Abstract
The Escherichia coli Pst system belongs to the family of ABC transporters. It is part of a phosphate (PHO) regulon which is regulated by extracellular phosphate. Under conditions of phosphate limitation, the response regulator PhoB is phosphorylated by the histidine kinase PhoR and binds to promoters that share a consensus PHO box. Under conditions of phosphate excess, PhoR, Pst, and PhoU downregulate the PHO regulon. Screening of a library of pneumococcal mutants with defects in exported proteins revealed a putative two-component regulatory system, PnpR-PnpS, and a downstream ABC transporter, similar to the Pst system in E. coli including a gene encoding a PhoU protein. Similar to E. coli, mutagenesis of the ATP-binding cassette gene, pstB, resulted in decreased uptake of phosphate. The effects of the loss of the pneumococcal Pst system extended to decreased transformation and lysis. Withdrawal of phosphate led to transformation deficiency in the parent strain R6x but not to penicillin tolerance, suggesting that reduced bacterial death was independent of phosphate. None of these phenotypes was observed in the pneumococcal loss-of-function mutant phoU. By using a lacZ reporter construct, it was demonstrated that expression of the two-component regulatory system PnpR-PnpS was not influenced by different concentrations of phosphate. These results suggest a more complex role of the Pst system in pneumococcal physiology than in that of E. coli.
Phosphate (P) uptake is of fundamental importance in the cell physiology of bacteria because P is required as a nutrient. The P acquisition system is best understood in Escherichia coli, which has evolved several gene clusters allowing the assimilation of P via a variety of systems. These different gene clusters are coregulated as members of the phosphate (PHO) regulon (42). For E. coli, two major inorganic phosphate (Pi) transporters, the Pst and the Pit systems, have been described (43). The expression of the PHO regulon is inhibited when Pi is in excess. Under such conditions, P is taken up by the low-affinity Pi transporter, Pit (33). Under P limitation, most PHO regulon genes are upregulated about 100-fold and the genes for the high-affinity Pi-specific transporter, Pst, are preferentially transcribed (42). Pst, encoded by the genes pstSCAB and phoU, belongs to the family of ATP-binding cassette (ABC) transporters. pstS encodes a phosphate-binding protein; the two following open reading frames (ORFs), pstC and pstA, encode transmembrane proteins; and pstB encodes an ATP-binding protein. phoU, the terminal gene, has no effect on Pi uptake by the Pst system (36). However, a phoU mutation has a deleterious effect on growth, indicating that the constitutive uptake of Pi by the Pst transporter in the absence of PhoU might be toxic (9).
The Pst system is subject to regulation by a two-component regulatory system, PhoBR, which monitors the external P concentration. Two-component systems are signal-transducing proteins that effect responses to changing environmental conditions (7, 28, 37). The histidine kinase has an autokinase activity that attaches the phosphate from ATP to a histidine residue. The product, phosphohistidine, serves as a high-energy intermediate for subsequent transfer of the phosphate to an aspartate residue in the response regulator (1). The phosphorylated form of the response regulator PhoB is required for the P-dependent control of transcription of the PHO regulon genes (19).
Since P is involved in many cellular functions of bacteria, it stands to reason that this substance also influences genetic transformation. Genetic transformation occurs in Streptococcus pneumoniae during a physiologically defined competent state (24, 40). The development of competence is cell density dependent and is mediated by the accumulation of an extracellular heptadecapeptide, competence-stimulating peptide (CSP) (10). During competence, different competence-induced loci are upregulated. All of these loci contain identical competence-induced promoter (CIP) sequences (4). One of the competence-induced loci is the rec operon, which includes the genes cogA, recA, dinF, and lytA (20, 29). The product of the recA gene facilitates homologous recombination between single- and double-stranded DNAs (14). The lytA gene encodes the pneumococcal amidase LytA. This amidase is responsible for remodeling the cell wall, cell death in stationary phase, and penicillin-induced cell lysis. Transcription of the lytA gene is strongly induced in competent cells from the competence-inducible promoter upstream of the rec operon, which cotranscribes recA and lytA (25). This links transformation and lysis on a genetic level; nevertheless, a physiological correlate remains unknown. Autolysis due to activation of the amidase is characteristic of most pneumococci. The ability of the organism to avoid lysis in the presence of antibiotics is defined as antibiotic tolerance. Tolerance arises if either the pneumococcal autolysin, which lyses the cell wall, is not triggered or the autolysin itself is not present.
In this report, we describe the identification and DNA sequence of the putative pneumococcal Pst locus, encoding an ABC transporter which participates in Pi uptake by S. pneumoniae. A two-component system, PnpR-PnpS, located directly upstream of the pneumococcal system did not seem to be functionally associated with the Pst system. In contrast to that of E. coli, the pneumococcal Pst system affected autolysis and transformation. In this study, we demonstrated that the pneumococcal Pst system is most likely involved in a signaling pathway regulating the activity of the major pneumococcal autolysin. Furthermore, Pi may control genetic transformation via this system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. The autolysin-deficient strain Lyt-4-4 is a stable point mutant that was created by chemical mutagenesis (39). Pneumococci were routinely grown on tryptic soy agar (Difco, Detroit, Mich.) supplemented with sheep blood to a final concentration of 3% (vol/vol). For growth in liquid culture, the bacteria were grown in a semisynthetic casein hydrolysate medium, supplemented with yeast extract, in the presence of 30 mM P (C+Y medium) (15). For experiments using various concentrations of P (0, 3, 10, 30, and 60 mM final concentrations), bacteria were grown in C+Y medium (pH 8.0). The buffer capacity of the medium was maintained by substituting an adequate amount of Tris-HCl. For the selection and maintenance of pneumococci containing chromosomally integrated plasmids, bacteria were grown in the presence of 1 μg of erythromycin (Sigma, St. Louis, Mo.)/ml. A library of translational fusions of random pneumococcal chromosomal DNA to a truncated gene for alkaline phosphatase (phoA), created by using the vector pHRM104, has been described elsewhere (31). Since pHRM104 does not contain an intrinsic promoter upstream of phoA, blue colonies indicate plasmid-derived production of fusion proteins generated from pneumococcal DNA that contains a promoter, a translational start site, and a functional signal sequence. Fifty-one independent pneumococcal mutants that survived exposure to penicillin at 10× its MIC were isolated from the first library of 3,000 clones.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source |
|---|---|---|
| R6x | Wild type | Rockefeller University collection |
| Lyt-4-4 | R6x with a point mutation in lytA | 39 |
| SPRU1001 | R6x with an insertion duplication in the locus pnpS-pstS | This study |
| SPRU1002 | R6x with lacZ fused to pnpR-pnpS | This study |
| SPRU589 | R6x with lacZ fused to recA | 29 |
| SPRU559 | R6x deficient in lacZ | 4 |
| ΔpnpR | R6x with chromosomally integrated construct pJDC9 containing a 646-bp fragment from nucleotide 32 to 678 of pnpR | This study |
| ΔpnpS | R6x with chromosomally integrated construct pJDC9 containing a 882-bp fragment from nucleotide 749 to 1631 of pnpS | This study |
| ΔpstB | R6x with chromosomally integrated construct pJDC9 containing a 330-bp fragment from nucleotide 2749 to 3079 of pstB | This study |
| ΔphoU | R6x with chromosomally integrated construct pJDC9 containing a 291-bp fragment from nucleotide 3556 to 3847 of phoU | This study |
Recombinant DNA methods.
DNA ligations, restriction endonuclease digestions, agarose gel electrophoresis, and DNA amplification by PCR were performed according to standard techniques (34). DNA purification and plasmid preparations were performed with kits from Qiagen (Santa Clarita, Calif.) and Promega/Wizard (Promega, Madison, Wis.) according to the manufacturer’s instructions. Transformation of E. coli with plasmid DNA was carried out with CaCl2-treated cells as described previously (3). Transformation of S. pneumoniae was performed according to standard protocols (31).
Oligonucleotides.
The following synthetic oligonucleotides were used for insertion duplication mutagenesis and inverse PCR: INVup, 5′-GCT CCC GTT TTT AGC CCC ACC C-3′ (forward); INVdown, 5′-ACC CAA AAG GTG AAC GAA GG-3′ (reverse); epnpR, 5′-TTA GCC GAA TTC TGA TTG CAG GGA TGG C-3′ (forward) (where underlining indicates restriction sites); bpnpR, 5′-AGC TGA GGA TCC AAG CTT CGA GC AAA TCC-3′ (reverse); epnpS, 5′-TTA GCC GAA TCC AAG CTT CGA GTC AAA TCC-3′ (forward); bpnpS, 5′-AGC TGA GGA TCC AAG CTT CGA GTC AAA TCC-3′ (reverse); bpstB, 5′-AAG ACA GGA TCC TGC CTT GAT AGG CC-3′ (forward); epstB, 5′-ATC TTT GAA TTC TTC CCA AAT GGC-3′ (reverse); bpstS, 5′-GAA AAT GGA TCC GGG ACA CGG GGG TGC-3′ (forward); epstS, 5′-CAG CGG GAA TTC CTG CTG AAG ACC C-3′ (reverse); ephoU, 5′-GAC TTA GGA TCC ATG AAT TAG AA-3′ (forward); bphoU, 5′-TGA CAA Ul;2GAA TTC GTC CCC GAT C-3′ (reverse); elacz, 5′-GCC TTT GGA TCC CAG CCG CTT ACA AAC TTC C-3′ (forward); and blacz, 5′-AGA CTT GAT AAG AAT TCT AGA GTT AGC TCC C-3′ (reverse).
Inverse PCR.
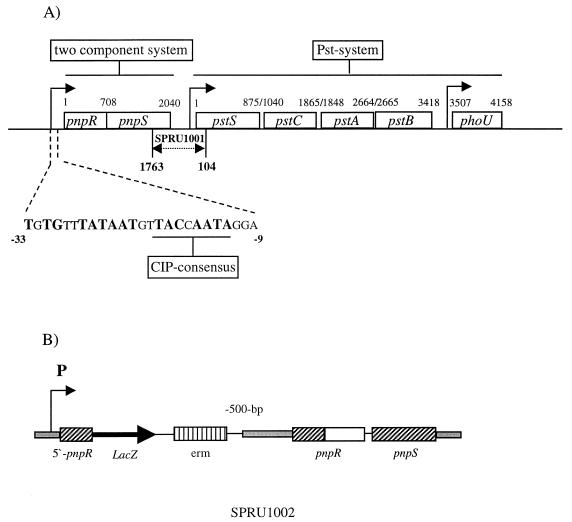
To analyze the mutation in SPRU1001, we sequenced the insert of a spontaneously excised plasmid (pHRM104) from this strain, revealing that it consisted of 647 bp. The inverse PCR technique was used to amplify the nucleotide sequence adjacent to the known sequence of the chromosomal 647-bp duplication (Fig. 1A). For this purpose, genomic DNA of S. pneumoniae strain R6x was digested with HincII, since this restriction enzyme had no cleavage site in the 647-bp target sequence. The digested DNA was ligated, and the region flanking the target sequence was amplified by using the oligonucleotide primers INVup and INVdown, which are complementary to the 5′ termini of the 647-bp insert. The amplification reaction gave a 3,712-bp fragment, which was gel purified and sequenced at the Protein/DNA Technology Center at Rockefeller University, New York, N.Y. Further sequence analysis demonstrated that plasmid pHRM104 disrupted the 3′ end of the pnpS gene, which encodes a putative histidine kinase, and the 5′ region of the pstS gene, which encodes a putative phosphate binding protein (Fig. 1A).
FIG. 1.
(A) Organization of the pnpR-pnpS ORF and the pst locus in S. pneumoniae. The nucleotides indicated are part of the upstream region including the putative CIP consensus sequence. The position of the insertion duplication mutagenesis in the mutant SPRU1001 is indicated by arrows (dashed line). (B) The strain SPRU1002 harboring the pnpR-lacZ-pnpS fusion was constructed by site-directed insertion duplication mutagenesis. A PCR fragment including the entire promoter region and the 5′ end of pnpR was cloned into the pEC2a vector and used to transform the lacZ-deficient strain SPRU559. Putative promoters are indicated by arrows.
Insertional inactivation of pnpR, pnpS, pstB, and phoU.
To create the knockout mutants, the method of insertional duplication mutagenesis, which is a homology-directed insertion of foreign DNA, was used (8, 22). For insertional duplication mutagenesis of the response regulator gene pnpR, an internal 646-bp fragment (bp 32 to 678) was amplified by using the total DNA of strain R6x and the primers epnpR and bpnpR. The PCR product was digested with EcoRI and BamHI. An identical strategy was used to knock out the other genes. The insertional inactivation of the histidine kinase gene, pnpS, was performed with the primers epnpS and bpnpS. The resulting fragment was 882 bp long (bp 749 to 1631). For the insertional inactivation of the ATP-binding cassette gene, pstB, an integral 330-bp fragment (bp 2749 to 3079) was amplified by using the primers epstB and bpstB. The insertional inactivation of phoU was performed with the primers ephoU and bphoU. The resulting fragment was 291 bp long (bp 3556 to 3847). Each amplified fragment was cloned in pJDC9 (5). The resulting recombinant plasmids were then transformed into strain R6x.
Northern blotting.
Total RNA was prepared according to the manufacturer’s instructions (Qiagen). Approximately 10 to 20 μg of total RNA was separated in a 1.2% formaldehyde gel. The gel was rinsed in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the RNA was transferred to nylon membranes (Hybond-N; Amersham, Little Chalfont, Buckinghamshire, England) by capillary blotting (34). A 550-bp PCR fragment generated by using primers bpstS and epstS was employed as the pstS-specific probe. The probe was labeled with [α-32P]dCTP (Amersham). Hybridization under stringent conditions was performed according to standard protocols (34).
Construction of pnpR-lacZ-pnpS fusion.
A strain harboring a pnpR-lacZ-pnpS fusion (SPRU1002 [Fig. 1B]) was constructed by cloning a 770-bp promoter region fragment upstream of pnpR-pnpS into the nonreplicating lacZ fusion vector pEC2A (6). The 770-bp fragment was amplified by PCR, using the primers elacZ and blacZ; gel purified; digested with EcoRI and BamHI; and then ligated to linearized vector pEC2A. The construct was introduced by transformation into SPRU559, a lacZ-deficient pneumococcal strain (4). Integration of the construct into the chromosome was confirmed by PCR and sequencing.
β-Galactosidase activity was measured by using a standard protocol (23). Bacteria were grown in C+Y medium with different concentrations of P to an optical density at 620 nm (OD620) of 0.2 or under conditions that either promote (pH 8.0) or repress (pH 6.8) the development of competence. After lysis with 0.1% deoxycholate, the resulting lysate was assayed for the hydrolysis of o-nitrophenyl-β-d-galactoside (ONPG; Sigma) at 30°C. As a positive control, SPRU589, a pneumococcal strain harboring a recA-lacZ fusion and known to be competence induced, was included (4).
Transformation assay.
Scoring for acquisition of a streptomycin resistance marker as described previously (30) assessed competence for DNA transformation. Pneumococci were grown to an OD620 of 0.1 to 0.2 or, alternatively 0.1, 0.3, 0.5, or 0.8 and incubated with the selectable marker Strr DNA (1 ng/ml, final concentration). Transformation efficiency was calculated as the percentage of Strr transformants among the total bacteria and compared to that of the parent strain, R6x. Transformations were performed with and without adding exogenous CSP (5 ng/ml, final concentration). For this purpose, bacteria were incubated with Strr DNA, with or without CSP, for 30 min at 30°C and for 60 min at 37°C.
To assess the influence of P on the transformation efficiency, pneumococci were grown in C+Y medium with a final P concentration of 0, 3, 10, 30, or 60 mM.
Preparation of autolysin and assay for autolytic activity.
Purification of the pneumococcal autolysin and the biological assay for autolytic activity were performed on the basis of established protocols (26, 27).
Penicillin susceptibility and autolysis rates.
Autolysis rates of the strains were determined with 10-ml cultures of S. pneumoniae exposed to benzylpenicillin at its 10× MIC (0.1 mg/ml; Sigma) when the OD620 reached 0.25 to 0.3. Autolysis rates were calculated as the first-order rate constant K = ln (A0/A120) × min−1, where A0 represents the peak of absorbance reading at 620 nm and A120 represents the reading after a further 120 min of incubation (17). The effect of penicillin treatment on the viability was determined by exposing 10-ml cultures in the early exponential phase of growth (OD620 = 0.3, corresponding to 5 × 107 CFU/ml) to benzylpenicillin at 10× its MIC. After various periods of exposure, 100-μl portions were removed, serially diluted in C+Y medium supplemented with 100 U of penicillinase (Sigma)/ml, and plated on tryptic soy agar supplemented with 3% (vol/vol) sheep blood.
Analytical and immunological methods.
Autolysin was analyzed by performing sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and by Western blotting, using Immobilon-P membranes (Millipore Corporation, Bedford, Mass.). A 30-μg portion of protein were loaded on the sodium dodecyl sulfate gel; this quantity was within the linear range of the assay. The membranes were incubated with polyclonal rabbit antiautolysin antibody (1:1,000) and developed by using a goat anti-rabbit antibody-horseradish peroxidase conjugate (Bio-Rad, Hercules, Calif.) and an enhanced chemiluminescence kit (ECL; Amersham).
Phosphate uptake measurement.
Measurement of Pi uptake was performed by growing the strains in C+Y to an OD620 of 0.4. A 1-ml volume of cells (4 × 108 bacteria) was withdrawn and centrifuged for 10 min at 5,000 × g. Cells were washed twice with prewarmed P-free C+Y medium (37°C) by centrifugation at 5,000 × g for 5 min and finally resuspended in P-free C+Y medium buffered with 30 mM Tris-HCl. After addition of 5 mM 32Pi (106 to 107 cpm/mol; Amersham) at 37°C, five samples were withdrawn over a period of 5 min. Those samples were washed once with phosphate-buffered saline by centrifugation at 5,000 × g for 5 min at 4°C, transferred to a 1-m Eppendorf tube, and layered onto 200 μl of Nyosil oil. To separate the bacteria from the C+Y medium containing free 32Pi, the tube was spun for 1 min at 14,000 × g. The residual medium at the top of the oil layer was discarded together with the oil. The bacterial pellet was resuspended in 1 ml of phosphate-buffered saline plus 9 ml of scintillation fluid before assessment of counts in a scintillation counter (Beckman model LS6000IC).
Computer-assisted sequence analysis.
Sequence analysis and alignments were conducted with the program DNA-Star and with the Genetics Computer Group sequence analysis software package. The computer program BLAST was used to search for amino acid sequences that were homologous to those of the two-component regulatory system and the pneumococcal Pst system.
Nucleotide sequence accession number.
The DNA sequence of the pneumococcal two-component regulatory system, PnpR-PnpS, and the Pst system has been assigned the GenBank accession no. AF118229.
RESULTS
Genetic organization of the pneumococcal Pst system and the pnpR-pnpS locus.
From a panel of 51 mutants of exported proteins, one mutant, SPRU1001, was selected for detailed study because it displayed both penicillin tolerance and a >90% reduction in transformation efficiency compared with the parent strain, R6x. To obtain the nucleotide sequence of the disrupted locus, spontaneously excised plasmids from the pneumococcal mutant were recovered and propagated in E. coli. An intact plasmid spontaneously excises at a low frequency and contaminates chromosomal DNA preparations from the mutants since the target fragments of the disrupted gene are duplicated. The insert of the recovered plasmid, pHRM104, was sequenced. Inverse PCR performed on chromosomal DNA of R6x with the primers INVup and INVdown, designed against an internal region of the insert, yielded 3,712 bp adjacent to the locus duplicated in SPRU1001 (Fig. 1A). Two complete ORFs which showed striking similarities to a two-component regulatory system, PhoP-PhoR, in Bacillus subtilis were revealed (Fig. 1A). The first coding sequence, pnpR, of the two-component system was 708 bp long and encoded a protein 51% identical to the B. subtilis response regulator PhoP (16). The response regulator PnpR contained a region characteristic of the OmpR helix-turn-helix motif involved in DNA binding (21). The N terminus of PnpR is highly conserved, including several aspartate residues (D9–10, D48, D84, and D98–99) which possibly form the acidic pocket representing the phosphorylation site of the protein. Immediately downstream of the putative response regulator was a second sequence, pnpS, of 1,335 bp, encoding a protein with 31% identity to the B. subtilis histidine kinase PhoR (35). The amino acid sequence of the putative histidine kinase, PnpS, was particularly well conserved in the transmitter region; the input domain did not show any similarity to known proteins. Histidine kinases demonstrate conserved motifs (H, N, D, F, and G boxes) (37, 38). Examination of the deduced amino acid sequence of PnpS revealed an H box, HEX3P, at amino acid positions 227 to 232. An N box, NX3N, was located at amino acid positions 338 to 342, followed by a D-F box, DXGXGX9F, at amino acid positions 370 to 384. Amino acid positions 402 to 405 comprised a G box, GXGL.
A sequence located downstream of the two-component system encoded a protein 42% identical to the phosphate binding protein PstS in Methanobacterium autotrophicum (Fig. 1A). Phosphate binding proteins vary in size and sequence but exhibit some similarities. The pneumococcal phosphate binding protein, PstS, contained the typical lipoprotein motif (LxxC) located at amino acid positions 19 to 22. In addition, the crystal structure of phosphate binding proteins has been described and shows a series of hydrogen bonds that contribute to the specificity of phosphate binding proteins and keep the P in position (18). The pneumococcal PstS protein contains similar putative hydrogen bond partners: threonine at amino acid positions 8 and 161 (10 and 141 in E. coli), arginine at amino acid position 130 (135 in E. coli), serine at amino acid positions 41 and 137 (38 and 139 in E. coli), glycine at amino acid position 144 (140 in E. coli), and phenylalanine at amino acid position 17 (11 in E. coli).
Scanning the pneumococcal genome sequence released by TIGR (http://www.tigr.org/tdb/mdb/mdb.html) for the pstS sequence revealed a contiguous putative phosphate ABC transporter with a high level of similarity to the Pst systems in M. autotrophicum and E. coli (Fig. 1A). The pneumococcal Pst system included two putative membrane proteins, PstC and PstA, each with six transmembrane helices. The ORF pstB, downstream of pstA, encoded a putative ABC protein with a high degree of similarity to the ABC protein PstB of E. coli. The pneumococcal PstB has two ATP binding domains, which include the Walker A motif, GX4GK(S/T) (41), at amino acid positions 36 to 44 and the Walker B motif, (R/K)X6–8hyd4D (where hyd is any hydrophobic residue) (2, 13, 41), at amino acid positions 159 to 171. An ABC signature sequence, (L/Y)SGG(Q/M) (12), at amino acid positions 147 to 151 possibly functions as a peptide linker domain, joining different domains of the protein. A fourth motif is a conserved histidine, located 33 amino acids downstream of the aspartic acid of the Walker B motif, which is preceded by four hydrophobic residues and followed by a charged residue. A gene, phoU, with 30% similarity to the phoU genes of Enterobacter cloacae and E. coli was located directly downstream of pstB.
Phosphate uptake and growth of the pstB mutant.
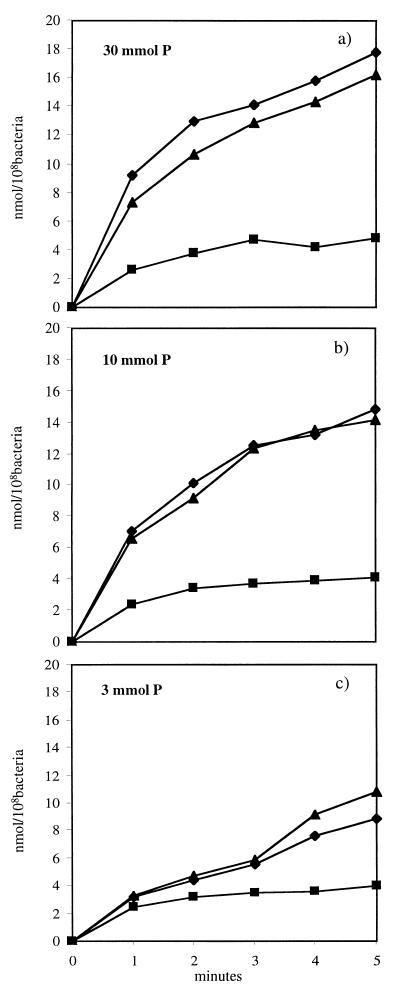
To investigate the role of the pneumococcal Pst system in P metabolism, the rate of Pi uptake by the parent strain, R6x, was compared to those of the pstB and phoU loss-of-function mutants during growth at different P concentrations. When the bacteria were grown in medium with 30 mM P, the amount of 32Pi taken up by strain R6x increased constantly over a period of 5 min, with the fastest rate of uptake being observed during the first 2 min (Fig. 2a). After 5 min, strain R6x accumulated about 18 nmol of 32Pi per 108 bacteria. Growing strain R6x with 3 and 10 mM P resulted in a decrease in Pi uptake to 8 and 14 nmol, respectively (Fig. 2b and c). Growing the pstB mutant with 30 mM P resulted in a 70% decrease in uptake of 32Pi compared to that of the parent strain, R6x, reaching an accumulation of 32Pi of about 6 nmol per 108 bacteria after 5 min (Fig. 2a). Performing the Pi uptake experiments with the pstB mutant grown under P-limited concentration conditions (3 and 10 mM) revealed only a slight decrease in Pi uptake compared to the uptake at 30 mM P. The phoU mutant behaved like the parent strain R6x (Fig. 2), indicating that PhoU in S. pneumoniae may not be directly involved in Pi uptake. The Pi uptake of the pnpR and pnpS mutants was also similar to that of the parent strain, R6x (data not shown).
FIG. 2.
Rates of uptake of 32Pi, expressed in nanomoles per 108 bacteria. Shown are Pi uptake measurements at 30 mM P, (a), 10 mM P (b), and 3 mM P (c). Parent strain R6x (⧫) and phoU mutant (▴) show comparable uptake kinetics. The pstB mutant (■) reached only 30% of the peak Pi uptake of strain R6x at 30 mM P. The results were expressed as the means of three independent measurements; the standard deviation was in each case <10%.
To address the impact of decreased Pi uptake by the pstB mutant on growth, growth rates were assessed for the parent strain, R6x, and the pstB mutant at different concentrations of P in the C+Y medium. The growth rate of the parent strain, R6x, was clearly dependent on the P concentration. Optimal growth conditions were achieved by supplementing the medium with 30 mM of P; the generation time was about 25 min. Decreasing the concentration of P to 3 mM led to an increase in the generation time to 60 min. The complete absence of P resulted in an almost complete growth arrest of strain R6x (data not shown). For the pstB mutant, supplementing the medium with 30 mM P led to a doubling time of about 50 min. Like with strain R6x, decreasing the P concentration to 3 mM led to an increase in the generation time to 60 min.
Genetic transformation in the pneumococcal Pst system and the PnpR-PnpS system.
In response to an as-yet-unknown cell density-dependent signal, pneumococci produce the pheromone CSP (10, 39). This extracellular polypeptide binds to its cognate receptor, probably the ComD histidine kinase, which in turn activates the response regulator ComE (6, 11, 32). The activation of ComE results in the transcription of a variety of genes preceded by the CIP consensus sequence; their gene products are required for the binding, uptake, and incorporation of exogenous DNA (4). To determine whether Pi uptake is critical for transformation, the efficiency of transformation of a streptomycin resistance marker, Strr DNA, was quantified for strain R6x and the pstB and phoU mutants. The efficiency of transformation of the parent strain, R6x, was dependent on the concentration of P in the medium, with maximal transformation being observed at 10 and 30 mM P (Table 2). This indicated a link between uptake or sensing of P and transformation. The pstB ABC mutant grown in the presence of 30 mM P to the density of peak transformation of the parent strain, R6x, showed a reduction of transformation of >95% (Table 2). Although the addition of CSP resulted in a slight increase in the transformation efficiency, the general transformation deficiency of the pstB mutant could not be restored (Table 2). The levels of transformation of the pstB mutant correlated with the reduced peak efficiency of transformation of the parent strain at low P concentrations of between 0 and 3 mM P. The phoU mutant displayed a wild-type transformation rate (Table 2). To assess whether the reduced transformation efficiencies of the pstB mutant and the parent strain R6x were due to competence development or transformation itself, transformation assays were performed at different P concentrations and at different growth stages (ODs of 0.1, 0.3, 0.5, and 0.8). The highest transformation rates for R6x and the pstB mutant, independent of P concentrations, occurred at an OD620 of 0.1 (data not shown). These findings suggested that the observed transformation deficiencies were due not to an impact on competence development but rather to transformation itself.
TABLE 2.
Transformation efficiencies of parent strain R6x and the pnpR, pnpS, pstB, and phoU mutants
| P concn (mM) | Transformation
efficiency for straina:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R6x
|
ΔpnpR
|
ΔpnpS
|
ΔpstB
|
ΔphoU
|
||||||
| −csp | +csp | −csp | +csp | −csp | +csp | −csp | +csp | −csp | +csp | |
| 0 | 0.002 | 0.09 | 0.001 | 0.08 | 0.002 | 0.08 | 0.001 | 0.006 | 0.003 | 0.11 |
| 3 | 0.09 | 0.91 | 0.05 | 0.78 | 0.045 | 0.52 | 0.006 | 0.012 | 0.12 | 0.86 |
| 10 | 0.19 | 2.56 | 0.11 | 2.43 | 0.12 | 2.23 | 0.008 | 0.067 | 0.179 | 2.34 |
| 30 | 0.24 | 2.79 | 0.14 | 2.81 | 0.16 | 3.11 | 0.005 | 0.072 | 0.21 | 2.38 |
| 60 | 0.04 | 1.32 | 0.009 | 1.55 | 0.008 | 1.08 | 0.007 | 0.075 | 0.06 | 1.56 |
Transformation was scored by acquisition of a streptomycin resistance marker in the absence (−) and the presence (+) of synthetic CSP (5 ng/ml). Numbers indicate percentages of transformed bacteria. Four independent experiments resulted in standard deviations of <15%.
To assess the role of the PnpR-PnpS two-component system in P dependence of genetic transformation, the pnpR and pnpS mutants were tested for their transformation efficiencies through a complete natural competence cycle. No shift of timing in competence in the pnpR and pnpS mutants occurred, as assessed at OD620s of 0.1, 0.3, 0.5, and 0.8 (data not shown). The pnpR and pnpS mutants grown to an OD620 of 0.1 showed a minor reduction in transformation efficiency of about 30% of that of the parent strain, R6x (Table 2). This partial defect was corrected by the addition of exogenous CSP (Table 2). These results indicated that the absence of a functional PnpS histidine kinase or PnpR response regulator does not have a major impact on transformation and has no effect on the response to CSP.
Effect of the pneumococcal Pst system and the PnpR-PnpS system on autolysis.
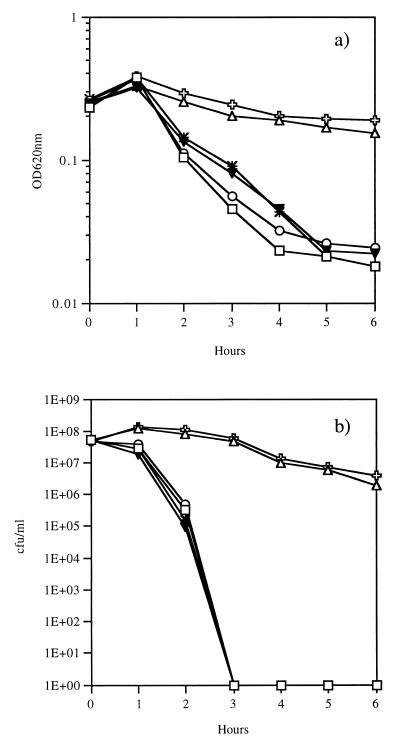
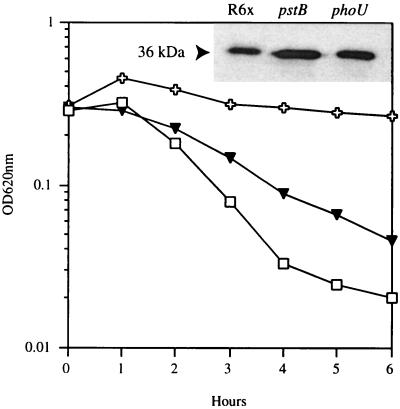
To determine the role of P in autolysis, cultures of strain R6x and its pnpR, pnpS, pstB, phoU, and Lyt-4-4 mutants at OD620s of 0.2 to 0.3 were exposed to penicillin at 10× its MIC. The effects on turbidity and viability were monitored. The parent strain, R6x, and the autolysin-deficient mutant Lyt-4-4 served as the positive control and the negative control, respectively. After 1 h, R6x and the pnpR, pnpS, and phoU mutants underwent antibiotic-induced lysis and killing (Fig. 3). In contrast, the pstB mutant and Lyt-4-4 demonstrated no significant autolysis and markedly reduced killing (Fig. 3). The reduced response to penicillin of the pstB mutant could be due to changes in either the expression or the activity of the autolysin, LytA. Western blot analysis of an autolysin preparation of the pstB mutant showed a normal translational LytA product (Fig. 4). However, the function of the autolysin appeared to be compromised. Exogenous autolysin from R6x reconstituted lysis in the autolysin-deficient strain Lyt-4-4, while the autolysin preparation of the pstB mutant failed to render Lyt-4-4 sensitive to penicillin (Fig. 4). These results suggest that the lack of autolysis in the pstB mutant is due to an alteration in amidase activity. However, growth of strain R6x in medium with a reduced P concentration did not result in reduced autolysis (data not shown), suggesting that either PstB has a direct effect on the autolysin independent of P transport or other P transport systems are compensatory in the wild-type background.
FIG. 3.
Effect of loss-of-function mutations on penicillin-induced lysis and loss of viability. Wild-type strain R6x (□), LytA-defective strain Lyt-4-4 (▵), a pnpR mutant (○), a pnpS mutant (∗), a pstB mutant ( ), and a phoU mutant (▾) were studied. Cultures in the early logarithmic phase of growth (107 CFU/ml) were treated with penicillin at 10× its MIC (0.1 mg/ml), and both the OD620 (a) and bacterial viability (b) were monitored for 6 h.
FIG. 4.
Functional assay of autolytic activity. Crude autolysin preparations of parent strain R6x (□), a pstB mutant ( ), and a phoU mutant (▾) were added to cultures of the autolysis-defective strain Lyt-4-4 at an OD620 of 0.25. Lysis was monitored after addition of penicillin at 10× its MIC (0.1 mg/ml) at an OD620 of 0.3. The results of immunoblot analysis of crude autolysin preparations of the parent strain R6x, the pstB mutant, and the phoU mutant are shown in the inset. The arrow indicates the size of LytA (36 kDa).
Expression of the two-component system PnpR-PnpS is independent of P concentration and transformation.
A specific CIP consensus sequence was recently detected directly upstream of the likely translational start of the response regulatory pnpR (4) (Fig. 1a). Since this promoter characterizes genetic elements induced during transformation in S. pneumoniae, the regulation of the two-component regulatory system under competence-inducing conditions was assessed. Pneumococci were transformed with a pnpR-lacZ construct to produce the strain SPRU1002, containing a pnpR-lacZ gene fusion, without disrupting the pnpR-pnpS locus (Fig. 1b). The next ORF upstream of pnpR (similar to an arginine tRNA synthetase) terminated 448 bp upstream of the start codon of pnpR. In order to include all possible regulatory elements, the entire intergenic region was included in the lacZ fusion. SPRU1002 was assessed for β-galactosidase activity under competence-inducing and competence-suppressing growth conditions. The pneumococcal strain SPRU589, harboring a recA-lacZ gene fusion which is strictly competence induced and driven by a CIP consensus promoter (4, 29), was used as a positive control. Expression of β-galactosidase activity by SPRU1002 was the same under both competence-inducing and -suppressing growth conditions, suggesting that transcription of the two-component system was not influenced by these conditions (data not shown). SPRU1002 was also assessed for variation of β-galactosidase activity under growth conditions in which the P concentration differed. At concentrations of 0, 3, 10, 30, and 60 mM P, the β-galactosidase activity remained unchanged (data not shown), suggesting that the transcription of pnpR and pnpS was not influenced by P. Northern blot analysis of the Pst locus in the parent strain R6x and the pnpS and pnpR mutants, using a probe specific for the pstS gene, showed no difference in levels of expression of that locus (data not shown).
DISCUSSION
In E. coli, P. assimilation is under the control of the PHO regulon, which includes 38 different genes. Phosphorylation and dephosphorylation of the response regulator PhoB are needed for the control of this regulon. The major Pi transporters of the PHO regulon are the Pst and the Pit systems, which function under conditions of P limitation and excess, respectively.
We describe a locus comprising a two-component system, PnpR-PnpS, and a putative pneumococcal Pst system which includes PstSACB and PhoU. The order of the pneumococcal genes corresponds to that of the E. coli Pst operon. The deduced amino acid sequences of the proteins encoded by these genes exhibit sequence similarities, ranging from 29 to 55%, to the corresponding genes of various bacteria, such as M. autotrophicum and E. coli. Insertion duplication mutagenesis of the gene encoding the ABC protein PstB revealed a pleiotropic phenotype of reduced Pi uptake, transformation deficiency, and penicillin tolerance.
In accordance with the high level of sequence similarities to the E. coli Pst locus, the reduced Pi uptake of the pstB mutant, but not of the phoU mutant, supported the hypothesis that the putative pneumococcal Pst system is involved in Pi uptake. Although reduction of the P concentration of the medium led to decreased Pi uptake. Although reduction of the P concentration of the medium led to decreased Pi uptake in the parent strain, R6x, the relationship was not linear. A 10-fold change in P concentration (30 to 3 mM) resulted in only a 50% decrease in Pi uptake, consistent with the described induction of the Pst system to compensate for decreased Pi uptake (42). The loss-of-function pstB mutation would be expected to interrupt this mechanism. The finding that Pi uptake in the pstB mutant was low and remained low during P limitation supports this hypothesis. Residual Pi uptake in the pstB mutant might be explained by the existence of alternative Pi transport systems.
Two models can be proposed to explain the reduced genetic transformation efficiency of the pstB mutant. In the first model, the reduced Pi uptake is directly responsible for the effect on transformation. P is an essential component for innumerable biomolecules and is also incorporated into many proteins posttranslationally. Genetic transformation involves a very complex interaction of several proteins; therefore, it is reasonable that a reduction in the intracellular level of P might affect those proteins. This possibility is supported by the fact that a reduction of the extracellular P concentration to ≤3 mM led to an almost complete loss of transformation in the wild-type pneumococcus. A possible link between the transformability of pneumococcal cultures and P was suggested previously by the observation that pneumococci are competent if grown in a medium (mal-C) in which maleate buffer replaces the P buffer. Since the mal-C medium was supplemented with dipotassium phosphate, a definitive conclusion about the role of P could not be drawn (40).
A second model hypothesizes that the pneumococcal Pst system is part of a signaling pathway independent of Pi uptake. The observation that the pstB mutant demonstrated penicillin tolerance regardless of the P concentration and of a lack an effect of P on wild-type autolysis are consistent with a non-Pi-related signaling role. This duality is similar to the recently described ABC-type permease complex Psa, which transports Mn2+ and Zn2+ but also is part of a signaling pathway indirectly affecting adhesion, lysis, transformation, and virulence of S. pneumoniae (27). The demonstration of a normal translational product of lytA indicates that the links between the pneumococcal Pst system, transformation, and autolysis do not arise at the level of transcription from the common promoter upstream of recA and lytA (25). Thus, the pneumococcal Pst system, like other ABC transporters (27), may function both in Pi uptake and in signaling autolytic activity. Loss of function of the two-component system, PnpR-PnpS, failed to recapitulate the phenotype of the Pst ABC mutant. Although the two-component system is located upstream of the pneumococcal Pst locus, no evidence for a functional relationship between the two loci was found. This finding is distinct from the coupling of the two corresponding loci in E. coli.
In summary, a putative pneumococcal Pst system was identified in S. pneumoniae. The pneumococcal Pst system is comparable to the Pst system in E. coli and contains the first locus described in S. pneumoniae that is involved in P assimilation. Insertion duplication mutagenesis of the ABC gene pstB revealed coincident decreases in Pi uptake, genetic transformation, and autolysis. Mutagenesis of the phoU gene did not lead to this phenotype. A transformation deficiency in the pstB mutant was most likely linked to the decreased Pi uptake. However, the inactivity of the autolysin was independent of the P concentration, most reasonably suggesting that the Pst system might be involved in a P-independent signaling pathway, comparable to the ABC transporter Psa (27). Although similar in sequence to the P-responsive two-component systems of E. coli and B. subtilis, the two-component regulatory system PnpR-PnpS, located upstream of the pneumococcal Pst system, did not seem to be responsive to different P concentrations and might be not linked to the pneumococcal Pst system.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grants AI27913 and AI39482, Cancer Center Support CORE grant P30 CA 21765, and American Lebanese Syrian Associated Charities grant CA21765.
We acknowledge the excellent technical assistance of Juan Li and John Winestone.
REFERENCES
- 1.Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 2.Ames G F, Mimura C S, Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia colito human: traffic ATPases. FEMS Microbiol Rev. 1990;6:429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown M C M, Weston A, Saunders J R, Humphreys G O. Transformation of E. coliC600 by plasmid DNA at different phases of growth. FEMS Microbiol Lett. 1979;5:219–222. [Google Scholar]
- 4.Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniaerevealed by genomic analysis. Mol Microbiol. 1998;27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen J D, Morrison D A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia colirequires vectors protected by strong transcriptional terminators. Gene. 1987;55:179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Q, Campbell E A, Naughton A M, Johnson S, Masure H R. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol Microbiol. 1997;23:687–692. doi: 10.1046/j.1365-2958.1997.2481617.x. [DOI] [PubMed] [Google Scholar]
- 7.Hakenbeck R, Stock J B. Analysis of two-component signal transduction systems involved in transcriptional regulation. Methods Enzymol. 1996;273:281–300. doi: 10.1016/s0076-6879(96)73026-4. [DOI] [PubMed] [Google Scholar]
- 8.Haldenwang W G, Banner C D B, Ollington J F, Losick R, Hoch J A, O’Connor M B, Sonenshein A L. Mapping a cloned gene under sporulation control by insertion of a drug resistance marker into the Bacillus subtilischromosome. J Bacteriol. 1980;142:90–98. doi: 10.1128/jb.142.1.90-98.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haldimann A, Daniels L L, Wanner B L. Use of new methods for construction of tightly regulated arabinose and rhamnose promotor fusions in studies of the Escherichia coliphosphate regulon. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havarstein L S, Gaustad P, Nes I F, Morrison D A. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol. 1996;21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 12.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 13.Hyde S C, Emsley P, Hartshorn M J, Mimmack M M, Gileadi U, Pearce S R, Gallagher M P, Gill D R, Hubbard R E, Higgins C F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 14.Kowalczykowski S C. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu Rev Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- 15.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 16.Lee J W, Hulett F M. Nucleotide sequence of phoP, the gene encoding PhoP, the response regulator of the phosphate regulon of in Bacillus subtilis. Nucleic Acids Res. 1992;20:5848. doi: 10.1093/nar/20.21.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H H, Tomasz A. Penicillin tolerance in multiply drug-resistant natural isolates of Streptococcus pneumoniae. J Infect Dis. 1985;152:365–372. doi: 10.1093/infdis/152.2.365. [DOI] [PubMed] [Google Scholar]
- 18.Luecke H, Quiocho F A. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature. 1990;347:402–406. doi: 10.1038/347402a0. [DOI] [PubMed] [Google Scholar]
- 19.Makino K, Shinagawa H, Amemura M, Kawamato T, Yamada M, Nakate A. Signal transduction in the phosphate regulon of Escherichia coliinvolves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 20.Martin B, Garcia P, Castanie M P, Claverys J P. The recA gene of Streptococcus pneumoniaeis part of a competence-induced operon and controls lysogenic induction. Mol Microbiol. 1995;15:367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 22.Mejean V, Claverys J P, Vasseghi H, Sicard A M. Rapid cloning of specific DNA fragments of Streptococcus pneumoniaeby vector integration into the chromosome followed by endonucleolytic excision. Gene. 1981;15:289–293. doi: 10.1016/0378-1119(81)90139-6. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.Morrison D A, Baker M F. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature. 1979;282:215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- 25.Mortier-Barriere I, de Saizieu A, Claverys J P, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 26.Mosser J L, Tomasz A. Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J Biol Chem. 1970;245:287–298. [PubMed] [Google Scholar]
- 27.Novak R, Braun J S, Charpentier E, Tuomanen E. Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese complex Psa. Mol Microbiol. 1998;29:1285–1296. doi: 10.1046/j.1365-2958.1998.01016.x. [DOI] [PubMed] [Google Scholar]
- 28.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 29.Pearce B J, Naughton A M, Campbell E A, Masure H R. The rec locus, a competence-induced operon in Streptococcus pneumoniae. J Bacteriol. 1995;177:86–93. doi: 10.1128/jb.177.1.86-93.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearce B J, Naughton A M, Masure H R. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol Microbiol. 1994;12:881–892. doi: 10.1111/j.1365-2958.1994.tb01076.x. [DOI] [PubMed] [Google Scholar]
- 31.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 32.Pestova E V, Havarstein L S, Morrison D A. Regulation of the transformability in Streptococcus pneumoniaeby an auto-induced peptide pheromone and a two component regulatory system. Mol Microbiol. 1996;9:1037–1050. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg H. Phosphate transport in prokaryotes. San Diego, Calif: Academic Press, Inc.; 1987. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 35.Seki T, Yoshikawa H, Takahashi H, Saito H. Nucleotide sequence of the Bacillus subtilis phoRgene. J Bacteriol. 1988;170:5935–5938. doi: 10.1128/jb.170.12.5935-5938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steed P M, Wanner B L. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoUoperon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol. 1993;175:6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 39.Tomasz A. Control of the competent state in pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature. 1965;208:155–159. doi: 10.1038/208155a0. [DOI] [PubMed] [Google Scholar]
- 40.Tomasz A, Hotchkiss R D. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker J E, Saraste M, Runswich M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wanner B L. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. . [Review.] [DOI] [PubMed] [Google Scholar]
- 43.Willsky G R, Malamy M H. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]