Abstract
Harsh environments provide opportunities to study how different species adapt, at the molecular level, to similar environmental stressors. High hydrostatic pressure, low temperature, and absence of sunlight in the deep-sea environment are challenging conditions for gene expression, cell morphology and vision. Adaptation of fish to this environment appears independently in at least 22 orders of fish, but it remains uncertain whether these adaptations represent convergent evolution. In this study, we performed comparative genomic analysis of 80 fish species to determine genetic evidences for adaptations to the deep-sea environment. The 80 fishes were divided into six groups according to their order. Positive selection and convergent evolutionary analysis were performed and functional enrichment analysis of candidate genes was performed. Positively selected genes (pik3ca, pik3cg, vcl and sphk2) were identified to be associated with the cytoskeletal response to mechanical forces and gene expression. Consistent signs of molecular convergence genes (grk1, ednrb, and nox1) in dark vision, skin color, and bone rarefaction were revealed. Functional assays of Grk1 showed that the convergent sites improved dark vision in deep-sea fish. By identifying candidate genes and functional profiles potentially involved in cold, dark, and high-pressure responses, the results of this study further enrich the understanding of fish adaptations to deep-sea environments.
Keywords: convergent evolution, positive selection, bathypelagic and abyssopelagic fishes, GRK1
Significance.
Deep-sea fish adaptations appear independently in fish at least 22 orders but have not been systematically studied. In this study, comparative genomic analysis was conducted on 36 bathypelagic and abyssopelagic fish species and 44 shallow-sea fish species to examine genetic evidence of adaptation by the fish to the deep sea. Many genes involved in deep-sea adaptations are positively selected or show convergent amino acid changes in deep-sea fishes. Functional assays of Grk1 were performed to confirm that the convergent sites improved dark vision in deep-sea fishes.
Introduction
The ocean occupies 70.55% of the Earth's surface area, which constitutes the largest area of the natural environment (Costello et al. 2010). Shallow seas have been extensively studied because of their moderate temperatures and pressures as well as abundant food sources dependent and enriched by sunlight. These factors make shallow seas the most hospitable regions in the ocean. In contrast, fewer species inhabit deep-sea areas because of the extremely harsh living conditions, such as tremendous pressure, reduced oxygen levels, scant food sources, constant darkness, and low temperatures (Sanders and Hessler 1969; Randall and Farrell 1997; Priede and Froese 2013). The deep sea refers to the bathypelagic zone below 1000 m that is devoid of daylight (Warrant and Adam Locket 2004). In the thermocline, the seawater temperature decreases with depth and the rate of change decreases after about 1000 m (Stewart 2018). As depth increases, dissolved oxygen decreases until the minimum at a few hundred meters to 1000 m is reached. At this point, the dissolved oxygen gradually increases with an increase in depth (Webb 2019).
Many deep-sea fish species possess physiological adaptations to high pressure, low temperatures, and darkness. In shallow-sea species, tens of atmospheres of pressure can disrupt enzyme ligand binding, kinetics and cytoskeletal protein assembly. In contrast, deep-sea fish are more resistant to high pressure (Lan et al. 2018). The ability of cells or tissues in an organism to respond to mechanical stimuli is important for normal functions, such as the formation and maintenance of bones, blood vessels, muscles, and other tissues, as well as blood pressure regulation, cell movement, and cell proliferation (Orr et al. 2006). The basis for many of these functions involves mechanosensitive adhesions mediated by membrane protein integrins. The force exerted by a cell's own cytoskeleton or external sources enhances adhesions, resulting in various intracellular signals (Del Rio et al. 2009; Friedland et al. 2009). For instance, the expansion of two adhesion gene families (syndecan and protocadherin) occurs in deep-sea mussels (Sun et al. 2017). The helical structure of DNA and RNA is slightly affected by hydrostatic pressure, while other nucleic acid structures, such as hairpin loops and gene transcription, can be more sensitive to hydrostatic pressure (Macgregor Jr 1998; Munro et al. 2015; Patra et al. 2018). At low temperatures, DNA and RNA tend to tighten their structures, hindering the involvement of enzymes in DNA replication, transcription, and translation, as well as disrupting the transcription and translation processes (Lan et al. 2018). In a dimly light environment, rods are used for vision. Studies on rod opsins, photosensitive pigment, and the retina have been conducted on the vision of deep-sea fishes (Denton and Warren 1957; Wagner et al. 1998; Musilova et al. 2019). In low-light environments, the color of deep-sea fish is light compared to that of shallow-sea fish. Species living below 200 m are either red or black; the color of most benthic species is light yellow to red (Johnsen 2005). The bones of deep-sea fish have different levels of reduction (dimensional reductions, loss of skeletal elements, and reductions in bone density) that reach neutral buoyancy. For example, Gonostoma elongatum (Isospondyli) and Xenodermichthys copei (Isospondyli) have fragile, lightly ossified skeletons (Denton and Marshall 1958). Research on snailfishes indicates that, with an increases in depth, a decrease in light levels, and changes in temperature, the density and size of bones are affected (Gerringer et al. 2021). With increasing depth, a progressive decrease in skeletal ash content occurs (Childress and Nygaard 1973).
The deep sea is a less-explored part of the ocean; additionally, deep-sea fish adaptations independently appear in at least 22 orders (Randall and Farrell 1997). Due to the difficulty in obtaining deep-sea fish samples, most studies on deep-sea adaptations are based on the adaptation of single species, and the convergence of species has only been studied at the level of mitochondrial and opsin genes (Musilova et al. 2019; Shen et al. 2019; Lupse et al. 2021). As samples of deep-sea fish become more available and complete genomes are mapped, studies on the convergence of deep-sea adaptation will be conducted on a larger scale.
We analyzed 80 genomes of different bathypelagic and abyssopelagic fishes in six taxonomic groups. To identify candidate genes associated with deep-sea adaptations, single-copy genes were searched for positively selected genes associated with deep-sea stresses. To identify patterns of molecular convergence, convergent amino acid substitutions were identified.
Results
Final Alignment Construction and Phylogeny
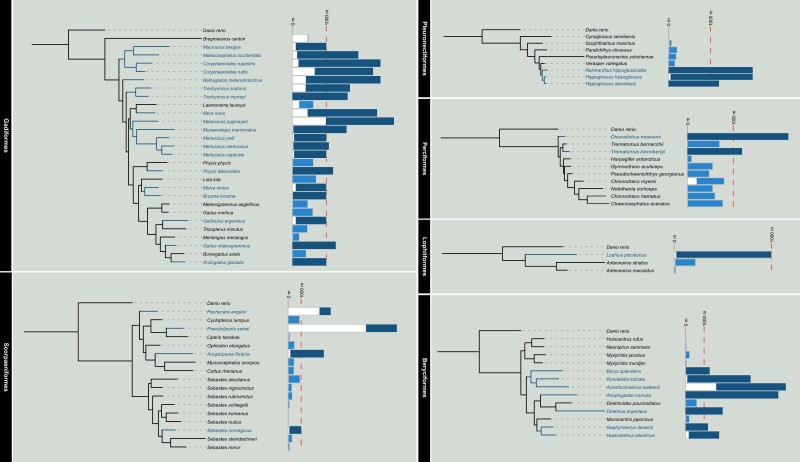
We obtained genome-wide single-copy sequences from 80 fish species, including 36 bathypelagic and abyssopelagic fishes, 43 shallow-sea fishes and zebrafish (supplementary table 1, Supplementary Material online). The fish represent six orders. Gadiformes had 19 deep-sea fish, Beryciformes had seven deep-sea fish, Scorpaeniformes and Pleronectiformes each had three deep-sea fish, Perciformes had two deep-sea fish and Lophiiformes had one. The orthologous genes of the six orders were identified for construction of a phylogenetic tree (fig. 1).
Fig. 1.
Phylogenetic relationships of six groups of fishes. The bars on the right represent the depth range inhabited by the fish.
Positive Selection on Six Groups and Kyoto Encyclopedia of Genes and Genomes Function Enrichment
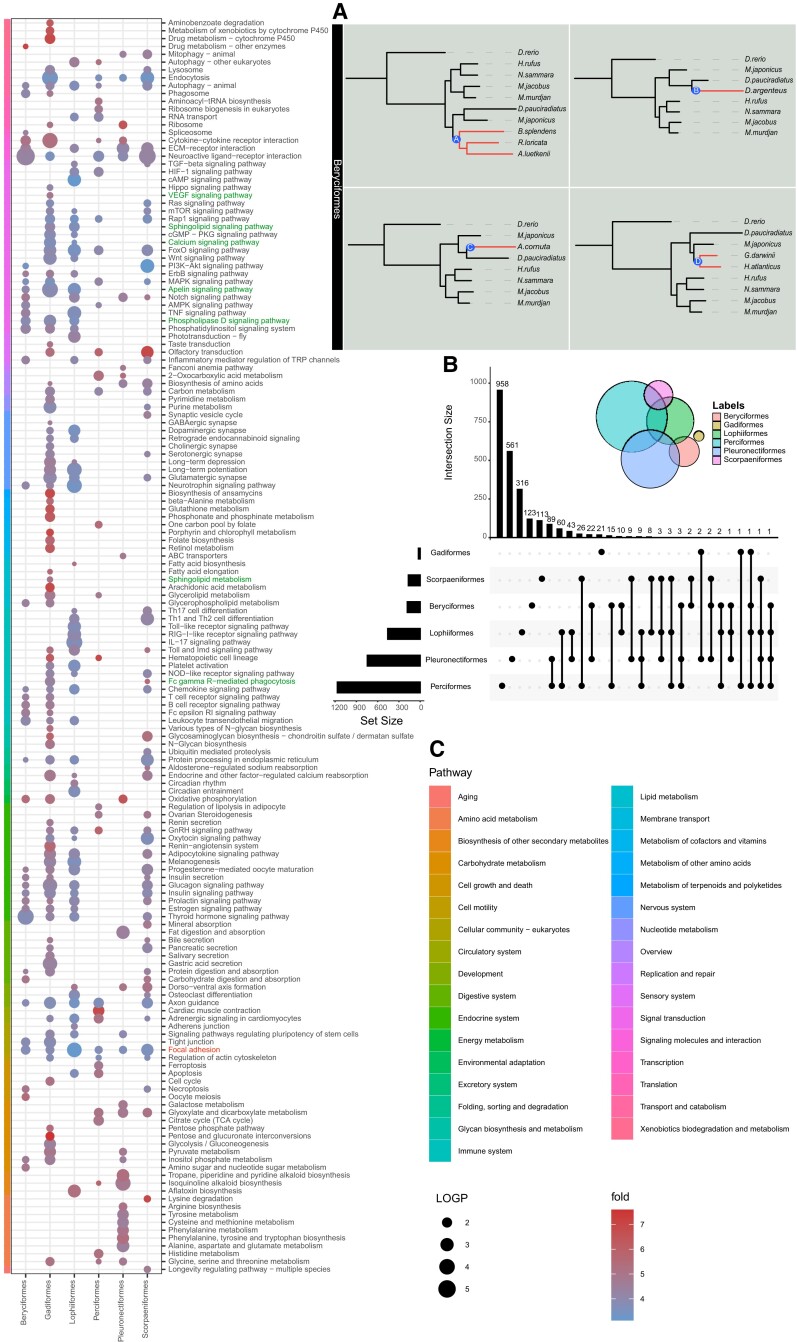
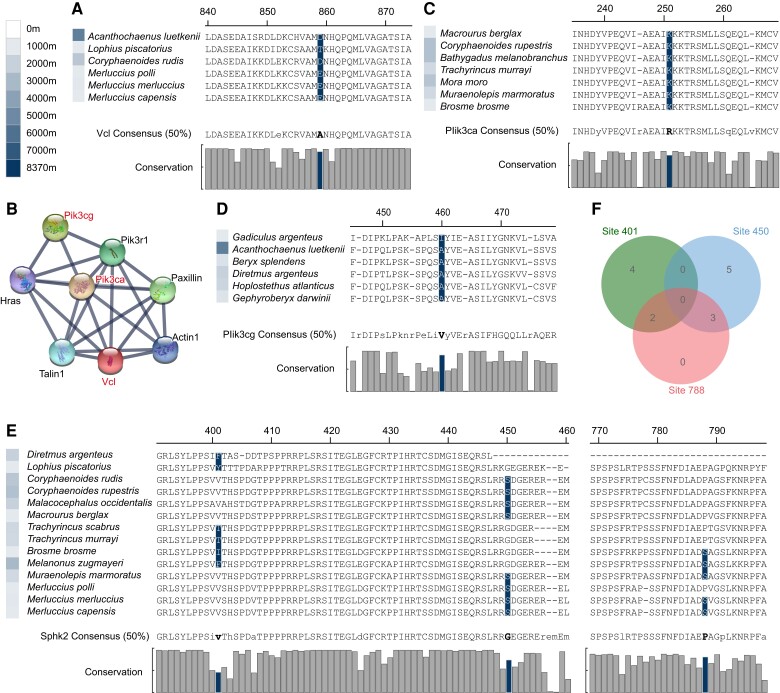
To detect genes that evolving under positive selection in the branches leading to the deep-sea species, branch-site tests were run separately for the six groups. Positive selection was calculated for each deep-sea fish, with only one deep-sea fish in each group for each calculation. See Beryciformes (fig. 2A) as an example. Many positive selection genes were found in all orders (fig. 2B). Positive selection genes were enriched by kyoto encyclopedia of genes and genomes (KEGG) and plotted according to P-values showing that there were many pathways enriched in each order (fig. 2C). Several interesting enriched pathways were revealed. All six orders were enriched in “Focal adhesion” pathway, and we found that Vinculin (vcl) was under positive selection in five species fish belonging to three orders (Beryciformes, Gadiformes, and Lophiiformes) (figs. 2B and 3A). Protein interaction networks linked two other positive selection genes, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha (pik3ca) and phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma (pik3cg), through ras, paxillin, and actin (fig. 3B). Pik3ca had one positively selected site in seven deep-sea fishes in the Gadiformes (fig. 3C) and pik3cg had one positively selected site in six deep-sea fishes belonging to Gadiformes and Beryciformes (fig. 3D). The positive-selection site of Pik3ca was located in the Ras-binding domain and that of Pik3cg was located in the C2 domain. Phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that can phosphorylating the 3’OH of the inositol ring of phosphoinositides. Our results revealed that seven deep-sea fishes of pik3ca and six fishes of pik3cg did not overlap. The PI3K-related genes were positively selected in 13 deep-sea fish. “Sphingolipid metabolism,” “Calcium signaling pathway,” “Sphingolipid signaling pathway,” “Phospholipase D signaling pathway,” “VEGF signaling pathway,” “Apelin signaling pathway,” “Fc gamma R-mediated phagocytosis”; among the seven pathways, the Sphingosine kinase 2 (sphk2) gene was positively selected (fig. 2C). Three positively selected sites in Sphk2 involving 14 deep-sea fishes and three groups (Gadiformes, Beryciformes, and Lophiiformes) were identified (fig. 3E). No species in which Sphk2 showed all three positively selected sites, and five possessed two positively selected sites (fig. 3F).
Fig. 2.
Results of positive selection analysis were presented. (A) Using Beryciformes as an example, positively selected genes were calculated by labeling branches A, B, C, and D respectively. (B) Positively selected genes statistic of six groups. The left bar graph shows the number of positively selected genes in each group, and the upper bar graph shows the number of statistical units, which are shown by dots and lines. (C) In the KEGG functional enrichment pathway diagram of positively selected genes, the bubble size represents -log10P-value, and the color represents the enrichment fold, which is the ratio of the probability of actual enrichment to the probability of random enrichment. See supplementary table 4, Supplementary Material online for abbreviations of species names.
Fig. 3.
(A) The positively selected site of Vcl. (B) STRING shows Vcl, Pik3ca, and Pik3cg interacting through Ras, Paxillin, and Actin. (C) The positively selected site of Pik3ca. (D) The positively selected site of Pik3cg. (E) The positively selected sites of Sphk2. (F) Venn diagram of gene distribution at three positively selected sites of Sphk2. The color of the square in front of the name of a species represents the maximum depth at which the species occurs. Consensus (50%) represents a consensus sequence at 50% conservation. The consensus sequence greater than 50% of the threshold is uppercase, and the consensus sequence less than 50% is lowercase. The histogram represents the conservation of each site in all species of the six orders.
Convergent Evolution in Six Groups and KEGG Function Enrichment
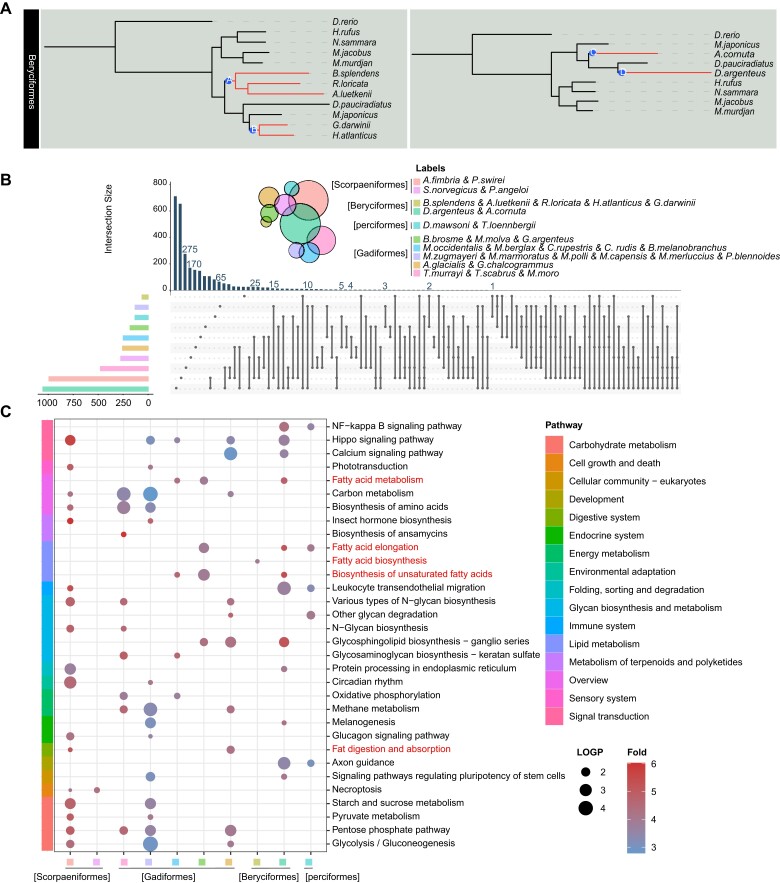
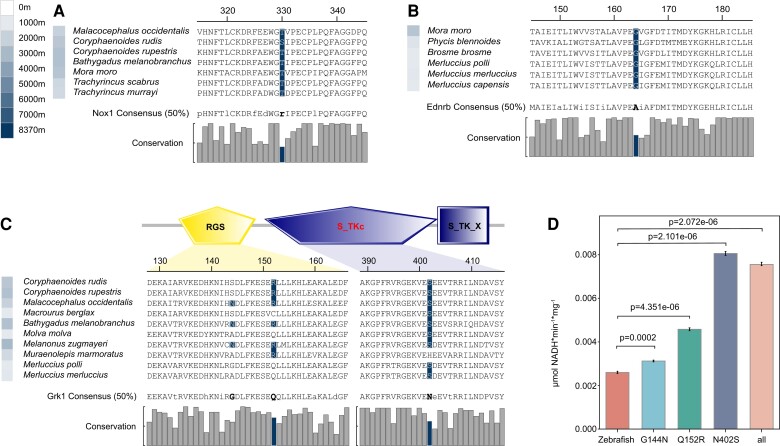
To estimate molecular convergence at the amino acid level, posterior expected numbers of convergent and divergent substitutions between pairs of deep-sea branches for the six groups were calculated separately. In each order, we first carried out the convergence between two branches, and then made statistics of all the convergent genes. Beryciformes is used as an example and shown in fig. 4A. Because Lophiiformes has only one deep-sea fish species, we were unable to do intra-order convergence calculations. In addition, we did not discover any convergent genes in Pleuronectiformes. All the convergent genes were analyzed statistically, and the vast majority of the convergent genes only converged in two branches (fig. 4B). KEGG enrichment was carried out for all the convergent genes, and the results showed that the convergent genes involved multiple pathways. There were not many overlapping genes among branches, but there were many overlapping pathways. KEGG pathway analysis showed that convergent genes were enriched in multiple adipose-related metabolic pathways, especially the “biosynthesis of unsaturated fatty acids” (fig. 4C). Previous studies demonstrated the positive role of unsaturated fatty acids in the deep-sea environment (Bell et al. 1986; Kamimura et al. 1993; Petursdottir et al. 2008; Saito and Hashimoto 2010). Gadiformes contained the largest number of deep-sea fish species in this study. We found several Gadiformes-specific convergent genes involving many fish. One site of NADPH oxidase 1 (nox1) was detected that showed a sign of convergent evolution in six species of Gadiformes (fig. 5A). The endothelin receptor-B gene (ednrb) was identified at one convergence site in six deep-sea species in the Gadiformes (fig. 5B). Rhodopsin kinase (grk1) was identified at three convergence sites in 10 deep-sea species in the Gadiformes. Domain prediction revealed two convergence sites in the regulator of the G-protein signaling domain, and one convergence site in the serine/threonine protein kinases catalytic domain, which added a phosphorylation site (fig. 5C and supplementary table 2, Supplementary Material online). The effect of convergence site mutation on Grk1 was examined. A kinase assay showed that the enzyme activity of Grk1 was more active than that of wild zebrafish after the mutation of the converging sites, either separately or together. In particular, mutation of site 402 N (Asn) to S (Ser) significantly increased kinase activity (fig. 5D).
Fig. 4.
Results of convergent evolutionary analysis were presented. (A) Using Beryciformes as an example, the A, B and C, D branches were labeled, and the gene convergence of the two branches was calculated. (B) Statistics of convergent genes per two branches. The left bar graph shows the number of convergence genes in each two branches, and the upper bar graph shows the number of statistical units, which are illustrated by dots and lines. (C) In the KEGG functional enrichment pathway diagram of convergence genes, the bubble size represents -log10P-value, and the color represents enrichment fold, which is the ratio of the probability of actual enrichment to the probability of random enrichment. See supplementary table 4 for abbreviations of species names.
Fig. 5.
(A) Convergence site of Nox1. (B) Convergence site of Ednrb. (C) Convergence sites of Grk1. Two convergence sites are located in RGS and S_TKc domain respectively. RGS: Regulator of G protein signaling domain; S_TKc: Serine/Threonine protein kinases, catalytic domain; S_TK_X: Extension to Ser/Thr-type protein kinases. (D) Kinase activities of zebrafish Grk1, zebrafish G144N Grk1, zebrafish Q152R Grk1, and zebrafish N402S Grk1 and all three sites were mutated Grk1. The error bars are the 95% confidence interval. The color of the square in front of the name of a species represents the maximum depth that the species inhabits. Consensus (50%) represents a consensus sequence at 50% conservation. The consensus sequence greater than 50% of the threshold is uppercase, and the consensus sequence less than 50% is lowercase. The histogram represents the conservation of each site in all species of six orders.
Discussion
Visual Adaptation to the Dark Environment
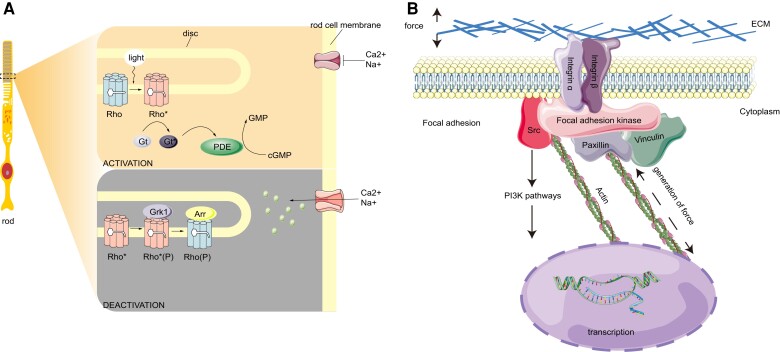
The vision of deep-sea fish vision is a topic of considerable interest. We found that convergent genes and positively selected genes were enriched in phototransduction signaling pathways. Grk1 was identified at three convergence sites in 10 deep-sea species in the Gadiformes. Additionally the N402S site mutation due to the introduction of a phosphorylation site in the structure of catalytic domain significantly improved Grk1 kinase activity. Grk1 is a member of G protein-coupled receptor kinase family, and specifically phosphorylates photo-activated rhodopsin (Rho*) in retinal photoreceptor cells. Phosphorylation of Rho* prevents activation of the G-protein transducin and promotes the binding of the regulatory protein arrestin to phosphorylated Rho*, thereby terminating the activation pathway of visual transduction (Wilden et al. 1986; Bennett and Sitaramayya 1988). This process is usually called photoreceptor desensitization, and is physiologically related to adaptation to darkness (Kühn and Dreyer 1972; Ohguro et al. 1995; Hurley et al. 1998; Qingming et al. 1998) (fig. 6A). Deficiency or mutation of grk1 leads to impairment in scotopic vision (Yamamoto et al. 1997; Dryja 2000). Grk1 is also under positive selection in nocturnally active birds (Wu 2019). Some deep-sea demersal fishes migrate vertically or horizontally day and night (Suetsugu and Ohta 2005). Changes in grk1 may have enhanced dark vision in deep-sea fishes and improved their predation and predation-avoidance capabilities.
Fig. 6.
(A) Schematic diagram of a rod, and Grk1 phosphorylates rhodopsin in the rod out segments. Light activates Rho, and the chromophore changes from 11-cis-retinal to all-trans-retinal. Activated Rho (Rho*) binds Gt to hydrolyze cGMP to GMP, reducing the concentration of cGMP in rod cytoplasm and closing the cGMP gated channel. To terminate the phototransduction, Rho* is phosphorylated by Grk1 and the subsequent binding of Arr. (B) Schematic showing a cell responding to mechanical force. The actin cell skeleton is connected to the ECM and Vcl, and the focal adhesion is formed. The focal adhesion can regulate cellular shape and motility. Focal adhesion kinase is responsible for the binding of integrin-associated protein, such as paxillin and Scr. Scr can activate the expression of downstream genes through the PI3K signaling pathway. Rho, rhodopsin; Gt, transducing; Arr, arrestin; ECM, extracellular matrix.
Other Adaptations to the Deep-Sea Environment
Hundreds of atmospheres of hydrostatic pressure can destroy the cytoskeleton organization and change cell morphology. The special connection between cells can maintain the tissue structure composition of the cells. Vcl, is an important component of focal adhesion, mediates the connection between the extracellular matrix and cytoskeleton, and participates in integrin-mediated cytomechanic-chemical signal transduction (Hu et al. 2009). Focal adhesions recognize mechanical forces generated either by either an external source or the cell itself and then translate them into a biological response. Vcl is a key protein regulating the transmission of contractile forces (Yang et al. 2010). The strength of the connection between the integrin adhesion receptors and the actomyosin cytoskeleton mediated by focal adhesion proteins determines the ability to generate and transmit forces. Contractile force generation is reduced when vinculin is absent and enhanced when vinculin is present (Mierke 2009). When pressure increases, vinculin is reduced and cell contraction is influenced (Yang et al. 2010). Focal adhesion complexes can connect the cytoskeleton with the adhesion site and trigger the downstream PI3K signaling pathway. Pik3ca and pik3cg regulate cell proliferation, differentiation, apoptosis, migration, and other physiological processes. This is accomplished by producing second messenger phosphatidylinositol 3,4,5 trisphosphate (PIP3) and acting on downstream molecules, such as mechanistic target of rapamycin complex 1 (mTORC1), Glycogen synthase kinase-3 (GSK3), and B-cell lymphoma 2 (Bcl-2) (König and Ostendorf 2015) (fig. 6B). Vcl is involved in sensing mechanical forces, and the cytoskeleton responds to changes in cell morphology. Pik3ca and pik3cg, as downstream genes, are involved in regulating various physiological processes and responding to them. We hypothesized that vcl, pik3ca, and pik3cg may contribute to a fish's resistance to deep-sea pressure.
In addition, the KEGG pathway showed that nox1 is involved in osteoclast differentiation. Nox1 and mitochondria are involved in receptor activator of nuclear factor-κB ligand (RANKL)-mediated osteoclastogenesis. This is because Reactive oxygen species (ROS) produced by Nox1 acts as an intracellular signal mediator for osteoclast differentiation (Lee et al. 2005; Chen et al. 2016). Osteocytes are considered to be mechanosensory cells. The mechanical forces acting on tissues inform osteoblasts and osteoclasts to adapt to them by adjusting the amount and space of the tissue (Burger et al. 1995). The balance between osteoclasts and osteogenesis is the key to maintaining normal bone mass (Xu and Teitelbaum 2013). A deceleration of bone mineralization occurs due to an increase in the number of osteoclasts or a decrease in the number of osteoblasts (Roy et al. 2002). The bones of deep-sea fish have different levels of reduction and nox1 may provide a breakthrough.
Sphk2 catalyzes the phosphorylation of sphingosine to form sphingosine-1-phosphate (S1P), this could indicate an interaction with histone and inhibit its deacetylase 1/2 (HDAC1/2) activity, resulting in an increase in histone acetylation and transcription of downstream target genes (Hait et al. 2009). Core histone acetylation and deacetylation determine chromatin transcriptional activity and are also closely related to gene regulation. Both low temperatures and high pressure inhibit the early stage of translation and reduce translation efficiency (Simonato et al. 2006). High pressure is stressful to life, because it forces a decrease in cell volume (Somero 1992). DNA is wrapped by histones on the chromosomes of eukaryotes. Histone acetylation is an epigenetic modification. Acetylation eliminates the positive charge of histone lysine and reduces its binding force to DNA (negatively charged), so that it can untwist the original tight chromosome structure and transform it into a looser form which is conducive to transcription and enhanced gene expression.
Ednrb is a G-protein-coupled seven-transmembrane receptor that interacts with a family of ligands, such as endothelins. Endothelin-1 is a major regulator of vascular function, and Ednrb is important for the control of vascular reactivity and blood pressure (Mazzuca and Khalil 2012). KEGG pathway analysis showed that ednrb is involved in melanogenesis. When the ednrb gene is deleted or suppressed, an almost complete loss of pigmentation occurs (Hosoda et al. 1994; Lee et al. 2003). The ednrb mutant shows a less dramatic phenotype with white spotting limited to about 20% of the coat (Hosoda et al. 1994). By studying mutations and polymorphisms affecting pigmentation and patterning, ednrb mutations are found to manifest in the white frame overo markings of horses, the white spots or fully white phenotype in mice, and the reduced melanophores in zebrafish (Mills and Patterson 2009). The expression levels of Ednrb from four color skins tones in goldfish skin, ranging from red > cyan > black, and the rare white skin (Gan et al. 2021). Ednrb was convergent in six fishes of Gadiformes, but there was no obvious convergent body color in these six fishes, and its function needs further study.
Conclusion
Determining the genetic basis of adaptive traits can be challenging. Through the positively selection and convergence analysis, to find a lot of interest genes, function enrichment shows lots different pathway. Adaptation to deep-sea environmental may differ in the evolution of each species, but there are some common adaptive approaches, such as unsaturated fatty acid metabolism, reactions to mechanical force, histone acetylation, reduced bone content, and enhanced dark vision. The 80 fishes we studied included 28 fishes in the Gadiformes, a total of 36 deep-sea fishes including 19 fishes in the Gadiformes. The high proportion of Gadiformes also contributed to the majority of the convergent genes involved in Gadiformes to a certain extent. Gadiformes have also emerged as a unique lineage of all deep-sea fishes in the study of the evolution of the immune system in deep-sea fishes (Malmstrøm et al. 2016). Andriashev divided the deep-sea fish fauna into ancient deep-sea forms and secondary deep-sea forms 1953. Gadiformes correspond to ancient deep-sea forms and are characterized by worldwide distribution, specialized morphology, and occupation of the deepest parts of the ocean (Priede and Froese 2013). Our results support the hypothesis that there are many specific sites in Gadiformes that may be functionally involved in deep-sea adaptation. Deep-sea invasive families, as secondary forms, such as Ophidiidae and Liparidae, make the greatest contribution to the fish fauna at depths >6000 m. With advances in ultra-abyss sampling technology, more abyss species will be discovered in the future, and these will allow Andriashev's hypothesis to be tested at the molecular level.
Materials and Methods
Source of Data
A total of 337 fish genomes were available at NCBI (https://www.ncbi.nlm.nih.gov/) as of July 2020. Information on the depth-ranges of these fish was collected from FishBase (https://www.fishbase.de/), OBIS (https://obis.org/), and Shen et al. (2019). Deep-sea fishes are usually considered those living at depths below 1,000 m (Angel 1997; Pradillon and Gaill 2007). The 337 species were classified by order. If no deep-sea fish species were found, the entire order was not studied further, leaving 180 species for consideration. The depth ranges of 34 species of the 172 fish species fell below 1,000 m (supplementary table 3, Supplementary Material online). For Perciformes, many shallow-water-dwelling sister taxa were available, and 10 species were selected according to the families of deep-sea fishes. In addition, four deep-sea fish genomes from the laboratory of this study were added. Considering that the zebrafish genome is well known, the zebrafish genome was used as the reference species. Finally, the remaining 36 deep-sea fishes and 44 shallow-living fishes were used in this study, and were placed into six groups according to the orders: 1) Beryciformes, 2) Gadiformes, 3) Lophiiformes, 4) Perciformes, 5) Pleuronectiformes, and 6) Scorpaeniformes.
Identification of Single-Copy Genes
The homologous genes in Fugu, Japanese medaka, Stickleback, Atlantic cod, Turbot, and Platyfish were researched in ENSEMBL (https://www.ensembl.org/) using BioMart tools with the Zebrafish genome as a reference based on one-to-one orthologs. Single-copy genes of these species were extracted based on one-to-one orthologs. The 10,058 single-copy homologous genes in at least six species were taken as the single-copy homologous gene set.
The genomes of 76 species from NCBI were downloaded. Our laboratory possesses the genomes of Coryphaenoides rudis, Pseudoliparis swirei, Liparis tanakae, and Pachycara angeloi sp.nov, and the genomes of these four species were included in this study (supplementary table 1, Supplementary Material online). Zebrafish protein sequences were extracted from protein data (Danio_rerio.GRCz11.pep.all.fa downloaded from NCBI) using a single-copy homologous gene set. TBLASTN (E-value cut-off 1e-3) (Altschul 2012) was used for the genomes of 80 species to determine the positional information of corresponding homologous genes. Scattered alignments were merged for each gene using Sorting Out Local Alignment Results v0.9.3 (Yu et al. 2006). The merged results with a cut-off <50 (cut-off being the aligned protein length/total protein length) were removed. After the cut-off selection, GENEWISE (Birney et al. 2004) was used to compare each zebrafish protein sequence with its optimal merged DNA segment sequences separately. GENEWISE scores >35 were used, and coding sequences were the resultant output. After the implementation of GENEWISE, homolog coding sequences of 80 species were obtained (supplementary table 4, Supplementary Material online).
Phylogenetic Inference
Phylogenetic trees were constructed of the six groups with zebrafish using single-copy orthologous genes. Each single-copy coding sequence of a group was translated into protein sequences and aligned against the zebrafish protein sequences using MAFFT v7.453 (L-INS-I) (Katoh and Standley 2013). Gblocks version 0.91b (Castresana 2000) extracted the conserved sites of multi-sequence alignment results of orthologous genes with option (−t = P, −b4 = 5, −b5 = h). A maximum likelihood (ML) tree was computed with IQ-TREE v2.0.3 (Lam-Tung et al. 2015) and RAxML v8.1.24 (raxmlHPC-HYBRID-SSE3 program) (Stamatakis 2014). ML trees of eight groups were constructed using the same method as previously described. ProtTest (Darriba et al. 2011) calculated the best amino acid substitution model required by RAxML (supplementary table 5, Supplementary Material online). These trees were visualized, rooted, and annotated using iTOL (Letunic and Bork 1988).
Positive Selection Analyses
The topologies of the six groups were used as the guide tree for positive selection analyses. Each group was separately tested for positive selection. The base-substitution mutation rate of non-synonymous mutation was dN, and the base-substitution mutation rate of synonymous mutation was dS. If dN/dS was > 1, also called positive selection, this signified that most non-synonymous mutations are favorable and retained in long-term evolution, and positive selection is the main driving force of biological evolution. The branch-site model of Codeml implemented in the PAML package (Yang 2007) was used to test for positive selection on individual codons along the lineage leading to each of the deep-sea species. PRANK v.170427 (Löytynoja 2014) was used to conduct multiple nucleotide alignments for coding sequences of single-copy orthologous genes. In order to keep the positively selected sites conservative, Gblocks was used to filtered unreliable blocks (−t = c, −b5 = h). The dN/dS ratios of filtered reliable codons were calculated by setting each deep-sea fish as the foreground branch and other shallow-sea fishes of the same group as background branches. To detect whether the positive selection signs were significant, likelihood ratio test statistics were calculated and P-values were tested by the false discovery rate (FDR) method in R version 4.1.0 (Team 2019). The FDR used a P-value ≤ 0.05 as the significance level. Meanwhile, check the result file after “Positive sites for foreground lineages Prob(W>1)” for positively selected sites, if there is more than one positively selected site is considered as positive selection gene. Unfiltered original sequences containing positively selected genes of six orders were extracted from all species and MEGA (Kumar et al. 2016) was used to check for conserved and consistency of positively selected sites in all deep-sea fishes.
Amino-Acid Convergence Analyses
PRANK v.170427 (Löytynoja 2014) was used to conduct multiple nucleotide alignments for coding sequences of single-copy orthologous genes and Gblocks to filtered unreliable blocks (−t = c, −b5 = h). In order to keep the convergence sites conservative, Gblocks was used to filtered the aligned bad blocks. Because the qualities of downloaded genomic data are different, if the number of species is too large, the filtered sequences will be too short, and many sites will not get convergence calculation. To ensure that convergence can be calculated for as many genes and sites as possible, each of the six orders was separately tested for convergent evolution. Grand Convergence (Castoe et al. 2009) was used to calculate the expected posterior numbers of convergent substitutions across all branch pairs. Since the Grand Convergence software was set up to mark only two branches at a time, we performed convergence calculations for every two branches of deep-sea species in each order. The program was run with the setting –free-bl = 1 to automatically estimate branch length by using the LG amino acid substitution model. A P-value (convergence) > 0.80 was considered to be a convergent site. Genes containing at least one convergent site were considered convergent genes. The convergent genes of all species were extracted and aligned. Unfiltered original sequences containing convergence genes of six orders were extracted from all species and MEGA (Kumar et al. 2016) was used to check for conserved and consistency of convergence sites in all deep-sea fishes.
Gene Annotations and Functional Analysis
SMART (Letunic et al. 2011) and InterPro (Hunter et al. 2012) facilitated the identification and annotation of domains of positive selection genes and convergence evolution genes. The protein-protein interaction database STRING (https://version11.string-db.org/) was applied to explore the interactions among candidate genes. The KEGG (https://www.genome.jp/kegg/) was used to ascertain the functions of candidate genes. The NetPhos 3.1 server predicted serine, threonine, or tyrosine phosphorylation sites in eukaryotic proteins using ensembles of neural networks (Blom et al. 2004).
Gene Synthesis, Plasmid Constructs, Cell Culture, and Transfection
To determine whether the convergence sites affected the Grk1 kinase activity, the grk1 gene and the mutant sites (G144N, Q152R, N402S and all three sites were mutated) gene from zebrafish were synthesized. The lentiviral vectors pCDH-CMV-MCS-EF1-copGFP-T2A-Puro were double-digested with NheI and BamHI endonuclease, and the T4 ligase was used to insert the synthesized genes into a plasmid. The recombinant plasmid was subsequently transfected to human embryonic kidney 293T (HEK293T) cells using Lipofectamine 2000 (Invitrogen). The 293T cells were homogenized for 20 s and immediately placed on ice, incubated for 5 min, centrifuged at 12,000 r/min for 10 min. We then removed the supernatant, added 200 μL chloroform, let stand at room temperature for 2 min, and centrifuged at 12,000 r/min at 4°C for 10 min. We then moved the supernatant into a new 1.5 mL centrifuge tube, added 600 μL isopropyl alcohol, let stand at room temperature for 15 min, centrifuged at 12,000 r/min at 4°C for 15 min, and discarded the supernatant. The precipitate was rinsed with 1 ml 75% ethanol, centrifuged at 12,000 r/min at 4°C for 5 min, and discarded the supernatant. We added 1 mL anhydrous ethanol, centrifuged at 12,000 r/min at 4°C for 5 min, discarded the supernatant, and dried the collection tube at room temperature for 10 min. We then added 40 μL of water to dissolve the RNA.
Cell RNA Extraction and qPCR Detection
At 48 h after transfection, RNA was extracted from the cells. Cells were taken and homogenized in 1 mL Trizol, homogenized for 20 s, and immediately placed on ice. They were then placed in a superclean platform, incubated for 5 min, centrifuged at 12,000 r/min for 10 min, and transferred the supernatant into a new 1.5 mL centrifuge tube, added 200 μL chloroform, shaken well. They were allowed to stand at room temperature for 2 min and then centrifuged at 12,000 r/min at 4°C for 10 min. We transferred the supernatant into a new 1.5 mL centrifuge tube, added 600 μL isopropyl alcohol, mixed well, allowed to stand at room temperature for 15 min, then centrifuged at 12,000 r/min at 4°C for 15 min and discarded the supernatant. The precipitate was rinsed with 1 ml 75% ethanol (750 μL ethanol and 250 μL DEPC water), centrifuged at 12,000 r/min at 4°C for 5 min, and then the supernatant was discarded. We added 1 mL anhydrous ethanol, rinsed the precipitate, centrifuged the solution at 12,000 r/min at 4°C for 5 min, and then discarded supernatant. We allowed the precipitate to dry at room temperature for 10 min. We then add 40 μL DEPC water to dissolve the RNA and stored this solution at −80°C for later use. To remove of DNA from the total RNA, 1 μL DnaseI, 1 μL 10×DNaseI Buffer, and DEPC-H2O were added to the total RNA to make a total volume of 10 μL. The mixture was subjected to 37°C for 30 min before 1 μL EDTA was added and the mixture was subjected to 65°C for10 min. RNA was reverse transcribed into first stand cDNA using M-MLV reverse transcriptase and random primers. Grk1 and Grk1 N402S primers were designed using Primer 5 (Lalitha 2000). Primers for upstream and downstream 0.5 µL, SYBRGreen Mix 12.5 µL, and ddH2O 9.5 µL were added to the cDNA template to achieve a volume of 25 µL. The internal reference gene used GAPDH. Reaction conditions were 95°C,10 min (95°C, 15 s; 60°C, 45 s)×40; 95°C,15 s; 60°C, 1 min; 95°C, 15 s; and 60°C, 15 s.
The expression intensity of cells was detected by qPCR (Real-time quantitative PCR). The primers used for loading control GAPDH were F: TCCGTCTTGAGAAACCTGCC, R: CAACCTGGTGCTCCGTGTAT, while those used for grk1 were F: TCCATGGCAGGAGGAGATGA, R: CACATCCCCCTCCTTTTGCT. The qPCR results were tabulated and calculated with the software LightCycler® 96 SW 1.1.
Protein Extraction and Concentration Determination
We used the appropriate amount of cracking fluid and added phenylmethylsulfonyl fluoride (PMSF) within a few minutes before use so that the final PMSF concentration was 1 mM. We removed the culture medium and washed it again with phosphate buffered saline (PBS), normal saline, or serum-free culture medium. We added cracking fluid at the rate of 100–200 ul cracking fluid to each well of a 6-well plate. We blow several times to make full contact between lysate and cells. Cells were usually lysed 1 to 2 seconds after the lysate came into contact with them. Protein concentration was determined using a bicinchoninic acid kit for protein determination.
Kinase Activity Test
A kinase activity assay kit of Grk1 (GENMED Scientifics Inc., Wilmington, DE, USA) was used to measure the kinase activity according to the manufacturer's instructions. The phosphorylation target of Grk1 was RRREEEEESAAA. In the presence or absence of Grk1 kinase sensitivity inhibitor gradient, the substrate RRREEEEESAAA was phosphorylated by Grk1 to generate the product phosphorylated polypeptide. Then, through the pyruvate kinase and lactate dehydrogenase reaction system, NADH (reduced nicotinamide adenine dinucleotide) was converted to NAD (nicotinamide adenine dinucleotide). Changes in peak absorbance (340 nm) were used to quantitatively analyze the specific activity of Grk1. The experimental procedure was carried out using product number is GMS50160.7.3 v.A according to manufacturer instructions (GENMED SCIENTIFICS INC. U.S.A). A Student's t-test was used to analyze the data.
Supplementary Material
Acknowledgments
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant Nos. XDB31000000 and XDB42000000), the National Natural Science Foundation of China (Grant Nos. 41876179, 32170480, and 31972866), the Major Scientific and Technological Projects of Hainan Province (Grant No. 2019PT03), Chinese Academy of Sciences (Youth Innovation Promotion Association, Chinese Academy of Sciences [http://www.yicas.cn]), State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences (Grant No. GREKF21-04), and the Young Top-notch Talent Cultivation Program of Hubei Province. This research was also supported by the Wuhan Branch of Supercomputing Center, Chinese Academy of Sciences, and the Institute of Deep-sea Science and Engineering, Supercomputing Center, Chinese Academy of Sciences.
Contributor Information
Jing Bo, Institute of Deep-sea Science and Engineering, Chinese Academy of Sciences, Sanya 572000, China; State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China; Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Han Xu, Institute of Deep-sea Science and Engineering, Chinese Academy of Sciences, Sanya 572000, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wenqi Lv, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Cheng Wang, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Shunping He, Institute of Deep-sea Science and Engineering, Chinese Academy of Sciences, Sanya 572000, China; State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China; Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China; Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, 650223, China.
Liandong Yang, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China; Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China.
Supplementary Material
Supplementary materials are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Authors Contributions
S.H. led the project. L.Y. conceived and designed the project. J.B. performed the computational analyses. H.X. provided data. W.L. and C.W. assisted in the experiments.
Data Availability
All data used in this study are provided in supplementary tables 1 and 3, Supplementary Material online.
Literature Cited
- Altschul SF. 2012. Basic local alignment search tool (BLAST). J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Andriashev A P. 1953. Ancient deep-water and secondary deep-water fishes and their importance in a zoogeographical analysis. Notes on special problems in ichthyology. [Google Scholar]
- Angel MV. 1997. 1 What is the deep sea? Fish Physiol. 16:1–41. [Google Scholar]
- Bell M, Henderson R, Sargent J. 1986. The role of polyunsaturated fatty acids in fish. Comp Biochem Physiol B 83:711–719. [DOI] [PubMed] [Google Scholar]
- Bennett N, Sitaramayya A. 1988. Inactivation of photoexcited rhodopsin in retinal rods: the roles of rhodopsin kinase and 48-kDa protein (arrestin). Biochemistry 27:1710–1715. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. Genewise and genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, et al. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649. [DOI] [PubMed] [Google Scholar]
- Burger EH, Klein-Nulend J, Van Der Plas A, Nijweide PJ. 1995. Function of osteocytes in bone—their role in mechanotransduction. J Nutr. 125:2020S–2023S. [DOI] [PubMed] [Google Scholar]
- Castoe TA, et al. 2009. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci. 106:8986–8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chen Y, et al. 2016. Alliin attenuated RANKL-induced osteoclastogenesis by scavenging reactive oxygen species through inhibiting Nox1. Int J Mol Sci. 17:1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress JJ, Nygaard MH. 1973. The chemical composition of midwater fishes as a function of depth of occurence off southern California. Deep Sea Research and Oceanographic Abstracts 20(12):1093–1109. [Google Scholar]
- Costello MJ, Cheung A, Hauwere ND. 2010. Surface area and the seabed area, volume, depth, slope, and topographic variation for the world's seas, oceans, and countries. Environ Sci Technol. 44:8821. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio A, et al. 2009. Stretching single talin rod molecules activates vinculin binding. Science 323:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton E, Marshall N. 1958. The buoyancy of bathypelagic fishes without a gas-filled swimbladder. J Mar Biol Assoc UK 37:753–767. [Google Scholar]
- Denton E, Warren F. 1957. The photosensitive pigments in the retinae of deep-sea fish. J Mar Biol Assoc UK 36:651–662. [Google Scholar]
- Dryja TP. 2000. Molecular genetics of Oguchi disease, fundus albipunctatus, and other forms of stationary night blindness: LVII Edward Jackson Memorial Lecture. Am J Ophthalmol. 130:547–563. [DOI] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. 2009. Mechanically activated integrin switch controls α5β1 function. Science 323:642–644. [DOI] [PubMed] [Google Scholar]
- Gan W, et al. 2021. Global tissue transcriptomic analysis to improve genome annotation and unravel skin pigmentation in goldfish. Sci Rep-Uk 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerringer M, et al. 2021. Habitat influences skeletal morphology and density in the snailfishes (family Liparidae). Front Zool. 18:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, et al. 2009. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325:1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, et al. 1994. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell 79:1267–1276. [DOI] [PubMed] [Google Scholar]
- Hu L-F, Qian A-R, Shang P. 2009. Role of vinculin in cell response to mechanical stimuli. Chin J Biochem Mol Biol. 9. [Google Scholar]
- Hunter S, et al. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40:D306–D312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JB, Spencer M, Niemi GA. 1998. Rhodopsin phosphorylation and its role in photoreceptor function. Vision Res. 38:1341–1352. [DOI] [PubMed] [Google Scholar]
- Johnsen S. 2005. The red and the black: Bioluminescence and the color of animals in the deep sea1. Integr Comp Biol. 45:234–246. [DOI] [PubMed] [Google Scholar]
- Kamimura K, Fuse H, Takimura O, Yamaoka Y. 1993. Effects of growth pressure and temperature on fatty acid composition of a barotolerant deep-sea bacterium. Appl Environ Microbiol. 59:924–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König K, Ostendorf A. 2015. Optically induced nanostructures: biomedical and technical applications. [PubMed] [Google Scholar]
- Kühn H, Dreyer WJ. 1972. Light dependent phosphorylation of rhodopsin by ATP. FEBS Lett. 20:1–6. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalitha S. 2000. Primer premier 5. Biotech Software & Internet Report: The Computer Software Journal for Scient 1:270–272. [Google Scholar]
- Lam-Tung N, Schmidt HA, Arndt VH, Quang MB. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, et al. 2018. De novo transcriptome assembly and positive selection analysis of an individual deep-sea fish. BMC Genomics 19:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK, et al. 2005. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 106:852–859. [DOI] [PubMed] [Google Scholar]
- Lee H-O, Levorse JM, Shin MK. 2003. The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 259:162–175. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 1988. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Febs Lett. 232:78–82. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2011. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40:D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. Multiple sequence alignment methods. 155–170. [DOI] [PubMed] [Google Scholar]
- Lupse N, et al. 2021. Visual gene expression reveals a cone-to-rod developmental progression in deep-sea fishes. Mol Biol Evol. 38:5664–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor RB Jr. 1998. Effect of hydrostatic pressure on nucleic acids. Biopolymers: Orig Res Biomolecules 48:253–263. [DOI] [PubMed] [Google Scholar]
- Malmstrøm M, et al. 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nature Genetics 48:1204–1210. [DOI] [PubMed] [Google Scholar]
- Mazzuca MQ, Khalil RA. 2012. Vascular endothelin receptor type B: structure, function and dysregulation in vascular disease. Biochem Pharmacol. 84:147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT. 2009. The role of vinculin in the regulation of the mechanical properties of cells. Cell Biochem Biophys. 53:115–126. [DOI] [PubMed] [Google Scholar]
- Mills MG, Patterson LB. 2009. Seminars in cell & developmental biology. [Google Scholar]
- Munro C, et al. 2015. The role of ontogeny in physiological tolerance: decreasing hydrostatic pressure tolerance with development in the northern stone crab Lithodes maja. Proc R Soc B: Biol Sci. 282:20150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova Z, et al. 2019. Vision using multiple distinct rod opsins in deep-sea fishes. Science 364:588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohguro H, Van Hooser JP, Milam AH, Palczewski K. 1995. Rhodopsin phosphorylation and dephosphorylation in vivo∗. J Biol Chem. 270:14259–14262. [DOI] [PubMed] [Google Scholar]
- Orr AW, Helmke BP, Blackman BR, Schwartz MA. 2006. Mechanisms of mechanotransduction. Dev Cell 10:11–20. [DOI] [PubMed] [Google Scholar]
- Patra S, Anders C, Schummel PH, Winter R. 2018. Antagonistic effects of natural osmolyte mixtures and hydrostatic pressure on the conformational dynamics of a DNA hairpin probed at the single-molecule level. Phys Chem Chem Phys. 20:13159–13170. [DOI] [PubMed] [Google Scholar]
- Petursdottir H, Gislason A, Falk-Petersen S. 2008. Lipid classes and fatty acid compositions of muscle, liver and skull oil in deep-sea redfish Sebastes mentella over the Reykjanes Ridge. J Fish Biol. 73:2485–2496. [Google Scholar]
- Pradillon F, Gaill F. 2007. Pressure and life: some biological strategies: Kluwer Academic Publishers. [Google Scholar]
- Priede I, Froese R. 2013. Colonization of the deep sea by fishes. J Fish Biol. 83:1528–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qingming Y, Jie Z, Jian C, Tianhua Z. 1998. Carboxyl terminal of rhodopsin kinase is required for the phosphorylation of photo-activated rhodopsin. Cell Res. 04:303–310. [DOI] [PubMed] [Google Scholar]
- Randall DJ, Farrell AP. 1997. Deep-sea fishes. Academic Press.
- Roy P, Witten P, Hall B, Lall S. 2002. Effects of dietary phosphorus on bone growth and mineralisation of vertebrae in haddock (Melanogrammus aeglefinus L.). Fish Physiol Biochem. 27:35–48. [Google Scholar]
- Saito H, Hashimoto J. 2010. Characteristics of the fatty acid composition of a deep-sea vent gastropod, Ifremeria nautilei. Lipids 45:537–548. [DOI] [PubMed] [Google Scholar]
- Sanders HL, Hessler RR. 1969. Ecology of the deep-sea benthos: more detailed recent sampling has altered our concepts about the animals living on the deep-ocean floor. Science 163:1419–1424. [DOI] [PubMed] [Google Scholar]
- Shen X, et al. 2019. Convergent evolution of mitochondrial genes in deep-sea fishes. Front Genet 10:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonato F, et al. 2006. Piezophilic adaptation: a genomic point of view. J Biotechnol. 126:11–25. [DOI] [PubMed] [Google Scholar]
- Somero GN. 1992. Adaptations to high hydrostatic pressure. Annu Rev Physiol. 54:557–577. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RH. 2018. Introduction to physical oceanography: introduction to physical oceanography. [Google Scholar]
- Suetsugu K, Ohta S. 2005. Day and nighttime changes in species composition of deep-sea demersal fishes. J Oceanogr. 61:187–196. [Google Scholar]
- Sun J, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 1:1–7. [DOI] [PubMed] [Google Scholar]
- Team RDC. 2019. R: a language and environment for statistical computing. [Google Scholar]
- Wagner H-J, Fröhlich E, Negishi K, Collin S. 1998. The eyes of deep-sea fish II. Functional morphology of the retina. Prog Retin Eye Res. 17:637–685. [DOI] [PubMed] [Google Scholar]
- Warrant EJ, Adam Locket N. 2004. Vision in the deep sea. Biol Rev. 79:671–712. [DOI] [PubMed] [Google Scholar]
- Webb P. 2019. Introduction to oceanography. [Google Scholar]
- Wilden U, Hall SW, Kuhn H. 1986. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci USA 83:1174–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. 2019. Widespread nocturnality of living birds stemming from their common ancestor. BMC Evol Biol. 19:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Teitelbaum SL. 2013. Osteoclasts: new insights. Bone Res. 1:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Sippel KC, Berson EL, Dryja TP. 1997. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 15:175–178. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. 2010. Elevated pressure downregulates ZO-1 expression and disrupts cytoskeleton and focal adhesion in human trabecular meshwork cells. Mol Vis. 17:2978–2985. [PMC free article] [PubMed] [Google Scholar]
- Yu X-J, et al. 2006. Detecting lineage-specific adaptive evolution of brain-expressed genes in human using rhesus macaque as outgroup. Genomics 88:745–751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are provided in supplementary tables 1 and 3, Supplementary Material online.