Abstract
The clinical significance of small TP53 clones detected with next generation sequencing (NGS) in chronic lymphocytic leukemia is an issue of active debate. According to the official guidelines, treatment decisions should be guided only by variants with variant allele frequency (VAF) ≥10%. We present data on 325 consecutive patients with chronic lymphocytic leukemia analyzed with NGS. In total 47 pathogenic/likely pathogenic (P/LP), TP53 variants were detected in 26 patients (8%). Eleven of these (23%) were in the 5% to 10% VAF range and reported according to our institutional policy. All TP53 variants in the 5% to 10% VAF range were confirmed (100% concordance) with a second NGS panel. Our results where further validated with the performance of Sanger sequencing and digital droplet PCR (ddPCR). In 12 patients with available fluorescence in situ hybridization data and TP53 mutations within 5% to 10% VAF, deletion of chromosome 17p (del(17p)) was detectable in only 1 patient. We propose a robust diagnostic algorithm, which allows the safe detection and reporting of TP53 variants with VAF down to 5% in the clinical setting. Our study provides evidence that NGS is equally potent to detect variants with VAF 5% to 10% compared to those with VAF 10% to 15%, highlighting the urgent need for harmonization of NGS methodologies across diagnostic laboratories.
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is an ideal example where the concept of precision medicine applies in the clinical setting, as a number of disease- and patient-related features dictate treatment choice.1,2 Moreover, numerous prognostic biomarkers have been suggested.3–17 Aberrations of the TP53 gene (deletions and mutations, TP53abs) and the somatic hypermutation status (SHM) of the immunoglobulin heavy variable (IGHV) gene expressed by the clonotypic B-cell receptor (BCR), are the main disease-related traits that predict treatment outcome.4–6,8,18,19 Therefore, they are taken into consideration before treatment initiation. In particular, patients carrying TP53abs, as well as those with unmutated BCRs (unmutated CLL, U-CLL), should be considered candidates for treatment with novel agents, such as B-cell signaling kinase inhibitors or the BCL-2 inhibitor venetoclax.1,20–24
Traditionally, TP53 gene evaluation was based on the detection of the deletion of the short arm of chromosome 17 [del(17p)] with either fluorescence in situ hybridization (FISH) or microarrays, whereas the mutation analysis was performed with Sanger sequencing.19,25–30 The majority of the cases with reported TP53abs (60–80% depending on the analyzed cohort) exhibit double-allelic abnormalities with 1 allele being deleted and the second being mutated.31,32 That said, a significant proportion of TP53 aberrant cases carry isolated deletions or mutations, with the second ones being far more common, reaching up to 20% TO 30% of all TP53 aberrant cases.33–36 According to the International Workshop on CLL (iwCLL) guidelines, isolated aberrations are considered equally unfavorable as the double-allelic ones and should be treated similarly.37
The advent of more sensitive next generation sequencing (NGS) technologies has revolutionized the characterization of cancer genomes, as it allowed the detection of variants present in minor subclones whose allelic burden is below the detection limit of Sanger sequencing.11,38 Recent ultradeep NGS analysis of TP53 in CLL revealed subclonal TP53 variants in ~5% TO 15% of all CLL cases.13,17,33,39 More importantly, in retrospective studies, the presence of subclonal TP53 aberrations was associated with inferior survival indicating that their prognostic impact may be equivalent to clonal aberrations.13,26,33,40,41 Interestingly, recent retrospective data suggest that even extremely small clones with VAF down to 0.1% may be clinically relevant.39,42 These findings remain to be prospectively validated in the context of clinical trials.
The latest European Research Initiative on CLL (ERIC) recommendations regarding the TP53abs suggest that only pathogenic/likely pathogenic (P/LP) variants with variant allele frequency (VAF) of at least 10% (which is theoretically equivalent to the limit of detection of Sanger sequencing) should be taken into consideration for treatment decision making, when NGS methods are applied.19 This circumspect approach is mostly advocated by (i) the risk that low TP53 clones identified by NGS may be technical artifacts rather than real clonal populations; (ii) the lack of harmonization across different NGS platforms used in the various laboratories and; (iii) the lack of prospective validation of a lower than Sanger detection limit within clinical trials. The same guidelines acknowledge however, that this 10% “Sanger-like” threshold is arbitrary and that variants with VAF 5% to 10% may be reported. Based on that, many institutions and CLL consortiums, among them the Swedish CLL working group,43 allow the reporting of P/LP variants with VAF ≥5% in the clinical setting, under the condition that the method is validated for detecting variants within that frequency range.
Here, we present our experience in the detection and reporting of low TP53 clones, which we define as those with VAF around 10% (VAF 5–10%) in a cohort of 325 patients with CLL. We propose a validated NGS-based diagnostic algorithm, which allows the safe reporting of TP53 variants with VAF ≥5%.
PATIENTS-METHODS
Patient cohort-variant calling
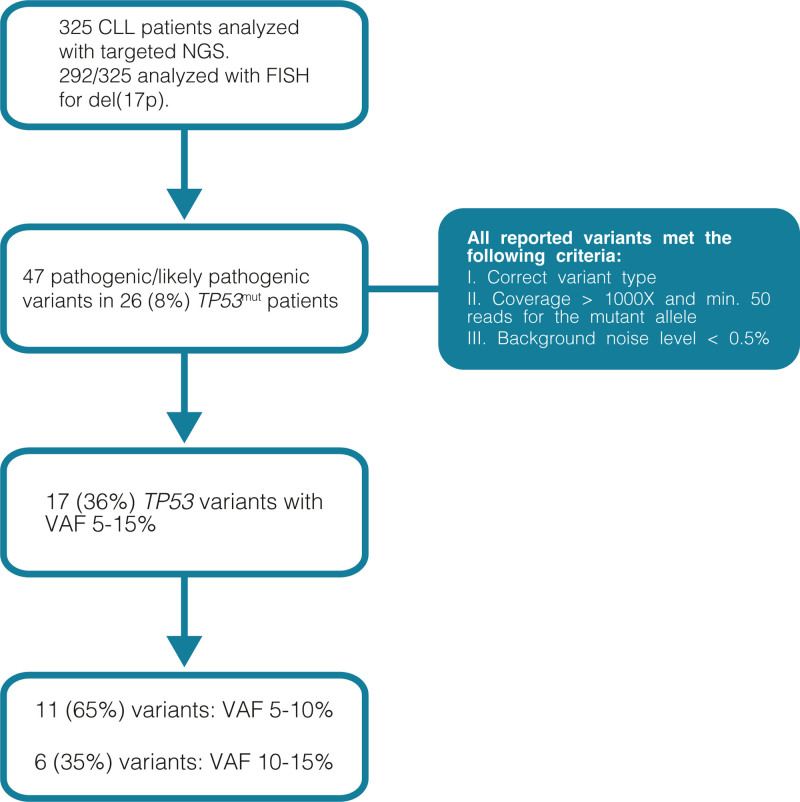
Between October 2014 and April 2020, 2952 patients with hematological malignancies, mainly myeloid neoplasms, were investigated with NGS (targeted sequencing, panel A, see below) at our department. In September 2016, we launched the same NGS panel for the analysis of TP53, NOTCH1, and SF3B1 genes in patients with CLL, replacing Sanger sequencing. Only TP53, SF3B1, and NOTCH1 genes are analyzed and reported for CLL patients. Data on the remaining 51 genes of the panel are not routinely further processed. A total of 325 patients with CLL, diagnosed according to the iwCLL 2018 criteria37 and analyzed between September 2016 and June 2019, are included in the present study (Figure 1). FISH data were available for 292/325 (90%) of the included patients.
Figure 1.
Flow chart of the methodology applied in the present cohort.
The diagnostic algorithm for reporting P/LP TP53 variants with VAF above 5% is illustrated in Figure 1. We report (i) missense variants located in known hotspot positions or truncating/frameshift/splice variants; (ii) coverage for the position of the variant should be ≥1000× with minimum 50 reads for the mutant allele; (iii) background noise level for each variant position ≤0.5% (median value +2 standard deviations). The known hotspot position is defined as the position where (i) the same amino acid change has been reported in at least 10 different cancer patients44; and (ii) a different amino acid change is reported at the same residue in at least 50 different cancer patients.44 For samples that show suboptimal coverage or high background due to poor DNA quality reanalysis or new sampling is recommended. Variants of unknown significance (VUS) with VAF <10% are not reported. The same approach is followed in our institution for all patients that are analyzed with NGS in clinically relevant genes. The study was approved by the Regional Ethical Committee (Dnr 2014/233) with the study protocol being in accordance with the declaration of Helsinki.
Next generation sequencing
NGS was performed with 2 independent panels: (i) TruSight Myeloid Sequencing panel, Illumina (panel A) and (ii) Archer VariantPlex Myeloid kit (panel B). Further details on the design and characteristics of panels A and B are provided in the Suppl. Material. Panel A covers exons 2–10 of the TP53 gene and is sequenced to a read-depth of at least 1000× for all positions (Suppl. Figure S1). The median background VAF with Panel A for all P/LP TP53 variants with VAF 5% to 15%, calculated in 774 samples, was 0.22% (range 0.07–0.94%) (Suppl. Table S1).
Panel B covers exons 1–11 of the TP53 gene and takes advantage of unique molecular identifiers (UMIs) to improve specificity (Suppl. Figure S1). After applying the UMIs, panel B provides an error corrected coverage of at least 100×. Panel A is the one that is used in the clinical setting while panel B was used as validation.
Fluorescence in situ hybridization
Interphase FISH analysis was performed in 292/325 patients (90%) using the probes for the 13q14 (LSI D13S319, LSI 13q34), 11q22 (LSI ATM), 17p13 (LSI TP53) regions, and trisomy 12 (CEP 12) with a detection limit of 5% according to our institutional policy.
Validation of NGS findings
Seven TP53 variants with VAF 5% to 15% were assessed with Sanger sequencing. Two specialized geneticists (TP and PB) evaluated the chromatograms without knowing the NGS results, providing reports as positive (identified variant), normal (no identified variant) or inconclusive. Finally, droplet digital PCR (ddPCR) was performed for validation of 2 TP53 variants with VAF in the range 1% to 5% (Suppl. Material). All the above-mentioned methodologies (NGS, Sanger Sequencing and ddPCR) were performed on genomic DNA extracted from whole blood using EZ1 DNA Blood Kit. Sanger sequencing chromatograms were generated in R (v4.0.3) using the sangerseqR (v1.26.0) software with standard parameters.45
Statistical analysis
Background noise level for individual positions across the panel A was calculated by estimating means, medians and standard deviations from 774 individuals. Pearson correlation was used to assess the relationship between VAF estimates from panel A and panel B. Student’s t-test was used to investigate if differences between VAF estimates were deviating from zero and if differences between VAF estimates were different in variants above and below 10% VAF. Stata v15.1 was used for statistical calculations. The r and r2 values for VAF correlation between NGS panel A and B were calculated using Microsoft Excel v16.41.
RESULTS
Detections of TP53 variants with panels A and validation with panel B
Among the patients with CLL analyzed with panel A, 47 P/LP TP53 variants were detected in 26 patients (8%). In total, 17 TP53 P/LP variants with VAF 5% to 15% (VAF 5–10%: n = 11 and 10–15%: n = 6, 36% of all reported TP53 mutations) were observed in 14 patients, as 2 patients carried more than 1 mutation (Table 1). Interestingly, among 12 patients with solely low VAF TP53 clones (VAF <10%) and available FISH data, only 1 (8%) carried del(17p) (Table 1, Suppl. Figure S2).
Table 1.
Overview of All TP53 Pathogenic/Likely Pathogenic Variants With VA 5% to 15%
| Patient | gDNA | cDNA | Protein | VAF Panel A (%) | VAF Panel B (%) | Sanger Sequencing | (FISH)del(17p) |
|---|---|---|---|---|---|---|---|
| 1 | 7578413 | c.517G>T | p.V173L | 14.7 | 20.8 | Positive | NA |
| 2 | 7577610 | c.673-2A>T | p.? | 12.1 | 11.6 | Positive | Trisomy 12 |
| 2 | 7579311 | c.375 + 1G>T | p.? | 13.6 | 14.9 | NA | Trisomy 12 |
| 3 | 7577086 | c.851_852del | p.T284fs*21 | 10.1 | 11.9 | NA | del(13q) |
| 3 | 7578263 | c.586C>T | p.R196* | 11.2 | 11.7 | Inconclusive | del(13q) |
| 3 | 7579575 | c.112del | p.Q38fs*6 | 9.8 | 9.6 | NA | del(13q) |
| 4 | 7578206 | c.643A>G | p.S215G | 6.1 | 5.9 | NA | del(13q) |
| 5 | 7577559 | c.722C>A | p.S241Y | 6.6 | 5.8 | Negative | NA |
| 6 | 7577114 | c.824G>A | p.C275Y | 7.9 | 9.1 | NA | del(17p) |
| 7 | 7577121 | c.817C>T | p.R273C | 5.0 | 6.8 | NA | del(11q) |
| 8 | 7577538 | c.743G>A | p.R248Q | 5.5 | 8.1 | NA | Trisomy 12 |
| 9 | 7577538 | c.743G>A | p.R248Q | 7.2 | 8.4 | Negative | Trisomy 12 |
| 10 | 7578394 | c.536A>G | p.H179R | 9.2 | 9.8 | Inconclusive | Normal |
| 11 | 7578212 | c.637C>T | p.R213* | 8.7 | 7.1 | Negative | NA |
| 12 | 7577121 | c.580C>T | p.L194F | 11.0 | 11.2 | NA | del(13q) |
| 13 | 7577538 | c.743G>T | p.R248L | 7.7 | 6.4 | NA | Normal |
| 14 | 7577108 | c.830G>T | p.C277F | 5.7 | 6.8 | NA | del(13q)/del(11q) |
NA = not analyzed; VAF = variant allele frequency.
Two variants with VAF 5% to 10% (c.11C>T; p.P4L [VAF 6.4%] and c.495_496del; p.Q165fs*15 [VAF 6.6%]) that were initially detected with panel A were not clinically reported. Variant p.P4L was interpreted as VUS and was not reported according to our institutional policy. Variant p.Q165fs*15 was detected in 1 sample with low DNA quality with 2 other pathogenic variants with VAF 20% and 36% being detected. Due to the low DNA quality the reporting VAF-threshold was raised to 10%, which resulted in omitting this variant from the report.
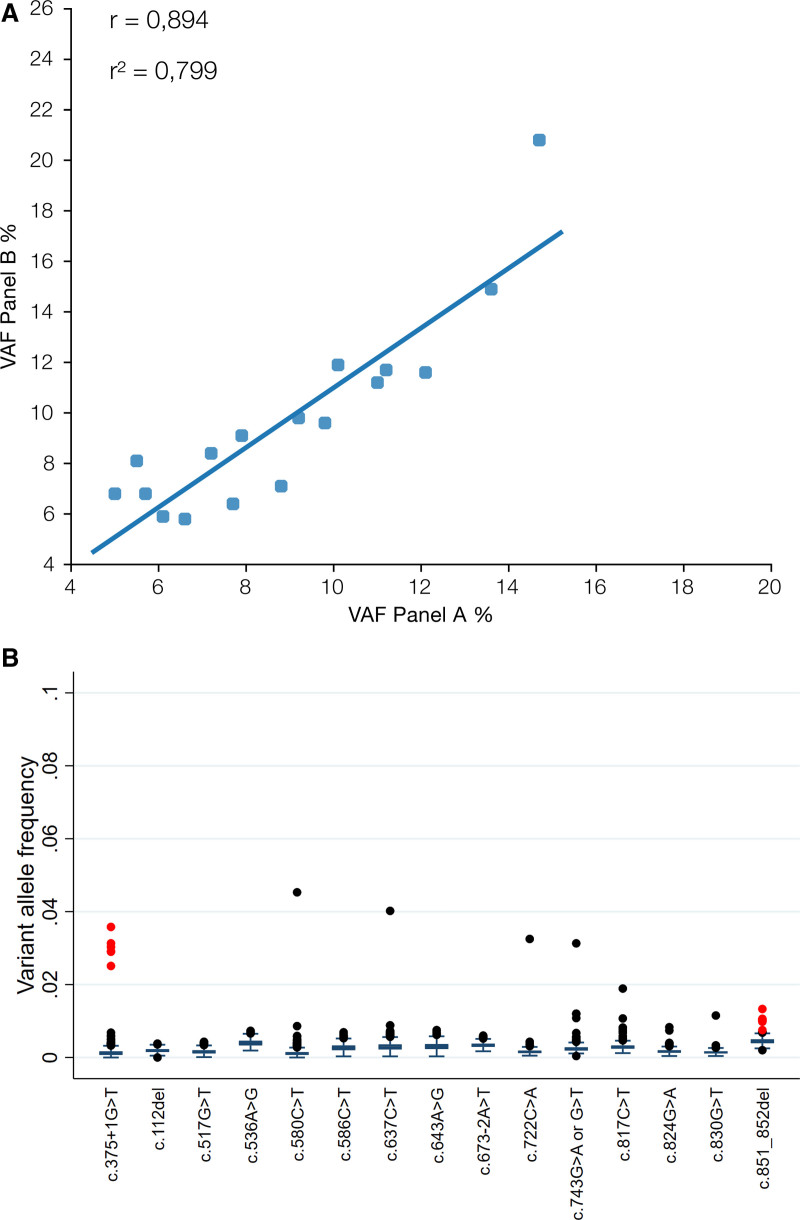
The presence of all 17 reported P/LP TP53 variants with VAF 5% to 15% detected by panel A was confirmed by panel B resulting in 100% concordance (Table 1). VAFs estimated by panel A and panel B were strongly correlated (r = 0.894, P < 0.00001) (Figure 2A), and there was no systematic variation in VAF estimation between the 2 panels (mean difference = –0,8%, P = 0.08) (Table 1). Furthermore, there was no systematic variation in VAF differences between variants that were above or below the 10% threshold (P = 0.21).
Figure 2.
(A) Correlation of VAFs between the 2 NGS panels. (B) Boxplot of background VAF levels of variants that were detected in the VAF 5%–15% range. Outliers in red represent samples that were included in the same library preparation and the same sequencing run (Suppl. Table S2). NGS = next generation sequencing; VAF = variant allele frequency.
No additional P/LP variant in TP53 gene with VAF > 5% was detected with panel B. Interestingly, the variant p.Q165fs*15, which was detected with panel A but omitted from the report due to poor quality of DNA, was also detected with panel B with a VAF of 7.5%, indicating that it was a true finding rather than an artifact. As mentioned above, the patient with the p.Q165fs*15 variant also carried 2 additional pathogenic variants with VAFs of 20% and 36%, therefore in this particular case, not reporting p.Q165fs*15 did not influence the treatment decision. However, in case the patient carried only this variant, nonreporting could have influenced the treatment choice. As mentioned above, the variant was not reported when performing the analysis with panel A due to lower quality of the sequencing data. In order to avoid similar errors when dealing with poor DNA quality samples, we have adjusted our analytic pipeline. Whenever a sample has lower coverage than expected, or if the number of detected variants is unusually high in the 3% to 5% range, we resequence the sample. If this is not possible, we advise the treating physician for repeating the analysis in a new sample.
Validation of TP53 variants with Sanger sequencing
Seven of the 14 patients harboring clinically reported low frequency TP53 clones (VAF: 5–9.99%, n = 4 and VAF: 10–15%, n = 3) were further analyzed with Sanger sequencing (Table 1, Suppl. Figure S3). Interpretation of the chromatograms was consistent between the 2 geneticists who evaluated the results. Two of the variants (VAF 14.7%, and 12.1%, patients 1 and 2, respectively) could be detected and thus Sanger sequencing was reported as positive, while 3 could not be detected (VAF 6.6%, 7.2% and 8.7%, patients 5, 9, and 11, respectively) and the sequencing was reported as normal (Suppl. Figure S3). For the remaining 2 variants (VAF 11% and 9%, patients 3 and 10, respectively), the results were reported as inconclusive and would motivate a recommendation of a new follow-up sample or evaluation with another method (Suppl. Figure S3). In summary, two-third of the variants with VAF above the 10% could be verified with Sanger sequencing, whereas for variants with VAF 5% to 9.99% Sanger sequencing was either inconclusive or negative.
Variants with VAF 1% to 5%: validation with ddPCR
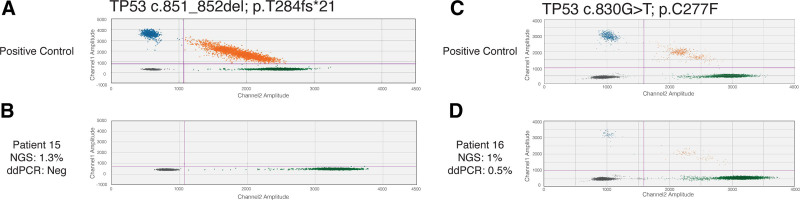
To further evaluate the performance of our NGS method as well as our reporting algorithm, we retrospectively sought for TP53 variants with VAF 1% to 5% in a cohort of 774 patients analyzed with panel A, focusing on the 17 TP53 variants that are included in this study and reported in the clinical setting. In total, 21 variants were identified in 19 patients (Figure 2B, Suppl. Table S2) with 11 variants located in either position c.375 + 1 (n = 6) or position c.851_852del (n = 5). The remaining 10 variants were found in 6 different positions. Re-evaluation of the results for positions with recurrent variants (c.375 + 1G<A and c.851_852del) revealed that all cases carrying the same aberration were included in the same library preparation and the same sequencing run, indicating that these findings may be technical artifacts rather than true variants (Suppl. Table S2). Indeed, ddPCR analysis failed to detect the presence of c.851_852del in the sample that initially was positive for the variant with VAF of 1.3% (Figure 3A,B), confirming that the low frequency variants observed in that same sequencing run were artifacts. In contrast, ddPCR analysis of the variant c.830G>T (p.C277F) in the sample that was shown to be a single positive case per NGS sequencing run and had VAF of 1% confirmed the presence of the variant (VAF = 0.5%) (Figure 3C,D).
Figure 3.
Variant specific ddPCR analysis of c.851_852del (p.T284fs*21) and c.830G>T (p.C277F). (A) 56565-positive control for c.851_852del; (B) patient 15: sample that showed VAF of 1% in NGS analysis; (C) positive control for c.830G>T; (D) patient 16: sample that showed VAF of 1% in NGS analysis. Green droplets indicate the presence of the wild-type allele, blue droplets indicate the mutant allele whereas orange show both wild type and mutant alleles in the same droplet.
DISCUSSION
The implementation of NGS in the clinical setting has revolutionized cancer-diagnostics, offering the possibility of detecting numerous genomic prognostic/predictive markers, present only in small subclonal populations, nondetectable with the traditional techniques.8,11,38,46–50 At the same time, many challenges emerged as there is no consensus on a universal NGS approach or a standard methodology regarding the reporting of NGS findings with potential clinical significance. Pathogenic variants in the TP53 gene in CLL are a typical example where the actual reporting threshold is a matter of active debate. According to the latest ERIC recommendation, only TP53 variants with VAF ≥10% should dictate the treatment choice justifying the use of other treatment options than chemo-based regimens, as this threshold is considered to better represent the sensitivity of Sanger sequencing.19 In the present study, we present our experience on the reporting of TP53 variants with VAF ≥5% in the clinical setting.
Our argument against the 10% as threshold stems mainly from the lack of reproducibility of NGS technologies in distinguishing variants with slight VAF differences, that is, a variant with VAF of 9.9% from a variant with a VAF of 10.2%. Moreover, several technical parameters both sample- and methodology-dependent such as DNA quality, cancer-cell fraction, experimental conditions, and amplification artifacts may have an impact on the sensitivity of each method, even of each run. In other words, the result of each analysis is both method but also variant specific.
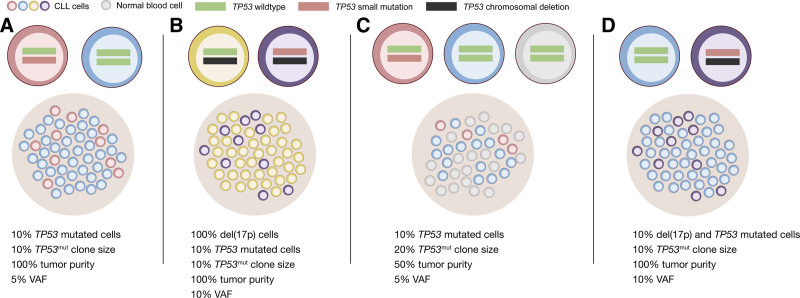
Another caveat of the 10% threshold is the fact that the resulting VAF of a specific variant may vary significantly due to factors such as focal copy number variations (CNVs) and tumor purity. A tumor with a TP53 variant in 10% of the tumor cells without concomitant del(17p) would in a sample with 100% tumor purity result in a VAF of 5% (Figure 4A). In a sample with 100% tumor purity, clonal del(17p) and a small TP53 variant in 10% of the tumor cells, the VAF would increase to 10% due to lack of wild-type allele sequence (Figure 4B).
Figure 4.
VAF and proportion of cells. (A) In a sample with 100% tumor purity and no del(17p), a TP53 mutation with VAF 5% (red cells) will correspond to a clone size of 10%. (B) A sample with 100% purity and a del(17p) (yellow cells) will lead to loss of the wild-type allele and increase the TP53 mutation VAF to 10% with a clone size of 10% (purple). (C) In a less pure sample with 50% tumor cells (blue and red cells), a TP53 mutation VAF of 5% will correspond to 10% of all cells being mutated but a clone size of 20%. (D) A sample with 100% tumor purity with 10% of the tumor cells having coexisting TP53 mutation and del(17p) (purple) will correspond to a clone size and VAF of 10%. CLL cells are depicted in blue (no TP53ab), yellow [del(17p)], no small TP53 mutation) red [small TP53 mutation, no del(17p)] or purple [small TP53 mutation and del(17p)]. Normal cells are depicted in gray.
An additional significant parameter is the actual sample source. In our institution, we use DNA extracted from whole blood without any sorting of the clonal cells, as the samples that we analyze are taken at the time of need for treatment and therefore by definition contain high tumor load. As a result, the detection of a variant with a VAF of 5% in a sample with a tumor purity of 50% corresponds to 20% of cancer cells carrying this variant within the tumor population (Figure 4C).
Having this in mind, the 10% threshold becomes even more questionable, in particular if we take into account the fact that there are no official recommendations regarding a relevant threshold for the analysis of del(17p) by FISH or microarrays. In the case of FISH, each lab may set its own threshold based on the background observed during the validation process. This leads to significant variations across different labs. In a retrospective study by ERIC, evaluating the prognostic impact of various genomic aberrations in CLL, the threshold for a positive result for del(17p) ranged from 2.5% to 20% among the included institutions.10 Interestingly, a TP53 deletion is considered equally unfavorable to a P/LP variant. Therefore, it is quite striking that the presence of a population of 5% of cells carrying del(17p) detected by FISH could dictate the treatment choice, whereas a population of 18% of cells carrying a P/LP TP53 variant (corresponding to a VAF of 9%, Figure 4), should not be taken into consideration. In our cohort, 23% of the TP53 P/LP variants exhibited a low VAF (5–10%). Of note, in 11 of 12 patients with available FISH data, no del(17p) was detected (Table 1, Suppl. Figure S2). In case we had followed the 10% threshold, the patients carrying TP53 variants with VAF up to 9.8% would have been reported as TP53 wild type and consequently treated accordingly.
We do acknowledge that NGS is a rather novel methodology not devoid of limitations. One of the major drawbacks of short read NGS, especially when it is amplicon-based and lacks unique molecular identifiers (UMIs), is the unequal amplification of different regions and the plethora of bioinformatic tools to align reads and call variants, which may lead to misinterpretation of the results and the reporting of artifacts instead of actual variants. In our suggested algorithm apart from considering the variant specific features, we evaluate the background of each position and take advantage of the large number of patients that we have analyzed thus far. With this approach, we have observed that the background noise level in panel A, especially for known hotspots in the TP53 gene, is usually very low (below 0.5%).
One could evidently argue in favor of reporting TP53 variants with even lower VAF than 5%. Various reports based on retrospective studies favor the concept of reporting variants with VAF down to 0.1% as these may be selected by chemo-based regimens and drive disease relapse.39 However, as revealed by ddPCR analysis of 2 variants with VAF <5% (1 considered as sequencing artifact and 1 considered as “true variant”), a comprehensive evaluation of the performance of the method for detecting low frequency variants should be conducted in order to avoid run-specific or data-quality related errors.
In conclusion, we suggest that VAF in the range of 5% to 10% represents an eligible threshold for P/LP variants in TP53 in CLL provided that the method has been extensively validated for VAFs in that frequency range. Our suggested diagnostic algorithm ensures the detection of relevant TP53 variants with VAF around 10% that failed to overcome the 10% threshold because of methodological factors rather than disease-related features. Finally, our results have further implications as NGS technologies today are the gold-standard for the diagnostic work-up for the majority of hematological malignancies, highlighting the urgent need for harmonization of NGS methodologies across diagnostic laboratories.
ACKNOWLEDGMENTS
We would like to acknowledge Clinical Genomics Uppsala, Science for Life Laboratory, Department of Immunology, Genetics and Pathology, Uppsala University, Sweden, for providing assistance in sequencing and analysis.
AUTHOR CONTRIBUTIONS
TP and VL performed research, analyzed the data, and wrote the article. CL, ME, MM, MH and LC performed research and analyzed the data. PB designed the study and wrote the article.
DISCLOSURES
PB has received honoraria from Abbvie, Gilead and Janssen and research funding from Gilead. All the other authors have no conflicts of interest to disclose.
SOURCE OF FUNDING
This study was supported in part by the Lion’s Cancer Research Foundation, Uppsala.
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
REFERENCES
- 1.Wierda WG, Byrd JC, Abramson JS, et al. Chronic lymphocytic leukemia/small lymphocytic lymphoma, version 4.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2020;18:185–217. [DOI] [PubMed] [Google Scholar]
- 2.Eichhorst B, Robak T, Montserrat E, et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2021;32:23–33. [DOI] [PubMed] [Google Scholar]
- 3.Baliakas P, Puiggros A, Xochelli A, et al. Additional trisomies amongst patients with chronic lymphocytic leukemia carrying trisomy 12: the accompanying chromosome makes a difference. Haematologica. 2016;101:e299–e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baliakas P, Iskas M, Gardiner A, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89:249–255. [DOI] [PubMed] [Google Scholar]
- 5.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Clinical effect of stereotyped B-cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multicentre study. Lancet Haematol. 2014;1:e74–e84. [DOI] [PubMed] [Google Scholar]
- 6.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133:1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco G, Puiggros A, Baliakas P, et al. Karyotypic complexity rather than chromosome 8 abnormalities aggravates the outcome of chronic lymphocytic leukemia patients with TP53 aberrations. Oncotarget. 2016;7:80916–80924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ljungström V, Baliakas P. Prognostic and predictive implications of cytogenetics and genomics. Hematol Oncol Clin North Am. 2021;35:703–713. [DOI] [PubMed] [Google Scholar]
- 9.Baliakas P, Mattsson M, Stamatopoulos K, et al. Prognostic indices in chronic lymphocytic leukaemia: where do we stand how do we proceed? J Intern Med. 2016;279:347–357. [DOI] [PubMed] [Google Scholar]
- 10.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29:329–336. [DOI] [PubMed] [Google Scholar]
- 11.Ljungström V, Cortese D, Young E, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2016;127:1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigolin GM, del Giudice I, Formigaro L, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia: Clinical and biologic correlations. Genes Chromosomes Cancer. 2015;54:818–826. [DOI] [PubMed] [Google Scholar]
- 13.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herling CD, Klaumünzer M, Rocha CK, et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128:395–404. [DOI] [PubMed] [Google Scholar]
- 15.Rossi D, Rasi S, Fabbri G, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatzikonstantinou T, Demosthenous C, Baliakas P. Biology and treatment of high-risk CLL: significance of complex karyotype. Front Oncol. 2021;11:788761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soussi T, Baliakas P. Landscape of TP53 alterations in chronic lymphocytic leukemia via data mining mutation databases. Front Oncol. 2022;12:808886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenquist R, Ghia P, Hadzidimitriou A, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: updated ERIC recommendations. Leukemia. 2017;31:1477–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malcikova J, Tausch E, Rossi D, et al. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia. 2018;32:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai KR, Jain P. Chronic lymphocytic leukemia (CLL)-Then and now. Am J Hematol. 2016;91:330–340. [DOI] [PubMed] [Google Scholar]
- 21.Brown JR, Hillmen P, O’Brien S, et al. Extended follow-up and impact of high-risk prognostic factors from the phase 3 RESONATE study in patients with previously treated CLL/SLL. Leukemia. 2018;32:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woyach JA, Ruppert AS, Heerema NA, et al. Ibrutinib regimens versus chemoimmunotherapy in older patients with untreated CLL. N Engl J Med. 2018;379:2517–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien S, Furman RR, Coutre S, et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: a 5-year experience. Blood. 2018;131:1910–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kater AP, Wu JQ, Kipps T, et al. Venetoclax plus rituximab in relapsed chronic lymphocytic leukemia: 4-year results and evaluation of impact of genomic complexity and gene mutations from the MURANO Phase III study. J Clin Oncol. 2020;38:4042–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Döhner H, Stilgenbauer S, Döhner K, et al. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J Mol Med (Berl). 1999;77:266–281. [DOI] [PubMed] [Google Scholar]
- 26.Malcikova J, Stano-Kozubik K, Tichy B, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dicker F, Herholz H, Schnittger S, et al. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23:117–124. [DOI] [PubMed] [Google Scholar]
- 28.Gunnarsson R, Isaksson A, Mansouri M, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2010;24:211–215. [DOI] [PubMed] [Google Scholar]
- 29.Schoumans J, Suela J, Hastings R, et al. Guidelines for genomic array analysis in acquired haematological neoplastic disorders. Genes Chromosomes Cancer. 2016;55:480–491. [DOI] [PubMed] [Google Scholar]
- 30.Leeksma AC, Baliakas P, Moysiadis T, et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: a multi-center study. Haematologica. 2021;106:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarian G, Tausch E, Eclache V, et al. TP53 mutations are early events in chronic lymphocytic leukemia disease progression and precede evolution to complex karyotypes. Int J Cancer. 2016;139:1759–1763. [DOI] [PubMed] [Google Scholar]
- 32.Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35:672–688. [DOI] [PubMed] [Google Scholar]
- 33.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi D, Cerri M, Deambrogi C, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15:995–1004. [DOI] [PubMed] [Google Scholar]
- 35.Trbusek M, Malcikova J. TP53 aberrations in chronic lymphocytic leukemia. Adv Exp Med Biol. 2013;792:109–131. [DOI] [PubMed] [Google Scholar]
- 36.Zenz T, Vollmer D, Trbusek M, et al. TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia. 2010;24:2072–2079. [DOI] [PubMed] [Google Scholar]
- 37.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131:2745–2760. [DOI] [PubMed] [Google Scholar]
- 38.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malcikova J, Pavlova S, Kunt Vonkova B, et al. Low-burden TP53 mutations in CLL: clinical impact and clonal evolution within the context of different treatment options. Blood. 2021;138:2670–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brieghel C, Kinalis S, Yde CW, et al. Deep targeted sequencing of TP53 in chronic lymphocytic leukemia: clinical impact at diagnosis and at time of treatment. Haematologica. 2019;104:789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bomben R, Rossi FM, Vit F, et al. TP53 mutations with low variant allele frequency predict short survival in chronic lymphocytic leukemia. Clin Cancer Res. 2021;27:5566–5575. [DOI] [PubMed] [Google Scholar]
- 43.Regional Cancer Centrum. 2022. Available at: https://kunskapsbanken.cancercentrum.se/globalassets/cancerdiagnoser/blod-lymfom-myelom/kll/vardprogram/nationellt-vardprogram-kronisk-lymfatisk-leukemi-kll.pdf. [in Swedish]. Accessed July 25, 2022.
- 44.COSMIC, Catalogue of Somatic Mutations in Cancer. 2022. Available at: https://cancer.sanger.ac.uk/cosmic. Accessed July 25, 2022.
- 45.Hill JT, Demarest BL, Bisgrove BW, et al. Poly peak parser: Method and software for identification of unknown indels using Sanger sequencing of polymerase chain reaction products. Dev Dyn. 2014;243:1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navrkalova V, Young E, Baliakas P, et al. ATM mutations in major stereotyped subsets of chronic lymphocytic leukemia: enrichment in subset #2 is associated with markedly short telomeres. Haematologica. 2016;101:e369–e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansouri L, Sutton LA, Ljungström V, et al. Feasibility of targeted next-generation sequencing of the TP53 and ATM genes in chronic lymphocytic leukemia. Leukemia. 2014;28:694–696. [DOI] [PubMed] [Google Scholar]
- 48.Sutton LA, Ljungström V, Mansouri L, et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach will facilitate implementation in a clinical setting. Haematologica. 2015;100:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28:108–117. [DOI] [PubMed] [Google Scholar]
- 50.Brieghel C, Aarup K, Torp MH, et al. Clinical outcomes in patients with multi-Hit TP53 chronic lymphocytic leukemia treated with Ibrutinib. Clin Cancer Res. 2021;27:4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.