Abstract
The epithelial-to-mesenchymal transition (EMT) is frequently co-opted by cancer cells to enhance migratory and invasive cell traits. It is a key contributor to heterogeneity, chemoresistance, and metastasis in many carcinoma types, where the intermediate EMT state plays a critical tumor-initiating role. We isolate multiple distinct single-cell clones from the SUM149PT human breast cell line spanning the EMT spectrum having diverse migratory, tumor-initiating, and metastatic qualities, including three unique intermediates. Using a multiomics approach, we identify CBFβ as a key regulator of metastatic ability in the intermediate state. To quantify epithelial-mesenchymal heterogeneity within tumors, we develop an advanced multiplexed immunostaining approach using SUM149-derived orthotopic tumors and find that the EMT state and epithelial-mesenchymal heterogeneity are predictive of overall survival in a cohort of stage III breast cancer. Our model reveals previously unidentified insights into the complex EMT spectrum and its regulatory networks, as well as the contributions of epithelial-mesenchymal plasticity (EMP) in tumor heterogeneity in breast cancer.
Intermediate EMT states, regulated by CBFβ, drive phenotypic heterogeneity and plasticity in breast cancer.
INTRODUCTION
The epithelial-to-mesenchymal transition (EMT) is a developmental cellular program frequently co-opted by cancer cells (1) and is a key contributor to intratumoral heterogeneity (2–4), chemoresistance, and metastasis (5, 6). Rather than being a switch from an epithelial to a mesenchymal state, increasing evidence points to the existence of intermediate EMT states, wherein cells coexpress both epithelial and mesenchymal traits (7–13). These robust stable and metastable transition states have unique characteristics (7, 8, 14, 15) and contribute to the complex heterogeneity of tumors and their overall metastatic behavior (16, 17). While much work has been carried out identifying and characterizing EMT-inducing transcription factors (18–21), the transcriptional and epigenetic networks responsible for the stability and maintenance of the midpoints along the EMT spectrum are poorly defined. In addition, there are currently no approaches to identifying and quantifying intermediate EMT subpopulations within patient tumors to evaluate their prognostic significance. Using single-cell clonally isolated derivatives of the SUM149PT breast cancer cell line, we systematically interrogate how each EMT state independently contributes to heterogeneity and influences metastatic progression, uncovering the role of CBFβ in stabilizing and maintaining metastatic capability in certain intermediate states. We develop an entropy-based model to quantify phenotypic heterogeneity and EMT status and primary patient tumors using SUM149PT-derived tumors stained with a panel of six EMT markers as a training set. The cell states captured in the SUM149PT model are represented in a cohort of patient tumors and are predictive of overall survival in these patients, laying the foundation for the quantification of epithelial-mesenchymal heterogeneity (EMH) and understanding the role of the intermediate EMT state in tumor progression.
RESULTS
Generation of a model to study EMT
Single-cell clones reside in multiple distinct EMT states
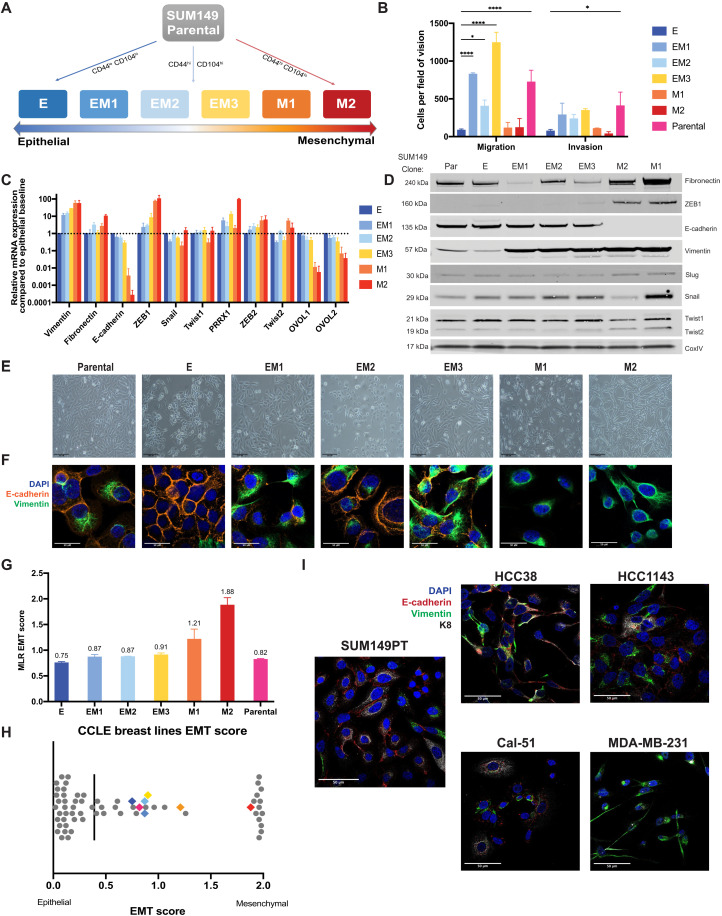
We derived single-cell clones from the SUM149PT estrogen and progesterone receptor-negative (ER−/PR−) inflammatory breast cancer cell line (22, 23) stratified by expression of CD44 and CD104 (Integrin β 4) (fig. S1A) (14). Six single cell–derived clonal populations were isolated, ranging from epithelial-like (E) to mesenchymal (M1 and M2), including three distinct intermediate states (EM1, EM2, and EM3)—hereafter referred to as “EMT clones” (Fig. 1A and fig. S1A). Briefly, the SUM149PT cell line was sorted into three populations stratifying the EMT spectrum (fig. S1A), and single cells were sorted into 96-well plates from which 14 single cell–derived clonal populations were chosen and isolated on the basis of morphological characteristics. Of these 14, 6 were chosen to best represent the spectrum of states within the SUM149PT parental cell line. These clones, which stably retained their EMT states in vitro, were ranked along the EMT spectrum relative to one another based on expression of hallmark epithelial and mesenchymal markers such as Vim (vimentin), CDH1 (E-cadherin), ZEB1, and SNAI1 (Snail) (Fig. 1, C and D), as well as by variable migratory and invasive characteristics in vitro (Fig. 1B). While the mesenchymal clones exhibited greater migratory and invasive ability than the epithelial clone, the intermediate clones displayed 2- to 10-fold higher migratory and invasive potential than the mesenchymal clones. These data suggest that earlier EMT studies did not discern intermediate states from mesenchymal ones when assessing these specific characteristics. Classic flow cytometry approaches using two to three markers were insufficient to distinguish between the intermediate states (fig. S1D). By other measures, however, the intermediate clones exhibit differences in cell migration (Fig. 1B), EMT marker expression (Fig. 1, C and D), cell morphology (Fig. 1E), and expression of vimentin and E-cadherin (Fig. 1F). The three intermediate clones most closely resemble the characteristics of the parental line in migratory and invasive ability in vitro (Fig. 1B) and coexpression of E-cadherin and vimentin (Fig. 1F), as well as their overall transcriptional profiles determined by unsupervised hierarchical clustering of Pearson’s correlation coefficients following RNA sequencing (RNA-seq) (fig. S1E). Notably, there were no substantial genetic differences between clones E, EM1, EM2, EM3, and M1, with M2 exhibiting some SNP and INDEL variations (fig. S1, B and C), indicating that the phenotypic and functional differences between these intermediate clones are likely driven by nongenetic mechanisms (24).
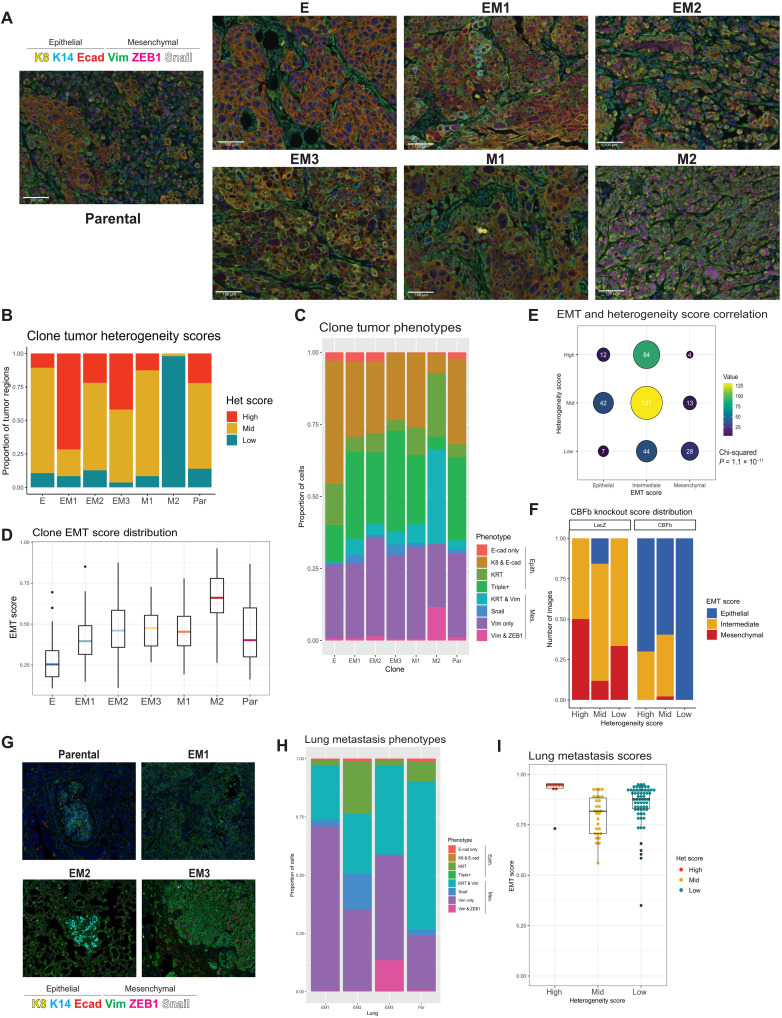
Fig. 1. The heterogeneous cell line SUM149PT contains multiple distinct EMT states that can be isolated as single-cell clones.
(A) A schematic of the flow cytometry method used to isolate single-cell clones that present as an epithelial (E), three distinct intermediate (EM1, EM2, and EM3), and two mesenchymal (M1 and M2) EMT states. (B) In vitro assessment of clonal migratory and invasive characteristics as measured in a standard transwell assay (n = 3, SD, ****P < 0.0001, and *P < 0.05). Canonical EMT marker expression levels as determined by (C) quantitative RT-PCR (SD, n = 4) or (D) immunoblotting to rank SUM149 clones along the EMT spectrum. (E) Bright-field and (F) immunofluorescent images of EMT clones in vitro stained with vimentin and E-cadherin displaying cell morphology and marker expression and localization, respectively. (G) EMT signature of EMT clones and parental line generated from the ordinal multinomial logistic regression method of gene scoring and (H) distribution of EMT score of the EMT clones among other breast cancer cell lines from the CCLE. (I) Immunofluorescent staining for E-cadherin (red), vimentin (green), and KRT8 (white) of four triple-negative breast cancer lines (two intermediate, HCC38 and Cal-51; one epithelial, HCC1143; and one mesenchymal, MDA-MB-231) from the CCLE displaying heterogeneous phenotypes.
Various methods have been developed to quantify the extent to which cells undergo an EMT and determine an absolute comparable EMT score (25–27), which have been reviewed recently (28). When applied to our model, all three methods—multinomial logistic regression-based scoring (MLR) (27), Kolmogorov–Smirnov (KS) (26), and 76 gene signature (76GS) (25)—predict that E through EM3, i.e., the epithelial and three intermediate clones, fall within the intermediate state, while M1 and M2 are mesenchymal (Fig. 1G and fig. S1, F and G). When plotted among the 59 breast cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE), these clones fall along the intermediate and more mesenchymal end of the spectrum (Fig. 1H; fig. S1, F and G; and table S1). This spread of EMT states is most relevant in the context of studying metastasis as many highly epithelial breast cancer cell lines such as those seen in this comparison (Fig. 1H) exhibit less migratory and invasive characteristics (29), which contribute significantly to metastatic potential. Notably, the parental line scored closer to the epithelial (E) clone in most methods (Fig. 1G and fig. S1, F and G).
Box 1. A list of qPCR primers for EMT-related genes.
| ZEB2 F | CAAGAGGCGCAAACAAGC | Zeb1 F | TGCACTGAGTGTGGAAAAGC |
| ZEB2 R | GGTTGGCAATACCGTCATCC | Zeb1 R | TGGTGATGCTGAAAGAGACG |
| PRRX1 F | CTGATGCTTTTGTGCGAGAA | Ecad F | TTGCACCGGTCGACAAAGGAC |
| PRRX1 R | ACTTGGCTCTTCGGTTCTGA | Ecad R | TGGATTCCAGAAACGGAGGCC |
| Twist2 F | GCAAGAAGTCGAGCGAAGAT | Fibronectin1 F | GAGAATGGACCTGCAAGCCCA |
| Twist2 R | GCTCTGCAGCTCCTCGAA | Fibronectin1 R | AGTGCAAGTGATGCGTCCGC |
| OVOL1 F | CCGTGCGTCTCCACGTGCAA | Vimentin F | ACCCGCACCAACGAGAAGGT |
| OVOL1 R | GGCTGTGGTGGGCAGAAGCC | Vimentin R | ATTCTGCTGCTCCAGGAAGCG |
| OVOL2 F | CCGATGGACACCTGGCGACC | RUNX1 F | CAGCTGCGGCGCACA |
| OVOL2 R | GACGGTTCAGCATGCGCTGC | RUNX1 R | GGATCTGCCTTGTATCCTGCAT |
| Twist1 F | TGCGGAAGATCATCCCCACG | RUNX2 F | AGCCCTCGGAGAGGTACCA |
| Twist1 R | GCTGCAGCTTGCCATCTTGGA | RUNX2 R | CGGAGCTCAGCAGAATAATTTTC |
| Snai1 F | CTGGGTGCCCTCAAGATGCA | RUNX3 F | GTTCAACGACCTTCGCTTC |
| Snai1 R | CCGGACATGGCCTTGTAGCA | RUNX3 R | GTCCACGGTCACCTTGATG |
| Snai2 F | TACCGCTGCTCCATTCCACG | CBFB F | CTTAGAAAGAGAAGCAGGCAAGG |
| Snai2 R | CATGGGGGTCTGAAAGCTTGG | CBFB R | AACTCCAGACAGCCCATACCA |
Heterogeneity in breast cancer cell lines and the presence of intermediate states have been validated across many breast cancer cell lines and EMT models (8, 30). To further corroborate these findings in the SUM149PT model, we validated the presence of multiple EMT states in four canonical breast cancer cell lines, which range from intermediate to mesenchymal in their EMT scores—HCC1143, HCC38, Cal-51, and MDA-MB-231 (MLR EMT score: 0.26, 0.76, 1.06, and 1.93, respectively; Fig. 1H). All four cell lines are composed of heterogeneous subpopulations along the epithelial-mesenchymal spectrum to varying degrees, as revealed by their coexpression of E-cadherin, vimentin, and Keratin 8 (KRT8), matching those observed in the SUM149PT parental cell line (Fig. 1I and fig. S1H). Our model, thus, highlights the presence of multiple distinct intermediate EMT states that are also found in other breast cancer cell lines, validating the suitability of this model and further investigation of each state’s role in tumor development and metastasis.
Characterizing in vivo roles of multiple EMT states
Intermediate clones possess high tumor-initiating cell frequencies
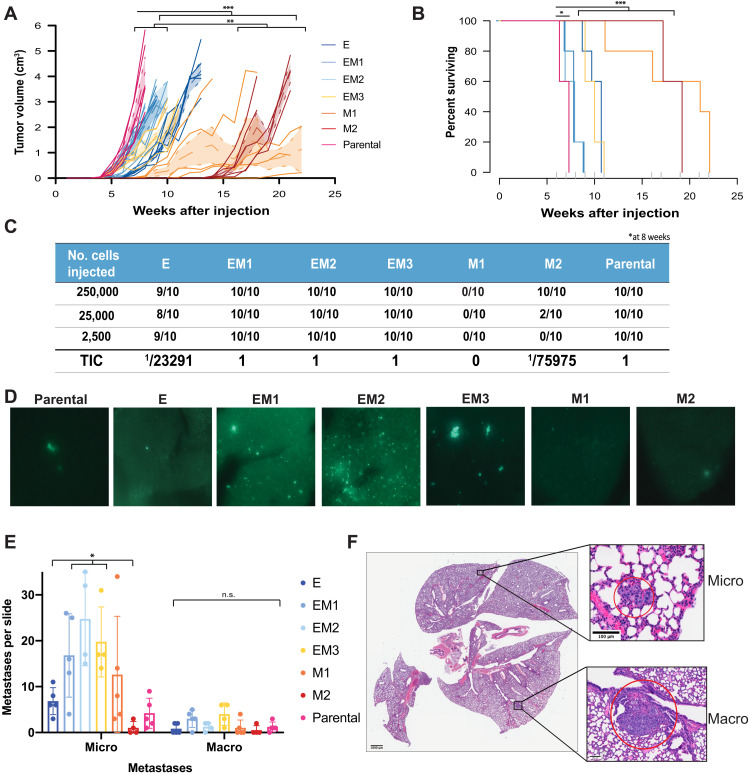
In vivo tumor initiation and growth further highlighted the individuality of these EMT clones. Upon orthotopic injection, the parental cell line was able to initiate and form tumors more rapidly than the other clones (Fig. 2A). Tumor growth analysis with a bimodal linear mixed model (31) revealed that the three intermediate clones were able to initiate tumors at the same rate as the parental line (initial-phase Holm adjusted P value <0.05) (Fig. 2, A and C) but exhibited a lag in growth (exponential-phase Holm adjusted P value <0.003). The epithelial (E) and two mesenchymal (M1 and M2) clones both failed to initiate tumors as readily or, in the case of the mesenchymal clones, grow as rapidly as the parental and intermediate clones (Holm adjusted P value <0.01). Increased tumor growth corresponded with decreased survival, with the parental line exhibiting shortest survival after injection and the mesenchymal clones exhibiting the longest (Fig. 2B). Limiting dilution analyses revealed a high tumor-initiating cell (TIC) frequency in the parental and intermediate clones, with all mice forming tumors by 8 weeks (Fig. 2C). The two mesenchymal clones (M1and M2) had the lowest TIC frequencies at 8 weeks (0 and 1 of 75,975, respectively) despite expressing high levels of CD44 expression, a marker frequently correlated with increased stemness and tumor-initiating ability (figs. S1D and S2B) (11, 12, 32). All clones generated high-grade, poorly differentiated, invasive ductal carcinomas of no special type (ductal). Clone M1 tumors notably exhibited 50% squamous differentiation, and clone M2 showed abundant spindle-cell morphology (fig. S2A). Tumor growth and TIC frequencies indicate that, while the intermediate EMT clones may represent a population with high tumor-initiating potential, a heterogeneous population such as the parental line is able to enter an exponential growth phase more rapidly. Notably, as with the clonal cell lines, flow cytometry was not able to distinguish between tumors of different clonal origin despite significant differences in growth and survival (fig. S2B).
Fig. 2. Differences in primary tumor growth and metastatic potential between EMT clones.
(A) Tumor growth curves measured weekly following orthotopic injection of clonal and parental cell lines at 2500 cells exhibit exponential growth differences between EMT states [TumGrowth (31) piecewise regression model breakpoint = 6 weeks, ***Holm adjusted P < 0.0001 and **Holm adjusted P < 0.01, n = 10]. (B) Survival curve of EMT clones and parental line displaying differences in survival across the parental line and early intermediate EMT states. Cox regression analysis (***Holm adjusted P < 0.0001 and *Holm adjusted P < 0.015). (C) Tumor-initiating cell frequency calculated by limiting dilution assay with cells injected at 250,000, 25,000, and 2500 cells per flank. TIC calculated at 8 weeks after injection. (D) Lung sections collected at the time of maximum tumor burden (2 cm3), with GFP-labeled tumor cells, following orthotopic injections as in (A). (E) Lungs fixed and stained from (A) with H&E and enumeration of micrometastatic (<10 adjacent cells) and macrometastatic (10+ adjacent cells) regions (SD, n = 5, micro P < 0.02 and macro P = n.s.). n.s., not significant. (F) Representative bright-field images of micro- and macrometastases from one mouse lung (EM1).
EMT state affects outgrowth of metastasis
For the purposes of monitoring metastatic outgrowth in vivo, derivatives of each EMT cell line stably expressing a luciferase-IRES-ZsGreen construct were generated, which allowed for both live and postmortem detection of tumor cells. As has been observed previously for SUM149PT (33), the parental line and all clones metastasized to the lung, with varying success, as seen by fluorescence (Fig. 2D). No other metastatic lesions could be detected by luciferase or fluorescence.
To precisely delineate the propensity of the clones to form micro- and macrometastasis, lungs from animals bearing orthotopic tumors were fixed and stained with hematoxylin and eosin (H&E) and counted for micrometastases (<10 adjacent cells) and macrometastases (>10 adjacent cells) (Fig. 2, E and F). The three intermediate (EM1 to EM3) clones seeded higher numbers of micrometastatic lesions per lung, compared to the most epithelial (E) and most mesenchymal (M2) (P value <0.05). Within this group, clones EM1 and EM3 seeded higher numbers of macrometastases compared to EM2 (Fig. 2E), although differences between the intermediate clones were not statistically significant. While exhibiting the highest rate of tumor growth and poorest survival, the parental line seeded fewer lung metastases than the intermediate clones, suggesting that other mechanisms could be contributing to mortality.
Identification of transcriptional networks that sustain EMT states
Transcriptomic profiles reveal shared intermediate gene signature
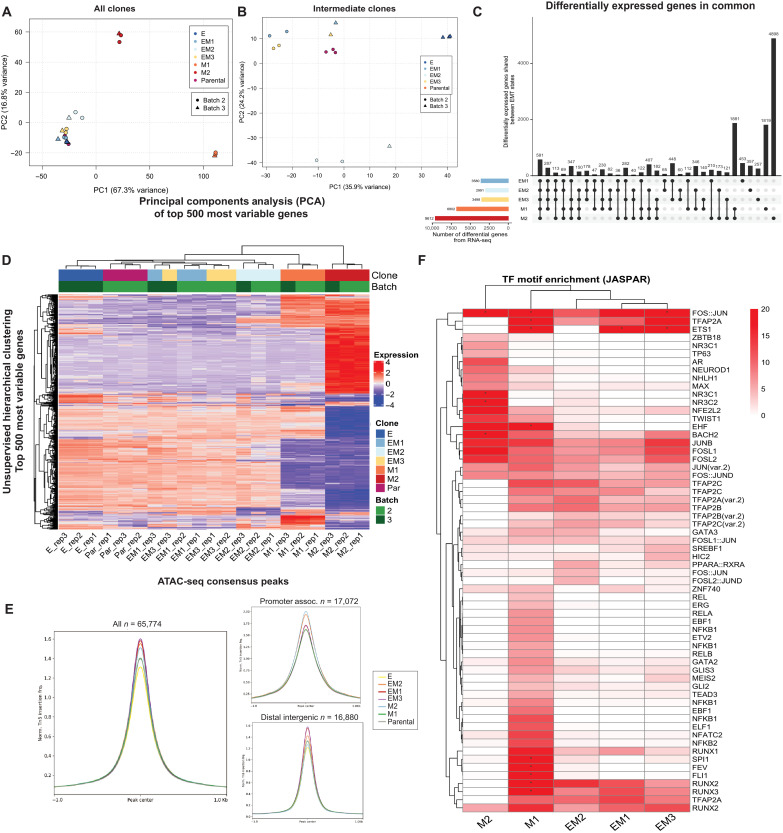
Given the lack of genetic differences between the clones, we hypothesized that the clonal variations in the EMT state were driven by alterations in their transcriptional profiles. Using the bulk RNA-seq of each clone, alterations in the expression levels of various transcription factors were analyzed using the epithelial clone E as a benchmark. Principal components analysis (PCA) demonstrates clustering of the parental line, intermediate EM, and E clones (Fig. 3, A and B), whereas the two mesenchymal clones (M1 and M2) share no overlap between themselves or any other cluster. This indicates that the EMT clones do not reside in a linear spectrum but rather embark upon multiple distinct trajectories. Unsupervised hierarchical clustering of the 500 most variable genes across all clones reveals distinct transcriptional programs separating the three intermediate clones from the epithelial (E) and mesenchymal (M1 and M2) ones (Fig. 3D). Within this intermediate cluster, EM2 and E are again distinct from the remaining intermediates and parental line (Fig. 3, B and D). Differential expression analysis revealed that 581 shared genes were significantly differentially expressed (adjusted P < 0.05) in all clones when compared to E, with 1881 genes shared between the two mesenchymal clones and 178 shared between the three intermediate clones (Fig. 3C, fig. S3A, and table S2). In comparison to the epithelial baseline clone E, more differentially expressed genes are exclusive to each clone than are shared between two or more clones (Fig. 3C), further corroborating the unique EMT states represented by this model. Gene set enrichment analysis (GSEA) (34) of the differentially expressed genes confirms activation of the hallmark EMT gene set in all intermediate and mesenchymal clones, as well as other gene sets corresponding to cell division and chromatin remodeling, as expected (fig. S3B).
Fig. 3. Identification of stabilizing transcription factors in the intermediate EMT state by transcriptional and chromatin analysis.
(A) PCA of the 500 most variable genes between all EMT clones and SUM149 parental line and (B) intermediate clones only from RNA-seq. (C) UpSet plot of all differentially expressed genes (referenced to clone E) that are shared and unique to each EMT clone. (D) Unsupervised hierarchical clustering of the top 500 differentially expressed genes in all comparisons to clone E (P value of <0.05). (E) ATAC-seq peak accessibility measured as counts per million (CPM) normalized Tn5 insertions surrounding consensus peaks, promoter-associated, and distal-intergenic peaks, respectively, for each EMT clone. (F) Unsupervised hierarchical clustering of transcription factor motif enrichment (−log10 adjusted P value, hypergeometric test) among accessible chromatin peaks unique to each clone, relative to (E). Motifs obtained from the JASPAR database were identified using motifmatchr (P < 0.05) and tested for enrichment against the background set of all peaks identified in the respective clone using the hypergeometric test. Asterisk indicates a −log10 P-value enrichment threshold greater than 20, scaled to fit.
Box 2. A list of CRISPR-Cas9 guides targeting RUNX family members.
| LacZ | CACCGTGCGAATACGCCCACGCGAT |
| RUNX2 #1 | CACCGGCGGACGAGTTCGGCCGGG |
| RUNX2 #2 | CACCGATGAGCGACGTGAGCCCGG |
| CBFb #1 | CACCGTCCAGAACGCCTGCCGCGA |
| CBFb #2 | CACCGAGTCGACATACTCTCGGCT |
To further explore the epigenetic landscape of these clones, we used Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) (35) to determine the differential chromatin accessibility across EMT clones in comparison to the epithelial clone E, which exhibits the most closed chromatin profile (Fig. 3E). Notably, the chromatin landscape is similarly diverse between mesenchymal and intermediate clones (fig. S3, C and D), as seen in the RNA-seq (Fig. 3D). To identify transcription factors (TFs) with a significant enrichment of motifs among peaks that were uniquely accessible in each clone (relative to E), the presence of transcription factor binding motifs was scanned for using motifmatchr (36) and tested for enrichment against the background set of all identified peaks (hypergeometric test, adjusted P < 0.05) (Fig. 3F). The three intermediate and early mesenchymal clones were highly enriched for motifs of the Runt-related transcription factor (RUNX) family (Fig. 3F). The Transcription factor AP-2 (TFAP2) family also exhibit enriched binding accessibility in these intermediate and early mesenchymal clones, albeit less significantly than the RUNX family. All three RUNX transcription factors and their cofactor CBFβ have been previously implicated in various cancers (37–39) as well as in metastatic progression of lung adenocarcinoma (40) and triple-negative breast cancer (TNBC) (41, 42). Enrichment of these RUNX TF motifs is unique to the intermediate (EM1, EM2, and EM3) and early mesenchymal (M1) clones, those that seeded the highest number of lung metastases (Fig. 2E).
Combined RNA-seq and ATAC-seq identifies EMT and MET networks
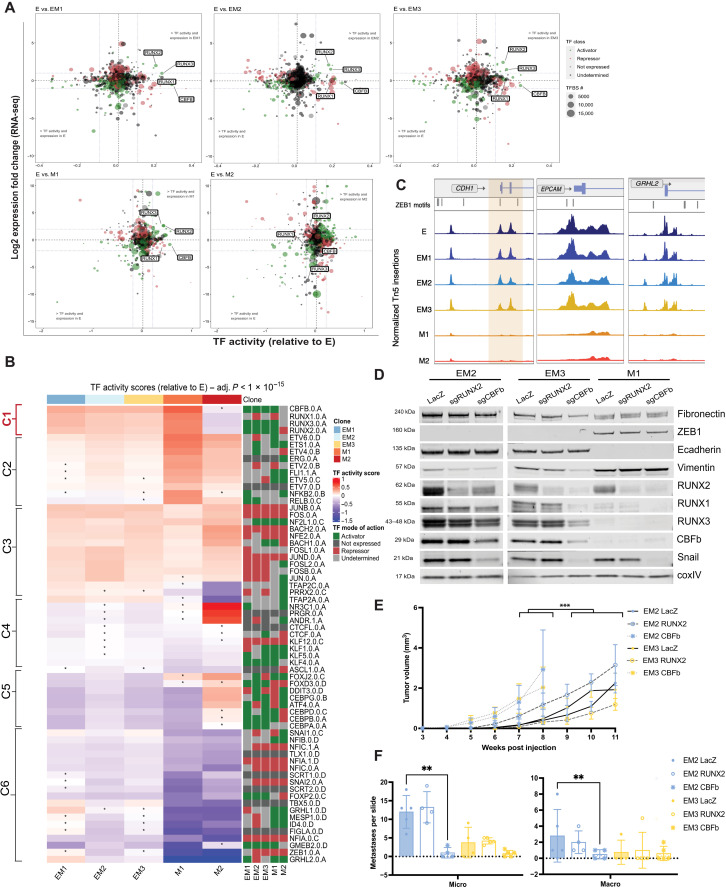
To provide a more comprehensive picture of the epigenetic and regulatory landscape of EMT, we used a new multiomics approach, DiffTF (43), to quantify TF activity and regulatory state by integrating RNA-seq and ATAC-seq data for each clone. Similar to motifmatchr, TF activity was inferred by computing the fold change in chromatin accessibility between each clone at each binding site of a given TF, once again using clone E as a baseline reference. Inferred TF activity was visualized against RNA-seq log2 fold change for each transcription factor (Fig. 4A and fig. S4A), facilitating simultaneous detection of TFs with increased expression and TF activity (top right quadrant). Analysis of the directional changes of TF expression from RNA-seq also indicated whether the TF in question acts as an activator (green) or repressor (red). The two mesenchymal clones revealed much larger shifts in both fold-change expression and TF activity, indicative of larger changes in their overall transcriptional and chromatin profiles relative to E. In the three intermediate and early mesenchymal clones, RUNX2 and RUNX3 are both among the most highly expressed and exhibit the highest TF activity of all significant transcription factors (Fig. 4A and fig. S4A). A list of all significant transcription factors, their relative TF activity, and fold-change expression are included in table S3. In addition, of the transcription factors that are more accessible in the intermediate state, the expression of RUNX2 positively correlated and that of TFAP2C negatively correlated with CCLE breast line EMT scores with high significance (fig. S4, B and C).
Fig. 4. Identification of stabilizing transcription factors in the intermediate EMT state by multiomics analysis.
(A) Advanced volcano plot of highly significant transcription factors, highlighting the RUNX family transcription factor activity, relative to clone E determined by diffTF from ATAC-seq along the x axis (label cutoffs at 0.1, −0.1 TF activity), plotted against log2 fold gene expression values of transcription factors on the y axis (label cutoffs at 1, −1 log2 fold). Transcription factor classification, determined by transcription factor expression, displayed in bubble color, and number of transcription factor binding sites used to determine TF activity plotted as bubble size. (B) Unsupervised hierarchical clustering of TF activity scores (z-score–transformed) compared to clone E (adjusted P value <1 × 10−15, asterisk indicates n.s. in that comparison). Right-hand side displays TF classification, determined by changes in TF expression (diffTF), as an activator, repressor, not expressed, or undetermined. (C) Peak accessibility of Tn5-normalized, merged coverage of three canonical ZEB1 target genes, CDH1, EPCAM, and Grainyhead like 2 (GRHL2), across all clones from ATAC-seq. ZEB1 TF motifs highlighted above signal tracks. (D) Protein levels of canonical EMT markers determined by Western blot following CRISPR-Cas9–mediated knockout of LacZ, RUNX2, and CBFβ in late intermediate and early mesenchymal clones. (E) Tumor growth curves measured weekly following orthotopic injection of CBFb knockout at 2500 cells [TumGrowth (31) piecewise regression model breakpoint = 6 weeks, ***initial-phase Holm adjusted P value <0.0001, n = 5]. (F) Lungs fixed and stained from (E) with H&E and enumeration of micrometastatic (<10 adjacent cells) and macrometastatic (10+ adjacent cells) regions (SD, n = 5, EM2 P < 0.001 and EM3 P = n.s.).
Box 3. Primary and secondary antibodies and OPALs in order for TSA staining.
HRP, horseradish peroxidase.
| Primary antibody | Secondary antibody | OPAL (TSA) |
| Snai1,1:400; CST, #3895 | Goat anti-mouse HRP, 1:1500 |
OPAL 620, 1:500 |
| K8, 1:300; Invitrogen, PA5-29607 |
Goat anti-rabbit HRP, 1:1500 |
OPAL 540, 1:1,000 |
| K14, 1:1000; Invitrogen, MA5-11599 |
Goat anti-mouse HRP, 1:1500 |
OPAL 520, 1:150 |
| Vimentin, 1:300; CST, #5741 |
Goat anti-rabbit HRP, 1:1500 |
OPAL 690, 1:150 |
| E-cadherin, 1:500; BD Biosciences, #610182 |
Goat anti-mouse HRP, 1:1500 |
OPAL 650, 1:500 |
| ZEB1, 1:1000; Invitrogen, PA5-82982 |
Goat anti-rabbit HRP, 1:1500 |
OPAL 570, 1:600 |
Unsupervised clustering analysis presents a clearer picture of how these TFs act in a transcriptional network, highlighting TFs that exhibit significant changes in their activity in at least one clone (adjusted P value <1 × 10−15) (Fig. 4B). Cluster 1 is composed of transcription factors that are up-regulated and function as activators in the clones that exhibit increased metastatic potential (EM1 to EM3 and M1)—the RUNX TF family and cofactor CBFβ. Cluster 2 includes factors that increase in activity with the progression of EMT, e.g., ETS1 and related factors, nuclear factor κB, RelB, and Friend leukemia integration 1 (FLI1). Cluster 3 identifies transcription factors that are activated and remain consistently active after entrance into the EMT, e.g., members and regulators of the Activator protein 1 (AP1) complex. Multiple members of the Krüppel-like factor (KLF) family of transcription factors and CCCTC-binding factor (CTCF) are found in cluster 4, which exhibit a consistent decrease in TF activity following entrance into an EMT. Cluster 5 highlights transcription factors that may be uniquely active in M2 while remaining inactive in the other EMT clones, including Forkhead box protein J2 (FOXJ2) and Forkhead box protein D3, CCAAT-enhancer-binding protein (CEBP) complex members, and Activating transcription factor 4 (ATF4). Last, cluster 6 delineates the transcription factors that have an overall and graded decrease in activity as the EMT progresses. This cluster contains multiple EMT TFs such as Snail, Slug, and ZEB1 as well as known mesenchymal-to-epithelial transition (MET)-promoting TFs such as Grainyhead like transcription factor 1 and 2 (GRHL1/2). Although these and other canonical EMT transcription factors such as Twist and Paired related homeobox 1 (PRRX1) are expressed at significantly high levels (P < 0.05), nonsignificant or decreased TF activity indicates no change in chromatin accessibility compared to clone E (table S3 and fig. S4A). These TFs decrease in activity, while their overall expression remains high (fig. S4A) likely resulting from strong repression of chromatin accessibility at their epithelial target genes, potentially through histone deacetylase recruitment (44, 45) or other mechanisms at ZEB1 target promoters CDH1, EPCAM, and GRHL2 (Fig. 4C).
CBFβ knockdown destabilizes EMT states
RUNX2 and its coactivator CBFβ have been implicated in mammary development (46), mammary stemness (47), and increased metastatic capacity in breast cancer (41, 48). To further examine the roles that these transcription factors play in EMT and the propagation of a metastatic state, we tested the effects of CRISPR-Cas9–mediated knockout of RUNX2 and CBFβ, which are uniquely expressed and active in the intermediate and early mesenchymal clones, as well as a nontargeting LacZ control (Figs. 3F and 4, A and B). Knockout of RUNX2 in the intermediate clones did not result in any significant alterations to the levels of canonical EMT markers, except a minor reduction in VIM expression in EM2 (Fig. 4D). On the other hand, knockout of CBFβ led to a down-regulation of SNAI1 and VIM expression in EM2 and EM3, as well as a more subtle reduction in FN1 levels, indicating a shift to a more epithelial state. The expression of all three RUNX TFs was reduced upon loss of CBFβ, likely due to its role as an essential cofactor (Fig. 4D).
To determine the effect of destabilizing EMT on tumor formation and metastasis, clone EM2 and EM3 bearing CBFβ knockout or LacZ nontargeting guide RNAs were injected orthotopically into NOD scid gamma (NSG) mice. These cell lines express high levels of both RUNX2 and CBFβ in comparison to clone E (Fig. 4B) and form predominantly either micro- or macrometastases, respectively, in the lung (Fig. 2E). In EM2 and, to a lesser extent, EM3, CBFβ knockout accelerated the time point at which tumors were first observed (initial-phase Holm adjusted P < 0.001) and time to tumor burden (Fig. 4E). However, these tumors had a near-complete absence of metastasis to the lung (Fig. 4F). In contrast, RUNX2 knockout did not appear to have any significant effects on tumor formation or metastasis, likely due to compensation from the other RUNX transcription factors. Thus, we conclude that loss of CBFβ destabilizes the intermediate EMT state in clones EM2 and EM3, leading them to acquire a more epithelial state that, despite being more proliferative, lacks lung-metastatic capacity.
We then leveraged data from The Cancer Genome Atlas (TCGA) to determine whether CBFβ expression could, on its own, serve as a single gene biomarker for patient prognosis. However, like many other single markers that have not been strong predictors of outcome (49, 50), CBFβ expression alone is not associated with any differences in patient overall survival in the TCGA-BRCA cohort, adjusting for age, stage, and PAM50 molecular subtype [(Hazard Ratio (HR): 1.03, P = 0.834] (fig. S4E). Other biomarkers such as ZEB1, Snail, and Twist have been tested as prognostic biomarkers across multiple cohorts with varying success and predictive power (51); however, no single biomarker has proven robust enough to be adopted into the clinic to predict metastatic disease outcome. This further indicates the need for a more nuanced multimodal metric to describe and identify intermediate EMT states within patient samples if EMT is to be leveraged as a diagnostic and prognostic tool.
Exploration of tumor heterogeneity in EMT clone-derived tumors
To better understand the role that EMH plays on tumor progression and metastasis, we used a multiround, multiplexed tyramide signal amplification (TSA) approach (52) to assess protein levels of EMT markers, segregating out stromal cells that could obfuscate EMT scoring. To capture the full spectrum of EMT states, we designed a panel of six EMT markers, containing three intermediate filament proteins (KRT8, KRT14, and Vimentin), two EMT transcription factors (ZEB1 and Snail), and an adherens junction protein that serves as a hallmark epithelial marker (E-cadherin). Tumors from each of the clones were stained and divided into 50 regions of interest (ROIs) and processed with the inForm analysis software (Akoya Biosciences) to generate composite images (Fig. 5A). inForm image training algorithms were used to discern tumor and stromal composition, as well as conduct cell segmentation (fig. S5A). Normalized percentile distributions of the arbitrary fluorescent units of each EMT marker, per cell, across all images provided an overview of the overall composition of these tumors (fig. S5B), indicating that no one unique marker signature defined any individual tumor. Overall, clone M2 tumors expressed less E-cadherin and more ZEB1, while clone E tumors contain less Vimentin, indicative of their initial EMT states in vitro. To determine the extent of EMH within the stained images, a scoring metric was developed using the SUM149PT clones as a training set. Tumor images were scored on the basis of a rubric of low (one major cell trait with up to one minor trait), mid (two major cell traits with up to three minor traits), and high (three or more major cell traits present with two or more minor traits) (Fig. 5C). Scoring based on these criteria revealed that the intermediate EMT clones form tumors that contain more regions of higher heterogeneity than the parental cell line (Fig. 5C). Thus, increasing levels of heterogeneity may not linearly correlate with tumor growth or metastasis, but rather, there exists an optimal ratio of cell traits within the tumor that determines its growth and metastatic potential. The requirement for this optimal ratio may explain the growth lag observed in the intermediate clones when compared to the parental line (Fig. 2A), which likely results from constraints in the generation of a requisite level of heterogeneity from a homogeneous cell culture.
Fig. 5. Multiplexed staining of SUM149 tumors and lungs identifies phenotypes and quantifies tumor heterogeneity and overall EMT state in clone-derived and CBFb-depleted tumors.
(A) EMT clone-derived tumors resected at 1.5 cm3 and stained with a six-marker EMT panel using multiplexed immunostaining (n = ~50 images per tumor). (B) Empirically determined heterogeneity scores of EMT clone-derived tumors. Rubric: low (one major cell trait with up to one minor trait), mid (two major cell traits with up to three minor traits), and high (three or more major cell traits present with two or more minor traits). (C) Boxplot of EMT phenotypes generated from inForm cell phenotype analysis displaying the composition of each clonally derived tumor (n = ~50 images per tumor). (D) EMT score distribution in clonally derived tumors generated from weighted multivariable logistic regression of the phenotypes in (C) present in each tumor. (E) Correlation of EMT (tertile; epithelial: 0 to 0.29, intermediate: 0.3 to 0.69, and mesenchymal: 0.7 to 1) and heterogeneity score in EMT clone-derived tumor images (n = 365, chi-squared P = 1.1 × 10−11). (F) Distribution of EMT (tertile) and heterogeneity scores in EM2 and EM3 clone-derived tumors following CBFb knockout, compared to LacZ control tumors. (G) Outgrowth lung metastases of EM1, EM2, EM3, and parental tumors stained with the six-marker EMT panel using multiplexed immunostaining to determine (H) EMT phenotypes present and (I) EMT and heterogeneity scores.
Composition of EMT phenotypes and EMH varies by EMT clone
In a parallel method to assess tumor diversity and phenotypic composition, the inForm analysis software was trained on a subset of tumor images to recognize eight distinct cell phenotypes encompassing the majority cell states present in all tumor images (fig. S5A). These phenotypes, spanning from most epithelial to most mesenchymal, are E-cadherin only, KRT8 and E-cadherin, KRT8 and KRT14 (KRT), triple positive (KRT8 + E-cadherin + vimentin), KRT8/14 and vimentin, Snail only, vimentin only, and vimentin and ZEB1. Phenotypes were validated manually and by fluorescent marker distribution (fig. S5C). Similar to EMT marker distribution, there were no phenotypes unique to any individual state, indicating that optimal tumor progression may be determined by global ratios of cell states rather than the presence or absence of an individual cell type. On either end of the spectrum, clone E– and clone M2–derived tumors are made up of more than 75% epithelial (E-cad only, KRT8 + E-cad, KRT, or Triple positive) or mesenchymal (KRT & vimentin, Snail only, vimentin only, or vimentin + ZEB1) phenotypes, respectively. Tumors derived from the intermediate clone and the parental line, meanwhile, all contained a roughly equal distribution of epithelial and mesenchymal phenotypes (Fig. 5C). These data further support the notion that the ratio of EMT states (phenotypes), rather than the presence of any particular state, is more reflective of tumor growth and metastatic potential.
To develop a scoring algorithm to assess EMH, three feature extraction methods were tested using the segmented cell data files (fig. S5A). These three approaches sought to determine the best method to assess heterogeneity: (i) an entropy-based approach (53, 54) using mean marker expression of cells per image, (ii) a nearest neighbor analysis approach (55) using cell phenotypes as determined above (Fig. 4C), and (iii) a hybrid approach combining approaches 1 and 2 (fig. S5D). All three approaches were developed and evaluated using a randomized subset of clone tumor images and their corresponding heterogeneity scores according to the rubric outlined in Fig. 5B as ground truths. Logistic regression performance on unseen test data indicated a 78% accuracy for entropy-based features of mean marker cell expressions. Fivefold cross-validation determined that the entropy-based approach 1 (F1 score = 0.78, Wilcoxon ranking test P = 0.004) proved to be the most robust at correctly assessing tumor heterogeneity (fig. S5D). This shows that E and M2 clones consisted largely of areas of low- or mid-level heterogeneity, whereas all intermediate clones were composed of regions of high heterogeneity (Fig. 5B).
In addition to scoring EMH, we sought to assign EMT scores to tumors based on the ratios of epithelial and mesenchymal traits they exhibit. EMT scores were generated by calculating the composition of cell phenotypes per image (50 images per clone) using the linearly weighted average of cell ratios expressing each phenotype from epithelial to mesenchymal. EMT scores range from 0 to 1, with zero being composed of all epithelial phenotypes and one composed of all mesenchymal phenotypes. Rather than attaining an equilibrated EMT state, clonal tumors held true to the EMT state of their starting populations, with clone E tumors being predominantly epithelial (mean = 0.25), intermediate (EM1, EM2, and EM3) and parental tumors maintaining an intermediate EMT score (mean = 0.4 to 0.6), and the most mesenchymal (M2) scoring >0.7 (mean = 0.7) (Fig. 5D). Clone M1 tumors also scored as intermediate despite starting as a quasi-mesenchymal, further validating this clone as a late intermediate (Fig. 5D). The variation in EMT scores between images in the intermediate clones (EM1, EM2, EM3, and M1) and the parental line was the highest of all of the groups, indicating higher intratumoral heterogeneity among tumor regions (Fig. 5D). In exploring the connection between heterogeneity and EMT scores, we found a correlation between the two scores upon splitting the EMT score into terciles (epithelial = 0 to 0.29, intermediate = 0.3 to 0.69, and mesenchymal = 0.7 to 1; chi-squared P = 1.1 × 10−11). Low-heterogeneity tumor regions correlated with more mesenchymal EMT scores, as can be seen in M2 tumors, and epithelial EMT scores correlate with mostly mid heterogeneity, seen in E tumors (Fig. 5A and fig. S5E). Mid- and high-heterogeneity regions were characterized by intermediate EMT scores despite encompassing a more diverse array of possible EMT states (Fig. 5E and fig. S5E).
Knockout of CBFβ in intermediate SUM149 clones reduces EMH and EMT score
Upon developing a metric to calculate EMH and EMT score, we questioned whether the decrease in metastasis observed upon loss of CBFβ (Fig. 4, E and F) resulted from alterations to overall EMT state or from a change in EMH. When tumors generated from EM2 and EM3 CBFβ knockout clones were stained with the multiplexed immunofluorescence panel, loss of CBFβ led to the formation of tumors with overall lower, more epithelial EMT scores (EM2 Fisher’s P = 3.5 × 10−7 and EM2 Fisher’s P = 4.0 × 10−5; Fig. 5F and fig. S5F). Clone EM2 additionally had decreased EMH in knockout conditions (EM2 Fisher’s P = 0.05 and EM3 Fisher’s P = 0.5; fig. S5G). This reduction arises from an increased presence of more epithelial phenotypes such as E-cadherin only and KRT8 and E-cadherin coexpressing cells, which are associated with a more proliferative state (56, 57), and concurrent decrease in mesenchymal phenotypes such as vimentin, which are more invasive (Fig. 5F and fig. S5H).
Lung metastases contain predominately late intermediate and mesenchymal phenotypes
To further understand the roles of tumor cell EMH and EMT score on the metastatic cascade, we resected the primary tumors to allow for maximal metastatic outgrowth beyond the primary tumor burden. Briefly, intermediate and parental cell lines were orthotopically injected and allowed to grow to a volume of 1 cm3 before surgical resection. Lungs were harvested, and metastases were quantified at 2.5 months after resection, determined by the burden of the relapsed ipsilateral tumor. Micro- and macrometastasis trends were maintained in this later growth model, with EM1 forming a mixture of micro- and macrometastases, EM2 forming predominantly micrometastases, and EM3 predominantly macrometastases.
We subsequently used the multiplexed immunofluorescence approach to stain these lung metastases to determine their heterogeneity and EMT phenotypes (Fig. 5G). To our surprise, all metastases, regardless of size or clone, were predominantly mesenchymal in composition, expressing very little E-cadherin or KRT8 (Fig. 5, G and H). EMT and heterogeneity scores calculated from this staining revealed overall low heterogeneity and high EMT scores, features that were infrequently exhibited in the primary tumors (Fig. 5I). These results suggest that, in contrast to trends observed in other models (58), the SUM149PT tumors give rise to metastases that are predominantly mesenchymal in nature.
High heterogeneity score and intermediate EMT states are predictive of poor survival outcomes in patients
To assess whether the EMT phenotypes observed in our model were representative of those observed in human breast cancer specimens, we carried out TSA immunostaining of a tumor microarray of the Cancer Diagnosis Program (CDP) Breast Cancer Stage III Prognostic Tissue Microarray collected and indexed by the Cooperative Human Tissue Network (CHTN) (Fig. 6A). Following staining with our six-marker EMT panel, we recovered and analyzed 124 cores from both hormone-positive (n = 86) and hormone-negative (n = 38) patients with long-term survival follow-up spanning 250 months. All eight phenotypes were reproducibly represented across all patient tumors (Fig. 6A and fig. S6A), and ratios of epithelial and mesenchymal phenotypes follow similar trends to the SUM149PT clonal model (fig. S6B). These data serve to validate the SUM149PT clones as a model to study various states along the EMT spectrum.
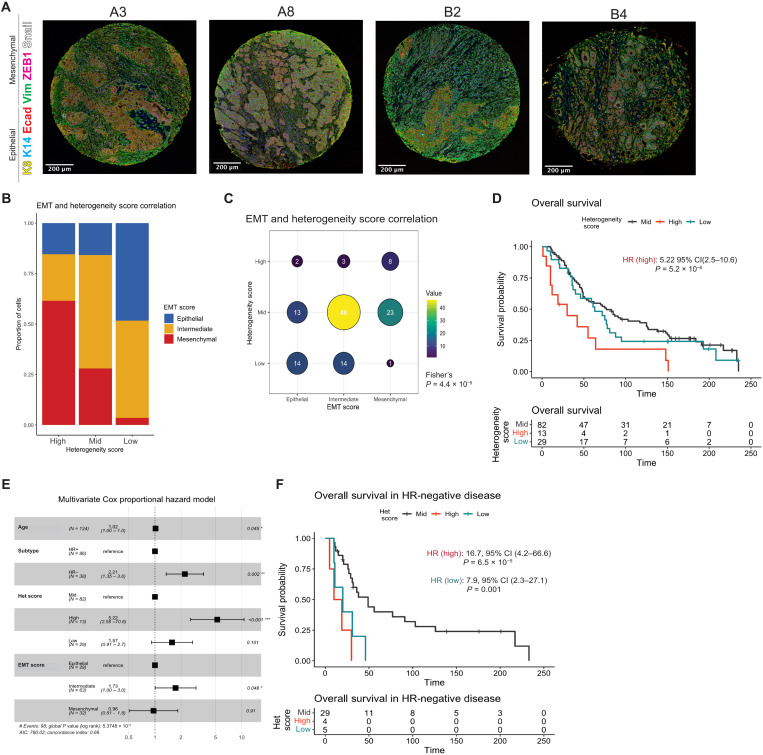
Fig. 6. High tumor heterogeneity and intermediate EMT states are associated with poor prognosis in patient tumors.
(A) Stage III breast cancer patient tumors (n = 124) stained with the six-marker EMT panel using multiplexed immunostaining. (B) Stage III breast cancer cohort tumor EMH score, determined by an entropy-based model of marker distribution at a single-cell level per image, trained and validated by the SUM149 clone tumors, plotted with EMT score (tertile; epithelial: 0 to 0.29, intermediate: 0.3 to 0.69, and mesenchymal: 0.7 to 1) generated from weighted multivariable logistic regression of the phenotypes present in each tumor for each patient sample. (C) Correlation of EMT and heterogeneity scores from (B) (Fisher’s exact test P = 4.4 × 10−5). (D) Kaplan-Meier plot of overall survival stratified by heterogeneity score. Hazard ratio and P value reported from (E) a forest plot of multivariate Cox proportional hazard model of overall survival for heterogeneity score and EMT score accounting for age and subtype. (F) Kaplan-Meier plot of overall survival in hormone-negative patient samples stratified by heterogeneity score. Hazard ratios and P values reported from multivariate Cox proportional hazard model.
Cores were phenotyped and analyzed as previously described to generate a heterogeneity score and EMT score, again segregated into terciles, for each patient (Fig. 6B and fig. S6C). Overall, high heterogeneity is associated with a more mesenchymal EMT score, while low heterogeneity is associated with an epithelial EMT score (Fisher’s exact test P = 4.4 × 10−5; Fig. 6, B and C, and fig. S6C). While this differs from trends seen in the SUM149PT model (Fig. 5E and fig. S5E), the samples in this cohort better represent the population distribution of stage III breast tumors. In survival models adjusting for patient age and hormone receptor status with all samples that passed quality control (QC) (n = 124), patients with a high heterogeneity score had significantly worse overall survival [HR: 5.2, 95% confidence interval (CI): 2.6 to 10.6, P = 5.2 × 10−6], as did patients with intermediate EMT scores (HR: 1.7, 95% CI: 1.0 to 3.0, P = 0.05) (Fig. 6, D and E). While high heterogeneity score on its own remained predictive of poor patient outcome, EMT score alone was not associated with survival [fig. S6D; HR (high): 3.9, 95% CI: 2.00 to 7.61, P = 6.04 × 10−5]. As these metrics describe associated yet independent metrics of tumor complexity and EMT status, our results further demonstrate nuances of tumor heterogeneity in disease progression. When subsetting for hormone-negative disease (n = 36), which is known to exhibit increased intratumoral heterogeneity (2), high and low heterogeneity were both associated with significantly worse overall survival [HR (high): 16.7, 95% CI: 4.2 to 66.6, P = 6.5 × 10−5; HR (low): 7.9, 95% CI: 2.3 to 27.1, P = 0.001] (Fig. 6F and fig. S6E), as was an intermediate EMT score (HR: 3.43, 95% CI: 1.04 to 11.3, P = 0.042). While the correlation between low heterogeneity and outcomes was unexpected, it could be a result of a subset of HR− patients whose tumors score as low heterogeneity/mesenchymal EMT (fig. S6F), similar to the M2 clone in our HR− SUM149 clonal model (fig. S5E). In the context of these patient tumors, poor survival outcomes could be a result of a bottlenecking event following chemotherapy administered to patients, which typically results in the outgrowth of more homogeneous resistant clones that are more aggressive and difficult to treat (59). Hormone-positive disease showed similar trends to the overall model (fig. S6G).
We then sought to explore whether the CBFβ cofactor, which we identify as a key regulator of the intermediate state, could hold predictive value for survival outcomes as a single marker. Sequential slides matching those used for the multiplexed immunofluorescence approach above were stained for CBFβ using immunohistochemistry and scored as negative, weak, moderate, or strong for CBFβ expression and penetrance within the sample core, noting nuclear or cytoplasmic localization (fig. S6H). An H score such as those used to define ER expression (60) was calculated using the strength of expression (0 to 3) multiplied by percentage of positively stained cells. The presence or absence of CBFβ (negative versus any staining) was not found to be associated with overall survival in these patient samples [HR (CBFβ+): 1.42, 95% CI: 0.89 to 2.25, P = 0.132; fig. S6I], nor was H score, adjusting for age and hormone receptor status (HR: 0.1, 95% CI: 0.1 to 1.0, P = 0.845). While CBFβ plays an important role in metastasis through stabilization of the intermediate EMT state and may represent a potential molecular target for future therapeutics, its expression alone is not predictive as a biomarker for patient survival.
Overall, these analyses demonstrate a novel method for scoring patient samples, displaying an increased risk of death in patients with highly heterogeneous tumors that are composed of intermediate EMT phenotypes. Moreover, they display the power of a multiplexed marker panel over a single biomarker, which was unable to correlate with patient survival. Together, this highlights the importance of tumor heterogeneity and EMT state in understanding and predicting patient prognosis as well as the benefit of combinatorial approaches in describing tumor heterogeneity.
DISCUSSION
Our study encompasses a comprehensive analysis of the spectrum of EMT states represented within a breast cancer cell line to interrogate the nuances of each state, their respective contributions to tumor initiation and metastasis, and their epigenetic regulatory networks. We uncover the presence of multiple unique EMT states within the intermediate EMT category, as well as two distinct mesenchymal-like states, suggesting multiple, nonlinear trajectories for EMT as has been previously shown (30, 61). These states were verified by flow cytometry (CD44 and CD104) and ranked by EMT scores among other CCLE breast cancer cell lines, where they fell between an early intermediate and mesenchymal. While this model does not span from one extreme state to the other, it represents the EMT spectrum within SUM149PT, a cell line that is classified as Basal-like 2 molecular subtype of TNBC (62), reflecting the heterogeneous nature of this subtype. We used the relative EMT states between the clones to study the epigenetic heterogeneity of metastatic breast cancer within an isogenic background and interrogate the fitness of each individual state. While the three intermediate EMT clones have the highest migratory and invasive potential in vitro, they are outperformed in tumor-initiating ability and growth by the parental cell line, which, by RNA-seq clustering, most closely resembles the epithelial clone.
A concern when studying heterogeneous cell subpopulations is the retention of their initial EMT state following their isolation and culture. All six of our clones retained their morphological and phenotypic traits through multiple passages in culture, although we did observe clone E drifting to acquire a more spindle-shaped morphology upon extended periods of culture. All single-cell clones retained their initial CD44/CD104 expression profiles except for the most mesenchymal clone, M2, which gained the expression of CD104 following its culture as an isolated single-cell clone. Given that the maintenance of EMT state is regulated by a complex set of paracrine and autocrine signals (63), it is plausible, albeit speculative, that the loss of specific paracrine signals upon isolated culture could have resulted in an altered state in the M2 clone that allowed expression of CD104. Nevertheless, to ensure that all clones retain their original EMT states upon culture, their passage numbers were restricted to below 20.
We further confirm that the intermediate EMT state exhibits higher levels of cellular plasticity, manifesting in tumors that grow more quickly than solely epithelial or mesenchymal cell states and exhibit high levels of EMH. Moreover, this plasticity-induced heterogeneity plays a key role in the metastatic propensity and tumor-initiating potential of these clones. However, no one individual EMT state is capable of recapitulating the aggressive growth and decrease in survival of the parental line. The intermediate clones, upon xenotransplantation, experience a growth lag followed by an overdiversification, resulting in high EMH-scored tumors compared to the parental line, indicating that their high levels of plasticity enable them to attain higher levels of heterogeneity that propel tumor growth and metastasis. In contrast, the extreme clones exhibit less plasticity, taking longer periods of time to generate tumors that are less heterogeneous with weaker metastatic potential. The intermediate population in isolation appears to be the most potent tumor initiator; however, the presence of multiple states (i.e., heterogeneity) within the parental line imparts additional fitness that provides robust and exponential tumor growth. This suggests that the presence of heterogeneous subpopulations within a tumor confer a greater tumor growth advantage than the presence of more homogeneous subpopulations that exhibit higher levels of plasticity. A corollary assessment would be that a tumor benefits from harboring heterogeneous subpopulations, only a small subpopulation of which are required to exhibit high levels of plasticity. Future studies investigating the dynamics of expansion of this intratumoral heterogeneity may elucidate how EMT phenotypes work cooperatively to support tumor growth and progression to metastasis.
Through a multiomics approach, we identify distinct transcriptional programs across the EMT spectrum. The intermediate state was found to be maintained and stabilized by a subset of transcription factors, including the RUNX family. Knockout of the coactivator of this RUNX family of transcription factors, CBFβ, results in decreased expression of all RUNX TFs in the intermediate EMT clones that leads to tumors that metastasize at lower rates as a result of reduced heterogeneity and increased presence of epithelial cells within the tumor. These observations are in line with other studies that have outlined a role for RUNX2 in metastasis (40) but provide additional granularity to this work by identifying specific cell states that benefit from the presence of active RUNX-CBFβ. This study also underscores the importance of combining the study of the transcriptional and chromatin state of cells as a means of uncovering their underlying regulatory networks, which particularly enabled the delineation between similar EMT states; the transcriptional analysis of these clones alone was not sensitive enough to identify differences in many of the canonical EMT transcription factors, let alone other less variable TFs. This is likely because TF gene expression is tightly regulated by multiple factors (64), and differences in TF activity would benefit from a higher-resolution study at the chromatin level.
Last, we develop an approach to quantifying EMH and “EMTness” within human tumors, the former showing promise in its ability to inform disease prognosis. A previous work has sought to elucidate phenotypic intratumoral heterogeneity in a manner of different contexts and analyses using an array of multiplexed staining methods (65, 66) as well as through single-cell approaches (67). Here, we sought to specifically delineate intratumoral heterogeneity across the EMT spectrum, which has strong prognostic value for predicting invasion and metastasis in our model. A significant challenge in the quantification of EMH has been the ability to discern carcinoma cells that exhibit mesenchymal traits from stromal cell types such as fibroblasts that express similar markers. Previous approaches to quantifying EMH have considered morphological features (68) and analyzed gene expression profiles from publicly available datasets that identify cells that have undergone EMT (26, 27). These approaches, while being highly useful to study tumor cell EMT status, have been unable to segregate stromal infiltrates and their contributions to aggregate EMT scoring. Our multiplexed immunostaining approach, which uses a set of six EMT markers to assess EMH and EMT score, incorporates a segmentation step, which ensures that the quantification excludes stromal elements. We were able to distinguish eight phenotypes within these EMT clone-derived tumors with high reproducibility, which were all present in a cohort of patient tumors, validating our approach to quantifying EMT in patient samples. We found that patients with high heterogeneity and overall intermediate EMT state had significantly worse overall survival than any other group, independent of patient age and clinical subtype. Notably, CBFβ expression alone was not a successful predictor of overall survival in this patient sample cohort, emphasizing the need for a more nuanced metric, such as the one presented here, that uses a combination of markers to assess tumor diversity and complexity. Moreover, the inability of a key regulator of tumorigenic and metastatic potential such as CBFβ to predict survival is in line with studies that identify many such proteins that play key roles in oncogenesis and remain important drug targets but are not associated with shorter survival times (69). Thus, identifying novel ways of assessing heterogeneity and EMT parameters within a patient tumor through the use of combinatorial predictive biomarkers could prove useful in the clinical assessment these features and inform therapeutic decision-making.
MATERIALS AND METHODS
Cell culture
The human-derived SUM149PT cell line was obtained from the Weinberg laboratory, which, in turn acquired it from S. P. Ethier (Michigan). All derivative cell lines were maintained in an F12 medium (Gibco, #11765-054) supplemented with 5% fetal bovine serum (FBS) (Gibco, #10438-026), insulin (1 mg/ml) (Gibco, #12585-014), hydrocortisone (1 mg/ml) (Sigma-Aldrich, #H4001), and 5% penicillin-streptomycin (Corning, #10-002-cl). All cell lines were incubated at 37°C with 5% CO2-air atmosphere with constant humidity. Cells were passaged with 0.25% trypsin (Corning, 25-053-Cl); passage number was kept on all cell lines, and cultures were discarded past a total of 20 passages to maintain their respective EMT phenotypes. The 293T cell line was maintained in Dulbecco’s modified Eagle’s medium + 10% FBS (Gibco, #10438-026) + 5% penicillin-streptomycin (Corning, #10-002-cl).
Lentiviral vectors
Lentivirus was made with 293T cells plated at 60% confluency in 10-cm tissue culture–treated plates and transfected using X-tremeGENE HP DNA Transfection Reagent (Sigma-Aldrich, # 6365779001) with lentiviral vectors, psPAX2 (1.5 μg; Addgene, #12260), pCMV-VSV-G (1.5 μg; Addgene, #8454), and pcDNA3–enhanced green fluorescent protein (0.5 μg; Addgene, #13031) plus the lentiviral vector of interest (3 μg). The supernatant was collected at 48 and 72 hours after transfection, concentrated using a Lenti-X concentrator (TakaraBio, #631232), and titer was determined with Lenti-X GoStix (TakaraBio, #631243).
Cell line generation
The parental cell line SUM149PT was maintained in standard media. To generate single-cell clones, fluorescence-activated cell sorting (FACS) was performed on SUM149PT with the FACSAria III Cell Sorter. Cells were stained with CD44-PeCy7 (1:100; BioLegend, 103030), CD104-APC (1:200; Invitrogen, #50-1049-82), or Epcam-BV510 (1:100; BioLegend, #324235) for 30 min on ice before the addition of 4′,6-diamidino-2-phenylindole (DAPI) (1:1000; Sigma-Aldrich 10236276001; 10 mM stock). Gating and compensation were done on single-stained controls, and cells were sorted into collection tubes and immediately plated at a dilution of 0.5 cells per well into a 96-well plate. Single-cell clones were then expanded and assessed for EMT characteristics.
ZsGreen-expressing cells
All SUM149PT clones and the parental line were infected with high-titer pHIV-Luc-ZsGreen (Addgene, #39196) virus so as to generate ZsGreen and luciferase-expressing tumor cells for metastasis tracking in mouse. A total of 6 × 105 cells were infected in six-well plates with 125 μl of high-titer virus in standard media with Polybrene (5 μg/ml) (Sigma-Aldrich). Media were changed after 24 hours, and cells were allowed to expand for 48 hours before sorting for ZsGreen-positive population on the FACSAria III Cell Sorter, as above.
Flow cytometry
Flow experiments were performed in the same manner as above, on a 10-color Gallios FACS cytometer (Beckman Coulter). Compensation, file analysis, and plot generation were conducted using FlowJo (BD Biosciences).
Reverse transcription quantitative polymerase chain reaction
RNA was harvested from six-well plates of cells at confluency, extracted using the Qiagen RNeasy Plus Kit (Qiagen, 74034) and quantified using a NanoDrop (Thermo Fisher Scientific, ND-2000-US-CAN). Reverse transcription polymerase chain reaction (RT-PCR) (Applied Biosystems, #4368814) was performed to generate complementary DNA (cDNA), and Power SYBR Green PCR Master Mix (Applied Biosystems) was used for quantitative polymerase chain reaction (qPCR) (Box 1).
Western blot
For Western blot, lysates were collected on-plate with 1× radioimmunoprecipitation assay buffer (EMD Millipore, #20-188) with protease and phosphatase inhibitors (Thermo Fisher Scientific, #1861280). Lysates were sonicated and cleared before quantification with a Bradford Protein assay (Bio-Rad) and loaded at 50 μg per lane and run on a NuPage bis-tris gel (Thermo Fisher Scientific) and transferred to nitrocellulose membrane with the iBlot semidry transfer system (Thermo Fisher Scientific) and blocked in 5% milk in Tris Buffered Saline + Tween 20 (TBST) before staining with fibronectin (BD Biosciences, #610078; 1:10,000), ZEB1 (LSBio, #LS-C288694; 1:2000), E-cadherin (BD Biosciences, #610182; 1:1000), vimentin [Cell Signaling Technology (CST), #5741; 1:2000], RUNX1 (CST, #4336; 1:2000), RUNX2 (CST, #12556; 1:2000), RUNX3 (CST, #9647; 1:2000), CBFb (Abcam, ab33516; 1:2000), Snail (CST, #3879; 1:1000), Twist1/2 (Abcam, ab50887: 1:50), and CoxIV (CST, #11967; 1:2000) overnight in 5% milk in TBST. LI-COR secondary antibodies, IRDye goat anti-rabbit and goat anti-mouse, 800CW (LI-COR, #925-32219), and 680RD (LI-COR, #925-68076), were applied at 1:10,000 for 1 hour at room temperature in 5% milk in TBST before imaging on the LI-COR Odyssey CLx Digital Imager.
Transwell assay
Transwell assays were conducted using Costar Transwell plates (#3422, 8.0 μm) in triplicate. For migration assays, 2.5 × 105 cells were added to the top of each well in 10% complete medium, with 100% complete medium beneath the transwell. Cells were incubated for 16 to 18 hours at 37°C. Media were aspirated, and cells were permeabilized with 100% methanol for 5 min at room temperature, followed by staining with crystal violet (0.5% crystal violet in 20% methanol). Transwells were imaged on a Nikon Eclipse TS100 microscope, and migrated cells were counted using ImageJ. Invasion assays were conducted as above on plates that were coated with 100 μl of 5% Matrigel to the top of each transwell and allowed to set at 37°C for 2 hours before seeding.
RUNX2 and CBFβ knockout
CRISPR-Cas9–mediated knockout was achieved through lentiviral infection of lentiCRISPR V2 (Addgene, #52961) containing guides targeting RUNX2, CBFβ, or a LacZ nontargeting control guide (Box 2, sequence acquired from Sigma predesigned CRISPR guide RNA). Constructs were confirmed by Sanger sequencing and used as vectors in lentivirus production as stated above. Cells were infected with 1/8th lentiviral product from a 10-cm plate in six-well plates. Media were changed after 24 hours, and puromycin selection (2 μg/ml) was applied at 48 hours until stable cell lines were generated. Cell lines were checked for knockout by Western blot, and LacZ #1, RUNX2 #2, and CBFb #2 were used for further analysis based on the level of knockout. Cell lines used for Western blotting are as described above, and orthotopic injection into the mammary fat pad of NSG mice is as described below.
Cell imaging
Bright field
Images were taken on a Nikon Eclipse TS100 microscope under ×20 magnification to determine cell morphology.
Immunofluorescence
Cells were grown to 60% confluency in chamber slides (Falcon 354118) with standard media. Cells were fixed and permeabilized before staining with primary antibody (vimentin, CST; 1:100; and E-cadherin, BD Biosciences; 1:100) overnight, followed by secondary antibody (anti-rabbit, Thermo Fisher Scientific, #31466; 1:10,000; anti-mouse, Thermo Fisher Scientific, #31431; 1:10,000) for 1 hour. Slides were washed and mounted using ProLong Diamond (Invitrogen, P36961) before imaging on Zeiss LSM 800 with Airyscan (63×).
In vivo studies
Cell lines were resuspended in 30% Matrigel (VWR, 47743-706) and injected in limiting dilutions (250,000, 25,000, and 2500 cells per flank) orthotopically into the inguinal mammary fat pat (no. 4) of NOD scid interleukin-2Rγnull (stock no. 005557, the Jackson Laboratory). Tumor growth was monitored weekly, and tumor volume was measured along three axes with calipers (VWR, 62379-531). Tumors and lungs were harvested at the time of tumor burden (total tumor volume of 2 cm3) and fixed overnight with 10% neutral-buffered formalin. Tumor growth curves and survival were statistically analyzed using TumGrowth (31). Tumor-initiating potential was calculated with extreme limiting dilution analysis (70).
Late metastasis models were obtained through primary tumor resection at 1 cm3. Mice were allowed to recover, and lungs were harvested at 2.5 months after surgery. This interval was determined by noticeable recurrence at the ipsilateral site and deteriorating health in the mice.
Tumor and lung staining
Tumors and lungs were extracted at the time of tumor burden (total tumor volume of 2 cm3) and fixed overnight with 10% neutral-buffered formalin. All samples were then processed and stained for H&E by the Dartmouth Hitchcock Pathology Shared Resource. Lungs bearing ZsGreen-positive EMT clones were counted by eye on a Nikon Eclipse TS100 microscope, and select images were taken at ×74 magnification.
Metastasis counting
H&E-stained lung slides were scanned on a PerkinElmer Vectra3 slide scanner at 10× and counted by eye for micrometastatic (>10 adjacent cells) and macrometastatic (10+ adjacent cells) tumors.
Whole-exome sequencing
Whole-exome sequencing and subsequent single-nucleotide polymorphism (SNP) and insertion and deletion (INDEL) alignment and discovery were performed by BGISeq on all EMT clones. They obtained 9242.46 Mb raw bases. After removing low-quality reads, we obtained, on average, 91,977,846 clean reads (9197.79 Mb). The clean reads of each sample had high Q20 and Q30, which showed high-sequencing quality. The average GC content was 50.63%. Reads were aligned with a Burrows-Wheeler Aligner. The HaploTypeCaller of GATK (v3.6) was used to call and identified 190,503 SNPs and 33,385 INDELs between all samples. SNPs and INDELs for each clone were tested against the consensus set for clone E to determine possible genetic mutations with a Fisher’s exact test and plotted as odds ratios.
TCGA survival analysis
The results shown here are in whole based on data generated by the TCGA Research Network: https://cancer.gov/tcga using the TCGA-BRCA cohort of patient samples. CBFβ raw counts were normalized using variance-stabilizing transformation, and subjects were stratified into high- versus low-expression groups based on 50th percentile CBFb (ENSG00000067955). Expression was modeled as a continuous variable in the Cox proportional hazards model adjusting for age, stage (low versus high), and molecular subtype (PAM50).
RNA-seq data processing
RNA was collected using a Qiagen RNeasy plus kit (Qiagen, 74034) and quantified using a NanoDrop (Thermo Fisher Scientific, ND-2000-US-CAN). Library preparation was performed with the Kapa mRNA HyperPrep Kit.
The quality of raw single-end RNA-seq data was confirmed using FastQC (v0.11.8) (71) before read trimming of polyA sequences and low-quality bases using Cutadapt (v2.4) (72). Reads were aligned to human genome hg38 using STAR (v2.7.2b) (73) with parameters “--outSAMattributes NH HI AS NM MD --outFilterMultimapNmax 10 --outFilterMismatchNmax 999 --outFilterMismatchNoverReadLmax 0.04 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1.” The quality of the alignments was assessed using CollectRNASeqMetrics [Picard Tools (74)], and duplicate reads were identified (but retained) with MarkDuplicates [Picard Tools (74)]. Gene-level abundance estimates were generated using RSEM (v1.3.2) (75) using the rsem-calculate-expression command with the parameters “--strandedness reverse --fragment-length-mean 313 --fragment-length-sd 91.”
Downstream RNA-seq data analysis
Gene-level abundance estimates generated with RSEM were imported into R and analyzed using the R package DESeq2 (76). To perform exploratory analysis of global transcriptional profiles, abundance was transformed using the regularized logarithm approach (76) implemented in the R package DESeq2, and the top 500 most variable genes across all clones were supplied to the prcomp() command in R to perform PCA. The 500 most variable genes were also used to perform unsupervised hierarchical clustering with the R package ComplexHeatmap. A differential expression analysis was performed on the raw gene-level abundance estimates assuming a negative binomial distribution, with clone E used as the reference group in all comparisons. Gene-wise dispersion estimates were reviewed in all analyses to confirm that the selected model was an appropriate fit for the data. Genes with a Benjamini-Hochberg adjusted P value <0.05 (Wald test) were considered statistically significant. GSEA was done on differential gene lists for each clone relative to clone E using the clusterProfiler (32) to determine overlaps with the hallmark gene sets at a threshold of 0.05.
ATAC-seq data processing
Tagmented DNA and library prep for ATAC sequencing was performed according to the protocol detailed in Buenrostro et al. (35). Before analysis, the quality of raw DNA sequences (in FASTQ format) was confirmed using FastQC (v0.11.8) (71). ATAC-seq data were then processed using the publicly available ENCODE ATAC-seq pipeline (https://encodeproject.org/pipelines/ENCPL792NWO/), and relevant commands and options used are described in detail below. Illumina adapter and transposase sequences were trimmed using Cutadapt (v1.9.1) (72) with parameters “--minimum-length 5 -e 0.1.” Trimmed reads were then aligned to human genome hg38 using Bowtie2 (v2.2.6) (77) in “--local” mode with parameters “-X 2000 -k 2.” Duplicate reads were identified using MarkDuplicates [Picard Tools (74)] and filtered from final alignments, in addition to unmapped reads and reads aligning to mitochondrial DNA, retaining only alignments formed by properly paired reads. For multimapping reads, one paired-end alignment was randomly selected as the primary alignment, while the remaining alignments were discarded. Alignments (in BAM format) were converted to tagAlign files and shifted +4 base pairs (bp) and −5 bp on the + and – strands, respectively, to account for insertion of adapter sequences by Tn5 transposase. Peaks were called for each replicate using the MACS2 (v2.1.1) (78) callpeak command with parameters “--shift -75 --extsize 150 --nomodel --keep-dup all --call-summits -p 1.0E-10” and filtered against the ENCODE hg38 blacklist. The irreproducible discovery rate (IDR) method was used to identify a set of reproducible peaks across biological replicates using an IDR threshold of 0.05. For visualization purposes, position-shifted (to account for Tn5 insertion) BAM files for biological replicates were merged MergeSamFiles [Picard Tools; (74)] and used to generate counts per million (CPM)–normalized signal tracks (in BigWig format) via the deepTools (v3.4.3) (79) bamCoverage command with parameters “--binSize 5 --normalizeUsing CPM --effectiveGenomeSize 2913022398 --ignoreForNormalization chrX.” Heatmaps of normalized Tn5 insertions in peak regions or specific regions were generated using deepTools commands computeMatrix and plotHeatmap.
Several metrics were used to confirm ATAC-seq data quality. To confirm whether sequencing libraries were of sufficient complexity, three specific quality control metrics were evaluated: nonredundant fraction (number of uniquely mapping reads/total read number), PCR bottlenecking coefficient 1 (PBC1, number of genomic positions with at least one read mapped/number of distinct genomics position to which a read maps uniquely), and PCR bottlenecking coefficient 2 (PBC2, number of locations where one read maps uniquely/number of genomic regions where two reads map uniquely). Fragment length distributions were generated and reviewed in R using the “Rsamtools” package. The fraction of reads in nucleosome-free regions (NFRs) was calculated to confirm that a sufficient fraction of reads were located in NFRs. Fraction of reads in peak regions (FRiP score) was calculated to assess the quality of the final IDR peak set. Enrichment of accessibility over transcriptional start sites (TSSs), calculated as the maximum number of normalized Tn5 insertions across a ±2-kb region flanking hg38 TSS regions, was used to confirm data quality in a peak agnostic fashion.
Downstream ATAC-seq data analysis
Basic peak annotation was performed using the annotatePeak() function from the ChIPseeker (80) package, using a range of ±3 kb to define promoter-associated regions. The R-package TxDb.Hsapiens.UCSC.hg38.knownGene was used to define gene models and coordinates of genomic features. To create a set of consensus peaks, the IDR peak sets for each sample groups were merged using the R package GenomicRanges. Tn5 insertions occurring in each peak of the consensus peak set were counted from position-shifted BAM files using the featureCounts() function (from the Rsubread package) with options “isPairedEnd = TRUE, countMultiMappingReads = FALSE.” To perform exploratory analyses of global chromatin accessibility profiles, raw counts were transformed using the regularized logarithm approach (76) implemented in the R package DESeq2. PCA was performed on the 3000 most variable consensus peak regions using the prcomp() function in R. The most variable peaks were defined as those with the greatest SD across all samples. Unsupervised hierarchical clustering was performed with the R package ComplexHeatmap, also using the 3000 most variable consensus peaks. Differential accessibility analysis of the consensus peak was also performed using the R package DESeq2, modeling raw counts using a negative binomial distribution, with clone E used as the reference group in all comparisons. Peaks with a Benjamini-Hochberg–corrected P value <0.05 (Wald test) were considered statistically significant.
Enrichment of Transcription Factor Binding Sites in clone-specific peak sets
To identify potential TFs responsible for mediating clone-specific phenotypes, we tested the differentially accessible peaks between clone E and each clone for overrepresentation of TF binding site motifs. We first restricted each peak set to regions that demonstrated statistically significant increases in chromatin accessibility compared to clone E (see description of differential accessibility analyses above) and scanned these peaks for TF motif occurrences using the R package motifmatchr (36). Position frequency matrices for human TF motifs used as input to motifmatchr were downloaded from the JASPAR database (81) using R packages JASPAR2018 and TFBSTools (82). Overrepresented TF motifs in each peak set were identified through hypergeometric testing using the R function phyper(), with all peaks identified in that clone used as the background set. TF motifs with a Bonferroni-corrected hypergeometric P value <0.05 were deemed as overrepresented. To identify potential groups of coordinately regulated TFs across the respective clones, −log10-transformed P values from hypergeometric testing were subjected to hierarchical clustering and visualized using the R package pheatmap. To prevent extreme motif enrichments from dominating the heatmap scale, −log10-transformed P values were capped at a maximum value of 20 (highlighted with an asterisk).
Differential TF activity analyses
Differential TF activity between single cell–derived clones, as well as TF mode of action (i.e., activator and repressor), was estimated using diffTF (43). Briefly, when used with ATAC-seq data, diffTF computes the fold change in chromatin accessibility between two conditions at each binding site of a given TF, and the distribution of fold changes is compared to a set of background fold-change values to assess statistical significance of differences in TF activity between the conditions. DiffTF was used in conjunction with matched RNA-seq (classification mode) to classify each TF into one of the four modes of action (activator, repressor, not expressed, and undetermined) through correlation of TF expression levels with target site accessibility. DiffTF was used with options “pairedEnd” and “RNASeqIntegration” set to “true,” with all remaining options using default settings. In silico–predicted binding sites based on the HOCOMOCO v11 database (83) and PWMScan (84) for hg38 across 768 human TFs was used to define the atlas of TFBS for diffTF analyses. To concentrate on the TFs with the most confidently estimated TF activity scores (weighted_meanDifference), we restricted our downstream analysis to TFs that achieved an adjusted P value of 1 × 10−15. To identify modules of cooperatively regulated TFs, unsupervised hierarchical clustering was performed on the diffTF activity scores using R package ComplexHeatmap.
Multiplexed TSA staining
Tumors were selected from each EMT clone at approximately 1 cm3 and stained with (in order) Snail (CST, #3895; 1:400), KRT8 (Invitrogen, PA5-29607; 1:300), KRT14 (Invitrogen, MA5-11599; 1:1000), vimentin (CST, #5741; 1:500), E-cadherin (BD Biosciences, #610182; 1:500), and ZEB1 (Invitrogen, PA5-82982; 1:1000). Antibody optimization and multiplexed staining were done according to the PerkinElmer OPAL Assay Development Guide (August 2017) and previous literature (52, 85). Briefly, slides were baked to remove paraffin wax and then sequentially washed with xylene and rehydrated with decreasing concentrations of ethanol and, finally, ddH2O before blocking. Then, slides were incubated with primary antibody and then secondary antibodies for 30 min at room temperature. Following washes, the selected OPAL fluorophore was applied to slides for precisely 6 min at room temperature in the dark and washed off, and slides were microwaved at 20% power for 15 min to affix OPAL to target regions and remove primary and secondary antibodies. Slides were blocked again, the staining process was repeated for each marker (Box 3), and, last, spectral DAPI (PerkinElmer, two drops/ml) was added before mounting on coverslips with ProLong Diamond (Invitrogen, P36961).
Image analysis
Whole-slide scans were captured at 10× with the PerkinElmer Vectra3 Slide Scanner, and ~50 ROIs per tumor were chosen manually with PhenoChart (PerkinElmer). ROIs were imaged at 20× resolution and imported into InForm analysis software (PerkinElmer). Spectral unmixing single stains and background fluorescence slides were generated from the parental tumor according to the OPAL Assay Development Guide. ROIs were spectrally unmixed and assigned colors and exported as composite images (Fig. 4A). Tissue segmentation (trainable to 98% accuracy) and cell segmentation were performed (nuclear compartment —DAPI; cytoplasm—vimentin and KRT8; and membrane—E-cadherin), and cells were phenotyped on the basis of expression of one or multiple markers [E-cadherin only, KRT8/14 and E-cadherin, KRT8 and/or KRT14, triple positive (KRT8 + E-cadherin + vimentin), KRT8/14 and vimentin, Snail only, vimentin only, and vimentin + ZEB1] and validated by marker distribution (fig. S5, A and B). Entire cell mean fluorescent units were extracted for each marker and normalized as a percentile of maximum and minimum fluorescence across all cells in all images.
Heterogeneity and EMT scores
Approach 1: Heterogeneity scores were generated using penalized logistic regression based on entropies of mean marker cell expressions to identify markers and cellular compartments (nucleus, cytoplasm, and membrane) that contributed most to the variability in the ranked tumor images (fig. S5C). In total, 134 entropy-based features were extracted, and 13 of them were selected by recursive feature elimination (86) as the most relevant. Logistic regression classified sample heterogeneity into levels mid, low, and high. Ground truths were determined from the rubric: low (one major cell trait with up to one minor trait), mid (two major cell traits with up to three minor traits), and high (three or more major cell traits present with two or more minor traits). These were used to train and validate the algorithm using 70% training and 30% test images (n = 409) in a fivefold cross-validation.
Approach 2: Nearest neighbor analysis was conducted with the scikit-learn Python package (55) using cell phenotypes determined from InForm. Similar feature selection methods were applied to nearest neighbors, with 26 of 49 features selected.
Approach 3: A hybrid approach used combined the 134 and 49 features from approaches 1 and 2 and selected 18 of 183 features.
To generate the EMT score, the seven derived phenotypes were weighted from epithelial to mesenchymal (E-cadherin only −3, KRT8 and E-cadherin −2, KRT14 only −1, triple positive +1, Snail only +2, vimentin only +3, and vimentin and ZEB1 + 4) and applied to a multivariate logistic regression. The code is available in GitHub (https://github.com/BMIRDS/cell-heterogeneity-emtscore).
Human patient tumors
Human patient samples were obtained from the CHTN CPD Breast Cancer Stage III Prognostic Tissue Microarray. Details on this tissue microarray can be found at https://chtn.org/. TMAs were stained as detailed above and unmixed as described. After QC, 124 sample cores were used for further analysis including phenotyping, as well as heterogeneity and EMT score calculation, both described above. Hazard ratios were calculated with a multivariate Cox proportional hazard model, adjusting for patient age and tumor hormone status (HR+ or HR−).
CBFb staining was conducted on sequential CHTN TMAs, as above, by immunohistochemical methods (Abcam, ab33516; 1:2000). Following staining, TMAs were scored by a licensed pathologist and marked as negative, weak, moderate, or strong for nuclear and/or cytoplasmic staining, as well as percentage of cells per core. H scores, such as those used to define ER expression (60), were calculated using the strength of expression (negative = 0, weak = 1, moderate = 2, and strong = 3) multiplied by percentage of positively stained cells (top estimate). Hazard ratios are calculated as above.
Survival analysis
Survival analysis was performed on patient data gathered from the CHTN. Overall survival was used to plot Kaplan-Meier and Cox proportional hazard models adjusting for patient age and patient subtype (HR+ or HR−). HR status was determined from ER and PR score (negative or positive) based on ASCO CAP Guidelines for ER and PR scoring.
Research animals
All animal experiments were carried out under ethical regulations approved by the Dartmouth College Institutional Animal Care and Use Committee.
Acknowledgments
We thank J. Fields, R. O’Meara, R V. Stan, W. Croteau, and F. W. Kolling IV for technical assistance; Molecular Biology Shared Resource at the Norris Cotton Center for assistance with RNA-seq and ATAC-seq assays; and Dartlab (flow cytometry) microscopy and pathology shared resources at the Norris Cotton Cancer Center.
Funding: Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (under grant number P20GM104416), a Prouty Pilot Grant from Friends of the Norris Cotton Cancer Center, funding from The Elmer R. Pfefferkorn & Allan U. Munck Education and Research Fund at the Geisel School of Medicine at Dartmouth, and an NCI Cancer Center Support Grant (5P30CA023108-40). O.M.W. was supported in part by a Cancer Center Core Grant (P30CA023108) from the National Cancer Institute and the National Institutes of Health–funded Center for Quantitative Biology at Dartmouth (COBRE, 5P20GM130454-03). This work was also supported by a Ramanujan Fellowship (SB/S2/RJN-049/2018) awarded by the Science and Engineering Research Board (SERB), the Department of Science and Technology (DST), the Government of India (to M.K.J.), and funding from the NIH 5R00CA201574-05 and 1R01CA267691-01 (to D.R.P).
Author contributions: This project was conceived by M.S.B., N.B.O., and D.R.P. N.B.O. and M.S.B. created the single-cell EMT clones, which was the starting point for the project. H.L. contributed to the generation of knockout cell lines. M.S.B. carried out all experiments except for generation of the entropy-based model for deriving heterogeneity scores and EMT scores, which was carried out by B.A. and S.H. RNA-seq and ATAC-seq data analysis was carried out by O.M.W. The EMT scoring was performed by P.C. and M.K.J. Patient survival and hazard model was performed under consultation with B.C.C. The manuscript was written by M.S.B. and D.R.P. with input and edits from all coauthors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data from RNA-seq and ATAC-seq experiments that support the findings in this study have been deposited and can be accessed in GEO (accession number GSE172613).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Thiery J. P., Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Easwaran H., Tsai H. C., Baylin S. B., Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 54, 716–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattabiraman D. R., Weinberg R. A., Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 13, 497–512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ognjenovic N. B., Bagheri M., Mohamed G. A., Xu K., Chen Y., Saleem M. A. M., Brown M. S., Nagaraj S. H., Muller K. E., Gerber S. A., Christensen B. C., Pattabiraman D. R., Limiting self-renewal of the basal compartment by pka activation induces differentiation and alters the evolution of mammary tumors. Dev. Cell 55, 544–557.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibue T., Weinberg R. A., EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Huang R. Y.-J., Wong M. K., Tan T. Z., Kuay K. T., Ng A. H. C., Chung V. Y., Chu Y. S., Matsumura N., Lai H. C., Lee Y. F., Sim W. J., Chai C., Pietschmann E., Mori S., Low J. J. H., Choolani M., Thiery J. P., An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to E-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis. 4, e915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., De Clercq S., Minguijón E., Balsat C., Sokolow Y., Dubois C., De Cock F., Scozzaro S., Sopena F., Lanas A., D’Haene N., Salmon I., Marine J.-C., Voet T., Sotiropoulou P. A., Blanpain C., Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jolly M. K., Boareto M., Huang B., Jia D., Lu M., Ben-Jacob E., Onuchic J. N., Levine H., Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front. Oncol. 5, 155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrix M. J. C., Seftor E. A., Seftor R. E. B., Trevor K. T., Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am. J. Pathol. 150, 483–495 (1997). [PMC free article] [PubMed] [Google Scholar]
- 11.Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A., The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morel A.-P., Lièvre M., Thomas C., Hinkal G., Ansieau S., Puisieux A., Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLOS ONE 3, e2888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosse-Wilde A., d’Hérouël A. F., McIntosh E., Ertaylan G., Skupin A., Kuestner R. E., del Sol A., Walters K. A., Huang S., Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLOS ONE 10, e0126522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bierie B., Pierce S. E., Kroeger C., Stover D. G., Pattabiraman D. R., Thiru P., Liu Donaher J., Reinhardt F., Chaffer C. L., Keckesova Z., Weinberg R. A., Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 114, E2337–E2346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolly M. K., Somarelli J. A., Sheth M., Biddle A., Tripathi S. C., Armstrong A. J., Hanash S. M., Bapat S. A., Rangarajan A., Levine H., Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 194, 161–184 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Kröger C., Afeyan A., Mraz J., Eaton E. N., Reinhardt F., Khodor Y. L., Thiru P., Bierie B., Ye X., Burge C. B., Weinberg R. A., Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116, 7353–7632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüönd F., Sugiyama N., Bill R., Bornes L., Hager C., Tang F., Santacroce N., Beisel C., Ivanek R., Bürglin T., Tiede S., Rheenen J., Christofori G., Distinct contributions of partial and full EMT to breast cancer malignancy. Dev. Cell 56, 3203–3221.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Grande M. T., Sánchez-Laorden B., López-Blau C., de Frutos C. A., Boutet A., Arévalo M., Rowe R. G., Weiss S. J., López-Novoa J. M., Nieto M. A., Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 21, 989–997 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Krebs A. M., Mitschke J., Lasierra Losada M., Schmalhofer O., Boerries M., Busch H., Boettcher M., Mougiakakos D., Reichardt W., Bronsert P., Brunton V. G., Pilarsky C., Winkler T. H., Brabletz S., Stemmler M. P., Brabletz T., The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19, 518–529 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Li Q.-Q., Xu J. D., Wang W. J., Cao X. X., Chen Q., Tang F., Chen Z. Q., Liu X. P., Xu Z. D., Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin. Cancer Res. 15, 2657–2665 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Guo W., Keckesova Z., Donaher J. L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zürrer-Härdi U., Bell G., Tam W. L., Mani S. A., van Oudenaarden A., Weinberg R. A., Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forozan F., Veldman R., Ammerman C. A., Parsa N. Z., Kallioniemi A., Kallioniemi O. P., Ethier S. P., Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br. J. Cancer 81, 1328–1334 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.F. M. Robertson, K. Chu, Genomic profiling of pre-clinical models of inflammatory breast cancer identifies a signature of epithelial plasticity and suppression of TGFβ signaling. J. Clin. Exp. Pathol. 2, 119 (2012).
- 24.Jolly M. K., Celià-Terrassa T., Dynamics of phenotypic heterogeneity associated with EMT and stemness during cancer progression. J. Clin. Med. 8, 1542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byers L. A., Diao L., Wang J., Saintigny P., Girard L., Peyton M., Shen L., Fan Y., Giri U., Tumula P. K., Nilsson M. B., Gudikote J., Tran H., Cardnell R. J. G., Bearss D. J., Warner S. L., Foulks J. M., Kanner S. B., Gandhi V., Krett N., Rosen S. T., Kim E. S., Herbst R. S., Blumenschein G. R., Lee J. J., Lippman S. M., Ang K. K., Mills G. B., Hong W. K., Weinstein J. N., Wistuba I. I., Coombes K. R., Minna J. D., Heymach J. V., An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 19, 279–290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan T. Z., Miow Q. H., Miki Y., Noda T., Mori S., Huang R. Y. J., Thiery J. P., Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 6, 1279–1293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George J. T., Jolly M. K., Xu S., Somarelli J. A., Levine H., Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 77, 6415–6428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty P., George J. T., Tripathi S., Levine H., Jolly M. K., Comparative study of transcriptomics-based scoring metrics for the epithelial-hybrid-mesenchymal spectrum. Front. Bioeng. Biotechnol. 8, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blick T., Widodo E., Hugo H., Waltham M., Lenburg M. E., Neve R. M., Thompson E. W., Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin. Exp. Metastasis 25, 629–642 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Cook D. P., Vanderhyden B. C., Context specificity of the EMT transcriptional response. Nat. Commun. 11, 2142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enot D. P., Vacchelli E., Jacquelot N., Zitvogel L., Kroemer G., TumGrowth: An open-access web tool for the statistical analysis of tumor growth curves. Onco. Targets. Ther. 7, e1462431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F., Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Gazzar A., Cai X., Reeves R. S., Dai Z., Caballero-Benitez A., McDonald D. L., Vazquez J., Gooley T. A., Sale G. E., Spies T., Groh V., Effects on tumor development and metastatic dissemination by the NKG2D lymphocyte receptor expressed on cancer cells. Oncogene 33, 4932–4940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G., Wang L. G., Han Y., He Q. Y., ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Schep, motifmatchr: Fast Motif Matching in R. R package version 1.12.0. (2020).
- 37.Ito Y., Bae S.-C., Shyue L., Chuang H., The RUNX family: Developmental regulators in cancer. Nat. Publ. Gr. 15, 81–95 (2015). [DOI] [PubMed] [Google Scholar]
- 38.O.-Otálora B. A., Henríquez B., L.-Kleine L., Rojas A., RUNX family: Oncogenes or tumor suppressors (Review). Oncol. Rep. 42, 3–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blyth K., Cameron E. R., Neil J. C., The RUNX genes: Gain or loss of function in cancer. Nat. Rev. Cancer 5, 376–387 (2005). [DOI] [PubMed] [Google Scholar]
- 40.LaFave L. M., Kartha V. K., Ma S., Meli K., Priore I. D., Lareau C., Naranjo S., Westcott P. M. K., Duarte F. M., Sankar V., Chiang Z., Brack A., Law T., Hauck H., Okimoto A., Regev A., Buenrostro J. D., Jacks T., Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell 38, 212–228.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran R., Harrison H., Ariffin N. S., Ayub R., Pegg H. J., Deng W., Mastro A., Ottewell P. D., Mason S. M., Blyth K., Holen I., Shore P., A role for CBFβ in maintaining the metastatic phenotype of breast cancer cells. Oncogene 39, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaraja G., Othman M., Fox B. P., Alsaber R., Pellegrino C. M., Zeng Y., Khanna R., Tamburini P., Swaroop A., Kandpal R. P., Gene expression signatures and biomarkers of noninvasive and invasive breast cancer cells: Comprehensive profiles by representational difference analysis, microarrays and proteomics. Oncogene 25, 2328–2338 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Berest I., Arnold C., R.-Palomares A., Palla G., Rasmussen K. D., Giles H., Bruch P.-M., Huber W., Dietrich S., Helin K., Zaugg J. B., Quantification of differential transcription factor activity and multiomics-based classification into activators and repressors: DiffTF. Cell Rep. 29, 3147–3159.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Peinado H., Ballestar E., Esteller M., Cano A., Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24, 306–319 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aghdassi A., Sendler M., Guenther A., Mayerle J., Behn C. O., Heidecke C. D., Friess H., Büchler M., Evert M., Lerch M. M., Weiss F. U., Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 61, 439–448 (2012). [DOI] [PubMed] [Google Scholar]
- 46.McDonald L., Ferrari N., Terry A., Bell M., Mohammed Z. M., Orange C., Jenkins A., Muller W. J., Gusterson B. A., Neil J. C., Edwards J., Morris J. S., Cameron E. R., Blyth K., RUNX2 correlates with subtype-specific breast cancer in a human tissue microarray and ectopic expression of Runx2 perturbs differentiation in the mouse mammary gland. DMM Dis. Model. Mech. 7, 525–534 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fritz A. J., Hong D., Boyd J., Kost J., Finstaad K. H., Fitzgerald M. P., Hanna S., Abuarqoub A. H., Malik M., Bushweller J., Tye C., Ghule P., Gordon J., Frietze S., Zaidi S. K., Lian J. B., Stein J. L., Stein G. S., RUNX1 and RUNX2 transcription factors function in opposing roles to regulate breast cancer stem cells. J. Cell. Physiol. 235, 7261–7272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chimge N.-O., Baniwal S. K., Little G. H., Chen Y.-B., Kahn M., Tripathy D., Borok Z., Frenkel B., Regulation of breast cancer metastasis by Runx2 and estrogen signaling: The role of SNAI2. Breast Cancer Res. 13, 1–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busch E. L., Keku T. O., Richardson D. B., Cohen S. M., Eberhard D. A., Avery C. L., Sandler R. S., Evaluating markers of epithelial-mesenchymal transition to identify cancer patients at risk for metastatic disease. Clin. Exp. Metastasis 33, 53–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen B., Chen B., Zhu Z., Ye W., Zeng J., Liu G., Wang S., Gao J., Xu G., Huang Z., Prognostic value of ZEB-1 in solid tumors: A meta-analysis. BMC Cancer 19, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matysiak M., Kapka-Skrzypczak L., Jodłowska-Jędrych B., Kruszewski M., EMT promoting transcription factors as prognostic markers in human breast cancer. Arch. Gynecol. Obstet. 295, 817–825 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Roy S., Axelrod H. D., Valkenburg K. C., Amend S., Pienta K. J., Optimization of prostate cancer cell detection using multiplex tyramide signal amplification. J. Cell. Biochem. 120, 4804–4812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu B., Li C., Li Z., Wang D., Ren X., Zhang Z., An entropy-based metric for assessing the purity of single cell populations. Nat. Commun. 11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo M., Bao E. L., Wagner M., Whitsett J. A., Xu Y., SLICE: Determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res. 45, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay E., Scikit-learn: Machine Learning in Python. J. Mach. Learn Res. 12, 2825–2830 (2011). [Google Scholar]
- 56.Vega S., Morales A. V., Ocaña O. H., Valdés F., Fabregat I., Nieto M. A., Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 18, 1131–1143 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mejlvang J., Kriajevska M., Vandewalle C., Chernova T., Sayan A. E., Berx G., Mellon J. K., Tulchinsky E., Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Mol. Biol. Cell 18, 4615–4624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai J. H., Donaher J. L., Murphy D. A., Chau S., Yang J., Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim C., Gao R., Sei E., Brandt R., Hartman J., Hatschek T., Crosetto N., Foukakis T., Navin N. E., Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell , 879–893.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kinsel L. B., Szabo E., Greene G. L., Konrath J., Leight G. S., McCarty K. S. Jr., Immunocytochemical analysis of estrogen receptors as a predictor of prognosis in breast cancer patients: Comparison with quantitative biochemical methods. Cancer Res. 49, 1052–1056 (1989). [PubMed] [Google Scholar]
- 61.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A. J., Burdziak C., Moon K. R., Chaffer C. L., Pattabiraman D., Bierie B., Mazutis L., Wolf G., Krishnaswamy S., Pe’er D., Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espinosa Fernandez J. R., Eckhardt B. L., Lee J., Lim B., Pearson T., Seitz R. S., Hout D. R., Schweitzer B. L., Nielsen T. J., Lawrence O. R., Wang Y., Rao A., Ueno N. T., Identification of triple-negative breast cancer cell lines classified under the same molecular subtype using different molecular characterization techniques: Implications for translational research. PLOS ONE 15, e0231953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheel C., Eaton E. N., Li S. H. J., Chaffer C. L., Reinhardt F., Kah K. J., Bell G., Guo W., Rubin J., Richardson A. L., Weinberg R. A., Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145, 926–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.González A. J., Setty M., Leslie C. S., Early enhancer establishment and regulatory locus complexity shape transcriptional programs in hematopoietic differentiation. Nat. Genet. 47, 1249–1259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin J. R., Izar B., Wang S., Yapp C., Mei S., Shah P. M., Santagata S., Sorger P. K., Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackson H. W., Fischer J. R., Zanotelli V. R. T., Ali H. R., Mechera R., Soysal S. D., Moch H., Muenst S., Varga Z., Weber W. P., Bodenmiller B., The single-cell pathology landscape of breast cancer. Nature , 615–620 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Karaayvaz M., Cristea S., Gillespie S. M., Patel A. P., Mylvaganam R., Luo C. C., Specht M. C., Bernstein B. E., Michor F., Ellisen L. W., Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 9, 3588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leggett S. E., Sim J. Y., Rubins J. E., Neronha Z. J., Williams E. K., Wong I. Y., Morphological single cell profiling of the epithelial-mesenchymal transition. Integr. Biol. (United Kingdom) 8, 1133–1144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith J. C., Sheltzer J. M., Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep. 38, 110569 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu Y., Smyth G. K., ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 71.S. Andrews, FastQC: A quality control tool for high throughput sequence data [Online]. (2010).
- 72.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011). [Google Scholar]
- 73.Dobin A. et. al, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Broad Institute. Picard Toolkit. GitHub Repository (2019).
- 75.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y. et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramírez F. et al., deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu G., Wang L.-G., He Q.-Y., ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Khan A. et al., JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan G., Lenhard B., TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32, 1555–1556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kulakovskiy I. V. et al., HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ambrosini G., Groux R., Bucher P., PWMScan: a fast tool for scanning entire genomes with a position-specific weight matrix. Bioinformatics 34, 2483–2484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lazarus J. et al., Optimization, design and avoiding pitfalls in manual multiplex fluorescent immunohistochemistry. J. Vis. Exp. 2019, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guyon I., Weston J., Barnhill S., Vapnik V., Gene selection for cancer classification using support vector machines. Mach. Learn. 46, 389–422 (2022). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S3