Abstract
Cancer remains one of the leading causes of death, and early detection of this disease is crucial for increasing survival rates. Although cancer can be diagnosed following tissue biopsy, the biopsy procedure is invasive; liquid biopsy provides an alternative that is more comfortable for the patient. While blood, urine, and cerebral spinal fluid can all be used as a source of liquid biopsy, saliva is an ideal source of body fluid that is readily available and easily collected in the most noninvasive manner. Characterization of salivary constituents in the disease setting provides critical data for understanding pathophysiology and the evaluation of diagnostic potential. The aim of saliva diagnostics is therefore to develop a rapid and noninvasive detection of oral and systemic diseases that could be used together with compact analysis systems in the clinic to facilitate point-of-care diagnostics.
Keywords: saliva liquid biopsy, salivaomics, saliva exosomics, salivary exosomes, circulating biomarkers
1. INTRODUCTION
Saliva is a critical bodily fluid required for the digestion of food and maintenance of good oral health. It contains secreted enzymes, hormones, cytokines, and antibodies that act as mediators of salivary functions. In addition, saliva contains microorganisms and cellular debris (1). There are three pairs of major salivary glands (parotid, submandibular, and sublingual) and many minor salivary glands dispersed throughout the oral mucosa (2). Owing to their proximity to blood vessels, the salivary glands are a rich source of metabolite exchange between the oral cavity and the circulatory system (3). Indeed, many proteins found in human serum can also be detected in saliva; this suggests that saliva could be used as a proxy for the measurement of disease-related circulating biomarkers (4). In the following review we provide a comprehensive overview of salivary biomarkers and the salivary exosomes in which they are encapsulated. We also describe a novel electrochemical sensing technology (electric field–induced release and measurement, or EFIRM) that can be used for biomarker detection and disease monitoring; this technology is pioneered to implement future point-of-care saliva testing methodologies.
2. SALIVAOMICS
Rising to prominence over the last decade, salivaomics refers to the integrated analysis of multiple large-scale molecular readouts from this important biological fluid (5, 6). Such readouts include genomics, epigenomics, transcriptomics, proteomics, microbiomics, and metabolomics. There are several challenges associated with obtaining a clear and disease-relevant signal from salivary samples. For example, components of food and the presence of bacteria in the oral cavity can contribute to noise in salivary data sets (7). The composition of saliva is also affected by natural circadian rhythms (8), the physical action of mastication (9), and the activity of amylase, which is abundant in saliva. RNAs (and many proteins, including histatins, statherin, and acidic prolinerich polypeptides) are also highly labile when taken out of the buffered environment of the saliva (10). Thus, methods to stabilize these molecular markers are paramount in order to preserve a representative snapshot of the true physiological state; thus, many protocols include protease and RNase inhibitors in extraction buffers. Finally, it is increasingly clear that analysis of bulk saliva samples may mask physiologically relevant signals from small subpopulations of cells or metabolites. Therefore, single-cell technologies or ultrasensitive methods of detection will ultimately be required to obtain more accurate information from saliva.
2.1. Salivary Genomics
Analysis of direct tumor biopsy is perhaps the most accurate approach for molecular diagnostics in cancer. However, taking tumor samples can be extremely uncomfortable for the patient, and repeat biopsy for disease monitoring may not always be possible. Furthermore, some deep-seated tumors may not be amenable to this approach. These disadvantages prompted the search for less-invasive methods for cancer diagnosis and surveillance. Circulating tumor DNA (ctDNA) is found in the serum and is composed of genomic DNA that is shed from the original tumor (11, 12). The cancer-specific mutational signatures of ctDNA can be differentiated from those of DNA from noncancerous tissues. Multiple studies have shown a high concordance between mutational profiles in ctDNA and those in the primary tumor (13-17). Furthermore, the relative abundance of ctDNA, or of specific mutations therein, can be used to monitor disease response to therapeutic intervention, and ctDNA samples can be taken repeatedly, which facilitates real-time surveillance of treatment (18-21). This ctDNA-based liquid biopsy approach is rapidly being adopted in many preclinical and clinical settings.
Although most liquid biopsies are taken by needle aspiration from the blood, it is possible that salivary fluid may offer an even less-invasive means of disease monitoring. The stability and relatively high quality of salivary DNA makes this an even more attractive possibility (22-24). Although diagnostics and monitoring using salivary ctDNA are in their infancy, a recent study indicated their utility in head and neck squamous cell carcinoma (HNSCC) (25, 26). Key cancer-associated somatic mutations and the presence of human papillomaviruses (HPV16 and 18) were evaluated among a cohort of 93 HNSCC patients (including 20 patients with early disease; see Table 1 for details). While plasma ctDNA analysis was associated with higher sensitivity for oropharynx, hypopharynx, and larynx cancers (plasma ctDNA: 86–100% versus salivary ctDNA: 47–70%), salivary ctDNA analysis had greater sensitivity for the detection of oral cancer (100% versus 80%). The latter result is likely attributable to the close physical proximity of salivary fluid to the actual tumor. Combined analysis of plasma and salivary ctDNA yielded a 96% detection rate, irrespective of tumor location or stage. Together, these data indicate that the optimal combination of bodily fluids used for ctDNA analysis should be chosen on a tumor type–specific basis.
Table 1.
Summary of saliva and plasma ctDNA biomarkers identified in HNSCC. Clinical and laboratory data were retrieved from a database published by Wang et al. (25)
| Site | ctDNA | % of positivity (number detected/examined) | ||
|---|---|---|---|---|
| Saliva | Plasma | Saliva and plasma | ||
| Oral cavity | TP53 | 100 (36/36) | 85 (11/13) | 100 (13/13) |
| PIK3CA | 100 (2/2) | 50 (1/2) | 100 (2/2) | |
| NOTCH1 | 100 (3/3) | NA | NA | |
| CDKN2A | 100 (2/2) | NA | NA | |
| Translocation | 100 (2/2) | NA | NA | |
| HPV16 DNA | 100 (1/1) | NA | NA | |
| Total | 100 (46/46) | 80 (12/15) | 100 (15/15) | |
| Oropharynx | TP53 | 80 (4/5) | 100 (1/1) | 100 (1/1) |
| PIK3CA | 25 (2/8) | 100 (5/5) | 100 (5/5) | |
| FBXW7 | 67 (2/3) | 100 (3/3) | 100 (3/3) | |
| HPV16 DNA | 41 (7/17) | 92 (11/12) | 92 (11/12) | |
| NRAS | 0 (0/1) | 0 (0/1) | 0 (0/1) | |
| Total | 44 (15/34) | 91 (20/22) | 91 (20/22) | |
| Larynx | TP53 | 70 (7/10) | 86 (6/7) | 100 (7/7) |
| Hypopharynx | TP53 | 67 (2/3) | 100 (3/3) | 100 (3/3) |
| Overall | 75 (70/93) | 87 (41/47) | 96 (45/47) | |
Abbreviations: ctDNA, circulating tumor DNA; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; NA, not applicable.
2.2. Salivary Transcriptomics
Saliva contains diverse types of RNA transcripts, including messenger RNA (mRNA), piwi-interacting RNA (piRNA), and micro RNA (miRNA) (27, 28). Although the value of piRNA as a biomarker remains to be determined, both mRNA and miRNA within the saliva have been used to detect several cancers, including pancreatic (29), breast (30), ovarian (31), and lung malignancies (32). We highlight some examples below and in Table 2.
Table 2.
Salivary RNA biomarkers in cancers
| Cancer | RNA type | Salivary RNA biomarker | Reference |
|---|---|---|---|
| Breast | mRNA | CSTA, TPT1, IGF2BP1, GRM1, GRIK1, H6PD, MDM4, S100A8 | 30 |
| Esophageal | miRNA | miR-10b, miR-98, miR-144, miR-363, miR-451 | 81 |
| Head and neck | mRNA | DUSP1, H3F3A, IL1B, IL8, OAZ1, S100P, SAT | 33 |
| miRNA | miR-125a, miR-200a | 36 | |
| Lung | mRNA | CCNI, FGF19, GREB1, FRS2, EGFR | 32 |
| Ovarian | mRNA | AGPAT1, B2M, IER3, IL1B, BASP1 | 31 |
Abbreviations: mRNA, messenger RNA; miRNA/miR-, micro RNA.
Profiling of saliva samples from patients with oral squamous cell carcinoma identified multiple mRNA biomarkers (33). A subset of four mRNAs (IL1B, OAZ1, SAT, and IL8) was sufficient for use in a logistic regression model to provide 91% sensitivity and 91% specificity for the detection of cancer.
Salivary miRNAs are packaged in salivary exosomes, where they are protected from RNase-dependent degradation (34, 35). Consistent with the general dysregulation of miRNAs in tumor cells themselves, the levels of specific miRNAs in the saliva of cancer patients are altered in comparison to those of healthy individuals. For example, the levels of miR-125a and miR-200a were significantly lower in saliva from oral cancer patients than healthy patients (36). Conversely, the levels of miR-27b and miR-31 were significantly higher in the saliva of oral cancer patients (37, 38). miR-139 and miR-31 reverted to baseline levels following excision of the malignant lesions, suggesting that these miRNAs could serve as prognostic biomarkers (37, 39).
Despite these intriguing findings, further preclinical and clinical studies are required to validate the roles of salivary miRNAs as disease biomarkers. It will be especially important to standardize the way salivary exosome miRNAs are detected and analyzed. Furthermore, researchers must find ways to deconvolute salivary miRNA signals that originated in immune cells versus tumor or salivary cells. This is critical, because systemic or local inflammation may perturb miRNA expression and generate variability, even within the same individual (40). It will be useful to crossreference data from future studies with the miRNA database, miRandola, which is a large catalog of extracellular noncoding RNAs found in a variety of diseases (http://mirandola.iit.cnr.it/) (41).
2.3. Salivary Proteomics
To the best of our knowledge, the first attempt at cancer diagnosis using salivary protein was made by Hoerman et al. (42) more than 60 years ago; the group showed that prostate cancer patients had elevated acid phosphatase enzymatic activity in parotid saliva. Since then, the advent of high-throughput mass spectrometry combined with bioinformatics has given rise to the field of proteomics, which holds great promise for disease detection and monitoring.
Although in its infancy, there are clear signs that salivary proteomics will prove extremely useful. For example, a US-based consortium has generated a comprehensive catalog of the salivary proteome of healthy individuals, identifying 1,166 proteins in parotid and submandibular/sublingual gland ductal saliva (43). The data are publicly available via the Human Salivary Proteome Wiki (https://salivaryproteome.nidcr.nih.gov). Between 20% and 30% of the salivary proteome overlaps with the plasma proteome, indicating that many salivary constituents are derived from the blood (4, 44). This observation, together with the close physical proximity of saliva and blood, suggests that saliva could be used as a proxy to detect disease. Unlike serum proteins, salivary proteins appear to be more susceptible to degradation (10, 45). Indeed, they degrade rapidly even during saliva collection and handling, which may compromise downstream experiments and limit the application of saliva-based methods (46). Protease inhibitors can be used to stabilize salivary proteins, thereby enabling the storage of saliva samples for up to two weeks without significant degradation (47). Table 3 summarizes salivary proteins that may have potential utility as biomarkers for cancer detection or disease monitoring.
Table 3.
Salivary protein biomarkers for cancers
| Cancer | Sample | Salivary protein biomarker | Reference |
|---|---|---|---|
| Breast | Whole saliva | EGF | 82 |
| ERBB2 | 83 | ||
| CA15–3, ERBB2 | 84 | ||
| VEGF, EGF, CEA | 85 | ||
| CA6 | 30 | ||
| LRP | 86 | ||
| Gastric | Whole saliva | CSTB, TPI1, DMBT1, CALML3, IGH, IL1RA | 87 |
| Head and neck | Whole saliva | A1BG, CFB | 88 |
| M2BP, MRP14, CD59, CAT, PFN | 89 | ||
| FGB, S100, TF, IGHG, CFL1 | 90 | ||
| ADA | 91 | ||
| IL-8, M2BP, IL-1B | 92 | ||
| Salivary EVs | A2M, HPa, MUC5B, LGALS3BP, IGHA1, PIP, PKM1/M2, GAPDH | 93 | |
| Lung | Whole saliva | HP, AZGP1, CALPR | 94 |
| Salivary EVs | Annexin A1, A2, A3, A5, A6, A11, NPRL2, CEACAM1, HIST1H4A, MUC1, PROM1, TNFAIP3 | 95 | |
| Ovarian | Whole saliva | CA125 | 96 |
Abbreviation: EV, extracellular vesicle.
3. SALIVA EXOSOMICS: NEXT-GENERATION SALIVAOMICS
Exosomes are nanosized extracellular vesicles with a diameter between 30 and 100 nm that have been isolated from virtually all types of body fluid, including saliva (48, 49). They are derived from endosomal membranes and are shuttled to the extracellular space during exocytosis. They are critical transporters of cell type–specific cargos that are delivered locally to the microenvironment and systemically via the vasculature. By relaying molecular information from their parental cell of origin to recipient cells, they play important roles in intercellular signaling and cellular homeostasis. Given their biological role in cancer pathogenesis, exosomes may harbor biomarkers that can be harnessed for detecting and monitoring cancer (50).
While exosomes are present in the saliva of healthy individuals, they may contain disease-related biomarkers from tumor cells that have been packaged and transported to the salivary glands (51). The use of these small but information-rich nanovesicles reduces the overall complexity of saliva (52). The term saliva exosomics defines the study of the genomic, transcriptomic, and proteomic features of exosomes and how they impact biological functions in oral and systemic diseases (7). Saliva exosomics is therefore considered next-generation salivaomics. Although the field is still in its infancy, it will undoubtedly reveal many novel facets of saliva biology as research gathers pace.
3.1. Salivary Exosomes
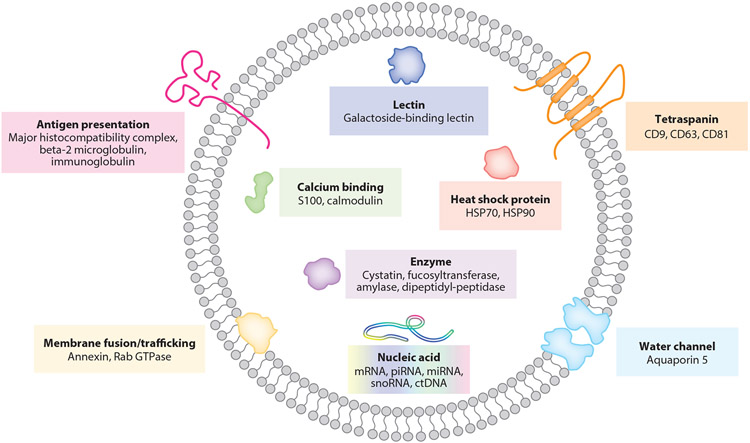
Salivary exosomes are nanoscale extracellular vesicles secreted by the salivary glands and oral epithelial cells (35, 53). Surrounded by a phospholipid bilayer, they carry many cell type–specific cargos (Figure 1). Prominent examples include tetraspanins, calcium-binding proteins, heat shock proteins, water channels, major histocompatibility complexes, and proteins associated with membrane fusion/trafficking (e.g., annexin, Rab GTPases) (54, 55). Almost half of salivary proteins are extracellular (e.g., immunoglobulin chains) or secretory (e.g., serum albumin), suggesting that they are derived from vesicles that originate from circulating lymphocytes and intravascular fluid (4, 44, 55).
Figure 1.
Structure and contents of typical salivary exosomes. The exosome is surrounded by a phospholipid bilayer, carrying many cell type–specific cargos. Abbreviations: ctDNA, circulating tumor DNA; mRNA, messenger RNA; miRNA, micro RNA; piRNA, piwi-interacting RNA; snoRNA, small nucleolar RNA.
Intriguingly, salivary exosomes play a role in the initiation of blood clotting (56). This is because they contain tissue factor, which works in concert with factor VII in the plasma to elicit coagulation. Salivary exosomes accelerate clotting in exosome-depleted plasma, and this can be attenuated by addition of anti-factor VII. These data highlight the importance of these exosomes in a critical physiological process.
Multiple types of RNA are found in salivary exosomes, where they are protected from RNase-dependent degradation (57). The exosomes therefore serve as an enriched source of RNA signaling mediators, primarily composed of piRNA (7.48%), miRNA (6.02%), and small nucleolar RNA (snoRNA; 0.02%) in descending order of abundance (27, 58). mRNA from salivary exosomes can be taken up and translated by recipient cells; this underscores the functional relevance of salivary exosome-mediated RNA transfer (57, 59). The Vesiclepedia (http://www.microvesicles.org) (60) and ExoCarta (http://www.exocarta.org) (61) databases are comprehensive resources for the types of molecular cargos found in extracellular vesicles.
3.2. Structure of Salivary Exosomes
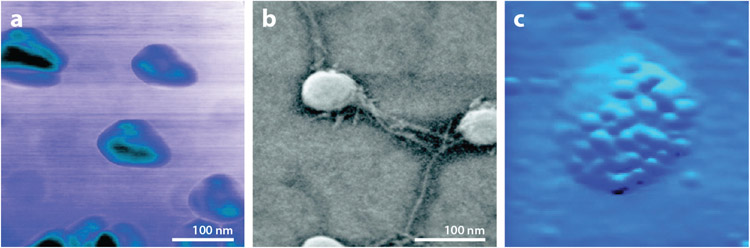
Atomic force microscopy (AFM) and field emission scanning electron microscopy (FESEM) have revealed that salivary exosomes have reversible elastic mechanical properties (57, 62). Specifically, exertion of an outside force causes these exosomes to transition from a spherical morphology to a trilobular structure (Figure 2a,b). Heterogeneity at the surface of salivary exosomes may be due to the presence of CD63 protein in the dense lipid membrane (Figure 2c).
Figure 2.
Nanostructure of salivary exosomes observed under atomic force microscopy (AFM) and field emission scanning electron microscopy (FESEM). (a) AFM phase image of salivary exosomes exhibits a trilobular substructure. Surface contrast is presumably attributed to variable constitutive elements on exosomal membrane (e.g., protein and lipid). (b) FESEM reveals round-shaped salivary exosomes with intervesicular connections. (c) Electron microscopy with anti-CD63 antibody-conjugated gold beads identifies dense tetraspanin molecules on the exosome surface. Figure adapted with permission from Reference 62; copyright 2010 American Chemical Society.
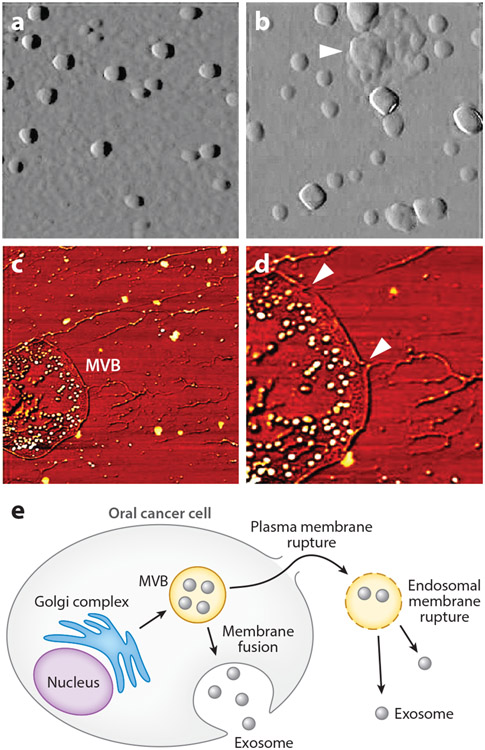
Variations in the nanostructure of salivary exosomes of oral cancer patients compared to healthy individuals may have disease relevance (63). Indeed, cancer-associated salivary exosomes are larger than their normal counterparts (98.3 ± 4.6 nm versus 67.4 ± 2.9 nm; P < 0.05) and appear to be derived from multivesicular bodies (MVBs) (Figure 3a,b). Membrane ruptures and elongated nanofilaments surrounding the lumen of MVBs likely contribute to the release of exosomes (Figure 3c-e). There is also a higher density of CD63 on the surface of cancer salivary exosomes.
Figure 3.
Exosomes and multivesicular bodies (MVBs) seen in saliva of oral cancer patients. (a) Salivary exosomes from healthy donors appear as homogeneous circular structures. (b) Salivary exosomes from oral cancer patients show irregular morphology with varying sizes and vesicle aggregation (arrow). (c) Elongated intervesicular filaments and exosome-like vesicles in MVBs are observed in cancer saliva. (d) At higher resolution, membrane ruptures are observed in cancer salivary MVBs (arrows). (e) Schematic of MVB endosomal membrane rupture and exosome release from oral cancer cell. Figure adapted with permission from Reference 63; copyright 2011 American Chemical Society.
3.3. Mechanistic Link Between Salivary Exosomes and Systemic Cancer
The discovery of the cancer-specific mutant EGFRvIII mRNA in circulating extracellular vesicles of glioblastoma patients suggested that tumor-derived microvesicles may harbor disease biomarkers (64). Indeed, cancer-derived exosomes are now known to be a rich source of omic information that reflects the genetic composition and status of their parent tumors. For example, miRNAs 21 and 141 are upregulated in serum exosomes of esophageal squamous cell carcinoma and prostate cancer, respectively (65, 66). Several biomarkers have also been detected in serum exosomes associated with pancreatic cancer, including mutant KRAS and TP53 (67) and the membrane-anchored exosomal protein glypican-1 (68). These findings demonstrate that the utility of tumor-derived exosomes in disease monitoring extends well beyond oral cancer and is likely to be broadly applicable.
Murine models have recapitulated some of the features of human pancreatic cancer and salivary exosome production. For example, mRNAs that originated in orthotopic pancreatic tumor xenografts were found in salivary exosomes (69). Furthermore, the biogenesis of tumor exosomes was suppressed by the introduction of a dominant-negative RAB11 GTPase (DN-RAB11), and this correlated with a reduction in biomarkers that were present in salivary exosomes. This study demonstrated that tumor-derived mRNAs are the cargo of exosomes and reach the salivary gland via the circulation, providing a mechanistic link between salivary exosomes and distal tumors. In subsequent studies using this model, the saliva from tumor-bearing mice was able to suppress the expression of genes associated with the activation of natural killer cells (70). Consistent with its effect on salivary biomarkers, the expression of DN-RAB11 reversed this effect. Thus, salivary exosomes also appear to dampen the immune response to their parental tumors through gastrointestinal tract.
The migration of exosomes from the primary tumor to the salivary glands is not confined to pancreatic cancer models. Orthotopically injected human lung cancer cells expressing a green fluorescent protein (GFP)-tagged cell surface marker (CD63) also gave rise to GFP-positive vesicles in mouse saliva (71). The cargo delivery role of these exosomes was confirmed by the presence of human GAPDH mRNA.
4. ELECTROCHEMICAL BIOSENSORS
The current gold standard methods for isolating exosomes involve ultracentrifugation through a density gradient or sucrose cushion (72). However, these approaches are expensive and laborious. Furthermore, detection of exosome-associated ctDNA in saliva using conventional polymerase chain reaction (PCR)-based methods has largely failed due to its short fragment length and low quantity (73). Therefore, there is a need to develop more practical and efficient methods for the isolation of exosomes, and the quantification of their molecular cargos.
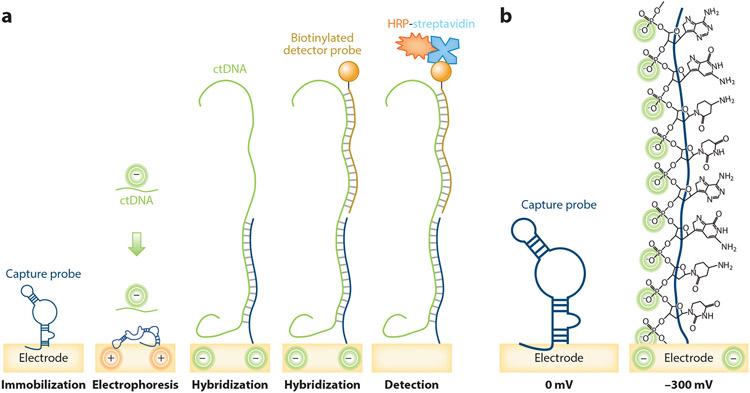
EFIRM is a technology that may meet this need. EFIRM allows quantification of ctDNA via an electrochemical sensor that is activated by capture and detector probes complementary to the ctDNA target (74, 75) (Figure 4). In the first step, pyrrole coating of gold electrodes facilitates the attachment of a single-stranded oligonucleotide capture probe at a surface density of 3.41 molecules/cm2 (74, 76). The saliva sample is then placed on the electrode in the presence of a cyclic square wave, which opens the hairpin structure (−300 mV, 9 s) of the capture probe and aids hybridization of the negatively charged DNA (+200 mV, 1 s). A complementary biotinylated single-stranded detector probe is then added, and binding to targets is measured by addition of horseradish peroxidase (HRP) and the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate. Redox of the HRP generates a current, which is amplified by TMB-dependent regeneration of HRP. The amount of current is proportional to the concentration of target immobilized on the electrode (77).
Figure 4.
Schematic of the EFIRM assay. (a) Surface preparation: The gold electrodes are precoated with pyrrole and DNA capture probe is immobilized onto the pyrrole-coated electrodes. Electrophoresis and target hybridization: The surface is incubated with the target ctDNA-containing saliva sample and a cyclic square wave electric field is applied at 30 cycles of +200 mV for 1 s and −300 mV for 9 s during hybridization. Detector probe hybridization: A complementary biotinylated single-stranded oligonucleotide detector probe hybridizes with the ctDNA target. Electrochemical detection: HRP-conjugated streptavidin and 3,3′,5,5′-tetramethylbenzidine substrate generate electrical current, which is detected by an electric sensor. (b) Steric effect: The negative potential makes a closed hairpin structure of DNA capture probe stretch and form an open structure required for highly efficient intermolecular hybridization. Abbreviations: ctDNA, circulating tumor DNA; EFIRM, electric field–induced release and measurement; HRP, horseradish peroxidase.
EFIRM has been used successfully to detect oncogenic mutations of the epidermal growth factor receptor (EGFR) gene in the saliva and plasma of patients with non-small cell lung cancer (73, 78, 79) (Figure 5). Receiver operating characteristic curve analysis demonstrated area under the curve values of 0.94 and 0.96 for EGFR exon 19 deletion and EGFP L858R, respectively. Capturing and analyzing EGFR exon 19 deletion and L858R mutation in saliva are emerging as a complementary technique in liquid biopsy and relevant in early cancer detection, as well as in guiding and managing patients on chemotherapy (80). These findings confirm that EFIRM has sufficient sensitivity to meet the demands of point-of-care testing.
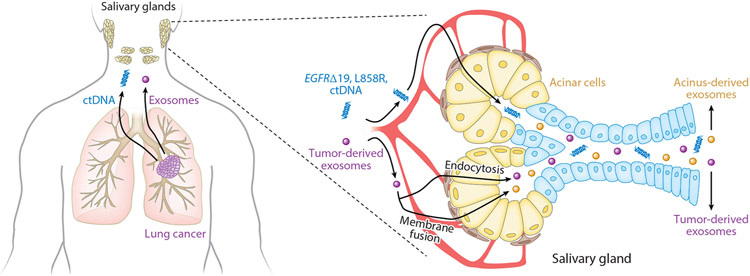
Figure 5.
Saliva-based liquid biopsy for non-small cell lung cancer. ctDNA and tumor-derived exosomes enter the circulation and reach the salivary glands. ctDNA and exosomes are uptaken by salivary gland acinar cells via endocytosis or membrane fusion. Central to saliva liquid biopsy techniques is the capture and analysis of ctDNA, which includes EGFR exon 19 deletion and L858R mutation. Combining salivary ctDNA and exosome analyses can provide more comprehensive panels of molecular markers for precision medicine application in a minimally invasive manner. Abbreviations: ctDNA, circulating tumor DNA; EGFR, epidermal growth factor receptor.
5. FUTURE PERSPECTIVE
The use of salivaomics for cancer detection, diagnosis, and disease monitoring is an exciting prospect. Indeed, the ease with which a biopsy from saliva can be obtained would have a positive impact on patients’ quality of life. However, before salivaomics can be successfully adopted in the clinical setting, more work is required to understand how exosomes mediate communication between distal tumors and organs such as the salivary glands. Furthermore, there must be a robust evaluation of the validity of salivary exosome-associated biomarkers. It is also currently unclear whether using salivary exosomes would be more effective than current methods of analysis in oncology such as those involving ctDNA, circulating tumor cells, or exosomal miRNA. On the one hand, mutational analysis of ctDNA does not reveal detailed information about signaling pathways that are active in a particular tumor. In this case, additional salivaomic analysis may provide a more information-rich basis upon which decisions regarding treatment could be made. Analysis of salivary exosomes may also be more representative of the whole tumor when compared to circulating tumor cells, which make up a very small fraction of the malignancy. On the other hand, analysis of exosomal miRNA may be desirable. This is because each exosome will contain a subset of the total cellular miRNA complement due to the randomized encapsulation of miRNAs at the point of vesicle formation.
Techniques for the isolation of salivary exosomes and the quantification of their cargo require further optimization. We suggest that EFIRM technology represents a significant step forward in this regard, because it offers a rapid, robust, and cost-effective way to perform salivary biomarker detection. Continued improvement of EFIRM, together with the development of other rapid biomarker isolation techniques, will lead to earlier detection of disease, more rapid treatment, and reduced morbidity and mortality.
ACKNOWLEDGMENTS
The work presented in this review was supported by NIH grants U18 TR003778, UG3/UH3 TR002978, UH2/UH3 CA206126, U01 CA233370, and U01 DE017790 to D.T.W.W. and R03 DE027759 and R03 DE29272 to T.N.
Footnotes
DISCLOSURE STATEMENT
D.T.W.W. is the cofounder of RNAmeTRIX Inc., a molecular diagnostic company; holds an equity in Liquid Diagnostics LLC; is a consultant to GlaxoSmithKline, PeriRx, Inc., Wrigley Co., and Colgate-Palmolive; and receives research grants from the US National Institutes of Health (NIH), US Department of Defense, UCLA, Colgate-Palmolive, and the Hirshberg Foundation. T.N. is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Chiappin S, Antonelli G, Gatti R, De Palo EF. 2007. Saliva specimen: a new laboratory tool for diagnostic and basic investigation. Clin. Chim. Acta 383:30–40 [DOI] [PubMed] [Google Scholar]
- 2.Lamy E, Mau M. 2012. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteom 75:4251–58 [DOI] [PubMed] [Google Scholar]
- 3.Carpenter GH. 2013. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol 4:267–76 [DOI] [PubMed] [Google Scholar]
- 4.Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, et al. 2009. Systematic comparison of the human saliva and plasma proteomes. Proteom. Clin. Appl 3:116–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai J, Smith B, Wong DT. 2010. Saliva ontology: an ontology-based framework for a Salivaomics Knowledge Base. BMC Bioinform. 11:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong DT. 2012. Salivaomics. J. Am. Dent. Assoc 143:19S–24S [DOI] [PubMed] [Google Scholar]
- 7.Nonaka T, Wong DTW. 2017. Saliva-exosomics in cancer: molecular characterization of cancer-derived exosomes in saliva. Enzymes 42:125–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson DB, Botchway CA. 1979. Circadian variations in the flow rate and composition of whole saliva stimulated by mastication. Arch. Oral. Biol 24:877–81 [DOI] [PubMed] [Google Scholar]
- 9.Mackie DA, Pangborn RM. 1990. Mastication and its influence on human salivary flow and alpha-amylase secretion. Physiol. Behav 47:593–95 [DOI] [PubMed] [Google Scholar]
- 10.Helmerhorst EJ, Oppenheim FG. 2007. Saliva: a dynamic proteome. J. Dent. Res 86:680–93 [DOI] [PubMed] [Google Scholar]
- 11.Diaz LA Jr., Bardelli A. 2014. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol 32:579–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, et al. 2001. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61:1659–65 [PubMed] [Google Scholar]
- 13.Beaver JA, Jelovac D, Balukrishna S, Cochran R, Croessmann S, et al. 2014. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res 20:2643–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, et al. 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, et al. 2008. Circulating mutant DNA to assess tumor dynamics. Nat. Med 14:985–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, et al. 2014. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med 20:548–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, et al. 2014. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med 20:430–35 [DOI] [PubMed] [Google Scholar]
- 18.Aarthy R, Mani S, Velusami S, Sundarsingh S, Rajkumar T. 2015. Role of circulating cell-free DNA in cancers. Mol. Diagn. Ther 19:339–50 [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri AA, Binkley MS, Osmundson EC, Alizadeh AA, Diehn M. 2015. Predicting radiotherapy responses and treatment outcomes through analysis of circulating tumor DNA. Semin. Radiat. Oncol 25:305–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ignatiadis M, Lee M, Jeffrey SS. 2015. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin. Cancer Res 21:4786–800 [DOI] [PubMed] [Google Scholar]
- 21.Polivka J Jr., Pesta M, Janku F. 2015.Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: Are we there yet? Expert Rev. Mol. Diagn 15:1631–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonne NJ, Wong DT. 2012. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 4:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen TV, Simonsen MK, Nielsen FC, Hundrup YA. 2007. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol. Biomarkers Prev 16:2072–76 [DOI] [PubMed] [Google Scholar]
- 24.Looi ML, Zakaria H, Osman J, Jamal R. 2012. Quantity and quality assessment of DNA extracted from saliva and blood. Clin. Lab 58:307–12 [PubMed] [Google Scholar]
- 25.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, et al. 2015. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med 7:293ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonaka T, Wong DTW. 2018. Liquid biopsy in head and neck cancer: promises and challenges. J. Dent. Res 97:701–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, et al. 2015. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem 61:221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park NJ, Li Y, Yu T, Brinkman BM, Wong DT. 2006. Characterization of RNA in saliva. Clin. Chem 52:988–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, et al. 2010. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138:949–57.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Xiao H, Karlan S, Zhou H, Gross J, et al. 2010. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLOS ONE 5:e15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YH, Kim JH, Zhou H, Kim BW, Wong DT. 2012. Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J. Mol. Med 90:427–34 [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, et al. 2012. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell. Mol. Life Sci 69:3341–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, St. John MAR, Zhou X, Kim Y, Sinha U, et al. 2004. Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res 10:8442–50 [DOI] [PubMed] [Google Scholar]
- 34.Gallo A, Tandon M, Alevizos I, Illei GG. 2012. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLOS ONE 7:e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, et al. 2010. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 16:34–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, et al. 2009. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res 15:5473–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu CJ, Lin SC, Yang CC, Cheng HW, Chang KW. 2012. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 34:219–24 [DOI] [PubMed] [Google Scholar]
- 38.Momen-Heravi F, Trachtenberg AJ, Kuo WP, Cheng YS. 2014. Genomewide study of salivary microRNAs for detection of oral cancer. J. Dent. Res 93:86S–93S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duz MB, Karatas OF, Guzel E, Turgut NF, Yilmaz M,et al. 2016. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: a pilot study. Cell. Oncol 39:187–93 [DOI] [PubMed] [Google Scholar]
- 40.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, et al. 2012. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res 5:492–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo F, Di Bella S, Vannini F, Berti G, Scoyni F, et al. 2018. miRandola 2017: a curated knowledge base of non-invasive biomarkers. Nucleic Acids Res. 46:D354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoerman KC, Chauncey HH, Herrold RD. 1959. Parotid saliva acid phosphatase in prostatic cancer. Cancer 12:359–63 [DOI] [PubMed] [Google Scholar]
- 43.Denny P, Hagen FK, Hardt M, Liao L, Yan W, et al. 2008. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res 7:1994–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. 2009. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J. Proteome Res 8:5590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulz BL, Cooper-White J, Punyadeera CK. 2013. Saliva proteome research: current status and future outlook. Crit. Rev. Biotechnol 33:246–59 [DOI] [PubMed] [Google Scholar]
- 46.Esser D, Alvarez-Llamas G, de Vries MP, Weening D, Vonk RJ, Roelofsen H. 2008. Sample stability and protein composition of saliva: implications for its use as a diagnostic fluid. Biomarker Insights 3:25–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao H, Wong DT. 2012. Method development for proteome stabilization in human saliva. Anal. Chim. Acta 722:63–69 [DOI] [PubMed] [Google Scholar]
- 48.Colombo M, Raposo G, Thery C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol 30:255–89 [DOI] [PubMed] [Google Scholar]
- 49.Kourembanas S 2015. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol 77:13–27 [DOI] [PubMed] [Google Scholar]
- 50.El Andaloussi S, Mäger I, Breakefield XO, Wood MJ. 2013. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov 12:347–57 [DOI] [PubMed] [Google Scholar]
- 51.Cheng J, Nonaka T, Wong DTW. 2019. Salivary exosomes as nanocarriers for cancer biomarker delivery. Materials 12:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Tarawneh SK, Border MB, Dibble CF, Bencharit S. 2011. Defining salivary biomarkers using mass spectrometry-based proteomics: a systematic review. OMICS 15:353–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. 2008. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull 31:1059–62 [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, et al. 2009. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J. Proteome Res 8:1304–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, et al. 2011. Proteomic analysis of two types of exosomes in human whole saliva. Biol. Pharm. Bull 34:13–23 [DOI] [PubMed] [Google Scholar]
- 56.Berckmans RJ, Sturk A, van Tienen LM, Schaap MC, Nieuwland R. 2011. Cell-derived vesicles exposing coagulant tissue factor in saliva. Blood 117:3172–80 [DOI] [PubMed] [Google Scholar]
- 57.Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. 2010. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLOS ONE 5:e8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. 2013. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol. Pharm. Bull 36:66–75 [DOI] [PubMed] [Google Scholar]
- 59.Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, et al. 2011. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, et al. 2012. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLOS Biol. 10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simpson RJ, Kalra H, Mathivanan S. 2012. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 1:18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, et al. 2010. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 4:1921–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma S, Gillespie BM, Palanisamy V, Gimzewski JK. 2011. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir 27:14394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, et al. 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol 10:1470–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Ma YY, Wang J, Zeng XF, Li R, et al. 2016. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 9:139–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, et al. 2013. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 119:1159–67 [DOI] [PubMed] [Google Scholar]
- 67.Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, et al. 2014. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem 289:3869–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, et al. 2015. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523:177–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, et al. 2013. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem 288:26888–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katsiougiannis S, Chia D, Kim Y, Singh RP, Wong DT. 2017. Saliva exosomes from pancreatic tumor-bearing mice modulate NK cell phenotype and antitumor cytotoxicity. FASEB J. 31:998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Wei F, Schafer C, Wong DT. 2014. Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLOS ONE 9:e110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thery C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol 2006:3.22.1–29 [DOI] [PubMed] [Google Scholar]
- 73.Li F, Wei F, Huang WL, Lin CC, Li L, et al. 2020. Ultra-short circulating tumor DNA (usctDNA) in plasma and saliva of non-small cell lung cancer (NSCLC) patients. Cancers 12:2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei F, Wang J, Liao W, Zimmermann BG, Wong DT, Ho CM. 2008. Electrochemical detection of low-copy number salivary RNA based on specific signal amplification with a hairpin probe. Nucleic Acids Res. 36:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei F, Yang J, Wong DT. 2013. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens. Bioelectron 44:115–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su X, Wu YJ, Robelek R, Knoll W. 2005. Surface plasmon resonance spectroscopy and quartz crystal microbalance study of streptavidin film structure effects on biotinylated DNA assembly and target DNA hybridization. Langmuir 21:348–53 [DOI] [PubMed] [Google Scholar]
- 77.Gau V, Ma SC, Wang H, Tsukuda J, Kibler J, Haake DA. 2005. Electrochemical molecular analysis without nucleic acid amplification. Methods 37:73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pu D, Liang H, Wei F, Akin D, Feng Z, et al. 2016. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: a pilot study. Thorac. Cancer 7:428–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei F, Lin CC, Joon A, Feng Z, Troche G, et al. 2014. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am. J. Respir. Crit. Care Med 190:1117–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim C, Xi L, Cultraro CM, Wei F, Jones G, et al. 2021. Longitudinal circulating tumor DNA analysis in blood and saliva for prediction of response to Osimertinib and disease progression in EGFR-mutant lung adenocarcinoma. Cancers 13:3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Du J, Zhang L. 2017. Analysis of salivary microRNA expression profiles and identification of novel biomarkers in esophageal cancer. Oncol. Lett 14:1387–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Navarro MA, Mesia R, Diez-Gibert O, Rueda A, Ojeda B, Alonso MC. 1997. Epidermal growth factor in plasma and saliva of patients with active breast cancer and breast cancer patients in follow-up compared with healthy women. Breast Cancer Res. Treat 42:83–86 [DOI] [PubMed] [Google Scholar]
- 83.Streckfus C, Bigler L, Dellinger T, Dai X, Kingman A, Thigpen JT. 2000. The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: a preliminary study. Clin. Cancer Res 6:2363–70 [PubMed] [Google Scholar]
- 84.Streckfus C, Bigler L, Tucci M, Thigpen JT. 2000. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Investig. 18:101–9 [DOI] [PubMed] [Google Scholar]
- 85.Brooks MN, Wang J, Li Y, Zhang R, Elashoff D, Wong DT. 2008. Salivary protein factors are elevated in breast cancer patients. Mol. Med. Rep 1:375–78 [PMC free article] [PubMed] [Google Scholar]
- 86.Wood N, Streckfus CF. 2015. The expression of lung resistance protein in saliva: a novel prognostic indicator protein for carcinoma of the breast. Cancer Investig. 33:510–15 [DOI] [PubMed] [Google Scholar]
- 87.Xiao H, Zhang Y, Kim Y, Kim S, Kim JJ, et al. 2016. Differential proteomic analysis of human saliva using tandem mass tags quantification for gastric cancer detection. Sci. Rep 6:22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohshiro K, Rosenthal DI, Koomen JM, Streckfus CF, Chambers M, et al. 2007. Pre-analytic saliva processing affect proteomic results and biomarker screening of head and neck squamous carcinoma. Int. J. Oncol 30:743–49 [PubMed] [Google Scholar]
- 89.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, et al. 2008. Salivary proteomics for oral cancer biomarker discovery. Clin. Cancer Res 14:6246–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dowling P, Wormald R, Meleady P, Henry M, Curran A, Clynes M. 2008. Analysis of the saliva proteome from patients with head and neck squamous cell carcinoma reveals differences in abundance levels of proteins associated with tumour progression and metastasis. J. Proteom 71:168–75 [DOI] [PubMed] [Google Scholar]
- 91.Rai B, Kaur J, Jacobs R, Anand SC. 2011. Adenosine deaminase in saliva as a diagnostic marker of squamous cell carcinoma of tongue. Clin. Oral Investig 15:347–49 [DOI] [PubMed] [Google Scholar]
- 92.Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, et al. 2012. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol. Biomarkers Prev 21:664–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Winck FV, Prado Ribeiro AC, Ramos Domingues R, Ling LY, Riano-Pachon DM, et al. 2015. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci. Rep 5:16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao H, Zhang L, Zhou H, Lee JM, Garon EB, Wong DTW. 2012. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell. Proteom 11. 10.1074/mcp.M111.012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun Y, Xia Z, Shang Z, Sun K, Niu X, et al. 2016. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci. Rep 6:24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen DX, Schwartz PE, Li FQ. 1990. Saliva and serum CA 125 assays for detecting malignant ovarian tumors. Obstet. Gynecol 75:701–4 [PubMed] [Google Scholar]