Abstract
Transcriptional variability facilitates stochastic cell diversification and can in turn underpin adaptation to stress or injury. We hypothesize that it may analogously facilitate progression of premalignancy to cancer. To investigate this, we initiated preleukemia in mouse cells with enhanced transcriptional variability due to conditional disruption of the histone lysine acetyltransferase gene Kat2a. By combining single-cell RNA sequencing of preleukemia with functional analysis of transformation, we show that Kat2a loss results in global variegation of cell identity and accumulation of preleukemic cells. Leukemia progression is subsequently facilitated by destabilization of ribosome biogenesis and protein synthesis, which confer a transient transformation advantage. The contribution of transcriptional variability to early cancer evolution reflects a generic role in promoting cell fate transitions, which, in the case of well-adapted malignancies, contrastingly differentiates and depletes cancer stem cells. That is, transcriptional variability confers forward momentum to cell fate systems, with differential multistage impact throughout cancer evolution.
Loss of Kat2a enhances transcriptional variability of ribosome biosynthetic programs and transiently accelerates preleukemia.
INTRODUCTION
Tumors evolve by genetic drift and natural selection (1, 2). Acquisition of new mutations confers a probability of adaptation to new environmental pressures (3), facilitates progression and transformation of premalignant lesions, promotes metastasis, and drives treatment resistance (4). In recent years, it became apparent that nongenetic instability, in particular variability in methylation epialleles, can confer adaptive advantages to tumor growth and survival irrespective of mutations and function as driver of therapy resistance and disease relapse in hematological malignancies (5, 6). Hematological malignancies, and, in particular, acute myeloid leukemia (AML), are strongly dependent on epigenetic regulation, both through mutation of chromatin factors and by co-option of unmutated chromatin regulators into maintenance of leukemogenic programs (7–9). Notably, AML has lower levels of mutations than solid tumors, supporting the notion that nongenetic events may be especially important in the former (7). Akin to genetic instability, epigenetic variability is increased in leukemia initiation and relapse but low in leukemia maintenance (10, 11), suggesting that reconfiguration of molecular/transcriptional programs may perturb the identity or survival of well-adapted leukemia cells by disrupting pro-oncogenic molecular signatures. We have recently captured this phenomenon upon loss of KAT2A (lysine acetyltransferase 2A), a histone acetyltransferase that promotes gene transcription through activation of transcriptional bursting and stabilization of gene expression levels. Kat2a loss (NULL) results in enhanced cell-to-cell transcriptional variability and progressive loss of leukemia stem cells (LSCs) transformed with the KMT2A-MLLT3 (MLL-AF9) gene fusion (12). Accordingly, KAT2A is required for maintenance of AML cell lines and in vitro self-renewal of patient AML blasts (13). At a cellular level, loss of Kat2a results in perturbation of leukemia lineage trajectories, with emergence of multiple incongruent differentiation pathways that deplete LSC but fail to uniformly differentiate leukemia cells (12). A similar pattern of incongruous exit from the stem cell state was observed upon KAT2A inhibition in mouse embryonic stem (ES) cells (14). MLL-AF9 results in an aggressive leukemia, both in mice and in humans, and requires minimal cooperativity from additional mutational events (7, 15). Hence, it provides a good representation of a well-adapted leukemia, with minimal genetic and epigenetic variability. However, it does not reflect what is observed with more common forms of AML such as those associated with RUNX1-RUNX1T1 (AML1-ETO), where progression in mouse models is slow and infrequent (7, 16), or clonal hematopoiesis, in which the associated mutations (e.g., in IDH1/2, TET2, DNMT3A) convey a self-renewal advantage but require additional genetic events for leukemia (7, 16). In these cases, we postulate that malignant progression may be facilitated by nongenetic instability, which can be promoted through loss of Kat2a. We tested this hypothesis through investigation of two preleukemia mouse models Idh1R132H and RUNX1-RUNX1T1[RT1(9a)] (17), which together represent up to 25% of human AML disease (7, 16, 18). We compared the effects of the respective mutations in the presence and absence of Kat2a and integrated functional in vitro and in vivo transformation assays with single-cell RNA sequencing (scRNA-seq) analysis, to illuminate consequences on transcriptional variability and differentiation trajectories and explain differential transformation progression.
RESULTS
Loss of Kat2a facilitates IDH1R132H preleukemia transformation
First, we developed a new inducible Idh1R132H allele (fig. S1, A to C) and crossed it into an Mx1-Cre background (fig. S1D), to activate the mutation in hematopoietic tissues. We verified the functionality of the Idh1R132H allele by accumulation of the oncometabolite 2-hydroxyglutarate (fig. S1, E and F). Idh1R132H mice develop leukemia rarely, with long latency and low penetrance, with no significant effects on overall survival (fig. S1G). In contrast, combination of Idh1R132H with other leukemogenic mutations, namely, NRas and Npm1c (triple-mutant), results in short-latency high-penetrance leukemia development (fig. S1G), confirming the preleukemic nature of the Idh1R132H model. Accordingly, triple-mutant bone marrow (BM) cells, but not cells with Idh1R132H alone, have enhanced colony-forming cell (CFC) assay–replating ability, an in vitro measure of transformation (fig. S1H). Comparison of RNA-seq from triple-mutant leukemias versus triple-mutant preleukemias, or versus Idh1R132H alone, revealed a gene signature that was specific to the leukemia state and in which down-regulated genes were enriched for KAT2A chromatin targets (fig. S1I). This association suggests that loss of KAT2A activity may contribute to progression of preleukemia to overt AML.
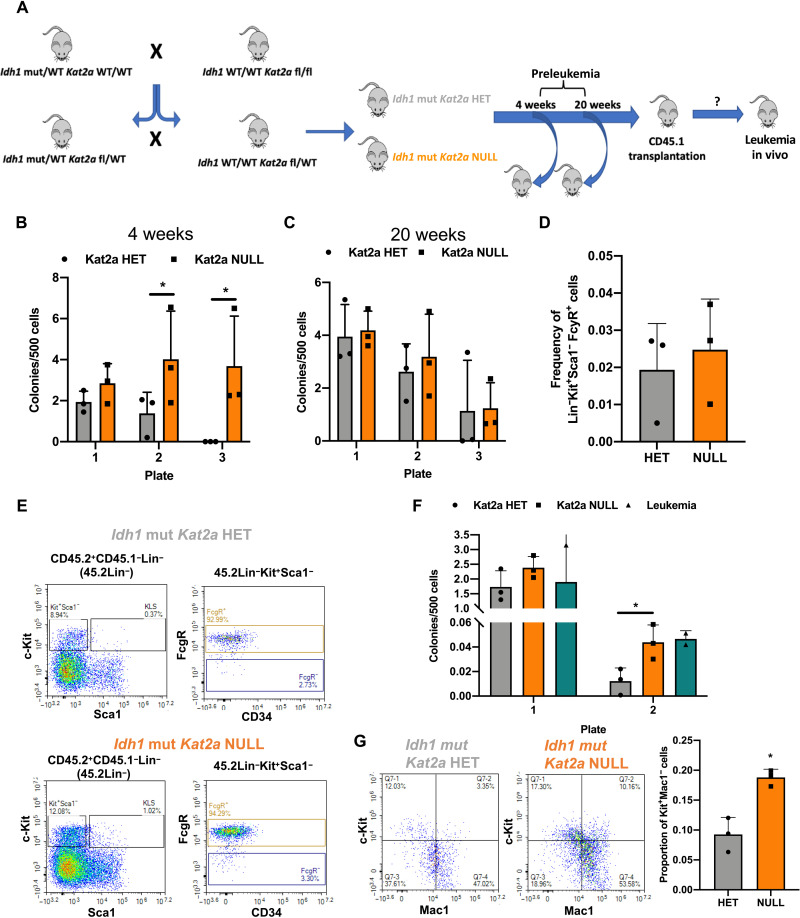
To investigate this putative contribution of Kat2a loss to preleukemia progression, we crossed conditional Idh1R132H and Kat2aFlox/Flox mice, into the Mx1-Cre background (Fig. 1A), to generate Idh1R132H animals that were heterozygous (HET) or NULL for Kat2a (fig. S2, A and B). We analyzed Idh1R132H Kat2aFlox/WT (Idhmut Kat2aHET) and Idh1R132H Kat2aFlox/Flox (IdhmutKat2aNULL) animals 4 and 20 weeks after Cre induction, to identify early and progressed Idh1R132H preleukemia states. Analysis of BM stem and progenitor composition revealed no differences between genotypes or time points (fig. S2, C to G). We did not observe differences in spleen or liver preleukemia burden (fig. S2, H and I). However, Idhmut Kat2aNULL samples had a significant advantage in CFC replating in early preleukemia (4 weeks) (Fig. 1B), which was not sustained at the 20-week time point. This could be compatible with earlier selection of preleukemia cells upon Kat2a loss, which is achieved later in Idhmut Kat2aHET animals as the Idh1mut phenotype progresses (Fig. 1C).
Fig. 1. Kat2a loss facilitates development of Idh1R132H preleukemia.
(A) Diagram of Idh1R132H (Idh1 mut) and Kat2afl/fl mouse crosses to generate Idh1 mut Kat2a HET and Idh1 mut Kat2aNULL cells used in preleukemia studies. WT, wild-type. (B) CFC assays of Idh1 mut Kat2a HET and NULL BM cells 4 weeks after polyinosinic:polycytidylic acid (pIpC) treatment. Mean ± SD, n = 3. (C) CFC assays of Idh1 mut Kat2a HET and NULL BM cells 20 weeks after pIpC treatment; mean ± SD, n = 3. (D) Quantification of GMP-like BM cells obtained from Idh1 mut CD45.2+ grafts; mean ± SD, n = 3 irradiated recipients (CD45.1). (E) Representative flow cytometry plots of BM cells in (D). Top: Idh1 mut Kat2a HET. Bottom: Idh1 mut Kat2aNULL. KLS, Lin-Kit+Sca1+. (F) Serial replating CFC assays of Idh1 mut BM grafts. Mean ± SD, n = 3 Idh1 mut Kat2a HET and NULL and n = 2 Idh1 mut leukemia. (G) Flow cytometry of colonies in (F). Left: representative plots. Right: Kit+Mac1− progenitor quantification. Mean ± SD, n = 3. All analyses two-tailed t test, *P < 0.05.
In an attempt to understand whether the early replating advantage in vitro could lead to accelerated leukemia development in vivo in the absence of other genetic events, we transplanted BM cells from Idhmut Kat2aHET and Idhmut Kat2aNULL mice, into irradiated CD45.1 recipients and followed them up for 1 year. Similar to single Idh1mut animals, we could not detect signs of leukemia development in transplanted mice (fig. S3A). Transplants showed accumulation of granulocyte-monocyte progenitor (GMP)–like (Lin-Kit+Sca1−FcγR+) donor cells, compatible with myeloproliferation (Fig. 1, D and E), which was identical between genotypes. Peripheral blood counts (fig. S3, B to D) and spleen and liver weights (fig. S3, E and F) were also similar. However, we observed the infiltration of the spleen and liver in one of three Idhmut Kat2aNULL recipients, which was not present in Idhmut Kat2aHET grafts (fig. S3G). Notably, Idhmut Kat2aNULL cells showed enhanced colony-replating potential relative to Idhmut Kat2aHET, which was comparable to that of BM from rare Idhmut leukemic animals (Fig. 1F). Idhmut Kat2aNULL cells in CFC assays were enriched in c-Kit+Mac1− cells (Fig. 1G) compatible with hindered differentiation and/or expansion of self-renewing cells. Overall, the results suggest that loss of Kat2a imparts leukemogenic properties to Idh1mut cells but is in itself not sufficient to drive leukemogenesis in the absence of additional cooperating genetic events.
Loss of Kat2a accelerates RUNX1-RUNX1T1 preleukemia–to-leukemia progression
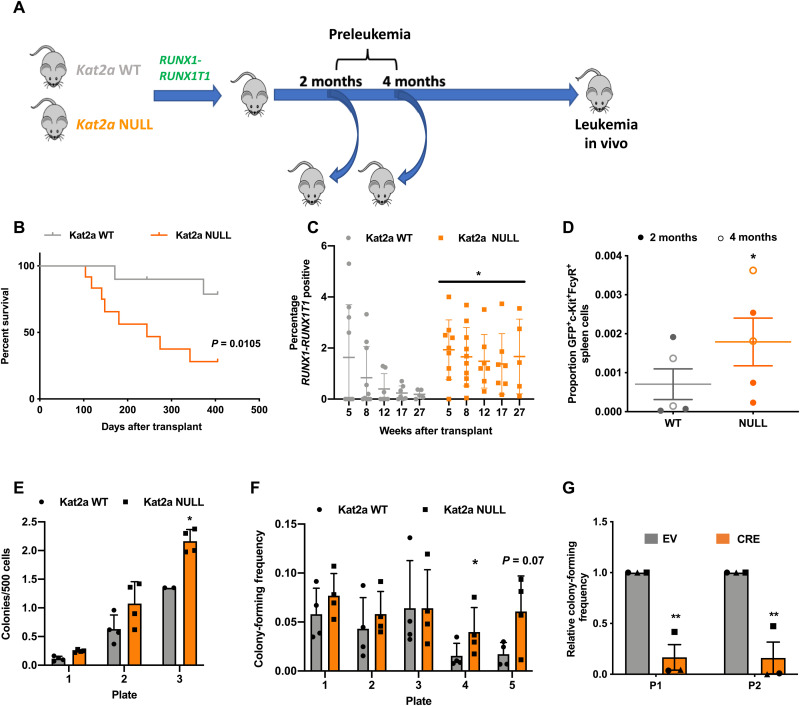
We next tested the impact of Kat2a loss on the preleukemia model driven by the exon 9a splicing variant of the RUNX1-RUNX1T1[RT1(9a)] fusion gene (17), which, when retrovirally delivered to adult BM cells, leads to long-latency, incomplete-penetrance leukemia in irradiated recipients (19–20). Using our previously described Kat2aFlox/Flox Mx1-Cre mice (12), we isolated progenitor-enriched BM cells after pIpC-induced locus excision (fig. S4A) and delivered the RT1(9a) construct by retroviral transduction, as described (20). In all experiments, Kat2aFlox/FloxMx1-Cre+/− (Kat2aNULL) cells were compared with Kat2aFlox/Flox Mx1-Cre−/− (Kat2aWT) cells. We started by evaluating leukemia development after transplantation of RT1(9a) Kat2aNULL and Kat2aWT BM cells (Fig. 2A). Loss of Kat2a led to a marked decrease in survival of RT1(9a) recipient animals, compatible with accelerated leukemia progression (Fig. 2B). Kat2aNULL leukemias had a nonsignificant trend toward higher white blood cell counts (fig. S4, B to D) and spleen leukemia burden, with minimal infiltration of other organs (fig. S4, E to G). The surface phenotype of the leukemias was not different between genotypes, with the majority of Kit+ progenitor cells also Sca1−/lowCD34− (fig. S4H), as described (17). Analysis of early time points after transplantation showed that RT1(9a) engraftment became quickly fixed in the absence of Kat2a (Fig. 2C). Kat2aNULL/RT1(9a) cells obtained from healthy presymptomatic recipients were mildly enriched for Kit+FcgR+ cells (Fig. 2D) and displayed enhanced colony formation (Fig. 2E), compatible with accelerated preleukemia development. Similarly, Kat2aNULL cells directly tested in CFC assays upon retroviral transduction displayed enhanced replating potential. (Fig. 2F). In contrast, excision of Kat2a in RT1(9a) cells after in vitro transformation by three rounds of serial replating led to a reduction in colony formation (Fig. 2G), suggesting that Kat2a loss favors leukemia development only at a preleukemia stage. These latest observations mirror our previously identified role for Kat2a in maintenance of established leukemia stem-like cells and suggest that Kat2a plays stage-specific roles during leukemogenesis, which are preserved across leukemia models.
Fig. 2. Kat2a loss accelerates RT1(9a) preleukemia to leukemia progression.
(A) Experimental design. (B) Survival curve of RT1(9a) Kat2aWT and Kat2aNULL Kit+ BM recipients. n = 12 animals per genotype. *P < 0.05, log-rank test. (C) Quantification of peripheral blood green fluorescent protein (GFP) for animals in (A); GFP reports RT1(9a). Mean ± SD, n = 10 animals/genotype (8 weeks). *P < 0.05, two-way analysis of variance (ANOVA). (D) Flow cytometry analysis of RT1(9a) Kat2aWT and Kat2aNULL graft spleen cells 2 and 4 months after transplantation. Mean ± SD, n = 5. (E) CFC assay of RT1(9a) Kat2aWT and Kat2aNULL graft BM cells 4 months after transplantation. Mean ± SD, n = 4. (F) In vitro transformation of Kat2aWT and Kat2aNULL Lin−/Kit+ BM cells transduced with RT1(9a) retrovirus tested in CFC serial replating. Mean ± SD, n = 4. (G) CFC replating (plate = P1 and P2) analysis of RT1(9a) Kat2aFlox/Flox Cre−/−Kit+/Lin− BM cells excised in vitro by lentiviral-delivered Cre recombinase [versus EV (empty vector)] after three rounds of colony replating. Mean ± SD, n = 3. All other analyses two-tailed t test, *P < 0.05 and **P < 0.01.
Loss of Kat2a results in preleukemia cellular diversification
We had previously associated Kat2a function in LSC maintenance with stability of transcriptional programs (12). Using scRNA-seq, we showed that Kat2a loss resulted in diversification and branching of differentiation trajectories and associated with enhanced transcriptional noise, particularly in biosynthetic programs (e.g., ribosomal biogenesis and translation). We asked whether similar mechanisms were at play in preleukemia progression facilitated by Kat2a loss. We hypothesized that enhanced transcriptional variability leading to program diversification might increase the probability of accessing or seeding leukemia programs, resulting in the observed acceleration in leukemia progression.
We performed scRNA-seq analysis of preleukemia cells on the 10X platform, comparing transcriptional landscapes of Kat2aNULL and Kat2aWT RT1(9a) asymptomatic animals obtained 2 and 4 months after transplantation. We sequenced a total of 1767 cells sorted as RT1(9a)/GFP+Kit+ stem/progenitor and retrieved an average of 174,770 aligned reads per cell, corresponding to medians of 5939 unique molecular identifiers (UMIs) and 1575 genes per cell (Supplementary File 1). Less than 0.2% of reads aligned to mitochondrial DNA, denoting successful sequencing. Preprocessing steps are detailed in Materials and Methods.
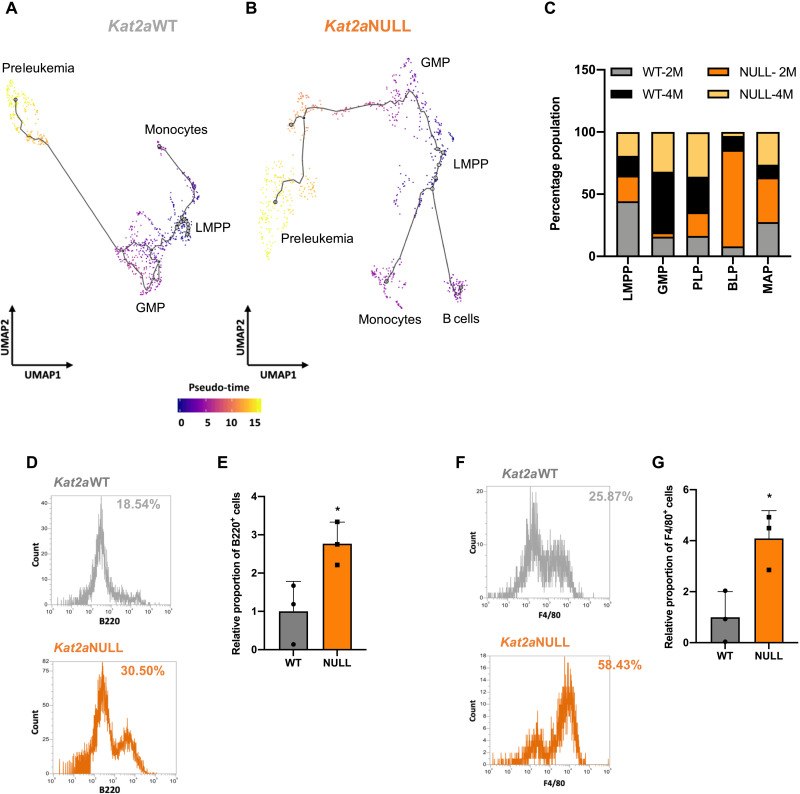
We used the pseudo-time alignment algorithm Monocle (21) to understand the presence of different subpopulations of preleukemia cells and to infer relationships between those subpopulations along putative transformation trajectories, within and across early time points of transformation. Monocle uses a reverse graph embedding algorithm to align individual cells along a differentiation trajectory defined by progressive changes in their gene expression profile. Upon learning overall gene expression–based trajectory, each cell is sequentially positioned along the trajectory path to infer dynamic mechanisms of cell state transitions. We considered Kat2aWT and Kat2aNULL separately (Fig. 3, A and B, and fig. S5, A and B) to identify differences in transcriptional states that accompany loss of Kat2a and may help explain Kat2aNULL advantage in leukemia progression. We used transcripts of cell surface markers routinely used (12, 22–23) for hematopoietic cell immunophenotyping to map the identity of cells along the pseudo-temporal trajectories (fig. S5, C and D). All cells were sorted as RT1(9a)+Kit+, thus capturing progenitor compartments. Cells at the origin of the trajectory expressed high Ly6e (Sca1), Cd34, and Flt3, compatible with lymphoid-myeloid primed progenitors (LMPPs) (22). LMPP-like cells were relatively enriched at 2 months after engraftment (Fig. 3E and fig. S5E), aligning temporal with pseudo-temporal trajectories. Accordingly, we did not find phenotypic LMPP-like cells in fully developed leukemias (fig. S6, A and B). LMPP-like cells were adjacent to an Ly6elowCd34+Fcgr3+ state (fig. S5, C and D), compatible with GMPs (23). Unlike LMPP-like, GMP-like cells are more abundant at 4 months (Fig. 3C), suggesting cellular and temporal trajectory progression. Kit+Sca1−/lowCD34+FcgR+ cells are variably represented in full-blown RT1(9a) leukemia (figs. S4H and S6, A and C). Additional states were represented in the trajectories. Two states, (1) and (2), diverged from LMPP-like cells in a distinct direction to GMP-like cells, with similar pseudo-times. The third state (3) denotes a later step in differentiation pseudo-time and follows the GMP-like path of the trajectory, with greater abundance of intermediate states upon Kat2a loss.
Fig. 3. Loss of Kat2a diversifies cell fates and promotes RT1(9a) preleukemia progression.
(A and B) Pseudo-time single-cell trajectory of (A) Kat2aWT cells (B) Kat2aNULL RT1(9a) cells 2 and 4 months after transplantation. Trajectories were inferred using Monocle3 (21); compartments were labeled as per hematopoietic markers in (fig. S5, C and D). (C) Proportion of candidate progenitor cell compartments in the RT1(9a) pseudo-time trajectory contributed by individual Kat2aWT or Kat2aNULL, 2-month or 4-month samples. (D) Representative flow cytometry histograms of B220 B cell marker detection in plate 2 CFC of RT1(9a)-transduced Kat2aWT and Kat2aNULL cells during in vitro transformation. (E) Aggregate results of B220 staining as in (D). Mean ± SD, n = 3. (F) Representative flow cytometry histograms of F4/80 monocyte marker detection in plate 2 CFC of RT1(9a)-transduced Kat2aWT and Kat2aNULL cells during in vitro transformation. (G) Aggregate results of F4/80 staining as in (F). Mean ± SD, n = 3.
State (1) comprised Ly6e+CD79a+Cd14− cells, which exhibit B lymphocyte–associated signatures (Supplementary File 3 and fig. S6D) and were designated B cell–affiliated progenitors (BAPs). These cells were more abundant in Kat2aNULL 2 months after transplantation but could also be observed at 4 months, in both genotypes (Fig. 3C and fig. S5E). Surface phenotyping of Kat2aNULL versus Kat2aWT cells undergoing in vitro transformation upon RT1(9a) transduction and serial replating independently confirmed enhanced specification of B220+ B lymphoid cells in Kat2aNULL samples (Fig. 3, D and E). Enhanced B lymphoid specification was transient, both transcriptionally (Fig. 3C) and cellularly (Fig. 3, D and E), and the phenotype also had reduced representation in fully developed leukemias (fig. S5E). State (2) comprised Ly6e+Fcgr3+Cd14+ cells, which have a monocytic/macrophage transcriptional affiliation (Supplementary File 4 and fig. S6E) and were dubbed monocyte-affiliated progenitors (MAPs). They could be observed in both genotypes, at both time points, with an enrichment in Kat2a-depleted samples (Fig. 3C and fig. S5E). Similar to B cell–affiliated cells, we captured more frequent emergence of F4/80+ monocyte-macrophages in Kat2aNULL samples undergoing in vitro transformation (Fig. 3, F and G), functionally confirming lineage diversification upon Kat2a knockout. MAP-like cells are minimally represented in fully developed RT1(9a) leukemia (fig. S6, A and B).
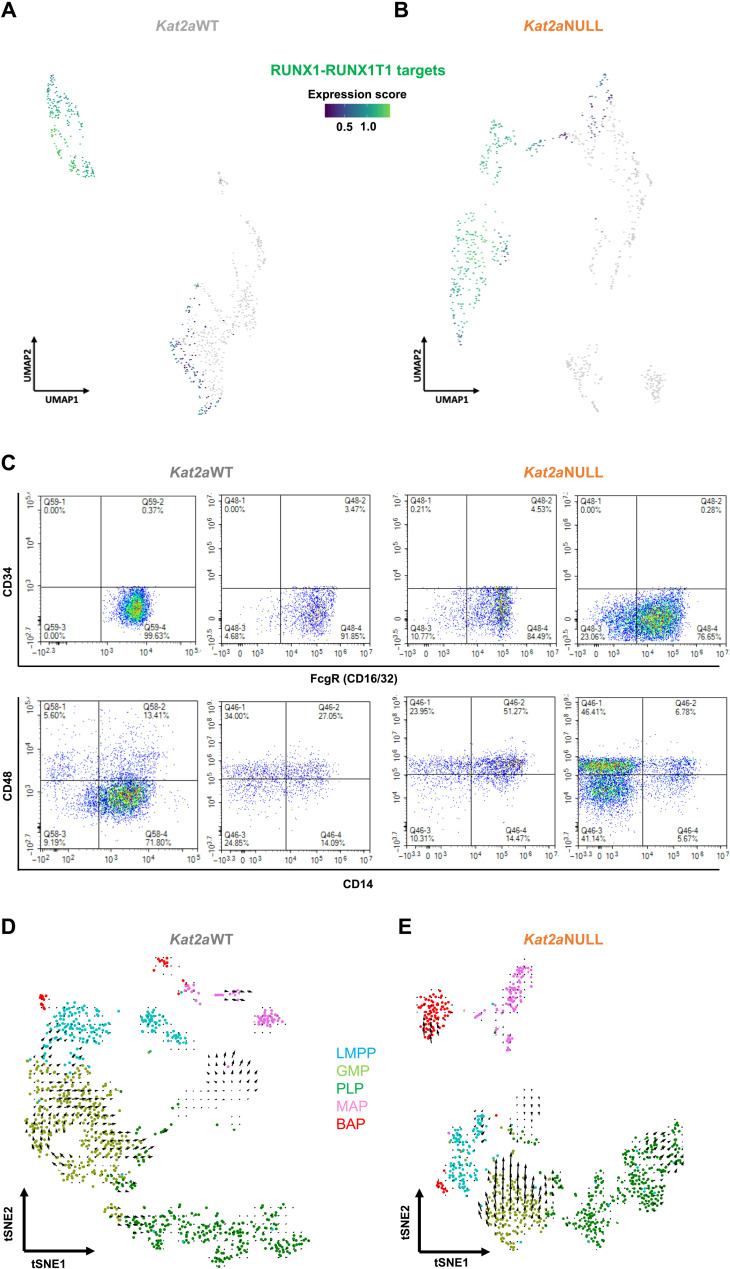
The last cell state (3) was characterized as Ly6elow/−Cd34−, with variable levels of Fcgr and Cd14 and rare detection of Cd48 (Fig. 3, C and D). RT1(9a) leukemic progenitors were originally described as Kit+Sca1−/lowCD34−FcgRlow (17), compatible with this last cell state. Accordingly, we detected expression of RT1 gene targets (24) specifically in this compartment (Fig. 4, A and B), suggesting development of a leukemogenic program. Inspection of the surface profile of RT1(9a) AML samples confirmed that most Kit+ cells were negative for Sca1 and CD34 (fig. S4H). RT1(9a) Kit+Sca1−/lowCD34− AML cells exhibited variable combinations of FcgR, CD48, and CD14 (Fig. 4C), suggesting that leukemias may have developed from different subclones within that compartment. We refer to this Ly6elow/−Cd34− state as preleukemia progenitors (PLPs). We sought to confirm the differentiation alignment of the different cell states using RNA velocity (Fig. 4, D and E) (25). The algorithm infers differentiation trajectories on the basis of relative representation of unspliced and spliced transcript variants, and the latter inferred as estimates of the future status of the cells over a relatively fast time frame. The greater distances observed in Monocle to BAP and MAP, and in opposite direction to PLP, are captured by scarcity of intermediate velocities that nevertheless recapitulate the Monocle directionality. Directionalities within the GMP-like and PLP states are better defined, particularly in Kat2aNULL cells. Significantly, Kat2aNULL cells (Fig. 4E) show increased velocity, which is unique within the PLP compartment, and may correspond to the more evenly populated trajectories within the Kat2aNULL Monocle trajectory (Fig. 3B).
Fig. 4. The PLP compartment captures RT1(9a) early transformed cells.
(A and B) Expression of RUNX1-RUNX1T1 chromatin immunoprecipitation sequencing targets (18) in (A) Kat2aWT cells and (B) Kat2aNULL RT1(9a) single-cell trajectories. (C) Flow cytometry analysis of RT1(9a) pseudo-time–associated hematopoietic cell surface markers in representative Kat2a WT and Kat2a NULL AML. Plots are gated on RT1(9a)/GFP+Kit+Sca1−CD34−PLP-like cells (see fig. S4H). (D and E) RNA velocity (25) plots of RT1(9a) preleukemia in the presence (D) and in the absence (E) of Kat2a. Color coding reflects the progenitor compartments defined in Fig. 3. tSNE, t-distributed stochastic neighbor embedding.
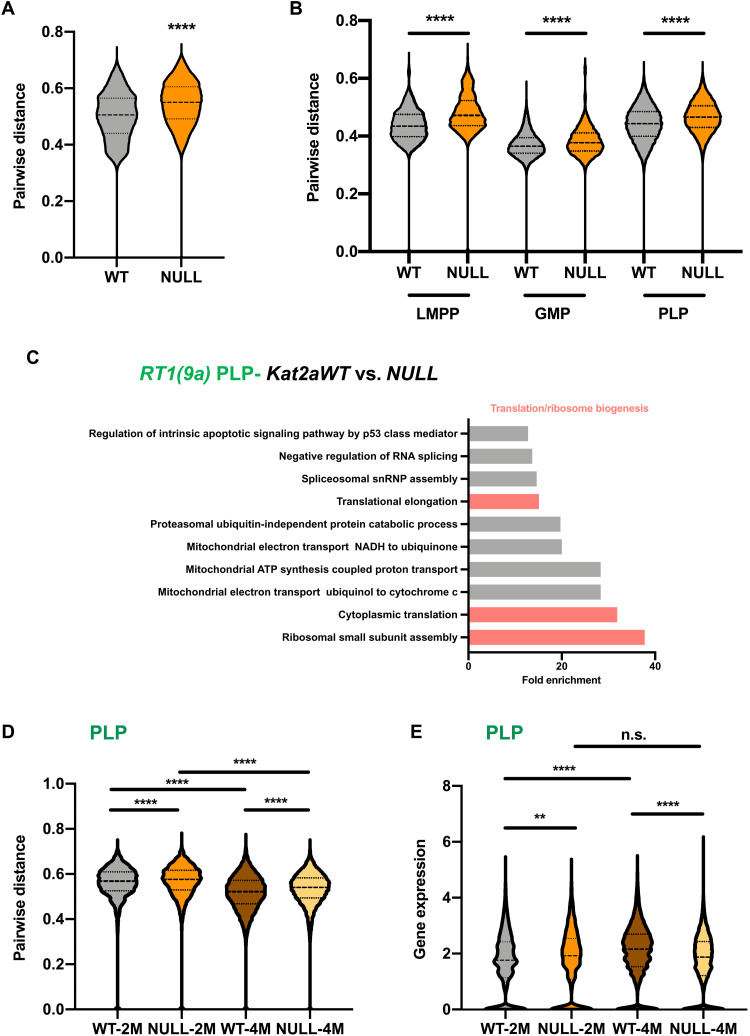
Loss of Kat2a increases transcriptional variability and destabilizes ribosomal biogenesis programs
Given the previously established association between KAT2A and transcriptional noise regulation (12, 14, 26), we asked whether the cellular diversification and higher trajectory velocities observed upon Kat2aNULL loss were accompanied by, and putatively attributable to, enhanced variability in transcription. Pairwise distance (27) defines highly variable genes on the basis of a mean expression-corrected coefficient of variation or distance to the median (DM) (28) and inverts gene-to-gene correlations to estimate dispersion or distance in gene expression programs. Pairwise distance has been used as a measure of global transcriptional variability, or noise (27). Perhaps expectedly, given the differential diversity of cell types observed between Kat2aNULL and Kat2aWT RT1(9a) preleukemias, global pairwise distance was increased in Kat2aNULL cells (Fig. 5A), putatively capturing cellular heterogeneity. However, the same gain in pairwise distance was observed in the individual cell states (Fig. 5B), suggesting that loss of Kat2a may affect transcriptional variability. GMP-to-PLP transition is also accompanied by enhanced transcriptional variability (Fig. 5B), supporting the notion that Kat2a loss may facilitate preleukemia progression through enhanced transcriptional noise. To understand the nature of the transcriptional programs perturbed upon (i) Kat2a loss and (ii) preleukemia progression, we performed differential gene expression analysis of the scRNA-seq dataset. Comparison of Kat2aNULL to Kat2aWT cells revealed minimal changes in gene expression levels (fig. S7A), which were of down-regulation, as previously observed upon Kat2a loss (12). Consistent with our published data (12), differentially expressed genes between genotypes predominantly associated with ribosomal assembly and translation ontologies (fig. S7B and Supplementary File 5), a pattern particularly prominent within PLP (Fig. 5C and Supplementary File 6). The same ontologies were specifically down-regulated in Kat2aWT RT1(9a) PLPs compared to other cell states (fig. S7C and Supplementary File 7), capturing a reported decrease in protein synthesis in RT1 leukemia (29). Ribosomal and translation ontologies (fig. S7, D and E, and Supplementary File 8) were also down-regulated in Idh1R132H mice. Together, our findings suggest a specific association of attenuated ribosomal programs with preleukemia progression, which may be further facilitated by Kat2a loss. Kat2a loss increases variability of ribosomal biogenesis programs in PLPs (Fig. 5D), which are themselves more variable than GMPs for the same programs (fig. S7F), suggesting enhanced noise at the transition (Supplementary File 9). The gene expression range in Kat2aNULL PLPs favors lower mean values (Fig. 5E), specifically at 4 months. In support of the functional impact of the transcriptional perturbation, Kat2a loss results in decreased protein synthesis (fig. S7, G and H).
Fig. 5. Loss of Kat2a destabilizes expression of ribosomal biogenesis and translation-associated genes.
(A) Pairwise distance transcriptional variability measure (27) of Kat2aWT and Kat2aNULL RT1(9a) cells; top 500 most variable genes/genotype calculated by distance to the median CV (DM). ****P < 0.0001, nonparametric Kolmogorov-Smirnov (KS) test of cumulative distributions. (B) Comparison of RT1(9a) Kat2aWT and Kat2aNULL genotype-specific pairwise distances within individual LMPP, GMP, and PLP compartments. NULL up in all comparisons, with ****P < 0.0001, KS test. (C) Overrepresented gene ontology categories for genes down-regulated in RT1(9a) Kat2aNULL versus Kat2aWT PLP. *P-adj < 0.05. snRNP, small nuclear ribonucleoprotein; NADH, reduced form of nicotinamide adenine dinucleotide; ATP, adenosine 5′-triphosphate. (D) Pairwise distance of RT1(9a) PLPs. Comparisons consider correlations between ribosomal biogenesis genes, ****P < 0.0001, KS test. NULL up in all comparisons with WT; 2 months up in comparison with 4 months. (E) Distribution of expression levels for gene signatures in (C). **P-adj < 0.01, ****P-adj < 0.0001, two-tailed t test; n.s., not significant. NULL up at 2 months and down at 4 months in comparison with WT; WT up at 4 months relative to 2 months; comparison of NULL time points not statistically significant.
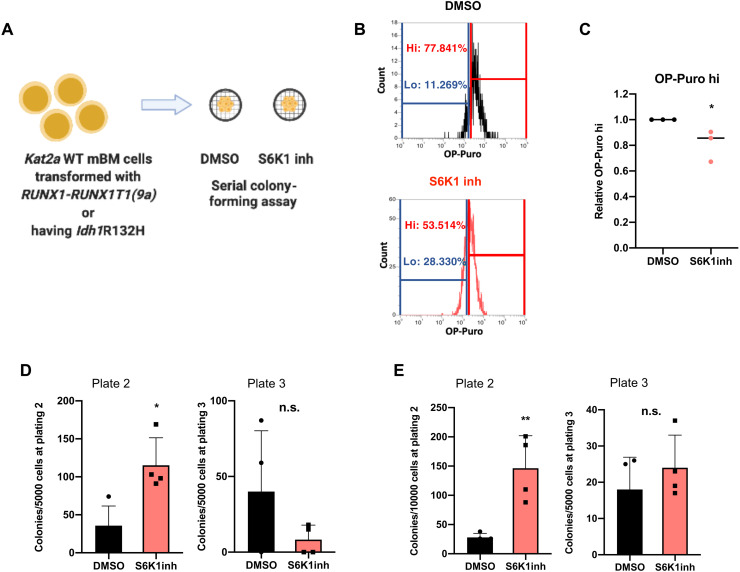
Reduced protein synthesis activity transiently facilitates preleukemia progression
We tested the contribution of reduced protein synthesis activity to preleukemia progression by treatment with the S6K1 inhibitor (S6K1inh) PF4708671 (Fig. 6A), which impairs protein synthesis activity confirmed by reduced O-propargyl-puromycin (OP-Puro) incorporation in nascent peptide chains (Fig. 6, B and C). We treated Kat2aWT RT1(9a) cells with S6K1inh and tested their leukemia transformation potential in vitro through CFC assay replating. S6K1-inhibited cells displayed enhanced colony formation upon replating (Fig. 6D), suggesting a contribution to leukemia transformation. However, the increase in colony formation was transient and eventually lost upon subsequent replating (Fig. 6D). This suggests that the effects of reduced protein synthesis on leukemia cells may vary with progression of transformation, reconciling our data with prior analysis of established MLL-AF9 cells, in which reduced OP-Puro incorporation associated with Kat2aNULL-mediated extinction of LSCs (12). We observed a similar pattern of transient increase in colony formation of Idh1R132H preleukemia cells treated with S6K1inh (Fig. 6E). Together, the data suggest that reduced ribosomal assembly and protein synthesis facilitate preleukemia progression. Exploration of lower levels of expression of translation-associated genes as a consequence of enhanced transcriptional variability may be instrumental in the acceleration of preleukemia to AML transition upon Kat2a loss. As leukemia progresses, variability in ribosomal biosynthesis programs may become attenuated with deviation from an optimal level no longer favorable to transformation.
Fig. 6. Inhibition of protein synthesis phenocopies effects of Kat2a loss facilitating preleukemia transformation.
(A) Schematic of S6K1 inhibition assays. mBM, mouse BM. (B) Representative OP-Puro incorporation flow cytometry of S6K1inh-treated RT1(9a) Kat2aWT cells. OP-Puro high cells, hi; OP-puro; low cells, lo. (C) Quantification of OP-Puro high (hi) cells in (B), relative to dimethyl sulfoxide (DMSO). Mean ± SD, n = 3. *P < 0.05, two-tailed t test. (D) CFC replating of Kat2aWT RT1(9a) in vitro transformation in the presence of S6K1inh (control, DMSO). Plate 2 (left): means ± SD, n = 4, *P < 0.05. Plate 3 (right): mean ± SD, n = 4, n.s., two-tailed t test. (E) (D) CFC replating of Idh1R132H Kat2aWT cells in the presence of S6K1inh (or DMSO), 4 weeks after locus activation. Plate 2 (left): mean ± SD, n = 4, **P < 0.01. Plate 3 (right): mean ± SD, n = 4, n.s., two-tailed t test.
DISCUSSION
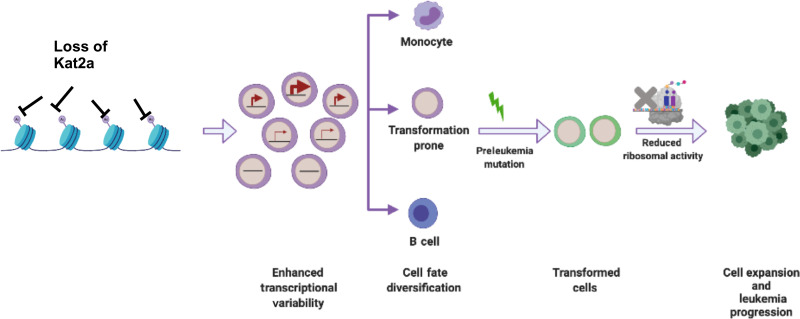
In this study, we have shown that Kat2a loss facilitates preleukemia progression in Idh1R132H and RUNX1-RUNX1T1(9a) mouse models of human disease, with acceleration of frank leukemia onset in the case of RT1(9a). Loss of Kat2a resulted in enhanced variability of transcription, leading to diversification of cell fates, including accumulation of PLP cells. In the context of an early genetic event such as RT1(9a) or Idh1R132H, which do not allow for full leukemia transformation, the cellular heterogeneity that ensues creates the opportunity for specification and expansion of transformation-prone cells, on which additional molecular events may act to progress the leukemic process (Fig. 7).
Fig. 7. Working model of the role of Kat2a loss in preleukemia progression.
Kat2a loss facilitates generation or selection of cell types susceptible to transformation. Preleukemia progression is further aided by variability in ribosomal protein gene transcription and reduced protein synthesis.
RT1 progenitor cells have been variably characterized as Kit+Sca1+ cells (17) and Kit+Sca1−FcgR+ GMP-like cells (19), with the (9a) variant denoting a Kit+Sca1−CD34−/lowFcgR−/low phenotype (17). The variability in cellular composition is notable between individual animals (fig. S5) and likely denotes the contribution of additional mutations to the establishment of full-blown leukemia (30), which may lose dependence on the presence of RUNX1-RUNX1T1 (29) and become sensitive to its level of expression (31, 32). Cellular variability may emerge as a downstream consequence of different additional genetic events or reflect differential upstream vulnerability to specific mutations. Loss of Kat2a may facilitate the latter process. By destabilizing transcription and diversifying cellular output as one consequence of moving stem and progenitor cells out of their status quo, Kat2aNULL animals may generate additional types of RT1(9a) translocation-carrying cells able to respond to downstream mutations and/or be transformed by them. It is unlikely that Kat2a loss itself contributes to the genetic load. Kat2aNULL animals are not at a risk of myeloproliferation (12), and no recurrent KAT2A mutations have been described to date in association with hematological or solid cancers. In contrast, Kat2a ablation consistently changes cellular composition (12–14, 33), making it a more likely facilitator event to generate “second hit”–responsive preleukemia cells. Future studies combining genetic barcoding and phenotyping on a time course of transformation should provide definitive evidence of such an effect.
Further to or concomitantly with cellular diversification and putative differential susceptibility to secondary genetic hits, the molecular programs affected by Kat2a loss can also contribute to the leukemogenic process. Ribosome biosynthetic and translation factor genes are pervasive targets of KAT2A (12, 33), and our data suggest that destabilization of translation acts to facilitate transformation at least transiently and down-regulation of translation-associated genes may accompany preleukemia to leukemia progression. Enhanced transformation may be achieved by surveying and selection of biosynthetically quiescent cell states, which evade further diversification and respond to additional mutations with disease propagation and progression. Inspection of noise-responsive programs in chronic lymphocytic leukemia has captured ribosome biogenesis and translation as a significant prognostic module (34), and loss of ribosome biosynthetic activities plays a role in T acute lymphoblastic leukemia progression (35). The latter study also implicated decreased mitochondrial metabolic activity, which we have shown to be responsive to Kat2a loss, in leukemia development.
However, established leukemia cells can be dependent on active translation for their maintenance (36), and AML subtypes, namely, those with RUNX1 mutations (37), are therapeutically sensitive to inhibition of protein synthesis. Despite the contribution of reduced or perturbed translational activity to transformation, our results suggest that the effect is transient, and we had previously observed that the inhibition of protein synthesis reduced colony formation in established MLL-AF9 leukemia cells (12). Accordingly, MLL-AF9 leukemia knockout for Kat2a displayed enhanced noise specifically in translation-associated genes, which was accompanied by reduced protein synthesis and associated with depletion of leukemia stem-like cells (12). It is possible that fully transformed, well-adapted leukemia cells buffer transcriptional variability to maintain stable self-renewal signatures and optimal biosynthetic, translation rates. In this context, instability of transcriptional programs may shift biosynthetic homeostasis and perturb cellular identity and mal-adapt leukemia stem-like cells, with antileukemia effects. Thus, stage-specific tuning and untuning of transcription and translation may be used to modulate cancer progression, a principle that can be extended to other cancer state transitions such as metastasis or drug resistance with prognostic and therapeutic potential.
MATERIALS AND METHODS
Preleukemia mouse models
Mice were kept in a specific pathogen–free animal facility, and all experimental work was carried out under U.K. Home Office regulations. Animal research was regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body. Peripheral blood was collected by saphenous vein, and differential blood cells counts were determined using a Vet abc automated counter (Scil Animal Care, Viernheim, Germany).
Generation of an Idh1R132H mouse model
Targeting vector was generated as follows using methods described previously (38). The WT Idh1 locus (endogenous Idh1 sequence, including arms of homology, was captured by gap repair as previously described). The following primer pairs were used to amplify the “U” cassette containing attR1 and attR2 gateway cloning sites containing a Zeo/PheoR selection cassette with the appropriate overhangs to allow insertion of this cassette between exons 2 and 3 of the Idh1 locus by recombineering (Table 1).
Table 1. Primers for genotyping.
| Target | Forward primer (5′-3′) | Reverse primer 1 (5′-3′) | Reverse primer 2 (5′-3′) |
| Mx1-Cre | CGTACTGACGGTGGGA GAAT | TGCATGATCTCCGGT ATTGA | – |
| Kat2a | CACAGAGCTTCTTGGA GACC | GGCTTGATTCCTGTA CCTCC | – |
| Idh1 | GTTGGTGGATTCCATTGCTT | TGTTAGTCCCAACCC CTTCC | GACAAACTGACAGGCTG CAA |
| Amplification of U cassette |
AAGTCCAACCTTATTGTCCCATCATAAGTTTTATA CTCTGTAAGTAATGACCGCCTACTGCGACTATAGA |
AGGTTCACCCTATGACTAACTGGCTCTAACAAAAG AGTTCTCAGCTCTTTAAGGCGCATAACGATACCAC |
– |
| Amplification of G cassette |
GCAATAGGAACCCTTTGCCATACTTAATTTTACTTCC ATAAATCTCAAGTTCCTGTGTGAAATTGTTATCCGC |
ACAAACTAGCTAACCTGATGGATGCAGTAATGAGT AACACAGGAGATCCTCCACTGGCCGTCGTTTTACA |
– |
| WT control assay | GGGCTAGGGGAAGCGCCATC | TGCGCAGGCCAAAAGCCCAT | – |
| 5′ integration | TGGCTGGAAAACAAAAAGATCGG | CGTTATGCGCCTTAAAGAGCTGA | – |
| 3′ integration | TGGATCCGGGAAGTTCCTATTCC | TGGCACAGGCACAGAGGGA | – |
| 3′ internal probe | GGAGTGTTGTATCGCAGCAA | GCGCTAGGATTAAAGGCACA | – |
| 5′ internal probe | TCAGCATTCCCTAGGCACAA | TCTCTTGAGTGTGAGGCCAG | – |
The following primers were used to amplify the “G” cassette that contains attR3 and attR4 gateway sites flanking an AmpR cassette. Appropriate overhangs were incorporated into these primers to allow “gap repair” subcloning and retrieval of arms of homology 5′ and 3′ to exons 2 and 4 (5.9 and 3.6 kb, respectively) from the Idh1 containing bacterial artificial chromosome (Table 1).
A custom gene block (GeneArt, Thermo Fisher Scientific) containing sequence encoding the mutant R132H substitution in exon 3 was cloned into the subsequent U/G-captured intermediate—replacing the WT exon3 by standard restriction enzyme cloning using Sna BI and Csp CI to generate an R132H mutant U/G vector. A custom cDNA flanked by Afl II and Asc I sites, containing AttL1-loxP-En2SA-Idh1 cDNA exons3 to 9–SV40 pA-loxP-FRT was synthesized (GeneArt, Thermo Fisher Scientific) and cloned into the PL1PL2 containing an FRT-flanked NeoR cassette. This generated the “SA-Idh1 exon 3-9 cDNA” cassette.
These vectors and the PL3L4 vector were combined in the downstream L/R clonase reaction to successfully generate the Idh1R132H-NeoRTV vector. All intermediate and final vectors were sequence-verified. The Idh1R132H-NeoRTV targeting vector was electroporated into mouse ES cells, and genotyping was performed for on target integration at the endogenous Idh1 locus using a series of long-range polymerase chain reaction (PCR) reactions (Table 1):
Genomic DNA (gDNA) extracted from heterozygous-targeted single-cell mouse ES cell clones were subjected to Southern blot to confirm the structural integrity and confirmation of site-directed recombination of FRT and LoxP recombination sites, before microinjection. FRT recombination and removal of the NeoR cassette were mediated by expression of flippase via transient transfection of the pCAG-FlpO plasmid (Addgene, #89574). LoxP recombination and deletion of the “SA-Idh1 exons 3 to 9 cDNA” cassette was mediated by expression of Cre via transient transfection of the pCAG-Cre plasmid (Addgene, #13775). gDNA extracted from single-cell clones were subjected to Southern blot to confirm the successful removal of the NeoR resistance cassette and LoxP recombination to generate the Idh1R132H-TV and Idh1R132H-KI alleles, respectively (fig. S1A). The primers that were used to generate Southern blot hybridization probes for the respective 3′ internal (FLP assay) and 5′ internal (Cre assay) are mentioned in Table 1.
Positively targeted heterozygous clones were selected for microinjection. Chimeric offspring were then selected for downstream breeding and germ line transmission of the Idh1R132H-NeoRTV-targeted allele.
The FRT-flanked neomycin-resistant cassette, used for positive enrichment of targeted mouse ES cells, was removed by breeding to FLPe mice [as previously described (39)]. F1 mice were backcrossed to WT C57Bl6 mice, and mice negative for the presence of the RosaFLPe transgene and positive for the inducible Idh1Knock-InR132H allele were selected for downstream cohort generation by subsequent crosses with the inducible Mx1-Cre transgenic mouse model (fig. S1C). Standard PCR genotyping for the WT and mutant alleles was performed (using the primers detailed in Table 1). Subsequent crosses to homozygous NrasG12D/G12D and Npm1cA/cA cohorts were used to generate experimental model cohorts, as described previously (40).
To generate a Mx1-Cre-inducible mouse with Idh1mut/WT- and Kat2a-floxed alleles, Idh1mut/WTKat2aWT/WTMx1-Cre+/− males were crossed with Idh1WT/WTKat2a fl/flCre−/− females. The first generation carried a Kat2a fl/WT genotype (referred as Kat2a HET) with either Idh1WT/WT or Idh1mut/WT and Mx1-Cre+/−. Idh1WT/WTKat2a fl/WT and Idh1mut/WTKat2a fl/fl offsprings were crossed to obtain experimental genotypes referred to as Kat2a HET (Idh1mut/WTKat2a fl/WT) and Kat2aNULL (Idh1mut/WTKat2a fl/fl) maintaining an heterozygous Mx1-Cre allele.
Kat2a conditional knockout model
Kat2afl/fl conditional knockout mice have been previously described (12).
Genotyping
Ear notch biopsies were digested overnight in lysis buffer [50 mM sodium chloride, 50 mM tris(hydroxymethyl)aminomethane hydrochloride, 5 mM ethylenediaminetetraacetic acid, 20% SDS, and proteinase K (0.5 mg/ml)] at 55°C and 750 rpm using a Thermo-Shaker (BioSan). DNA extraction used isopropanol-based precipitation. Mice were genotyped using the primers in Table 1, following the PCR protocol: 95°C, 3 min, 40× [94°C, 30 s; 60°C, 30 s (57°C, 30 s for Idh1); 72°C, 90 s (30 s for Idh1)]; 72°C, 10 min. DNA products were run on a 1% agarose gel in TAE (Tris-acetate-EDTA) (1×), at 100 V and visualized using an AlphaImager ultraviolet transilluminator (Protein Simple). Cre-mediated recombination was induced in 8-week-old mice by administration of five–alternate day intraperitoneal injections of polyinosinic:polycytidylic acid (pIpC), 300 μg per dose. Idh1 recombination was confirmed by PCR (Table 1, reverse primer 2) following the same PCR protocol.
Preleukemia cell engraftment
BM cells were isolated from long bones of Kat2aWT and Kat2aNULL animals as described (12). Briefly, following red blood cell lysis, BM-nucleated cells were depleted of differentiated cells using a cocktail of biotinylated lineage (Lin) antibodies and streptavidin-labeled magnetic nanobeads (BioLegend), according to the manufacturer’s instructions. Lin-depleted cells pooled from four Kat2aWT or Kat2aNULL animals were cultured separately overnight at 37°C and 5% CO2 in RPMI supplemented with 20% heat-inactivated fetal bovine serum (FBS) (R20), l-glutamine (2 mg/ml), 1% prostate-specific antigen, murine interleukin-3 (mIL-3; 10 ng/ml), mIL-6 (10 ng/ml), and murine stem cell factor (mSCF; 20 ng/ml) (cytokines from PeproTech) (supplemented R20), followed by retroviral transduction.
Retroviral construct MSCV-AML1/ETO-IRES-GFP was previously described (20). Viral particle production and transduction of the genotype-specific pools were done as described previously (12). Green fluorescent protein (GFP) levels were assessed by flow cytometry. One million BM cells obtained after transduction of Kat2aWT and Kat2aNULL pools were injected per animal into >8-week-old, C57/BL6 mice, which were lethally irradiated [2 × 5.5 gray (Gy)]. Seventeen mice per group were injected for preleukemia and leukemia studies. Leukemia survival studies were performed as two independent experiments. Leukemic mice were collected on the basis of symptoms of hunched posture, inappetence, and lethargy.
BM cells obtained from Idh1R132H-transformed Kat2aHET and Kat2aNULL animals 20 weeks after pIpC were injected into CD45.1, C57/BL6 mice (n = 8 per group), which were sublethally irradiated (1 × 8 Gy). Bones and spleens were collected after 1 year of transplantation and analyzed by flow cytometry.
CFC assays
For analysis of preleukemia samples from RT1(9a) and Idh1R132H, 50,000 BM cells were plated in MethoCult M3434 (STEMCELL Technologies), following the manufacturer’s protocols. Colonies were scored 7 to 10 days after plating. Cells were collected from plates, washed, dispersed to a single-cell suspension, and serially replated for transformation analysis.
In S6K1 inhibition studies, 10,000 Kat2aWT BM cells transduced with RT1(9a) or carrying the recombined Idh1R132H allele, were plated in MethoCult M3434 containing freshly added dimethyl sulfoxide (DMSO) (vehicle) or 10 μM PF4708671 (Tocris) with a final concentration of 0.1% DMSO. Colonies were scored as above.
Leukemia maintenance in vitro
Pooled BM cells collected from two to three 12-week-old Kat2a floxed Mx1-Cre−/− animals without pIpC treatment were retrovirally transduced with RT1(9a) and serially replated in MethoCult M3434 for a total of three platings (4000 cells per plating). At plate 3, cells were collected, transduced with a MIGR-Cre-OP-Puro (Cre+) or a MIGR-OP-Puro (empty) retrovirus, and cultured for 48 hours under puromycin selection, as described (20). Transduced and antibiotic-selected cells were assessed for colony formation over two rounds of plating in MethoCult M3434 (4000 cells per plating per condition) in the presence of puromycin. Colonies were scored 7 to 10 days after plating.
Flow cytometry analysis
Cell surface analysis of mouse BM and spleen was performed as described (12), using the antibodies in Table 2. Where indicated in the text, the following gating strategies were used for quantification of hematopoietic stem and progenitor cell compartments: hematopoietic stem cell (HSC), Lin−cKit+Sca1+CD34−Flt3−; multipotent progenitor (MPP), Lin−cKit+Sca1+CD34+Flt3−; lympho-myeloid primed progenitor (LMPP), Lin−cKit+Sca1+CD34+Flt3+; common myeloid progenitor (CMP), Lin−cKit+Sca1−CD34+/lowCD16/32low; granulocyte-monocyte progenitor (GMP), Lin−cKit+Sca1+CD34+CD16/32high; megakaryocyte–erythroid progenitor (MEP), Lin−cKit+Sca1+CD34−CD16/32−; Lin−, CD3e−B220−Gr1−CD11b−Ter119−.
Table 2. Antibodies used in flow cytometry analysis, cell sorting and lineage selection.
| Antibody | Fluorochrome | Catalog ID | Clone | Dilution | Supplier |
| CD45R/ B220 | APC-Cy7 | 103223 | RA3-6B2 | 1:50 | BioLegend |
| CD45R/ B220 | PerCP-Cy5.5 | 103235 | RA3-6B2 | 1:100 | BioLegend |
| CD117/c-Kit | APC-Cy7 | 105826 | 2B8 | 1:50 | BioLegend |
| CD117/c-Kit | BV785 | 105841 | 2B8 | 1:100 | BioLegend |
| CD11b/Mac1 | AF700 | 101222 | M1/70 | 1:200 | BioLegend |
| CD16/32/FcγR | PE | 101308 | 93 | 1:100 | BioLegend |
| CD16/32/FcγR | BV421 | 101331 | 93 | 1:200 | BioLegend |
| CD34 | APC | 128612 | HM34 | 1:100 | BioLegend |
| F4/80 | PE | 123109 | BM8 | 1:100 | BioLegend |
| Gr1 | PB | 108430 | RB6-8C5 | 1:100 | BioLegend |
| CD14 | PE-Cy7 | 123315 | Sa14–2 | 1:200 | BioLegend |
| CD48 | BV510 | 103443 | HM48–1 | 1:200 | BioLegend |
| Sca1 | PE-Cy7 | 108114 | D7 | 1:100 | BioLegend |
| Sca1 | PerCP | 108121 | D7 | 1:100 | BioLegend |
| CD45R/B220 (Lin) | Biotin | 103204 | RA3-6B2 | 1:300 | BioLegend |
| Ter119 (Lin) | Biotin | 116204 | Ter119 | 1:300 | BioLegend |
| Gr1 (Lin) | Biotin | 108404 | RB6-8C5 | 1:300 | BioLegend |
| CD3e (Lin) | Biotin | 100304 | 145-2 C11 | 1:300 | BioLegend |
| CD11b (Lin) | Biotin | 101204 | M1/70 | 1:300 | BioLegend |
| CD48 | Biotin | 103409 | HM48-1 | 1:100 | BioLegend |
| Streptavidin | BV421 | 405226 | – | 1:200 | BioLegend |
| Streptavidin | BV605 | 405229 | – | 1:200 | BioLegend |
| Streptavidin | APC-Cy7 | 405208 | – | 1:200 | BioLegend |
| Nanobeads | Streptavidin | 76447 | – | 1:10 | BioLegend |
| Hoechst 33258 | – | H3569 | – | 1:10,000 | Invitrogen |
| Click-iT Cell Reaction Buffer Kit |
AF647 azide | A10277 | – | 1:500 | Invitrogen |
scRNA-seq preparation and analysis
Preleukemia BM samples were collected from individual animals engrafted with RT1(9a)-transduced Kat2aWT or Kat2aNULL pooled cells, 2 and 4 months after transplantation, and stored at −150°C. Cells were thawed, recovered in R20 medium, and sorted on an Influx sorter (BD) as Hoechst 33258-negative (live), GFP+ [RT1(9a) reporter], and cKit+ (early progenitors) singlets. Sorted cells were immediately used for library preparation with the Chromium Next GEM Single-Cell 3′ GEM, Library, and Gel Bead Kit v2 (10X Genomics). Libraries were quality-controlled and underwent paired-end sequencing on an Illumina NextSeq 500 Sequencer. Library preparation and sequencing were performed at Cancer Research UK (CRUK) Cambridge Research Institute. Raw single-cell RNA-seq fastq reads were analyzed using Cell Ranger software (v2.2) to obtain the cell-gene count matrix (Table 3). Preprocessing analysis yielded a gene-count matrix with 1675 cells in total with a median UMI count of 5939 and 1575 median genes per cell. The count-matrix data were preprocessed with Seurat v2.4 (41) as described (12). Each cell that expressed less than 500 genes was considered to be of poor quality and was filtered out. Differential gene expression was obtained with DESeq2 (42), as per the implementation in Seurat v2.4, for pairwise comparisons between genotypes, globally or at individual time points, between two individual cell compartments, or between one individual compartment and the remaining cells, e.g., for BAP- and MAP-associated signatures. For genes with adjusted P < 0.05, differential expression calculated using log2 fold change (FC) was deemed as significant where |log2 FC| > 0.26 (20% fold difference in averages). Gene ontology analysis was performed using Panther 14.0 (43), selecting Fisher’s exact test with Bonferroni correction. Transcriptional variability analysis used pairwise distance between gene correlations as a measure of cellular heterogeneity, by identifying the top 500 highly variable genes based on distance-to-median (DM) and calculating Spearman correlation coefficients between all gene pairs (27). Pseudo-time analysis was performed using Monocle v3.0 (21) separately for Kat2aWT and Kat2aNULL cells. Cell identities were attributed by inspection of the presence and level of transcripts of lineage-associated markers and transcription factors commonly used in classification of hematopoietic progenitors (see fig. S5, C and D) (22, 23) and manual annotation of contiguous or discrete regions on the basis of dominant combinatorial marker expression. LMPP-like space, Ly6e+CD34+Flt3+CD79a−CD14− (also Gata2+Myb+), was observed at the origin of the trajectories. Ly6e+CD34+Flt3−CD79a−CD14−FcgR+ (also Cebpa+), Ly6e+CD34−Flt3−CD79a+CD14−FcgR− (also CD19+Il7r+Ebf+), and Ly6e+CD34−Flt3−CD79a−CD14+FcgR+ (also Mafb+) were GMP-like, B cell–affiliated, and monocyte/macrophage-affiliated, respectively. Uniform manifold approximation and projection (UMAP) plot regions were consistent between genotype-specific and global pseudo-time trajectories. RNA velocity analysis was performed using velocyto.py analysis pipeline (25). Raw fastq files were used to generate .loom files that were used for velocity calculation for individual genotypes and individual time points.
Table 3. Cell-gene count matrix specification.
| Total number of cells sequenced | 1767 |
| Median number of genes per cell | 1575 |
| Kat2aWT 2 months | 379 |
| Kat2aNULL 2 months | 369 |
| Kat2aWT 4 months | 518 |
| Kat2aNULL 4 months | 501 |
Bulk RNA-seq analysis
Total RNA was extracted from mouse BM aspirates, following enucleated cell lysis. Paired-end RNA-seq reads were mapped to the mouse genome (UCSC mm10) using STAR with default parameters (44). The total number of reads aligning to the exons of each gene [as per GENCODE vM4 (45)] were counted using HTSeq count (46). Read counts were used for FPKM (Fragments Per Kilobase of transcript per Million mapped reads) computation and differential expression analyses using DESeq2 between sex-matched preleukemia and AML samples (42). Preleukemia samples analyzed were: Idh1R132H (n = 2), Idh1R132H Npm1c (n = 2), Npm1c N-RasG12D Idh1R132H (n = 2), and Idh1WT (n = 6 female, 4 male); AML samples analyzed were: N-RasG12D Idh1R132H (n = 3) and Npm1c N-RasG12D Idh1R132H (n = 3).
Measurement of protein synthesis
Protein synthesis rates were estimated using OP-Puro (Thermo Fisher Scientific) incorporation, as described (12). In detail, 1 million RT1(9a) Kat2aWT or Idh1R132H Kat2aHET cells treated with 10 μM S6K1inh PF4708671 versus DMSO (vehicle) were collected from successive replating of colony-forming assays and cultured for 2 hours in the presence of cytokines, mSCF (20 ng/ml), mIL-3 (10 ng/ml), and mIL-6 (10 ng/ml) in R20 medium. In other assays, Idh1R132H Kat2aHET versus Kat2aNULL BM cells collected 20 weeks after pIpC treatment for locus activation/excision were thawed and cultured overnight in the same conditions. A final concentration of 12.5 μM OP-Puro was added directly to 80% of each culture for the last hour of the culture period; the remainder 20% cells were treated with phosphate-buffered saline (PBS) and processed in parallel as control. After incubation, cells were washed with ice-cold PBS without Ca2+ or Mg2+ (Sigma-Aldrich) and resuspended in PBS/10% FBS for cell surface staining with c-Kit–APCC7, CD11b-biotin, and Gr1-biotin (all from BioLegend; see Table 2), followed by Streptavidin Brilliant Violet 605 (also from BioLegend), both staining steps for 30 min on ice. After washing, cells were fixed in 1% paraformaldehyde in PBS for 15 min on ice protected from light, washed, and permeabilized in PBS/3% FBS/0.1% saponin (permeabilization buffer) at room temperature, in the dark, for 5 min. Cells were washed and used immediately in the azide-alkyne cyclo-addition reaction with the Click-iT Cell Reaction Buffer Kit (Thermo Fisher Scientific, C10269) and Alexa Fluor 647–Azide (Thermo Fisher Scientific, A10277) with a master reaction solution freshly prepared for immediate use, as per the manufacturer’s instructions. Alexa Fluor 647–Azide was used at a final concentration of 5 μM. The reaction proceeded in the dark at room temperature for 30 min; cells were washed twice in permeabilization buffer and resuspended in PBS, 5 min before flow cytometry analysis.
Statistical analysis
Experiments were performed in triplicate, with any exceptions specifically indicated in the text or figure legends. Data are plotted as mean ± SD with statistical tests described in the respective figure legends. Statistical analysis was performed using GraphPad Prism 8.0 software. R language was used for analysis of single-cell RNA-seq data.
Acknowledgments
We would like to thank the following: Central Biomedical Services of the University of Cambridge for expert animal husbandry; CRUK Genomics Core Facility at the Cambridge Research Institute and the Wellcome Trust Sanger Research Institute Genomics Core Facility for library preparation and next-generation sequencing; the Flow Cytometry facilities at the Cambridge Institute for Medical Research, the NIHR Cambridge BRC Cell Phenotyping Hub, and the Department of Pathology of the University of Cambridge (J. Cerveira) for cell sorting; R. Bandiera for discussions and reagent sharing; and M. Wayland for assistance with Cell Ranger installation and 10X Genomics data matrix generation.
Funding: This study was funded by a Lady Tata Memorial Trust International PhD Scholarship to S.G. (2017-2021), a Kay Kendall Leukaemia Fund Intermediate Fellowship to C.P. (KKL888), Cancer Research UK (C22324/A23015) and Wellcome Trust (WT098051) Senior Fellowships to G.S.V., and Cancer Research UK (C18680/A25508) and European Research Council (647685) grants to B.J.H. This research was also supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014) and was funded in part by the Wellcome Trust, who supported the Wellcome–MRC Cambridge Stem Cell Institute (203151/Z/16/Z). C.P. was funded by a Leuka John Goldman Fellowship for Future Science (2017-2019), a Wellcome Trust/University of Cambridge ISSF Grant (2019), and a Start-up Grant from Brunel University London CHMLS (2019-2021). Work in G.S.V. laboratory is also funded by the European Research Council, Kay Kendall Leukaemia Fund, Blood Cancer UK, and the Wellcome Trust. S.G. received partial PhD studentships from the Trinity Henry Barlow and the Cambridge Commonwealth, European and International Trusts, and additional support from Murray Edwards College and the University of Cambridge Lundgren Award. L.D. is a student in the University of Groningen Masters’ Program in Molecular Medicine and Innovative Treatment (MMIT), University Medical Center Groningen, The Netherlands.
Author contributions: Study conception: C.P. Experimental design: S.G., O.M.D., G.S.V., and C.P. Data collection: S.G., O.M.D., A.F.D., O.W.C., C.M.C., G.G., J.R., J.C., M.G., L.D., R.J.A., N.A.-J., and V.H.-H. Data analysis and interpretation: S.G., O.M.D., A.F.D., O.W.C., C.M.C., G.S.V., and C.P. Critical reagents: S.P. and B.J.H. Writing: S.G. and C.P., with contributions from O.M.D. and G.S.V. All authors approved the final version of the manuscript.
Competing interests: V.H.-H. is cofounder and CSO of Axovia Therapeutics. S.P. is the CEO of NonExomics Inc. G.S.V. is consultant to STRM.BIO and has a research grant from AstraZeneca. Axovia, NonExomics, STRM.BIO, and AstraZeneca did not provide funding to this study and did not influence study design, execution, data analysis, or interpretation. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. scRNA-seq and bulk RNA-seq data were deposited in ArrayExpress with accession number E-MTAB-10853 and ERP006862, respectively. Code used for single-cell RNA-seq data analysis was deposited in Zenodo and can be accessed as G. Shikha and P. Cristina (2022). scRNA-seq analysis of RUNX1-RUNX1T1(9a) preleukemia, Zenodo; https://doi.org/10.5281/zenodo.6584118.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
References
Other Supplementary Material for this manuscript includes the following:
Supplementary Files S1 to S9
REFERENCES AND NOTES
- 1.Greaves M., Maley C. C., Clonal evolution in cancer. Nature 481, 306–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGranahan N., Swanton C., Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Zahir N., Sun R., Gallahan D., Gatenby R. A., Curtis C., Characterizing the ecological and evolutionary dynamics of cancer. Nat. Genet. 52, 759–767 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Persi E., Wolf Y. I., Horn D., Ruppin E., Demichelis F., Gatenby R. A., Gillies R. J., Koonin E. V., Mutation–Selection balance and compensatory mechanisms in tumour evolution. Nat. Rev. Genet. 22, 251–262 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Landau D. A., Clement K., Ziller M. J., Boyle P., Fan J., Gu H., Stevenson K., Sougnez C., Wang L., Li S., Kotliar D., Zhang W., Ghandi M., Garraway L., Fernandes S. M., Livak K. J., Gabriel S., Gnirke A., Lander E. S., Brown J. R., Neuberg D., Kharchenko P. V., Hacohen N., Getz G., Meissner A., Wu C. J., Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan H., Jiang Y., Boi M., Tabbò F., Redmond D., Nie K., Ladetto M., Chiappella A., Cerchietti L., Shaknovich R., Melnick A. M., Inghirami G. G., Tam W., Elemento O., Epigenomic evolution in diffuse large B-cell lymphomas. Nat. Commun. 6, 6921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network, Ley T. J., Miller C., Ding L., Raphael B. J., Mungall A. J., Robertson A. G., Hoadley K., Triche T. J. Jr., Laird P. W., Baty J. D., Fulton L. L., Fulton R., Heath S. E., Kalicki-Veizer J., Kandoth C., Klco J. M., Koboldt D. C., Kanchi K.-L., Kulkarni S., Lamprecht T. L., Larson D. E., Lin L., Lu C., McLellan M. D., McMichael J. F., Payton J., Schmidt H., Spencer D. H., Tomasson M. H., Wallis J. W., Wartman L. D., Watson M. A., Welch J., Wendl M. C., Ally A., Balasundaram M., Birol I., Butterfield Y., Chiu R., Chu A., Chuah E., Chun H.-J., Corbett R., Dhalla N., Guin R., He A., Hirst C., Hirst M., Holt R. A., Jones S., Karsan A., Lee D., Li H. I., Marra M. A., Mayo M., Moore R. A., Mungall K., Parker J., Pleasance E., Plettner P., Schein J., Stoll D., Swanson L., Tam A., Thiessen N., Varhol R., Wye N., Zhao Y., Gabriel S., Getz G., Sougnez C., Zou L., Leiserson M. D. M., Vandin F., Wu H.-T., Applebaum F., Baylin S. B., Akbani R., Broom B. M., Chen K., Motter T. C., Nguyen K., Weinstein J. N., Zhang N., Ferguson M. L., Adams C., Black A., Bowen J., Gastier-Foster J., Grossman T., Lichtenberg T., Wise L., Davidsen T., Demchok J. A., Shaw K. R. M., Sheth M., Sofia H. J., Yang L., Downing J. R., Eley G., Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson M. A., Prinjha R. K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.-I., Robson S. C., Chung C.-w., Hopf C., Savitski M. M., Huthmacher C., Gudgin E., Lugo D., Beinke S., Chapman T. D., Roberts E. J., Soden P. E., Auger K. R., Mirguet O., Doehner K., Delwel R., Burnett A. K., Jeffrey P., Drewes G., Lee K., Huntly B. J. P., Kouzarides T., Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernt K. M., Zhu N., Sinha A. U., Vempati S., Faber J., Krivtsov A. V., Feng Z., Punt N., Daigle A., Bullinger L., Pollock R. M., Richon V. M., Kung A. L., Armstrong S. A., MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shlush L. I., Zandi S., Mitchell A., Chen W. C., Brandwein J. M., Gupta V., Kennedy J. A., Schimmer A. D., Schuh A. C., Yee K. W., McLeod J. L., Doedens M., Medeiros J. J. F., Marke R., Kim H. J., Lee K., McPherson J. D., Hudson T. J.; HALT Pan-Leukemia Gene Panel Consortium, Brown A. M. K., Yousif F., Trinh Q. M., Stein L. D., Minden M. D., Wang J. C. Y., Dick J. E., Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmerman M. R. C., Hong W.-J., Weissman I. L., Medeiros B. C., Majeti R., Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. U.S.A. 111, 2548–2553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingues A. F., Kulkarni R., Giotopoulos G., Gupta S., Vinnenberg L., Arede L., Foerner E., Khalili M., Adao R. R., Johns A., Tan S., Zeka K., Huntly B. J., Prabakaran S., Pina C., Loss of KAT2A enhances transcriptional noise and depletes acute myeloid leukemia stem-like cells. eLife 9, e51754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzelepis K., Koike-Yusa H., De Braekeleer E., Li Y., Metzakopian E., Dovey O. M., Mupo A., Grinkevich V., Li M., Mazan M., Gozdecka M., Ohnishi S., Cooper J., Patel M., Kerrell T. M., Chen B., Domingues A. F., Gallipoli P., Teichmann S., Ponstingl H., Dermott U. M., Saez-Rodriguez J., Huntly B. J. P., Iorio F., Pina C., Vassiliou G. S., Yusa K., A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moris N., Edri S., Seyres D., Kulkarni R., Domingues A. F., Balayo T., Frontini M., Pina C., Histone acetyltransferase KAT2A stabilizes pluripotency with control of transcriptional heterogeneity. Stem Cells 36, 1828–1838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somervaille T. C. P., Cleary M. L., Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 10, 257–268 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V. I., Paschka P., Roberts N. D., Potter N. E., Heuser M., Thol F., Bolli N., Gundem G., Van Loo P., Martincorena I., Ganly P., Mudie L., Laren S. M., Meara S. O.’, Raine K., Jones D. R., Teague J. W., Butler A. P., Greaves M. F., Ganser A., Döhner K., Schlenk R. F., Döhner H., Campbell P. J., Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan M., Kanbe E., Peterson L. F., Boyapati A., Miao Y., Wang Y., Chen I.-M., Chen Z., Rowley J. D., Willman C. L., Zhang D.-E., A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 12, 945–949 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Paschka P., Schlenk R. F., Gaidzik V. I., Habdank M., Krönke J., Bullinger L., Späth D., Kayser S., Zucknick M., Götze K., Horst H.-A., Germing U., Döhner H., Döhner K., IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 28, 3636–3643 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Rasmussen K. D., Jia G., Johansen J. V., Pedersen M. T., Rapin N., Bagger F. O., Porse B. T., Bernard O. A., Christensen J., Helin K., Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 29, 910–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basheer F., Giotopoulos G., Meduri E., Yun H., Mazan M., Sasca D., Gallipoli P., Marando L., Gozdecka M., Asby R., Sheppard O., Dudek M., Bullinger L., Döhner H., Dillon R., Freeman S., Ottmann O., Burnett A., Russell N., Papaemmanuil E., Hills R., Campbell P., Vassiliou G. S., Huntly B. J. P., Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J. Exp. Med. 216, 966–981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C. T., Bryder D., Yang L., Borge O.-J., Thoren L. A. M., Anderson K., Sitnicka E., Sasaki Y., Sigvardsson M., Eirik S., Jacobsen W., Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Akashi K., Traver D., Miyamoto T., Weissman I. L., A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Ptasinska A., Assi S. A., Mannari D., James S. R., Williamson D., Dunne J., Hoogenkamp M., Wu M., Care M., McNeill H., Cauchy P., Cullen M., Tooze R. M., Tenen D. G., Young B. D., Cockerill P. N., Westhead D. R., Heidenreich O., Bonifer C., Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia 26, 1829–1841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manno G. L., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A., Fan J., Borm L. E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P. V., RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raser J. M., O’Shea E. K., Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1814 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammed H., Hernando-Herraez I., Savino A., Nichols J., Marioni J. C., Reik W., Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 20, 1215–1228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolodziejczyk A. A., Kim J. K., Tsang J. C. H., Ilicic T., Henriksson J., Natarajan K. N., Tuck A. C., Gao X., Bühler M., Liu P., Marioni J. C., Teichmann S. A., Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17, 471–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai X., Gao L., Teng L., Ge J., Oo Z. M., Kumar A. R., Gilliland D. G., Mason P. J., Tan K., Speck N. A., Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17, 165–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christen F., Hoyer K., Yoshida K., Hou H.-A., Waldhueter N., Heuser M., Hills R. K., Chan W., Hablesreiter R., Blau O., Ochi Y., Klement P., Chou W.-C., Blau I.-W., Tang J.-L., Zemojtel T., Shiraishi Y., Shiozawa Y., Thol F., Ganser A., Löwenberg B., Linch D. C., Bullinger L., Valk P. J. M., Tien H.-F., Gale R. E., Ogawa S., Damm F., Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): An international study on 331 patients. Blood 133, 1140–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Mandoli A., Singh A. A., Prange K. H. M., Tijchon E., Oerlemans M., Dirks R., Huurne M. T., Wierenga A. T. J., Jannsen-Megens E. M., Berentsen K., Sharifi N., Kim B., Matarese F., Nguyen L. N., Hubner N. C., Rao N. A., van den Akker E., Altucci L., Vellenga E., Stunnenberg H. G., Martens J. H. A., The hematopoietic transcription factors RUNX1 and ERG prevent AML1-ETO oncogene overexpression and onset of the apoptosis program in t(8;21) AMLs. Cell Rep. 17, 2087–2100 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Wichmann C., Coco I. Q.-L., Yildiz Ö., Chen-Wichmann L., Weber H., Syzonenko T., Döring C., Brendel C., Ponnusamy K., Kinner A., Brandts C., Henschler R., Grez M., Activating c-KIT mutations confer oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors. Leukemia 29, 279–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arede L., Foerner E., Wind S., Kulkarni R., Domingues A. F., Giotopoulos G., Kleinwaechter S., Mollenhauer-Starkl M., Davison H., Chandru A., Asby R., Samarista R., Gupta S., Forte D., Curti A., Scheer E., Huntly B. J. P., Tora L., Pina C., KAT2A complexes ATAC and SAGA play unique roles in cell maintenance and identity in hematopoiesis and leukemia. Blood Adv. 6, 165–180 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ecker S., Pancaldi V., Rico D., Valencia A., Higher gene expression variability in the more aggressive subtype of chronic lymphocytic leukemia. Genome Med. 7, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gachet S., el-Chaar T., Avran D., Genesca E., Catez F., Quentin S., Delord M., Thérizols G., Briot D., Meunier G., Hernandez L., Pla M., Smits W. K., Buijs-Gladdines J. G., van Loocke W., Menschaert G., André-Schmutz I., Taghon T., van Vlierberghe P., Meijerink J. P., Baruchel A., Dombret H., Clappier E., Diaz J. J., Gazin C., de Thé H., Sigaux F., Soulier J., Deletion 6q drives T-cell leukemia progression by ribosome modulation. Cancer Discov. 8, 1614–1631 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Signer R. A. J., Magee J. A., Salic A., Morrison S. J., Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 508, 49–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mill C. P., Fiskus W., DiNardo C. D., Birdwell C., Davis J. A., Kadia T. M., Takahashi K., Short N., Daver N., Ohanian M., Borthakur G., Kornblau S. M., Green M. R., Qi Y., Su X., Khoury J. D., Bhalla K. N., Effective therapy for AML with RUNX1 mutation by cotreatment with inhibitors of protein translation and BCL2. Blood 139, 907–921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., de Jong P. J., Stewart A. F., Bradley A., A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M., High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139–140 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Dovey O. M., Cooper J. L., Mupo A., Grove C. S., Lynn C., Conte N., Andrews R. M., Pacharne S., Tzelepis K., Vijayabaskar M. S., Green P., Rad R., Arends M., Wright P., Yusa K., Bradley A., Varela I., Vassiliou G. S., Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood 130, 1911–1922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., Kokocinski F., Aken B. L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., Mukherjee G., Rajan J., Despacio-Reyes G., Saunders G., Steward C., Harte R., Lin M., Howald C., Tanzer A., Derrien T., Chrast J., Walters N., Balasubramanian S., Pei B., Tress M., Rodriguez J. M., Ezkurdia I., van Baren J., Brent M., Haussler D., Kellis M., Valencia A., Reymond A., Gerstein M., Guigó R., Hubbard T. J., GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S., Pyl P. T., Huber W., HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., Lachmann A., Wojciechowicz M. L., Kropiwnicki E., Jagodnik K. M., Jeon M., Ma’ayan A., Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
References
Supplementary Files S1 to S9