Abstract
Eukaryotes initiate autophagy when facing environmental changes such as a lack of external nutrients. However, the mechanisms of autophagy initiation are still not fully elucidated. Here, we showed that deacetylation of ATG4B plays a key role in starvation-induced autophagy initiation. Specifically, we demonstrated that ATG4B is activated during starvation through deacetylation at K39 by the deacetylase SIRT2. Moreover, starvation triggers SIRT2 dephosphorylation and activation in a cyclin E/CDK2 suppression–dependent manner. Meanwhile, starvation down-regulates p300, leading to a decrease in ATG4B acetylation at K39. K39 deacetylation also enhances the interaction of ATG4B with pro-LC3, which promotes LC3-II formation. Furthermore, an in vivo experiment using Sirt2 knockout mice also confirmed that SIRT2-mediated ATG4B deacetylation at K39 promotes starvation-induced autophagy initiation. In summary, this study reveals an acetylation-dependent regulatory mechanism that controls the role of ATG4B in autophagy initiation in response to nutritional deficiency.
The deacetylation of ATG4B accelerates its action on LC3 and promotes starvation-induced autophagy.
INTRODUCTION
Macroautophagy/autophagy is a highly conserved protein or organelle degradation system in species ranging from yeast to humans. It involves the autophagosome delivering needed degradable molecules into the vacuole/lysosome (1). Autophagy is a multistep process that includes commencement, membrane expansion and extension, fusion of the autophagosome with the vacuole/lysosome, and destruction of components in the autolysosomes (2). Each stage is governed by distinct proteins (2). So far, around 40 autophagy-related (ATG) genes have been identified as being involved in the regulation of these intricate membrane dynamics during autophagy (3). Recent advances in yeast and human genetics have greatly enhanced the knowledge of autophagy′s molecular mechanisms and physiological effects (4). Autophagy dysfunction is associated with various physiological and pathological processes, including development, differentiation, metabolism, immunology, inflammation, cancer, and so on (5–7).
Autophagy is initiated by the development and maturation of double-membraned autophagosomes, which ingest and transport cargo to lysosomes for degradation (1). ATG proteins are the central players in autophagy by modulating the initiation and maturation of autophagosomes (8). It is well known that the functions of some ATG proteins are regulated by posttranslational modifications (PTMs), such as phosphorylation, methylation, acetylation, and ubiquitination (9, 10). Fine regulation of autophagy in cells occurs through multiple strategies, including PTMs of ATG proteins. Therefore, it is crucial to explore the previously unidentified PTMs of ATG proteins, especially some key ATG proteins such as autophagy-related 4B cysteine peptidase (ATG4B).
ATG4 cysteine proteases cut freshly generated ATG8 to expose a glycine residue that is conjugated with membrane-bound phosphatidylethanolamine (PE) during the process of lipidation at the beginning of autophagy. ATG4 can also remove PE from ATG8 in the process of delipidation (8, 11). In humans, the best-characterized ATG8 isoform is microtubule-associated protein 1 light chain 3 (LC3) (12, 13). Moreover, ATG4B (but not ATG4A, ATG4C, and ATG4D) shows a very selective preference for LC3 (ATG4B is 1500-fold more catalytically effective than the other three ATG4 isoforms in activating LC3) (14). The role of ATG4B in physiology and pathology has been examined using genetically altered mouse models, and it has been demonstrated that ATG4B deficiency can result in the decrease of systemic autophagic activity (15–17). In addition, ATG4B has been linked as a biomarker and a therapeutic target in cancer (18). The level or activity of ATG4B can be regulated at transcriptional, translational, and posttranslational levels. Our previous study has shown that heat shock transcription factor 1 (HSF1) enhances the transcription of ATG4B and promotes autophagy, which causes epirubicin resistance in hepatocellular carcinoma cells (19). In addition, our team and another group have revealed that the expression of ATG4B is suppressed by miR-34c-5p and miR-34a (20, 21). Furthermore, it has been reported that phosphorylation, ubiquitination, O-GlcNAcylation, S-nitrosylation, and oxidation modification of ATG4B are important for its function and activity (22–24). For instance, the phosphorylation of ATG4B at S316, S383, or S392 is critical for its activity (25, 26). Nevertheless, it is still unclear whether the activity and function of ATG4B could be modulated by acetylation.
Acetylation of lysine residues on the ε-amino group has lately emerged as a critical PTM for protein function regulation, which influences a variety of physiological and pathological processes such as development, cell proliferation, metabolism, inflammation, and cancer (27–29). Recently, mounting evidence suggested that protein acetylation may play an important role in autophagy regulation (9, 10). Acetyl–coenzyme A (Ac-CoA), the only acetyl group donor for protein acetylation, has been shown to prevent autophagy caused by hunger and aging (30, 31). Numerous regulators and autophagic machinery key components, including unc-51–like kinase 1 (ULK1), ATG5, ATG7, ATG8, ATG12, sequestosome 1 (SQSTM1), and LC3, have been shown to undergo changes in their acetylation levels and govern autophagy (32, 33). However, it is unknown whether ATG4B, a key “scissor” for processing pro-LC3 and lipidated LC3 to drive the autophagy progress, is regulated by acetylation.
In the present study, we found that the level of ATG4B acetylation was negatively correlated with autophagy under nutrient deprivation. Mechanistically, p300 promoted ATG4B acetylation at K39, while silence information regulator 2 (SIRT2) enhanced ATG4B deacetylation at the same site. Starvation down-regulated p300 protein level and increased SIRT2 activity, leading to the deacetylation of ATG4B at K39 and the subsequent up-regulation of ATG4B activity and autophagy initiation. The in vivo study in Sirt2 knockout (Sirt2−/−) mice also showed that SIRT2 promoted autophagy by reducing ATG4B acetylation at K39. Together, our results elucidate a previously unknown molecular mechanism by which acetylation controls ATG4B activity and function in autophagy initiation.
RESULTS
ATG4B is deacetylated under starvation stimulation
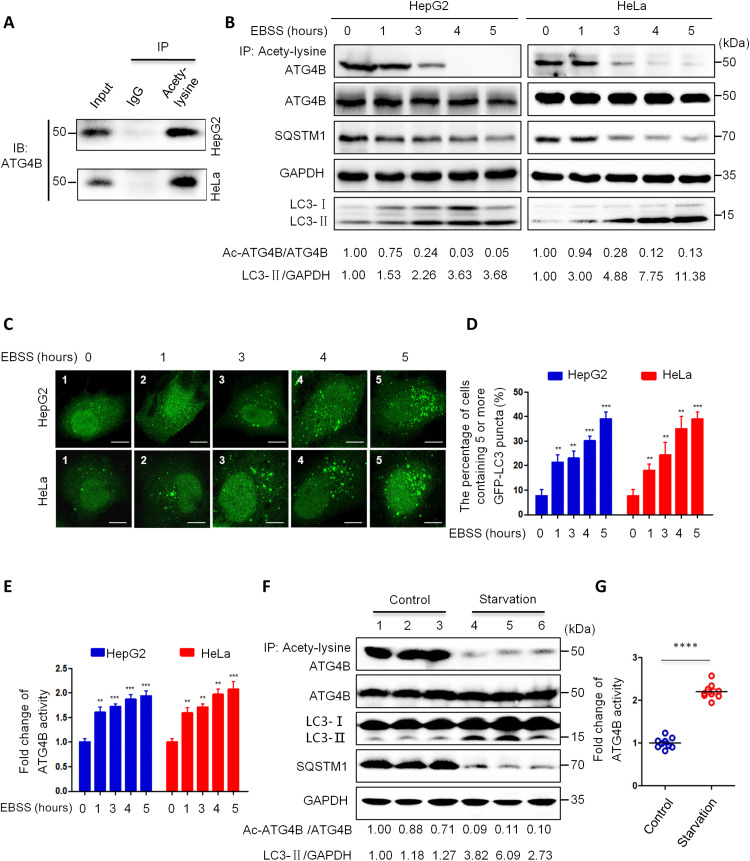
Because acetylation is a general and key PTM that plays important roles in autophagy regulation, we explored whether ATG4B could be acetylated in HepG2 and HeLa cells. As shown in Fig. 1A, our results showed that ATG4B was definitely acetylated in the cells. We found that starvation notably reduced acetylated ATG4B (Ac-ATG4B) and induced autophagy in HepG2 and HeLa cells (Fig. 1B). Subsequently, we investigated whether the other ATG4 family members (ATG4A, ATG4C, and ATG4D) could be deacetylated after starvation. As shown in fig. S1, starvation just slightly reduced ATG4A acetylation but had no significant influence on the acetylation of ATG4C and ATG4D. Next, we validated the effect of starvation on autophagy induction and showed that starvation stimulation remarkably enhanced LC3-II processing and SQSTM1 degradation, and increased green fluorescent protein (GFP)–LC3 puncta numbers in HepG2 and HeLa cells (Fig. 1, B to D). Meanwhile, starvation also markedly elevated the activity of ATG4B (Fig. 1E), which was consistent with a previous report (34). Moreover, knockdown of ATG4B markedly reduced LC3-II conversion, SQSTM1 degradation, GFP-LC3 puncta formation, and cell viability under starvation conditions (fig. S2, A to D). The ATG4B inhibitor NSC 185058 (18) had a similar effect with its small interfering RNA (siRNA) on LC3-II conversion, SQSTM1 degradation, cell viability, and autophagy induction (fig. S2, E to G). Furthermore, we examined whether the level of Ac-ATG4B could be regulated by starvation in vivo in mouse model. As shown in Fig. 1 (F and G), we detected liver tissues of starvation mice and found that starvation obviously decreased Ac-ATG4B and enhanced autophagy and ATG4B activity in vivo in mice. The findings above imply that starvation-induced autophagy in vitro and in vivo is associated with a considerable drop in ATG4B acetylation and an increase in ATG4B activity.
Fig. 1. Starvation induces the deacetylation of ATG4B.
(A) Acetylated proteins were immunoprecipitated (IP) from cell lysates using an antibody against acetylated lysine, and the acetylation level of endogenous ATG4B was determined by immunoblotting (IB). (B) HepG2 and HeLa cells were treated with EBSS for the indicated time, the acetylation level of ATG4B was assessed as in (A), and the levels of listed proteins were evaluated by Western blot. (C) Cells with stable GFP-LC3 expression were treated with EBSS for the indicated time and then observed under a fluorescence microscope. Representative images were shown. Scale bar, 5 μm. (D) The findings from (C) were quantified as the proportion of cells with five or more GFP-LC3 puncta. (E) After treatment as in (B), cells were lysed and the activity of ATG4B was determined using the AU4S substrate. (F) Six C57BL/6N mice were randomly separated into two groups and either given conventional laboratory chow (mice 1, 2, and 3) or deprived (mice 4, 5, and 6) for 48 hours. Mice were then slaughtered, and their livers were harvested and lysed. Following that, the acetylation level of ATG4B and the protein levels of ATG4B, SQSTM1, and LC3-I/II were detected. (G) The activity of ATG4B in mouse liver tissue lysates was measured as in (E). The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (B) and (F). Data are means ± SD (n = 3). **P < 0.01, ***P < 0.001, and ****P < 0.0001. Ac, acetylated; IgG, immunoglobulin G; EBSS, Earle’s balanced salt solution; GFP, green fluorescent protein.
ATG4B is acetylated at K39
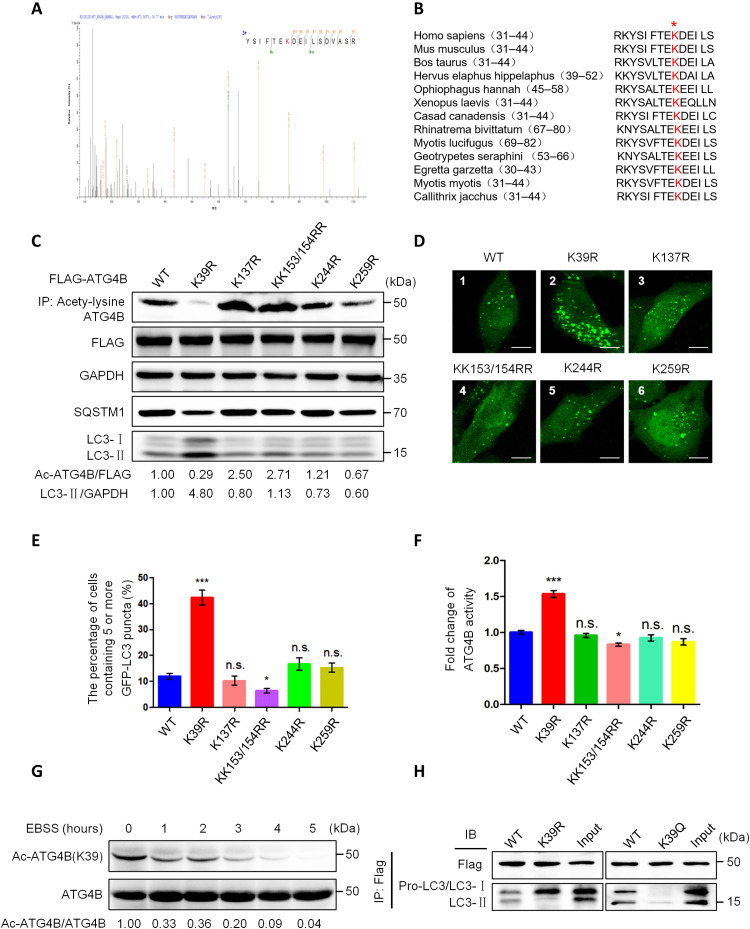
To determine the acetylation sites in ATG4B, we analyzed the six lysine (K) acetylation sites (K39, K137, K153, K154, K244, and K259) predicted by bioinformatics on ATG4B. By isolating ectopically expressed ATG4B and examining ATG4B acetylation using mass spectrometry, we found an acetyl-lysine–containing peptide (YSIFTEKAcDEILSDVASR) in three different tests, which was mapped to the K39-containing region on human ATG4B (Fig. 2A). Homology analysis of protein sequences showed that K39 in ATG4B is highly conserved among different species (Fig. 2B), and K42 in ATG4A has a similar conservation (fig. S3A). To study the function of the potential acetylation sites, we separately mutated each corresponding K site to an arginine (R) (KK153/154RR was in one mutant construct), which mimicked the deacetylation state on the lysine(s). Then, we tested the effects of these mutations on ATG4B activity and autophagy regulation in ATG4BCRISPR HepG2 cells (ATG4B hemizygous knockout cells) (24). As shown in Fig. 2 (C to F), the K-to-R mutation of K39 (ATG4BK39R) resulted in a marked reduction in ATG4B acetylation and a significant elevation of autophagy induction and ATG4B activity. On the other side, the ATG4BK39Q mutation [K39 was mutated to glutamine (Q), which mimics an acetylated state] could obviously decrease ATG4B activity and decrease GFP-LC3 puncta numbers in ATG4BCRISPR HepG2 cells (fig. S3, B to E). In addition, we investigated whether the K39 acetylation of ATG4B (Ac-ATG4BK39) could influence the lipidation of γ-aminobutyric acid type A (GABAA) receptor–associated protein–like 2 (GABARAPL2), another member of the ATG8 family. As shown in fig. S3 (F and G), ATG4BK39R mutation slightly increased the lipidation of GABARAPL2, while the ATG4BK39Q mutation had little effect on GABARAPL2 lipidation. These results indicated that the K39 deacetylation of ATG4B preferably processed LC3 rather than GABARAPL2, which is consistent with the previous study (35). Subsequently, we generated the antibody against acetylated K39 of ATG4B using N-SIFTE(Ac-K)DE-C as an immunogen, and the antibody′s titer was over 1:32,000 (fig. S4A). Then, we verified the specificity of the antibody with Western blot (fig. S4, B and C) and demonstrated that starvation markedly reduced Ac-ATG4BK39 using this antibody (Fig. 2G).
Fig. 2. The K39 acetylation of ATG4B regulates ATG4B activity and autophagy initiation.
(A) The acetylation sites of ATG4B were detected using mass spectrometry after ATG4B protein was purified by IP and SDS-PAGE. Schematic representation of the acetylated sites on ATG4B. (B) Sequence analysis of the conservation degree of Lys39 in different species of ATG4B. (C) ATG4BCRISPR HepG2 cells (ATG4B hemizygous knockout cells) were transfected with the indicated expression plasmids with point mutation. Cell lysates were used for IP and Western blot assays with the corresponding antibody. (D) The ATG4BCRISPR HepG2 cells with stable GFP-LC3 expression were transfected with the indicated expression plasmids following observation under a fluorescence microscope. Representative images were shown. Scale bar, 5 μm. (E) Quantification of the data from (D); the proportion of cells harboring five or more GFP-LC3 puncta was calculated. (F) The activity of ATG4B in (C) was measured. (G) HepG2 cells were treated with EBSS for various times, and ATG4B acetylation was assessed by Western blot using the K39-acetylated ATG4B antibody. (H) ATG4BCRISPR HepG2 cells were transfected with the wild-type (WT) or K39R mutated or K39Q mutated ATG4B plasmids for 24 hours, following treatment with EBSS for 5 hours, and then whole-cell lysates were immunoprecipitated using an antibody against FLAG tag and analyzed by IB with LC3 antibody. The lysates containing K39R mutation ATG4B or K39Q mutation ATG4B were used as input control. The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (C) and (G). Data are means ± SD (n = 3). n.s., no significance; *P < 0.05 and ***P < 0.001.
Since Earle’s balanced salt solution (EBSS) contains neither contain amino acids nor glucose or growth factors, we further investigated which of the deprivations triggers ATG4B K39 deacetylation. As shown in fig. S5 (A to C), glucose deprivation only led to a slight down-regulation of Ac-ATG4BK39 (fig. S5A), while deprivation of amino acids could rapidly decrease Ac-ATG4BK39 at 1-hour point (fig. S5B), and serum deprivation (mimicking growth factor deficiency) also markedly reduced Ac-ATG4BK39 at 5-hour point (fig. S5C). These results indicated that deficiency of either amino acids or glucose or growth factors could trigger ATG4B deacetylation at K39, and amino acid starvation had the strongest effect on it. Moreover, we assayed whether the K39 deacetylation of ATG4B is specific for macroautophagy. As shown in fig. S6A, ATG4BK39R mutation obviously up-regulated the expression of PTEN-induced putative kinase 1 (PINK1; a marker of mitophagy), while the ATG4BK39Q mutation mildly down-regulated PINK1, suggesting that the K39 deacetylation of ATG4B might play a role in the process of mitophagy. Despite the up-regulation effect of mitophagy inducer cccp on PINK1 expression at different doses, the low doses (2.5 and 5.0 μM) of cccp slightly decreased Ac-ATG4BK39, while the high dose (10 μM) of cccp even slightly increased Ac-ATG4BK39 (fig. S6B), which suggests that the K39 deacetylation of ATG4B does not play a key role in the mitophagy induction. Next, we checked the role of K39 deacetylation on the interaction of ATG4B with LC3. As shown in Fig. 2H, wild-type (WT) ATG4B interacted with both pro-LC3 and LC3-II, which was consistent with a previous report (36). However, ATG4BK39R mainly interacted with pro-LC3, while ATG4BK39Q had no obvious interaction with pro-LC3. In addition, we found that the K39 (de)acetylation of ATG4B had little effect on ATG4B distribution in the cytoplasm and nucleus (fig. S7, A and B). These results above indicate that the acetylation of ATG4B at K39 is a key PTM that suppresses ATG4B activity and macroautophagy initiation.
p300 is an acetyltransferase of ATG4B
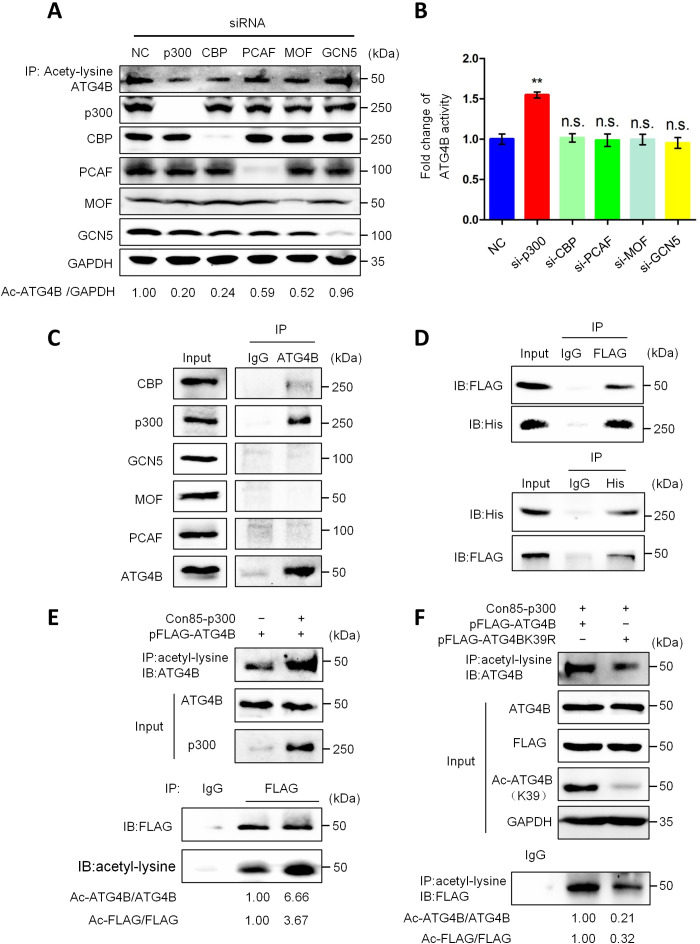
To identify the acetyltransferase that regulates ATG4B acetylation, we separately knocked down five acetyltransferases, which are reported to be closely related to autophagy in mammals, including E1A-binding protein/300 kDa (p300) (37), CREB (adenosine 3′,5′-monophosphate response element–binding protein)–binding protein (CBP) (38), p300/CBP-associated factor (PCAF) (39), males absent on the first (MOF; also known as MYST1) (40), and general control nondepressible 5 (GCN5; also known as KAT2A) (41). As shown in Fig. 3A, knockdown of p300 or CBP markedly decreased the acetylation level of ATG4B, whereas knockdown of the other three acetyltransferases had no obvious influence on ATG4B acetylation. Meanwhile, we observed that knockdown of p300 (but not the other four acetyltransferases) significantly enhanced the activity of ATG4B (Fig. 3B). Furthermore, the coimmunoprecipitation (co-IP) result showed that ATG4B was physically associated with p300 (Fig. 3, C and D). In addition, the ectopic expression of p300 enriched the acetylation of WT ATG4B (but not mutant ATG4BK39R) (Fig. 3, E and F). Together, these data demonstrate that p300 is an acetyltransferase of ATG4B and promotes the acetylation of ATG4B at K39.
Fig. 3. p300 is the acetyltransferase of ATG4B.
(A) HepG2 cells were transfected with indicated siRNAs of five acetyltransferases for 24 hours, and then cell lysates were used for IP and Western blot assays with the corresponding antibody. (B) The activity of ATG4B in (A) was measured. (C) The association of endogenous ATG4B and different acetyltransferases was assessed by IP and Western blot. (D) HepG2 cells were cotransfected with pHIS-p300 and pFLAG-ATG4B plasmids, and then cell lysates were used for IP and IB assays with the corresponding antibodies. (E) HepG2 cells were transfected with pFLAG-ATG4B with or without pHIS-p300 plasmids, and then the acetylation level of ATG4B was detected by IP and IB. (F) HepG2 cells were transfected with indicated plasmids, and then the acetylation level of ATG4B was analyzed as in (E). The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (A), (E), and (F). Data are means ± SD (n = 3). **P < 0.01.
Starvation down-regulates the expression of p300
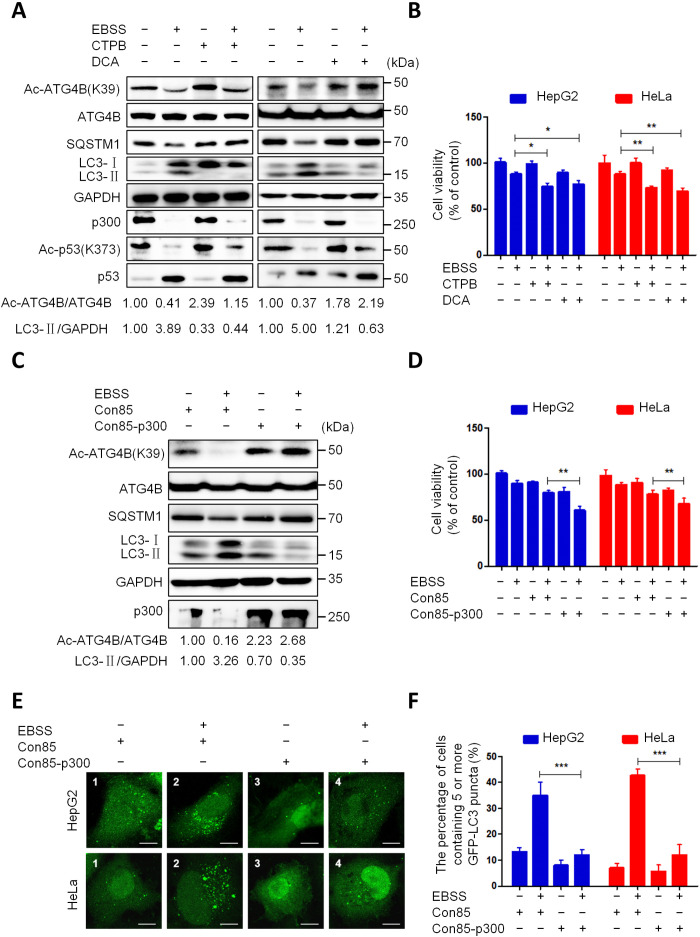
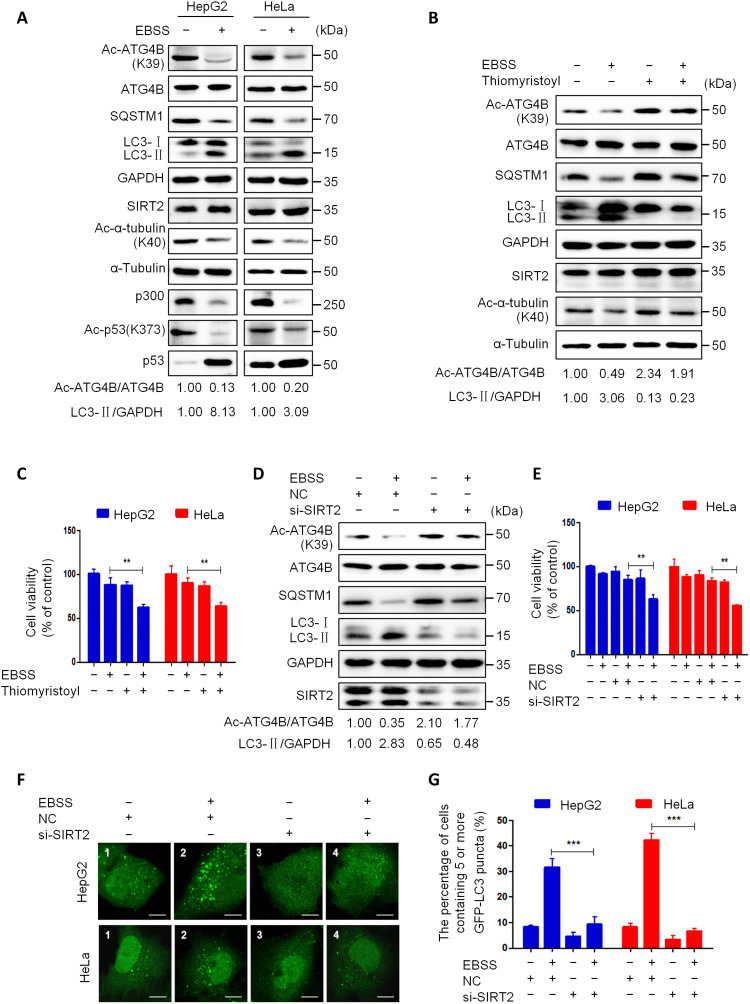
We next investigate the changes of p300 expression and activity under starvation conditions. As shown in Fig. 4A, starvation stimulation markedly decreased the protein level of p300. Meanwhile, starvation markedly reduced the acetylation levels of ATG4BK39 and p53K373 (a known substrate of p300) (42). A previous study has shown that Ac-CoA is an important factor in maintaining p300 activity (30). Therefore, we used CTPB (a direct activator of p300) or DCA (a drug that increases Ac-CoA by promoting aerobic respiration in cells) to treat cells under starvation conditions. As shown in Fig. 4A, both CTPB and DCA substantially attenuated the starvation-induced deacetylation of ATG4B and p53 and autophagy. Meanwhile, CTPB or DCA remarkably enhanced the suppression effect of starvation on cell survival (Fig. 4B). Moreover, ectopic expression of p300 obviously weakened the starvation-induced ATG4B deacetylation, autophagy, and repression of cell survival (Fig. 4, C to F). These data indicate that starvation reduces ATG4B acetylation and induces autophagy, at least in part by down-regulating the expression of p300.
Fig. 4. Starvation down-regulates the expression of p300.
(A) HepG2 cells were treated with EBSS alone, EBSS combined with CTPB (100 μM), or EBSS combined with DCA (20 mM) for 24 hours. The levels of Ac-ATG4BK39, Ac-p53K373, and listed proteins were analyzed by Western blot. (B) Cells were treated as in (A), and cell viability was detected using the CCK-8 kit. (C) HepG2 cells were transfected with a control vector or pHIS-p300 for 36 hours, following treatment with EBSS or normal medium for 5 hours, and then the levels of Ac-ATG4BK39 and listed proteins were analyzed as in (A). (D) Cells were treated as in (C), and cell viability was measured using the CCK-8 kit. (E) Cells with stable GFP-LC3 expression were treated as in (C) and then observed under a fluorescence microscope. Representative images were shown. Scale bar, 5 μm. (F) Quantification of the data from (E); the proportion of cells carrying five or more GFP-LC3 puncta were calculated. The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (A) and (C). Data are means ± SD (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001.
SIRT2 is the deacetylase of ATG4B
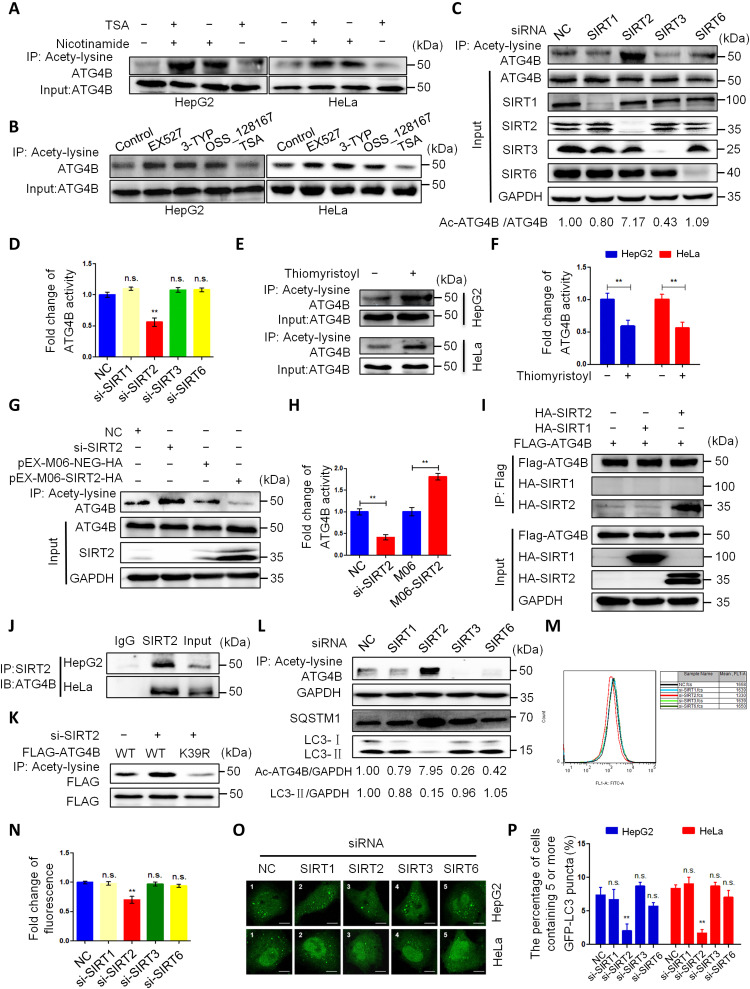
As acetylation modification can be regulated by acetyltransferase and deacetylase, we next investigate which deacetylase regulates ATG4B acetylation. First, we treated cells with trichostatin A (TSA) [an inhibitor of histone deacetylase (HDAC) classes I, II, and IV] and/or nicotinamide (NAM; an inhibitor of the SIRT family deacetylases). As shown in Fig. 5A, NAM alone or NAM combined with TSA markedly promoted ATG4B acetylation, whereas TSA alone had no obvious influence on ATG4B acetylation, suggesting that the deacetylase of ATG4B may be a member of the SIRT family. Therefore, we used three inhibitors separately targeting SIRT1 (selisistat, EX527), SIRT3 (3-TYP), and SIRT6 (OSS_128167) to figure out the deacetylase involved in ATG4B deacetylation. Each of the three inhibitors increased ATG4B acetylation in cells (Fig. 5B). Considering the off-target effect of the pharmacological inhibitors, we further synthesized the siRNAs separately targeted to SIRT1, SIRT2, SIRT3, or SIRT6. As shown in Fig. 5C, knockdown of SIRT2 (but not SIRT1, SIRT3, and SIRT6) with siRNA remarkably increased the acetylation level of ATG4B. Meanwhile, knockdown of SIRT2 also markedly decreased the activity of ATG4B (Fig. 5D). Moreover, an effective SIRT2 inhibitor, thiomyristoyl (43), could notably enhance ATG4B acetylation and inhibit ATG4B activity (Fig. 5, E and F). Next, we found that knockdown of SIRT2 increased Ac-ATG4B (Fig. 5G) and decreased its activity (Fig. 5H), while overexpression of SIRT2 reduced Ac-ATG4B (Fig. 5G) and elevated its activity (Fig. 5H). Furthermore, the co-IP experiment showed that ATG4B was physically associated with SIRT2 at both endogenous and exogenous levels (Fig. 5, I and J). In addition, knockdown of SIRT2 promoted the acetylation of WT ATG4B (but not mutant ATG4BK39R) (Fig. 5K), indicating that SIRT2 deacetylated ATG4B at K39. Last, we determined the effect of SIRT2 on autophagy. The results showed that knockdown of SIRT2 decreased LC3-II and GFP-LC3 puncta and increased Ac-ATG4B and SQSTM1 (Fig. 5, L to P), indicating that SIRT2 strengthened autophagy. Collectively, these data demonstrate that SIRT2 is the deacetylase of ATG4B, and it can enhance autophagy by promoting the deacetylation of ATG4B at K39.
Fig. 5. SIRT2 is the deacetylase of ATG4B.
(A and B) Cells were treated with indicated reagents for 24 hours, and then cell lysates were used for IP and Western blot. (C and D) HepG2 cells were transfected with the indicated siRNAs for 24 hours, and cell lysates were used for IP, Western blot (C), and ATG4B activity (D) assays. (E and F) Cells were treated with thiomyristoyl for 24 hours, and then the level of Ac-ATG4B (E) and ATG4B activity (F) was measured. (G to I) HepG2 cells were transfected with the indicated siRNAs or plasmids for 24 hours, and then cell lysates were used for IP, Western blot (G and I), and ATG4B activity (H) analysis. (J) The association of endogenous ATG4B and SIRT2 was assessed by IP and IB. (K) ATG4BCRISPR HepG2 cells were transfected with the indicated plasmids or siRNAs, and then the level of Ac-ATG4B was determined. (L and M) HepG2 cells were treated as in (C), and cell lysates were used for IP and Western blot (L). Meanwhile, the treated cells were stained with CYTO-ID Green, fluorescence intensity of autophagosomes was detected with a flow cytometer, and autophagic cells were calculated (M). (N) Quantification of the data from (M). (O) Cells with stable GFP-LC3 expression were treated as in (C) and then observed under a fluorescence microscope. Representative images were shown. Scale bar, 5 μm. (P) Quantifying the data from (O) and showing as the percentage of cells containing five or more GFP-LC3 puncta. The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (C) and (L). Data are means ± SD (n = 3). **P < 0.01.
Starvation activates SIRT2 in cells
To explore the role of SIRT2 in starvation-induced autophagy, we examined the expression and activity of SIRT2 after starvation treatment in cells. As shown in Fig. 6A, a short-time nutritional deficiency had no obvious influence on the level of SIRT2, but it could substantially reduce the acetylation levels of ATG4BK39 and α-tubulinK40 (a known substrate of SIRT2) (44), suggesting that starvation may activate SIRT2 in cells. Moreover, the SIRT2 inhibitor thiomyristoyl notably attenuated the starvation-induced autophagy and deacetylation of ATG4BK39 and α-tubulinK40 in cells (Fig. 6B). Meanwhile, thiomyristoyl markedly enhanced the suppression effect of starvation on cell survival (Fig. 6C). Furthermore, knockdown of SIRT2 markedly alleviated the starvation-induced ATG4B deacetylation, autophagy, and inhibition of cell survival (Fig. 6, D to G). These data reveal that starvation activates SIRT2 and subsequently promotes the deacetylation of ATG4B, which leads to autophagy induction.
Fig. 6. Starvation activates SIRT2 in cells.
(A) Cells were treated with EBSS for 5 hours; the levels of Ac-ATG4BK39, Ac-α-tubulinK40, Ac-p53K373, and listed proteins were analyzed by Western blot using the corresponding antibodies. (B) HepG2 cells were treated with EBSS, thiomyristoyl (50 μM), or EBSS combined with thiomyristoyl for 24 hours, and then the protein levels were detected as in (A). (C) Cells were treated as in (B); cell viability was detected using the CCK-8 kit. (D) HepG2 cells were transfected with a control siRNA or SIRT2 siRNA for 24 hours following treatment with EBSS or normal medium for 5 hours, and then protein levels were analyzed as in (A). (E) Cells were treated as described in (D), and the vitality of cells was determined using the CCK-8 kit. (F) Cells expressing GFP-LC3 were treated as described in (D) and then viewed under a fluorescence microscope. Representative images were shown. Scale bar, 5 μm. (G) Quantification of the data from (F); data were expressed as the percentage of cells containing five or more GFP-LC3 puncta. The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (A), (B), and (D). Data are means ± SD (n = 3). **P < 0.01 and ***P < 0.001.
Starvation activates SIRT2 by suppressing cyclin E/CDK2
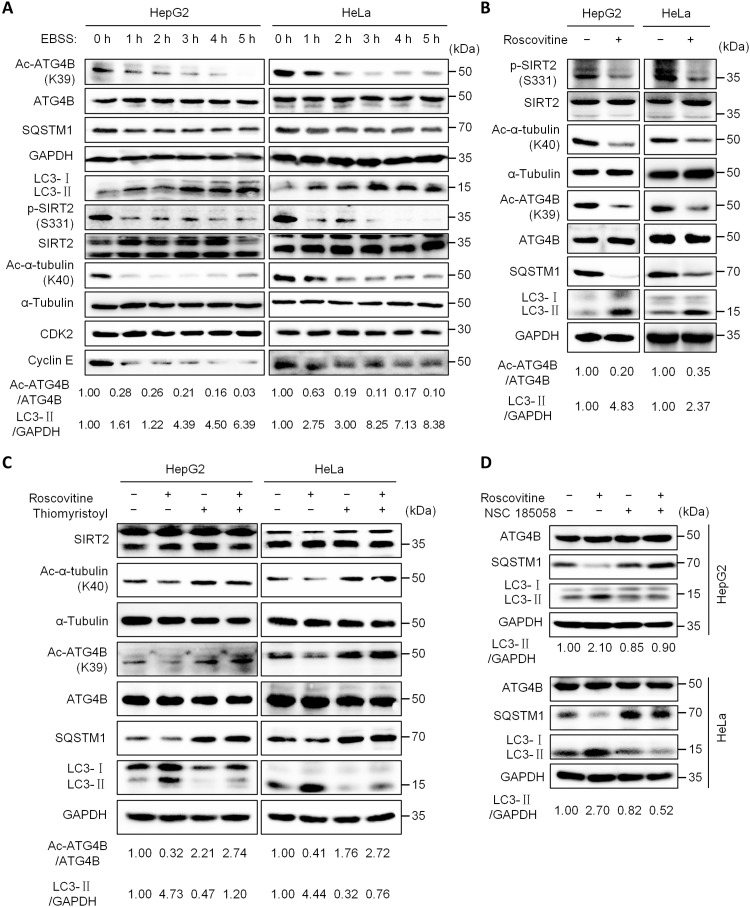
A previous study has demonstrated that cyclin E/cyclin-dependent kinase 2 (CDK2) complex–mediated phosphorylation of SIRT2 at serine-331 (SIRT2S331) plays a vital role in regulating SIRT2 activity (45). Therefore, we detected the levels of phosphorylated SIRT2S331 (p-SIRT2S331), cyclin E, and CDK2 under starvation conditions. The results showed that starvation markedly reduced the levels of p-SIRT2S331, Ac-ATG4BK39, cyclin E, SQSTM1, and Ac-α-tubulinK40 but increased the protein level of LC3-II (Fig. 7A). As it has been demonstrated that CDK2 activation requires the formation of the cyclin E/CDK2 complex (46), we speculated that the starvation-induced cyclin E down-regulation may decrease the formation of the cyclin E/CDK2 complex, leading to the inhibition of CDK2 activity. Consistent with this hypothesis, we found that the CDK2 inhibitor roscovitine notably reduced the levels of p-SIRT2S331, Ac-ATG4BK39, SQSTM1, and Ac-α-tubulinK40 but elevated the protein level of LC3-II (Fig. 7B), which was blocked by the SIRT2 activator thiomyristoyl (Fig. 7C). In addition, the ATG4B inhibitor NSC 185058 markedly attenuated roscovitine-mediated SQSTM1 down-regulation and LC3-II up-regulation (Fig. 7D), indicating that ATG4B is involved in CDK2-mediated autophagy regulation. In all, these data reveal that starvation activates SIRT2 by suppressing cyclin E/CDK2.
Fig. 7. Starvation activates SIRT2 by suppressing cyclin E/CDK2.
(A) Cells were treated with EBSS for the indicated time; the levels of Ac-ATG4BK39, Ac-α-tubulinK40, p-SIRT2S331, and listed proteins were analyzed by Western blot using the corresponding antibodies. (B) Cells were treated with roscovitine (100 μM) for 24 hours, and then the protein levels were analyzed as in (A). (C) Cells were treated with roscovitine (100 μM), thiomyristoyl (50 μM), or their combination for 24 hours, and then the protein levels were analyzed as in (A). (D) Cells were treated with roscovitine (100 μM), NSC 185058 (50 μM), or their combination for 24 hours, and then the protein levels were analyzed as in (A). The relative intensity of listed proteins in each lane was calculated and normalized with control (lane 1) after quantification and shown in (A) to (D). The experiments were repeated three times, with similar results obtained.
SIRT2 is a key modulator of ATG4B deacetylation and autophagy in vivo
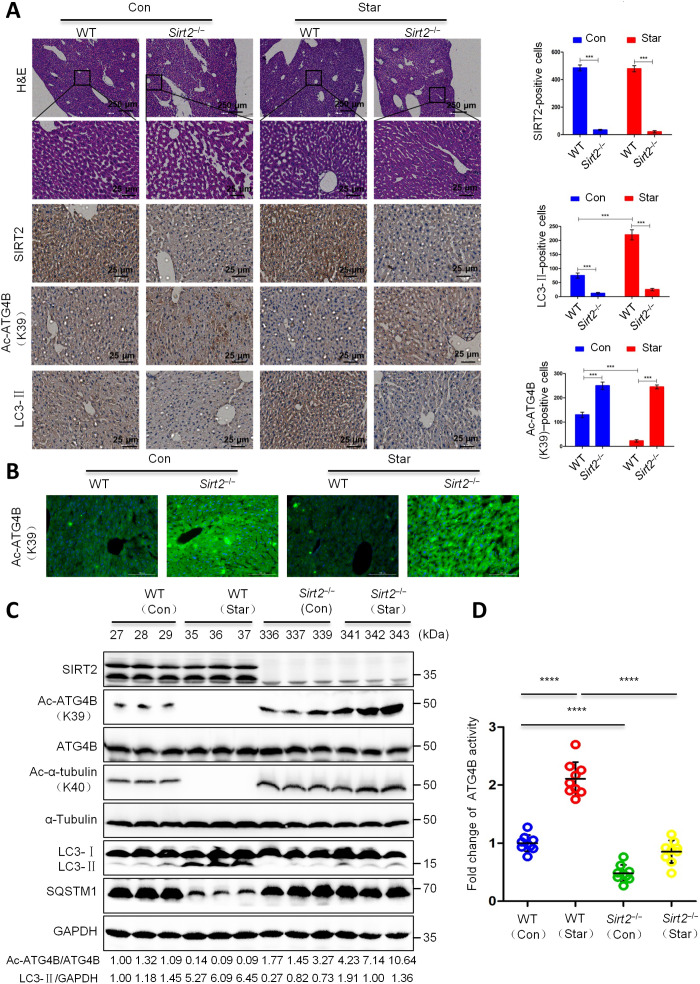
To further verify the importance of SIRT2 on ATG4B deacetylation and autophagy, we used Sirt2 knockout (Sirt2−/−) mice as a starvation animal model in the experiment. The WT or Sirt2−/− mice were randomly separated into two groups and starved or not for 2 days, and then liver tissues were collected for further analysis. First, we did not observe an obvious difference in hematoxylin and eosin (H&E) staining in liver tissues between WT and Sirt2−/− mice, whether starved or not (Fig. 8A). However, compared with WT mice, Sirt2−/− mice exhibited substantially higher levels of Ac-ATG4BK39 and lower levels of LC3-II, with a defective autophagy phenotype [the phenotype is consistent with the previous report (47)] (Fig. 8, A and B). Next, we found that starvation markedly down-regulated Ac-ATG4BK39 and SQSTM1 but up-regulated LC3-II in WT mice (but not in Sirt2−/− mice) (Fig. 8, A to C), indicating that SIRT2 played an important role in starvation-mediated promotion of ATG4BK39 deacetylation and autophagy in vivo. In addition, starvation remarkably enhanced ATG4B activity in liver tissues of WT mice (but not Sirt2−/− mice) (Fig. 8D). Together, these data indicate that SIRT2 plays a vital role in promoting the deacetylation and activity of ATG4B and autophagy in vivo.
Fig. 8. SIRT2 is a key modulator of ATG4B deacetylation and autophagy in vivo.
WT C57BL/6N mice or Sirt2 knockout mice (Sirt2−/−) were starved or not for 2 days, and then liver tissues were used for further analysis. (A) The levels of SIRT2, Ac-ATG4BK39, and LC3 were detected by immunohistochemistry. Positive cells were counted among a total of 1000 cells on average. Images were obtained at ×4 or ×20 magnification. Scale bar, 250 or 25 μm. ***P < 0.001. (B) The level of Ac-ATG4BK39 was assessed by immunofluorescent staining. Scale bar, 25 μm. (C) The levels of Ac-ATG4BK39, Ac-α-tubulinK40, and listed proteins were evaluated by Western blot. Relative intensity of Ac-ATG4B and LC3-II in each lane was calculated and normalized with control (lane 1) after quantification and shown. The experiments were repeated three times, with similar results obtained. (D) The activity of ATG4B was measured. ****P < 0.0001.
DISCUSSION
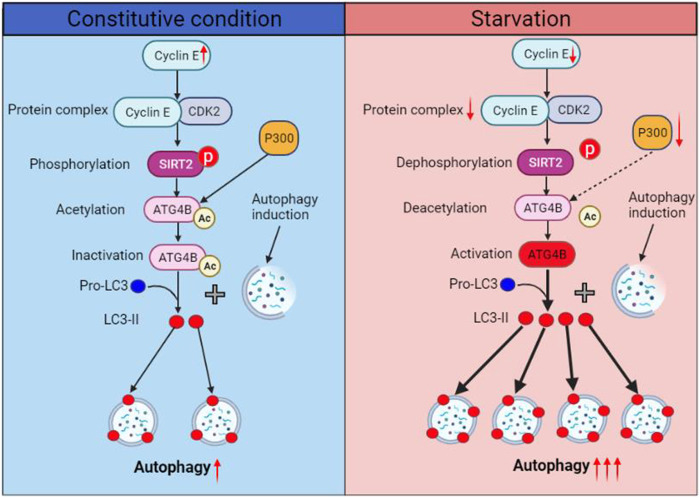
This study presents evidence that acetylation modulates ATG4B activity and autophagy initiation under starvation conditions. Specifically, we found that the deacetylation of ATG4B at K39 plays an important role in autophagy initiation, which advances the molecular mechanisms of starvation-induced autophagy (Fig. 9). In a sufficient nutrient environment, the amount of cyclin E is enough to form the cyclin E/CDK2 complex with CDK2 and maintain the activity of CDK2. The active CDK2 phosphorylates SIRT2 at S331 and inhibits its activity. The low activity of SIRT2 and the high level of p300 cause a high acetylation level of ATG4B at K39, resulting in a low ATG4B activity and a low level of autophagy initiation. However, in a nutrient deficiency environment, cyclin E and p300 are down-regulated in cells. The amount of the cyclin E/CDK2 complex is not enough to keep the activity of CDK2. As a result, the phosphorylation of SIRT2 is suppressed and the activity of SIRT2 is increased. The high activity of SIRT2 and the low level of p300 bring a low acetylation level of ATG4B at K39, resulting in a high ATG4B activity and a high level of autophagy initiation.
Fig. 9. Schematic illustration for the ATG4B deacetylation–induced autophagy initiation.
In a sufficient nutrient environment (left), cyclin E and p300 are in relative high levels in cells. Cyclin E is sufficient to form the cyclin E/CDK2 complex with CDK2 and maintain the activity of CDK2. The active CDK2 phosphorylates SIRT2 at S331 and inhibits its activity. The low activity of SIRT2 and the high level of p300 cause a high acetylation level of ATG4B at K39, leading to low levels of ATG4B activity and autophagy initiation. In a nutrient deficiency (starvation) environment (right), cyclin E and p300 are down-regulated in cells. The amount of the cyclin E/CDK2 complex is not enough to keep the activity of CDK2. Subsequently, the phosphorylation of SIRT2 is suppressed and the activity of SIRT2 is increased. The high activity of SIRT2 and the low level of p300 bring a low acetylation level of ATG4B at K39, resulting in high levels of ATG4B activity and autophagy initiation. Ac, acetylation; p, phosphorylation; ↑, up-regulation; ↓, down-regulation.
Acetylation (here means acetylation of the ε-amino group of lysine residues) is a key PTM for histone and nonhistone proteins, which plays an important role in autophagy regulation (9). Ac-CoA, the sole acetyl group donor for protein acetylation, has been proven to suppress age-related and starvation-induced autophagy. The acetyltransferase p300 has been shown to inhibit autophagy by acetylating ATG5, ATG7, ATG8, and ATG12 (48). Our study revealed that deacetylation of ATG4B promoted ATG4B activity and autophagy initiation, consistent with previous studies on other ATGs (10). We further ascertained the specific K39 acetylation site on ATG4B and demonstrated that it is vital for ATG4B activity and autophagy initiation. In addition, the K39 of ATG4B is an evolutionally conserved site among different species, suggesting that the acetylation of ATG4B may be a general phenomenon across species.
Acetylation and deacetylation are a conversed and dynamic process, which is catalyzed by lysine acetyltransferases (KATs; also known as histone acetylases) and lysine deacetylases (KDACs; also known as HDACs), respectively (27). In the current study, we found that p300 acts as the KAT of ATG4B acetylation and SIRT2 exerts the role of KDAC. p300, a KAT that belongs to the P300/CBP subfamily, has been reported to inhibit autophagy by acetylizing several substrates. Here, we found that ATG4B is a previously unidentified substrate of p300. The activity of ATG4B is suppressed by p300-medicated acetylation in normal cells. Consistent with previous studies (48), reduced p300 levels alleviated ATG4B acetylation and boosted its activity under hunger conditions. On the other hand, a member of the NAM adenine dinucleotide (NAD)–dependent deacetylase family, SIRT2 has been reported to play important roles in numerous biological processes, including genome integrity, mitosis regulation, cell homeostasis, cell differentiation, infection, aging, inflammation, autophagy, and oxidative stress. In recent years, increasing substrate numbers of SIRT2 have been validated to mediate a wide range of biological processes (49). In this study, we verified that SIRT2 is the HDAC of ATG4B deacetylation. Selisistat, 3-TYP, or OSS_128167 cotreatment could increase ATG4B acetylation. Selisistat (EX527) can effectively inhibit the activity of SIRT1 deacetylase (50), accompanied by partial inhibition on the activity of SIRT2 and SIRT3. The median inhibitory concentration (IC50) of 3-TYP for SIRT1, SIRT2, and SIRT3 is 88, 92, and 16 nM, respectively. The SIRT6 inhibitor OSS_128167 can also partially inhibit the activity of SIRT1 and SIRT2. Therefore, the effect of selisistat, 3-TYP, or OSS_128167 on ATG4B acetylation may attribute to its action on SIRT2. The results of SIRT1, SIRT2, SIRT3, and SIRT6 knockdown further supported this point. Furthermore, the role of SIRT2 in autophagy regulation is paradoxical (51). An impaired autophagy process has been observed in Sirt2−/− mice (47). However, SIRT2 reduction promotes autophagy in normal HCT116 cells (52). Our study revealed that SIRT2 promotes autophagy by accelerating ATG4B deacetylation when facing nutrient deprivation. We guess that the discrepancy of SIRT2 on autophagy regulation may attribute to the different treatment conditions, different cells, or tissue backgrounds.
Although several pathways such as MTOR (mechanistic target of rapamycin) (53), AMPK (AMP-dependent protein kinase) (54), and AKT/protein kinase B (55) have been reported to regulate autophagy under nutrient and energy deprivation, the molecular mechanisms of the rapid initiation of autophagy are far from elucidation. In this study, we observed that the acetylation of ATG4B at K39 is markedly decreased, accompanied by autophagy induction even at 1 hour after starvation. Although deficiency of either amino acids or glucose or growth factors could trigger ATG4B deacetylation at K39, we preliminarily speculate that amino acid deficiency is more important in triggering ATG4B deacetylation. Amino acid deficiency may rapidly activate SIRT2, which is consistent with the report of SIRT2 in amino acid–related lipid metabolism (56, 57). These findings may supply a previously unidentified perspective on the rapid initiation of autophagy because posttranslational regulation is more convenient and faster than transcription regulation. We further observed that short-term starvation promoted the deacetylation of ATG4B by enhancing the activity of SIRT2, which was demonstrated by the fact that short-term starvation has little effect on the protein level of SIRT2 but notably reduces the acetylation level of α-tubulin (a recognized substrate of SIRT2 that indirectly reflects the activity of SIRT2). Short-term starvation down-regulates the protein but not the mRNA level of cyclin E and inhibits the activity of CDK2, which leads to the dephosphorylation of SIRT2 at the S331 site. Previous studies have demonstrated that starvation can block the cell cycle and keep cells in the G0-G1 phase. Therefore, we speculate that the down-regulation of cyclin E may lead to cell cycle arrest and autophagy induction to maintain cell survival in a nutrient-deficient environment.
In our model, starvation induces the deacetylation of ATG4B and its activation, which promotes the conversion of pro-LC3 to LC3-I and accelerates autophagy initiation. Our data show that the deacetylated ATG4B mainly interacts with pro-LC3, while the acetylated ATG4B has no interaction with pro-LC3. We also found that the acetylated ATG4B level is closely negative relative to the level of LC3-II in cells. Therefore, we speculate that activated ATG4B promotes the generation of LC3-I by cleaving pro-LC3, and then LC3-I is further turned into LC3-II rapidly. According to Nguyen et al.’s report (35), ATG4B can also promote mitophagy independent of its activity. Our data showed that the acetylation of ATG4B does not play a key role in the mitophagy induction. Therefore, we presume that the acetylation of ATG4B mainly regulates autophagy by changing its activity.
In conclusion, our research revealed that the acetylation of ATG4B regulates its activity and autophagy initiation. Moreover, we verified that p300 acetylates ATG4B at K39, which the SIRT2 can antagonize. Specifically, nutrient deprivation, on one side, down-regulates p300 and, on another side, activates SIRT2 by suppressing cyclin E/CDK2. Consequently, ATG4B was deacetylated and activated. Last, autophagy was induced. These findings not only reveal a previously unknown posttranslational modification of ATG4B but also may supply a clinical opportunity for treating ATG4B and ATG diseases. It will be interesting to find out whether the above results apply to the treatment of other ATG4B and ATG diseases.
MATERIALS AND METHODS
Cell culture and reagents
HepG2 and HeLa cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (GE Healthcare Life Sciences), streptomycin (100 mg/ml), and penicillin (100 U/ml) (Gibco). The cell lines were originally evaluated by ATCC and passaged for less than 6 months in the laboratory. Cells were rinsed with phosphate-buffered saline (PBS) and grown for 1 to 5 hours in EBSS (Sigma-Aldrich) to induce autophagy and starvation. Cells were incubated with 100 μM CTPB, 20 mM DCA, 0.5 μM TSA, 100 μM NAM, 1 μM EX527, 100 μM 3-TYP, 100 μM OSS_128167, 5 μM thiomyristoyl, 50 μM NSC 185058, or 100 μM roscovitine at 37°C under 5% CO2. The small-molecule inhibitors listed above were obtained from Selleck Chemicals LLC. Transient transfection of HepG2 and HeLa cells with expression vectors was performed using Lipofectamine 3000 (Invitrogen) according to the package recommendations.
Antibodies
The antibodies against acetyl-lysine (ab22550), p53 (acetyl K373, ab62376), p53 (ab131442), ATG4A (ab108322), ATG4C (ab183516), ATG4D (ab237751), and SIRT2 (ab211033) were from Abcam. The antibodies against SQSTM1 (P0067), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (G9545), LC3 (L7543), GABARAPL2 (HPA036726), FLAG (F3165), His (SAB1305538), and hemagglutinin (HA) (H9658) were purchased from Sigma-Aldrich. The antibodies against ATG4B (#13507), p300 (#86377), CBP (#7389), PCAF (#3378), MOF (#46862), GCN5 (#3305), SIRT1 (#2493), SIRT3 (#2627), SIRT6 (#12486), acetyl-α-tubulin (Lys40, #5335), α-tubulin (#3873), CDK2 (#18048), PINK1 (#6946), and cyclin E (#20808) were obtained from Cell Signaling Technology. The antibody against SIRT2 phospho-Ser331 (61363) was from Active Motif.
RNA interference
All siRNAs were obtained from GeneCopoeia. Transfection of siRNA was performed using Lipofectamine 3000 (Invitrogen) following the manufacturer’s protocol. siGENOME SMARTpool reagents (Dharmacon) were diluted with Opti-MEM reduced serum medium (Invitrogen). The siRNA sequences for the target genes were as follows: p300 (5′-GCAAAGGAAUUGCCUUAUUTT-3′), CBP (5′-GCGUGUGUACAUUUCUUAUTT-3′), PCAF (5′-GGAGCCACUUUAAUGGGAUTT-3′), MOF (5′-CCCUGCAAUCCCUCAAUAUTT-3′), GCN5 (5′-GCUACCUACAAGGUCAAUUTT-3′), SIRT1 (5′-GGAUAGAGCCUCACAUGCA-3′), SIRT2 (5′-GCCAACCAUCUGUCACUACUU-3′), SIRT3 (5′-GAAACUACAAGCCCAACGU-3′), SIRT6 (5′-GAAUGUGCCAAGUGUAAGA-3′), and ATG4B (5′-GGUGUGGACAGAUGAUCUUUG-3′).
Plasmids and transfection
The plasmids pcDNA3.1, pcDNA3.1-SIRT1-HA, pEX-M06, pEX-M06-SIRT2-HA, pCMV-ATG4BK39R, pCMV-ATG4BK39Q, pCMV-ATG4BK137R, pCMV-ATG4BKK153/154RR, pCMV-ATG4BK244R, and pCMV-ATG4BK259R were obtained from GeneCopoeia. The plasmids CON85-p300 and CON85 were purchased from Genechem (Shanghai, China). The plasmids pCMV-ATG4B and pCMV were products of Lab Cell Biotechnology (Chongqing, China). Lipofectamine 3000 (Invitrogen) was used to transfect plasmids according to the manufacturer’s instructions.
Western blot
Whole-cell extracts were prepared by directly lysing cells with cell lysis buffer (Thermo Fisher Scientific). Lysates were boiled with 6× SDS loading buffer for 10 min and electrophoresed on SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels. A total of 25 μg of protein in each sample was analyzed. Then, protein samples were electrophoretically transferred to a polyvinylidene difluoride membrane (Bio-Rad). Membranes were blocked with 5% skimmed milk for 1 hour at room temperature and then incubated with primary antibodies overnight at 4°C. After washing three times with PBST (phosphate-buffered saline, 0.1% Tween 20), membranes were incubated with horseradish peroxidase–labeled secondary antibodies for 1 hour at room temperature. The protein blots were detected and imaged with ChemiDoc Touch System (Bio-Rad).
Immunoprecipitation
For IP, we used the Pierce Cross-link Magnetic IP/Co-IP Kit (Thermo Fisher Scientific). Cells were collected by a cell scraper and centrifuged at 15,000g for 10 min. For immunoprecipitation, 1.5 mg of protein in 500 μl of cell lysis buffer solution (pH 7.4, 25 mM tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol) was added to each sample, using 5 μg of the antibody against the protein of interest. First, the target antibody was bound to Protein A/G magnetic beads and cross-linked with disuccinimidyl suberate. Second, 500 μl of the diluted cell lysate from each of the lysed samples was added to a tube containing the cross-linked magnetic beads and incubated overnight at 4°C on a rotator or mixer. The final eluate was neutralized, and a nonreducing electrophoresis loading buffer was added to boil, denature, and perform immunoblotting. The membranes were blocked with 5% nonfat milk in TBS-T (tris-buffered saline–Tween 20) and probed for the proteins of interest. The protein band density was analyzed using Image Lab software. On the basis of three replicated experiments, the ratios relative to the control were calculated.
Immunocytochemistry
Liver tissue was fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with H&E. Immunohistochemical staining of paraffin-embedded tissues was performed using SIRT2 (Abcam, ab211033), Ac-ATG4B (Lys39), and LC3 (Sigma-Aldrich, L7543) primary antibodies and an ABC Elite immunoperoxidase kit according to the manufacturer’s instructions.
Immunofluorescent staining
Liver tissue was fixed with 4% formaldehyde (Thermo Fisher Scientific) and then dehydrated with 30% sucrose solution (Sangon Biotech). The frozen liver tissue sections were dried at room temperature for 15 min, permeabilized with 0.5% Triton X-100 for 10 min, then blocked with 3% bovine serum albumin (BSA) + 0.1% Triton X-100 for 30 min, diluted 1:100 with primary antibody, and incubated at 4°C overnight. Tissue sections were incubated with Alexa 488–labeled secondary antibody (dilution 1:200), washed several times, mounted with 4′,6-diamidino-2-phenylindole (DAPI)–containing mounting solution Vectashield (Vector Laboratories), and observed using a Nikon inverted microscope equipped with an Eclipse Ti-U digital camera.
Cell viability assay
In 96-well plates, cells were plated at a density of 2000 cells per well and grown overnight (specific density depends on cell type). After 24 hours, cells were treated following the prescribed protocol. Then, cells were cultured in the dark for 1 to 3 hours with CCK-8 reagent (Dojindo Molecular Technologies) according to the manufacturer’s procedure. Last, absorbance values were measured at 450 nm using a microplate reader.
GFP-LC3 analysis
Cells with stable GFP-LC3 expression were plated into six-well plates at appropriate densities, treated with different reagents, and finally fixed for 10 min with 4% formaldehyde. Following that, five random views of the cells were taken with a fluorescence microscope (Olympus IX81). The number of positive autophagic cells containing five or more GFP-LC3 puncta was counted from 50 cells per view. Representative cell pictures were captured by laser confocal microscopy (Carl Zeiss AG).
ATG4B activity assay
Following our previous study (34), cells or tissues were lysed with lysis buffer containing 2 mM dithiothreitol. The lysates were incubated with 0.2 μM AU4S for 40 min. The reaction solution without cell lysates was used as background control and incubated with AU4S. Then, fluorescence intensity was analyzed with a fluorescence microplate reader. The value (the difference between AU4S and background fluorescence intensity) was calculated and normalized by the amount of protein. Data are means ± SD from three experiments.
Fluorescent assay of autophagosomes
Autophagy in living cells was examined using the Cyto-ID Autophagy Detection Kit (Enzo) and a proprietary probe to selectively detect autophagosomes according to the manufacturer’s procedure. Cells were collected, resuspended in 500 μl of freshly diluted Cyto-ID Green Detection Solution, and incubated for 30 min at room temperature in the dark. Then, fluorescence intensity was measured using a flow cytometer (Beckman).
Mass spectrometry
HepG2 cells were transfected with the ATG4B expression plasmid for 36 hours and then treated with 0.5 μM TSA and 100 μM NAM for 24 hours. Subsequently, cells were harvested and lysed with IP lysis buffer supplemented with 2 mM TSA and 10 mM NAM. Next, cell lysates were immunoprecipitated using ATG4B antibody–coupled protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology), and immunoprecipitates were separated by SDS-PAGE and stained with Coomassie blue. After treatment with destaining solution, the ATG4B band was extracted from the gel, followed by tryptic digestion and mass spectrometry. Database searches were used to identify proteins and modifications, and peptide identifications were confirmed using the Peptide Prophet.
Mice
Sirt2 knockout (Sirt2−/−) mice and WT litters were used to analyze liver autophagy in mice induced by starvation. This research used only male mice. Mice were obtained from Cyagen Biosciences Inc. Mouse breeding was conducted at the Army Medical University’s Animal Center’s specific pathogen–free mouse facility. Army Medical University’s Institutional Animal Care and Use Committee authorized all mouse trials.
Statistical analysis
Data are reported as means ± SD of at least three independent experiments. Comparisons between multiple groups were performed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons tests using GraphPad Prism 6.0 software (GraphPad Software). For comparison between two groups, paired t test was used. In all cases, a difference of P < 0.05 was judged statistically significant. For in vivo experiments, sample size was determined, taking into account experimental feasibility and statistical significance. Animals and samples were randomized, and investigators were not blinded to allocation during studies and result evaluation.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81872024 to F.H., 82073300 to J.L., and 31900449 to H.X.), the Chongqing Natural Science Foundation (cstc2018jcyjA2018 to J.L.), and Post-doctor Program of Natural Science Foundation of Chongqing (cstc2021jcyj-bshX0200 to W.J.).
Author contributions: Conceptualization: L.S. and H.X. Methodology: H.X., X.D., X.Y., Y.W., M.Y., M.S., T.L., and J.Y. Investigation: L.S., H.X., L.C., W.J., and H.H. Supervision: F.H. and J.L. Writing—original draft: L.S. and H.X. Writing—review and editing: F.H. and J.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
REFERENCES AND NOTES
- 1.Ohsumi Y., Historical landmarks of autophagy research. Cell Res. 24, 9–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakatogawa H., Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Fukuda T., Kanki T., Atg43, a novel autophagy-related protein, serves as a mitophagy receptor to bridge mitochondria with phagophores in fission yeast. Autophagy 17, 826–827 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B., Kroemer G., Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadwell K., Crosstalk between autophagy and inflammatory signalling pathways: Balancing defence and homeostasis. Nat. Rev. Immunol. 16, 661–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dikic I., Elazar Z., Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Eskelinen E. L., Autophagy: Supporting cellular and organismal homeostasis by self-eating. Int. J. Biochem. Cell Biol. 111, 1–10 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N., Yoshimori T., Ohsumi Y., The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Banreti A., Sass M., Graba Y., The emerging role of acetylation in the regulation of autophagy. Autophagy 9, 819–829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Y., Kang R., Sun X., Zhong M., Huang J., Klionsky D. J., Tang D., Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White E., The role for autophagy in cancer. J. Clin. Invest. 125, 42–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T., LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Xie Z., Klionsky D. J., Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Li M., Hou Y., Wang J., Chen X., Shao Z. M., Yin X. M., Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 286, 7327–7338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabrera S., Fernandez A. F., Marino G., Aguirre A., Suarez M. F., Espanol Y., Vega J. A., Laura R., Fueyo A., Fernandez-Garcia M. S., Freije J. M., Kroemer G., Lopez-Otin C., ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy 9, 1188–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marino G., Fernandez A. F., Cabrera S., Lundberg Y. W., Cabanillas R., Rodriguez F., Salvador-Montoliu N., Vega J. A., Germana A., Fueyo A., Freije J. M., Lopez-Otin C., Autophagy is essential for mouse sense of balance. J. Clin. Invest. 120, 2331–2344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Read R., Savelieva K., Baker K., Hansen G., Vogel P., Histopathological and neurological features of Atg4b knockout mice. Vet. Pathol. 48, 486–494 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Akin D., Wang S. K., Habibzadegah-Tari P., Law B., Ostrov D., Li M., Yin X. M., Kim J. S., Horenstein N., Dunn Jr. W. J., A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 10, 2021–2035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N., Wu Y., Lyu X., Li B., Yan X., Xiong H., Li X., Huang G., Zeng Y., Zhang Y., Lian J., Ni Z., He F., HSF1 upregulates ATG4B expression and enhances epirubicin-induced protective autophagy in hepatocellular carcinoma cells. Cancer Lett. 409, 81–90 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Wu Y., Dai X., Ni Z., Yan X., He F., Lian J., The downregulation of ATG4B mediated by microRNA-34a/34c-5p suppresses rapamycin-induced autophagy. Iran. J. Basic Med. Sci. 20, 1125–1130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y., Ni Z., Yan X., Dai X., Hu C., Zheng Y., He F., Lian J., Targeting the MIR34C-5p-ATG4B-autophagy axis enhances the sensitivity of cervical cancer cells to pirarubicin. Autophagy 12, 1105–1117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T., Kim C. K., Alvarez A. A., Pangeni R. P., Wan X., Song X., Shi T., Yang Y., Sastry N., Horbinski C. M., Lu S., Stupp R., Kessler J. A., Nishikawa R., Nakano I., Sulman E. P., Lu X., James C. D., Yin X. M., Hu B., Cheng S. Y., MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell 32, 840–855.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng X., Yang Z., Gu Q., Xia F., Fu Y., Liu P., Yin X. M., Li M., The protease activity of human ATG4B is regulated by reversible oxidative modification. Autophagy 16, 1838–1850 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni Z., He J., Wu Y., Hu C., Dai X., Yan X., Li B., Li X., Xiong H., Li Y., Li S., Xu L., Li Y., Lian J., He F., AKT-mediated phosphorylation of ATG4B impairs mitochondrial activity and enhances the Warburg effect in hepatocellular carcinoma cells. Autophagy 14, 685–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z., Wilkie-Grantham R. P., Yanagi T., Shu C. W., Matsuzawa S., Reed J. C., ATG4B (Autophagin-1) phosphorylation modulates autophagy. J. Biol. Chem. 290, 26549–26561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pengo N., Agrotis A., Prak K., Jones J., Ketteler R., A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat. Commun. 8, 294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narita T., Weinert B. T., Choudhary C., Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Verdin E., Ott M., 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Guan K. L., Xiong Y., Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 36, 108–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marino G., Pietrocola F., Eisenberg T., Kong Y., Malik S. A., Andryushkova A., Schroeder S., Pendl T., Harger A., Niso-Santano M., Zamzami N., Scoazec M., Durand S., Enot D. P., Fernandez A. F., Martins I., Kepp O., Senovilla L., Bauvy C., Morselli E., Vacchelli E., Bennetzen M., Magnes C., Sinner F., Pieber T., Lopez-Otin C., Maiuri M. C., Codogno P., Andersen J. S., Hill J. A., Madeo F., Kroemer G., Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell 53, 710–725 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Schroeder S., Pendl T., Zimmermann A., Eisenberg T., Carmona-Gutierrez D., Ruckenstuhl C., Marino G., Pietrocola F., Harger A., Magnes C., Sinner F., Pieber T. R., Dengjel J., Sigrist S. J., Kroemer G., Madeo F., Acetyl-coenzyme a. Autophagy 10, 1335–1337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You Z., Jiang W. X., Qin L. Y., Gong Z., Wan W., Li J., Wang Y., Zhang H., Peng C., Zhou T., Tang C., Liu W., Requirement for p62 acetylation in the aggregation of ubiquitylated proteins under nutrient stress. Nat. Commun. 10, 5792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang R., Liu W., Identifying an essential role of nuclear LC3 for autophagy. Autophagy 11, 852–853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni Z., Gong Y., Dai X., Ding W., Wang B., Gong H., Qin L., Cheng P., Li S., Lian J., He F., AU4S: A novel synthetic peptide to measure the activity of ATG4 in living cells. Autophagy 11, 403–415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen T. N., Padman B. S., Zellner S., Khuu G., Uoselis L., Lam W. K., Skulsuppaisarn M., Lindblom R., Watts E. M., Behrends C., Lazarou M., ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. Mol. Cell 81, 2013–2030.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Kauffman K. J., Yu S., Jin J., Mugo B., Nguyen N., O’Brien A., Nag S., Lystad A. H., Melia T. J., Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy 14, 992–1010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan W., You Z., Xu Y., Zhou L., Guan Z., Peng C., Wong C., Su H., Zhou T., Xia H., Liu W., mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol. Cell 68, 323–335.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Chen X., Li Y., Wang C., Tang Y., Mok S. A., Tsai R. M., Rojas J. C., Karydas A., Miller B. L., Boxer A. L., Gestwicki J. E., Arkin M., Cuervo A. M., Gan L., Promoting tau secretion and propagation by hyperactive p300/CBP via autophagy-lysosomal pathway in tauopathy. Mol. Neurodegener. 15, 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong D., Ying B., Zhang J., Ying H., PCAF regulates H3 phosphorylation and promotes autophagy in osteosarcoma cells. Biomed. Pharmacother. 118, 109395 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Zehender A., Li Y. N., Lin N. Y., Stefanica A., Nuchel J., Chen C. W., Hsu H. H., Zhu H., Ding X., Huang J., Shen L., Gyorfi A. H., Soare A., Rauber S., Bergmann C., Ramming A., Plomann M., Eckes B., Schett G., Distler J., TGFβ promotes fibrosis by MYST1-dependent epigenetic regulation of autophagy. Nat. Commun. 12, 4404 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S., Liang M., Naqvi N. I., Lin C., Qian W., Zhang L. H., Deng Y. Z., Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in Magnaporthe oryzae. Autophagy 13, 1318–1330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., Zhu W. G., Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol. Cell. Biol. 26, 2782–2790 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jing H., Hu J., He B., Negron A. Y., Stupinski J., Weiser K., Carbonaro M., Chiang Y. L., Southard T., Giannakakou P., Weiss R. S., Lin H., A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and rxhibits broad anticancer activity. Cancer Cell 29, 297–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E., The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Pandithage R., Lilischkis R., Harting K., Wolf A., Jedamzik B., Luscher-Firzlaff J., Vervoorts J., Lasonder E., Kremmer E., Knoll B., Luscher B., The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J. Cell Biol. 180, 915–929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fagundes R., Teixeira L. K., Cyclin E/CDK2: DNA replication, replication stress and genomic instability. Front. Cell Dev. Biol. 9, 774845 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu G., Park S. H., Imbesi M., Nathan W. J., Zou X., Zhu Y., Jiang H., Parisiadou L., Gius D., Loss of NAD-dependent protein deacetylase sirtuin-2 alters mitochondrial protein acetylation and dysregulates mitophagy. Antioxid. Redox Signal. 26, 849–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee I. H., Finkel T., Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 284, 6322–6328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Yang J., Hong T., Chen X., Cui L., SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res. Rev. 55, 100961 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Broussy S., Laaroussi H., Vidal M., Biochemical mechanism and biological effects of the inhibition of silent information regulator 1 (SIRT1) by EX-527 (SEN0014196 or selisistat). J. Enzyme Inhib. Med. Chem. 35, 1124–1136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aventaggiato M., Vernucci E., Barreca F., Russo M. A., Tafani M., Sirtuins’ control of autophagy and mitophagy in cancer. Pharmacol. Ther. 221, 107748 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Inoue T., Nakayama Y., Li Y., Matsumori H., Takahashi H., Kojima H., Wanibuchi H., Katoh M., Oshimura M., SIRT2 knockdown increases basal autophagy and prevents postslippage death by abnormally prolonging the mitotic arrest that is induced by microtubule inhibitors. FEBS J. 281, 2623–2637 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Feng Z., Zhang H., Levine A. J., Jin S., The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. U.S.A. 102, 8204–8209 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Y., Chen H., Zhang L., Lin X., Li X., Zhuang H., Fan H., Meng T., He Z., Huang H., Gong Q., Zhu D., Xu Y., He P., Li L., Feng D., The AMPK-MFN2 axis regulates MAM dynamics and autophagy induced by energy stresses. Autophagy 17, 1142–1156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raina K., Agarwal C., Wadhwa R., Serkova N. J., Agarwal R., Energy deprivation by silibinin in colorectal cancer cells: A double-edged sword targeting both apoptotic and autophagic machineries. Autophagy 9, 697–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reis T., Van Gilst M. R., Hariharan I. K., A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLOS Genet. 6, e1001206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banerjee K. K., Ayyub C., Sengupta S., Kolthur-Seetharam U., dSir2 deficiency in the fatbody, but not muscles, affects systemic insulin signaling, fat mobilization and starvation survival in flies. Aging 4, 206–223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7