Abstract
Type 1 regulatory T (Tr1) cells represent a subset of IL-10–producing CD4+Foxp3− T cells and play key roles in promoting transplant tolerance. However, no effective pharmacological approaches have been able to induce Tr1 cells in vivo. We herein report the combined use of a CD28 superagonist (D665) and anti–glucocorticoid-induced tumor necrosis factor receptor–related protein monoclonal antibody (G3c) to induce Tr1 cells in vivo. Large amounts of IL-10/interferon-γ–co-producing CD4+Foxp3− Tr1 cells were generated by D665-G3c sequential treatment in mice. Mechanistic studies suggested that D665-G3c induced Tr1 cells via transcription factors Prdm1 and Maf. G3c contributed to Tr1 cell generation via the activation of mitogen-activated protein kinase–signal transducer and activator of transcription 3 signaling. Tr1 cells suppressed dendritic cell maturation and T cell responses and mediated permanent allograft acceptance in fully major histocompatibility complex–mismatched mice in an IL-10–dependent manner. In vivo Tr1 cell induction is a promising strategy for achieving transplant tolerance.
In vivo generation of IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 induced permanent allograft acceptance.
INTRODUCTION
Organ transplantation is considered the optimal treatment for a variety of end-stage organ diseases. However, the life-long, systemic immune suppression required after transplantation severely compromises host immune defense and is associated with adverse effects, including organ toxicity, infections, and malignancies. Operational tolerance has long been the ultimate goal in the field of transplantation, which would enable transplant recipients to maintain a stable and acceptable graft function without the need for immunosuppression therapy, thus avoiding undesirable side effects (1).
Allograft immunity is a complex process that results from the interplay of multiple different cell types, including lymphocytes, monocytes, macrophages, and dendritic cells (DCs). Recipient alloreactive T cells recognize non–self-donor alloantigens presented by donor or recipient antigen-presenting cells and initiate the adaptive inflammatory immune response, leading to allograft rejection. Regulatory immune cells regulate or suppress immune responses of other cells and help prevent anti-donor immune responses. Regulatory immune cell–based therapies, via inducing or adoptively transferring regulatory immune cells, are emerging as promising strategies for achieving permanent donor-specific immune tolerance, thus minimizing or even obviating immunosuppressants after organ transplantation.
CD4+ T cells coordinate immune responses by helping to activate and regulate other immune cells and are critical in determining transplantation rejection or tolerance. Type 1 regulatory T (Tr1) cells represent a subset of CD4+Foxp3− T cells and secrete high amounts of interleukin-10 (IL-10), their signature cytokine, with potent immunosuppressive properties. The potent immunosuppressive function of Tr1 cells has been implicated both in vitro and in vivo (2). Tr1 cells have been proven to play key regulatory roles in peripheral immune tolerance and are considered an emerging therapeutic target for improving transplant tolerance (3). However, as with Foxp3+ regulatory T (Treg) cells, in vivo effective pharmacological approaches to induce Tr1 cells are largely lacking.
CD28 superagonist is a monoclonal antibody (mAb) that engages CD28 costimulatory receptors, which laterally bind to the CD28 homodimer, thus allowing its clustering via lattice formation (4, 5). The particular binding topology of CD28 superagonist confers its superagonist properties, resulting in potent T cell activation and expansion independent of concomitant T cell receptor (TCR) engagement (6). CD28 superagonist was shown to expand Treg cells preferentially over effector T (Teff) cells both in vitro and in vivo, a characteristic that was used to prevent autoimmune disease, graft-versus-host disease (GvHD), and allograft rejection (6–8). Glucocorticoid-induced tumor necrosis factor receptor–related protein (GITR), also referred to as TNFRSF18, is a type I transmembrane protein of the tumor necrosis factor receptor (TNFR) superfamily, characterized by three cysteine-rich domain pseudorepeats in its extracellular domain (9). GITR is constitutively expressed at high levels on Treg cells and at low levels on naïve and memory T cells (9–11). Its expression is rapidly up-regulated upon TCR activation on both Treg and Teff cells (11, 12). GITR serves as a costimulatory molecule that exerts multiple distinct effects on T cells. GITR engagement results in the proliferation and cytokine production of activated T cells but abrogates the suppressive activity of Treg cells (10, 11, 13, 14). Targeting GITR has been evaluated for its utility in the treatment of cancers, autoimmune diseases, and organ rejection in both animal models and clinical trials (15–19). However, the effects of GITR agonism on T cells, which is cell specific and context dependent, remain controversial (20, 21).
We herein report the combined use of CD28 superagonist D665 and anti-GITR mAb G3c to induce Tr1 cells in vivo. Large amounts of IL-10/interferon-γ (IFN-γ)–co-producing CD4+Foxp3− Tr1 cells were generated by D665-G3c sequential treatment in mice. Tr1 cells suppressed DC maturation and T cell responses in an IL-10–dependent manner. Further mechanical studies suggested that D665-G3c induced Tr1 cells via the positive regulatory domain zinc finger protein 1 (Prdm1), and musculoaponeurotic fibrosarcoma (Maf) pathways. G3c contributed to Tr1 cell generation via the activation of mitogen-activated protein kinase–signal transducer and activator of transcription 3 (MAPK-STAT3) signaling. The combined use of D665 and G3c induced permanent allograft acceptance in a Tr1-dependent manner. Our study presented an effective pharmacological approach to generate Tr1 cells in vivo by D665-G3c sequential treatment and demonstrated their therapeutic potential in transplantation.
RESULTS
Induction of Tr1 cells in vivo
Foxp3-GFP mice [expressing green fluorescent protein (GFP) under the control of the mouse Foxp3 promoter] and IL-10–Venus mice [expressing yellow fluorescent protein (Venus) under the control of the mouse IL-10 promoter] were treated with D665 (250 μg per mouse) and G3c (250 μg per mouse) on days −3 and 0 sequentially (Fig. 1A). On day 0, before G3c administration, D665 induced a robust expansion of Treg cells over Teff cells (Fig. 1B), consistent with previous reports (22–25). The GITR expression was significantly up-regulated on the surface of both Treg and Teff cells (Fig. 1, C and D). When targeting GITR with G3c, large amounts of CD4+Foxp3−IL-10+ Tr1 cells were generated on days 3 and 7 (Fig. 1, E and F). Although Treg cells were also potently expanded, they were obviously decreased on day 7 in the combination treatment group (Fig. 1, E and G). On day 7, most of the CD4+ IL-10+ T cells were IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 cells (Fig. 1E and fig. S1A). The gene expression of IL-10 and Ifng exhibited a consistent trend with flow cytometry (FCM) results (fig. S1B). The numbers of splenocytes obtained in each group are shown in fig. S1C.
Fig. 1. Protocol for the induction of IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 cells.
(A) D665 (250 μg per mouse) and G3c (250 μg per mouse) were intraperitoneally injected on days −3 and 0 sequentially. (B) Representative flow cytometry (FCM) analysis and frequency of Treg cells in the splenocytes of D665- and immunoglobulin G (IgG)–treated mice on day 0 (n = 6 for each group). ****P < 0.0001. (C and D) The median fluorescence intensity (MFI) of the GITR on CD4+Foxp3+ Treg cells and CD4+Foxp3− Teff cells in the splenocytes of D665- and IgG-treated mice on day 0 (n = 5 for each group). **P < 0.01 and ****P < 0.0001. (E) Representative FCM analysis of CD4+Foxp3+ Treg cells and CD4+Foxp3−IL-10+ Tr1 cells in the splenocytes of each treatment group day 3 (D3) and D7 (n = 4 for each group). (F) Frequency of CD4+Foxp3−IL-10+ Tr1 cells in the splenocytes of each treatment group on day 3 and day 7 (n = 4 for each group). *P < 0.05, **P < 0.01, and ****P < 0.0001. (G) Frequency of CD4+Foxp3+ Treg cells in the splenocytes of each treatment group on day 3 and day 7 (n = 4 for each group). *P < 0.05, ***P < 0.001, and ****P < 0.0001. Values are shown as the mean ± SEM.
We further analyzed the phenotype of Treg and Tr1 cells. Treg cells showed a central memory phenotype and strongly expressed the activation markers CD25 and CD69, coinhibitory markers programmed cell death protein 1 (PD-1), inducible costimulatory molecule (ICOS), T cell immunoglobulin and mucin-domain containing-3 (TIM-3), T cell protein cytotoxic T lymphocyte antigen 4 (CTLA-4), lymphocyte activation gene 3 (LAG3), and T cell immunoreceptor with immunoglobulin (Ig) and ITIM domains (TIGIT). The detected GITR expression was slightly lower compared to that in naïve CD4+ T cells because of blockade of FCM antibody binding to GITR by G3c (Fig. 2A and fig. S2A). Tr1 cells comprised both central and effector memory phenotypes and strongly expressed coinhibitory markers PD-1, ICOS, CTLA-4, and LAG3 but weakly expressed activation markers CD25 and CD69 (Fig. 2B and fig. S2B). The coexpression of CD49b and LAG3, identified as biomarkers for a population of murine and human memory Tr1 cells, was not observed (26).
Fig. 2. The phenotype of Treg and Tr1 cells.
(A) Representative histogram of molecule markers on CD4+Foxp3(GFP)+ Treg cells as evaluated by FCM (n = 4 for each group). (B) Representative histogram of molecule markers on CD4+Foxp3−IL-10(Venus)+ Tr1 cells as evaluated by FCM (n = 4 for each group).
Function of induced Tr1 cells in vitro
Tr1 cells are characterized by the production of high amounts of IL-10, which has a potent immunosuppressive function. IL-10 exerts its major suppressive effects on the DC maturation and accessory functions (27, 28). To determine the suppressive function of Tr1 cells on DCs, we isolated CD4+IL-10+ T cell, CD4+IL-10− T cells, and naïve CD4+ T cells and cocultured them with bone marrow–derived DCs (BMDCs) stimulated with lipopolysaccharide (LPS) on day 5. CD11b+CD11c+ population cells were identified as BMDCs to assess maturation state on day 7 (fig. S3). Tr1 cells prevented LPS-mediated BMDC maturation, as measured by the decreased median fluorescence intensity of surface CD40, CD80, CD86, and major histocompatibility complex class II (MHC-II) molecules (Fig. 3, A and B). Furthermore, the addition of anti–IL-10–neutralizing antibody diminished the Tr1 cell–mediated prevention of BMDC maturation.
Fig. 3. Tr1 cells suppress immune responses via IL-10 signaling.
(A) Representative histogram of surface MHC-II, CD40, CD80, and CD86 on BMDCs in each group (n = 4 for each group). (B) Quantitative data of the MFI of surface MHC-II, CD40, CD80, and CD86 on BMDCs in each group (n = 4 for each group). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (C) Representative histogram of CFSE dilution of CD8+ T cells in each group (n = 4 for each group). (D) Quantitative data of the proportion of CFSEdim proliferating CD8+ T cells in each group (n = 4 for each group). Values are shown as the mean ± SEM; ***P < 0.001 and ****P < 0.0001.
IL-10 also exerts inhibitory effects by suppressing T cell proliferation (29, 30). Cytotoxic CD8+ T cells are the principal driving force of allograft destruction and regulated by helper CD4+ T cells. We therefore next investigated the suppressive effects of Tr1 cells on CD8+ T cells using a one-way mixed lymphocyte reaction (MLR) system, where purified B6/J mice CD8+ T cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) as responders and cocultured with BALB/c BMDC stimulators. FCM-sorted CD4+IL-10+ T cells, CD4+IL-10− T cells, and naïve CD4+ T cells were added to the MLR systems as regulators and cocultured for 3 days. As shown in Fig. 3 (C and D), CD8+ T cell proliferation was significantly suppressed in the presence of CD4+IL-10+ Tr1 cells, whereas the addition of an anti–IL-10 antibody restored the CD8+ T cell proliferation. These data indicated that Tr1 cells mediated the suppressive function mainly dependent on IL-10 signaling.
Mechanism underlying Tr1 cell generation
To investigate the mechanism underlying the generation of Tr1 cells by D665-G3c sequential treatment, we isolated CD4+IL-10+ Tr1 cells and CD4+IL-10− T cells from D665-G3c–treated IL-10–Venus mice on day 7 and conducted a transcriptome RNA sequencing (RNA-seq) analysis (Fig. 4A). The gene expression of IL-10 and Ifng in both cell populations was confirmed by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 4B). Following the RNA-seq analysis, a magnitude component (principal component #1) separated samples by cell type, explaining 83% of the total variation (fig. S4). Compared to CD4+IL-10− T cells, 1963 differentially expressed genes (DEGs) were obtained in CD4+IL-10+ Tr1 cells, with a threshold of absolute log2 fold change (log2FC) > 0.5 and adjusted P value of <0.05, including 525 genes that were up-regulated and 1438 that were down-regulated (Fig. 4, C and D). Consistent with the cell-sorting strategy, IL-10 was the most up-regulated gene compared to the CD4+IL-10− T cells (Fig. 4C).
Fig. 4. Results of an RNA-seq analysis of CD4+IL-10+ and CD4+IL-10− T cells.
(A) Total CD4+ T cells were purified by magnetic-activated cell sorting (MACS) from the splenocytes of D665-G3c–treated IL-10–Venus mice on day 7 and then subjected to CD4+IL-10+ and CD4+IL-10− T cell sorting based on the Venus expression. The isolated CD4+IL-10+ and CD4+IL-10− T cell samples were prepared in three duplicates for the subsequent transcriptome RNA-seq analysis. (B) The relative mRNA expression of IL-10 and Ifng in CD4+IL-10+ and CD4+IL-10− T cells, detected by qRT-PCR, normalized with 18S for each sample (n = 4 for each group). Paired Student’s t test; *P < 0.05 and **P < 0.01. (C) The volcano plot shows the DEGs, with a threshold of absolute log2FC of >0.5 and adjusted P value of <0.05, between CD4+IL-10+ T cells versus CD4+IL-10− T cells. ns, not significant. (D) The heatmap shows 1963 DEGs, among which 525 were up-regulated and 1438 were down-regulated, between CD4+IL-10+ cells versus CD4+IL-10− cells. (E) GO cluster plot displaying a circular dendrogram of the clustering of the DEGs with color-coded log2FC (inner ring) and the assigned functional terms (outer ring). (F) Heatmap of differentially expressed TFs known to regulate Tr1 differentiation. (G) The relative mRNA expression of differentially expressed TFs identified in (F), validated by qRT-PCR, and normalized with 18S for each sample (n = 4 for each group). Paired Student’s t test; *P < 0.05 and **P < 0.01. Values are shown as the mean ± SEM.
We then performed a gene ontology (GO) enrichment analysis to gain insight into the molecular functions of the DEGs that were enriched in CD4+IL-10+ Tr1 cells (Fig. 4E). The most enriched GO terms were those involving signaling receptor activity, molecular transducer activity, transmembrane signaling activity, cell adhesion molecular binding, immune receptor activity, cytokine binding, G protein–coupled receptor activity, growth factor binding, cytokine receptor activity, and cytokine activity. To identify transcription factors (TFs) that might be responsible for IL-10 production, we extracted TFs known to regulate Tr1 differentiation from DEGs, including interferon regulatory factor 4 (Irf4), early growth response 2 (Egr2), aryl hydrocarbon receptors (Ahr), T-box transcription factor 21 (Tbx21), Eomesodermin (Eomes), Prdm1, and Maf. Of these, Prdm1, Maf, Eomes, and Tbx21 were up-regulated with IL-10, whereas Irf4, Egr2, and Ahr were down-regulated in CD4+IL-10+ Tr1 cells (Fig. 4F). The gene expression of the TFs was further validated by qRT-PCR. Prdm1, Maf, Eomes, and Tbx21 were verified to be significantly up-regulated in CD4+IL-10+ Tr1 cells (Fig. 4G).
To further explore the role of D665 and G3c in Tr1 cell generation, we examined the expression of TFs in different treatment groups on days 3 and 7 (Fig. 5A). Single-D665 treatment induced Prdm1 and Maf up-regulation, and the combination treatment further increased their expression. We also observed an elevated Prdm1 expression in the single-G3c treatment group compared to the naïve group on day 7. The Eomes expression was only up-regulated in the combination treatment group. Next, we detected the Blimp1 (encoded by the Prdm1 gene) and c-Maf (encoded by the Maf gene) protein expression by Western blotting (Fig. 5, B and C). The expression of both Blimp1 and c-Maf protein was significantly up-regulated in the combination treatment group. It has been shown that STAT3 plays a critical role in Tr1 cell generation and is a potent inducer of both Blimp1 and c-Maf expression during Tr1 cell differentiation (31–33). In our study, high STAT3 phosphorylation was observed in the single-G3c treatment group as well as the combination treatment group (Fig. 5, B and C), possibly hinting at the essential role of GITR signaling in Tr1 cell generation. Extracellular signal–related kinase (ERK) MAPK, as downstream signaling of GITR activation, have been shown to induce and maintain IL-10 production in Tr1 cells (34). Our results revealed that the phosphorylation of ERK was significantly increased in the single-G3c treatment group as well as the combination treatment group (Fig. 5, B and C). In addition, although IL-27 was highlighted as an important driver of Tr1 cell in previous studies (35, 36), we only observed a slightly increased expression of IL-27p28 in the combination treatment group on day 7 (Fig. 5D). The level of Epstein-Barr virus–induced gene 3 (Ebi3), a subunit of IL-27 heterodimer, was comparable among groups.
Fig. 5. The mechanism underlying the Tr1 cell functions.
(A) The relative mRNA expression of differentially expressed TFs identified in Fig. 4F on day 3 and day 7, normalized with 18S for each sample (n = 4 for each group). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (B) Results of a Western blot analysis of the phosphorylated STAT3 (p-STAT3), STAT3, p-ERK, ERK, Blimp1, c-Maf, and β-actin protein expression in each group on day 7. Data are representative of three independent experiments. (C) Quantitative data for the expression of p-STAT3 relative to STAT3, p-ERK relative to ERK, and Blimp1 and c-Maf relative to β-actin in each group on day 7 (n = 3 for each group). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (D) The relative mRNA expression of IL-27p28 and Ebi3 on day 3 and day 7, normalized with 18S for each sample (n = 4 for each group). **P < 0.01. Values are shown as the mean ± SEM.
Generation of Tr1 cells for permanent allograft acceptance
Previous studies have shown that Tr1 cells play a critical role in promoting and maintaining tolerance (3). We therefore next determined whether or not Tr1 cells generated by the combination of D665 and G3c treatment could induce permanent allograft acceptance.
By using a fully MHC-mismatched (donor: BALB/c, H-2kd; recipient: B6/J, H-2kb) mouse model of heterotopic heart transplantation (Fig. 6A), we found that the use of D665 alone prolonged the survival of heart allograft, whereas the single use of G3c showed a minimal effect on the heart allograft survival. Unexpectedly, combinations of D665 and G3c treatments induced permanent allograft acceptance (Fig. 6B). A histological analysis of the heart allograft on postoperative day 7 (POD7) indicated pronounced inflammatory infiltration and severe myocyte damage in the no-treatment control transplantation group, effects that were markedly ameliorated in the D665-G3c–treated group (Fig. 6C and fig. S5A). D665 potently expanded Treg cells, and G3c further enforced the expansion of Treg cells on POD3. However, on POD7, Treg cells were largely diminished in the combined treatment group (Fig. 6D and fig. S5B). Large amounts of IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 cells were observed both in the cardiac graft-infiltrating lymphocytes (GILs) and splenocytes in the D665-G3c–treated group on POD7 (Fig. 6E and fig. S5C).
Fig. 6. D665-G3c treatment induced permanent allograft acceptance.
(A) Treatment protocol of D665 and G3c in mouse heart transplantation. D665 (250 μg per mouse) and G3c (250 μg per mouse) were intraperitoneally injected on days −3 and 0 sequentially. Heart transplantation was performed on day 0. (B) The survival of cardiac allografts in each group (n = 9 to 12 for each group). The graft survival of each group was evaluated using Kaplan-Meier curves and log-rank tests. ****P < 0.0001. (C) Representative hematoxylin and eosin staining of cardiac grafts in each treatment group on POD7 (n = 6 for each group). Scale bars, 100 μm. (D) A representative FCM analysis of CD4+CD25+Foxp3+ Treg cells in the splenocytes of each treatment group on POD3 and POD7. (E) A representative FCM analysis of IL-10/IFN-γ–co-producing CD4+ Tr1 cells in the splenocytes (SPs) and GILs of each treatment group on POD7 (n = 4 for each group). (F) A representative FCM analysis of CD4+ T and CD8+ T cells in the splenocytes and GILs of each treatment group on POD7 (n = 4 for each group). (G) Representative CD8 (blue), 5-bromo-2′-deoxyuridine (BrdU) (red), and type IV collagen (yellow) triple immunohistochemistry staining of heart grafts in each group on POD7 (n = 4 for each group). Scale bars, 100 μm. (H) Survival of cardiac allografts in each group (n = 6 for each group). The graft survival of each group was evaluated using Kaplan-Meier curves and log-rank tests. ***P < 0.001.
Further FCM analyses revealed that the CD4+/CD8+ T cell ratio were significantly increased in both the graft and spleen in the combination treatment group on POD7 (Fig. 6F and fig. S5D). The numbers of splenocytes and GILs obtained in each group are shown in fig. S5E. Consistently, myocardial infiltration of both of cytotoxic CD8+ T cell and proliferating cytotoxic CD8+ T cells, considered the major executor of transplantation rejection, were notably decreased in the combination treatment group on POD7 detected by immunohistochemical staining (Fig. 6G).
Because Foxp3+ Treg cells were also transiently expanded in our model, we next examined the contribution of Treg and Tr1 cells to the induction of heart allograft acceptance. As shown in Fig. 6H, depletion of Treg cells on POD−1, POD3, and POD7, respectively, using anti-CD25 treatment did not disrupt heart tolerance, whereas the neutralization of IL-10 by anti–IL-10 or IL-10 knockout resulted in heart rejection. These results indicated that the combination of D665 and G3c treatment induced permanent allograft acceptance in a Tr1 cell–dependent rather than Treg cell–dependent manner.
DISCUSSION
Tr1 cells are potent IL-10–producing cells capable of suppressing immune responses to self, foreign, and allogeneic antigens. We found in the present study that combinations of CD28 superagonist D665 and anti-GITR antibody G3c could generate large amounts of IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 cells in vivo. Mechanistic studies suggested that D665 and G3c treatment induced Tr1 cells via TFs Prdm1 and Maf. G3c contribute to Tr1 cell generation via the activation of MAPK-STAT3 signaling. Tr1 cells suppressed DC maturation and T cell proliferation in an IL-10–dependent manner. Furthermore, in a mouse heart transplantation model, combinations of D665 and G3c treatments induced permanent allograft acceptance in fully MHC-mismatched mice in a Tr1 cell–dependent manner rather than a Treg cell–dependent manner.
CD28 superagonist bivalently binds to the laterally exposed C″D loop of the extracellular Ig-like domains of the CD28 homodimer and forms a stable lattice on the T cell membrane, which provides strong activating signals and leads to potent polyclonal T cell expansion (4, 5, 37). The CD28 superagonist D665 has been shown to preferentially expand Treg cells over Teff cells in various rodent models for Treg-based interference with autoimmune and inflammatory disease (22–25). We also observed robust Treg cell expansion consistent with previously published data (Fig. 1). However, use of D665 alone resulted in a prolonged cardiac allograft survival rather than permanent allograft acceptance in the BALB/c to B6/J strain combination with strong rejection responses (Fig. 6). Following D665 treatment, the GITR expression was strongly up-regulated on both Treg and Teff cells. GITR signaling is complicated, and its functions are cell specific and context dependent (21). The conventional anti-GITR antibody DTA-1 has been reported to activate Teff lymphocytes while depleting GITR-expressing Treg cells, thereby promoting antitumor immune responses. G3c, another agonist anti-GITR antibody, showed a stronger costimulatory activity than DTA-1 for both Teff and Treg cells but failed to remove Treg cells in vivo and cure tumor-bearing mice (38). The application of G3c targeting GITR following D665 generated large amounts of Tr1 cells. G3c played an important role in the induction of Tr1 cells based on D665 treatment.
The strong synthesis of IL-10 is a hallmark of Tr1 cells. Over the past few years, many studies have explored the mechanism underlying the IL-10 expression in Tr1 cells. While the molecular mechanisms underlying the development of Tr1 cells are still unclear, a plethora of TFs have been identified as being involved in IL-10 production (2). Notably, several TFs have been identified that contribute to the development of Tr1 cells activated by IL-27 signaling (35). Prdm1 and Maf were identified as two central regulators that cooperatively drive the expression of IL-10 expression and Tr1 signature genes induced by IL-27 (36). IL-27 induced potent Egr2 expression via STAT3 signaling, which is required for IL-27–induced Prdm1-mediated IL-10 production in CD4+ T cells (32). Furthermore, Ahr and Maf, which are located downstream of IL-27 signaling, collaboratively promoted the development of Tr1 cells (39). In addition, Eomes and Prdm1 cooperated to generate Tr1 cells and mediated the expression of granzyme B, which is another feature of Tr1 cells (40). The induction of Eomes in Tr1 cells requires Tbx21 and IL-27 signaling. Recently, Eomes has been reported as a lineage-defining TF of the unique granzyme K+ Tr1 cells (41). In the present study, Prdm1, Maf, Tbx21, and Eomes were up-regulated in Tr1 cells (Fig. 4), accompanied by strong phosphorylation of STAT3 (Fig. 5), whereas Egr2 and Ahr were down-regulated in Tr1 cells (Fig. 4). Of these, Egr2 was reported to be essential for IL-27–induced Blimp-1–dependent IL-10 induction. A slightly elevated IL-27 expression was only observed in the combination treatment group on day 7, whereas a considerable amount of Tr1 cells had been generated on day 3. We suspect that the cell-intrinsic function of GITR signaling may be responsible for the development of Tr1 cells. GITR induced activation of ERK via TNFR-associated factor 5, which is an adaptor protein and signal transducer of TNFRs (42). The ERK/MAPK signaling pathways were able to induce IL-10 production through STAT3 activation (43). We therefore proposed that GITR signaling might contribute to Tr1 cell generation via MAPK-STAT3 signaling (Fig. 7). The high expression of Tbx21, the lineage-defining TF of T helper type 1 (TH1) cells, and IFN-γ, the signature TH1 cytokine, suggested that TH1 cells might be the origin of Tr1 cells. Switching of IFN-γ–secreting TH1 cells into IL-10/IFN-γ–co-producing Tr1 cells manifest the self-regulation of TH1 immune response, which represent a mechanism of Tr1 generation (44).
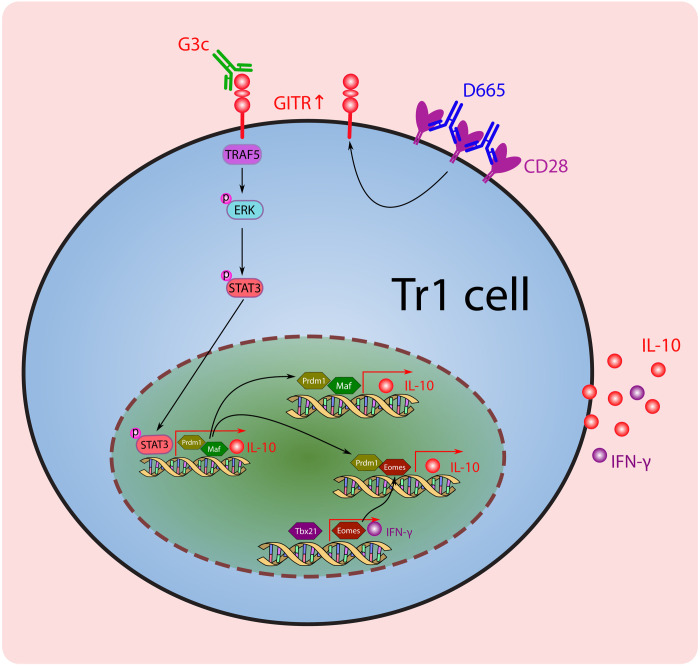
Fig. 7. The proposed cellular and transcriptional regulation of Tr1 cell development in D665-G3c–treated mice.
The CD28 superagonist D665 induced up-regulation of GITR along with activation and expansion of TH1 cells. The application of G3c targeting GITR contributes the conversion of TH1 cells into IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 cells via the activation of MAPK-STAT3 signaling. TF Blimp1, c-Maf, Eomes, and Tbx21 are responsible for IL-10/IFN-γ production in D665-G3c–induced Tr1 cells. TRAF5, TNFR-associated factor 5.
IL-10 is an important pleiotropic cytokine with broad immunomodulatory functions. Both immunosuppressive and immunostimulatory effects of IL-10 have been reported, the discrepancy in which may be due to differences in diseases, microenvironments, cell status, or other factors in these studies (45). In general, the potent anti-inflammatory and immunosuppressive effects represent the primary roles of IL-10 in immune regulation (46). One key function of Tr1 cells is to inhibit the DC maturation and function, which is critical for the initiation and determining the magnitude of an immune response (47). Coculture of BMDCs with Tr1 cells down-regulated the expression of MHC-II and costimulatory molecules, thereby reducing the magnitude of adaptive immune responses. Tr1 cells also inhibit the DC function by directly killing DCs via granzyme B and perforin secretion (48). Tr1 cells can regulate adaptive immune responses by directly suppressing T cells. We showed in the present study that Tr1 cells suppressed alloantigen-driven T cell proliferation in an IL-10–dependent manner (Fig. 3). Furthermore, Tr1 can also suppress T cells via cell contact–dependent mechanisms mediated by CTLA-4 or PD-1 and CD39-mediated metabolic disruption (49, 50).
The therapeutic effects of Tr1 cells have been investigated in many preclinical models of immune-mediated diseases, including autoimmune diseases, allergic diseases, GvHD, and transplantation (40, 51–54). Tr1 cells, via de novo induction or adoptive transfer, promote pancreatic islet graft tolerance in mouse transplantation models (51, 52). An increased number of circulating Tr1 cells was associated with a stable graft function and operational tolerance in renal transplant patients (55). Several good manufacturing practices and clinical-grade compatible protocols have been established to generate human antigen-specific Tr1 cells in vitro (56, 57). We demonstrated that D665-G3c–induced Tr1 cells mediated permanent cardiac allograft acceptance. D665-G3c induced both Treg and Tr1 cells, but the Treg cells were only transiently elevated, and the heart transplantation tolerance was dependent on Tr1 rather than Treg cells (Fig. 6). Further translational studies concerning donor specificity and the safety of Tr1 cells generated by the D665-G3c induction strategy are needed.
In the present study, we found that the combination of D665 and G3c treatment generated large amounts of Tr1 cells, resulting in permanent cardiac allograft acceptance. We proposed that D665-G3c induced Tr1 cells via TFs Prdm1 and Maf. G3c contributed to Tr1 cell generation via the activation of MAPK-STAT3 signaling. Tr1 cells suppressed both innate and adaptive immunity, thus causing permanent cardiac allograft acceptance in an IL-10–dependent manner. Further studies should focus on the associated molecular mechanisms and clinical translation of Tr1 cells. We believe that the in vivo pharmacological induction of Tr1 cells opens up possibilities for transplantation tolerance and can be successfully translated to the clinical setting.
MATERIALS AND METHODS
Animals
Specific pathogen–free inbred male C57BL/6J (B6/J; H-2kb) and BALB/c mice (H-2kd), 8 to 12 weeks old, were purchased from Japan SLC Inc. (Shizuoka, Japan). Foxp3-GFP mice were purchased from the Jackson Laboratory (Bar Harbor, ME). IL-10–Venus mice were provided by K. Takeda (Graduate School of Medicine, Osaka University, Osaka, Japan) (58), and IL-10−/− mice were provided by T. Yoshimoto (Tokyo Medical University, Tokyo, Japan). All mice received humane care in accordance with the guidelines of the Animal Use and Care Committee of the National Research Institute for Child Health and Development, Tokyo, Japan (permission number: A2008-004-C10). All mouse experiments conformed to the National Institutes of Health guidelines for the care and use of laboratory animals.
Heterotopic heart transplantation
Mouse heterotopic heart transplantation was performed as previously described (59). Fully vascularized hearts from donor BALB/c mice were heterotopically transplanted to the abdomen of recipient B6/J mice. The graft survival was monitored by daily abdominal palpation of the heart impulse. Total cessation of heartbeat was considered to indicate heart allograft rejection and confirmed by direct laparotomy. D665 and G3c antibodies were purified from supernatant of hybridomas, gifts from T. Hunig (University of Würzburg, Würzburg, Germany) and J. Shimizu (Kyoto University, Kyoto, Japan), respectively. For IL-10 neutralization, mice were intraperitoneally injected with 250 μg of anti–IL-10 (Bio X Cell, Lebanon, NH, catalog no. BE0049) on pre- and posttransplantation days −1, 1, 3, 6, 9, and 12. For Treg cell depletion, mice were intraperitoneally injected with 250 μg of anti-CD25 (Bio X Cell, catalog no. BE0012) on pre- and posttransplantation days −1, 3, and 7, respectively. For the in situ immunoproliferative response analysis, recipient mice received a single intravenous injection of 5-bromo-2′-deoxyuridine (BrdU) (0.6 mg per mouse; Sigma-Aldrich, St. Louis, MO) 1 hour before sampling.
Cell preparation and cell sorting
Total CD4+ and CD8+ T cells were purified from the mouse splenocytes using magnetic-activated cell sorting (MACS) mouse CD4+ (Miltenyi Biotec, Bergisch Gladbach, Germany, catalog no. 130-104-454) and CD8+ (Miltenyi Biotec, catalog no. 130-095-236) T cell isolation kits according to the manufacturer’s instructions. For CD4+IL-10+ cells and CD4+IL-10− cell preparation, CD4+ T cells were isolated from D665- and G3c-treated IL-10–Venus mice on day 7 using MACS and then subjected to cell sorting for CD4+Venus+ and CD4+Venus− populations using a FACSAria (BD Biosciences, Franklin Lakes, NJ). Because most CD4+Venus+ T cells (>95%) were IL-10/IFN-γ–co-producing CD4+Foxp3− Tr1 cells (Fig. 1E and fig. S1A), the sorted CD4+Venus+ T cells were considered Tr1 cells.
BMDC culture
Bone marrow cells were obtained from the femur and tibia of male BALB/c mice, and then, erythrocytes were removed by lysis. To generate BMDCs, bone marrow cells (1 × 106 per well) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) (Thermo Fisher Scientific, Waltham, MA), granulocyte macrophage colony-stimulating factor (10 ng/ml; PeproTech, Cranbury, NJ), IL-4 (10 ng/ml; PeproTech), and 50 μM β-mercaptoethanol (Wako, Osaka, Japan) in 24-well tissue culture plates. On day 2, floating granulocyte clusters were drained away, and half of the medium was refreshed. On day 5, for maturation, cells were collected and reseeded (1 × 105 per well) in a 96-well flat-bottom plate with 100 μl per well of fresh medium in the presence of LPS (10 ng/ml; Sigma-Aldrich) and stimulated for 2 days. Different-sorted CD4+ T cells (1 × 105 per well) were also added to the medium to test their effect on BMDC maturation. On day 7, different groups of BMDCs were harvested for subsequent analyses.
Mixed lymphocyte reaction
In the one-way MLR, MACS-isolated CD8+ T cells from B6/J mouse splenocytes were labeled with CellTrace CFSE (Thermo Fisher Scientific) as responders. BALB/c-derived BMDCs were irradiated with 20-Gy x-ray and used as stimulators. Naïve CD4+ T cells, CD4+IL-10+ cells, and CD4+IL-10− cells were seeded to serve as regulators. Stimulator BMDCs (1 × 104 per well), responder CD8+ T cells (1 × 105 per well), and regulators CD4+ T cells (1 × 105 per well) were cocultured in a 96-well U-bottom plate with 100 μl per well of complete RPMI 1640 medium at 37°C for 3 days. For neutralization of secreted cytokines, anti–IL-10 (20 ng/ml; Bio X Cell, catalog no. BE0049) mAb was added at the start of the culture. At the end of the assay, cells were collected for measurement of CFSE dilution by FCM.
Isolation of mouse cardiac GILs
Cardiac grafts were harvested on POD7 and cut into 1- to 2-mm pieces on ice. The tissue was then mechanically disrupted and digested at 37°C for 20 min in 10 ml of digestion solution, which included collagenase IV (0.5 mg/ml; Sigma-Aldrich) and deoxyribonuclease I (50 U/ml; Thermo Fisher Scientific) in phosphate-buffered saline (PBS). Subsequently, 10 ml of iced RPMI 1640 with 5% FCS was added to stop digestion. The digested material was filtered through a nylon mesh (100 μm) to remove aggregates and centrifuged at 200g for 10 min to pellet the cells. The pellet was then suspended in 5 ml of PBS, loaded onto 5 ml of Lympholyte-M (Cedarlane, Ontario, Canada), and centrifuged at 1500g for 25 min at room temperature. GILs were collected from the Lympholyte-M interface and washed twice in PBS before staining.
Flow cytometry
Cells were stained with LIVE/DEAD staining (Thermo Fisher Scientific) for labeling dead cells and blocked with anti-CD16/CD32 (BioLegend, San Diego, CA; catalog no. 101302) Fc Block antibody to prevent nonspecific antibody binding. For cell surface staining, the cells were incubated with different combinations of fluorochrome-conjugated antibodies against mouse CD45 (BioLegend, catalog no. 103136), CD3 (BioLegend, catalog no. 100328), CD4 (BioLegend, catalog no. 100526), CD8α (BioLegend, catalog no. 100730), CD11b (BioLegend, catalog no. 101216), CD11c (BioLegend, catalog no. 117310), CD40 (BioLegend, catalog no. 124610), CD80 (BioLegend, catalog no. 104722), CD86 (BioLegend, catalog no. 105030), MHC-II (BioLegend, catalog no. 107606), CD44 (BioLegend, catalog no. 103012), CD62L (BioLegend, catalog no. 104408), GITR (BioLegend, catalog no. 126310), CD39 (BioLegend, catalog no. 143804), CD25 (BioLegend, catalog no. 102012), ICOS (BioLegend, catalog no. 313508), TIGIT (BioLegend, catalog no. 142104), CD49b (BioLegend, catalog no. 103515), PD-1 (BioLegend, catalog no. 109104), TIM-3 (BioLegend, catalog no. 119718), Neuropilin-1 (BioLegend, catalog no. 145212), and CD69 (BioLegend, catalog no. 104512). For intracellular staining, cells were fixed and permeabilized using the Transcription Factor Staining Buffer Set (eBioscience, San Diego, CA) according to the manufacturer’s instructions and then stained with fluorochrome-conjugated antibodies against the following: IL-10 (BioLegend, catalog no. 505008), IFN-γ (BioLegend, catalog no. 505826), CTLA-4 (BioLegend, catalog no. 106306), and Foxp3 (eBioscience, catalog no. 11-5773-82). For intracellular cytokine staining, cells were stimulated with phorbol 12-myristate 13-acetate (50 ng/ml; BD Golgi Plug), 1 mM ionomycin (Sigma-Aldrich), and brefeldin A (eBioscience) in complete medium for 4 hours, followed by surface and intracellular staining. The isotype- and fluorochrome-matched IgG was used as a negative control. Flow data were acquired with an LSRFortessa Cell Analyzer (BD Biosciences) and analyzed using the FlowJo v10 software program (BD Biosciences).
RNA-seq analyses
CD4+IL-10+ and CD4+IL-10− T cell samples were prepared in three duplicates. Total RNA extraction and DNA removal were performed as described above. These RNA samples were then subjected to preprocessing, DNA nanoball-based library construction, and high-throughput sequencing by BGI Japan (Kobe, Japan) on the DNBseq sequencing platform. The raw sequencing reads were checked for their quality with FastQC (v0.11.8), trimmed with TrimGalore (v0.5.0), and aligned to the GRCm38 reference genome using the RNA-seq aligner HISAT2 (v2.1.0). Read counts for each gene were calculated with Feature Counts. DEGs were identified by DESeq2 (v1.32.0) with the following criteria: absolute log2FC of ≥0.5 and adjusted P value of ≤0.05. Variance stabilization–transformed expression data were Z-transferred and used to generate a heatmap by pheatmap (v1.0.12). A GO enrichment analysis was performed using ClusterProfiler (v4.0.5). The top 10 enriched clusters were visualized with GOplot (v1.0.2).
RNA purification and qRT-PCR
Total RNA from heart grafts was extracted using reagent Sepasol-RNA I Super G (Nacalai Tesque, Kyoto, Japan). Total RNA from the cell suspension was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. After treating samples with a DNA-free kit (Ambion, Life Technologies, Carlsbad, CA), total RNA was reverse-transcribed to generate complementary DNA using a PrimeScript RT Reagent Kit (Takara Bio, Shiga, Japan). qRT-PCR was performed in an Applied Biosystem PRISM 7700 instrument (Applied Biosystems, Foster City, CA) using the SYBR Green system. The threshold cycle (Ct) values of the target genes were normalized to the Ct value of 18S ribosomal RNA. The relative gene expression was calculated by the ΔΔCt calculation method. The sequences of 18S and target gene primers used in this research are shown in Table 1.
Table 1. Primer sequences for qRT-PCR.
| Gene | Forward primer (5′ to 3′) | Reverse primer (3′ to 5′) |
| Prdm1 | CCCTCATCGGTGAAGTCTA | ACGTAGCGCATCCAGTTG |
| Maf | GCAGAGACACGTCCTGGAGTCG | CGAGCTTGGCCCTGCAACTAGC |
| Eomes | GCGCATGTTTCCTTTCTTGAG | GGTCGGCCAGAACCACTTC |
| Ahr | AGCCGGTGCAGAAAACAGTAA | AGGCGGTCTAACTCTGTGTTC |
| Egr2 | GCCAAGGCCGTAGACAAAATC | CCACTCCGTTCATCTGGTCA |
| Tbx21 | AGCAAGGACGGCGAATGTT | GGGTGGACATATAAGCGGTTC |
| Irf4 | TCCGACAGTGGTTGATCGAC | CCTCACGATTGTAGTCCTGCTT |
| Il-27p28 | CCTGACATGGGCCAGGTGACAGGAGACC | TCACTCGAGTTAGGAATCCCAGGCTGAG |
| Ebi3 | CTTACAGGCTCGGTGTGGC | GTGACATTTAGCATGTAGGGCA |
| IL-10 | GCTCTTACTGACTGGCATGAG | CGCAGCTCTAGGAGCATGTG |
| Ifng | AAGCGTCATTGAATCACACCTGA | ACCTGTGGGTTGTTGACCTCAA |
| 18S | ACATCGACCTCACCAAGAGG | TCCCATCCTTCACATCCTTC |
Western blotting
Total cell protein was extracted with radioimmunoprecipitation assay lysis buffer (Wako) containing 1% protease inhibitor cocktail, 1% phosphatase inhibitor cocktail 1, and 1% phosphatase inhibitor cocktail 2 (Sigma-Aldrich), measured concentrations with the BCA Protein Assay (Thermo Fisher Scientific). Twenty micrograms of protein was resolved on 10% SDS–polyacrylamide gel electrophoresis gels and transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). After blocking, the membranes were incubated with primary antibodies containing phosphorylated STAT3 (p-STAT3) (Cell Signaling Technology, Danvers, MA, catalog no. 9131), STAT3 (Cell Signaling Technology, catalog no. 4904), p-ERK (Cell Signaling Technology, catalog no. 4370), ERK (Cell Signaling Technology, catalog no. 4695), Blimp1 (Cell Signaling Technology, catalog no. 9115), c-Maf (Bethyl Laboratories, Montgomery, TX; catalog no. A300-613A-M), and β-actin (Cell Signaling Technology, catalog no. 4970) overnight at 4°C, followed by horseradish peroxidase–linked anti-rabbit IgG secondary antibody (Cell Signaling Technology, catalog no. 7074) probing for 1 hour at room temperature. The chemiluminescence signal intensity of protein bands were detected with enhanced chemiluminescence (GE Healthcare, Piscataway, NJ) and the ImageQuant LAS4000 System (GE Healthcare). The protein expression was quantitated with the ImageJ software program (National Institutes of Health, Bethesda, MD).
Histological analyses
Heart grafts were fixed in 10% neutral buffered formalin (Wako), dehydrated, and then embedded in paraffin. Paraffin sections (4 μm thick) were stained with hematoxylin and eosin stain. Heart allograft rejection was graded by two investigators blinded to the group and labeling allocation during the experiment, according to the International Society for Heart and Lung Transplantation criteria (60).
Immunohistochemistry
Heart grafts were frozen in Tissue-Tek optimal cutting temperature compound (Sakura Finetek, Torrance, CA) and then cut into 4-μm-thick cryosections. Cryosections were rehydrated, fixed with formaldehyde calcium solution, and then blocked using block ace for 10 min. For CD8α T cell staining, the sections were immunostained with CD8α (BioLegend, catalog no. 100777) as the primary antibody and then incubated with alkaline phosphatase (ALP)-conjugated donkey anti-rat IgG (Jackson ImmunoResearch, West Grove, PA; catalog no. 712-055-153) as the secondary antibody. The positive antigens were visualized with the Vector Blue Alkaline Phosphatase Substrate Kit (Vector Laboratories, Burlingame, CA) according to the manufacturer’s instructions. For type IV collagen staining, the sections were immunostained with rabbit–anti-mouse type IV collagen polyclonal antibody (Cosmo Bio, Tokyo, Japan, catalog no. LSL-LB-1403) and then incubated with POD-conjugated goat–anti-rabbit Ig (Jackson ImmunoResearch, catalog no. 111-036-144) as the secondary antibody. The positive antigens were visualized with diaminobenzidine (DAB) (Dojindo, Kumamoto, Japan) substrate reaction. For BrdU staining, the sections were digested with a pepsin (Sigma-Aldrich) solution, immunostained with anti-BrdU (Bio-Rad Laboratories, catalog no. OBT0030CX) as the primary antibody, and then incubated with ALP-conjugated donkey anti-rat IgG (Jackson ImmunoResearch, catalog no. 712-055-153) as the secondary antibody. The positive antigens were visualized with New Fuchsin (Dako, Santa Clara, CA) in a substrate reaction. Last, the sections were fixed in formaldehyde calcium solution and mounted with Aquatex (Merck, Whitehouse Station, NJ).
All of the sections were imaged with a DP70 camera (Olympus, Osaka, Japan). The images were processed and analyzed using the ImageJ software program. Positive cells were determined by counting nine random 400× high-power fields on each slide.
Statistical analyses
All data were analyzed using the GraphPad Prism software program (v7.0, GraphPad Software, San Diego, CA). The results were presented as the mean ± SEM. Student’s t test (normal distribution data) was used for comparisons between the two groups. A one-way analysis of variance (ANOVA), followed by a post hoc test was used for comparisons between multiple groups. A log-rank (Mantel-Cox) test was used for the survival data. In all experiments, differences were considered statistically significant at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We thank K. Takeda (Graduate School of Medicine, Osaka University, Osaka, Japan) for providing IL-10–Venus mice. We thank T. Yoshimoto (Tokyo Medical University, Tokyo, Japan) for providing IL-10−/− mice. We also thank T. Hunig (University of Würzburg, Würzburg, Germany) and J. Shimizu (Kyoto University, Kyoto, Japan) for providing D665- and G3c-producing hybridomas, respectively.
Funding: This study was supported, in part, by research grants from the National Center for Child Health and Development (30-20 and 2021B-18) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (21 K08634 and 17H04277).
Author contributions: Conceptualization: W.Q. and X.-K.L. Methodology: W.Q., K.M. and X.H. Investigation: W.Q., K.M., X.H., and W.-Z.G. Visualization: X.H. and W.-Z.G. Supervision: X.-K.L. Writing—original draft: W.Q. and X.-K.L. Writing—review and editing: W.Q. and X.-K.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.Leventhal J. R., Mathew J. M., Outstanding questions in transplantation: Tolerance. Am. J. Transplant 20, 348–354 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Roncarolo M. G., Gregori S., Bacchetta R., Battaglia M., Gagliani N., The biology of T regulatory type 1 cells and their therapeutic application in immune-mediated diseases. Immunity 49, 1004–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Song Y., Wang N., Chen L., Fang L., Tr1 cells as a key regulator for maintaining immune homeostasis in transplantation. Front. Immunol. 12, 671579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lühder F., Huang Y., Dennehy K. M., Guntermann C., Müller I., Winkler E., Kerkau T., Ikemizu S., Davis S. J., Hanke T., Hünig T., Topological requirements and signaling properties of T cell–Activating, anti-CD28 antibody superagonists. J. Exp. Med. 197, 955–966 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans E. J., Esnouf R. M., Manso-Sancho R., Gilbert R. J. C., James J. R., Yu C., Fennelly J. A., Vowles C., Hanke T., Walse B., Hünig T., Sørensen P., Stuart D. I., Davis S. J., Crystal structure of a soluble CD28-Fab complex. Nat. Immunol. 6, 271–279 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Beyersdorf N., Hanke T., Kerkau T., Hünig T., Superagonistic anti-CD28 antibodies: Potent activators of regulatory T cells for the therapy of autoimmune diseases. Ann. Rheum. Dis. 64, iv91–iv95 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyersdorf N., Werner S., Wolf N., Hünig T., Kerkau T., In vitro polyclonal activation of conventional T cells with a CD28 superagonist protects mice from acute graft versus host disease. Eur. J. Immunol. 45, 1997–2007 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Kitazawa Y., Fujino M., Li X.-K., Xie L., Ichimaru N., Okumi M., Nonomura N., Tsujimura A., Isaka Y., Kimura H., Hünig T., Takahara S., Superagonist CD28 antibody preferentially expanded Foxp3-expressing nTreg cells and prevented graft-versus-host diseases. Cell Transplant 18, 627–638 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Nocentini G., Giunchi L., Ronchetti S., Krausz L. T., Bartoli A., Moraca R., Migliorati G., Riccardi C., A new member of the tumor necrosis factor/nerve growth factor receptor family inhibits T cell receptor-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 94, 6216–6221 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McHugh R. S., Whitters M. J., Piccirillo C. A., Young D. A., Shevach E. M., Collins M., Byrne M. C., CD4+CD25+ immunoregulatory T cells: Gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity 16, 311–323 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Shimizu J., Yamazaki S., Takahashi T., Ishida Y., Sakaguchi S., Stimulation of CD25+CD4+ regulatory T cells through GITR breaks immunological self-tolerance. Nat. Immunol. 3, 135–142 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Kanamaru F., Youngnak P., Hashiguchi M., Nishioka T., Takahashi T., Sakaguchi S., Ishikawa I., Azuma M., Costimulation via glucocorticoid-induced TNF receptor in both conventional and CD25+ regulatory CD4+ T cells. J. Immunol. 172, 7306–7314 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Tone M., Tone Y., Adams E., Yates S. F., Frewin M. R., Cobbold S. P., Waldmann H., Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells. Proc. Natl. Acad. Sci. U.S.A. 100, 15059–15064 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens G. L., McHugh R. S., Whitters M. J., Young D. A., Luxenberg D., Carreno B. M., Collins M., Shevach E. M., Engagement of glucocorticoid-induced TNFR family-related receptor on effector T cells by its ligand mediates resistance to suppression by CD4+CD25+ T cells. J. Immunol. 173, 5008–5020 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Kim J. I., Sonawane S. B., Lee M. K., Lee S.-H., Duff P. E., Moore D. J., O’Connor M. R., Lian M.-M., Deng S., Choi Y., Yeh H., Caton A. J., Markmann J. F., Blockade of GITR–GITRL interaction maintains Treg function to prolong allograft survival. Eur. J. Immunol. 40, 1369–1374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushell A., Wood K., GITR ligation blocks allograft protection by induced CD25+CD4+ regulatory T cells without enhancing effector T-cell function. Am. J. Transplant. 7, 759–768 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Sukumar S., Wilson D. C., Yu Y., Wong J., Naravula S., Ermakov G., Riener R., Bhagwat B., Necheva A. S., Grein J., Churakova T., Mangadu R., Georgiev P., Manfra D., Pinheiro E. M., Sriram V., Bailey W. J., Herzyk D., McClanahan T. K., Willingham A., Beebe A. M., Sadekova S., Characterization of MK-4166, a clinical agonistic antibody that targets human GITR and inhibits the generation and suppressive effects of T regulatory cells. Cancer Res. 77, 4378–4388 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Petrillo M. G., Ronchetti S., Ricci E., Alunno A., Gerli R., Nocentini G., Riccardi C., GITR+ regulatory T cells in the treatment of autoimmune diseases. Autoimmun. Rev. 14, 117–126 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Mitsui J., Nishikawa H., Muraoka D., Wang L., Noguchi T., Sato E., Kondo S., Allison J. P., Sakaguchi S., Old L. J., Kato T., Shiku H., Two distinct mechanisms of augmented antitumor activity by modulation of immunostimulatory/inhibitory signals. Clin. Cancer Res. 16, 2781–2791 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Ephrem A., Epstein A. L., Stephens G. L., Thornton A. M., Glass D., Shevach E. M., Modulation of Treg cells/T effector function by GITR signaling is context-dependent. Eur. J. Immunol. 43, 2421–2429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clouthier D. L., Watts T. H., Cell-specific and context-dependent effects of GITR in cancer, autoimmunity, and infection. Cytokine Growth Factor Rev. 25, 91–106 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Chen J., Xie L., Toyama S., Hünig T., Takahara S., Li X.-K., Zhong L., The effects of Foxp3-expressing regulatory T cells expanded with CD28 superagonist antibody in DSS-induced mice colitis. Int. Immunopharmacol. 11, 610–617 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Wagner J. C., Leicht S., Hofmann M., Seifert F., Gahn S., Germer C.-T., Beyersdorf N., Otto C., Klein I., CD28 superagonist D665-mediated activation of mouse regulatory T cells maintains their phenotype without loss of suppressive quality. Immunobiology 226, 152144 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Copsel S., Wolf D., Komanduri K. V., Levy R. B., The promise of CD4+FoxP3+ regulatory T-cell manipulation in vivo: Applications for allogeneic hematopoietic stem cell transplantation. Haematologica 104, 1309–1321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilliams M., Bosschaerts T., Hérin M., Hünig T., Loi P., Flamand V., De Baetselier P., Beschin A., Experimental expansion of the regulatory T cell population increases resistance to african trypanosomiasis. J. Infect Dis. 198, 781–791 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Gagliani N., Magnani C. F., Huber S., Gianolini M. E., Pala M., Licona-Limon P., Guo B., Herbert D. R., Bulfone A., Trentini F., Di Serio C., Bacchetta R., Andreani M., Brockmann L., Gregori S., Flavell R. A., Roncarolo M.-G., Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat. Med. 19, 739–746 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Haase C., Jørgensen T. N., Michelsen B. K., Both exogenous and endogenous interleukin-10 affects the maturation of bone-marrow-derived dendritic cells in vitro and strongly influences T-cell priming in vivo. Immunology 107, 489–499 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corinti S., Albanesi C., la Sala A., Pastore S., Girolomoni G., Regulatory activity of autocrine IL-10 on dendritic cell functions. J. Immunol. 166, 4312–4318 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Smith L. K., Boukhaled G. M., Condotta S. A., Mazouz S., Guthmiller J. J., Vijay R., Butler N. S., Bruneau J., Shoukry N. H., Krawczyk C. M., Richer M. J., Interleukin-10 directly inhibits CD8+ T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groux H., Bigler M., de Vries J. E., Roncarolo M.-G., Inhibitory and stimulatory effects of IL-10 on human CD8+ T cells. J. Immunol. 160, 3188–3193 (1998). [PubMed] [Google Scholar]
- 31.Schmetterer K. G., Pickl W. F., The IL–10/STAT3 axis: Contributions to immune tolerance by thymus and peripherally derived regulatory T-cells. Eur. J. Immunol. 47, 1256–1265 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki Y., Fujio K., Okamura T., Yanai A., Sumitomo S., Shoda H., Tamura T., Yoshida H., Charnay P., Yamamoto K., Egr-2 transcription factor is required for Blimp-1-mediated IL-10 production in IL-27-stimulated CD4+ T cells. Eur. J. Immunol. 43, 1063–1073 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Yang Y., Ochando J., Yopp A., Bromberg J. S., Ding Y., IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J. Immunol. 174, 2720–2729 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Farez M. F., Mascanfroni I. D., Méndez-Huergo S. P., Yeste A., Murugaiyan G., Garo L. P., B. Aguirre M. E., Patel B., Ysrraelit M. C., Zhu C., Kuchroo V. K., Rabinovich G. A., Quintana F. J., Correale J., Melatonin contributes to the seasonality of multiple sclerosis relapses. Cell 162, 1338–1352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasanthakumar A., Kallies A., IL-27 paves different roads to Tr1. Eur. J. Immunol. 43, 882–885 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Zhang H., Madi A., Yosef N., Chihara N., Awasthi A., Pot C., Lambden C., Srivastava A., Burkett P. R., Nyman J., Christian E., Etminan Y., Lee A., Stroh H., Xia J., Karwacz K., Thakore P. I., Acharya N., Schnell A., Wang C., Apetoh L., Rozenblatt-Rosen O., Anderson A. C., Regev A., Kuchroo V. K., An IL-27-driven transcriptional network identifies regulators of IL-10 expression across T helper cell subsets. Cell Rep. 33, 108433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hünig T., Dennehy K., CD28 superagonists: Mode of action and therapeutic potential. Immunol. Lett. 100, 21–28 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Nishioka T., Nishida E., Iida R., Morita A., Shimizu J., In vivo expansion of CD4+Foxp3+ regulatory T cells mediated by GITR molecules. Immunol. Lett. 121, 97–104 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Apetoh L., Quintana F. J., Pot C., Joller N., Xiao S., Kumar D., Burns E. J., Sherr D. H., Weiner H. L., Kuchroo V. K., The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11, 854–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P., Lee J. S., Gartlan K. H., Schuster I. S., Comerford I., Varelias A., Ullah M. A., Vuckovic S., Koyama M., Kuns R. D., Locke K. R., Beckett K. J., Olver S. D., Samson L. D., de Oca M. M., de L. Rivera F., Clouston A. D., Belz G. T., Blazar B. R., MacDonald K. P., McColl S. R., Thomas R., Engwerda C. R., Degli-Esposti M. A., Kallies A., Tey S.-K., Hill G. R., Eomesodermin promotes the development of type 1 regulatory T (TR1) cells. Sci. Immunol. 2, eaah7152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruarin P., Maglie S., De Simone M., Häringer B., Vasco C., Ranzani V., Bosotti R., Noddings J. S., Larghi P., Facciotti F., Sarnicola M. L., Martinovic M., Crosti M., Moro M., Rossi R. L., Bernardo M. E., Caprioli F., Locatelli F., Rossetti G., Abrignani S., Pagani M., Geginat J., Eomesodermin controls a unique differentiation program in human IL-10 and IFN-γ coproducing regulatory T cells. Eur. J. Immunol. 49, 96–111 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Esparza E. M., Lindsten T., Stockhausen J. M., Arch R. H., Tumor necrosis factor receptor (TNFR)-associated factor 5 is a critical intermediate of costimulatory signaling pathways triggered by glucocorticoid-induced TNFR in T cells. J. Biol. Chem. 281, 8559–8564 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Lucas M., Zhang X., Prasanna V., Mosser D. M., ERK activation following macrophage FcγR ligation leads to chromatin modifications at the IL-10 locus. J. Immunol. 175, 469–477 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Cope A., Friec G. L., Cardone J., Kemper C., The Th1 life cycle: Molecular control of IFN-γ to IL-10 switching. Trends Immunol. 32, 278–286 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ouyang W., O’Garra A., IL-10 family cytokines IL-10 and IL-22: From basic science to clinical translation. Immunity 50, 871–891 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Saraiva M., Vieira P., O’Garra A., Biology and therapeutic potential of interleukin-10. J. Exp. Med. 217, e20190418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Smedt T., Van Mechelen M., De Becker G., Urbain J., Leo O., Moser M., Effect of interleukin-10 on dendritic cell maturation and function. Eur. J. Immunol. 27, 1229–1235 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Magnani C. F., Alberigo G., Bacchetta R., Serafini G., Andreani M., Roncarolo M. G., Gregori S., Killing of myeloid APCs via HLA class I, CD2 and CD226 defines a novel mechanism of suppression by human Tr1 cells. Eur. J. Immunol. 41, 1652–1662 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akdis M., Verhagen J., Taylor A., Karamloo F., Karagiannidis C., Crameri R., Thunberg S., Deniz G., Valenta R., Fiebig H., Kegel C., Disch R., Schmidt-Weber C. B., Blaser K., Akdis C. A., Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. J. Exp. Med. 199, 1567–1575 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mascanfroni I. D., Takenaka M. C., Yeste A., Patel B., Wu Y., Kenison J. E., Siddiqui S., Basso A. S., Otterbein L. E., Pardoll D. M., Pan F., Priel A., Clish C. B., Robson S. C., Quintana F. J., Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med. 21, 638–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jofra T., Di Fonte R., Galvani G., Kuka M., Iannacone M., Battaglia M., Fousteri G., Tr1 cell immunotherapy promotes transplant tolerance via de novo Tr1 cell induction in mice and is safe and effective during acute viral infection. Eur. J. Immunol. 48, 1389–1399 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Gagliani N., Jofra T., Stabilini A., Valle A., Atkinson M., Roncarolo M.-G., Battaglia M., Antigen-specific dependence of Tr1-cell therapy in preclinical models of islet transplant. Diabetes 59, 433–439 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H., Gagliani N., Ishigame H., Huber S., Zhu S., Esplugues E., Herold K. C., Wen L., Flavell R. A., Intestinal type 1 regulatory T cells migrate to periphery to suppress diabetogenic T cells and prevent diabetes development. Proc. Natl. Acad. Sci. U.S.A. 114, 10443–10448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuda M., Doi K., Tsutsumi T., Inaba M., Hamaguchi J., Terada T., Kawata R., Kitatani K., Nabe T., Adoptive transfer of type 1 regulatory T cells suppressed the development of airway hyperresponsiveness in ovalbumin-induced airway inflammation model mice. J. Pharmacol. Sci. 141, 139–145 (2019). [DOI] [PubMed] [Google Scholar]
- 55.van den Boogaardt D. E. M., van Miert P. P. M. C., de Vaal Y. J. H., de Fijter J. W., Claas F. H. J., Roelen D. L., The ratio of interferon-gamma and interleukin-10 producing donor-specific cells as an in vitro monitoring tool for renal transplant patients. Transplantation 82, 844–848 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Gregori S., Roncarolo M. G., Engineered T regulatory Type 1 cells for clinical application. Front. Immunol. 9, 233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mfarrej B., Tresoldi E., Stabilini A., Paganelli A., Caldara R., Secchi A., Battaglia M., Generation of donor-specific Tr1 cells to be used after kidney transplantation and definition of the timing of their in vivo infusion in the presence of immunosuppression. J. Transl. Med. 15, 40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., Taniguchi T., Takeda K., Hori S., Ivanov I. I., Umesaki Y., Itoh K., Honda K., Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Que W., Hu X., Fujino M., Terayama H., Sakabe K., Fukunishi N., Zhu P., Yi S.-Q., Yamada Y., Zhong L., Li X.-K., Prolonged cold ischemia time in mouse heart transplantation using supercooling preservation. Transplantation 104, 1879–1889 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Stewart S., Winters G. L., Fishbein M. C., Tazelaar H. D., Kobashigawa J., Abrams J., Andersen C. B., Angelini A., Berry G. J., Burke M. M., Demetris A. J., Hammond E., Itescu S., Marboe C. C., McManus B., Reed E. F., Reinsmoen N. L., Rodriguez E. R., Rose A. G., Rose M., Suciu-Focia N., Zeevi A., Billingham M. E., Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J. Heart Lung Transplant. 24, 1710–1720 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5