycin. Selected exconjugants were screened for plasmid integration at the correct site on the chromosome by PCR with the KAN4 primer, specific to the N-acetyltransferase gene sequence on pKAS46, and a 3′ primer specific to chromosomal sequences outside of the region of homology on the integrated plasmid. Plasmid loss from all confirmed C6706str2 and JM191 integrants was carried out in the presence of streptomycin (1 mg/ml) at 30°C as previously described (47). C6706str2 streptomycin-resistant recombinants were screened by PCR for insertion of TT500 upstream of yhdA, mshI, and mshB, using the appropriate primer pair flanking each insertion site, and resulted in the identification of strains JM282, JM225, and JM223, respectively. Similarly, insertion of TT500 upstream of yhdA, mshI, and mshB in JM191 resulted in JM242, JM265, and JM222, respectively.
Operon fusion construction.
PCR products carrying the promoter region upstream of mshI or mshB were generated by amplification of C6706str2 genomic DNA with primer pair EC1-EC2 or MSP5-MSP6, respectively. The resulting PCR products were blunt-end ligated into the unique SmaI site located upstream of the promoterless lacZ gene on plasmid pRS415. The lacZ deletion strain, MC4100, was electroporated with either ligation reaction mixture, with selection on LB agar containing ampicillin. Plasmids containing promoter inserts were identified by colony PCR using the original primer pair, EC1-EC2 or MSP5-MSP6. Orientation of the mshI promoter fragment was established by PCR amplification of plasmid DNA with MSP5 or MSP6 and the LAC1 primer, which is homologous to lacZ sequences on pRS415 and directed toward the cloned mshI promoter fragment. This resulted in the identification of operon fusion plasmid pJM307, carrying a 500-bp mshI promoter fragment in the forward orientation, and plasmid pJM318, containing the mshI promoter fragment in the opposite orientation [mshI(rev)]. A SalI site on the cloned mshB promoter fragment and in the plasmid backbone allowed the orientation of the mshB promoter with respect to lacZ. Restriction digest of plasmid DNA identified the operon fusion pJM246 carrying a 700-bp mshB promoter fragment in the forward orientation and plasmid pJM247, which contains the mshB promoter directed away from the lacZ sequence [mshB(rev)].
Chromosomal capture.
Chromosomal capture (49) was used to clone a region containing the 5′ portion of the msh locus and adjacent sequences. Genomic DNA from strain JM127, which carries a pGP704 plasmid insertion in the mshL gene, was digested with NcoI, self-ligated, and electroporated into strain S17-1λpir, with selection on LB agar containing ampicillin. The resulting plasmid, pJM133, carries a chromosomal NcoI fragment containing approximately 5 kb of sequence upstream of the yhdA gene. The chromosomal region encompassing the 3′ portion of the msh locus and beyond was similarly cloned from JM127 by ligation of SmaI-digested chromosomal DNA and resulted in plasmid pJM139, which carries approximately 4 kb of DNA downstream of mshC. Chromosomal capture of sequences downstream of mshQ was carried out by NsiI digestion of JM227 genomic DNA and resulted in the isolation of pJM245, which carries approximately 2 kb of DNA downstream of mshQ. Plasmid DNA from each construct was purified by anion-exchange chromatography (Qiagen, Inc.) and used in subsequent automated DNA sequence analysis.
Sequence analysis.
Plasmid DNA obtained by chromosomal capture was sequenced by using a Prism dye terminator ready reaction kit (Applied Biosystems, Inc. [ABI], Foster City, Calif.). Briefly, 1 μg of purified plasmid DNA and 3.2 pmol of oligonucleotide primer were mixed with 8 μl of ABI dye terminator mix and brought to a final 20-μl volume with distilled deionized water. The mixture was subjected to 25 cycles of 96°C for 30 s, 50°C for 15 s, and 60°C for 4 min, followed by Centrisep column purification (Princeton Separations) and gel analysis on an ABI model 373 Stretch automated DNA sequencer. Oligonucleotide primers homologous to known MSHA sequences included in the chromosomal capture were designed such that sequence analysis could proceed from both ends and on both strands of the captured DNA. Each primer was used in six separate sequencing reactions to ensure the fidelity of the sequence data. Subsequent oligonucleotide primers were designed in accordance with the resulting consensus sequence. DNAstar and ABI software packages were used for sequence analysis and alignment.
5′ RACE.
To identify transcription start sites, we used the 5′ RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Gibco BRL). Briefly, total RNA (1 μg) isolated from log-phase C6706str2 and JM225 cells by RNeasy silica gel membrane column (Qiagen) purification was reverse transcribed into cDNA with primer MSP2 or MSP8, specific to mshI or mshB, respectively. A control tube without reverse transcriptase was included for each primer to ensure that the resulting product was due to the amplification of cDNA and not contaminating chromosomal DNA. The cDNA was GlassMax column purified (Gibco BRL), poly(dC) tailed with terminal deoxynucleotidyltransferase, and subjected to PCR using mshI or mshB gene-specific nested primer MSH50 or MSH49, respectively, and the 5′ RACE abridged anchor primer, which contains 3′ sequence complementary to the homopolymeric poly(dC) tail. The resulting PCR product was reamplified with either MSH50 (for the mshI-specific product) or MSH49 (for the mshB-specific product) and the 5′ RACE universal amplification primer, which is complementary to the abridged anchor primer. The resulting products were silica gel membrane purified on Qiaquik columns (Qiagen), and 50 ng of the purified product was used as the template for automated sequence analysis with mshI or mshB gene-specific primer MSP3 or MSP6, respectively.
Hemagglutination assay.
Hemagglutination assays were carried out in 96-well round-bottom microtiter plates in a final volume of 200 μl. Bacteria (∼107) were serially diluted in Krebs-Ringer buffer (15). CD-1 mouse (Charles River Laboratories) erythrocytes were collected in the presence of heparin, washed three times, and resuspended at a final concentration of 1% in Krebs-Ringer buffer. An equal volume of serially diluted bacteria was mixed with the mouse erythrocytes and incubated at room temperature for 1 h. Hemagglutination titers are expressed as the reciprocal of the highest bacterial dilution that gave strong hemagglutination.
β-Galactosidase and alkaline phosphatase assays.
β-Galactosidase assays were performed on mid-logarithmic-phase cultures of MC4100 carrying lacZ operon fusion plasmid pJM246, pJM247, pJM307, and pJM318. Alkaline phosphatase assays were performed on overnight cultures of mshA-phoA gene fusion strains JM191, JM222, JM242, and JM265. Assays were performed as previously described (32).
Nucleotide sequence accession numbers.
The DNA sequence generated has been entered in the GenBank database under accession no. AF079406 (which corresponds to the partial coding sequence of SSB (single-stranded DNA-binding protein) and the complete coding sequence of YhdA) and AF079234 (which corresponds to the 3′ coding sequence of the MSHA operon including the partial coding sequence of MshD, the complete coding sequences of MshO, MshP, and MshQ, and the complete coding sequence of MreB). Accession no. AF079781 corresponds to the partial coding sequence of UvrA.
RESULTS
Identification and characterization of additional msh genes.
Previous studies identified 13 open reading frames required for MSHA pilus biogenesis (20, 26, 33). In the present study, three additional open reading frames downstream of mshD were identified by chromosomal capture and sequencing of V. cholerae El Tor genomic DNA (Fig. 1). These genes, designated mshO, mshP, and mshQ, are contiguous with and oriented in the same direction as the previously described msh genes. Predicted protein homologies and functions for the entire MSHA gene locus and nearby genes are listed in Table 3. The predicted gene product of mshO resembles a type 4 prepilin subunit in that it contains a consensus prepilin peptidase cleavage site and a C-terminal pair of cysteine residues which may form the characteristic pilin subunit disulfide hairpin (40). The predicted mshO gene product has a leader sequence that is only four amino acids long but contains invariant amino acids at positions −1, +1, +3, and +5 relative to the peptidase cleavage site and maintains overall N-terminal amino acid residue conservation characteristic of the type 4 prepilins. The mshO gene is the last of five consecutive open reading frames that encode type 4 prepilin or prepilin-like proteins. The deduced amino acid sequence of the mshP gene shows 52% homology to OutG of Erwinia chrysanthemi. This protein is a component of the E. chrysanthemi pectic enzyme secretion pathway and contains a type 4 prepilin leader sequence (31). While MshP does not appear to contain a consensus type 4a prepilin leader sequence, the presence of a charged amino terminus followed by a hydrophobic stretch of amino acids suggests that this protein is secreted across the bacterial inner membrane, potentially in a signal sequence-dependent manner. While the ultimate destination of the putative MshP protein to the periplasm or the outer membrane could not be predicted from its primary amino acid sequence, the OutG homology suggests that MshP may contribute to the secretion of pilin subunits and the assembly of the MSHA pilus fiber. Finally, the mshQ open reading frame encodes a product with a predicted molecular mass of 134.8 kDa that is likely to initiate with a valine residue, based on the location of the mshP stop codon. MshQ is likely to be an outer membrane protein, based on the presence of a potential signal sequence with a processing site at position 24 and the extensive predicted beta-sheet structure which occurs throughout the mature protein (36).
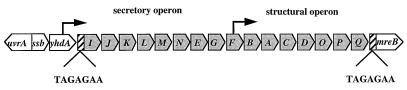
FIG. 1.
Schematic representation of the MSHA gene locus and flanking open reading frames. The MSHA gene locus is 16.7 kb in length and consists of 16 contiguous open reading frames which are flanked by a 7-bp direct repeat (▨). The three genes depicted upstream of the 5′ end (yhdA, ssb, and uvrA) and the one gene downstream of the 3′ end of the locus (mreB) are not involved in MSHA pilus biogenesis. The intragenic promoters required for the expression of MSHA are indicated by arrows.
TABLE 3.
Sizes, protein homologies, and predicted functions or locations of MSHA gene products
| Gene product | Predicted mol wt (103) | Homology
|
Predicted function or location | Reference(s) | |
|---|---|---|---|---|---|
| Protein | %a | ||||
| UvrA | 104 | H. influenzae UvrA | 93 | Excision repair | 9 |
| SSB | 16 | E. coli SSB | 98 | Single-stranded DNA binding | 43 |
| YhdA | 73.3 | E. coli YhdA | 60 | Hypothetical protein | 12 |
| MshI | 53.4 | None | Inner membraneb | 20 | |
| MshJ | 24.5 | E. carotovora OutM | 48 | Outer membrane | 20, 42 |
| MshK | 10.9 | None | Periplasmicb | 20 | |
| MshL | 60.1 | E. chrysanthemi OutD | 47 | Outer membrane | 31 |
| MshM | 31.7 | A. hydrophila ExeA | 56 | Cytoplasmic | 20, 24 |
| MshN | 42.6 | None | Inner membraneb | 20 | |
| MshE | 52.5 | V. cholerae EpsE | 63 | Cytoplasmic, ATP binding | 20, 44 |
| MshG | 44.8 | N. gonorrhoeae PilG | 55 | Inner membrane | 33, 55 |
| MshF | 17.9 | None | Periplasmicb | 26 | |
| MshB | 19.7 | Type 4 prepilin | Minor pilin, other | 26 | |
| MshA | 18.4 | Type 4 prepilin | Major pilin subunit | 26 | |
| MshC | 17.3 | Type 4 prepilin | Minor pilin, other | 26 | |
| MshD | 19.7 | Type 4 prepilin | Minor pilin, other | 26 | |
| MshO | 27.8 | Type 4 prepilin | Minor pilin, other | This study | |
| MshP | 15.2 | E. chrysanthemi OutG | 52 | Periplasm or outer membraneb | 31 |
| MshQ | 134.8 | None | Outer membraneb | This study | |
| MreB | 36.9 | E. coli MreB | 95 | Rod cell shape | 12 |
MSHA gene locus boundaries delineated.
Chromosomal capture and sequence analysis of C6706str2 El Tor genomic DNA upstream of the secretory genes at the 5′ region of the MSHA locus identified three open reading frames (Fig. 1). Immediately upstream of the mshI gene lies an open reading frame predicted to encode a gene product with 60% homology to YhdA of E. coli, a 73.3-kDa protein of unknown function encoded within the mreB-accB intergenic region (Table 3) (SWISS-PROT entry P13518). The predicted molecular mass of the presumed V. cholerae YhdA protein is 75 kDa. Upstream of yhdA and oriented in the same direction lies a gene that encodes a putative V. cholerae SSB (Fig. 1). The predicted amino acid sequence of this protein shows 96% homology to SSB (a protein involved in DNA replication, recombination, and repair) of many bacteria (Table 3). Therefore, this V. cholerae gene has been named ssb. An additional open reading frame discovered upstream of ssb was designated uvrA due to the significant amino acid homology of its predicted product with the Haemophilus influenzae excision repair protein (Table 3). The organization of the DNA repair genes observed in the V. cholerae chromosome, with the uvrA gene located upstream and adjacent to ssb but transcribed in the opposite direction, is evident in a number of bacterial genomes (11, 43). The location of these V. cholerae recombination-repair genes is indicative of the upstream MSHA gene locus boundary.
Downstream of mshQ at the 3′ region of the MSHA locus lies a gene predicted to encode a protein with 78% homology to MreB of a number of bacteria. The MreB protein is required for the formation of the characteristic rod shape in E. coli (12). This E. coli protein is encoded within a gene cluster which is involved in the regulation of bacterial cell division (57). Due to these observations and the significant homology to the E. coli MreB protein, the V. cholerae gene has been designated mreB. Interestingly, the yhdA and mreB genes, which are adjacent to one another on the E. coli chromosome, are separated by the MSHA gene locus on the V. cholerae chromosome. This observation is intriguing given the identification of a 7-bp direct repeat, TAGAGAA, located 5′ of mshI and 3′ of mshQ (Fig. 1). These findings suggest that the MSHA gene locus is delineated by these 7-bp direct repeats and raise the possibility that this locus was acquired by V. cholerae as a mobile genetic element that inserted between the yhdA and mreB genes of the ancestral V. cholerae chromosome.
Genetic confirmation of the MSHA gene locus boundaries.
To confirm the 5′ and 3′ boundaries of the MSHA gene locus, the genes surrounding the MSHA gene locus were subjected to mutational analysis. Plasmid disruption of yhdA and mreB gene sequence in the V. cholerae El Tor chromosome was performed to determine if the products of these genes are involved in MSHA pilus biogenesis. Hemagglutination of mouse erythrocytes was used to quantitate MSHA on the surface of the V. cholerae mutants. A hemagglutination assay performed with the C6706str2 yhdA and mreB mutant strains JM157 and JM278, respectively, demonstrated that neither of these genes is required for the MSHA phenotype. Both JM157 and JM278 displayed wild-type hemagglutination titers (Table 4). The wild-type hemagglutination titer observed for the yhdA mutant suggests that mshI is the first gene in the MSHA pilus biogenesis operon, since a TnphoA insertion in mshI abolishes hemagglutination in the El Tor strain N16961 (20).
TABLE 4.
Hemagglutination titers of C6706 derivatives
| Strain | Genotype | Titer |
|---|---|---|
| C6706str2 | Wild type | 128 |
| JM157 | yhdA::pGP704 | 128 |
| JM223 | mshF::TT500::mshB | <2 |
| JM225 | yhdA::TT500::mshI | <2 |
| JM227 | mshQ::pKAS32 | 32 |
| JM278 | mreB::pKAS32 | 128 |
| JM282 | ssb::TT500::yhdA | 128 |
Upstream of the mreB gene on the V. cholerae chromosome lies the mshQ gene. To determine if MshQ is the last protein encoded on the MSHA gene locus, plasmid disruption mutant JM227 was created and tested for its ability to hemagglutinate mouse erythrocytes. Disruption of MshQ function resulted in a 75% decrease in the hemagglutination titer compared to both the wild-type C6706str2 control strain and the mreB mutant (Table 4), indicating that mshQ is the last gene of the MSHA locus.
Transcriptional organization of the MSHA gene locus.
Type 4 pilus biogenesis operons are typically organized with the pilin gene located promoter proximal, followed immediately by a sequence that decreases transcription readthrough (transcriptional down sequence) and the pilus assembly and secretory genes. This organization allows for coordinate expression of all genes while maintaining proper stoichiometry of the pilin subunits relative to the assembly and secretory components of the pilus biogenesis operon. The organization of the MSHA pilus genes illustrated in Fig. 1 is unusual in that the secretory components are encoded upstream of the pilus structural components. This arrangement can best be explained if the MSHA secretory and structural components are in fact encoded on separate operons and regulated by individual promoters. Evidence for two operons is supported by the identification of potential consensus sigma 70 promoters located upstream of both mshI and mshB coding sequences (20, 26). To ascertain whether these promoters were necessary and sufficient for msh gene expression, the region encompassing either the mshI or the mshB putative promoter was deleted and replaced with the E. coli rrnB transcription terminator (TT500) in wild-type C6706str2 and isogenic mshA-phoA gene fusion strains. Deletion of the promoter region upstream of mshB and replacement with TT500 abolished mshA expression, as illustrated by the low level of alkaline phosphatase activity exhibited by strain JM222 compared to the control, JM191 (Table 5). Confirmation of this expression defect was provided by the hemagglutination-negative phenotype of the isogenic C6706str2 TT500 derivative, JM223 (Table 4). These data demonstrate that a promoter is not present immediately upstream of the mshA structural subunit gene and indicate that regulation of mshA expression must originate from a promoter located upstream of mshB.
TABLE 5.
Alkaline phosphatase activities of C6706 mshA-phoA derivatives
| Strain | Genotype | Activity (U)a |
|---|---|---|
| C6706str2 | Wild type | 20 ± 3 |
| JM191 | mshA-phoA | 1,985 ± 28 |
| JM222 | mshF::TT500::mshB | 37 ± 4 |
| JM265 | yhdA::TT500::mshI | 1,670 ± 158 |
| JM242 | ssb::TT500::yhdA | 1,847 ± 211 |
Average ± standard deviation of triplicate samples.
The possibility existed that one promoter located upstream of mshI was required to drive the expression of the entire MSHA gene locus. To address this possibility, the putative mshI promoter was replaced with TT500 in both C6706str2 and the JM191 mshA-phoA gene fusion strain. The TT500 mshA-phoA derivative, JM265, exhibited 80% of the control alkaline phosphatase activity, yet hemagglutination activity was completely abolished in the C6706str2 TT500 derivative, JM225 (Tables 4 and 5). This finding indicates that a promoter downstream of mshI is responsible for the majority of the transcriptional activity exhibited by the mshA-phoA gene fusions and that MSHA pilus biogenesis is completely dependent on expression from a promoter located upstream of mshI. Both JM282 and JM242, containing the TT500 terminator upstream of the yhdA gene, expressed wild-type levels of mshA, as reflected by their hemagglutination titers and alkaline phosphatase activities (Tables 4 and 5). These data indicate that a promoter upstream of mshI but downstream of yhdA is required for expression of the msh secretory genes, while a second promoter upstream of mshB but downstream of mshI is necessary for expression of the mshA structural subunit gene. These results support the two-promoter hypothesis for MSHA pilus expression, assembly, and secretion in V. cholerae.
Mapping the transcriptional start sites by 5′ RACE.
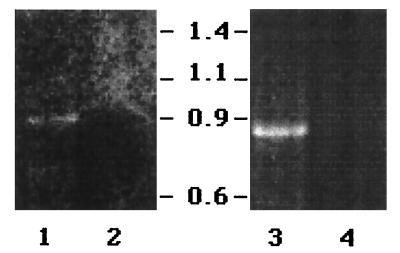
To establish the exact locations of the promoters driving msh gene expression, the 5′ ends of the corresponding mRNAs were determined by 5′ RACE. Primers specific to either the mshI or mshB transcript were used for subsequent cDNA synthesis and PCR amplification. The mshI-specific cDNA amplified a 900-bp product that was seen only in wild-type C6706str2 cells expressing the mshI transcript (Fig. 2, lane 1). No PCR product was observed with cDNA prepared from JM225, the C6706str2 TT500 derivative that does not express mshI (Fig. 2, lane 2). A 700-bp band was specifically amplified from mshB-specific cDNA prepared from C6706str2 total RNA (Fig. 2, lane 3). A control cDNA reaction without the addition of reverse transcriptase gave no product, indicating that the PCR product observed for mshB is specific to the mshB transcript and not contaminating DNA (Fig. 2, lane 4).
FIG. 2.
PCR amplification of gene-specific cDNAs. RNA isolated from C6706str2 or JM225 was reverse transcribed into cDNA followed by 5′ RACE with primers specific to mshI (lane 1 and 2) and mshB (lanes 3 and 4). Lane 1, C6706str2; lane 2, JM225; lane 3, C6706str2; lane 4, C6706str2, negative control cDNA reaction in the absence of reverse transcriptase. DNA standards are indicated in kilobase pairs.
The PCR products obtained from the 5′ RACE were subjected to sequence analysis to localize the start of transcription of each potential operon. Interestingly, both transcription start sites were found to be intragenic. The mshI transcription start site maps to the middle of the yhdA gene, while the start of mshB transcription lies within the mshF gene (Fig. 1). A sigma 70 consensus promoter was identified at an appropriate position upstream of each transcription start site (Fig. 3).
FIG. 3.
Comparison of MSHA and sigma 70 consensus promoters. Nucleotide sequences upstream of the 5′ ends of the mshI and mshB transcripts are depicted. The putative start of transcription is designated +1. The −35 and −10 promoter regions are indicated in boldface. The sigma 70 consensus sequence is indicated below the mshI and mshB promoter sequences.
Confirmation of promoter activity.
To demonstrate transcriptional activity, the putative promoters identified by 5′ RACE were cloned into the promoterless lacZ reporter plasmid pRS415. Activities of the resulting mshI and mshB operon fusions were determined by β-galactosidase assay of E. coli cells carrying pJM307 and pJM246, respectively. In addition, the activity of each promoter fragment in the reverse orientation was tested. As shown in Table 6, each promoter supported expression of the lacZ gene at a level consistently higher than that for the pRS415 control. The mshI promoter expressed on plasmid pJM307 demonstrated a greater level of transcription than the cloned mshB promoter on plasmid pJM246 (5,700 versus 1,500 U). The mshI promoter fragment cloned in the reverse orientation with respect to the lacZ gene on plasmid pJM318 expressed low-level β-galactosidase activity (Table 6). This result is in contrast to the relatively high level of β-galactosidase expression observed when an inverted mshB promoter fragment was placed upstream of the lacZ gene on plasmid pJM247 (Table 6). This level of lacZ gene expression indicates that there is promoter activity in the opposite direction and suggests a potential mechanism for negative regulation of pilin subunit gene expression. These data indicate that the two promoters required for MSHA type 4 pilus biogenesis are functional in vivo.
TABLE 6.
β-Galactosidase activities of msh promoter constructs
| Plasmid | Construct | Activity (U)a |
|---|---|---|
| pRS415 | Vector | 59 ± 5 |
| pJM307 | mshI::lacZ | 5,682 ± 87 |
| pJM318 | mshI(rev)::lacZ | 139 ± 8 |
| pJM246 | mshB::lacZ | 1,576 ± 233 |
| pJM247 | mshB(rev)::lacZ | 637 ± 89 |
Average ± standard deviation of triplicate samples.
DISCUSSION
In this study, the entire MSHA type 4 pilus biogenesis gene locus has been defined. Three additional open reading frames located downstream of the structural gene locus originally described by Jonson et al. (26) have been identified. The newly identified msh genes have been designated mshO, mshP, and mshQ. The amino acid sequence of the predicted mshO gene product contains a short, basic leader sequence and a conserved processing motif characteristic of the type 4 prepilins. This finding increases the total number of type 4 prepilins encoded in the MSHA gene locus to five. A similar cluster of genes encoding five type 4 prepilins has been described for the exe secretory locus of Aeromonas hydrophila (23). While a type 4 pilus is not associated with the exe gene locus, it is interesting that a similar pilin subunit gene arrangement exists at the MSHA gene locus and that several of the predicted MSHA gene products show significant homology to proteins involved in the bacterial general secretory pathway (Table 3). The Pseudomonas aeruginosa type 4 pilus responsible for colonization, twitching motility, and phage sensitivity requires the expression of seven type 4 prepilin and prepilin-like proteins which are encoded on three distinct loci of the bacterial chromosome (2). The MshA pilin subunit has previously been shown to be the major structural subunit of the MSHA pilus (26); however, the role of the additional pilin proteins in MSHA biogenesis remains unclear. The possibility exists that the MSHA pilus is a heteropolymer of pilin subunits or that accessory pilin subunits are required for the assembly of a pilus secretory apparatus. Recent evidence has demonstrated that the type 4 prepilin peptidase required for MshA prepilin processing is also required for processing of the Eps prepilin-like proteins associated with V. cholerae extracellular secretion (34). These observations demonstrate the similarities between extracellular secretion and type 4 pilus assembly and raise the possibility that the MSHA genetic element was at one time involved in extracellular secretion.
Many of the genes located upstream of the mshA pilin subunit gene encode homologs of general secretory pathway components (Table 3) (20, 41). These homologies predict that the region upstream of the mshA pilin structural gene is involved in the secretion and assembly of MSHA pilin subunits into a functional pilus on the bacterial cell surface. Previous studies have demonstrated that the genes encoding the secretory components are physically linked to the mshA structural locus on the V. cholerae chromosome (30). This observation suggested that the contiguous set of uniformly oriented genes required for MSHA pilus biogenesis may be organized into a single operon. This genetic arrangement would be unusual since type 4 pilus biogenesis operons are typically arranged such that the major pilin subunit gene is promoter proximal, providing a means to generate greater expression of pilin monomers relative to the secretory components. This report demonstrates that two promoters are required to drive expression of the MSHA gene locus. Analysis of various transcription terminator insertions in an mshA-phoA gene fusion strain demonstrated that mshA prepilin subunit gene expression is dependent on promoter activity generated upstream of mshB but downstream of mshI. This analysis indicates that the MSHA locus is organized as two operons. One operon encodes five type 4 prepilin subunits including the major structural subunit of the pilus, MshA; the second operon encodes proteins predicted to be involved in the assembly and secretion of the pilin structural subunits and is located immediately upstream of the operon encoding the pilin subunits.
The genetic organization of the MSHA gene locus more closely resembles that found for the family of type 4b pili than type 4a pili. The type 4b pili, including the bundle-forming pilus of enteropathogenic E. coli and TCP of V. cholerae, are encoded in a single locus which is located on a plasmid (bundle-forming pilus) or a pathogenicity island (TCP) (13, 28). Within these operons, the ancillary components essential for pilus biogenesis are typically encoded downstream of the major pilin subunit gene. However, the MSHA locus differs in that the genes required for assembly and secretion of the pilus are located directly upstream of the genes encoding the pilin subunit. This unusual genetic arrangement has been described for several fimbriae of E. coli whose structural subunit genes are thought to be regulated by independent promoters in order to maintain proper stoichiometry of pilin subunits relative to the secretion components (5, 39). A similar mechanism for pilin subunit regulation may exist for the MSHA locus.
5′ RACE analysis of msh transcripts indicated that the structural operon transcript initiating upstream of mshB was more abundant than the secretory operon transcript which initiates upstream of mshI. While 5′ RACE is not quantitative, these results are consistent with the possibility that increased levels of MshA pilin subunit could be regulated at the level of transcription or through increased message stability. Contrary to this hypothesis, the operon fusion analysis in E. coli found the mshI promoter to have more transcriptional activity than the mshB promoter. This finding may reflect the absence of a required V. cholerae regulatory protein in E. coli. The poor consensus homology for the −35 region of the mshB promoter further suggests that this promoter may require an additional V. cholerae factor for specific activation.
There is no apparent transcriptional terminator within the secretory operon, suggesting that one long transcript which is initiated from the mshI promoter and contributes to the structural gene operon expression may be generated. This hypothesis is supported by studies using the mshA-phoA gene fusion strain, JM265, which contains a transcription terminator within the secretory operon and shows less alkaline phosphatase activity than the isogenic control strain, JM191. These results indicate that expression from the secretory operon is essential for maximum mshA structural subunit gene expression. Alternatively, expression of the secretory components may stabilize the MshA-PhoA hybrid protein or increase its secretion across the inner membrane in this system. In either case, evidence of a second transcript beginning at a promoter located upstream of mshB and in the middle of the mshF gene is provided by 5′ RACE analysis and suggests that expression from a long mshI-initiated transcript is not sufficient for MSHA structural operon expression.
The MSHA gene locus consists of 16 open reading frames flanked by two genes whose predicted gene products show significant homology to YhdA and MreB of E. coli. Interestingly, the genes encoding these proteins are adjacent to one another on the E. coli chromosome. Neither protein has any function associated with pilus formation or extracellular secretion in E. coli. Furthermore, inactivation of these homologs in V. cholerae by plasmid insertion had no effect on hemagglutination titers. These results help define the MSHA gene locus boundary given that mshI or mshQ gene disruption results in hemagglutination defects. In addition, the identification of a 7-bp direct repeat flanking these genes suggests that the MSHA gene locus may have been acquired by V. cholerae as a transposable or otherwise mobilizable genetic element. While no integrase function can be attributed to any of the proteins encoded on the locus, it is possible that during genetic transfer to the V. cholerae chromosome the integrase gene, essential for MSHA gene locus transposition, was lost.
While the MSHA type 4 pilus has no function in colonization of the mammalian intestine (51), this finding does not exclude the possibility that MSHA is an important attachment factor in the environment, where V. cholerae is often found associated with a variety of aquatic organisms (7). Studies indicate that many bacteria form complex communities or biofilms on solid biotic and abiotic surfaces (8, 38). Recent evidence indicates that mshA mutants are unable to form biofilms (59). This finding suggests that MSHA may be involved in the initial stages of V. cholerae biofilm production in the aquatic environment. Although further studies are necessary, the MSHA locus may represent a genetic cassette that plays a significant role in survival of V. cholerae in the environment. We propose that the MSHA locus, analogous to bacterial pathogenicity islands, be termed an environmental persistence island.
ACKNOWLEDGMENTS
We gratefully acknowledge the helpful comments and suggestions of Nicholas Morris, discussions with Jean-François Tomb, and the expert technical assistance of Emily Cornell and the Dartmouth Molecular Biology Core Facility.
This work was supported by National Institutes of Health grant AI-25096 (R.K.T.). J.W.M. is the recipient of a predoctoral fellowship from the National Institutes of Health (training grant AI-07519).
REFERENCES
- 1.Albert M J. Vibrio cholerae O139 Bengal. J Clin Microbiol. 1994;32:2345–2349. doi: 10.1128/jcm.32.10.2345-2349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal pilC homologue. Mol Microbiol. 1996;22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attridge S R, Manning P A, Holmgren J, Jonson G. Relative significance of mannose-sensitive hemagglutinin and toxin-coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect Immun. 1996;64:3369–3373. doi: 10.1128/iai.64.8.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilge S S, Clausen C R, Lau W, Moseley S L. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. 1989;171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 7.Colwell R R. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 8.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.de la Morena M L, Hendrixson D R, St. Geme J W. Isolation and characterization of the Haemophilus influenzae uvrA gene. Gene. 1996;177:23–28. doi: 10.1016/0378-1119(96)00264-8. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 11.de Vries J, Wackernagel W. Cloning and sequencing of the Serratia marcescens gene encoding a single-stranded DNA-binding protein (SSB) and its promoter region. Gene. 1993;127:39–45. doi: 10.1016/0378-1119(93)90614-9. [DOI] [PubMed] [Google Scholar]
- 12.Doi M, Wachi M, Ishino F, Tomioka S, Ito M, Sakagami Y, Suzuki A, Matsuhashi M. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in the formation of the rod shape of Escherichia coli cells. J Bacteriol. 1988;170:4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnenberg M S, Giron J A, Nataro J P, Kaper J B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 14.Finkelstein R A, Mukerjee S. Haemagglutination. A rapid method for the differentiating Vibrio cholerae from El Tor vibrios. Proc Soc Exp Biol Med. 1963;112:355–359. [Google Scholar]
- 15.Freter R, Jones G W. Adhesive properties of Vibrio cholerae: nature of the interaction with intact mucosal surfaces. Infect Immun. 1976;14:2246–2256. doi: 10.1128/iai.14.1.246-256.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giron J A, Gomez-Duarte O G, Jarvis K G, Kaper J B. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili—a minireview. Gene. 1997;192:39–43. doi: 10.1016/s0378-1119(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 17.Giron J A, Levine M M, Kaper J B. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12:71–82. doi: 10.1111/j.1365-2958.1994.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 18.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall R H, Khambaty F M, Kothary M H, Keasler S P, Tall B D. Vibrio cholerae non-O1 serogroup associated with cholera gravis genetically and physiologically resembles O1 El Tor cholera strains. Infect Immun. 1994;62:3859–3863. doi: 10.1128/iai.62.9.3859-3863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hase C C, Bauer M E, Finkelstein R A. Genetic characterization of mannose-sensitive hemagglutinin (MSHA)-negative mutants of Vibrio cholerae derived by Tn5 mutagenesis. Gene. 1994;150:17–25. doi: 10.1016/0378-1119(94)90852-4. [DOI] [PubMed] [Google Scholar]
- 21.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 23.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahagirdar R, Howard S P. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J Bacteriol. 1994;176:6819–6826. doi: 10.1128/jb.176.22.6819-6826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonson G, Holmgren J, Svennerholm A M. Identification of a mannose-binding pilus on Vibrio cholerae El Tor. Microb Pathog. 1991;11:433–441. doi: 10.1016/0882-4010(91)90039-d. [DOI] [PubMed] [Google Scholar]
- 26.Jonson G, Lebens M, Holmgren J. Cloning and sequencing of Vibrio cholerae mannose-sensitive haemagglutinin pilin gene: localization of mshA within a cluster of type 4 pilin genes. Mol Microbiol. 1994;13:109–118. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 27.Jouravleva E A, McDonald G A, Marsh J W, Taylor R K, Boesman-Finkelstein M, Finkelstein R A. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect Immun. 1998;66:2535–2539. doi: 10.1128/iai.66.6.2535-2539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S R, Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 31.Lindeberg M, Collmer A. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J Bacteriol. 1992;174:7385–7397. doi: 10.1128/jb.174.22.7385-7397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 33.Marsh J W, Sun D, Taylor R K. Physical linkage of the Vibrio cholerae mannose-sensitive hemagglutinin secretory and structural subunit gene loci: identification of the mshG coding sequence. Infect Immun. 1996;64:460–465. doi: 10.1128/iai.64.2.460-465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsh J W, Taylor R K. Identification of the Vibrio cholerae type 4 prepilin peptidase required for toxin secretion and pilus formation. Mol Microbiol. 1998;29:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 35.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 37.O’Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 38.O’Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 39.Oudega B, De Graaf F K. Genetic organization and biogenesis of adhesive fimbriae of Escherichia coli. Antonie Leeuwenhoek. 1988;54:285–299. doi: 10.1007/BF00393521. [DOI] [PubMed] [Google Scholar]
- 40.Parge H E, Forest K T, Hickey M J, Christensen D A, Getzoff E D, Tainer J A. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 41.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves P J, Whitcombe D, Wharam S, Gibson M, Allison G, Bunce N, Barallon R, Douglas P, Mulholland V, Stevens S, et al. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 43.Sancar A, Williams K R, Chase J W, Rupp W D. Sequences of the ssb gene and protein. Proc Natl Acad Sci USA. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandkvist M, Morales V, Bagdasarian M. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene. 1993;123:81–86. doi: 10.1016/0378-1119(93)90543-c. [DOI] [PubMed] [Google Scholar]
- 45.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 46.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 47.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 48.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 49.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 51.Tacket C O, Taylor R K, Losonsky G, Lim Y, Nataro J P, Kaper J B, Levine M M. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thelin K H, Taylor R K. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tonjum T, Freitag N E, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 56.Tonjum T, Koomey M. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships. Gene. 1997;192:155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 57.Wachi M, Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 59.Watnick, P. I., and R. Kolter. Personal communication.