A combined inject-and-transfer system has been developed for sample delivery in serial femtosecond crystallography.
Keywords: serial crystallography, sample delivery, injection, fixed-target scanning, viscous media
Abstract
Serial femtosecond crystallography (SFX) enables the determination of room-temperature crystal structures of macromolecules with minimized radiation damage and provides time-resolved molecular dynamics by pump–probe or mix-and-inject experiments. In SFX, a variety of sample delivery methods with unique advantages have been developed and applied. The combination of existing sample delivery methods can enable a new approach to SFX data collection that combines the advantages of the individual methods. This study introduces a combined inject-and-transfer system (BITS) method for sample delivery in SFX experiments: a hybrid injection and fixed-target scanning method. BITS allows for solution samples to be reliably deposited on ultraviolet ozone (UVO)-treated polyimide films, at a minimum flow rate of 0.5 nl min−1, in both vertical and horizontal scanning modes. To utilize BITS in SFX experiments, lysozyme crystal samples were embedded in a viscous lard medium and injected at flow rates of 50–100 nl min−1 through a syringe needle onto a UVO-treated polyimide film, which was mounted on a fixed-target scan stage. The crystal samples deposited on the film were raster scanned with an X-ray free electron laser using a motion stage in both horizontal and vertical directions. Using the BITS method, the room-temperature structure of lysozyme was successfully determined at a resolution of 2.1 Å, and thus BITS could be utilized in future SFX experiments.
1. Introduction
Serial femtosecond crystallography (SFX) enables determination of crystal structures at room temperature while minimizing radiation damage (Chapman et al., 2011 ▸, 2014 ▸; Boutet et al., 2012 ▸; Liu et al., 2013 ▸; Pan et al., 2022 ▸; Ohmer et al., 2022 ▸). Moreover, SFX combined with pump–probe experiments enables visualization of time-resolved molecular dynamics of macromolecules (Kupitz et al., 2014 ▸; Tenboer et al., 2014 ▸; Pandey et al., 2020 ▸; Skopintsev et al., 2020 ▸; Ibrahim et al., 2020 ▸; Nass et al., 2020 ▸; Claesson et al., 2020 ▸). In serial crystallography (SX) experiments using X-ray free electron laser (XFEL) or synchrotron X-rays, many crystals are delivered to the X-ray interaction point and exposed to X-rays only once. Various sample delivery methods such as injectors (DePonte et al., 2008 ▸; Weierstall et al., 2014 ▸; Vakili et al., 2020 ▸), syringes with viscous media (Sugahara et al., 2015 ▸; Berntsen et al., 2019 ▸; Park & Nam, 2019 ▸; Nam, 2019 ▸, 2020a ▸), capillary methods (Stellato et al., 2014 ▸; Nam, 2020c ▸), fixed-target scanning (Hunter et al., 2014 ▸; Cohen et al., 2014 ▸; Mueller et al., 2015 ▸; Lee et al., 2019 ▸; Tolstikova et al., 2019 ▸), electrospinning (Sierra et al., 2012 ▸) and microfluidic devices (Monteiro et al., 2019 ▸, 2020 ▸; Echelmeier et al., 2020 ▸; Nam & Cho, 2021 ▸) have been applied to deliver microcrystals into X-ray interactions in a serial manner.
Among them, the liquid jet injector has the advantage of maintaining the crystal hydrate environment and providing fresh crystals from the injector to the X-ray beam with reduced exposure to the atmosphere (DePonte et al., 2008 ▸). However, since this method operates at a high flow rate to provide a stable injection flow, large numbers of crystals are not irradiated by X-rays due to the pulsed nature of the XFEL source (Echelmeier et al., 2020 ▸). Thus, this method is not suitable in XFELs with low repetition rates or synchrotrons in terms of sample consumption. Recently, to reduce sample consumption in the injector method, a segmented flow generator method, wherein droplets containing crystals are segmented with immiscible oil in a microfluidic device, has been developed (Echelmeier et al., 2020 ▸).
The method of delivering the crystals embedded in a viscous medium via a high-viscosity injector or syringe provides a stable injection stream even at low flow rates and is therefore widely applied in XFEL facilities with low repetition rates or synchrotrons (Weierstall et al., 2014 ▸; Conrad et al., 2015 ▸; Sugahara et al., 2015 ▸). Several types of viscous media, such as lipidic cubic phase and hydrophobic and hydrophilic injection matrices, have been developed and successfully applied to SX experiments (Nam, 2019 ▸). However, depending on the crystal characteristics and crystallization solution composition, the viscosity of the viscous medium is subject to change, which may produce an unstable injection stream (Nam, 2019 ▸). In such cases, stable injection conditions must be found, which lower the efficiency of data collection and increase sample consumption during optimization of the injection stream. To solve this problem, the delivery of crystals embedded in a viscous medium via capillary or single-channel microfluidics aligned to the X-ray interaction point was developed (Nam, 2020c ▸; Nam & Cho, 2021 ▸). Since the inner channel of the capillary or microfluidics is aligned to the X-ray beam path, all crystals continuously pass through the inner channel of the devices and are exposed to X-rays. Although these methods have been successfully demonstrated in serial synchrotron crystallography experiments, glass or polyimide channels may be physically damaged when exposed to intense XFELs.
The fixed-target scanning method has advantages in that it consumes fewer crystal samples compared with the sample delivery method using an injector, minimizes physical damage on the crystal during sample delivery, and can be programmed so that the sample is delivered to the desired location (Hunter et al., 2014 ▸; Mueller et al., 2015 ▸; Roedig et al., 2017 ▸; Lee et al., 2019 ▸, 2020 ▸; Nam et al., 2021 ▸). Although significant radiation damage has not yet been reported in crystal structures determined by the fixed-target scanning method, theoretically, radicals are generated after the X-rays have passed (Lee et al., 2019 ▸); therefore, diffusion of these radicals may affect the diffraction data of crystals collected later. In such cases, application of this method may be limited to time-resolved studies using an optical laser with caged molecules.
In addition, hybrid-type sample delivery methods that combine injection and motion have been developed. In the mix-and-diffuse (Beyerlein et al., 2017 ▸) or drop-on-drop (Butryn et al., 2021 ▸) method, crystal samples are placed on a polyimide tape by an injector, and these crystals are delivered to the X-ray interaction point by driving a conveyor belt. This method enables liquid-based time-resolved studies for various time delays by controlling the mixing speed and conveyor belt speed (Beyerlein et al., 2017 ▸; Butryn et al., 2021 ▸). In the liquid application method for time-resolved analyses (LAMA) (Mehrabi et al., 2019 ▸) approach, the crystals are dispensed, deposited or pipetted onto a fixed-target chip, and these crystals are delivered to the X-ray interaction point by translation stage motion. This method enables time-resolved SX experiments using the mix-and-inject approach at a synchrotron source. Taken together, these hybrid sample delivery systems are useful for application in time-resolved SFX experiments at XFEL or synchrotron facilities.
The sample delivery system will be selected according to the characteristics of each X-ray facility, the properties of the X-rays, the crystal sample and the desired target experimental setup. Different sample delivery methods have their own advantages, and the combination of multiple sample delivery methods enables creation of new experimental approaches.
In this study, we introduce a new sample delivery method referred to as the ‘combination of inject-and-transfer system’ (BITS), which combines the advantages of delivering a fresh sample every time (from the previously reported injection method) and programmed crystal sample delivery to the desired location in the XFEL (from the SFX fixed-target scanning method). Lysozyme crystals were embedded in lard and extruded on an ultraviolet ozone (UVO)-treated polyimide film through a syringe. The crystal samples were scanned by moving a translation stage in the horizontal and vertical directions. Using this method, we successfully determined the room-temperature structure of lysozyme, demonstrating that this sample delivery method can be applied to SFX data collection.
2. Methods and materials
2.1. Crystallization
Chicken egg white lysozyme was purchased from Sigma–Aldrich (catalogue No. L6876, St Louis, MO, USA). Crystal samples were obtained using the batch method described previously (Park et al., 2019 ▸). In brief, the lysozyme was dissolved in a solution containing 10 mM Tris–HCl pH 8.0 and 200 mM NaCl. Then the lysozyme solution (50 mg ml−1) was mixed with an equal volume of crystallization solution containing 0.1 M sodium acetate pH 4.0, 5%(w/v) PEG 8000 and 3.5 M NaCl in a 1.5 ml tube. Incubation at room temperature gave cube-shaped lysozyme crystals that grew to ≤10 × 10 × 10 µm within 1 h.
2.2. Ultraviolet ozone treatment
Polyimide film (50 µm) was purchased from Suzhou Kying Industrial Materials Co. Ltd (Suzhou, China). The UVO treatment was conducted on a 70 × 70 mm polyimide film with a UVO Cleaner (Ahtech LTS Co. Ltd, Anyang, Republic of Korea) for >10 min. The UVO-treated polyimide film was immediately packed under vacuum sealing to avoid the recovery of the polyimide film surface.
2.3. Injection experiment
The injection experiment was performed offline without the XFEL beam. The experiment used a crystallization solution, and the film was mounted on a translation stage and moved at a speed of 1.5 mm s−1 at 50 µm intervals in raster scan mode. For sample injection, a blunt-type syringe needle (length: 100 mm; inner diameter: 150 µm) was purchased from Solution Korea (Osan, Republic of Korea). In vertical scan mode for BITS, the crystallization solution was injected on a polyimide film at flow rates of 10 and 100 nl min−1, or on a UVO-treated polyimide film at flow rates of 0.5, 1, 2, 5, 10, 50, 100, 200, 500, 1000, 2000, 5000 and 10 000 nl min−1. The translation stage system consisted of a piezoelectric SLLV42 (SamartAct, dimension: 60 × 16 mm) and an SLL12 (SmartAct, dimension: 27 × 10 mm) actuator for motion in the horizontal and vertical directions, respectively. The translation pattern was implemented by user definition, according to the purpose of the experiment, and the translation code used in this experiment is given in Table S1 of the supporting information. In the combined vertical and horizontal scan mode for BITS, the crystallization solution was injected onto a UVO-treated polyimide film at flow rates of 0.5, 1, 5, 10, 50, 100, 200, 500 and 1000 nl min−1. Using the motion stage, the film was moved three times at 50 µm intervals from the bottom to the top, then moved 50 µm to the right, then moved three times at 50 µm intervals from the top to the bottom and then again to the right. By repeating this scan method, both horizontal and vertical measurements were made.
2.4. Crystal embedding in lard
The lard viscous medium was prepared as previously reported (Nam, 2020b ▸). In brief, lard was obtained from pork giblets and purified by heat-treatment and phase-separation methods. The lysozyme crystals and lard were mechanically mixed in a dual syringe setup, as previously reported (Nam, 2019 ▸, 2020b ▸). The lysozyme crystal suspension was transferred into a syringe. Then, the lard was dissolved by immersion in hot water (approximately 80°C) for 10 min, transferred into a syringe and left to stand until it solidified. The syringes containing the lysozyme crystals (200 µl) and lard (200 µl) were connected using a coupler and mixed by moving the plunger back and forth more than 30 times. Finally, the crystals embedded in the lard were transferred to a syringe connected to a needle with an inner diameter of 150 µm.
2.5. Data collection
SFX data collection was performed at the nano-crystallography and coherent imaging (NCI) experimental hutch in the Pohang Accelerator Laboratory X-ray Free Electron Laser (PAL-XFEL) (Ko et al., 2017 ▸; Kang et al., 2017 ▸). The photon energy and flux used in this experiment were 9500 eV and 2–5 × 1011 photons per pulse, respectively. The X-ray beam size was 2 × 3 µm (V × H FWHM) focused using a Kirkpatrick–Baez mirror (Kim et al., 2018 ▸). A 1000 µl syringe (BD Plastics Ltd, Sunderland, United Kingdom) containing the crystals embedded in the lard injection matrix (500 µl) was mounted on a Fusion 200 Touch syringe pump (Chemyx Inc., Houston, TX, USA; see Fig. S1 of the supporting information). The sample was transferred from the syringe to a needle with an inner diameter of 790 µm along polytetrafluoroethylene tubing (Precigenome LLC, San Jose, CA, USA). The position of the needle was adjusted using an XY manual motion stage, and the needle tip was tilted at an angle of 30–60° to contact the lower-left corner of the UVO-treated polyimide film (Fig. S2). Crystal samples were extruded at a 50–100 nl min−1 flow rate with a Fusion 200 Touch syringe pump (Chemyx Inc., Houston, TX, USA). The scanning stage was moved from left to right without synchronization with the arrival of an XFEL pulse, and then moved again to the left and then the right in a zigzag manner in the scanning direction. For the vertical mode, it was moved from the bottom to the top in three intervals, then to the right in one interval, then from the top to the bottom in three intervals and again to the right, and each interval was 50 µm. This type of raster scanning motion was repeatedly performed to obtain complete three-dimensional structural data. Diffraction data were collected in a helium atmosphere at 23–25°C using an MX225-HS detector (Rayonix, LLC, Evanston, IL, USA) with a 30 Hz readout.
2.6. Structure determination
The images containing the Bragg peaks were filtered using the Cheetah program (Barty et al., 2014 ▸) and were indexed and scaled using CrystFEL (White et al., 2016 ▸). The phasing was obtained by molecular replacement with the MOLREP program (Vagin & Teplyakov, 2010 ▸), with the lysozyme crystal structure (PDB entry 7wkr) as the search model. The model structure was built using Coot (Emsley & Cowtan, 2004 ▸) and refined using phenix.refine in PHENIX (Afonine et al., 2012 ▸). The geometries of the final model structures were verified using MolProbity (Williams et al., 2018 ▸). Structural figures were generated using PyMOL (https://pymol.org/). Data collection and refinement statistics are summarized in Table 1 ▸. Coordinates and structure factors have been deposited (PDB entry 7wuc) in the Protein Data Bank (https://www.rcsb.org/). Diffraction images were deposited (CIDB entry ID197; https://cxidb.org/id-197.html) in the Coherent X-ray Imaging Data Bank (Maia, 2012 ▸).
Table 1. Data collection and processing.
| Data collection | |
| Diffraction source | NCI beamline, PAL-XFEL |
| Wavelength (Å) | 1.305 |
| Temperature (K) | 296.15–298.15 |
| Photons per pulse | ∼5 × 1011 |
| Detector | MX225-HS |
| Crystal-to-detector distance (mm) | 123 |
| No. of collected images | 85901 |
| No. of hits | 27413 |
| No. of indexed images | 17411 |
| No. of indexed patterns | 22302 |
| Space group | P43212 |
| a, b, c (Å) | 79.48, 79.48, 38.48 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (Å) | 80.0–2.10 (2.17–2.10) |
| No. of unique reflections | 7521 (724) |
| Completeness (%) | 100 (100) |
| Redundancy | 590.7 (377.5) |
| SNR | 7.38 (1.57) |
| CC | 0.9866 (0.4766) |
| CC* | 0.9966 (0.8034) |
| R split (%)† | 10.44 (71.03) |
| Overall B factor from Wilson plot (Å2) | 46.9 |
| Refinement | |
| Resolution (Å) | 39.74–2.10 (2.26–2.10) |
| σ cutoff | F > 1.35σ(F) |
| Final R work ‡ | 0.220 (0.3563) |
| Final R free § | 0.249 (0.3833) |
| B factor (averaged) | |
| Protein | 43.87 |
| Water | 46.45 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.664 |
| Ramachandran plot (%) | |
| Favoured | 97.64 |
| Allowed | 2.36 |
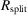

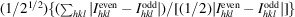
R
work
 ∑||F
obs| − |F
calc||/∑|F
obs|, where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively.
∑||F
obs| − |F
calc||/∑|F
obs|, where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively.
R free was calculated as R work using a randomly selected subset (9.47%) of unique reflections not used for structure refinement.
3. Results and discussion
3.1. Combination of inject-and-transfer system
We developed BITS from two perspectives: (i) It was intended to be used immediately with minimal effort to create a stable injection stream when the provided injection stream, including crystals delivered from the injector or syringe, is unstable. (ii) When the sample is delivered through an injector or syringe, since the XFEL beam impinges only at one location of the injection stream, unnecessary sample consumption occurs when the width of the injection stream is large. It was intended to scan the injection stream to reduce sample consumption.
We benchmarked a hybrid sample delivery method in which samples are transferred into the XFEL beam after being delivered to a specific location by an injector, as in the conveyor belt and LAMA approaches, and developed a new sample delivery system for SX data collection.
BITS is composed of injection and motion systems, and the sample chamber and motion stage utilized a previously developed fixed-target SFX chamber system (Lee et al., 2019 ▸, 2020 ▸). The main components of BITS can be divided into the injector, which delivers the crystal sample from the syringe and tube to the film via a syringe needle; and a motion stage, which moves the crystal sample that has been deposited on the film to the X-ray interaction point (Fig. 1 ▸). The position of the syringe needle tip was adjusted with a manual stage such that the needle contacts the film at the point where the crystals are deposited. The crystal sample forms a small drop at the needle tip and, when in contact with the film, is deposited on the film according to the flow rate (Fig. S3). However, when the needle tip does not directly contact the polyimide film, a larger sample drop is formed at the end of the needle tip by surface tension, and data collection cannot be performed until the drop has grown large enough to reach the polyimide film (Fig. S3). This causes an increase in the amount of unnecessary sample consumption and impairs data collection efficiency. Accordingly, the needle tip must make contact with the polyimide film during device installation and data acquisition. Therefore, it is important that the polyimide film is installed flat on the frame and that the translation stage is accurately aligned horizontally and vertically with the needle (Fig. S4). When the injected crystal sample was placed on the film, the movement of the translation stage transferred the sample to the X-ray interaction point and exposed it to the XFEL. In this experiment, we manually aligned the UVO-treated polyimide film and syringe needle in the plane. To make the system more efficient, it would be useful to apply the previously reported plane-levelling via xyz-centering (Sherrell et al., 2015 ▸).
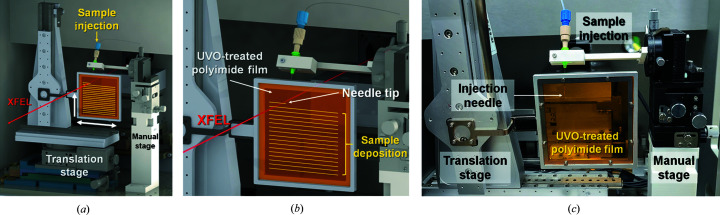
Figure 1.
Schematics of the experimental setup of BITS. (a) BITS is composed of an injection and translation system. (b) The protein crystal sample (yellow) is injected from the needle and deposited on the UVO-treated polyimide film; the needle position is fixed and the sample position is moved to the XFEL path (red line) by the motion stage. (c) Photograph of the BITS experimental setup.
Collectively, the BITS method is a combination of the conventional injection method and the fixed-target scanning method. It has the advantages of both an injector that always delivers a fresh crystal sample and fixed-target scanning that allows the sample to be freely delivered to the desired location. In addition, because the stability of the injection flow is not critical, it can be delivered at very low flow rates to reduce sample consumption.
3.2. Sample loading
To demonstrate the stable sample loading to the film, an injection experiment on a polyimide film was performed using a crystallization solution containing 0.1 M sodium acetate pH 4.0, 5%(w/v) PEG 8000 and 3.5 M NaCl, without crystal samples. The results show that, rather than the crystal solution being uniformly deposited on the polyimide film, a drop was formed at the syringe needle tip, and it often settled on the polyimide film with a random size [Fig. 2 ▸(a) and Movies S1 and S2 of the supporting information]. Droplet formation at the needle tip is favoured over deposition of the crystallization solution on the film because the polyimide film surface is very hydrophobic (Damaceanu et al., 2014 ▸). Moreover, it was considered that the crystallization solution will preferentially maintain the form of the drop on the polyimide film because the crystallization solution has no affinity for the polyimide film hydrophobic surface, even if the droplet becomes large and settles on the film.
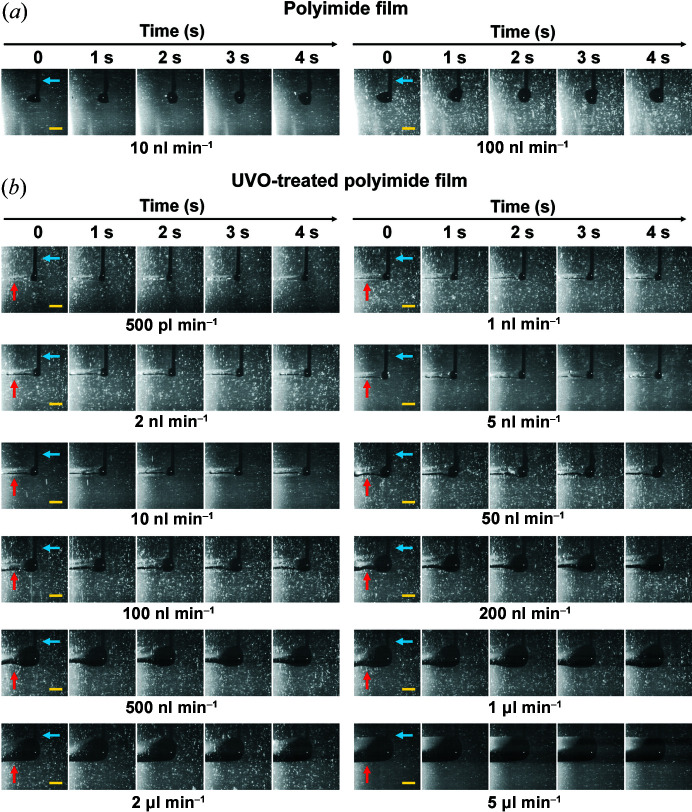
Figure 2.
Sample injection experiment on polyimide film. (a) Snapshots of sample injection on the polyimide film at flow rates of 10 and 100 nl min−1 during vertical scanning. (b) Snapshots of sample injection on the UVO-treated polyimide film at flow rates of 0.5, 1, 2, 5, 10, 50, 100, 200, 500, 1000, 2000 and 5000 nl min−1 during vertical scanning. The injection needle and deposited sample are indicated by blue and red arrows, respectively. Scale bar indicates 500 µm.
To solve this problem, the hydrophilicity of the polyimide film surface was increased by UVO surface pre-treatment. The results showed that the crystallization solution was deposited more evenly onto the UVO-treated polyimide film [Fig. 2 ▸(b)]. Therefore, the hydrophilic surface helped the sample settle on the film and was suitable for application in the BITS. However, the hydrophilic surface of the UVO-treated polyimide film can revert to its original hydrophobic form over time. Therefore, using the polyimide film immediately after UVO treatment is ideal.
The stability of the injected sample and the sample consumption vary depending on the injection flow rate and translation stage speed. Therefore, the target scanning was performed at 30 Hz at 50 µm intervals in the horizontal direction, and the crystallization solution was injected while moving the translation stage at a speed of 1.5 mm s−1. Then, the sample pattern on the UVO-treated polyimide film was analyzed according to various injection flow rates [Fig. 2 ▸(b) and Movies S3–S15]. The crystallization solution could be stably mounted on the UVO-treated polyimide film at an injection rate of 1–100 nl min−1. At 500 pl min−1, the sample was continuously deposited on the UVO polyimide film; however, in this case, it was expected that a dehydration problem could occur. The sample was uniformly deposited on the film, even at a flow rate of >200 nl min−1, but the deposit was unnecessarily wide and thick, and the solution not deposited formed droplets on the needle tip. This is an excessive injection flow rate, which temporarily causes unnecessary crystal consumption.
We also used a nylon mesh as the crystal settling plane material, and stably and uniformly mounted the crystal solution. However, the needle tip position was often changed slightly while the translation stage was moving. This is caused by a physical effect that occurs when the needle tip is in contact with a nylon mesh that has an uneven surface and moves. Although it is possible to collect data by injecting crystal samples onto nylon mesh, a UVO-treated polyimide film was used here to deliver samples in an optimized state.
3.3. XFEL data collection
To demonstrate the BITS application, we performed an SFX experiment using lysozyme crystals as a model sample. In the initial XFEL data collection, the lysozyme crystal suspension was loaded onto a UVO-treated polyimide film through a syringe needle for XFEL exposure. During SFX data collection, lysozyme diffraction data could be collected for several minutes; however, we observed a reduction in crystal diffraction as injection progressed. When the sample tubing and syringe needle were disassembled, we confirmed that the crystal sample was clogged at the needle inlet. We considered that, when the crystal suspension was delivered at a low flow rate, the crystals sank in the tubing and needle under gravity, and a large number of crystals clogged the needle inlet, with only the crystallization solution flowing after clogging. It may be possible to reduce the crystal clogging by gentle shaking using an anti-settler where the crystals are stored, or by minimizing the distance of the tubing through which the crystals pass.
On the other hand, our previous study investigated how the delivery of crystals mixed with viscous media such as agarose, gelatin or shortening prevented the crystals from sinking under gravity without clogging in quartz capillary and single-channel microfluidics (Nam, 2020c ▸; Nam & Cho, 2021 ▸). Therefore, lysozyme crystals were embedded in lard in the present experiment to prevent clogging of the crystals in the needle and tubing. Lard was used to produce a stable injection stream, at a low flow rate, in the previous SX experiment, and a stable stream was produced when the lard content was >80% in the mixture of lard and protein suspension (Nam, 2020b ▸). In the present experiment, the crystal suspension and lard ratio was 1:1; therefore, a directly measurable injection stream was not created, and the lard was used only to prevent the crystals from sinking. As a result, crystals embedded in lard could be stably delivered to UVO-treated polyimide films, without clogging the tubing or syringe needle. The lard used in this study showed background scattering at about 15 Å (Fig. S5), which had no effect on the structural determination; however, if the diffraction intensity of the target crystal is weak or it is necessary to minimize background noise, viscous materials with low background scattering should be used. In this experiment, a mixture of viscous material and crystals which cannot create a directly measurable injection stream was injected onto a film. Data collection was possible by utilizing the injection stream on the film, similarly to general sample delivery using a viscous medium in an SX experiment. When using a viscous material to prevent the crystals from sinking under gravity, it is necessary to select a viscous material that does not have a physical and chemical effect during the mixing and storage of the crystal and the viscous material. Lard was used in this experiment, but if lard is not an appropriate material for target crystals in terms of crystal stability, it may be possible to prevent or delay the sinking of the crystal sample by employing other viscous materials with different physical and chemical characteristics. On the other hand, there is no need to use a viscous material if the crystal suspension is used and there is no clogging problem.
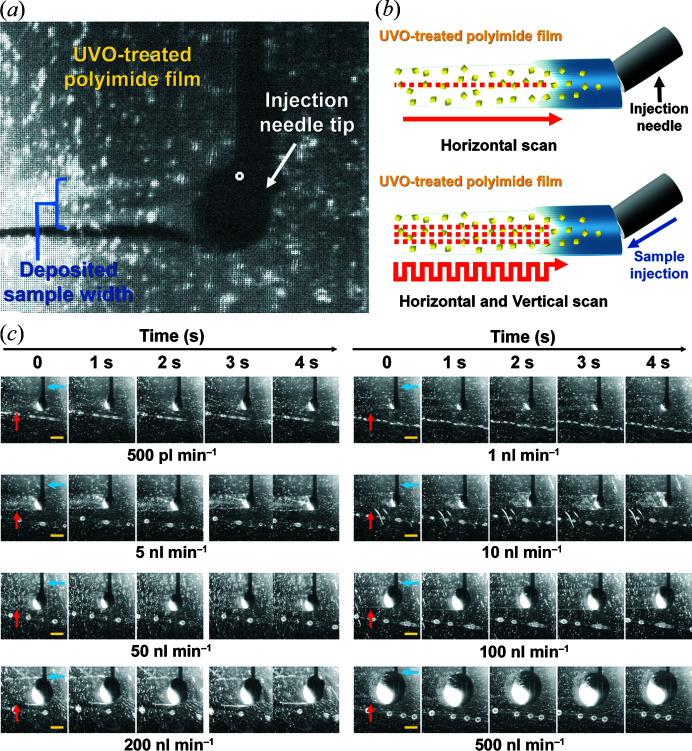
The width of the injected sample on the UVO-treated polyimide film was approximately 300 µm [Fig. 3 ▸(a)]. Since the size of the beam was within 5 µm (FWHM), most crystal samples were not exposed to X-rays when the translation stage solely moved in the horizontal direction during data collection [Fig. 3 ▸(b)]. Alternatively, BITS can be programmed to move the sample to the desired location. Therefore, the width of the injected sample is not an important factor in terms of sample consumption. In this experiment, to avoid radiation damage to the surrounding sample when the XFEL beam is transmitted, crystals were scanned at 50 µm intervals. During data collection, the process of scanning the injected sample vertically at 50 µm intervals for three scan points and moving it 50 µm to the right was repeated (Movie S16). The applied injection sample shows various injection sample widths on the film depending on the inner needle (or nozzle) diameter used and the crystallization solution viscosity when applying BITS. However, the injected sample width is not critical, as data collection can be accomplished by simply programming the scan interval according to the injected sample width. In the existing injector/syringe, X-rays can only irradiate one point of the injection stream, whereas in BITS, adjacent regions of the sample, both vertically and horizontally, can be exposed to X-rays at a lower flow rate, so that sample consumption can be significantly reduced. For example, if a viscous medium containing crystals is extruded – by an injector or syringe – into an injection stream with a 150 µm width, the XFEL beam impinges at one location regardless of the width of the injection stream. On the other hand, in BITS, when the sample is deposited on the UVO-treated polyimide film, the actual width becomes wider than 150 µm. In this case, when the deposited sample is scanned with an XFEL beam in the vertical direction at 50 µm intervals using a motion stage, the sample is exposed three times in the vertical direction of the injected sample, so the sample consumption in BITS can be reduced 3× compared with the injector/syringe-based sample delivery. In this experiment, scans were performed at 50 µm intervals to prevent radiation damage from XFEL exposure, but with a smaller XFEL, scanning at shorter intervals for distances where there is no radiation damage problem can further reduce sample consumption.
Figure 3.
Injection and data collection strategies in the BITS approach. (a) Snapshot of injection of the crystallization solution on UVO-treated polyimide film for data collection using BITS. (b) Data collection strategy in BITS. (Top) When the motion stage moves in one direction, only one position along the injected sample is exposed to X-rays, and most of the crystal sample is not. (Bottom) In BITS, more data can be collected from the injected sample because the motion stage moves both horizontally and vertically. (c) Snapshots of the sample injections on the UVO-treated polyimide film at flow rates of 0.5, 1, 5, 10, 50, 100, 200 and 500 nl min−1 during vertical and horizontal scanning. The injection needle and deposited sample are indicated by blue and red arrows, respectively. Scale bar indicates 500 µm.
We performed an injection experiment to investigate BITS, in which the vertical and horizontal directions were driven at various injection flow rates using a crystallization solution [Fig. 3 ▸(c) and Movies S17–S25]. When scanning a sample deposited on the film in both directions, the horizontal movement speed was found to be lower than when scanning only in the vertical direction in which the translation stage moves (Fig. 2 ▸). Therefore, even at the same injection flow rate, the bidirectional scan method in BITS increases the relative size of the droplets on the needle tip, and since they also move in the vertical direction, the width of the deposited sample is increased. It was consequently determined that with BITS, even if the injected sample was deposited on the film at a flow rate of 0.5 nl min−1, data could still be collected by scanning in both directions. Meanwhile, at a flow rate >100 nl min−1, more sample was found to be hanging from the needle tip than deposited on the film, indicating unnecessary sample consumption.
Our injection test showed that the sample injected was stably mounted on the film, even at 0.5 nl min−1 with both vertical and horizontal scanning [Fig. 3 ▸(b)]. However, the crystallization solution of lysozyme contains a 3.5 M NaCl concentration, and prolonged exposure to the atmosphere during data collection may cause salt crystal growth and changes in the crystal structure by dehydration (Lobley et al., 2016 ▸; Lee et al., 2019 ▸). Therefore, to prevent this in advance, crystals embedded in the lard were injected at 50–100 nl min−1 in the actual experiment. In BITS, if there were no dehydration issues of the crystallization solution, the crystal sample would be ejected at a lower flow rate. In previous studies, to avoid evaporation of the sample solution, a sample delivery system combined with a humidity stream (Mehrabi et al., 2019 ▸) or an environmental control box (Mehrabi et al., 2021 ▸) has been developed and successfully applied in SFX experiments. The combination of these systems with BITS will provide better sample environments to avoid sample evaporation.
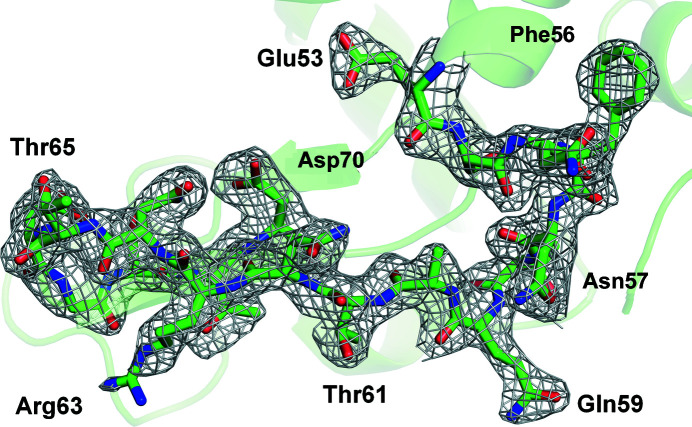
Finally, for the structure determination, XFEL data were collected using BITS with lysozyme crystals embedded in lard material (Movie S26). SFX data were collected for about 50 min using an XFEL beam of ∼1 × 1011 photons per pulse, and the amount of lysozyme crystal solution consumed was <1.88 µl. A total of 85 901 images were collected, and the number of hit images containing Bragg peaks and indexed images was 27 413 (hit rate: 31.91%) and 22 302 (indexing rate: 81.35%), respectively. The data were processed up to 2.1 Å. Relevant parameters are given in Table 1 ▸. Lysozyme showed a clear electron density map for Lys19–Leu147 (Fig. 4 ▸). The room-temperature structure of lysozyme obtained by BITS showed high similarity to the previously reported room-temperature structures of lysozyme delivered in polyacrylamide (PDB entry 6ig6; Park et al., 2019 ▸), shortening (PDB entry 6kcb; Nam, 2020d ▸), lard (PDB entry 7cjz; Nam, 2020b ▸) and beef tallow (PDB entry 7e02; Nam, 2022 ▸), with an RMSD of 0.111–0.128 Å.
Figure 4.
2mF o − DF c (grey mesh, 1.0σ) electron density map of room-temperature lysozyme embedded in lard delivered by BITS.
4. Conclusions
In this study, we introduced BITS, a hybrid method in which viscous injection is coupled with a fixed-target matrix. Using this approach, we successfully collected XFEL diffraction data and determined the room-temperature structure of lysozyme. This method does not require any effort to create a stable injection stream in the existing injector/syringe-based sample delivery method, and can thus deliver samples at very low flow rates. In addition, even if the width of the injection sample applied to the experiment varied, most of the injected samples could be scanned by programming the motion stage. As a result, compared with the sample delivery method using the existing injector/syringe, this requires less effort for sample injection and has the advantage of significantly reducing sample consumption. However, since the types of crystal solution or viscous medium used with BITS can be varied, the width of the sample deposited on the film and the characteristics of evaporation can be varied too. In addition, the data collection strategy may change depending on the size and repetition rate of the XFEL beam. Accordingly, depending on the sample injected and its environment, optimization may be required to minimize sample consumption. Overall, the BITS method was found to be suitable for SFX data collection.
Supplementary Material
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup1.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup2.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup3.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup4.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup5.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup6.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup7.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup8.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup9.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup10.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup11.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup12.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup13.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup14.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup15.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup16.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup17.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup18.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup19.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup20.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup21.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup22.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup23.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup24.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup25.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup26.mp4
Supporting figures and BITS code. DOI: 10.1107/S1600576722005556/te5095sup27.pdf
PDB reference: 7wkr
PDB reference: 7wuc
Acknowledgments
We thank the beamline staff at NCI bemline at Pohang Accelerator Laboratory X-ray Free Electron Laser (PAL-XFEL) for their assistance with data collection. The XFEL experiments were carried out at the NCI endstation at PAL-XFEL (proposal Nos. 2021-2nd-NCI-013). The authors thank Global Science experimental Data hub Center (GSDC) at Korea Institute of Science and Technology Information (KISTI) for computational support.
Funding Statement
The following funding is acknowledged: National Research Foundation of Korea (grant Nos. NRF-2017M3A9F6029736; NRF-2020M3H1A1075314; NRF-2021R1I1A1A01050838).
References
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Barty, A., Kirian, R. A., Maia, F. R. N. C., Hantke, M., Yoon, C. H., White, T. A. & Chapman, H. (2014). J. Appl. Cryst. 47, 1118–1131. [DOI] [PMC free article] [PubMed]
- Berntsen, P., Hadian Jazi, M., Kusel, M., Martin, A. V., Ericsson, T., Call, M. J., Trenker, R., Roque, F. G., Darmanin, C. & Abbey, B. (2019). Rev. Sci. Instrum. 90, 085110. [DOI] [PubMed]
- Beyerlein, K. R., Dierksmeyer, D., Mariani, V., Kuhn, M., Sarrou, I., Ottaviano, A., Awel, S., Knoska, J., Fuglerud, S., Jönsson, O., Stern, S., Wiedorn, M. O., Yefanov, O., Adriano, L., Bean, R., Burkhardt, A., Fischer, P., Heymann, M., Horke, D. A., Jungnickel, K. E. J., Kovaleva, E., Lorbeer, O., Metz, M., Meyer, J., Morgan, A., Pande, K., Panneerselvam, S., Seuring, C., Tolstikova, A., Lieske, J., Aplin, S., Roessle, M., White, T. A., Chapman, H. N., Meents, A. & Oberthuer, D. (2017). IUCrJ, 4, 769–777. [DOI] [PMC free article] [PubMed]
- Boutet, S., Lomb, L., Williams, G. J., Barends, T. R., Aquila, A., Doak, R. B., Weierstall, U., DePonte, D. P., Steinbrener, J., Shoeman, R. L., Messerschmidt, M., Barty, A., White, T. A., Kassemeyer, S., Kirian, R. A., Seibert, M. M., Montanez, P. A., Kenney, C., Herbst, R., Hart, P., Pines, J., Haller, G., Gruner, S. M., Philipp, H. T., Tate, M. W., Hromalik, M., Koerner, L. J., van Bakel, N., Morse, J., Ghonsalves, W., Arnlund, D., Bogan, M. J., Caleman, C., Fromme, R., Hampton, C. Y., Hunter, M. S., Johansson, L. C., Katona, G., Kupitz, C., Liang, M., Martin, A. V., Nass, K., Redecke, L., Stellato, F., Timneanu, N., Wang, D., Zatsepin, N. A., Schafer, D., Defever, J., Neutze, R., Fromme, P., Spence, J. C., Chapman, H. N. & Schlichting, I. (2012). Science, 337, 362–364.
- Butryn, A., Simon, P. S., Aller, P., Hinchliffe, P., Massad, R. N., Leen, G., Tooke, C. L., Bogacz, I., Kim, I. S., Bhowmick, A., Brewster, A. S., Devenish, N. E., Brem, J., Kamps, J., Lang, P. A., Rabe, P., Axford, D., Beale, J. H., Davy, B., Ebrahim, A., Orlans, J., Storm, S. L. S., Zhou, T., Owada, S., Tanaka, R., Tono, K., Evans, G., Owen, R. L., Houle, F. A., Sauter, N. K., Schofield, C. J., Spencer, J., Yachandra, V. K., Yano, J., Kern, J. F. & Orville, A. M. (2021). Nat. Commun. 12, 4461. [DOI] [PMC free article] [PubMed]
- Chapman, H. N., Caleman, C. & Timneanu, N. (2014). Phil. Trans. R. Soc. B, 369, 20130313. [DOI] [PMC free article] [PubMed]
- Chapman, H. N., Fromme, P., Barty, A., White, T. A., Kirian, R. A., Aquila, A., Hunter, M. S., Schulz, J., DePonte, D. P., Weierstall, U., Doak, R. B., Maia, F. R. N. C., Martin, A. V., Schlichting, I., Lomb, L., Coppola, N., Shoeman, R. L., Epp, S. W., Hartmann, R., Rolles, D., Rudenko, A., Foucar, L., Kimmel, N., Weidenspointner, G., Holl, P., Liang, M., Barthelmess, M., Caleman, C., Boutet, S., Bogan, M. J., Krzywinski, J., Bostedt, C., Bajt, S., Gumprecht, L., Rudek, B., Erk, B., Schmidt, C., Hömke, A., Reich, C., Pietschner, D., Strüder, L., Hauser, G., Gorke, H., Ullrich, J., Herrmann, S., Schaller, G., Schopper, F., Soltau, H., Kühnel, K.-U., Messerschmidt, M., Bozek, J. D., Hau-Riege, S. P., Frank, M., Hampton, C. Y., Sierra, R. G., Starodub, D., Williams, G. J., Hajdu, J., Timneanu, N., Seibert, M. M., Andreasson, J., Rocker, A., Jönsson, O., Svenda, M., Stern, S., Nass, K., Andritschke, R., Schröter, C.-D., Krasniqi, F., Bott, M., Schmidt, K. E., Wang, X., Grotjohann, I., Holton, J. M., Barends, T. R. M., Neutze, R., Marchesini, S., Fromme, R., Schorb, S., Rupp, D., Adolph, M., Gorkhover, T., Andersson, I., Hirsemann, H., Potdevin, G., Graafsma, H., Nilsson, B. & Spence, J. C. H. (2011). Nature, 470, 73–77.
- Claesson, E., Wahlgren, W. Y., Takala, H., Pandey, S., Castillon, L., Kuznetsova, V., Henry, L., Panman, M., Carrillo, M., Kübel, J., Nanekar, R., Isaksson, L., Nimmrich, A., Cellini, A., Morozov, D., Maj, M., Kurttila, M., Bosman, R., Nango, E., Tanaka, R., Tanaka, T., Fangjia, L., Iwata, S., Owada, S., Moffat, K., Groenhof, G., Stojković, E. A., Ihalainen, J. A., Schmidt, M. & Westenhoff, S. (2020). eLife, 9, e57105.
- Cohen, A. E., Soltis, S. M., González, A., Aguila, L., Alonso-Mori, R., Barnes, C. O., Baxter, E. L., Brehmer, W., Brewster, A. S., Brunger, A. T., Calero, G., Chang, J. F., Chollet, M., Ehrensberger, P., Eriksson, T. L., Feng, Y., Hattne, J., Hedman, B., Hollenbeck, M., Holton, J. M., Keable, S., Kobilka, B. K., Kovaleva, E. G., Kruse, A. C., Lemke, H. T., Lin, G., Lyubimov, A. Y., Manglik, A., Mathews, II, McPhillips, S. E., Nelson, S., Peters, J. W., Sauter, N. K., Smith, C. A., Song, J., Stevenson, H. P., Tsai, Y., Uervirojnangkoorn, M., Vinetsky, V., Wakatsuki, S., Weis, W. I., Zadvornyy, O. A., Zeldin, O. B., Zhu, D. & Hodgson, K. O. (2014). Proc. Natl Acad. Sci. USA, 111, 17122–17127.
- Conrad, C. E., Basu, S., James, D., Wang, D., Schaffer, A., Roy-Chowdhury, S., Zatsepin, N. A., Aquila, A., Coe, J., Gati, C., Hunter, M. S., Koglin, J. E., Kupitz, C., Nelson, G., Subramanian, G., White, T. A., Zhao, Y., Zook, J., Boutet, S., Cherezov, V., Spence, J. C. H., Fromme, R., Weierstall, U. & Fromme, P. (2015). IUCrJ, 2, 421–430. [DOI] [PMC free article] [PubMed]
- Damaceanu, M.-D., Constantin, C.-P., Nicolescu, A., Bruma, M., Belomoina, N. & Begunov, R. S. (2014). Eur. Polym. J. 50, 200–213.
- DePonte, D. P., Weierstall, U., Schmidt, K., Warner, J., Starodub, D., Spence, J. C. H. & Doak, R. B. (2008). J. Phys. D Appl. Phys. 41, 195505.
- Echelmeier, A., Cruz Villarreal, J., Messerschmidt, M., Kim, D., Coe, J. D., Thifault, D., Botha, S., Egatz-Gomez, A., Gandhi, S., Brehm, G., Conrad, C. E., Hansen, D. T., Madsen, C., Bajt, S., Meza-Aguilar, J. D., Oberthür, D., Wiedorn, M. O., Fleckenstein, H., Mendez, D., Knoška, J., Martin-Garcia, J. M., Hu, H., Lisova, S., Allahgholi, A., Gevorkov, Y., Ayyer, K., Aplin, S., Ginn, H. M., Graafsma, H., Morgan, A. J., Greiffenberg, D., Klujev, A., Laurus, T., Poehlsen, J., Trunk, U., Mezza, D., Schmidt, B., Kuhn, M., Fromme, R., Sztuk-Dambietz, J., Raab, N., Hauf, S., Silenzi, A., Michelat, T., Xu, C., Danilevski, C., Parenti, A., Mekinda, L., Weinhausen, B., Mills, G., Vagovic, P., Kim, Y., Kirkwood, H., Bean, R., Bielecki, J., Stern, S., Giewekemeyer, K., Round, A. R., Schulz, J., Dörner, K., Grant, T. D., Mariani, V., Barty, A., Mancuso, A. P., Weierstall, U., Spence, J. C. H., Chapman, H. N., Zatsepin, N., Fromme, P., Kirian, R. A. & Ros, A. (2020). Nat. Commun. 11, 4511.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hunter, M. S., Segelke, B., Messerschmidt, M., Williams, G. J., Zatsepin, N. A., Barty, A., Benner, W. H., Carlson, D. B., Coleman, M., Graf, A., Hau-Riege, S. P., Pardini, T., Seibert, M. M., Evans, J., Boutet, S. & Frank, M. (2014). Sci. Rep. 4, 6026. [DOI] [PMC free article] [PubMed]
- Ibrahim, M., Fransson, T., Chatterjee, R., Cheah, M. H., Hussein, R., Lassalle, L., Sutherlin, K. D., Young, I. D., Fuller, F. D., Gul, S., Kim, I.-S., Simon, P. S., de Lichtenberg, C., Chernev, P., Bogacz, I., Pham, C. C., Orville, A. M., Saichek, N., Northen, T., Batyuk, A., Carbajo, S., Alonso-Mori, R., Tono, K., Owada, S., Bhowmick, A., Bolotovsky, R., Mendez, D., Moriarty, N. W., Holton, J. M., Dobbek, H., Brewster, A. S., Adams, P. D., Sauter, N. K., Bergmann, U., Zouni, A., Messinger, J., Kern, J., Yachandra, V. K. & Yano, J. (2020). Proc. Natl Acad. Sci. USA, 117, 12624–12635.
- Kang, H. S., Min, C. K., Heo, H., Kim, C., Yang, H., Kim, G., Nam, I., Baek, S. Y., Choi, H. J., Mun, G., Park, B. R., Suh, Y. J., Shin, D. C., Hu, J., Hong, J., Jung, S., Kim, S. H., Kim, K., Na, D., Park, S. S., Park, Y. J., Han, J. H., Jung, Y. G., Jeong, S. H., Lee, H. G., Lee, S., Lee, S., Lee, W. W., Oh, B., Suh, H. S., Parc, Y. W., Park, S. J., Kim, M. H., Jung, N. S., Kim, Y. C., Lee, M. S., Lee, B. H., Sung, C. W., Mok, I. S., Yang, J. M., Lee, C. S., Shin, H., Kim, J. H., Kim, Y., Lee, J. H., Park, S. Y., Kim, J., Park, J., Eom, I., Rah, S., Kim, S., Nam, K. H., Park, J., Park, J., Kim, S., Kwon, S., Park, S. H., Kim, K. S., Hyun, H., Kim, S. N., Kim, S., Hwang, S. M., Kim, M. J., Lim, C. Y., Yu, C. J., Kim, B. S., Kang, T. H., Kim, K. W., Kim, S. H., Lee, H. S., Lee, H. S., Park, K. H., Koo, T. Y., Kim, D. E. & Ko, I. S. (2017). Nat. Photon. 11, 708–713.
- Kim, J., Kim, H.-Y., Park, J., Kim, S., Kim, S., Rah, S., Lim, J. & Nam, K. H. (2018). J. Synchrotron Rad. 25, 289–292. [DOI] [PMC free article] [PubMed]
- Ko, I. S., Kang, H. S., Heo, H., Kim, C., Kim, G., Min, C. K., Yang, H., Baek, S. Y., Choi, H. J., Mun, G., Park, B. R., Suh, Y. J., Shin, D. C., Hu, J., Hong, J., Jung, S., Kim, S. H., Kim, K., Na, D., Park, S. S., Park, Y. J., Jung, Y. G., Jeong, S. H., Lee, H. G., Lee, S., Lee, S., Oh, B., Suh, H. S., Han, J. H., Kim, M. H., Jung, N. S., Kim, Y. C., Lee, M. S., Lee, B. H., Sung, C. W., Mok, I. S., Yang, J. M., Parc, Y. W., Lee, W. W., Lee, C. S., Shin, H., Kim, J. H., Kim, Y., Lee, J. H., Park, S. Y., Kim, J., Park, J., Eom, I., Rah, S., Kim, S., Nam, K. H., Park, J., Park, J., Kim, S., Kwon, S., An, R., Park, S. H., Kim, K. S., Hyun, H., Kim, S. N., Kim, S., Yu, C. J., Kim, B. S., Kang, T. H., Kim, K. W., Kim, S. H., Lee, H. S., Lee, H. S., Park, K. H., Koo, T. Y., Kim, D. E. & Lee, K. B. (2017). Appl. Sci. 7, 479.
- Kupitz, C., Basu, S., Grotjohann, I., Fromme, R., Zatsepin, N. A., Rendek, K. N., Hunter, M. S., Shoeman, R. L., White, T. A., Wang, D., James, D., Yang, J.-H., Cobb, D. E., Reeder, B., Sierra, R. G., Liu, H., Barty, A., Aquila, A. L., Deponte, D., Kirian, R. A., Bari, S., Bergkamp, J. J., Beyerlein, K. R., Bogan, M. J., Caleman, C., Chao, T.-C., Conrad, C. E., Davis, K. M., Fleckenstein, H., Galli, L., Hau-Riege, S. P., Kassemeyer, S., Laksmono, H., Liang, M., Lomb, L., Marchesini, S., Martin, A. V., Messerschmidt, M., Milathianaki, D., Nass, K., Ros, A., Roy-Chowdhury, S., Schmidt, K., Seibert, M., Steinbrener, J., Stellato, F., Yan, L., Yoon, C., Moore, T. A., Moore, A. L., Pushkar, Y., Williams, G. J., Boutet, S., Doak, R. B., Weierstall, U., Frank, M., Chapman, H. N., Spence, J. C. H. & Fromme, P. (2014). Nature, 513, 261–265.
- Lee, D., Baek, S., Park, J., Lee, K., Kim, J., Lee, S. J., Chung, W. K., Lee, J. L., Cho, Y. & Nam, K. H. (2019). Sci. Rep. 9, 6971. [DOI] [PMC free article] [PubMed]
- Lee, K., Lee, D., Baek, S., Park, J., Lee, S. J., Park, S., Chung, W. K., Lee, J.-L., Cho, H.-S., Cho, Y. & Nam, K. H. (2020). J. Appl. Cryst. 53, 1051–1059.
- Liu, W., Wacker, D., Gati, C., Han, G. W., James, D., Wang, D., Nelson, G., Weierstall, U., Katritch, V., Barty, A., Zatsepin, N. A., Li, D., Messerschmidt, M., Boutet, S., Williams, G. J., Koglin, J. E., Seibert, M. M., Wang, C., Shah, S. T. A., Basu, S., Fromme, R., Kupitz, C., Rendek, K. N., Grotjohann, I., Fromme, P., Kirian, R. A., Beyerlein, K. R., White, T. A., Chapman, H. N., Caffrey, M., Spence, J. C. H., Stevens, R. C. & Cherezov, V. (2013). Science, 342, 1521–1524. [DOI] [PMC free article] [PubMed]
- Lobley, C. M. C., Sandy, J., Sanchez-Weatherby, J., Mazzorana, M., Krojer, T., Nowak, R. P. & Sorensen, T. L. (2016). Acta Cryst. D72, 629–640. [DOI] [PMC free article] [PubMed]
- Maia, F. R. N. C. (2012). Nat. Methods, 9, 854–855. [DOI] [PubMed]
- Mehrabi, P., Schulz, E. C., Agthe, M., Horrell, S., Bourenkov, G., von Stetten, D., Leimkohl, J. P., Schikora, H., Schneider, T. R., Pearson, A. R., Tellkamp, F. & Miller, R. J. D. (2019). Nat. Methods, 16, 979–982. [DOI] [PubMed]
- Mehrabi, P., von Stetten, D., Leimkohl, J.-P., Tellkamp, F. & Schulz, E. C. (2021). bioRxiv, 2021.11.07.467596.
- Monteiro, D. C. F., von Stetten, D., Stohrer, C., Sans, M., Pearson, A. R., Santoni, G., van der Linden, P. & Trebbin, M. (2020). IUCrJ, 7, 207–219. [DOI] [PMC free article] [PubMed]
- Monteiro, D. C. F., Vakili, M., Harich, J., Sztucki, M., Meier, S. M., Horrell, S., Josts, I. & Trebbin, M. (2019). J. Synchrotron Rad. 26, 406–412. [DOI] [PubMed]
- Mueller, C., Marx, A., Epp, S. W., Zhong, Y., Kuo, A., Balo, A. R., Soman, J., Schotte, F., Lemke, H. T., Owen, R. L., Pai, E. F., Pearson, A. R., Olson, J. S., Anfinrud, P. A., Ernst, O. P. & Dwayne Miller, R. J. (2015). Struct. Dyn. 2, 054302. [DOI] [PMC free article] [PubMed]
- Nam, K. H. (2019). Int. J. Mol. Sci. 20, 1094.
- Nam, K. H. (2020a). Int. J. Mol. Sci. 21, 3332.
- Nam, K. H. (2020b). Int. J. Mol. Sci. 21, 5977.
- Nam, K. H. (2020c). J. Appl. Cryst. 53, 45–50.
- Nam, K. H. (2020d). Sci. Rep. 10, 107. [DOI] [PMC free article] [PubMed]
- Nam, K. H. (2022). Sci. Rep. 12, 694. [DOI] [PMC free article] [PubMed]
- Nam, K. H. & Cho, Y. (2021). J. Appl. Cryst. 54, 1081–1087.
- Nam, K. H., Kim, J. & Cho, Y. (2021). Sci. Rep. 11, 13115. [DOI] [PMC free article] [PubMed]
- Nass, K., Gorel, A., Abdullah, M. M. V., Martin, A., Kloos, M., Marinelli, A., Aquila, A., Barends, T. R. M., Decker, F.-J., Bruce Doak, R., Foucar, L., Hartmann, E., Hilpert, M., Hunter, M. S., Jurek, Z., Koglin, J. E., Kozlov, A., Lutman, A. A., Kovacs, G. N., Roome, C. M., Shoeman, R. L., Santra, R., Quiney, H. M., Ziaja, B., Boutet, S. & Schlichting, I. (2020). Nat. Commun. 11, 1814. [DOI] [PMC free article] [PubMed]
- Ohmer, C. J., Dasgupta, M., Patwardhan, A., Bogacz, I., Kaminsky, C., Doyle, M. D., Chen, P. Y.-T., Keable, S. M., Makita, H., Simon, P. S., Massad, R., Fransson, T., Chatterjee, R., Bhowmick, A., Paley, D. W., Moriarty, N. W., Brewster, A. S., Gee, L. B., Alonso-Mori, R., Moss, F., Fuller, F. D., Batyuk, A., Sauter, N. K., Bergmann, U., Drennan, C. L., Yachandra, V. K., Yano, J., Kern, J. F. & Ragsdale, S. W. (2022). J. Inorg. Biochem. 230, 111768. [DOI] [PMC free article] [PubMed]
- Pan, D., Oyama, R., Sato, T., Nakane, T., Mizunuma, R., Matsuoka, K., Joti, Y., Tono, K., Nango, E., Iwata, S., Nakatsu, T. & Kato, H. (2022). IUCrJ, 9, 134–145. [DOI] [PMC free article] [PubMed]
- Pandey, S., Bean, R., Sato, T., Poudyal, I., Bielecki, J., Cruz Villarreal, J., Yefanov, O., Mariani, V., White, T. A., Kupitz, C., Hunter, M., Abdellatif, M. H., Bajt, S., Bondar, V., Echelmeier, A., Doppler, D., Emons, M., Frank, M., Fromme, R., Gevorkov, Y., Giovanetti, G., Jiang, M., Kim, D., Kim, Y., Kirkwood, H., Klimovskaia, A., Knoska, J., Koua, F. H. M., Letrun, R., Lisova, S., Maia, L., Mazalova, V., Meza, D., Michelat, T., Ourmazd, A., Palmer, G., Ramilli, M., Schubert, R., Schwander, P., Silenzi, A., Sztuk-Dambietz, J., Tolstikova, A., Chapman, H. N., Ros, A., Barty, A., Fromme, P., Mancuso, A. P. & Schmidt, M. (2020). Nat. Methods, 17, 73–78. [DOI] [PMC free article] [PubMed]
- Park, J., Park, S., Kim, J., Park, G., Cho, Y. & Nam, K. H. (2019). Sci. Rep. 9, 2525. [DOI] [PMC free article] [PubMed]
- Park, S.-Y. & Nam, K. H. (2019). J. Synchrotron Rad. 26, 1815–1819. [DOI] [PubMed]
- Roedig, P., Ginn, H. M., Pakendorf, T., Sutton, G., Harlos, K., Walter, T. S., Meyer, J., Fischer, P., Duman, R., Vartiainen, I., Reime, B., Warmer, M., Brewster, A. S., Young, I. D., Michels-Clark, T., Sauter, N. K., Kotecha, A., Kelly, J., Rowlands, D. J., Sikorsky, M., Nelson, S., Damiani, D. S., Alonso-Mori, R., Ren, J., Fry, E. E., David, C., Stuart, D. I., Wagner, A. & Meents, A. (2017). Nat. Methods, 14, 805–810. [DOI] [PMC free article] [PubMed]
- Sherrell, D. A., Foster, A. J., Hudson, L., Nutter, B., O’Hea, J., Nelson, S., Paré-Labrosse, O., Oghbaey, S., Miller, R. J. D. & Owen, R. L. (2015). J. Synchrotron Rad. 22, 1372–1378. [DOI] [PMC free article] [PubMed]
- Sierra, R. G., Laksmono, H., Kern, J., Tran, R., Hattne, J., Alonso-Mori, R., Lassalle-Kaiser, B., Glöckner, C., Hellmich, J., Schafer, D. W., Echols, N., Gildea, R. J., Grosse-Kunstleve, R. W., Sellberg, J., McQueen, T. A., Fry, A. R., Messerschmidt, M. M., Miahnahri, A., Seibert, M. M., Hampton, C. Y., Starodub, D., Loh, N. D., Sokaras, D., Weng, T.-C., Zwart, P. H., Glatzel, P., Milathianaki, D., White, W. E., Adams, P. D., Williams, G. J., Boutet, S., Zouni, A., Messinger, J., Sauter, N. K., Bergmann, U., Yano, J., Yachandra, V. K. & Bogan, M. J. (2012). Acta Cryst. D68, 1584–1587.
- Skopintsev, P., Ehrenberg, D., Weinert, T., James, D., Kar, R. K., Johnson, P. J. M., Ozerov, D., Furrer, A., Martiel, I., Dworkowski, F., Nass, K., Knopp, G., Cirelli, C., Arrell, C., Gashi, D., Mous, S., Wranik, M., Gruhl, T., Kekilli, D., Brünle, S., Deupi, X., Schertler, G. F. X., Benoit, R. M., Panneels, V., Nogly, P., Schapiro, I., Milne, C., Heberle, J. & Standfuss, J. (2020). Nature, 583, 314–318. [DOI] [PubMed]
- Stellato, F., Oberthür, D., Liang, M., Bean, R., Gati, C., Yefanov, O., Barty, A., Burkhardt, A., Fischer, P., Galli, L., Kirian, R. A., Meyer, J., Panneerselvam, S., Yoon, C. H., Chervinskii, F., Speller, E., White, T. A., Betzel, C., Meents, A. & Chapman, H. N. (2014). IUCrJ, 1, 204–212. [DOI] [PMC free article] [PubMed]
- Sugahara, M., Mizohata, E., Nango, E., Suzuki, M., Tanaka, T., Masuda, T., Tanaka, R., Shimamura, T., Tanaka, Y., Suno, C., Ihara, K., Pan, D., Kakinouchi, K., Sugiyama, S., Murata, M., Inoue, T., Tono, K., Song, C., Park, J., Kameshima, T., Hatsui, T., Joti, Y., Yabashi, M. & Iwata, S. (2015). Nat. Methods, 12, 61–63. [DOI] [PubMed]
- Tenboer, J., Basu, S., Zatsepin, N., Pande, K., Milathianaki, D., Frank, M., Hunter, M., Boutet, S., Williams, G. J., Koglin, J. E., Oberthuer, D., Heymann, M., Kupitz, C., Conrad, C., Coe, J., Roy-Chowdhury, S., Weierstall, U., James, D., Wang, D., Grant, T., Barty, A., Yefanov, O., Scales, J., Gati, C., Seuring, C., Srajer, V., Henning, R., Schwander, P., Fromme, R., Ourmazd, A., Moffat, K., Van Thor, J. J., Spence, J. C., Fromme, P., Chapman, H. N. & Schmidt, M. (2014). Science, 346, 1242–1246. [DOI] [PMC free article] [PubMed]
- Tolstikova, A., Levantino, M., Yefanov, O., Hennicke, V., Fischer, P., Meyer, J., Mozzanica, A., Redford, S., Crosas, E., Opara, N. L., Barthelmess, M., Lieske, J., Oberthuer, D., Wator, E., Mohacsi, I., Wulff, M., Schmitt, B., Chapman, H. N. & Meents, A. (2019). IUCrJ, 6, 927–937. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vakili, M., Vasireddi, R., Gwozdz, P. V., Monteiro, D. C. F., Heymann, M., Blick, R. H. & Trebbin, M. (2020). Rev. Sci. Instrum. 91, 085108. [DOI] [PubMed]
- Weierstall, U., James, D., Wang, C., White, T. A., Wang, D., Liu, W., Spence, J. C., Bruce Doak, R., Nelson, G., Fromme, P., Fromme, R., Grotjohann, I., Kupitz, C., Zatsepin, N. A., Liu, H., Basu, S., Wacker, D., Won Han, G., Katritch, V., Boutet, S., Messerschmidt, M., Williams, G. J., Koglin, J. E., Marvin Seibert, M., Klinker, M., Gati, C., Shoeman, R. L., Barty, A., Chapman, H. N., Kirian, R. A., Beyerlein, K. R., Stevens, R. C., Li, D., Shah, S. T., Howe, N., Caffrey, M. & Cherezov, V. (2014). Nat. Commun. 5, 3309. [DOI] [PMC free article] [PubMed]
- White, T. A., Mariani, V., Brehm, W., Yefanov, O., Barty, A., Beyerlein, K. R., Chervinskii, F., Galli, L., Gati, C., Nakane, T., Tolstikova, A., Yamashita, K., Yoon, C. H., Diederichs, K. & Chapman, H. N. (2016). J. Appl. Cryst. 49, 680–689. [DOI] [PMC free article] [PubMed]
- Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. & Richardson, J. S. (2018). Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup1.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup2.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup3.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup4.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup5.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup6.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup7.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup8.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup9.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup10.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup11.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup12.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup13.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup14.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup15.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup16.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup17.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup18.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup19.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup20.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup21.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup22.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup23.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup24.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup25.mp4
Supporting information file. DOI: 10.1107/S1600576722005556/te5095sup26.mp4
Supporting figures and BITS code. DOI: 10.1107/S1600576722005556/te5095sup27.pdf
PDB reference: 7wkr
PDB reference: 7wuc