Abstract
Attachment to surfaces by the prosthecate bacterium Caulobacter crescentus is mediated by an adhesive organelle, the holdfast, found at the tip of the stalk. Indirect evidence suggested that the holdfast first appears at the swarmer pole of the predivisional cell. We used fluorescently labeled lectin and transmission electron microscopy to detect the holdfast in different cell types. While the holdfast was readily detectable in stalked cells and at the stalked poles of predivisional cells, we were unable to detect the holdfast in swarmer cells or at the flagellated poles of predivisional cells. This suggests that exposure of the holdfast to the outside of the cell occurs during the differentiation of swarmer to stalked cells. To investigate the timing of holdfast synthesis and exposure to the outside of the cell, we have examined the regulation of a holdfast attachment gene, hfaA. The hfaA gene is part of a cluster of four genes (hfaABDC), identified in strain CB2A and involved in attachment of the holdfast to the polar region of the cell. We have identified the hfaA gene in the synchronizable C. crescentus strain CB15. The sequence of the CB2A hfaA promoter suggested that it was regulated by ς54. We show that the transcription of hfaA from either strain is not dependent on ς54. Using a hfaA-lacZ fusion, we show that the transcription of hfaA is temporally regulated during the cell cycle, with maximal expression in late-predivisional cells. This increase in expression is largely due to the preferential transcription of hfaA in the swarmer pole of the predivisional cell.
Cell division of the gram-negative bacterium Caulobacter crescentus gives rise to two distinct progeny cells, a sessile cell (containing a stalk) and a motile cell (containing a single polar flagellum) (5). Stalked cells are often found attached to surfaces by means of an adhesive holdfast found at the tip of the stalk. Staining properties and enzyme sensitivity studies indicate that the holdfast is a complex polysaccharide (20), with acidic components such as uronic acids (33). It has been proposed that the appearance of the holdfast is temporally regulated during the cell cycle and that the holdfast first appears at the base of the flagellum in the swarmer pole of the predivisional cell (26). The presence of the holdfast at the tip of the stalk is thought to result from the growth of the stalk at the site previously occupied by the flagellum during the differentiation of swarmer to stalked cells. How the spatial and temporal regulation of holdfast expression is achieved is unclear.
A cluster of four genes (hfaABDC) involved in the attachment of the holdfast to the cell was previously identified by Tn5 insertion mutagenesis (16). The exact role of each of these genes is unknown. The C-terminal region of HfaA is similar to the C termini of pilus tip proteins, such as the PapG adhesin from Escherichia coli and the SmfG adhesin from Serratia marcescens (16). These adhesins interact with host cell polysaccharides and with a protein anchor in the pilus, thus mediating the attachment of these bacteria to host cell surfaces. It is possible that HfaA functions analogously in C. crescentus (16). HfaB is similar to proteins that function in transcriptional activation and may activate the transcription of hfaC (16). The sequence of HfaD contains three putative membrane-spanning regions, and it has been suggested that it acts as the membrane-associated protein anchor between HfaA and the cell (17). HfaC is similar to ATP-binding transport-related proteins (17).
A sequence identical to the consensus for ς54-dependent promoters was found upstream of the hfaA transcription start site, suggesting that its transcription may be subject to cell cycle regulation like other known ς54 promoters in C. crescentus (16). ς54 is present in both gram-negative and gram-positive members of the Eubacteria and is required for the expression of a wide variety of genes, including those involved in nitrogen fixation, pilus production, dicarboxylic acid transport, and xylene catabolism in these bacteria (21). In C. crescentus, ς54 does not appear to be required for general metabolic functions but is needed for the biosynthesis of two polar organelles, the flagellum and the stalk (6).
The hfaA gene was initially characterized in C. crescentus CB2A (16, 23). In this study, we extend this work to investigate the transcriptional regulation of hfaA in strain CB15. We show that hfaA is not transcribed by a ς54 promoter. We find that the transcription of hfaA is cell cycle regulated and that hfaA is maximally expressed in the swarmer compartment of the predivisional cell. The use of fluorescently labeled lectin to detect the holdfast on live cells indicates that neither the flagellar poles of predivisional cells nor swarmer cells possess a holdfast. However, the holdfast was readily detectable at the tips of short stalks. This is consistent with the hypothesis that the holdfast appears at the tips of nascent stalks during the differentiation of swarmer to stalked cells.
MATERIALS AND METHODS
Materials, bacterial strains, and growth conditions.
Oligonucleotides hfaA Rev (5′GAACGAAGCCGAAAAGCTTGACATCGATTG3′), hfaA+135 (5′CCATTTTTTCGCTGCAGTGGGGCTACC3′), and hfaA+60 (5′GGGCTGGTCCCTGCAGTCTATCTAGGG3′) were obtained from either the Institute for Molecular and Cellular Biology at Indiana University or Operon Technologies, Inc. Ludox was obtained from Dupont, radionuclides were obtained from ICN Radiochemicals, and antibiotics were obtained from Sigma or Amresco. The strains and plasmids used in this study are described in Table 1. The strain YB1371 (NA1000 hfaA::pAA2) used in the analysis of the cell cycle expression of hfaA was constructed as follows. Plasmid pAA2 is a transcriptional fusion containing the CB15 hfaA promoter cloned upstream of the promoterless lacZ gene in the plasmid pGSZ. This plasmid was introduced into NA1000 by conjugation. As pAA2 does not replicate in C. crescentus, selection for gentamicin-resistant colonies selects for integrants that arise by homologous recombination.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or construction | Source or reference |
|---|---|---|
| E. coli | ||
| S17-1 | E. coli 294::RP4-2(Tc::Mu)(Km::Tn7) | 29 |
| DH5αF′ | φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (r− m+) deoR thi-1 supE44 λ− gyrA96 relA1 | 18 |
| C. crescentus | ||
| CB15 | Wild type | 25 |
| NA1000 | syn-1000, previously called CB15N, a synchronizable derivative of CB15 | 10 |
| CB2A | rsaA mutant of CB2 | 31 |
| CB2AG9 | hfaAB::Tn5 (Kmr) | 23 |
| SU213 | CB15 rpoN::Tn5 | 6 |
| SC1117 | flgH::Tn5 | 9 |
| YB1293 | NA1000 ORF203::Ω Spr | 14 |
| YB1306 | NA1000 ORF208::Ω Spr | 14 |
| YB1270 | NA1000 ORF159::pRJ23 | 14 |
| YB1369 | CB15 hfaA::pAA2 | This work |
| YB1370 | CB15 rpoN::Tn5 hfaA::pAA2 | This work |
| YB1371 | NA1000 hfaA::pAA2 | This work |
| Plasmids | ||
| pBB31 | 1.7-kb BclI-BglII hfaAB fragment from CB2A cloned into pUC19 | 16 |
| pIC-HfaA | 325-bp PstI-ClaI fragment from pBB31 containing the CB2A hfaA promoter cloned into PstI-ClaI sites of pIC20H | This work |
| pRJ38 | 325-bp PstI-HindIII fragment from pIC-HfaA cloned into PstI-HindIII sites of pRKlac290 | This work |
| pRJ41 | Cosmid 4-1, a pLAFR5-derived cosmid containing hfaA from CB15 | This work |
| pRJ39 | 3.0-kb PstI-SmaI fragment from pRJ41 containing hfaA from CB15 cloned into PstI-SmaI sites of pSKII− | This work |
| pRJ52 | 245-bp PCR product created by using hfaA+60 and hfaA Rev primers cut with PstI and HindIII and cloned into PstI-HindIII sites of pRKlac290 | This work |
| pRJ54 | PstI-HindIII sites of pRKlac290 177-bp PCR product created by using hfaA+135 and hfaA Rev primers cut with PstI and HindIII and cloned into PstI-HindIII sites of pRKlac290 | This work |
| pAA1 | 325-bp PstI-ClaI fragment from pRJ39 cloned into PstI-ClaI sites of pIC20R | This work |
| pAA2 | 325-bp BamHI-EcoRI fragment from pAA1 containing the hfaA promoter cloned into BamHI-EcoRI sites of pGSZ | This work |
| pUC19 | Ampr, lacZ | 35 |
| pSKII− | Phagemid, Ampr, ColE1 ori, f1− ori | 2 |
| pIC20R | 2.7-kb phagemid, Ampr, derivative of pUC18 | 19 |
| pIC20H | 2.7-kb phagemid, Ampr, derivative of pUC18 | 19 |
| pRKlac290 | lacZ transcriptional fusion vector, Tetr, IncP-1 replicon, mob+ | 12 |
| pGSZ | Sper, derivative of pGMTZ1 | 1 |
E. coli strains were grown at 37°C in Luria-Bertani medium with one or more of the following antibiotics: ampicillin (100 μg/ml), gentamicin (15 μg/ml), or tetracycline (12 μg/ml). C. crescentus strains were routinely grown at 30°C in peptone-yeast extract (PYE) medium (25) supplemented with nalidixic acid (20 μg/ml) and either tetracycline (2 μg/ml) or gentamicin (2.5 μg/ml). M2 minimal glucose (M2G) medium (15) was used when cells were synchronized by Ludox density gradient centrifugation.
General DNA manipulations, cloning, and sequencing.
General cloning procedures were done as described previously (3, 13). The hfaA gene from C. crescentus CB15 was identified by Southern blot hybridization as follows. A 400-bp SacII fragment of CB2A hfaA from pBB31 was used as a probe to screen a C. crescentus cosmid library, and a cosmid containing hfaA (pRJ41) was isolated. A 3.0-kb PstI-SmaI fragment from pRJ41 that hybridized to the CB2A hfaA probe was subcloned (pRJ39). Several overlapping subclones were generated from pRJ39 and used for sequencing CB15 hfaA. DNA sequencing was done by the dideoxynucleotide chain termination method (28) on double-stranded templates isolated with Qiagen Mini-Prep kits. Reactions were done by a modification of the Thermo Sequenase dye terminator cycle sequencing protocol (Amersham). The following program was used: 1 min at 96°C, followed by 25 cycles of 96°C for 30 s, 50°C for 15 s, and 60°C for 4 min in the presence of either M13 forward or reverse primers and labeled dideoxynucleoside triphosphates. Reactions were run in the Institute for Molecular and Cellular Biology at Indiana University on an ABI PRISM 377 DNA sequencer. The DNA sequence was analyzed with the package of the Genetics Computer Group of the University of Wisconsin (8) and Sequencher 3.0 (Gene Codes Corporation).
Detection of the holdfast with fluorescein-labeled lectin.
A fluorescent lectin-binding assay was used to detect the holdfast (20). A volume (2 μl) of fluorescein-conjugated wheat germ agglutinin (FITC-WGA; Molecular Probes) (5 mg/ml of stock) was added to 200 μl of cells growing exponentially. The mixture was incubated at room temperature for 20 min, diluted with 1 ml of water, and centrifuged. The cell pellet was resuspended in 30 μl of Slowfade antifading reagent (Molecular Probes), and 1 μl was examined by fluorescence microscopy. Epifluorescence photomicroscopy was performed on a Nikon Eclipse E800 light microscope equipped with a Nikon B-2E fluorescein isothiocyanate (FITC) filter cube for FITC and a 100× Plan Apo oil objective. Images were captured by a Princeton Instruments cooled charge-coupled device camera (model 1317) and the Metamorph imaging software package (v. 3.0).
Electron microscopy.
Cells were grown to mid-log phase in PYE medium, washed by centrifugation at 5,000 rpm for 5 min in an Eppendorf centrifuge, and resuspended in a one-fifth volume of phosphate-buffered saline. Cells (5 μl) were spotted onto carbon-coated grids and allowed to settle for 20 min. The grids were blotted dry and washed once with water. The cells were stained with 1% uranyl acetate for 30 s and washed four times with water. They were examined in a Philips model 300 electron microscope at 60 kV.
Analysis of the promoter.
The hfaA promoter and mutant versions of the hfaA promoter were cloned upstream of the lacZ gene in pRKlac290 or pGSZ and analyzed in wild-type and mutant backgrounds for promoter activity. Assays were done in duplicate on a minimum of two independent cultures in each case. β-Galactosidase activity conferred by these plasmids was measured at 30°C as described previously (22), except that cells were permeabilized with chloroform.
To assay the time of transcription of hfaA, late log-phase cultures grown in M2G medium containing tetracycline were synchronized by the Ludox density centrifugation method (10). Swarmer cells were collected and allowed to proceed through the cell cycle in fresh M2G medium at 30°C. At 15-min intervals, 1-ml culture samples were labeled with 15 μCi of [35S]methionine (Trans-Label) for 5 min, collected by centrifugation, and frozen at −20°C. Cells were lysed in wash buffer (50 mM Tris, pH 8.3–450 mM NaCl–0.5% Triton X-100). A small volume of each sample was precipitated with 10% trichloroacetic acid, collected on glass fiber filters, and counted in a scintillation counter cocktail. Equivalent counts of radiolabeled protein were then immunoprecipitated with an antiflagellin antibody (an internal control for the cell cycle) and an anti-β-galactosidase antibody (Boehringer Mannheim). The samples were processed as described previously (14).
Pole-specific expression was measured as described previously (34). Synchronized swarmer cells were allowed to proceed to the predivisional stage and were pulse-labeled for 10 min with 30 μCi of [35S]methionine. Unlabeled methionine (0.1 μM) was used to chase the label, and the cells were allowed to divide. The progeny swarmer and stalked cells were separated by Ludox density gradient centrifugation, and the transcription level from the hfaA promoter was determined as described above.
Nucleotide sequence accession number.
The DNA sequence of the hfaA gene was submitted to GenBank and has been given the accession no. AF058792.
RESULTS
ς54 is not required for holdfast synthesis or attachment.
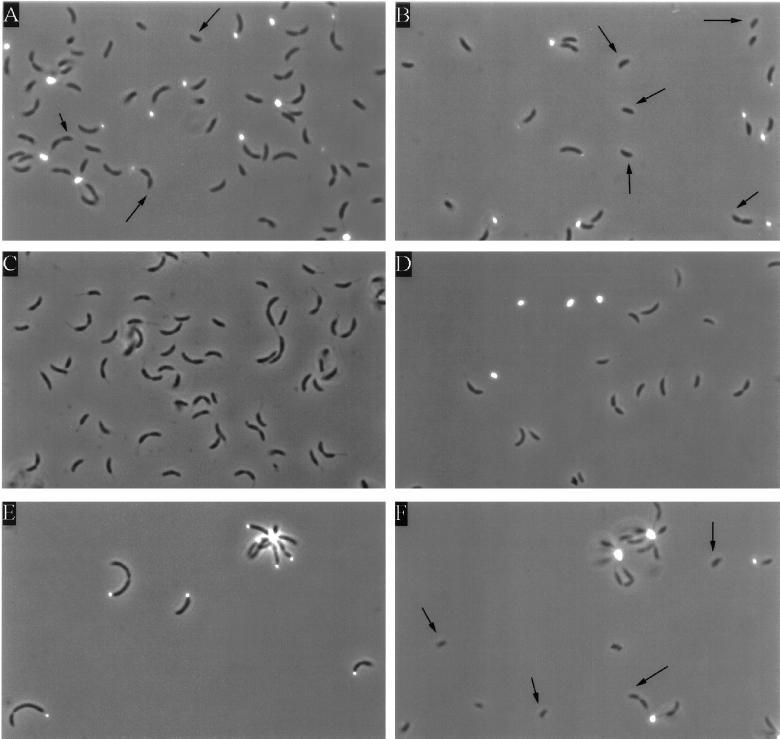
Since the CB2A hfaA gene contains sequences at nucleotides −24 and −12 (numbered in relation to the transcription initiation site) that are identical to those recognized by the ς54-RNA polymerase holoenzyme (16), we expected a strain that lacks ς54 (such as an rpoN::Tn5 mutant) to display the holdfast-shedding phenotype seen in the hfaAB::Tn5 mutant. To determine if this was the case, we labeled the holdfast in an rpoN::Tn5 mutant, SU213, with FITC-lectin. The wheat germ agglutinin lectin binds specifically to the holdfast, and its conjugation to FITC allows its visualization by epifluorescence microscopy (20). We assayed for the presence of the holdfast and quantitated this labeling in the wild-type strains CB15 and CB2A. Wild-type CB15, which forms rosettes and contains a normal holdfast, showed spots of FITC-lectin attached to the stalks in stalked cells and predivisional cells (Fig. 1A and B). No fluorescent labeling was seen in NA1000, a strain that lacks a holdfast, indicating that the background level of fluorescence from this technique is very low (Fig. 1C). Figure 1D shows labeling of the holdfast material in CB2AG9, an hfaAB::Tn5 mutant, which synthesizes a normal holdfast but sheds it into the medium (23). This shed holdfast was labeled, as indicated by the FITC-conjugated lectin spots which are not associated with cells. Only 10% of the predivisional cells in CB2AG9 were labeled, indicating that most of the predivisional cells had shed their holdfasts (Table 2). In the rpoN::Tn5 mutant, 75% of the predivisional cells were labeled with fluorescent lectin (Fig. 1E; Table 2). This is comparable to the percentages of predivisional cells labeled by FITC-lectin in the C. crescentus wild-type strains CB15 and CB2A (Table 2). In addition, shed holdfasts were not detected in the culture medium of the rpoN::Tn5 mutant. This suggests that ς54 is not required for hfaA expression. It is also possible that two promoters drive the expression of hfaA and only one of them is ς54 dependent or that CB15 hfaA is regulated differently than CB2A hfaA. Alternatively, shedding may not occur efficiently in the stalkless rpoN mutant, because the holdfast is not subject to the same shearing force as when it is at the tips of stalks in wild-type cells.
FIG. 1.
Fluorescein-conjugated lectin labeling of the holdfast in various strains of C. crescentus. Micrographs were taken in combined fluorescence and phase-contrast modes of wild-type CB15 (A and B), NA1000 (lacking a holdfast) (C), holdfast-shedding mutant CB2AG9 (D), SU213 (rpoN::Tn5) (E), and SC1117 (flgH::Tn5) (F). The arrows indicate swarmer cells and the swarmer poles of predivisional cells.
TABLE 2.
Quantitation of holdfast labeling in different strains with fluorescein-labeled lectin
| C. crescentus strain | % of cells labeleda
|
|
|---|---|---|
| Predivisional | Swarmer | |
| CB15 | 81 | 0 |
| CB2A | 87 | 0 |
| SU213 (rpoN::Tn5) | 75 | ND |
| SC1117 (flgH::Tn5) | 79 | 0 |
| CB2AG9 (hfaAB::Tn5) | 10 | ND |
A minimum of 112 predivisional cells and 45 swarmer cells were examined in each case. ND, not determined.
Identification of hfaA from C. crescentus CB15.
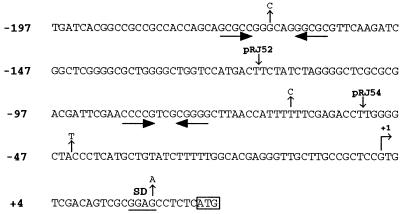
To determine whether hfaA from CB15 had regulatory sequences similar to those of the CB2A hfaA gene, we cloned hfaA from CB15 (see Materials and Methods). Analysis of the nucleotide sequence of the CB15 hfaA gene indicated that it was 98% identical to the CB2A hfaA gene, while the predicted product of CB15 hfaA (GenBank accession no. AF058792) is 95% identical to that of CB2A HfaA. Only 3 nucleotides differed between CB15 and CB2A in the 200 bp upstream of the transcription start site (Fig. 2). This is consistent with previous evidence which revealed that the freshwater C. crescentus strains CB15 and CB2A are highly similar (32). The high degree of identity between the CB15 and CB2A hfaA regulatory sequences makes it unlikely that the CB15 and CB2A hfaA genes are regulated differently; therefore, we studied the regulation of the CB2A hfaA promoter in CB15.
FIG. 2.
Sequence of the hfaA promoter region in CB2A and CB15. Positions where the CB15 hfaA promoter sequence differs are shown above the sequence. Inverted repeat sequences are shown by inverted horizontal arrows. The 5′ endpoints of plasmids pRJ52 and pRJ54, used for deletion analysis, are indicated. The transcription initiation site (+1), as mapped previously (16), is indicated by the bent arrow; the translation initiation codon is boxed, and the nucleotides are numbered relative to the transcription start site. The putative Shine-Dalgarno sequence is underlined.
The hfaA gene is not transcribed by a ς54 promoter.
The presence of a promoter proximal to hfaA was previously suggested by complementation and S1 mapping studies (16). To confirm the presence of a promoter for hfaA in this region, a 325-bp fragment containing the putative CB2A hfaA promoter and the first 30 codons of hfaA was cloned upstream of a promoterless lacZ gene to generate a transcriptional fusion. This fusion (pRJ38 [Table 3]) yielded approximately 2,500 Miller units of β-galactosidase activity in both CB15 (wild type) and NA1000 (a synchronizable derivative of CB15). Deleting sequences upstream of nucleotide −117 (pRJ52) reduced transcriptional activity to 1,090 Miller units. Removing bases upstream of nucleotide −52 (pRJ54) completely abolished transcription (Table 4). Thus, essential promoter elements are present upstream of nucleotide −52, and sequences sufficient for promoter activity are present downstream of nucleotide −117.
TABLE 3.
Expression of the hfaA promoter in different mutant backgrounds
| C. crescentus strain | Genotype | β-Galactosidase activitya |
|---|---|---|
| NA1000 | syn-1000 | 2,560 |
| CB15 | Wild type | 2,350 |
| CB2A | Wild type | 2,580 |
| SU213 | rpoN::Tn5 | 3,400 |
| YB1270 | ORF159 mutant | 1,730 |
| YB1293 | ORF203 mutant | 2,480 |
| YB1306 | ORF208 mutant | 2,360 |
| YB1369 | CB15 hfaA::pAA2 | 1,230 |
| YB1370 | rpoN::Tn5 hfaA::pAA2 | 2,350 |
All results are expressed in Miller units and have a standard deviation of less than 10%. Activities of pRJ38 were measured in each case, except in YB1369 and YB1370, for which the activity of the chromosomal fusion pAA2 was measured.
TABLE 4.
Expression of mutant hfaA promoters in wild-type and rpoN::Tn5 backgrounds
| Plasmid | Nucleotide rangea | β-Galactosidase activityb in:
|
|
|---|---|---|---|
| Wild type | rpoN::Tn5 | ||
| pRJ38 | −197 to +125 | 2,500 | 3,220 |
| pRJ52 | −117 to +125 | 1,090 | 1,540 |
| pRJ54 | −52 to +125 | 90 | 110 |
The numbers represent the endpoints of the fragments cloned upstream of lacZ, relative to the hfaA transcription start site, which has been designated +1.
All results are expressed in Miller units and have a standard deviation of less than 10%.
To determine if the transcription of the hfaA gene is controlled by a ς54 promoter, we studied its expression in an rpoN null mutant. When the pRJ38 hfaA-lacZ fusion was introduced into SU213, a strain that lacks ς54, it produced 3,400 Miller units of β-galactosidase activity (Table 3). Similarly, transcription of the pRJ52 fusion was approximately 1.5-fold higher in the rpoN mutant. These results demonstrate that ς54 is not required for promoter activity in the 325-bp fragment of hfaA. The increase in β-galactosidase activity of the hfaA-lacZ fusion in the rpoN::Tn5 mutant suggests that ς54 or a ς54-dependent gene has a negative effect on hfaA transcription. The same effect of an rpoN mutation on hfaA transcription was observed with an hfaA-lacZ transcriptional fusion integrated at the hfaA locus. The structure of the integrations is shown in Fig. 3A. The chromosomal hfaA-lacZ fusion produced 1,230 Miller units of β-galactosidase activity in CB15 (YB1369 [Table 3]) and 2,350 Miller units in the rpoN::Tn5 mutant (YB1370 [Table 3]).
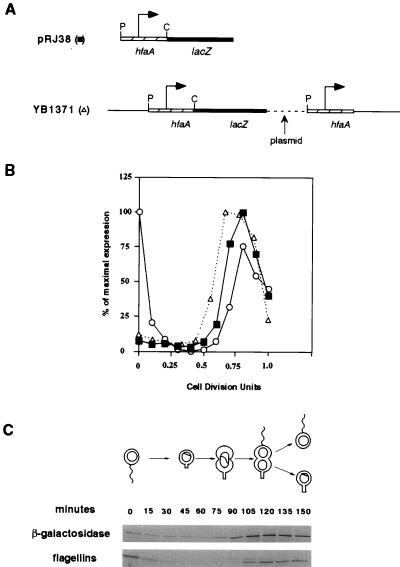
FIG. 3.
Cell cycle expression of the hfaA promoter. (A) The construct pRJ38, containing the hfaA promoter fused to a promoterless lacZ gene, is shown, and the relevant restriction sites are indicated (P, PstI; C, ClaI). The bent arrow indicates the position of the promoter. A diagram of the integrated hfaA-lacZ transcriptional fusion in YB1371 is also shown. The hatched box represents hfaA, and the black box represents lacZ. The plasmid sequences are represented by dashed lines, while the bent arrow indicates the location of the promoter driving hfaA. Integration of the fusion plasmid restores a wild-type hfaA gene downstream of the promoter. (B) Graph of the cell cycle expression of plasmid-borne hfaA (■) and the 25-kDa flagellin protein (○), compared to the temporal transcription of chromosomal hfaA (▵). The gel corresponding to the autoradiographs shown in panel C was quantitated by phosphorimaging, and the intensities of the bands were plotted as the percentages of the maximal intensity for the band corresponding to each protein. The time of division was 150 min, which is represented as 1 cell division unit. Each synchrony experiment was repeated twice with similar results. (C) Immunoprecipitation of β-galactosidase and flagellins from the strain containing pRJ38 throughout the cell cycle. The progression through the cell cycle is shown above the autoradiograph.
We determined that the effect of the rpoN::Tn5 mutation on hfaA transcription was not due to polar effects on downstream genes. The transcription of the hfaA-lacZ fusion was assayed in ORF208, ORF203, and ORF159 mutants (14). In both the ORF208 and ORF203 mutant backgrounds, the expression of hfaA was comparable to that in wild-type cells, whereas in the ORF159 mutant, hfaA expression decreased by 30% (Table 3). This result is similar to what is observed with the expression of the ς54-dependent flagellar gene fljK in these mutant backgrounds (14). Since the expression of hfaA is not increased in any of these mutant backgrounds, the open reading frames downstream of rpoN do not play a role in the ς54-mediated repression of the hfaA promoter. These results indicate that rpoN acts genetically like a negative regulator of hfaA transcription.
Cell cycle and compartment-specific expression of hfaA.
We used a transcriptional fusion of the hfaA promoter to lacZ to analyze the cell cycle transcription of hfaA (Fig. 3). Swarmer cells were isolated by Ludox density gradient centrifugation from a mixed culture of NA1000 cells harboring pRJ38. These swarmer cells were allowed to proceed synchronously through the cell cycle. At 15-min intervals, samples of the culture were pulse-labeled with [35S]methionine and cell extracts were immunoprecipitated with antiflagellin and anti-β-galactosidase antibodies. Figure 3C shows the results of this immunoprecipitation after the samples had been subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The synthesis of β-galactosidase was at a low level for the first half of the cell cycle, after which it increased approximately 10-fold (Fig. 3B). The peak level of expression from the hfaA promoter occurred at the predivisional cell stage coincident with the increase in flagellin expression. The expression from this promoter then decreased as the cells divided. This demonstrates that the transcription of hfaA is temporally controlled in C. crescentus, with the maximal level of expression occurring in the predivisional cells.
We also analyzed the temporal control of hfaA transcription in the NA1000::hfaA-lacZ integration strain, YB1371. In this strain, the lacZ gene is transcribed from the hfaA promoter in the 325-bp fragment and any upstream promoter(s) (Fig. 3A). The transcription of hfaA was assayed in synchronized cells as described above. As indicated in Fig. 3B, the cell cycle expression of hfaA is similar whether it is measured by using a plasmid-borne or a chromosomal fusion.
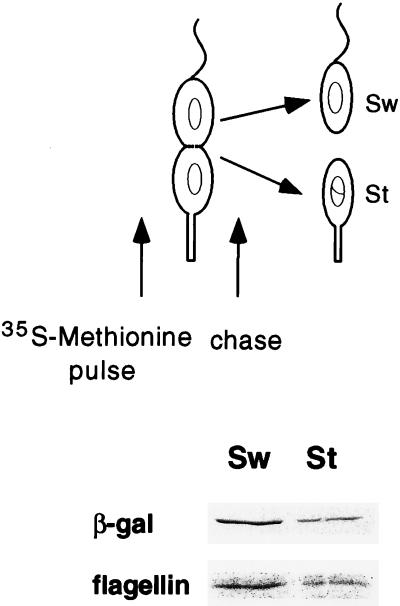
The swarmer and stalked compartments of the predivisional cell differ not only in morphology but also in their programs of gene expression. Because a holdfast is already present at the stalked pole, hfaA expression may not be required in the stalked compartment of the predivisional cell. To determine in which pole of the predivisional cell hfaA is transcribed, NA1000 cells containing the hfaA-lacZ fusion were synchronized, and the swarmer cells were allowed to proceed to the late-predivisional cell stage. At this stage, proteins were pulse- labeled for 10 min with [35S]methionine and then chased with an excess of nonradioactive methionine (Fig. 4). The cells were allowed to divide, and progeny cells were isolated. Cell extracts were immunoprecipitated with antiflagellin and anti-β-galactosidase antibodies and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. In this experiment, the amounts of labeled β-galactosidase in the swarmer and stalked cell fractions reflect the rate of transcription of hfaA from its promoter in the swarmer or stalked compartment of the predivisional cell (11). As seen in Fig. 4, the transcription of hfaA occurred preferentially in the swarmer pole of the predivisional cell.
FIG. 4.
Cell type-specific expression of hfaA. The transcriptional fusion containing the hfaA promoter region (pRJ38) is shown in Fig. 3A. Cells containing pRJ38 were synchronized, and the swarmer cells were allowed to proceed to the late-predivisional stage (135 min). Proteins were labeled with [35S]methionine for 10 min and then chased with unlabeled methionine as shown. After division, the stalked (St) and swarmer (Sw) cells were separated by density gradient centrifugation and then processed as shown in Fig. 3. The autoradiograph of immunoprecipitated β-galactosidase and flagellin proteins is shown. This experiment was repeated twice with reproducible results.
The holdfast is not detectable in swarmer cells.
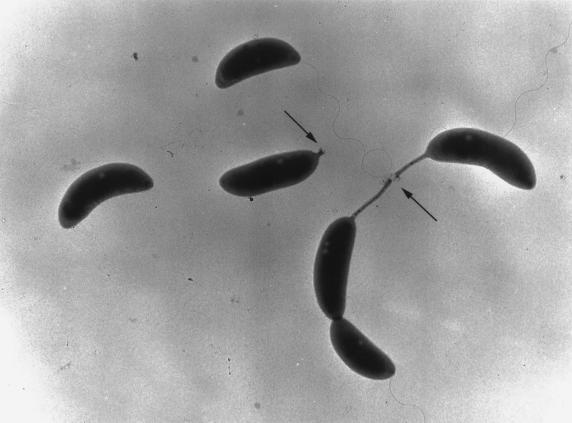
The ability of the swarmer poles of predivisional cells to attach to surfaces has suggested that the holdfast first appears at the base of the flagellum at the swarmer pole of the predivisional cell (25, 26). To determine if the holdfast is present at the swarmer pole, we used FITC-lectin to label the holdfast in various strains. Approximately 80% of the predivisional cells were labeled in the C. crescentus strains CB15 (Fig. 1A and B) and CB2A (data not shown), with labeling always occurring at the stalked pole (Table 2). Thus, lectin binds very efficiently to the stalked pole of the predivisional cell. In hundreds of predivisional cells examined in many cell cultures, we never observed FITC-lectin labeling at the swarmer pole of the predivisional cell (Fig. 1). In addition, we never observed FITC-lectin labeling of swarmer cells (Fig. 1) but readily observed labeling at the tips of stalked cells possessing short stalks. To eliminate the possibility that the fluorescein-conjugated lectin could not bind a putative holdfast at the flagellated pole of a predivisional cell because of the flagellum, we labeled the holdfast in a flagellar mutant (flgH::Tn5 [Table 2 and Fig. 1F]). A total of 79% of the predivisional cells were labeled in the flgH::Tn5 strain, and the FITC-conjugated lectin spots were always associated with the stalked poles of these predivisional cells. No labeling was observed in swarmer cells or at the flagellated poles of predivisional cells (Fig. 1), but labeling was readily observed at the tips of short stalks. All these observations are consistent with electron micrographs, in which the holdfast is clearly visible as an amorphous material at the tips of stalks but not at the base of the flagellum in predivisional or swarmer cells (Fig. 5). This suggests that the holdfast appears at the tips of stalks at the time of or shortly after the initiation of stalk synthesis.
FIG. 5.
The holdfast is visible after the initiation of stalk synthesis in this transmission electron micrograph of wild-type C. crescentus (CB15) grown in PYE medium. The holdfast material is indicated by arrows and is visible as amorphous material at the tips of stalks.
DISCUSSION
The holdfast is important for the attachment of C. crescentus cells to surfaces; however, little is known about the regulation of holdfast synthesis and holdfast attachment genes during the cell cycle. Furthermore, the time of holdfast appearance at the pole of the cell is not known. In this paper, we investigate two aspects of holdfast synthesis: the transcription of a holdfast attachment gene, hfaA, and the appearance of the holdfast at the pole of the cell. We demonstrate that hfaA transcription is temporally controlled during the cell cycle, with maximal transcription in the swarmer pole of the predivisional cell. We show that the holdfast is not present in swarmer cells or at the swarmer poles of predivisional cells. Our results suggest that the holdfast appears during the differentiation of swarmer to stalked cells.
The hfaA gene possesses a ς54 promoter sequence at the requisite distance from the transcription start site (16). However, our results indicate that hfaA is not transcribed by a ς54-dependent promoter. hfaA is still transcribed in a ς54 null mutant, and lectin labeling of the holdfast in an rpoN::Tn5 mutant demonstrates that ς54 is not required for holdfast attachment. It is possible that hfaA is transcribed by the ς54-RNA polymerase holoenzyme under certain conditions that are as yet unknown. Surprisingly, our data indicates that ς54 or a ς54-dependent event has a negative effect on the transcription of hfaA. The ability of ς54 to bind to certain promoters in the absence of core polymerase (7) raises the possibility that ς54 itself could bind to the sequences at nucleotides −24 and −12 of the hfaA promoter and repress the transcription of hfaA. A similar case that has recently been found is the Bradyrhizobium japonicum fixRnifA promoter that contains two overlapping promoters: one that is ς54 dependent and one that is dependent on a second unidentified form of RNA polymerase (4). Alternatively, the increase in hfaA transcription in the rpoN mutant could be an indirect consequence of the pleiotropic phenotype of rpoN mutants. The fact that some of the rpoN mutant cells have a holdfast at both poles suggests that rpoN mutants have an increased level of holdfast synthesis (Fig. 1E).
Using an hfaA-lacZ fusion integrated at the hfaA locus, we were able to show that the transcription of hfaA is temporally regulated during the cell cycle, with maximal levels of transcription occurring in predivisional cells. Because the holdfast does not seem to appear until the differentiation of the swarmer cell during the next cell cycle (see below), the reason for the preferential transcription of hfaA in the swarmer compartment of the predivisional cell is unclear. This burst in hfaA transcription may serve to load hfaA mRNA or HfaA itself in the swarmer compartment prior to the beginning of the next cell cycle. This would ensure that the holdfast attachment protein is present in the swarmer cell, ready to be used when the swarmer cell differentiates into a stalked cell.
Previous observations that the swarmer poles of predivisional cells can attach to surfaces suggested that the holdfast first appears at the flagellated poles of predivisional cells (25, 26). Electron micrographs fail to reveal the presence of a holdfast at the base of the flagellum, whereas it is clearly visible at the tips of short stalks. In our studies, we were unable to detect any binding of fluorescent lectin to swarmer cells or to the flagellated poles of predivisional cells, whereas the binding of lectin to stalked cells and to the stalked poles of predivisional cells was very efficient. Our failure to detect any binding of fluorescent lectin at the swarmer pole is not due to the presence of the flagellum, because we could not detect any binding to the swarmer pole in a flagellar mutant. These results suggest that the exposure of the holdfast to the outside of the cell occurs during the differentiation of swarmer to stalked cells. In addition to a single flagellum, the swarmer pole contains pili (30). Pili are involved in mediating attachment to surfaces in many bacteria (13) and have also been implicated in promoting the primary adhesion event in Hyphomonas, another prosthecate bacterium (27). Thus, perhaps the attachment of the swarmer pole of C. crescentus to surfaces is mediated by the pilus and not the holdfast. This is consistent with the observation that swarmer cells collide and stick more frequently to glass surfaces than nonmotile stalked and dividing cells (24). Based on our findings, we suggest that the holdfast is either not present or not accessible at the swarmer poles of predivisional cells and that other adhesive components of that pole, perhaps pili, facilitate its attachment to surfaces.
ACKNOWLEDGMENTS
We are particularly grateful to John Smit for generously providing many clones and strains and for helpful discussions. We thank Anahita Amiri for constructing pAA2 and members of the Brun lab for helpful suggestions on the manuscript.
This work was supported by National Institutes of Health predoctoral fellowship GM07757 (to R.S.J.) and National Institutes of Health grant GM51986 (to Y.V.B.).
REFERENCES
- 1.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altin-Mees M A, Short J M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley/Greene; 1989. [Google Scholar]
- 4.Barrios H, Grande R, Olvera L, Morett E. In vivo footprinting analysis reveals that the complex Bradyrhizobium japonicum fixRnifA promoter region is differently occupied by two distinct RNA polymerase holoenzymes. Proc Natl Acad Sci USA. 1998;95:1014–1019. doi: 10.1073/pnas.95.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brun Y, Marczynski G, Shapiro L. The expression of asymmetry during cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 6.Brun Y V, Shapiro L. A temporally controlled sigma factor is required for cell-cycle dependent polar morphogenesis in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 7.Buck M, Cannon W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature. 1992;358:422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- 8.Devereux D, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ely B, Ely T W. Use of pulsed field gel electrophoresis and transposon mutagenesis to estimate the minimal number of genes required for motility in Caulobacter crescentus. Genetics. 1989;123:649–654. doi: 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gober J W, Champer R, Reuter S, Shapiro L. Expression of positional information during cell differentiation in Caulobacter. Cell. 1991;64:381–391. doi: 10.1016/0092-8674(91)90646-g. [DOI] [PubMed] [Google Scholar]
- 12.Gober J W, Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol Biol Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hultgren S J, Abraham S, Caparon M, Falk P, St. Geme III J, Normark S. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 14.Janakiraman R S, Brun Y V. Transcriptional and mutational analyses of the rpoN operon in Caulobacter crescentus. J Bacteriol. 1997;179:5138–5147. doi: 10.1128/jb.179.16.5138-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson R C, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurtz H D, Jr, Smit J. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J Bacteriol. 1992;174:687–694. doi: 10.1128/jb.174.3.687-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurtz H D, Jr, Smit J. The Caulobacter crescentus holdfast: identification of holdfast attachment complex genes. FEMS Microbiol Lett. 1994;116:175–182. doi: 10.1111/j.1574-6968.1994.tb06697.x. [DOI] [PubMed] [Google Scholar]
- 18.Liss L R. New M13 host: DH5αF′ competent cells. Focus. 1987;9:3–13. [Google Scholar]
- 19.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 20.Merker R I, Smit J. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrick M J. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 23.Mitchell D, Smit J. Identification of genes affecting production of the adhesion organelle of Caulobacter crescentus CB2. J Bacteriol. 1990;172:5425–5431. doi: 10.1128/jb.172.9.5425-5431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton A. Role of transcription in the temporal control of development in Caulobacter crescentus. Proc Natl Acad Sci USA. 1972;69:447–451. doi: 10.1073/pnas.69.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poindexter J S. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poindexter J S. The caulobacters: ubiquitous unusual bacteria. Microbiol Rev. 1981;45:123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintero E J, Busch K, Weiner R M. Spatial and temporal deposition of adhesive extracellular polysaccharide capsule and fimbriae by Hyphomonas strain MHS-3. Appl Environ Microbiol. 1998;64:1246–1255. doi: 10.1128/aem.64.4.1246-1255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon R, Prieffer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–790. [Google Scholar]
- 30.Smit J. Localizing the subunit pool for the temporally regulated polar pili of Caulobacter crescentus. J Cell Biol. 1987;105:1821–1828. doi: 10.1083/jcb.105.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit J, Agabian N. Cloning of the major protein of the Caulobacter crescentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J Bacteriol. 1984;160:1137–1145. doi: 10.1128/jb.160.3.1137-1145.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl D A, Key R, Flesher B, Smit J. The phylogeny of marine and freshwater caulobacters reflects their habitat. J Bacteriol. 1992;174:2193–2198. doi: 10.1128/jb.174.7.2193-2198.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umbreit T H, Pate J L. Characterization of the holdfast region of wild-type cells and holdfast mutants of Asticcacaulis biprosthecum. Arch Microbiol. 1978;118:157–168. [Google Scholar]
- 34.Wingrove J A, Mangan E K, Gober J W. Spatial and temporal phosphorylation of a transcriptional activator regulates pole-specific gene expression in Caulobacter. Genes Dev. 1993;7:1979–1992. doi: 10.1101/gad.7.10.1979. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]