Abstract
INTRODUCTION
The subcellular compartmentalization of eukaryotic cells requires selective transport of folded proteins and protein/nucleic acid complexes. Embedded in nuclear envelope pores, generated by the circumscribed fusion of the inner and outer nuclear membranes, nuclear pore complexes (NPCs) are the sole bidirectional gateways for nucleocytoplasmic transport. The ~110 MDa human NPC is an ~1,000 protein assembly comprising multiple copies of ~34 different proteins, collectively termed nucleoporins. The symmetric core of the NPC is composed of an inner ring encircling the central transport channel, and outer rings formed by Y-shaped coat nucleoporin complexes (CNCs) anchored atop both sides of the nuclear envelope. The outer rings are decorated with compartment-specific asymmetric nuclear basket and cytoplasmic filament nucleoporins, which establish transport directionality and provide docking sites for transport factors and the small GTPase Ran. The cytoplasmic filament nucleoporins also play an essential role in the irreversible remodeling of messenger ribonucleoprotein particles (mRNPs) as they exit the central transport channel. Unsurprisingly, the NPC’s cytoplasmic face represents a hotspot for disease-associated mutations and is commonly targeted by viral virulence factors.
RATIONALE
Previous studies established a near-atomic composite structure of the human NPC’s symmetric core by combining biochemical reconstitution elucidating the interaction network between symmetric nucleoporins, crystal and single particle cryo-EM structure determination of nucleoporins and nucleoporin complexes revealing their three-dimensional shape and the molecular details of their interactions, quantitative docking in cryo-ET maps of the intact human NPC uncovering nucleoporin stoichiometry and positioning, and cell-based assays validating the physiological relevance of the biochemical and structural findings. Here, we extended our approach to the cytoplasmic filament nucleoporins to elucidate the near-atomic architecture of the cytoplasmic face of the human NPC.
RESULTS
Using biochemical reconstitution, we elucidated the protein-protein and protein-RNA interaction networks of the human and C. thermophilum cytoplasmic filament nucleoporins, establishing an evolutionarily conserved hetero-hexameric cytoplasmic filament nucleoporin complex (CFNC) held together by a central hetero-trimeric coiled-coil hub that tethers two separate mRNP-remodeling complexes. Further biochemical analysis and determination of a series of crystal structures revealed that the metazoan-specific cytoplasmic filament nucleoporin NUP358 is composed of 16 distinct domains, including an N-terminal S-shaped α-helical solenoid followed by a coiled-coil oligomerization element, numerous Ran-interacting domains, an E3 ligase domain, and a C-terminal prolyl-isomerase domain. Physiologically validated quantitative docking into cryo-ET maps of the intact human NPC revealed that pentameric NUP358 bundles, conjoined by the oligomerization element, are anchored through their N-terminal domains to the central stalk regions of the CNC, projecting flexibly attached domains as far as ~600Å into the cytoplasm. Using cell-based assays, we demonstrated that NUP358 is dispensable for the architectural integrity of the assembled interphase NPC and RNA export but is required for efficient translation. After NUP358 assignment, the remaining 4-shaped cryo-ET density matched the dimensions of the CFNC coiled-coil hub, in close proximity to an outer ring NUP93. Whereas the N-terminal NUP93 assembly sensor motif anchors the properly assembled related coiled-coil channel nucleoporin heterotrimer to the inner ring, biochemical reconstitution confirmed the NUP93 assembly sensor is reused in anchoring the CFNC to the cytoplasmic face of the human NPC. In contrast, two C. thermophilum CFNCs are anchored by a divergent mechanism involving assembly sensors located in unstructured portions of two CNC nucleoporins. Whereas unassigned cryo-ET density occupies the NUP358 and CFNC binding sites on the nuclear face, docking of the nuclear basket component ELYS established that the equivalent position on the cytoplasmic face is unoccupied, suggesting that mechanisms other than steric competition promote asymmetric distribution of nucleoporins.
CONCLUSION
We have substantially advanced the biochemical and structural characterization of the asymmetric nucleoporins’ architecture and attachment at the cytoplasmic and nuclear faces of the NPC. Our near-atomic composite structure of the human NPC’s cytoplasmic face provides a biochemical and structural framework for elucidating the molecular basis of mRNP remodeling, viral virulence factor interference with NPC function, and the underlying mechanisms of nucleoporin diseases at the cytoplasmic face of the NPC.
The nuclear pore complex (NPC) is the sole bidirectional gateway for nucleocytoplasmic transport. Despite recent progress in elucidating the NPC symmetric core architecture, the asymmetrically decorated cytoplasmic face, essential for mRNA export and a hotspot for nucleoporin-associated diseases, has remained elusive. Here, we report a composite structure of the human cytoplasmic face obtained by combining biochemical reconstitution, crystal structure determination, docking into cryo-electron tomographic reconstructions, and physiological validation. Whereas species-specific motifs anchor an evolutionarily conserved ~540kDa hetero-hexameric cytoplasmic filament nucleoporin complex above the central transport channel, attachment of the NUP358 pentameric bundles depends on the double-ring arrangement of the coat nucleoporin complex. Our composite structure and its predictive power provide a rich foundation for elucidating the molecular basis of mRNA export and nucleoporin diseases.
One-Sentence Summary:
An interdisciplinary analysis established the near-atomic molecular architecture of the cytoplasmic face of the human nuclear pore complex.
The sequestration of genetic material in the nucleus represents one of the great hallmarks of evolution but creates the necessity for selective bidirectional transport across the nuclear envelope (1–4). The nuclear pore complex (NPC) is the sole gateway through which folded proteins and protein/nucleic acid complexes cross the nuclear envelope, making this transport organelle an essential machine for all eukaryotic life. Besides its direct role as a transport channel, the NPC serves as an organizer for nuclear and cytoplasmic processes that are essential for the flow of genetic information from DNA to RNA to protein, including transcription, spliceosome assembly, mRNA export, and ribosome assembly (1–4). Dysfunction of the NPC or its components represents a major cause of human disease (2, 5, 6).
Architecturally, the NPC consists of a central core with an 8-fold rotational symmetry across a nucleocytoplasmic axis and a two-fold rotational symmetry across the plane of the nuclear envelope, which links to compartment-specific asymmetric “cytoplasmic filaments” (CF) and a “nuclear basket” structure (Fig.1A) (1, 2). The NPC is built from ~34 different proteins, termed nucleoporins (nups) that are organized into distinct subcomplexes. Multiple copies of each nup in the NPC add up to an assembly that reaches an extraordinary molecular mass of ~110MDa in vertebrates. The symmetric core of the NPC is composed of an inner ring and two spatially segregated outer rings. The inner ring is embedded in nuclear envelope pores generated by the circumscribed fusion of the double membrane of the nuclear envelope. The diffusion barrier is formed by unstructured phenylalanine-glycine (FG) repeats that fill the central transport channel, imposing a gradually increasing barrier to passive diffusion of macromolecules >40kDa (1–4). Transport factors collectively termed karyopherins overcome the diffusion barrier by binding to FG repeats, thereby transporting cargo across the nuclear envelope (7–9). A significant fraction of the FG repeats in the inner ring is contributed by a hetero-trimeric channel nup complex (CNT) which is anchored by a single assembly sensor motif (10–12). The outer rings sit atop the nuclear envelope, sandwiching the inner ring from both sides. They are primarily formed by the Y-shaped coat nup complex (CNC, also referred to as Y-complex or Nup107–160 complex), and serve as a platform for the asymmetric incorporation of the CF and nuclear basket nups.
Fig. 1. Reconstitution of a 16-protein C.thermophilum coat-cytoplasmic filament nup complex.
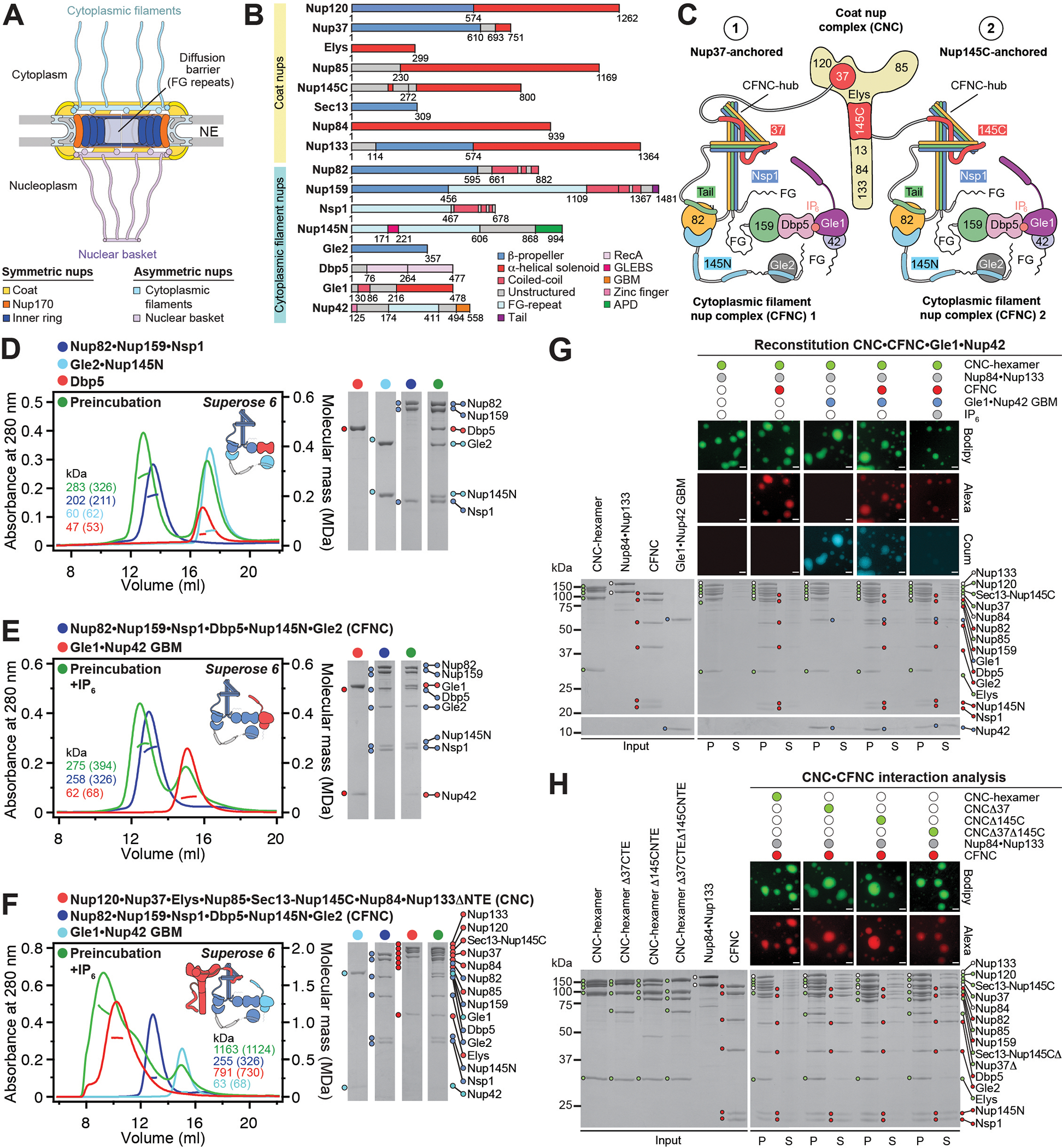
(A) Cross sectional schematic of the fungal NPC architecture. (B) Domain structures of the coat and cytoplasmic filament nups. (C) Schematic representation summarizing our biochemical reconstitution and dissection experiments with purified recombinant C.thermophilum nups, illustrating the cytoplasmic filament nup complex (CFNC) architecture and its attachment to the coat nup complex (CNC). The CNC harbors two assembly sensors, Nup37CTE and Nup145CNTE, each anchoring a CFNC via its central hub, with Nup37CTE exhibiting tighter binding than Nup145CNTE. (D-F) SEC-MALS interaction analyses, showing the stepwise biochemical reconstitution starting with (D) the CFNC (green) from Nup82•Nup159•Nsp1 (blue), Gle2•Nup145N (cyan), and Dbp5 (red), (E) CFNC•Gle1•Nup42GBM (green) from CFNC (blue) and Gle1•Nup42GBM (red), and culminating with (F) the 16-protein CNC•CFNC•Gle1•Nup42GBM complex (green) from CNC (red), CFNC (blue), and Gle1•Nup42GBM (cyan). SDS-PAGE gel strips of peak fractions are shown. Measured molecular masses are indicated, with respective theoretical masses in parentheses. (G, H) Liquid-liquid phase separation (LLPS) interaction assays, assessing (G) CFNC (red) and Gle1•Nup42GBM (cyan) incorporation into CNC-LLPS (green), and (H) CFNC incorporation into CNC-LLPS, lacking either one or both Nup37CTE and Nup145CNTE assembly sensors. N-terminally fluorescently labeled CNC (Bodipy), CFNC (Alexa Fluor 647), and Gle1•Nup42GBM (Coumarin) were visualized by fluorescence microscopy. Pelleted CNC condensate phase (P) and soluble (S) fractions were analyzed by SDS-PAGE and visualized by Coomassie brilliant blue staining. Scale bars are 10μm.
Two decades ago, the atomic level characterization of the NPC began with individual nup domains and progressed to nup complexes of increasing size and complexity, culminating in the ~400kDa hetero-heptameric CNC (11, 13–28). Simultaneously, advances in cryo-electron tomographic (cryo-ET) data acquisition and processing gradually increased the resolution of intact NPC 3D reconstructions (29). Docking of the CNC into a ~32Å cryo-ET map of the intact human NPC demonstrated that two reticulated eight-membered CNC rings, linked by head-to-tail interactions, are present on each side of the nuclear envelope (27, 30). Moreover, this advance established that the resolution gap between high- and low-resolution structural methods can be overcome by combining biochemical reconstitution and X-ray crystallographic characterization of nups with cryo-ET reconstruction of the intact NPC. Expansion of this approach to the nine nups constituting the inner ring rapidly led to the reconstitution of two distinct ~425kDa inner ring complexes (IRCs) and the elucidation of their components’ structures (10–12, 20, 31–38). In turn, this advance enabled the determination of the near-atomic composite structure of the entire ~56MDa symmetric core of the human NPC, establishing the stoichiometry and placement of all 17 symmetric nups within a ~23Å cryo-ET reconstruction (38, 39). Subsequently, the architecture of the Saccharomyces cerevisiae NPC was determined with a similar approach, utilizing high-resolution nup crystal structures and ~25Å cryo-ET maps of either detergent purified or in-situ NPCs (40, 41). Compared to the human NPC, the S.cerevisiae NPC lacks the distal CNC ring and associated nups on both sides of the nuclear envelope, but the relative nup arrangement within the rest of the symmetric core remains essentially identical (38, 39, 42).
Projecting from the cytoplasmic face of the NPC, the CF nups recruit cargo•transport factor complexes for nucleocytoplasmic transport and orchestrate the export and remodeling of messenger ribonucleoprotein particles (mRNPs) in preparation for translation (2, 43). The nine-component CF nup machinery represents a hotspot for human diseases ranging from degenerative brain disorders and cardiac diseases to cancer (2, 5, 6). Although linked to the human CF nups NUP358, NUP214, NUP62, NUP88, NUP98, GLE1, NUP42, RAE1, and DDX19, the pathophysiology and optimal therapeutic strategies for these conditions remain ill-defined.
Here, we present insight into the atomic and higher order architecture, function, and mechanism of action of the CF nups in the human and thermophilic fungus Chaetomium thermophilum NPCs. First, we uncover a conserved modular architecture within the hetero-hexameric CF nup complex (CFNC) of both species: Holding the CFNC together is a coiled-coil hub built like the CNT, but formed by NUP62 with the C-terminal regions of NUP88 and NUP214, while their N-terminal β-propeller domains link to the mRNA export factors NUP98•RAE1 and the DEAD-box RNA helicase DDX19, respectively, which in turn recruit the remaining complex components. We further uncover evolutionary divergent mechanisms for the attachment of the intact CFNC at the cytoplasmic face of the NPC, which in C.thermophilum involves two distinct assembly sensors in the CNC that do not exist in humans. We assemble the C.thermophilum CNC and CF nups into a ~1.1MDa 16-protein complex and find that it can be remodeled by inositol hexaphosphate (IP6). Towards dissecting the molecular mechanism of mRNA export, we systematically characterize the propensity of CF nups for RNA binding and find novel capabilities in two CFNC subcomplexes GLE1•NUP42 and NUP88•NUP214•NUP98 as well as different parts of the metazoan-specific NUP358. To build a composite structure of the human NPC cytoplasmic face, we determine crystal structures of the NUP88NTD•NUP98APD complex and all remaining structurally uncharacterized regions of NUP358, uncovering a hereto unobserved, S-shaped fold of three α-helical solenoids of the NUP358 N-terminal domain as well as a complex mechanism for NUP358 oligomerization. Docking of the novel structures along with previously characterized CF nups into a previously reported ~23Å and an ~12Å cryo-ET map of the intact human NPC (provided by the Beck group), as well as an ~8Å region of an anisotropic single particle cryo-EM composite map of the Xenopus laevis cytoplasmic NPC face accounts for all of the asymmetric density on the cytoplasmic NPC-side resolved in the maps (44–46). Validating our quantitative docking analysis in human cells engineered to enable rapid, inducible NUP358 depletion, we surprisingly find NUP358 to be dispensable for the architectural integrity of the assembled interphase NPC and mRNA export but having a general role in translation. The docking of the CFNC-hub in close proximity to a NUP93 fragment that, in the inner ring, acts as the assembly sensor for the CNT, allows us to predict and experimentally confirm that NUP93 also recruits the structurally related CFNC on the cytoplasmic face, thereby enabling identification of the elusive human CFNC NPC anchor. Thus, our near-atomic composite structure possesses predictive power, demonstrating its general utility for the mechanistic dissection of essential cellular events occurring on the cytoplasmic face of the human NPC.
RESULTS
Modular architecture of the evolutionarily conserved 6-protein cytoplasmic filament nup complex
Although pair-wise interactions between selected CF nups had previously been reported, comprehensive knowledge on the entire CF nup interaction network has remained unavailable (11, 47–64). Utilizing nups from the thermophilic fungus C.thermophilum, which exhibit superior biochemical stability, we previously elucidated the interaction network of the seventeen symmetric core nups (38). Therefore, we first sought to establish the protein-protein interaction network and complex stoichiometry of the eight evolutionarily conserved C.thermophilum CF nups Nup159, Nup82, Nsp1, Nup145N, Gle2, Dbp5, Gle1, and Nup42 (Fig.1B and fig.S1) (2). Most CF nups contain both structured and unstructured regions that can harbor multiple distinct binding sites and FG-repeats. We established expression and purification protocols for the C.thermophilum CF nups, omitting FG-repeat regions, as well as an unstructured linker region in Nup145N to improve solubility, and analyzed their binding by size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS), and a liquid-liquid phase separation (LLPS) interaction assay (Fig.1, figs.S1 to S26, and tables S1 to S6). For a detailed description of these experiments, see Supplementary Text.
Mixture of Nup82•Nup159•Nsp1 with Gle2•Nup145N and Dbp5 results in the formation of a stoichiometric hetero-hexameric CFNC (Fig.1, C and D and fig.S4A), which is held together by a parallel coiled-coil hetero-trimer formed by the C-terminal domains of Nup82•Nup159•Nsp1, termed the CFNC-hub (figs.S4 to S6). The CFNC is tethered to the NPC by two mutually exclusive assembly sensors targeting the CFNC-hub. These anchor points are located within primarily unstructured regions (NTEs, CTEs) present in the CNC constituents Nup37CTE and Nup145CNTE, which supply a strong and weak binding site, respectively, permitting two CFNCs to bind a single CNC (figs.S7 to S17). The Gle1•Nup42 complex has also been shown to locate at the cytoplasmic face of the NPC, forming an IP6 dependent interaction with Dbp5 (53, 58, 63, 65). We demonstrate stoichiometric incorporation of Gle1•Nup42 into both the CFNC and CNC•CFNC complexes in the presence of IP6 (Fig.1, E and F, and figs.S18 to 23). Additionally, we identify an interaction formed between Gle1•Nup42 and the CNC that is disrupted upon addition of IP6, establishing that the CNC-CF nup interaction network can be remodeled (figs.S23 to S25).
Given the special importance of the CF nups in human disease, we next tested whether the molecular architecture of the CFNC is evolutionarily conserved from C.thermophilum to humans. The human CFNC is comprised of NUP88, NUP214, NUP62, NUP98, RAE1, and DDX19. Apart from a rearrangement of the FG-repeat and coiled-coil regions in NUP214, the domain organization of the human CFNC nups is identical to that of the C.thermophilum orthologs (Fig.2 and fig.S27). Indeed, mixing the NUP88•NUP214•NUP62 hetero-trimer with RAE1•NUP98 and DDX19 resulted in a stoichiometric H.sapiens CFNC hetero-hexamer (Fig.2C and figs.S28 and S29). Similarly, a systematic pairwise interaction analysis, established that the modular CFNC architecture characterized in C.thermophilum is conserved in humans (Fig.2D and E, and figs.S30 to S39).
Fig. 2. Conserved modular architecture and RNA binding properties of the human CF nups.
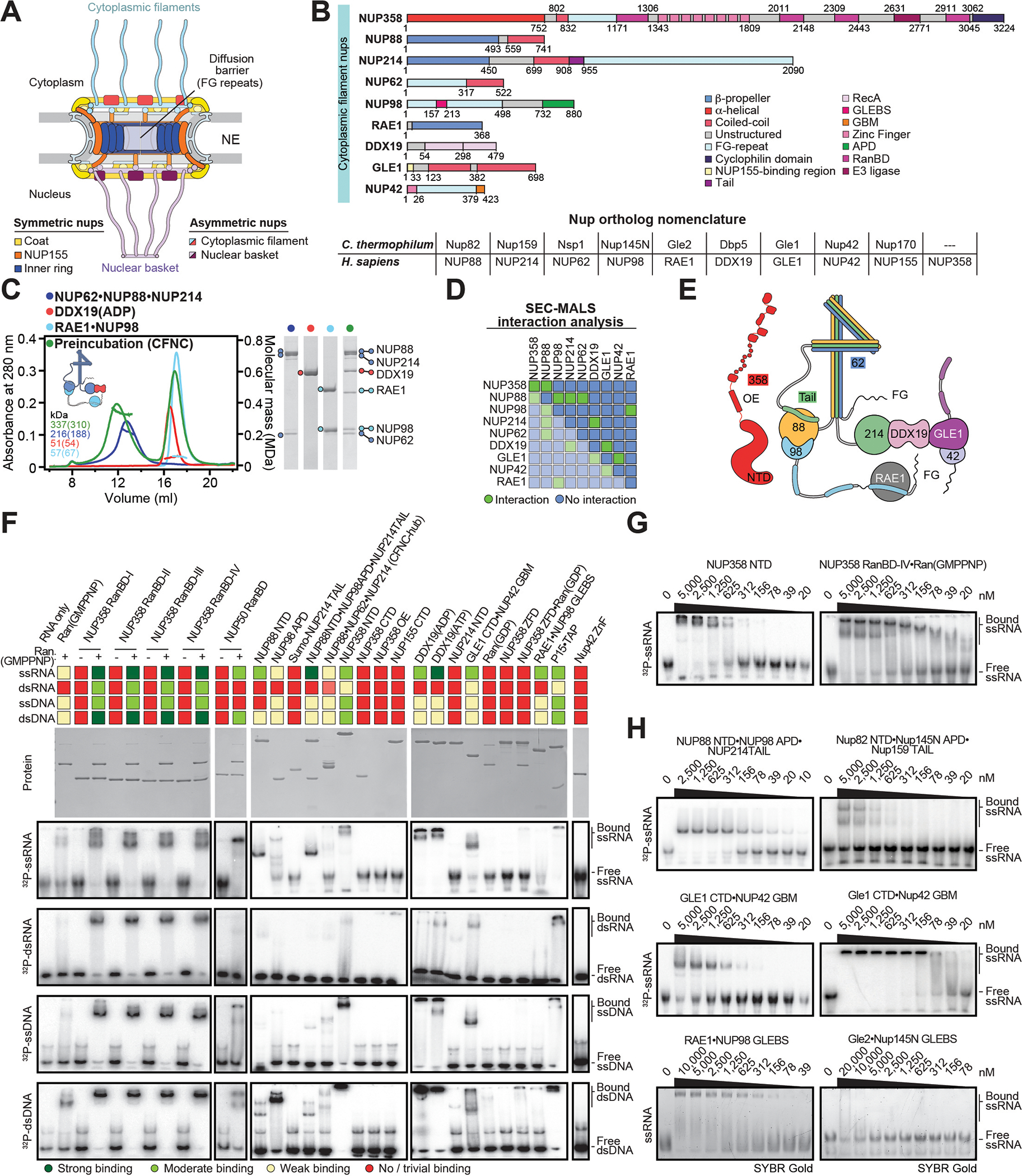
(A) Cross sectional schematic of the human NPC architecture. (B) Domain structures of human cytoplasmic filament nups. Nomenclature of H.sapiens and C.thermophilum nup orthologs is indicated. (C) Biochemical reconstitution of the ~310kDa hetero-hexameric human CFNC. SEC-MALS interaction analysis of NUP88•NUP214•NUP62 (blue), DDX19 (ADP) (red), RAE1•NUP98 (cyan), and their preincubation (green). Measured molecular masses are indicated, with theoretical masses in parentheses. SDS-PAGE gel strips of peak fractions visualized by Coomassie brilliant blue staining are shown. (D) Summary of pairwise SEC-MALS interaction analyses between human cytoplasmic filament nups. (E) Schematic summary of the human CFNC architecture and the cytoplasmic filament nup interaction network. (F) Human cytoplasmic filament nup domains and complexes were assayed for binding to single-stranded (ss) and double-stranded (ds) RNA and DNA probes by electrophoretic mobility shift assay (EMSA). Input proteins resolved by SDS-PAGE were visualized by Coomassie brilliant blue staining. Qualitative assessment of nucleic acid binding is denoted by color-coded boxes. (G, H) EMSAs with ssRNA titrated against (G) metazoan-specific NUP358NTD and NUP358RanBD-IV•Ran(GMPPNP), and (H) the indicated H.sapiens CFNC subcomplexes and their C.thermophilum orthologs.
Together, our data establish that the CF nups form an evolutionarily conserved six-protein complex that is held together by an extensive parallel coiled-coil hub generated by the C-terminal regions of Nup82/NUP88, Nup159/NUP214 and Nsp1/NUP62, which shares architectural similarities with the heterotrimeric Nsp1/NUP62•Nup49/NUP58•Nup57/NUP54 CNT (11). The Nup82/NUP88 N-terminal β-propeller domain is attached by an interaction between the C-terminal α-helical TAIL fragment of Nup159/NUP214 and provides a binding site for the Nup145N/NUP98 APD, which in turn recruits Gle2/RAE1 to the NPC. Analogously, the Nup159/NUP214 N-terminal β-propeller domain provides a binding site for the DEAD-box helicase Dbp5/DDX19. In C.thermophilum, the CFNC-hub is anchored to the CNC by two distinct assembly sensors in Nup37CTE and Nup145CNTE, similar to the anchoring of the CNT by the Nic96R1/NUP93R1 assembly sensor in the inner ring. In contrast, the human CNC lacks comparable assembly sensor motifs, suggesting alternative mechanisms for anchoring CF nups at the cytoplasmic face of the human NPC.
RNA interactions of human cytoplasmic filament nups
Given their essential roles in mRNA export, we next sought to identify which of the human CF nups possessed RNA-binding capabilities (50, 66–69). Previous work utilizing disparate methods and diverse but inconsistent probes had established DDX19 and RAE1•NUP98GLEBS binding to U10 single-stranded ssRNA, degenerate ssRNA, poly(A), poly(C), poly(G) RNA, as well as ssDNA and double-stranded dsDNA across a variety of assays (50, 56, 57, 63, 70).
Taking advantage of our complete set of purified human CF nup domains and sub-complexes, we carried out a comprehensive electrophoretic mobility shift assay (EMSA) screen to systematically assess binding against a consistent set of ss/dsRNA probes (Fig.2F). In addition to the established RNA binders DDX19 and RAE1•NUP98, we identified ssRNA binding by GLE1CTD•NUP42GBM and NUP88NTD, an activity enhanced in the context of the NUP88NTD•NUP98APD•NUP214TAIL complex. We tested their C.thermophilum orthologs and found these RNA binding activities to be conserved (Fig.2, G and H, and fig.S40). Next, we analyzed the metazoan-specific NUP358. We detected moderate RNA binding for NUP358 N-terminal domain (NTD) (Fig.2F and fig.S40G). Additionally, we also found that the four NUP358RanBD•Ran(GMPPNP) complexes preferentially bound ssRNA (Fig.2F and fig.S40E). Additional details of RNA binding can be found in Supplementary Text. Future studies need to delineate whether these RNA binding sites present sequence-specific RNA affinity, and what the implications of such specificity would be in the overall mRNA export pathway.
Structural and biochemical analyses of NUP358
NUP358 is a 3,224-residue metazoan-specific CF component and the largest constituent of the NPC (71–74). Previous studies established that its N-terminal ~900-residue α-helical region is necessary for nuclear envelope recruitment (75). Within this region, the first 145 residues have been biochemically and structurally characterized, shown to form three tetratricopeptide repeats (TPR) (76). Guided by secondary structure predictions, we systematically screened expression constructs for solubility, identifying three fragments: NUP358NTDΔTPR (residues 145–752), NUP358NTD (residues 1–752), and an extended region spanning residues 1–832 (NUP3581–832). Subsequent purifications revealed that the NUP358NTD and NUP3581–832 fragments behave differently, with the latter forming amorphous precipitates in buffer NaCl concentrations below 300mM. Therefore, we characterized these NUP358 fragments in both high salt (350mM NaCl) and low salt (100mM NaCl) buffers, wherever possible.
NUP358NTD exhibited concentration-dependent homo-dimerization in low salt buffer, with measured molecular masses between values corresponding to monomeric and dimeric species but existed as a monomeric species in high salt buffer (Fig.3, A and B and fig.S41). Conversely, NUP358NTDΔTPR was exclusively monomeric, suggesting that the TPR mediates homo-dimerization (Fig.3B and fig.S41). Furthermore, the extended NUP3581–832 fragment forms oligomers with measured molecular masses between a tetramer and a pentamer (Fig.3C and fig.S41). Subsequent C-terminal mapping revealed an oligomerization element (OE) between residues 802–832, forming salt-insensitive concentration-dependent oligomers between dimers and tetramers (Fig.3G and fig.S42). Thus, NUP358 oligomerization is mediated by the TPR and OE regions, located on opposite sides of the N-terminal α-helical region.
Fig. 3. Structural analysis and biochemical characterization of NUP358.
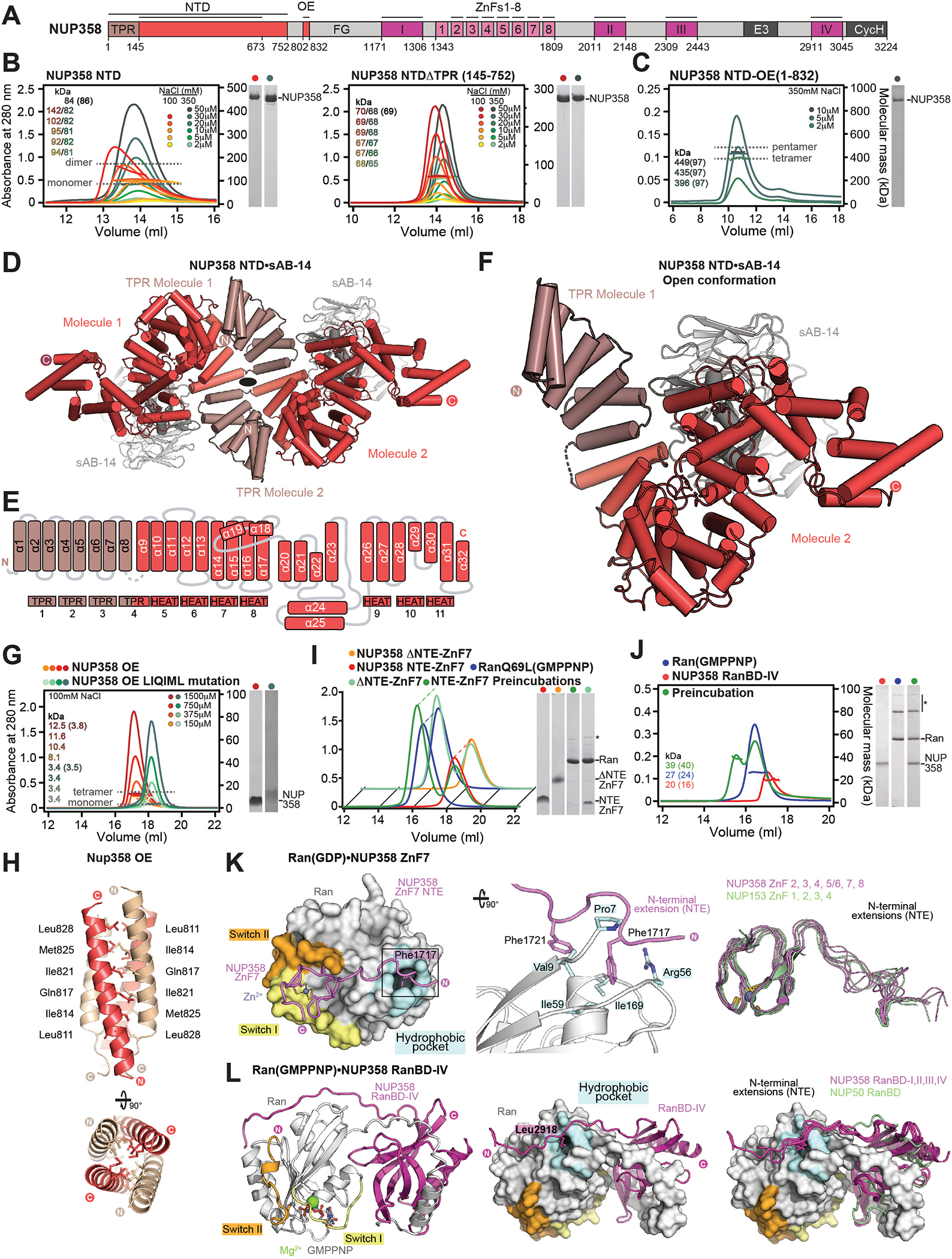
(A) Domain structure of NUP358. Black lines indicate the boundaries of the crystallized fragments. (B, C) SEC-MALS analysis of the oligomeric behavior of (B) NUP358NTD and NUP358NTDΔTPR, and (C) NUP358NTD-OE, performed at the indicated protein concentrations. Measured molecular masses are indicated, with theoretical masses in parentheses. SDS-PAGE gel strips of peak fractions are shown and visualized by Coomassie brilliant blue staining. (D) Cartoon representation of the NUP358NTD•sAB-14 co-crystal structure dyad of the P6522 lattice, illustrating the NUP358NTD dimer between symmetry-related molecules. (E) Schematic of the NUP358NTD structure and structural motifs. (F) TPR of molecule 1 complements α-helical stacking of the N-terminal solenoid of the symmetry-related molecule 2, generating the open conformation of NUP358NTD. (G) SEC-MALS analysis of the oligomeric behavior of NUP358OE and the NUP358OE LIQIML mutant performed at the indicated protein concentrations. (H) Cartoon representation of the homo-tetrameric NUP358OE crystal structure with hydrophobic core residues shown in ball-and-stick representation. (I) SEC interaction analysis of NUP358ZnF7 and NUP358ZnF7ΔNTE binding to Ran(GTP). (J) SEC-MALS interaction analysis of NUP358RanBD-IV binding to Ran(GMPPNP). (K) Co-crystal structure of NUP358ZnF7•Ran(GDP), shown in cartoon and surface representation (left). The inset indicates the location of the magnified and 90° rotated view of the Ran hydrophobic pocket (middle). Superposition of the six NUP358ZnF•Ran(GDP) and four NUP153ZnF•Ran(GDP) co-crystal structures with the Zn2+-coordinating cysteines and Ran-burying NTE hydrophobic residues shown as sticks (right). (L) Co-crystal structure of NUP358RanBD-IV•Ran(GMPPNP) with NUP358RanBD-IV shown in cartoon and Ran(GMPPNP) shown in cartoon (left) or surface (middle) representation. Superposition of Ran(GMPPNP) bound to NUP358RanBD-I, NUP358RanBD-II, NUP358RanBD-III, NUP358RanBD-IV, and NUP50RanBD (right).
To aid the crystallization of the entire NUP358NTD, we generated high-affinity, synthetic antibody fragments (sABs) by phage display selection (77). By systematically screening the generated 62 sABs as crystallization chaperones, we identified a NUP358NTD•sAB-14 complex that crystallized, enabling de novo structure determination of the entire NUP358NTD at 3.95Å resolution (tables S7 to S10). To unambiguously assign the Nup358NTD sequence register, we crystallized 17 Seleno-L-methionine mutants (fig.S43 and tables S11 and S12).
The asymmetric unit contained two copies of the NUP358NTD•sAB-14 complex, in one of which the first three and a half TPR repeats are not resolved. The second copy forms extensive interactions with a symmetry related molecule (Fig.3, D to F, and fig.S44). This NUP358NTD dimer reveals two alternative TPR conformations, in which the TPR either forms a continuous N-terminal solenoid (open), or folds back, separating TPR4 and forming electrostatic interactions with HEAT repeats 5–7 of the N-terminal solenoid (closed) (Fig.3F, fig.S44C, and Movie 1). Toggling between these two states provides a molecular explanation for the salt-sensitive, concentration-dependent, dimerization behavior of NUP358NTD (Fig.3B). Because the open confirmation is the one identified in the intact NPC (see below), we focus our description on this state.
Movie 1. Structure of NUP358NTD.
A 360° rotation of the NUP358NTD•sAB-14 cocrystal structure, illustrating the NUP358NTD dimer between symmetry-related molecules, followed by a comparison of the two possible TPR conformations giving rise to the open and closed states, concluding with a 360° rotation of the NUP358NTD open confirmation.
The open conformation of NUP358NTD can be divided in three sections: an N-terminal α-helical solenoid composed of four TPRs and four HEAT repeats, a central α-helical wedge domain, and a short C-terminal α-helical solenoid formed by three HEAT repeats (fig.S44D). The N- and C-terminal TPR and HEAT repeats are capped by solvating helices. Inserted between α17 of the N-terminal solenoid and α20 of the wedge domain is a ~50-residue loop that wraps around the convex face of the N-terminal solenoid. The N-terminal solenoid and wedge domain form a composite concave surface with a striking overall positive charge (figs.S45 and S46). The central wedge domain makes extensive hydrophobic contacts with the sides of the N- and C-terminal solenoids, generating a non-canonical S-shaped architecture (fig.S44D). Indeed, a Dali 3D search of the Protein Data Bank revealed that the NUP358NTD architecture is novel (78).
Our biochemical analysis revealed that NUP358NTD interacts weakly with NUP88NTD and possesses RNA-binding activity, both of which were salt-sensitive (figs.S47 and S48). By splitting NUP358NTD into two fragments, NUP358TPR and NUP358NTDΔTPR, we show that both halves are necessary, yet insufficient, for either NUP88NTD or RNA binding (figs.S47 and S48). To further map these interactions, we performed a saturating NUP358NTD surface mutagenesis, screening 106 mutants for NUP88NTD and RNA binding (fig.S49). We found that positively charged residues in the concave surface mediate binding to both NUP88NTD and RNA. Mutations that abolished NUP88NTD-binding clustered exclusively on the N-terminal solenoid, whereas RNA disruption required additional mutations in the wedge domain. By systematically combining individual alanine substitutions we identified a NUP358NTD 2R5K mutation, which abolished both interactions (fig.S50).
Next, we determined the crystal structure of NUP358OE at 1.1Å resolution (table S13). NUP358OE is a small α-helical element that homo-tetramerizes to form an anti-parallel bundle (Fig.3, G and H, and fig.S42A). The core of the α-helical bundle is lined with hydrophobic residues that coordinate oligomeric inter-helical packing, demonstrated by the monomeric form assumed by the NUP358OE LIQIML mutant (Fig.3G, fig.S42, B to E, and Movie 2). To validate our NUP358OE structure, we tested the effect of introducing the LIQIML mutation into the larger NUP358NTD-OE. Whereas wildtype NUP358NTD-OE formed higher-order oligomeric species, the oligomerization profile of the LIQIML NUP358NTD-OE mutant matched that of the OE-less NUP358NTD, presenting concentration-dependent dimerization in low salt buffer but persisting in a monodisperse monomeric state in high salt buffer (fig.S42, F and G).
Movie 2. Structure of NUP358OE.
A 360° rotation of the of the homotetrameric NUP358OE crystal structure, with hydrophobic core residues shown in ball-and-stick representation followed by an end-on view.
Our data show that NUP358NTD is composed of three distinct α-helical solenoids that interact in a novel manner, adopting a unique overall S-shaped architecture with a propensity to form domain-swapped homo-dimers. Connected to NUP358NTD by a ~50-residue linker is an oligomerization element that forms homo-tetramers/pentamers in solution. These dual modes of homo-oligomerization provide a plausible explanation for NUP358’s propensity to form phase separation, as observed during NPC assembly in Drosophila melanogaster oocytes (79).
Ran interactions with human asymmetric nups
Nucleocytoplasmic transport depends on Karyopherin transport factors (Kaps) with directionality imposed by a cellular gradient of the small GTPase Ran, nuclear Ran(GTP) is ~200 fold elevated compared with the primarily cytoplasmic Ran(GDP) (2, 7, 9). Multiple Ran-binding sites are distributed among the asymmetric nups at the cytoplasmic and nuclear sides of the NPC in the form of distinct Ran-binding domains (RanBDs) and Zn2+-finger (ZnF) modules. On the cytoplasmic face, NUP358 contains four dispersed RanBDs and a central zinc finger domain (ZFD) with a tandem array of eight ZnFs (Fig.3A) (80). On the nuclear side, NUP153 and NUP50 contain a central ZFD with four ZnFs and a solitary C-terminal RanBD, respectively (fig.S51) (81, 82).
Testing the Ran(GDP/GTP)-binding activity of all 17 domains by SEC-MALS, we confirmed that all domains bound to Ran, as expected, except for NUP358ZnF1 (Fig.3, I and J, and figs.S51 to S55). Consistent with previous reports, the RanBDs of NUP358 and NUP50 only bound Ran(GTP), whereas the ZnFs in NUP358 and NUP153 bound Ran in both nucleotide states but showed a preference for Ran(GDP) (figs.S52 and S54) (80, 83–85). To clarify the molecular basis for the differential binding behaviors, we determined the co-crystal structures of all 16 domains bound to Ran in their preferred nucleotide-bound state at 1.8Å-2.45Å resolutions (figs.S51 and S55, Movies 3 and 4, and tables S14 to S16). For expanded description of these structures, see Supplementary Text.
Movie 3. Comparison of NUP358 and Nup153 ZnF Ran(GDP) complexes.
Crystal structures of the six NUP358 ZnF•Ran(GDP) and four NUP153 ZnF•Ran(GDP) cocrystal structures are shown individually followed by a superposition. A 360° rotation of the NUP358 and NUP153 ZnF superposition and Ran(GDP) is provided, with a zoom-in view showing hydrophobic residues of the ZnF N-terminal extension buried in the Ran hydrophobic pocket. Colors are as in Fig. 3K.
Movie 4. Comparison of NUP358 and Nup50 RanBD Ran(GMPPNP) complexes.
A 360° rotation of the of NUP358RanBD-IV•Ran(GMPPNP) cocrystal structure, colored as in Fig. 3L. A zoom-in view is provided, transitioning between the four different Nup358 RanBD structures and the single NUP50RanBD structure, showing hydrophobic residues of the NUP358 RanBD N-terminal extension buried in the Ran hydrophobic pocket. Finally, the interaction of individual RanBD basic patches with the Ran acidic tail is shown, colored as in fig. S55D.
Together, our data establish that the human CF and nuclear basket nups NUP358, NUP153, and NUP50 harbor a total of 16 distinct Ran-binding sites that, given their stoichiometry in the NPC, could together recruit up to several hundred Ran molecules. Considering the substantial size difference between metazoan and S.cerevisiae cells, it is conceivable that additional Ran-binding sites provided by the metazoan-specific asymmetric nups NUP358 and NUP153 help ensure high enough Ran concentrations in the NPC vicinity to enable nucleocytoplasmic transport, as has been previously suggested (85).
Docking of NUP358NTD into the cytoplasmic unassigned density cluster I
NUP358 is known to reside on the cytoplasmic face of the NPC. Lacking a structure, its location in the NPC could previously be inferred only from the differential absence of unassigned density in an ~38Å cryo-ET map of a NUP358-depleted human NPC (44). In the accompanying manuscript, quantitative docking of residue-level resolution structures into the symmetric core of a ~12Å cryo-ET map of the intact human NPC led to a novel assignment of 16 copies of the symmetric nups NUP205 and NUP93 in the cytoplasmic outer ring, as well as the identification of two clusters (I and II) of unassigned density (Fig.4A), of which the first corresponds to the previously observed NUP358-dependent density (42). Because of the large size and unique fold of our newly determined NUP358NTD crystal structure, we sought to directly determine its position in the intact human NPC (Fig.4A).
Fig. 4. Docking of NUP358NTD on the cytoplasmic face of the NPC.
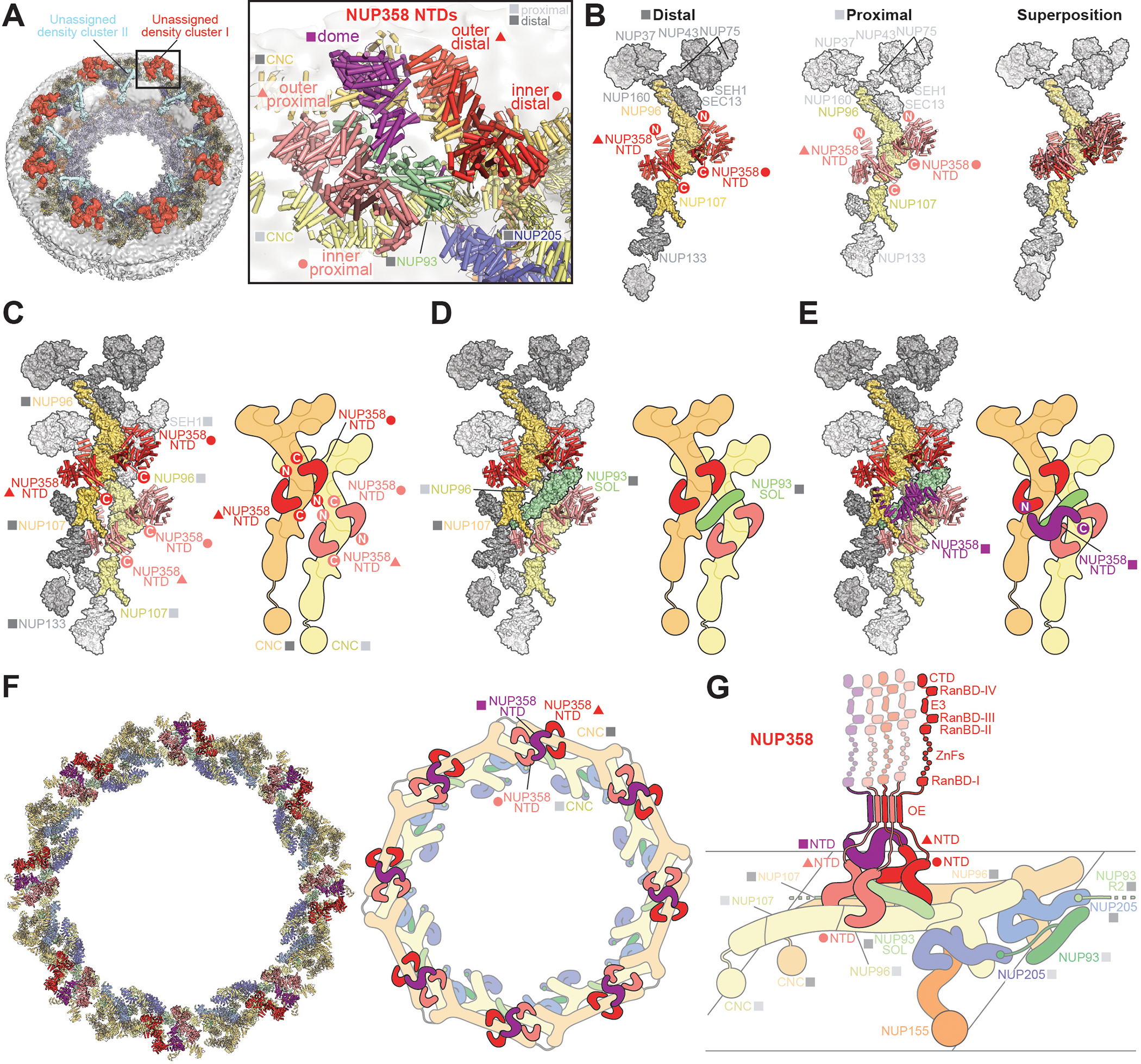
(A) Overview of the NPC cytoplasmic face with isosurface rendering of unexplained density clusters I (red) and II (cyan) of the ~12Å cryo-ET map of the intact human NPC. The inset indicates the location of the magnified view showing cartoon representations of five copies of NUP358NTD docked in unassigned density cluster I. (B) Comparison of the binding of two NUP358NTD copies (cartoon) to distal and proximal CNCs (surface). (C-E) Architecture of the pentameric NUP358NTD bundle attachment site on a cytoplasmic outer ring spoke with cartoon- and surface-represented structures (left) and schematics (right), sequentially illustrating the placement of (C) four copies of NUP358NTD around NUP96 and NUP107 interfaces on the stalks of tandem-arranged Y-shaped CNCs, (D) a distal copy of NUP93SOL collocated at the center of the NUP358NTD bundle, interfacing with both proximal and distal CNC stalks, and (E) the NUP358NTD dome copy interfacing with the stalk-attached NUP358NTD quartet beneath. (F) Overview of the entire cytoplasmic face of the NPC in cartoon representation and as schematic, illustrating the distribution of 40 NUP358NTD copies anchored as pentameric bundles across the eight NPC spokes. (G) Schematic of NUP358 attachment to the cytoplasmic outer ring spoke. The NUP358 pentameric bundle is linked together by interactions between oligomerization elements (OEs). Anchored by NUP358NTD, the rest of NUP358 domains are linked by unstructured linker sequences and are expected to freely project from the cytoplasmic face of the NPC. Distal and proximal positions are labeled according to the legend in (A).
In our docking analysis, we calculated correlations between a new ~12Å cryo-ET map of the intact human NPC (provided by the Beck group) and one million resolution-matched densities simulated from either open or closed conformation of the NUP358NTD crystal structure, randomly placed and locally fit-optimized in the asymmetric unit of the full ~12Å cryo-ET map (46). Unlike for the closed NUP358NTD conformation, docking scores for five placements of the open NUP358NTD conformation segregated to high confidence of placement and located to the previously unassigned density cluster I, leaving no unexplained density (fig.S56). We found four copies of NUP358NTD to be interfaced with the α-helical solenoid folds of the CNC components NUP96, NUP107, and the distal copy of NUP93SOL, wrapping around the stalks of the tandem arranged Y-shaped CNCs in pairs, at equivalent distal and proximal positions (Fig.4, B and C). As identified in the docking analysis of the symmetric core reported in the accompanying manuscript, the distal NUP93SOL bisects the stalks of the tandem-arranged Y-shaped CNCs by interfacing with the distal NUP107 and the proximal NUP96 α-helical solenoids, cloistered between the four NUP358NTD copies (Fig.4D) (42). Lastly, the fifth NUP358NTD referred to as the dome copy was docked above the other four NUP358NTD copies and the distal NUP93SOL, with its N- and C-termini oriented towards the C-termini of the outer distal and inner proximal copies of NUP358NTD, respectively (Fig.4E). Though unexpected, the placement of the dome NUP358NTD was the second most confident docking solution into both the current ~12Å and previously reported ~23Å cryo-ET maps of the intact human NPC (44) (fig.S57). The placement of 40 molecules of NUP358 per NPC is in agreement with previous experimental lower-bound stoichiometry estimates of 32 molecules of NUP358 (86). Finally, we successfully placed the composite structure of the entire cytoplasmic outer ring protomer, including all five NUP358NTD copies, into an anisotropic ~7Å region of a composite single particle cryo-EM map of the X.laevis NPC cytoplasmic outer ring protomer (figs.S58 and S59) (45).
The arrangement of five NUP358NTD copies in each spoke places their C-termini in proximity of each other, projecting the remaining domains towards the cytoplasm. Consequently, the oligomerization domains of the five NUP358NTD copies are constrained to form a homomeric assembly within the same spoke (Fig.4, F and G). The oligomerization of NUP358 observed in the NUP358OE crystal structure and SEC-MALS analysis would boost the avidity of NUP358 attachment to the cytoplasmic face of the NPC (Figs.3, G and H, and 4G).
NUP358 is dispensable for NPC integrity during interphase
Our quantitative docking showed that NUP358NTD is the primary attachment point for NUP358 at the cytoplasmic face of the NPC. To validate this result physiologically, we sought to determine the subcellular localization of structure-guided NUP358 fragments in intact cells. To prevent default localization of ectopically expressed fragments at the nuclear envelope because of homo-oligomerization with the NUP358OE of endogenous proteins, we generated an inducible NUP358 knockout cell line, in which an N-terminal auxin-inducible degron (AID) tag was inserted into both genomic NUP358 loci (AID::NUP358 HCT116) (fig.S60). Addition of auxin resulted in the rapid, selective, and complete degradation of endogenous NUP358 within three hours, confirmed by the loss of immunofluorescent nuclear envelope rim staining and western blot analysis of cellular NUP358 protein levels (Fig.5A and figs.S60 and S61).
Fig. 5. NUP358 plays a general role in translation of exported mRNA.
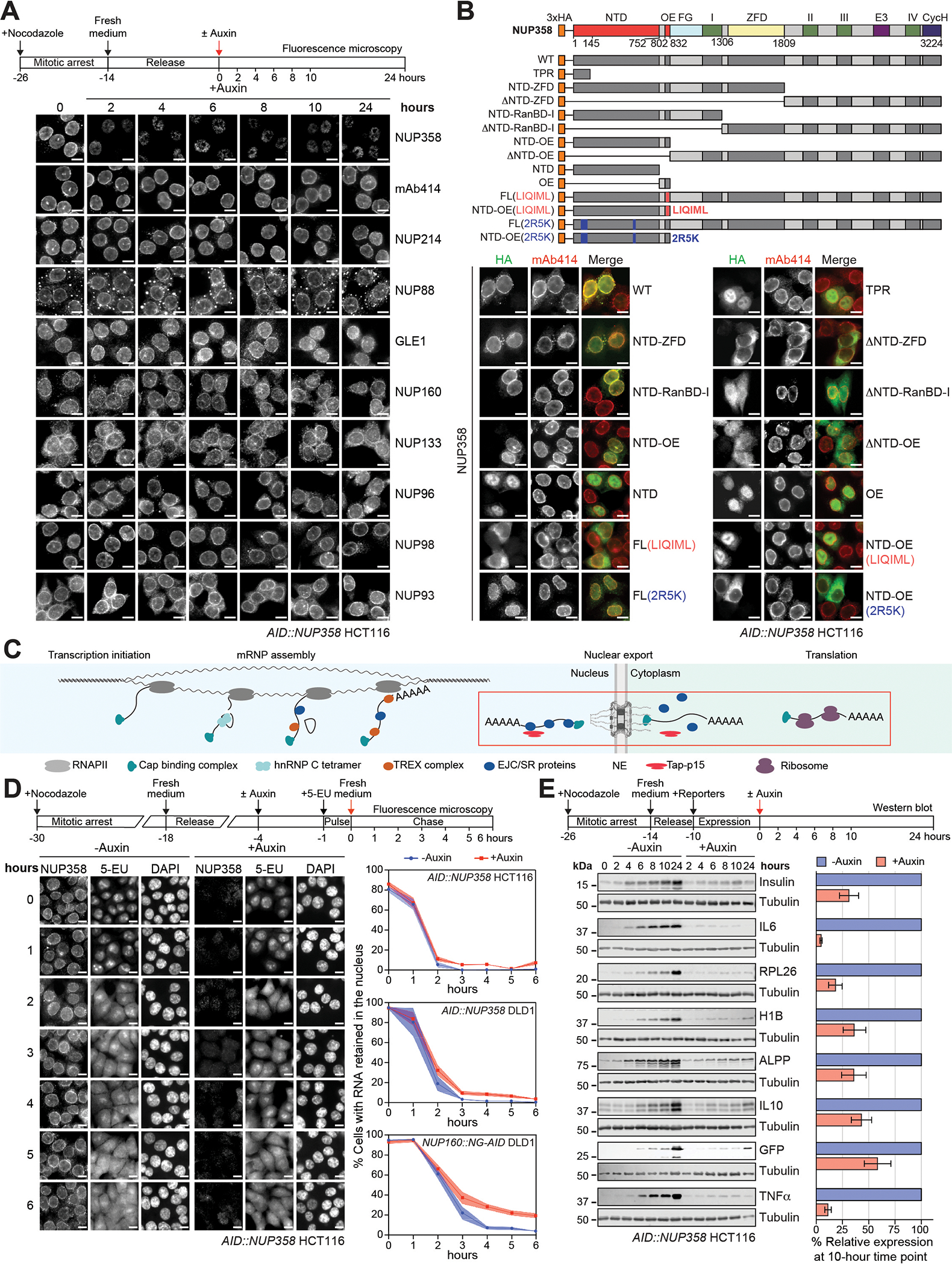
(A) Subcellular localization by immunofluorescent staining of endogenous nups in synchronized AID::NUP358 HCT116 cells at the indicated time points following auxin-induced NUP358 depletion. Cytoplasmic puncta arise from NUP88 overexpression inherent to HCT116 cells. (B) Subcellular localization of N-terminally 3×HA-tagged NUP358 variants in AID::NUP358 HCT116 cells by immunofluorescence microscopy. mAb414 staining marks nuclear envelope rim location. Domain structure of the transfected NUP358 variants is shown (above). (C) Schematic illustrating the life cycle of messenger ribonucleoprotein particles (mRNPs) from transcription, maturation, export, remodeling at the cytoplasmic face of the NPC, to translation. Steps associated with the NPC are highlighted by a red box. (D) Time-resolved analysis of RNA nuclear retention in synchronized AID::NUP358 HCT116 cells upon auxin-induced NUP358 depletion visualizing 5-EU metabolically pulse-labeled RNA. Representative images (left) and quantitation (right) of the proportion of cells (n>200/timepoint) with nuclear RNA retention are shown with mean values (squares) and the respective standard error (shaded area) of triplicate experiments. Quantitation from unsynchronized AID::NUP358 DLD1 and NUP160::NG-AID DLD1 cells are also shown. (E) Time-resolved western blot analysis of the expression of C-terminally 3×FLAG-tagged reporter proteins in synchronized AID::NUP358 HCT116 cells upon auxin-induced NUP358 depletion. Quantitated reporter expression in auxin-treated cells was normalized to expression in control cells, at the 10-hour timepoint. Experiments performed in triplicate, with mean and associated standard error shown. Scale bars are 10μm. Experimental timelines are shown above each experiment.
To identify the minimal NUP358 region necessary and sufficient for nuclear envelope targeting, we generated a systematic series of HA-tagged N- and C-terminal fragments, splitting the protein into two pieces after the NTD, OE, RanBD-I, or ZFD, and determined their subcellular localization by immunofluorescence microscopy. NUP358 targeting to the nuclear envelope in the absence of auxin required both NTD and OE regions, whereas all other domains were dispensable (Fig.5B and fig.S62). When NUP358NTD or NUP358OE were tested in isolation, neither was found to be sufficient, with both domains exhibiting strong nuclear staining (Fig.5B). Introduction of the NUP358 2R5K mutation, located on the NUP358NTD concave surface contacting the CNC, either abolished or severely reduced nuclear envelope rim staining when introduced into HA-NUP358NTD-OE or HA-NUP358FL, respectively (Fig.5B). Analogously, NUP358 oligomerization is required for localization to the nuclear envelope, with introduction of the oligomerization-deficient LIQIML mutation eliminating nuclear envelope rim staining of both HA-NUP358NTD-OE and HA-NUP358FL (Fig.5B). Crucially, we repeated the fluorescence microscopy analysis after auxin-induced depletion of endogenous NUP358 and obtained identical results (fig.S63).
The previous ~38Å cryo-ET map of the human NPC showed loss of the distal cytoplasmic CNC ring and potentially other CF nups in the absence of NUP358, leading to the conclusion that NUP358 is required for the integrity of the interphase NPC (44). To determine whether the architectural stability of the interphase NPC depends on NUP358, we sought to analyze the effect of auxin-induced NUP358 depletion on the subcellular localization of eight nups representative of all NPC sub-complexes by immunofluorescence microscopy (Fig.5A). Because previous studies showed NUP358 depletion results in cell cycle arrest at the G2/M transition (87), we first monitored the levels of cell cycle markers determining a cell cycle length of ~14-hours, consistent with previous reports for HCT cells (fig.S64) (88). We then induced NUP358-degradation in nocodazole-synchronized cells and imaged nups at various timepoints prior to cells entering mitosis and at 24 hours. Although NUP358 nuclear envelope rim staining was rapidly lost two hours after induction of degradation and remained absent throughout the remaining timepoints, all eight representative nups continued to display robust nuclear envelope rim staining (Fig.5A). This suggests that NPC integrity is not dependent on NUP358 attachment to the NPC and also demonstrates the specificity of the auxin-induced NUP358 knockout. To reconcile the apparent conflict between our results and the aforementioned cryo-ET study, we investigated whether NUP358 depletion led to release of nups from the nuclear fraction during cellular fractionation. Indeed, we observed an auxin-dependent leakage of NUP214, NUP88, and NUP160 from the nuclear to the cytoplasmic fraction (fig.S61C). Curiously, we also consistently observed a reduction of the nuclear basket nup ELYS in the nuclear fraction upon NUP358 depletion.
Together, these data confirm the quantitative docking of NUP358NTD, validate the physiological relevance of NUP358OE-mediated bundling, and establish that NUP358 is dispensable for the architectural integrity of the assembled interphase NPC, although its depletion made the structural integrity of the cytoplasmic face of the NPC susceptible to the biochemical stresses inherent to cell fractionation. Future studies need to establish the extent of NUP358’s role in the formation of the double CNC-ring architecture during NPC assembly.
NUP358 plays a general role in translation of exported mRNAs
Export of mRNA from the nucleus to the cytoplasm is an essential step in the expression of eukaryotic proteins (Fig.5C) (43). Our biochemical analysis revealed that NUP358 has multiple RNA-binding domains distributed throughout the protein, suggesting a potential role in RNA export and mRNP remodeling (Fig.2G). The docking of five NUP358NTD copies in the intact human NPC revealed occlusion of the RNA/NUP88NTD-binding surfaces of the dome and inner distal NUP358 copies by the CNC stalk, but exposure of the remaining copies’ binding sites (fig.S65). Thus, some NUP358NTD copies could potentially be simultaneously attached to the NPC and dynamically interacting with RNA/NUP88NTD.
Previous studies had found that efficient translation of secretory proteins requires NUP358 binding to the ~63-nucleotide GC-rich signal sequence coding region (SSCR) of mRNAs encoding secretory proteins (89). NUP358 knockdown by short hairpin (sh) RNAs was shown to prevent the translation of various secretory protein reporters, but had no effect on the distribution of mRNA in the cell (89). These experiments involved extended incubation periods and achieved only a partial NUP358 knockdown, potentially allowing secondary phenotypes to emerge from prolonged NPC disruption, non-NPC related NUP358 effects, or defective postmitotic NPC re-assembly. With the ability to rapidly deplete NUP358 in our AID cell line, we sought to examine whether NUP358 is directly involved in mRNA export or mRNP remodeling by monitoring the subcellular distribution of 5-ethynyl uridine (5-EU) pulse-labeled, newly synthesized RNA after NUP358 depletion. In situ labeled RNA was visualized by fluorescence microscopy at hourly intervals during the chase for up to 6 hours (Fig.5D). At early time points, 90–100% of cells displayed a strong nuclear 5-EU-labeled RNA signal that decreased over time with a concomitant increase in the cytoplasmic signal, indicative of RNA being exported. After six hours, only ~5% of NUP358-depleted cells exhibited nuclear retention of labeled RNA, compared to <1% of control cells (Fig.5D). Because of this subtle effect, we sought to confirm the result in AID::NUP358 DLD1 cells (Fig.5D and figs.S66 and S67). Similar to the AID::NUP358 HCT116 cell results, only ~3% of NUP358-depleted DLD1 cells exhibited nuclear RNA retention after a 6-hour chase (Fig.5D and fig.S68). By contrast, nuclear RNA retention was present in ~20% of NUP160::NG-AID DLD1 cells after depletion of NUP160, whose knockout causes RNA retention in S.cerevisiae, demonstrating the principle suitability of our experimental approach (Fig.5D and fig.S68) (90).
Next, we analyzed the fate of the genetic message downstream of mRNA export by examining the dependence of cellular protein expression on NUP358 in AID::NUP358 HCT116 cells using eight different reporter constructs. Synchronized cells were transfected with C-terminally FLAG-tagged reporter constructs prior to NUP358 depletion, and the amount of reporter in whole cell extracts was determined by western blot analysis (Fig.5E and fig.S69). First, we focused our analysis on representative secretory protein reporter constructs including insulin, interleukin-10 (IL-10), IL-6, tumor necrosis factor alpha (TNFα), and membrane-bound placental alkaline phosphatase (ALPP). NUP358 depletion significantly reduced the cellular levels of all five reporters. We also tested non-secretory protein reporters and contrary to the previous observation of secretory protein-effect specificity, we found that NUP358 depletion also significantly reduced the cellular levels of ribosomal protein L26 (RPL26), green fluorescence protein (GFP), and histone 1B (H1B) reporters (Fig.5E and fig.S69).
In summary, our data confirm that NUP358 depletion does not result in marked nuclear RNA accumulation, but nevertheless affects the efficient translation of secreted and membrane-bound proteins, as previously proposed (89). However, our findings also demonstrate that the observed translational defect is not restricted to secretory proteins, suggesting a more general role of NUP358 in mRNP remodeling events that occur at the cytoplasmic face of the NPC after mRNA export.
Characterization of NUP358 harboring ANE1 mutations
Acute necrotizing encephalopathy (ANE) is an autoimmune disease in which previously healthy children experience a cytokine storm following common viral infections, resulting in brain inflammation and rapid deterioration from seizures to coma that can ultimately be fatal (91). ANE1, the familial and recurring form of ANE, has been associated with four distinct NUP358 mutations: T585M, T653I, I656V, and W681C (91, 92). All four ANE1 mutations map to the C-terminal α-helical solenoid of NUP358NTD (fig.S70). Apart from T585, which is exposed on the surface, the ANE1 mutations locate in the closely packed hydrophobic core (fig.S70C). We determined co-crystal structures of NUP358NTD harboring the individual ANE1 mutations T585M, T653I, and I656V in complex with sAB-14 revealing no substantial structural changes, with root-mean square deviation (RMSD) calculated over 746 Cα atoms of ~0.5Å (fig.S70 and table S10). Moreover, we did not detect differences in nuclear envelope rim staining or binding to NUP88NTD or ALPP SSCR RNA (fig.S71). Notably, we found that in vitro thermosolubility of the W681C, T653I, and I656V mutants was reduced to levels below body temperature (fig.S72), but increase beyond wildtype levels by binding to sAB-14 in all three mutants (fig.S73).
Together, our results indicate that ANE1 mutations neither directly disturb the fold observed in the crystal structure, nor affect the known cellular functions of NUP358. Our observation of a substantially reduced thermosolubility of NUP358 ANE1 mutants is interesting, considering that the sudden onset of symptoms appears to require a fever-inducing trigger such as a viral infection (91). Future studies will need to systematically assess whether ANE1 mutations affect unknown cellular functions of NUP358.
Structural and biochemical analysis of NUP88NTD•NUP98APD•NUP214TAIL
Besides NUP358, the NUP88NTD•NUP98APD•NUP214TAIL complex had up to now been another CF component for which atomic level structural information had remained unavailable. Through extensive screening of crystallization fragments and conditions, we solved the structure of the hetero-dimeric NUP88NTD•NUP98APD at 2.0Å resolution (Fig.6H and table S17). Despite low sequence homology, the overall architecture of the NUP88NTD•NUP98APD complex is conserved from fungi to humans, although the orientation of NUP98APD relative to NUP88NTD varies between the co-crystal structures of human and fungal orthologs by as much as ~20° (figs.S74 to S77, and Movie 5) (11, 59). For a detailed description of the structure, see Supplementary Text (figs.S76 to S81).
Fig. 6. Docking analysis reveals that NUP93 anchors the CFNC to the cytoplasmic outer ring.
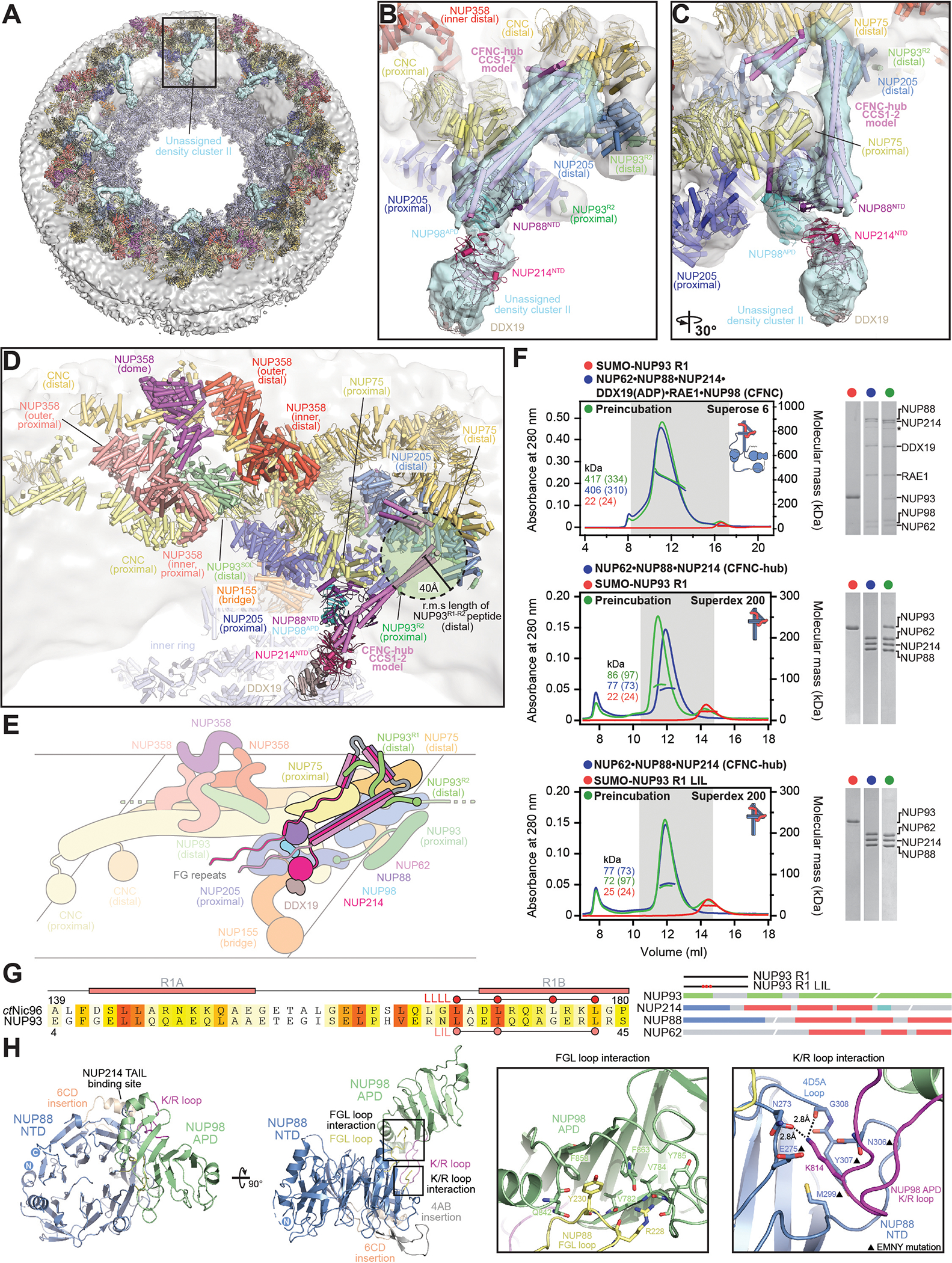
(A) Overview of the NPC cytoplasmic face with isosurface rendering of unexplained density cluster II (cyan) of the ~12Å cryo-ET map of the intact human NPC. The inset indicates the location of the magnified view in (B). (B, C) Two views of manually placed poly-alanine models of CFNC-hub segments CCS1 and CCS2, as well as of the tentatively placed NUP88NTD•NUP98APD and NUP214NTD•DDX19 co-crystal structures shown in cartoon representation. (D) Cartoon representation of a cytoplasmic face spoke. The root-mean square (r.m.s.) end-to-end length estimate for the NUP93R1-R2 linker defines a ~40Å radius for the expected location of the distal NUP93R1 region. (E) Schematic of a cytoplasmic face spoke illustrating CFNC-hub anchoring by the distal NUP93R1 positioned by the distal NUP205-bound NUP93R2. (F) SEC-MALS interaction analysis showing the binding of SUMO-NUP93R1 to the hetero-hexameric CFNC or CFNC-hub and illustrating the abolished binding of the SUMO-NUP93R1 LIL mutant to the CFNC-hub. Proteins were visualized by Coomassie brilliant blue staining. (G) An alignment of C.thermophilum Nic96 and H.sapiens NUP93 sequences shows the conservation of residues targeted by the LLLL and LIL mutations that abolish binding to the CNT. Residues are colored according to a multispecies sequence alignment from white (less than 55% similarity), to yellow (55% similarity), to red (100% identity), using the BLOSUM62 weighting algorithm. Domain architectures of NUP214, NUP88, NUP62, and NUP93. The location of the NUP93 LIL mutation is indicated (red dots). (H) Two views of the NUP88NTD•NUP98APD co-crystal structure in cartoon representation. Inset boxes indicate regions of magnified views (right) of NUP88NTD FGL-loop interaction with NUP98APD, and of NUP98APD K/R-loop interaction with NUP88NTD. Black triangles indicate alanine substitutions in the NUP88NTD EMNY mutant.
Movie 5. Evolutionary conservation of the NUP88NTD•NUP98APD architecture.
A 360° rotation of the NUP88NTD•NUP98APD cocrystal structure and the previously determined crystal structures of S. cerevisiae Nup82NTD•Nup116CTD•Nup159TAIL (PDB ID 3PBP) (59) and C. thermophilum Nup82NTD•Nup145NAPD•Nup159TAIL (PDB ID 5CWW) (11), colored as in fig. S77.
Because the NUP214TAIL-NUP88NTD interaction was crystallographically intractable, we mapped a minimal NUP88NTD-binding region spanning NUP214 residues 938–955 by systematic truncation (figs.S78 and S79). NUP214TAIL forms a hydrophobic interaction with NUP88NTD at the 6CD insertion, which was abolished by a combined NUP88NTD LLL mutation, analogous to a mutation we had previously shown to abolish the interaction between the S.cerevisiae orthologs Nup159TAIL and Nup82NTD (fig.S79) (59). Interestingly, this NUP88NTD LLL mutation straddles a naturally occurring D434Y mutation in NUP88 linked to a fatal disorder called fetal akinesia deformation sequence (FADS), which is associated with congenital malformations and impaired fetal movement (fig.S80) (93). Given its location, the D434Y mutation is expected to interfere with the NUP214TAIL interaction.
Combined, our structural and biochemical analysis of NUP88, NUP214, NUP98, and their interactions shows that their shape, mode of interaction, and the overall architecture of their complexes are evolutionarily conserved from fungi to humans despite primary sequence divergence.
Docking of the cytoplasmic filament nup complex into unassigned cluster II
Following the placement of five NUP358NTD copies into unassigned density cluster I, we reasoned that the remaining unassigned density cluster II would represent the CFNC. Unassigned density cluster II is composed of two near-perpendicular tube-like segments that bisect the NUP75 arms of the distal and proximal Y-shaped CNCs, a globular segment lodged between the base of the long tube-like segment and the proximal NUP75 arm, and a dumbbell-shaped globular density that projects toward the central transport channel (Fig.6A). Due to the small size and lack of distinctive shape features, the quantitative docking of NUP88NTD•NUP98APD, NUP214NTD•DDX19, GLE1CTD•NUP42GBM, GLE1CTD•NUP42GBM•DDX19, RAE1•NUP98GLEBS, NUP358RanBD•Ran, NUP358ZnF•Ran, and NUP358CTD, into the ~12Å cryo-ET map of the intact human NPC, from which all the hereto explained density had been subtracted, did not result in high confidence solutions (fig.S82). We therefore took the less objective approach of manual placement based on shape complementarity and biochemical restraints, followed by local rigid body refinement.
We used the C.thermophilum and X.laevis CNT crystal structures, the latter containing NUP62, as a template for the polyalanine model of the coiled-coil segments (CCS) 1 and 2 of the CFNC-hub (10, 11). Remarkably, the CCS1 and CCS2 models based on CNT structures matched the shape and dimensions of two near-perpendicular segments of tube-like density, suggesting that the CFNC-hub coiled-coil architecture is similar to the CNT (Fig.6, B and C, and fig.S83A). NUP88NTD•NUP98APD fit best at the base of the long CCS1 segment, interfacing with the NUP75 arm of the proximal CNC. The tentative placement would be consistent with the biochemically mapped interaction between NUP88NTD•NUP98APD and NUP214TAIL segment expected to emanate from the C-terminal base of the CCS3 segment and thus restrain NUP88NTD•NUP98APD near the CFNC-hub base (Fig.6, B to E). A dumbbell-shaped density interfacing with NUP88NTD•NUP98APD and extending towards the central transport channel is consistent with the shape and size of the NUP214NTD•DDX19 crystal structure, although it could also be explained by other more transiently tethered components of the nucleocytoplasmic transport machinery or cargo (Fig.6, B and C, and fig.S83A). However, the NUP214NTD•DDX19 complex forms tighter interactions when DDX19 is in its ADP-bound state and would therefore be expected to exist in the ATP-depleted environment of the purified nuclear envelope (50, 63). The placements of the CFNC-hub model and NUP88NTD•NUP98APD were further supported by manual docking into an anisotropic ~8Å region of a composite single particle cryo-EM map of the X.laevis cytoplasmic outer ring protomer, though the map masking excluded the region comprising the dumbbell-shaped density (fig.S83B) (45).
Placement of the CFNC-hub into the tube-like density puts CCS1, CCS2, and likely the unresolved CCS3, within reach of the ~40Å r.m.s length of the linker that tethers NUP93R1 to the NUP205-bound NUP93R2 (Fig.6D and fig.S84). In the accompanying paper, we demonstrated that the NUP93R1 fragment (residues 2–93), like the orthologous C.thermophilum Nic96R1 assembly sensor, binds to the CNT complex of the inner ring (42). The proximity of the CFNC-hub coiled-coil segments to the expected NUP93R1 location suggested that NUP93R1 might act as assembly sensor for the CFNC-hub, as well. Indeed, the NUP93R1 fragment formed stable complexes with the intact CFNC and CFNC-hub (Fig.6F and fig.S85). Strikingly, the NUP93R1 LIL mutation that abolished CNT binding (42) also abolished the interaction with the CFNC-hub (Fig.6, F and G). To ensure we did not miss an interaction of the C.thermophilum CFNC (ctCFNC), we tested whether Nic96R1 could bind the ctCFNC-hub. In fact, Nic96R1 did not bind to ctCFNC-hub, consistent with our reconstitution results that identified two distinct assembly sensors for the ctCFNC in the CNC (fig.S86). These data indicate that the long elusive assembly sensor anchoring point of the human CFNC is not provided by the Y-shaped CNC, but rather by the NUP205-positioned NUP93R1, corroborated by the recent finding that NUP93 depletion displaces the CFNC nups NUP214, NUP88, and NUP62 from the nuclear envelope (94).
A second NUP93R1 assembly sensor emanating from the proximal NUP205-positioned NUP93R2 represents a potential anchoring site for a second, flexibly attached proximal CFNC (fig.S84). The placement of 16 copies of the CFNC on the cytoplasmic face of the NPC, half of which are unresolved in the ~12Å cryo-ET map, is consistent with the previously established stoichiometry (86). Recent in situ ~37Å and ~34Å cryo-ET maps of the dilated human NPC (95, 96) present unexplained elongated density near the expected location of the proximal NUP93R1 but could not be further interpreted at the current solutions (fig.S87).
Finally, we tentatively placed the human CF nup GLE1CTD•NUP42GBM crystal structure into a region of unexplained density in the ~12Å cryo-ET map of the intact human NPC between the cytoplasmic bridge NUP155 and the cytoplasmic face of the nuclear envelope, consistent with our previous analysis (fig.S88) (63).
Steric occlusion is insufficient to explain asymmetric decoration of the NPC
Having assigned all cytoplasmic density of clusters I and II to NUP358 pentameric bundles and CFNCs, respectively, we next wondered whether any structural features prevent NUP358 or CFNC mis-localization at the nuclear face of the NPC. We found that unexplained nuclear density adjacent to the NUP160 arms of the Y-shaped CNCs could be assigned to 16 copies of the structured N-terminal domains of the nuclear basket nup ELYS (fig.S89) (25). The ELYS domains did not overlap with nuclear regions equivalent to the sites occupied by NUP358 and CFNC on the cytoplasmic face, thereby excluding that steric competition with NUP358 or CFNC prevents ELYS mis-localization (Fig.7, A and B). On the contrary, the ~12Å cryo-ET map revealed rod-shaped unassigned densities atop the nuclear outer ring in regions equivalent to NUP358 sites on the cytoplasmic face (Fig.7C). Analogously, we asked if recruitment of the CFNC to the nuclear face was prevented by steric hindrance from a nuclear basket component. Although the NUP205-NUP93R2 attachment site from which NUP93R1 is flexibly projected remains unencumbered, an unassigned rod-shaped cryo-ET density present on the nuclear face overlaps with an area equivalent to the CFNC-hub docking sites on the cytoplasmic face (Fig.7C). Together, these findings suggest that mechanisms other than steric competition alone, such as active nuclear transport of asymmetric nups, as previously indicated for NUP214 and NUP153 (81, 97), are key determinants of the asymmetric localization of NUP358, CFNC and ELYS.
Fig. 7. Comparison of cytoplasmic and nuclear faces of the human NPC.
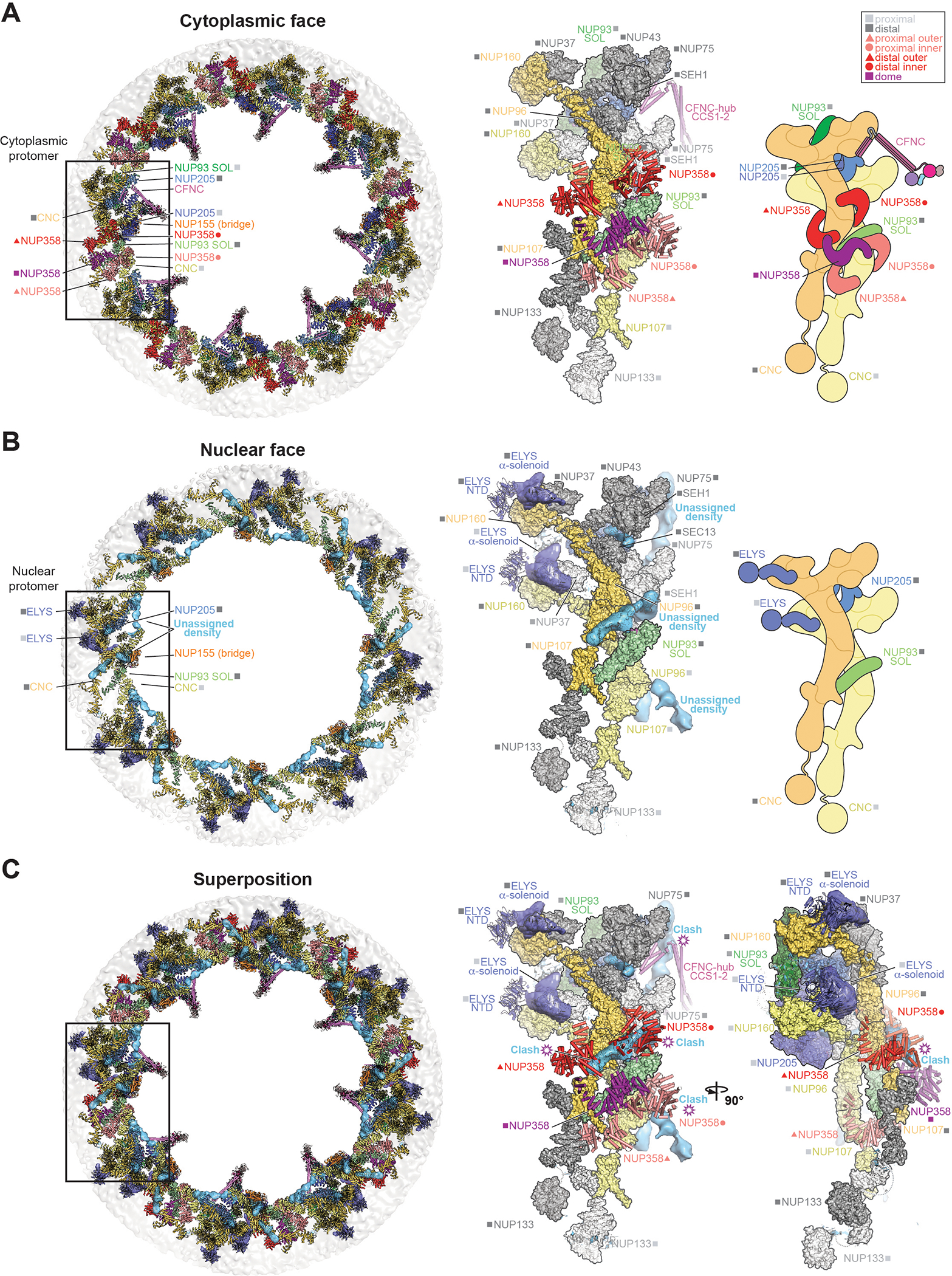
Overall top view (left), single spoke protomer with symmetric core nups in surface, docked asymmetric nups in cartoon, and unexplained density of the ~12Å cryo-ET map in isosurface representation (middle), and schematic (right) of the (A) cytoplasmic and (B) nuclear face of the intact human NPC. (C) Superpositions of the overall view (left) and two orthogonal views of single spoke protomers (middle and right) of the nuclear and cytoplasmic faces, with hypothetical steric clashes between the cytoplasmic filaments in cartoon representation, and the unassigned asymmetric nuclear density (cyan), indicated. Distal and proximal positions are labeled according to the legend. Inset boxes indicate regions of magnified protomer views (right).
Together, our data complete the near-atomic composite structure of the symmetric and cytoplasmic asymmetric portions of the human NPC (Fig.8 and Movie 6).
Fig. 8. Architecture of the human NPC cytoplasmic face.
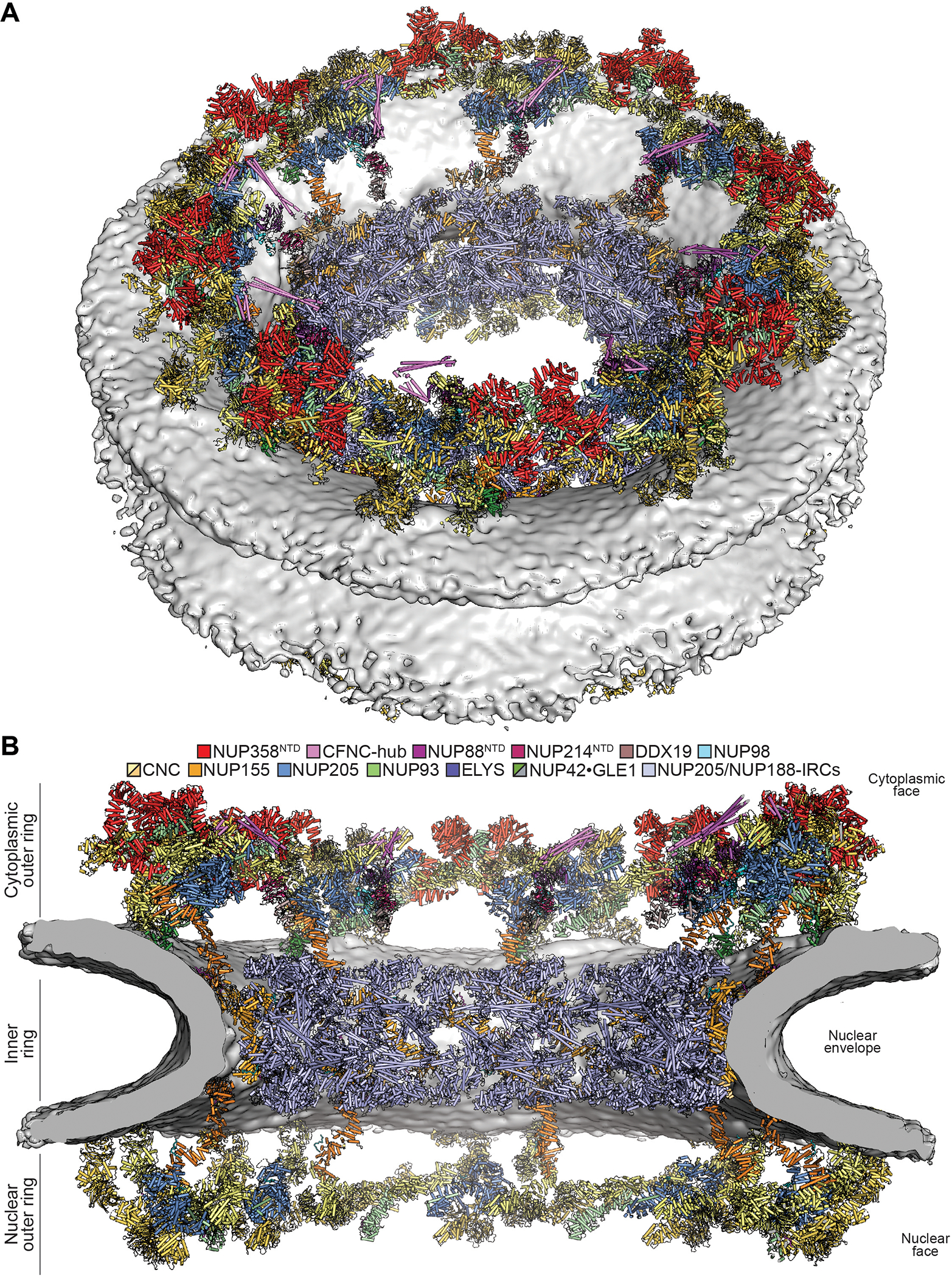
Near-atomic composite structure of the human NPC generated by docking individual nup and nup complex crystal and cryo-EM structures into a ~12Å cryo-ET map of the intact human NPC, viewed from (A) the cytoplasmic face, and (B) the central transport channel, as a cross-section. Newly placed structures include the quantitatively docked NUP358NTD and the manually docked NUP88NTD•NUP98APD, NUP214NTD•DDX19, GLE1CTD•NUP42GBM, ELYSNTD, and a CFNC-hub model. The nuclear envelope is rendered as a grey isosurface. Nups are shown in cartoon representation and colored according to the legend.
Movie 6. Overview of the composite structure of the human NPC cytoplasmic face.
The structures are shown docked into the cryo-ET reconstruction of the intact human NPC, colored according to Fig. 8. A pentameric bundle of NUP358NTD is docked followed by relative placement of NUP358OE and additional NUP358 domains, followed by the CFNC components. An overview of a single-spoke cytoplasmic face protomer is shown followed by a comparison cytoplasmic and nuclear face protomers.
Conclusions
Situated on the cytoplasmic face of the NPC, CF nups remodel mRNPs as they emerge from the central transport channel, ensuring directional transport of mRNA and preparing it for downstream translation. Given this essential cellular function, it is unsurprising that CF nups are a hotspot for mutations associated with currently incurable diseases, ranging from neurodegenerative and auto-immune disorders to aggressive cancers. Through a comprehensive analysis combining in vitro complex reconstitution, crystal structure determination, quantitative docking, and in vivo validation, we established a near-atomic composite structure of the cytoplasmic face of the human NPC.
Our biochemical reconstitution highlights the evolutionary conservation of the CFNC modular assembly, which consists of a central hetero-trimeric coiled-coil hub that tethers two separate mRNP remodeling complexes together. Despite the divergence in attachment mechanisms, the anchoring of two copies of the CFNC module to each of the eight NPC spokes appears to be an evolutionarily conserved architectural outcome: The C.thermophilum NPC presents two distinct assembly sensor motifs for the CFNC-hub in the Nup37 and Nup145C subunits of each CNC. The human NPC reuses the NUP93 sensor for the assembly and anchoring of the CNT in the inner ring as an anchor for the CFNC in the cytoplasmic outer ring by intercalating two NUP205•NUP93 copies among the tandem-arranged CNCs of each spoke. Previous studies have also shown that the S.cerevisiae NPC incorporates a P-shaped CFNC dimer (61) to a single site within each of the eight outer ring spokes (40, 41).
In addition to the CFNC, the asymmetric CF nup decoration of the human NPC cytoplasmic face includes NUP358. Conjoined by an oligomerization element, pentameric bundles of the uniquely folded NUP358NTD envelop the tandem-arrayed stalks of a CNC pair in each of the eight spokes. Each attached NUP358NTD anchors an extensive ~2,400-residue C-terminal region that harbors 14 different domains connected by unstructured linkers, thereby extending as much as ~60nm from the outer ring (98). Our placement of the CNC and CF nups explains the entirety of the observed cytoplasmic face cryo-ET density, accounting for ~23MDa of structured mass. The more flexibly attached regions of the CF nups that are not captured by the current sub-tomogram averaged cryo-ET map account for an additional ~19MDa of mass.
In addition to these flexibly attached structured domains, NUP358, NUP214, NUP98, and NUP42 contain extended FG-repeat regions emanating from various anchor points at the outer ring. The degree to which these regions contribute to the architectural integrity of the human NPC, as has been shown for the S.cerevisiae NPC (99), and the NPC’s diffusion barrier remain important future questions.
We found that the interactions between the C.thermophilum CF nups and the CNC are modulated by the small molecule IP6, the presence of which is required for mRNA export. Future studies need to address the concerted role of post-translational modifications, second messengers and other small molecules, and macromolecular factors in regulating the assembly and functions of the NPC cytoplasmic face in mRNA export.
The integral membrane proteins and nuclear basket portions of the NPC represent an outstanding challenge for structural determination. Nevertheless, our analysis has already identified that competition for binding sites could play a role in the segregation of CF and nuclear basket nups to opposite faces of the NPC. However, steric occlusion alone is insufficient to deterministically establish NPC polarity, whereby the correct asymmetric nups are segregated to either the cytoplasmic or the nuclear face, or the proximal NUP93 and NUP205 copies are excluded from the nuclear outer ring. Nuclear and cytoplasmic eviction mediated by the nucleocytoplasmic transport machinery is perhaps the most obvious candidate for a mechanism that maintains the polar subcellular segregation of asymmetric nups.
The data presented provide a comprehensive biochemical foundation and a structural framework for the design of future experiments aimed at elucidating the multiple mechanistic steps involved in mRNP export and remodeling. This mechanistic insight will be vital for illuminating disease mechanisms associated with CF nup genetic variants and mechanisms by which viral virulence factors, e.g. SARS-CoV2 ORF6, hijack the functions of the NPC (100).
Our results represent a significant step towards the complete in vitro reconstitution of the NPC and establish a near-atomic composite structure of the entire cytoplasmic face of the human NPC. More broadly, they illustrate the effectiveness of our divide-and-conquer approach in successfully elucidating the near-atomic architecture of an assembly as large and complex as the NPC, serving as a paradigm for studying similar macromolecular machines, which remains a major frontier in structural cell biology.
Methods Summary
Full details of the Materials and Methods are presented in the Supplementary Materials. Briefly, the source of materials and reagents are summarized in Table S1. Bacterial, insect, and mammalian cell expression constructs and conditions are described in Tables S2 to S4. Proteins were purified using standard chromatography techniques, with purification procedures summarized in Table S5. Purified proteins and complex formation were characterized by analytical size-exclusion chromatography coupled to inline multi-angle light scattering (SEC-MALS), summarized in Table S6. Liquid-liquid phase separation (LLPS) of purified protein mixtures was analyzed by centrifugal separation followed by SDS-PAGE and Coomassie staining, and by fluorescence microscopy subsequent to N-terminal amino labeling with fluorescent dyes. Nup-RNA binding interactions were assayed by electrophoretic mobility shift assays (EMSAs) employing either 32P-labeled or unlabeled nucleic acid probes, visualized by autoradiography or SybrGold-staining, respectively. Structures were determined by X-ray crystallography, with crystallization conditions and X-ray diffraction data collection, processing, and refinement statistics summarized in Tables S7 to S17. Quantitative docking was performed by randomly placing and scoring densities simulated from crystal structures into ~12Å and ~23Å cryo-ET maps of the intact human NPC (44, 46). Experimental structures used to generate the near-atomic composite structure of the intact human NPC are inventoried in Table S18. NUP358 localization, NPC integrity, RNA export, and reporter expression levels were assessed in auxin-inducible degron cell lines.
Supplementary Material
Fig. 0. Cytoplasmic face of the human nuclear pore complex (NPC).
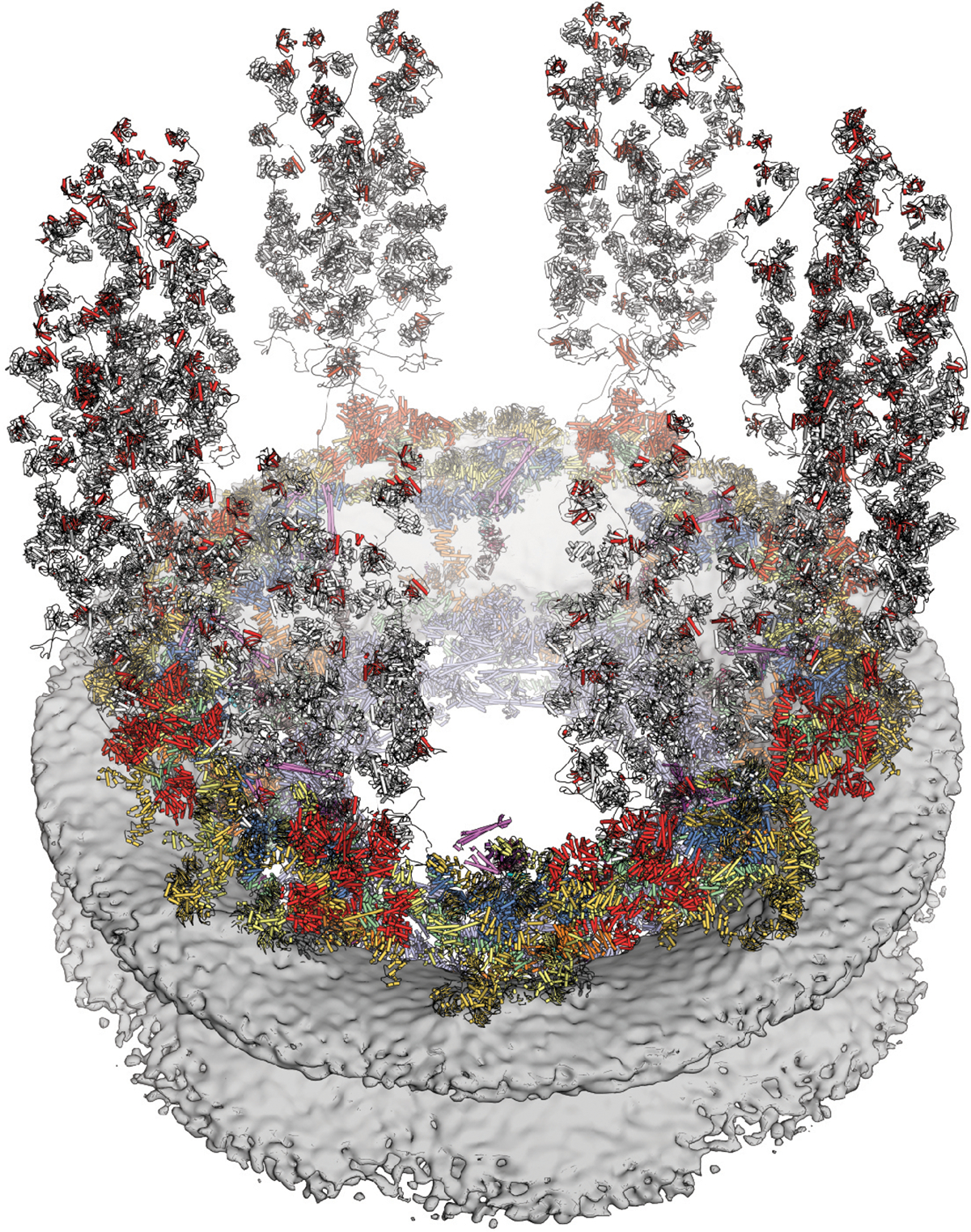
Near-atomic composite structure of the NPC generated by docking high-resolution crystal structures into a cryo-ET reconstruction of an intact human NPC. The symmetric core, embedded in the nuclear envelope, is decorated with NUP358 (red) domains bound to Ran (gray) flexibly projected into the cytoplasm and cytoplasmic filament nucleoporin complexes (pink) overlooking the central transport channel.
Acknowledgements:
We thank the members of the Hoelz laboratory, Mitchell Guttman, and Shu-ou Shan for insightful discussions, Alexander Cohen, Jacqueline Chou, Reeti Gulati, Young Jeon, Hannah Margolis, Evelyn Stuwe, Tobias Stuwe, and Jimmy Thai for experimental support, Martin Beck for sharing the ~12Å cryo-ET reconstruction of the intact human NPC prior to publication, and Valerie Doye, Ulrike Kutay, Ed Hurt, and Iain Mattaj for providing material. We acknowledge Jens Kaiser and the scientific staff of the Stanford Synchrotron Radiation Laboratory (SSRL) Beamline 12–2 and the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility (GM/CA) at the Advanced Photon Source (APS) for their support with X-ray diffraction measurements.
Funding:
The Molecular Observatory at the California Institute of Technology (Caltech) is supported by Donald and Judith Voet, the Gordon and Betty Moore Foundation, and the Beckman Institute. The operations at the SSRL and APS are supported by the U.S. Department of Energy and the National Institutes of Health (NIH). GM/CA has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). SP and FMH were supported by a Ph.D. fellowship of the Boehringer Ingelheim Fonds. DHL was supported by an NIH Research Service Award (5 T32 GM07616) and Amgen Graduate Fellowship through the Caltech-Amgen Research Collaboration. SC, SGR and MD were supported by National Institute of Child Health and Human Development Division of Intramural Research funding ZIAHD001902 and ZIAHD008954. AAK was supported by NIH grants R01-GM117372 and P50-GM082545. AH was supported by a Camille-Dreyfus Teacher Scholar Award (TC-15-082) and NIH grants R01-GM117360 and R01-GM111461, is an Investigator of the Heritage Medical Research Institute (HMRI-15-09-01), and a Faculty Scholar of the Howard Hughes Medical Institute (55108534).
Footnotes
Competing interests: The authors declare no financial conflicts of interest.
Supplementary Materials:
References
Data and materials availability:
Materials generated in this study are available on request from the corresponding author. The auxin-inducible AID::NUP358 HCT116, AID::NUP358 DLD1, and NUP160::NG AID DLD1 degron cell lines are subject to a materials transfer agreement, which is available upon request. The coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 7MNJ (NUP358145–673), 7MNK (NUP358OE), 7MNI (NUP88NTD•NUP98APD), 7MNL (NUP358NTD•sAB-14), 7MNM (NUP358NTD T585M•sAB-14), 7MNN (NUP358NTD T653I•sAB-14), 7MNO (NUP358NTD I656V•sAB-14), 7MNP (NUP358ZnF2•Ran(GDP)), 7MNQ (NUP358ZnF2•Ran(GDP)), 7MNR (NUP358ZnF3•Ran(GDP)), 7MNS (NUP358ZnF4•Ran(GDP)), 7MNT (NUP358ZnF5/6•Ran(GDP)), 7MNU (NUP358ZnF7•Ran(GDP)), 7MNV (NUP358ZnF8•Ran(GDP)), 7MNW (NUP358RanBD-I•Ran(GMPPNP)), 7MNX (Nup358RanBD-II•Ran(GMPPNP)), 7MNY (NUP358RanBD-III•Ran(GMPPNP)), 7MNZ (NUP358RanBD-IV•Ran(GMPPNP)), 7MO0 (NUP50RanBD•Ran(GMPPNP)), 7MO1 (NUP153ZnF1•Ran(GDP)), 7MO2 (NUP153ZnF2•Ran(GDP)), 7MO3 (NUP153ZnF3•Ran(GDP), 2.05Å), 7MO4 (NUP153ZnF3•Ran(GDP), 2.4Å), 7MO5 (NUP153ZnF4•Ran(GDP)). PyMol and Chimera sessions containing the composite structures of the constricted and dilated human NPCs can be obtained from our webpage (http://ahweb.caltech.edu) and coordinates are deposited in the PDB with accession numbers 7TBL and 7TBM, respectively. Quantitative docking data, workflow code, PyMol and Chimera sessions were deposited on CaltechDATA (101).
REFERENCES AND NOTES
- 1.Hoelz A, Debler EW, Blobel G, The structure of the nuclear pore complex. Annu. Rev. Biochem 80, 613–643 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Lin DH, Hoelz A, The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem 88, 725–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampoelz B, Andres-Pons A, Kastritis P, Beck M, Structure and Assembly of the Nuclear Pore Complex. Annu. Rev. Biophys 48, 515–536 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Knockenhauer KE, Schwartz TU, The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 164, 1162–1171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM, Viral subversion of nucleocytoplasmic trafficking. Traffic 15, 127–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohler A, Hurt E, Gene regulation by nucleoporins and links to cancer. Mol. Cell 38, 6–15 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Cook A, Bono F, Jinek M, Conti E, Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem 76, 647–671 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Pumroy RA, Cingolani G, Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem. J 466, 13–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chook YM, Suel KE, Nuclear import by karyopherin-betas: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D, Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science 350, 106–110 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, Huber FM, Lin DH, Paduch M, Koide A, Lu V, Fischer J, Hurt E, Koide S, Kossiakoff AA, Hoelz A, Architecture of the fungal nuclear pore inner ring complex. Science 350, 56–64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E, Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol 22, 774–781 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Hodel AE, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, Powers MA, The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell 10, 347–358 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Berke IC, Boehmer T, Blobel G, Schwartz TU, Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol 167, 591–597 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsia KC, Stavropoulos P, Blobel G, Hoelz A, Architecture of a coat for the nuclear pore membrane. Cell 131, 1313–1326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU, Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322, 1369–1373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehmer T, Jeudy S, Berke IC, Schwartz TU, Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol. Cell 30, 721–731 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A, A fence-like coat for the nuclear pore membrane. Mol. Cell 32, 815–826 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Leksa NC, Brohawn SG, Schwartz TU, The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 17, 1082–1091 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whittle JRR, Schwartz TU, Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J. Biol. Chem 284, 28442–28452 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A, Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc. Natl. Acad. Sci. U.S.A 106, 14281–14286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brohawn SG, Schwartz TU, Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat. Struct. Mol. Biol 16, 1173–1177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A, Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc. Natl. Acad. Sci. U.S.A 106, 17693–17698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilokapic S, Schwartz TU, Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A 109, 15241–15246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilokapic S, Schwartz TU, Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure 21, 572–580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C, Li Z, He H, Wernimont A, Li Y, Loppnau P, Min J, Crystal structure of human nuclear pore complex component NUP43. FEBS Lett 589, 3247–3253 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A, Nuclear pores. Architecture of the nuclear pore complex coat. Science 347, 1148–1152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU, Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol 22, 425–431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoelz A, Glavy JS, Beck M, Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat. Struct. Mol. Biol 23, 624–630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres-Pons A, Glavy JS, Beck M, Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Handa N, Kukimoto-Niino M, Akasaka R, Kishishita S, Murayama K, Terada T, Inoue M, Kigawa T, Kose S, Imamoto N, Tanaka A, Hayashizaki Y, Shirouzu M, Yokoyama S, The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain. J. Mol. Biol 363, 114–124 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Jeudy S, Schwartz TU, Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J. Biol. Chem 282, 34904–34912 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR, Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell 29, 46–55 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Seo HS, Blus BJ, Jankovic NZ, Blobel G, Structure and nucleic acid binding activity of the nucleoporin Nup157. Proc. Natl. Acad. Sci. U.S.A 110, 16450–16455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, Ketaren NE, Matsui T, Weiss TM, Stokes DL, Sauder JM, Burley SK, Sali A, Rout MP, Almo SC, Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 21, 560–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU, Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. Elife 2, e00745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuwe T, Lin DH, Collins LN, Hurt E, Hoelz A, Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. U.S.A 111, 2530–2535 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, Davenport AM, Jeon YE, Hoelz A, Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, Hurt E, Beck M, Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, Chemmama IE, Pellarin R, Echeverria I, Shivaraju M, Chaudhury AS, Wang J, Williams R, Unruh JR, Greenberg CH, Jacobs EY, Yu Z, de la Cruz MJ, Mironska R, Stokes DL, Aitchison JD, Jarrold MF, Gerton JL, Ludtke SJ, Akey CW, Chait BT, Sali A, Rout MP, Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turonova B, Karius K, Bormel M, Zhang X, Muller CW, Schwab Y, Mahamid J, Pfander B, Kosinski J, Beck M, In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586, 796–800 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A, Architecture of the linker-scaffold in the nuclear pore. Accompanying Manuscript, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart M, Polyadenylation and nuclear export of mRNAs. J. Biol. Chem 294, 2977–2987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, Beck M, In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang G, Zhang Y, Zhu X, Zeng C, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, Shi Y, Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis. Cell Res 30, 520–531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosalaganti S, Obarska-Kosinska A, Siggel M, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull M-T, Hagen W, Hummer G, Beck M, Kosinski J, Artificial intelligence reveals nuclear pore complexity. bioRxiv, 2021.2010.2026.465776 (2021). [Google Scholar]
- 47.Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC, A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J. Cell Biol 130, 1263–1273 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E, Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J 17, 1107–1119 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole CN, Doye V, Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell 9, 3475–3492 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E, Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 18, 4332–4347 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho AK, Shen TX, Ryan KJ, Kiseleva E, Levy MA, Allen TD, Wente SR, Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol 20, 5736–5748 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weirich CS, Erzberger JP, Berger JM, Weis K, The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16, 749–760 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR, Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol 8, 711–716 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Napetschnig J, Blobel G, Hoelz A, Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc. Natl. Acad. Sci. U.S.A 104, 1783–1788 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A, Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc. Natl. Acad. Sci. U.S.A 106, 3089–3094 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Moeller H, Basquin C, Conti E, The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol 16, 247–254 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Ren Y, Seo HS, Blobel G, Hoelz A, Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc. Natl. Acad. Sci. U.S.A 107, 10406–10411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K, A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A, Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A 108, 16571–16576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A, Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex. J. Mol. Biol 419, 330–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaik M, Flemming D, von Appen A, Kastritis P, Mucke N, Fischer J, Stelter P, Ori A, Bui KH, Bassler J, Barbar E, Beck M, Hurt E, Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol 208, 283–297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teimer R, Kosinski J, von Appen A, Beck M, Hurt E, A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat. Commun 8, 1107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin DH, Correia AR, Cai SW, Huber FM, Jette CA, Hoelz A, Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat. Commun 9, 2319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP, Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 167, 1215–1228 e1225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K, Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol 8, 668–676 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Enninga J, Levy DE, Blobel G, Fontoura BM, Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295, 1523–1525 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Forler D, Rabut G, Ciccarelli FD, Herold A, Kocher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E, RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol. Cell. Biol 24, 1155–1167 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutten S, Kehlenbach RH, Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol 26, 6772–6785 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Walde S, Joseph J, Kehlenbach RH, van Deursen JM, Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol 194, 597–612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quan B, Seo HS, Blobel G, Ren Y, Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 * Nup98). Proc. Natl. Acad. Sci. U.S.A 111, 9127–9132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E, Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem 270, 14209–14213 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. , A giant nucleopore protein that binds Ran/TC4. Nature 376, 184–188 (1995). [DOI] [PubMed] [Google Scholar]
- 73.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT, The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol 148, 635–651 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ, Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol 158, 915–927 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Joseph J, Dasso M, The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett 582, 190–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kassube SA, Stuwe T, Lin DH, Antonuk CD, Napetschnig J, Blobel G, Hoelz A, Crystal structure of the N-terminal domain of Nup358/RanBP2. J. Mol. Biol 423, 752–765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paduch M, Koide A, Uysal S, Rizk SS, Koide S, Kossiakoff AA, Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 60, 3–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holm L, Using Dali for Protein Structure Comparison. Methods Mol. Biol 2112, 29–42 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Hampoelz B, Schwarz A, Ronchi P, Bragulat-Teixidor H, Tischer C, Gaspar I, Ephrussi A, Schwab Y, Beck M, Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell 179, 671–686 e617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yaseen NR, Blobel G, Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc. Natl. Acad. Sci. U.S.A 96, 5516–5521 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakielny S, Shaikh S, Burke B, Dreyfuss G, Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 18, 1982–1995 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L, Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol 20, 5619–5630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higa MM, Alam SL, Sundquist WI, Ullman KS, Molecular characterization of the Ran-binding zinc finger domain of Nup153. J. Biol. Chem 282, 17090–17100 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Schrader N, Koerner C, Koessmeier K, Bangert JA, Wittinghofer A, Stoll R, Vetter IR, The crystal structure of the Ran-Nup153ZnF2 complex: a general Ran docking site at the nuclear pore complex. Structure 16, 1116–1125 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Partridge JR, Schwartz TU, Crystallographic and biochemical analysis of the Ran-binding zinc finger domain. J. Mol. Biol 391, 375–389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ori A, Banterle N, Iskar M, Andres-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, Lemke EA, Beck M, Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol 9, 648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salina D, Enarson P, Rattner JB, Burke B, Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol 162, 991–1001 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pereira PD, Serra-Caetano A, Cabrita M, Bekman E, Braga J, Rino J, Santus R, Filipe PL, Sousa AE, Ferreira JA, Quantification of cell cycle kinetics by EdU (5-ethynyl-2’-deoxyuridine)-coupled-fluorescence-intensity analysis. Oncotarget 8, 40514–40532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mahadevan K, Zhang H, Akef A, Cui XA, Gueroussov S, Cenik C, Roth FP, Palazzo AF, RanBP2/Nup358 potentiates the translation of a subset of mRNAs encoding secretory proteins. PLoS Biol 11, e1001545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN, Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol 131, 1677–1697 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neilson DE, Adams MD, Orr CM, Schelling DK, Eiben RM, Kerr DS, Anderson J, Bassuk AG, Bye AM, Childs AM, Clarke A, Crow YJ, Di Rocco M, Dohna-Schwake C, Dueckers G, Fasano AE, Gika AD, Gionnis D, Gorman MP, Grattan-Smith PJ, Hackenberg A, Kuster A, Lentschig MG, Lopez-Laso E, Marco EJ, Mastroyianni S, Perrier J, Schmitt-Mechelke T, Servidei S, Skardoutsou A, Uldall P, van der Knaap MS, Goglin KC, Tefft DL, Aubin C, de Jager P, Hafler D, Warman ML, Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am. J. Hum. Genet 84, 44–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sell K, Storch K, Hahn G, Lee-Kirsch MA, Ramantani G, Jackson S, Neilson D, von der Hagen M, Hehr U, Smitka M, Variable clinical course in acute necrotizing encephalopathy and identification of a novel RANBP2 mutation. Brain Dev 38, 777–780 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Bonnin E, Cabochette P, Filosa A, Juhlen R, Komatsuzaki S, Hezwani M, Dickmanns A, Martinelli V, Vermeersch M, Supply L, Martins N, Pirenne L, Ravenscroft G, Lombard M, Port S, Spillner C, Janssens S, Roets E, Van Dorpe J, Lammens M, Kehlenbach RH, Ficner R, Laing NG, Hoffmann K, Vanhollebeke B, Fahrenkrog B, Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet 14, e1007845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Regmi SG, Lee H, Kaufhold R, Fichtman B, Chen S, Aksenova V, Turcotte E, Harel A, Arnaoutov A, Dasso M, The Nuclear Pore Complex consists of two independent scaffolds. bioRxiv, 2020.2011.2013.381947 (2020). [Google Scholar]
- 95.Zila V, Margiotta E, Turonova B, Muller TG, Zimmerli CE, Mattei S, Allegretti M, Borner K, Rada J, Muller B, Lusic M, Krausslich HG, Beck M, Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032–1046 e1018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schuller AP, Wojtynek M, Mankus D, Tatli M, Kronenberg-Tenga R, Regmi SG, Dip PV, Lytton-Jean AKR, Brignole EJ, Dasso M, Weis K, Medalia O, Schwartz TU, The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamed M, Caspar B, Port SA, Kehlenbach RH, A nuclear export sequence promotes CRM1-dependent targeting of the nucleoporin Nup214 to the nuclear pore complex. J. Cell Sci 134, (2021). [DOI] [PubMed] [Google Scholar]
- 98.Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M, The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol 158, 63–77 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K, Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell 171, 904–917 e919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, Danziger O, White KM, Rathnasinghe R, Uccellini M, Gao S, Aydillo T, Mena I, Yin X, Martin-Sancho L, Krogan NJ, Chanda SK, Schotsaert M, Wozniak RW, Ren Y, Rosenberg BR, Fontoura BMA, Garcia-Sastre A, SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U.S.A 117, 28344–28354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SR, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A, Architecture of the cytoplasmic face of the nuclear pore, Version 1.0, CaltechDATA (2021). doi: 10.22002/D1.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kellner N, Schwarz J, Sturm M, Fernandez-Martinez J, Griesel S, Zhang W, Chait BT, Rout MP, Kuck U, Hurt E, Developing genetic tools to exploit Chaetomium thermophilum for biochemical analyses of eukaryotic macromolecular assemblies. Sci. Rep 6, 20937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fontoura BM, Blobel G, Matunis MJ, A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol 144, 1097–1112 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kendirgi F, Rexer DJ, Alcazar-Roman AR, Onishko HM, Wente SR, Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol. Biol. Cell 16, 4304–4315 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoelz A, Nairn AC, Kuriyan J, Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol. Cell 11, 1241–1251 (2003). [DOI] [PubMed] [Google Scholar]
- 106.Mossessova E, Lima CD, Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Romier C, Ben Jelloul M, Albeck S, Buchwald G, Busso D, Celie PHN, Christodoulou E, De Marco V, van Gerwen S, Knipscheer P, Lebbink JH, Notenboom V, Poterszman A, Rochel N, Cohen SX, Unger T, Sussman JL, Moras D, Sixma TK, Perrakis A, Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. D Biol. Crystallogr 62, 1232–1242 (2006). [DOI] [PubMed] [Google Scholar]
- 108.Mukherjee S, Erramilli SK, Ammirati M, Alvarez FJD, Fennell KF, Purdy MD, Skrobek BM, Radziwon K, Coukos J, Kang Y, Dutka P, Gao X, Qiu X, Yeager M, Eric Xu H, Han S, Kossiakoff AA, Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins. Nat. Commun 11, 1598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Doublie S, Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276, 523–530 (1997). [PubMed] [Google Scholar]
- 110.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A, Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46 (1999). [DOI] [PubMed] [Google Scholar]
- 111.Wyatt PJ, Multiangle light scattering: The basic tool for macromolecular characterization. Instrum. Sci. Technol 25, 1–18 (1997). [Google Scholar]
- 112.Sabath K, Staubli ML, Marti S, Leitner A, Moes M, Jonas S, INTS10-INTS13-INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module. Nat. Commun 11, 3422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miller KR, Koide A, Leung B, Fitzsimmons J, Yoder B, Yuan H, Jay M, Sidhu SS, Koide S, Collins EJ, T cell receptor-like recognition of tumor in vivo by synthetic antibody fragment. PLoS One 7, e43746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mukherjee S, Ura M, Hoey RJ, Kossiakoff AA, A New Versatile Immobilization Tag Based on the Ultra High Affinity and Reversibility of the Calmodulin-Calmodulin Binding Peptide Interaction. J. Mol. Biol 427, 2707–2725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 116.Kabsch W, Xds. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Winter G, Waterman DG, Parkhurst JM, Brewster AS, Gildea RJ, Gerstel M, Fuentes-Montero L, Vollmar M, Michels-Clark T, Young ID, Sauter NK, Evans G, DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol 74, 85–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Evans PR, Murshudov GN, How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vonrhein C, Blanc E, Roversi P, Bricogne G, Automated structure solution with autoSHARP. Methods Mol. Biol 364, 215–230 (2007). [DOI] [PubMed] [Google Scholar]
- 121.Sheldrick GM, A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Grosse-Kunstleve RW, Adams PD, Substructure search procedures for macromolecular structures. Acta Crystallogr. D Biol. Crystallogr 59, 1966–1973 (2003). [DOI] [PubMed] [Google Scholar]
- 123.Terwilliger TC, Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr 56, 965–972 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC, MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 129.Lin DH, Zimmermann S, Stuwe T, Stuwe E, Hoelz A, Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. J. Mol. Biol 425, 1318–1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Flory PJ, Volkenstein M, Statistical mechanics of chain molecules. Biopolymers 8, 699–700 (1969). [Google Scholar]
- 131.Evers TH, van Dongen EM, Faesen AC, Meijer EW, Merkx M, Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry 45, 13183–13192 (2006). [DOI] [PubMed] [Google Scholar]
- 132.Thierbach K, von Appen A, Thoms M, Beck M, Flemming D, Hurt E, Protein interfaces of the conserved Nup84 complex from Chaetomium thermophilum shown by crosslinking mass spectrometry and electron microscopy. Structure 21, 1672–1682 (2013). [DOI] [PubMed] [Google Scholar]
- 133.Waldman T, Kinzler KW, Vogelstein B, p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res 55, 5187–5190 (1995). [PubMed] [Google Scholar]
- 134.Knutsen T, Padilla-Nash HM, Wangsa D, Barenboim-Stapleton L, Camps J, McNeil N, Difilippantonio MJ, Ried T, Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines. Genes Chromosomes Cancer 49, 204–223 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yau KC, Arnaoutov A, Aksenova V, Kaufhold R, Chen S, Dasso M, RanBP1 controls the Ran pathway in mammalian cells through regulation of mitotic RCC1 dynamics. Cell Cycle 19, 1899–1916 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patke A, Murphy PJ, Onat OE, Krieger AC, Ozcelik T, Campbell SS, Young MW, Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell 169, 203–215 e213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ, Multiple sequence alignment with Clustal X. Trends Biochem. Sci 23, 403–405 (1998). [DOI] [PubMed] [Google Scholar]
- 140.Barton GJ, ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6, 37–40 (1993). [DOI] [PubMed] [Google Scholar]
- 141.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Grandi P, Schlaich N, Tekotte H, Hurt EC, Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J 14, 76–87 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Loughlin FE, Mansfield RE, Vaz PM, McGrath AP, Setiyaputra S, Gamsjaeger R, Chen ES, Morris BJ, Guss JM, Mackay JP, The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5’ splice site-like sequences. Proc. Natl. Acad. Sci. U.S.A 106, 5581–5586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Loughlin FE, Lukavsky PJ, Kazeeva T, Reber S, Hock EM, Colombo M, Von Schroetter C, Pauli P, Clery A, Muhlemann O, Polymenidou M, Ruepp MD, Allain FH, The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity. Mol Cell 73, 490–504 e496 (2019). [DOI] [PubMed] [Google Scholar]
- 145.Kuhlmann J, Macara I, Wittinghofer A, Dynamic and equilibrium studies on the interaction of Ran with its effector, RanBP1. Biochemistry 36, 12027–12035 (1997). [DOI] [PubMed] [Google Scholar]
- 146.Drozdetskiy A, Cole C, Procter J, Barton GJ, JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43, W389–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kobayashi W, Takizawa Y, Aihara M, Negishi L, Ishii H, Kurumizaka H, Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding. Commun Biol 2, 163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ, ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 17, 1657–1662 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, Doye V, The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J 26, 1853–1864 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, Kutay U, Antonin W, The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 22, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- 151.Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W, MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8, 165–172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E, Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289 (2011). [DOI] [PubMed] [Google Scholar]
- 153.Reverter D, Lima CD, Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials generated in this study are available on request from the corresponding author. The auxin-inducible AID::NUP358 HCT116, AID::NUP358 DLD1, and NUP160::NG AID DLD1 degron cell lines are subject to a materials transfer agreement, which is available upon request. The coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 7MNJ (NUP358145–673), 7MNK (NUP358OE), 7MNI (NUP88NTD•NUP98APD), 7MNL (NUP358NTD•sAB-14), 7MNM (NUP358NTD T585M•sAB-14), 7MNN (NUP358NTD T653I•sAB-14), 7MNO (NUP358NTD I656V•sAB-14), 7MNP (NUP358ZnF2•Ran(GDP)), 7MNQ (NUP358ZnF2•Ran(GDP)), 7MNR (NUP358ZnF3•Ran(GDP)), 7MNS (NUP358ZnF4•Ran(GDP)), 7MNT (NUP358ZnF5/6•Ran(GDP)), 7MNU (NUP358ZnF7•Ran(GDP)), 7MNV (NUP358ZnF8•Ran(GDP)), 7MNW (NUP358RanBD-I•Ran(GMPPNP)), 7MNX (Nup358RanBD-II•Ran(GMPPNP)), 7MNY (NUP358RanBD-III•Ran(GMPPNP)), 7MNZ (NUP358RanBD-IV•Ran(GMPPNP)), 7MO0 (NUP50RanBD•Ran(GMPPNP)), 7MO1 (NUP153ZnF1•Ran(GDP)), 7MO2 (NUP153ZnF2•Ran(GDP)), 7MO3 (NUP153ZnF3•Ran(GDP), 2.05Å), 7MO4 (NUP153ZnF3•Ran(GDP), 2.4Å), 7MO5 (NUP153ZnF4•Ran(GDP)). PyMol and Chimera sessions containing the composite structures of the constricted and dilated human NPCs can be obtained from our webpage (http://ahweb.caltech.edu) and coordinates are deposited in the PDB with accession numbers 7TBL and 7TBM, respectively. Quantitative docking data, workflow code, PyMol and Chimera sessions were deposited on CaltechDATA (101).


