Fig. 3. Structural analysis and biochemical characterization of NUP358.
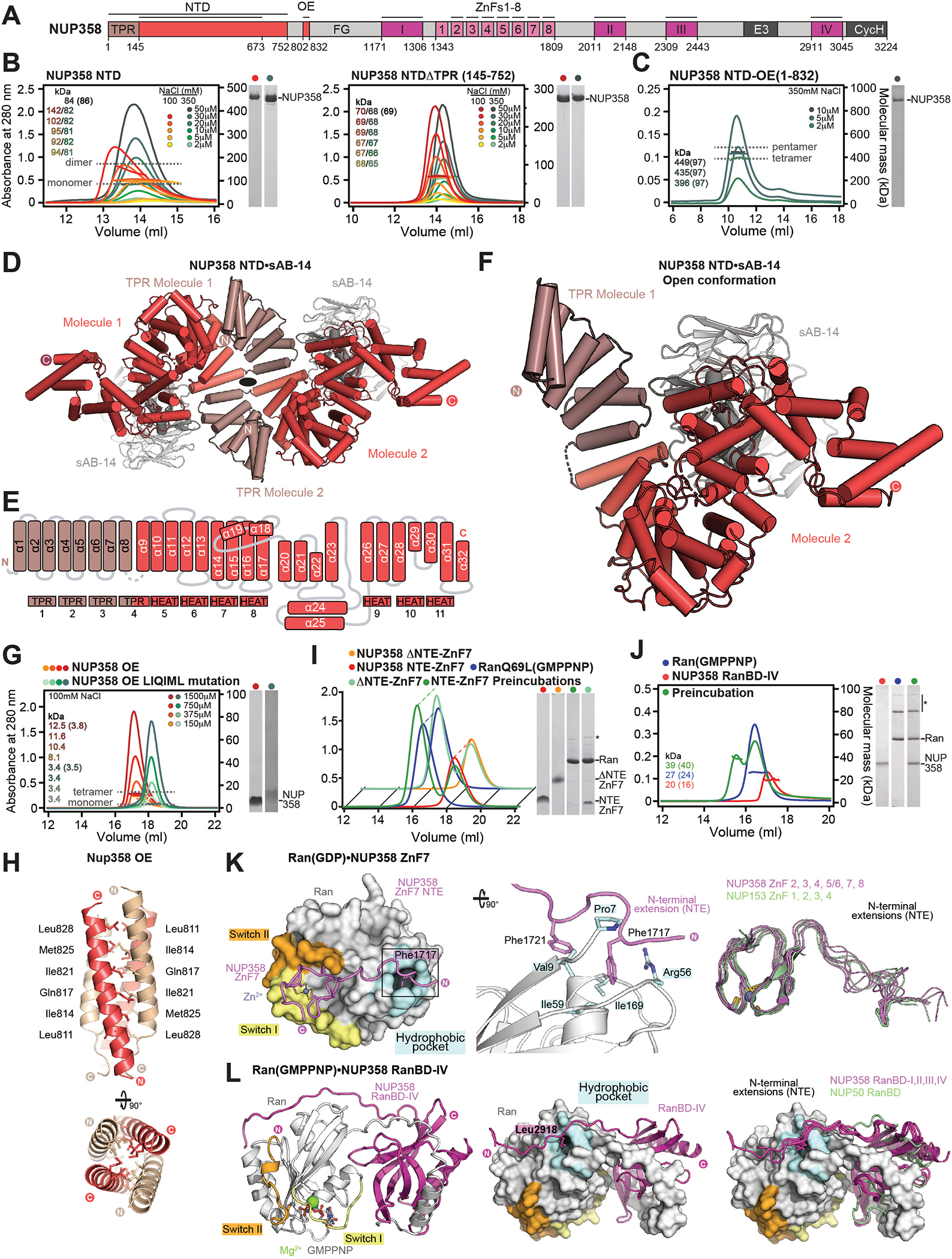
(A) Domain structure of NUP358. Black lines indicate the boundaries of the crystallized fragments. (B, C) SEC-MALS analysis of the oligomeric behavior of (B) NUP358NTD and NUP358NTDΔTPR, and (C) NUP358NTD-OE, performed at the indicated protein concentrations. Measured molecular masses are indicated, with theoretical masses in parentheses. SDS-PAGE gel strips of peak fractions are shown and visualized by Coomassie brilliant blue staining. (D) Cartoon representation of the NUP358NTD•sAB-14 co-crystal structure dyad of the P6522 lattice, illustrating the NUP358NTD dimer between symmetry-related molecules. (E) Schematic of the NUP358NTD structure and structural motifs. (F) TPR of molecule 1 complements α-helical stacking of the N-terminal solenoid of the symmetry-related molecule 2, generating the open conformation of NUP358NTD. (G) SEC-MALS analysis of the oligomeric behavior of NUP358OE and the NUP358OE LIQIML mutant performed at the indicated protein concentrations. (H) Cartoon representation of the homo-tetrameric NUP358OE crystal structure with hydrophobic core residues shown in ball-and-stick representation. (I) SEC interaction analysis of NUP358ZnF7 and NUP358ZnF7ΔNTE binding to Ran(GTP). (J) SEC-MALS interaction analysis of NUP358RanBD-IV binding to Ran(GMPPNP). (K) Co-crystal structure of NUP358ZnF7•Ran(GDP), shown in cartoon and surface representation (left). The inset indicates the location of the magnified and 90° rotated view of the Ran hydrophobic pocket (middle). Superposition of the six NUP358ZnF•Ran(GDP) and four NUP153ZnF•Ran(GDP) co-crystal structures with the Zn2+-coordinating cysteines and Ran-burying NTE hydrophobic residues shown as sticks (right). (L) Co-crystal structure of NUP358RanBD-IV•Ran(GMPPNP) with NUP358RanBD-IV shown in cartoon and Ran(GMPPNP) shown in cartoon (left) or surface (middle) representation. Superposition of Ran(GMPPNP) bound to NUP358RanBD-I, NUP358RanBD-II, NUP358RanBD-III, NUP358RanBD-IV, and NUP50RanBD (right).
