Abstract
A comparison of the spectra of spontaneous growth-dependent and adaptive mutations in ebgR shows that both spectra are dominated by insertion sequence (IS)-mediated mutations. The difference between growth-dependent mutations (61% IS mediated) and adaptive mutations (80% IS mediated) is highly significant (P < 0.0001). In contrast, the spectra of growth-dependent and adaptive non-IS-mediated mutations do not differ from each other and therefore do not provide support for the hypothesis that adaptive and growth-dependent mutations arise by substantially different mechanisms.
Adaptive mutations are spontaneous mutations that occur in microorganisms during periods of prolonged stress in nondividing (2) or very slowly dividing (8) populations. The key feature that distinguishes adaptive mutations from growth-dependent mutations is that the former are specific to the selective challenge that is imposed, i.e., the selective conditions are not generally mutagenic and the only mutations that are recovered are mutations in the site that is under selection (10, 12) (but see reference 3 for an exception to this rule). The phrase “only mutations that are recovered” was chosen carefully to avoid implying that other mutations do not occur. A variety of experiments have looked very hard for mutations at sites that are not under selection and have failed to find such mutations (9, 10), but that does not rule out the possibility that mutations occur but are lost before they can be recovered. One possibility that has gained increasing acceptance over the last few years is that mutations do occur at other sites, but that the cells that suffer those mutations die (because the mutations confer no advantage), and thus the mutations are not recovered (6).
Adaptive mutations, which have been shown to occur in both bacteria and yeast (reviewed in reference 12), are usually observed by subjecting populations to nonlethal selection for reversion of known mutations in genes for carbon source catabolism or for amino acid biosynthesis. The first revertants to appear are presumed to be the result of mutations that were present in the population prior to plating. Typically, additional revertant colonies continue to appear for periods ranging from a few days up to a month, and it is those late-appearing colonies that are said to result from adaptive mutations.
Some of the most important evidence that adaptive mutagenesis is a different process from growth-dependent mutagenesis comes from showing that the spectra of adaptive mutations differ significantly from the spectra of growth-dependent mutations. A series of F′-borne lacZ alleles that could revert only by specific base substitutions was used to show that the base substitution spectra were different (7), and both Foster and Trimarchi (4) and Rosenberg et al. (19) used the F′-borne lacI33 allele to show that the frameshift-reversion spectra were different. There are a number of reasons, reviewed in reference 12, to suspect that adaptive reversion of F′-borne alleles may be a special case. Because of concerns about the generality of conclusions based on studies of F′-borne reporter alleles, it is important to compare the mutagenic spectra of growth-dependent and adaptive mutations in a chromosomal gene.
Reversion systems are unsatisfactory for determining mutational spectra because, for any given mutation, there are a very limited number of sites and kinds of mutations that will produce a revertant phenotype. Some kinds of mutational events, such as insertion of insertion sequence (IS) elements, are completely excluded from studies that employ reversion systems. Because no constraints are placed on the specific nature of the mutations, it is much more useful to select for loss of function in the target gene than to select for reversion. The lacI gene has served as a very powerful system both for studying spontaneous mutagenesis and for helping to elucidate the functions of the methyl-directed mismatch repair system (14, 20–22). lacI mutants allow constitutive expression of lacZY and thus permit growth on phenyl-β-galactoside, which is a substrate for β-galactosidase, but is not an inducer of the lac operon. Aside from a four-base repeat hot spot for frameshifts that causes 70% of spontaneous lacI mutations, of the remainder about 35% are deletions, 39% are base substitutions, 3.9% result from insertion elements, 18% from single-base frameshifts, and 3.5% from duplications (14).
Over the long periods of selection required for adaptive mutagenesis studies, the basal level of lacZ-encoded β-galactosidase generates considerable background growth (data not shown); thus the lacI system is not suitable as a reporter for adaptive mutagenesis. Adaptive mutations have proven readily detectable in the Ebg repressor gene ebgR (8), and adaptive ebgR mutants accumulated continuously over a period of 14 days. The ebgAC-encoded β-galactosidase does not generate significant background growth. Like lacI, ebgR specifies a repressor that controls expression of an adjacent β-galactosidase gene, and the two genes have 25% homology at the amino acid level (11). Beyond the issue of adaptive mutations, a comparison of the spontaneous mutation spectra of these similar genes could provide insight into the generality of the very detailed spectra that have been generated for lacI. I have therefore compared the spectra of forward growth-dependent and adaptive mutations in ebgR.
MATERIALS AND METHODS
Escherichia coli strains.
Strain SJ134 (11) is F− ΔlacZ4680 ebgA51 ebgR+ rpsL. Strain SJ2 is F− ΔlacZ4680 lacY+ ebgR+ ebgA+ rpsL metC.
Transductions.
Transductions were mediated by bacteriophage P1vir as described by Miller (17).
Media.
Mineral salts (MS) medium consisted of 423 mg of sodium citrate, 100 mg of MgSO4 · 7H2O, 1 g of (NH4)2SO4, 540 μg of FeCl3, 1 mg of thiamine, 3 g of KH2PO4, and 7 g of K2HPO4 per liter, plus a carbon source. Lactulose selection medium was MS medium containing 1 g of lactulose per liter, 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to induce expression of the lacY-encoded β-galactoside permease and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (20 mg/liter), a noninducing substrate that produces an intense blue color (to facilitate the detection of colonies). Limiting glycerol medium was MS medium containing 0.01% (vol/vol) glycerol and 0.2 mM IPTG. X-Gal medium was MS medium containing 0.2% (vol/vol) glycerol, 0.1% (wt/vol) Casamino Acids (Difco), 20 mg of X-Gal/liter, and 0.2 mM IPTG.
L agar consisted of Luria-Bertani-agar (17) plus 1 g of glucose per liter. Solid media were solidified with Sigma Purified Agar. MacConkey lactulose indicator medium was prepared from MacConkey agar base (Difco) and lactulose according to the manufacturer’s instructions and included 0.2 mM IPTG. ebgR colonies of strain SJ134 are red on MacConkey lactulose plates, while wild-type colonies are white.
PCR amplification of ebgR.
Amplifications for initial characterization of mutants used primers 2 and 7 (Fig. 1) and 2.5 μl of crude genomic DNA in a 25-μl reaction that contained 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates (dNTP), 0.5 U of Taq polymerase (Gibco), and 2.5 μl of 10× PCR buffer (Gibco). Crude genomic DNA was prepared by suspending 600 μl of an overnight L-broth-grown culture in 50 μl of sterile water, heating the suspension in a sealed Microfuge tube at 100°C for 15 min, centrifuging at 4°C for 15 min in a Microfuge, and transferring the supernate to a fresh tube for storage. PCR products for DNA sequencing were amplified from genomic DNA prepared by the cetyltrimethylammonium bromide method (1) using either primers 2 and 7 or, when sequencing failed to reveal any differences from the canonical sequence, primers 1 and 8 in a 100-μl reaction that contained 100 ng of genomic DNA, 1.5 mM MgCl2, 200 μM each dNTP, 2 U of either AmpliTaq Gold (Perkin-Elmer) or Qiagen Taq polymerase, and 10 μl of corresponding 10× PCR buffer. PCR products used for DNA sequencing were purified by the QiaQuick (Qiagen) method.
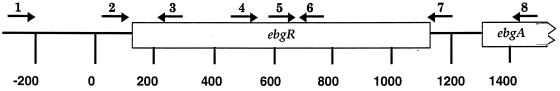
FIG. 1.
Oligonucleotide primers used for amplification and sequencing. Base numbering is according to the published sequence of the ebg operon (13) (GenBank accession no. M64441) except for primer 1, which is numbered according to GenBank accession no. AE000389. The ebgR coding region begins at bp 126 and ends at bp 1109 The locations of the primers are as follows: primer 1, bp 4501 to 4525; primer 2, bp 73 to 94; primer 3, complement of bp 219 to 243; primer 4, bp 473 to 497; primer 5, bp 624 to 660; primer 6, complement of bp 660 to 684; primer 7, complement of bp 1144 to 1168; primer 8, complement of bp 1435 to 1459.
DNA sequencing.
Purified PCR products were sequenced by cycle sequencing using the primers shown in Fig. 1 and an ABI kit according to the manufacturer’s instructions. Sequencing products were separated and analyzed on an ABI model 377 automated DNA sequencer.
RESULTS
Kinetics of adaptive mutations in ebgR.
Strain SJ134 carries the ebgA51 allele, which encodes a mutant Ebg β-galactosidase that hydrolyzes lactulose effectively, but that strain cannot utilize lactulose as a carbon source because lactulose is not an effective inducer of the ebg operon. Mutations in ebgR, which permit constitutive expression of the ebg operon, allow ebgA51 strains to utilize lactulose effectively.
To monitor the accumulation of ebgR mutants, on day 0 approximately 107 SJ134 cells were spread onto lactulose selection plates, and the plates were incubated at 30°C. On days 1 through 4 and on day 7, cells were washed from two plates, suitably diluted, and plated onto L agar to determine the number of viable cells. Once ebgR colonies began to appear, they were immediately eliminated from a subset of plates, with little disturbance of surrounding cells, through the use of a diathermy probe (Hyfrecator Plus model 7-796; Birtcher Medical Supplies), an electrosurgery device that delivers an intense spark which kills the cells in the colony. Those treated plates were then used to determine the number of viable cells. During the first few hours after plating, the populations grew from 2.4 × 107 to 2.3 × 108 cells at the expense of trace contaminants in the medium. After day 1, the population declined, with a death rate of −0.41 day−1.
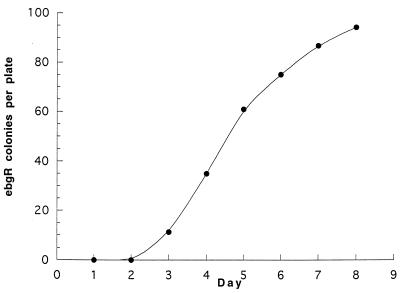
Figure 2 shows the accumulation of ebgR mutants over the course of 7 days. Reconstruction experiments have shown that >98% of preexisting ebgR mutants of strain SJ134 form visible colonies in 3 days under these conditions (11), which is consistent with the first appearance of ebgR colonies on day 3. Based on the assumption that colonies appeared 3 days after the ebgR mutations occurred, the adaptive mutatation rate on any given day is the average number of mutants that appeared 3 days later per plate divided by the average number of viable cells per plate. The average adaptive mutation rate to ebgR was (1.9 ± 0.24) × 10−7 mutations per cell per day.
FIG. 2.
Accumulation of ebgR mutants on lactulose selection medium. Data along the ordinate are the average numbers of mutant colonies per plate based on counting at least 10 plates each day.
Isolation of growth-dependent and adaptive ebgR mutants.
A total of 144 independent 0.2-ml cultures of SJ134 were grown from inocula of about 2 × 103 cells at 30°C in limiting glycerol medium to ensure that all cultures were at the same density (about 108 cells per ml). Next, 127 of the 0.2-ml cultures were spread onto lactulose selection plates, and the remainder were used to determine that the average number of cells per culture was 2.4 × 107. The lactulose selection plates were incubated at 30°C, and the colonies on each plate were counted and marked on day 3. Those early-arising colonies resulted from growth-dependent mutations that occurred either during the growth of the cultures prior to plating or during growth at the expense of trace contaminants to a final density of 2.7 × 108 cells per plate. This selection constituted a fluctuation test (16), and the growth-dependent mutation rate to ebgR was estimated from the distribution of the number of ebgR colonies per plate on day 3 according to the method of Stewart et al. (23) as implemented by Stewart’s DataFit program. That program estimates separately the average number of mutations that occurred prior to plating and the average number that occurred after plating. In this experiment, there was an average of 1.52 mutations per culture prior to plating and 1.3 mutations per culture after plating. Dividing the average number of mutations per culture by the number of cells per culture at the time of plating yielded the estimated growth-dependent mutation rate to ebgR of 6.3 × 10−8 per cell division.
To avoid isolating sibling mutants, one early-arising (day-3) mutant was isolated from each of 100 lactulose selection plates by restreaking onto X-Gal medium.
The incubation of lactulose selection plates was continued at 30°C, and late-arising colonies were marked on days 4, 5, and 6. One day-5 and one day-6 colony were isolated from each of 100 lactulose selection plates and restreaked onto X-Gal medium. Those were designated as late-arising mutants.
All mutants were grown in L broth from single colonies and stored at −80° in 7% dimethyl sulfoxide.
If the spectra of growth-dependent mutations (early-arising mutants) and adaptive mutations (late-arising mutants) are to be compared, it is important to be confident that the sample of late-arising mutants does not include growth-dependent mutations in which the ebgR individual either grew slowly or lagged before starting to grow. Bacteriophage P1vir lysates were prepared from four of the ebgR mutants, including two missense mutants, one nonsense mutant, and one frameshift mutant. The lysates were used to transduce strain SJ2 (ebgR+ ebgA+), and the transduced cells were spread onto lactulose selection plates that had previously been scavenged with about 108 strain SJ2 cells to eliminate trace contaminants that would allow the transduced population to grow at the expense of nutrients other than lactulose. Strain SJ2 cannot easily mutate to lactulose utilization because it makes both the functional ebg repressor, which does not respond to lactulose as an inducer, and the wild-type Ebg enzyme, which cannot hydrolyze lactulose effectively. Indeed, SJ2 control cells that were not transduced with P1vir produced no colonies on the lactulose selection plates over the course of this experiment. Transduction of an ebgR allele into an ebgR+ cytoplasm mimics the situation when a new ebgR allele arises by mutation. An average of 191 ebgR transductants were obtained from each of the four donors. During incubation at 30°C, no colonies appeared on day 1 after the transductions, <1% of the final total of colonies appeared on day 2, >98% appeared on day 3, <1% appeared on day 4, and no additional colonies appeared after day 4. This result shows that newly arisen ebgR mutations do produce colonies on the lactulose-selective plates within 3 days and indicates that the sample of late-arising mutants isolated on days 5 and 6 was unlikely to have included any growth-dependent mutations.
Initial characterization of mutants.
PCR amplification products generated by using primers 2 and 7 (Fig. 1) were resolved on 0.7% Tris-borate-EDTA agarose gels. Gels were stained with Syber Green, and the sizes of the PCR products were determined by comparison with a 1-kb ladder (Gibco) on the same gel. The wild-type PCR product of amplification with primers 2 and 7 is 1,096 bp. Mutants whose PCR products were detectably larger fell into two size classes, those whose PCR product was about 2 kb and those whose PCR product was about 2.5 kb. The 2-kb product was likely to have arisen from insertion of the 768-bp IS1 insertion element, while the larger PCR product was likely to have arisen from insertion of other IS elements, all of which are in the 1.2- to 1.5-kb range.
Distribution of IS-mediated mutations.
Several of the putative IS1 insertion mutants were sequenced with primers 2 and/or 7 and proved to contain authentic IS1 insertions. Putative IS1-mediated mutants were amplified using a three-primer cocktail consisting of primer 2 (Fig. 1) and primers corresponding to bp 685 to 709 and the complement of bp 53 to 77 of IS1. Because amplification was with primer 2 and one of the two outward-amplifying IS1 primers the position, but not the orientation, of the IS1 insertion element could be determined. Over 95% of the putative IS1 insertions were authentic, and the remainder proved to have been misidentified on the basis of anomalously migrating bands in the initial determinations.
Several of the mutants whose PCR products were about 2.5 kb were sequenced with primers 2 and/or 7 and all proved to contain IS30 insertions. The remainder of the putative IS30 insertion mutants were amplified with primer 2 and a primer corresponding to bp 1043 to 1067 of IS30. Those that failed to produce a PCR product in that reaction were amplified with the IS30 primer and primer 7. Those reactions allowed the positions of the remaining IS30 insertions to be determined.
The remainder of the putative insertion element mutants were sequenced with primer 2 and/or primer 7 to determine the identity and position of the element.
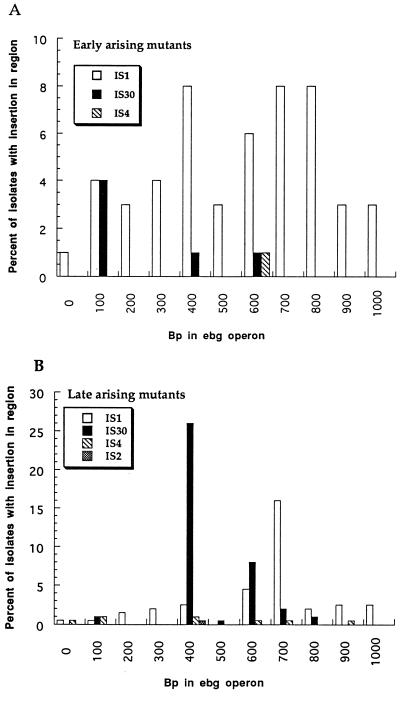
Among the 100 early-arising mutants there were 54 IS1, 1 IS4, and 6 IS30 insertions. Their positions are shown in Fig. 3.
FIG. 3.
Distribution of IS elements among early-arising and late-arising ebgR mutants. Data along the abscissa are positions in the ebg operon numbered according to reference 13 (GenBank accession no. M64441). Data along the ordinate are the percentages of total isolates with that insertion element in the indicated 100-bp interval. Note the difference in scale of the ordinates of panel A and panel B.
Among the 200 late-arising mutants, one which consistently produced two bands upon amplification with primers 2 and 7 and three which could not be amplified were not further considered. Among the remaining 196 late-arising mutants, there were 70 IS1, 78 IS30, 1 IS2, and 8 IS4 insertions. Over half of the late-arising IS30 insertions were in the 400- to 500-bp region (Fig. 3); therefore four of those mutants were sequenced. All proved to have IS30 inserted at bp 472. The region centered at bp 472 matches the consensus target for IS30 insertion (18) at 20 of the 24 bp, and that region is the best match to the consensus found within ebgR. Thus, it appears that for late-arising mutants that site is a hot spot for IS30 insertion.
Sequence changes in non-IS-mediated mutants.
The remaining sample of 39 early-arising and 39 late-arising mutants that were not mediated by insertion elements was not large enough for the comparison of mutational spectra; thus another 100 early-arising and 200 late-arising mutants were isolated as described above. As before, they were amplified with primers 2 and 7 to identify those that were caused by insertion of IS elements, but no attempt was made to identify those elements or the positions of the insertions. In total, an additional 34 early-arising and 44 late-arising non-IS-mediated mutants were isolated.
Sequence changes in those mutants were determined by sequencing with the primers shown in Fig. 1. Mutations were identified by aligning the sequences to bp 73 to 1168 of the wild-type sequence (GenBank accession no. M64441). Base changes were accepted as authentic only if they were found on both strands, but regions in which the sequence was unambiguous and did not differ from the wild-type sequence were not necessarily completely sequenced on both strands.
In a few cases, the sequence changes could not be identified, although the mutants were clearly phenotypically ebgR. In those cases, the region extending from the 234 bp 5′ to ebgR through the first 142 bp of ebgA was amplified with primers 1 and 8 and sequenced as above.
Table 1 and Fig. 4 show the distribution of non-IS-mediated growth-dependent (early-arising) and adaptive (late-arising mutations) in ebgR. These include two multiple mutations, a deletion of G393 and C397→A, and AA1211–1212→GC. There were four duplications ranging from 2 to 5 bp, one of which was an imperfect duplication (CCTA instead of CCTG).
TABLE 1.
Sequence analysis of spontaneous mutations in ebgR
| Growth-dependent mutations
|
Adaptive mutations
|
|||
|---|---|---|---|---|
| Mutationa | Result | Mutation | Result | |
| C130→A | Ala→Glu | A126→C | Met→Leu | |
| C130→A | Ala→Glu | Δ130–142 | ||
| A138→G | Lys→Glu | T136→G | Leu→Arg | |
| A144→T | Ile→Phe | C157→A | Ala→Asp | |
| C148→A | Ala→Glu | C157→A | Ala→Asp | |
| ΔC151 | Frameshift | C157→A | Ala→Asp | |
| G153→T | Ochre | T165→C | Ser→Pro | |
| T165→C | Ser→Pro | T177→A | Val→Glu | |
| C172→A | Ala→Glu | A183→T | Arg→Cys | |
| C172→A | Ala→Glu | A183→T | Arg→Cys | |
| T178→G | Val→Gly | G184→A | Arg→Lys | |
| T180→A | Ser→Thr | G184→A | Arg→Lys | |
| C181→A | Ser→Tyr | A199→C | Asp→Ala | |
| C181→T | Ser→Phe | T200→G | Asp→Glu | |
| G184→A | Arg→Lys | T200→G | Asp→Glu | |
| T187→G | Val→Gly | A210→G | Asn→Asp | |
| A191→C | Leu→Phe | A210→G | Asn→Asp | |
| A199→C | Asp→Ala | G213→A | Val→Met | |
| A210→G | Asn→Asp | T insertion after 220 | Frameshift | |
| G213→A | Val→Met | C226→A | Thr→Lys | |
| G219→T | Ochre | G243→T | Amber | |
| C250→A | Ala→Asp | G243→T | Amber | |
| ΔG260 | Frameshift | G243→T | Amber | |
| C266→A | Ochre | G252→T | Ochre | |
| C266→G | Amber | C266→A | Ochre | |
| C266→G | Amber | A285→T | Ochre | |
| C309→T | Ochre | C325→A | Ala→Asp | |
| C309→T | Ochre | C332→A | Ochre | |
| C338→G | Amber | G382→T | Arg→Leu | |
| G351→T | Amber | G403→T | Cys→Phe | |
| Δ368–381 | G403→T | Cys→Phe | ||
| C368→A | Ochre | C404→A | Opal | |
| ΔG393 and C397→A | Frameshift | A462→T | Ochre | |
| C404→A | Opal | ΔC520 | Frameshift | |
| G405→T | Ochre | ΔC520 | Frameshift | |
| ΔG413 | Frameshift | ΔC520 | Frameshift | |
| T451→A | Ochre | T543→C | Cys→Arg | |
| T451→A | Ochre | G582→A | Ala→Thr | |
| T451→G | Opal | G615→T | Ochre | |
| Δ454–463 | Δ622–632 | |||
| A462→T | Ochre | C639→T | Amber | |
| Δ485–495 | C639→T | Amber | ||
| C578→A | Ochre | C639→T | Amber | |
| ΔC583 | Frameshift | C639→T | Amber | |
| G615→T | Ochre | C639→T | Amber | |
| C639→T | Amber | 2-bp duplication after 647 | Frameshift | |
| C639→T | Amber | G678→T | Amber | |
| C639→T | Amber | T695→G | Asp→Glu | |
| T insertion after 686 | Frameshift | 5-bp duplication after 715 | Frameshift | |
| G702→T | Amber | ΔT722 | Frameshift | |
| G702→T | Amber | C735→T | Ochre | |
| G717→T | Ochre | T742→G | Val→Gly | |
| Duplication 760–811 | G750→T | Amber | ||
| G761→A | Opal | Δ760–809 | ||
| Δ767–818 | G761→A | Opal | ||
| G792→A | Ochre | G766→C | Gly→Ala | |
| ΔA872 | Frameshift | T insertion after 769 | Frameshift | |
| Δ904–918 | G769→T | Gly→Val | ||
| ΔT917 | Frameshift | C775→T | Ser→Phe | |
| C919→A | Amber | C784→A | Amber | |
| C919→A | Amber | G819→T | Ochre | |
| Δ936–1019 | T insertion after 824 | Frameshift | ||
| C951→T | Opal | T838→C | Leu→Pro | |
| G993→T | Amber | G852→T | Asp→Tyr | |
| G993→T | Amber | T865→C | Ile→Ser | |
| G1002→A | Gly→Arg | G868→A | Gly→Asp | |
| G1003→A | Gly→Glu | G868→T | Gly→Val | |
| G1012→A | Gly→Ser | C929→A | Ser→Arg | |
| ΔA1017 | Frameshift | Δ931–945 | ||
| 4-bp duplication after 1018 | A937→G | Asp→Gly | ||
| Δ1019–1022 | T956→A | Phe→Leu | ||
| Δ1047–1048 | G993→T | Amber | ||
| Δ1083–1092 | G993→T | Amber | ||
| G993→T | Amber | |||
| G993→T | Amber | |||
| G1003→A | Gly→Glu | |||
| A insertion after 1029 | Frameshift | |||
| ΔGG 1044–1045 | ||||
| A1080→T | Ochre | |||
| T1084→A | Ochre | |||
| ΔG1095 | Frameshift | |||
| ΔG1095 | Frameshift | |||
| C1099→G | Thr→Arg | |||
| AA1211,1212→GC | Ochre | |||
Numbers refer to positions in the ebg operon as numbered in GenBank accession no. M64441.
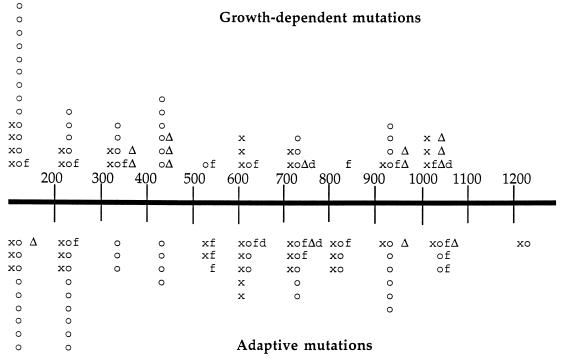
FIG. 4.
Distribution of non-IS-mediated mutations. The scale indicates base pair numbers according to reference 13 (GenBank accession no. M64441). Symbols above the scale indicate growth-dependent mutations, and those below the scale indicate adaptive mutations. Symbols: x, transition; o, transversion; f, single base insertion or deletion frameshifts; d, multibase duplication; Δ, multibase deletion. Positions of symbols indicate sites of mutations within 100-bp region.
This study adds to previous knowledge of the ebg operon. Three insertion mutations, involving IS1 at bp 57, IS1 at bp 76, and IS4 at bp 75, permitted identification of a regulatory region 70 bp upstream of the EbgR coding region.
In three of the mutants, instead of being constitutive for expression of Ebg, the repressor had become sensitive to lactulose as an inducer. Nine such mutants have previously been identified (5), and the mutations responsible have been identified as resulting in amino acid substitutions Asp190→Glu, Ala195→Thr, and Phe196→Cys (13). The two novel mutants resulted from Gly215→Val and Ser217→Phe replacements. This observation expands the sugar-binding domain to the 188- to 217-amino-acid region of the repressor.
One mutation, AA1211–1212→GC, is well outside of the ebgR coding region and is probably in the ebgA operator. The region from bp 1205 to 1225, half of which overlaps the previously identified −35 region of the promoter, forms a perfect palindrome and is probably the operator.
DISCUSSION
Both the spectra of growth-dependent (early-arising) and adaptive (late-arising) mutations in ebgR are clearly dominated by IS-mediated mutations. Although domination by IS-mediated mutations is not the case for spontaneous lacI mutations (14), where only about 4% are IS mediated, IS element domination of the mutation spectrum has also been reported for spontaneous mutations in tonB (15). There is a highly significant difference between the growth-dependent mutants, of which only 60% are IS mediated, and the adaptive mutants, of which 80% are IS mediated (P < 0.0001 by Fisher’s exact test of 2 × 2 contingency tables). There is also a highly significant difference between early-arising and late-arising mutations in terms of the relative frequencies with which the different IS elements cause those mutations (P < 0.0001 by the likelihood ratio [G test] of contingency tables).
The large contribution of IS30 insertions at the hot spot in late-arising mutants raises the question of whether that hot spot alone accounts for those highly significant differences. It does not. Elimination of those mutants with IS30 inserted at the hot spot still results in a highly significant difference (P = 0.0065) between early and late arising in terms of the relative frequencies with which the different IS elements cause those mutations, and a significant difference (P = 0.04) between growth-dependent and adaptive mutants in terms of the contributions of IS elements to the mutational spectrum. It is emphasized that there is neither a good statistical reason nor a good biological reason to eliminate those hot spot IS30 mutants from consideration.
Despite these differences, the distribution of IS-mediated mutations may tell us more about IS biology in starving cells relative to growing cells than it does about adaptive mutagenesis in general.
Table 2 compares the spectra of growth-dependent (early-arising) and adaptive (late-arising) mutations. There is no significant difference between the spectra of growth-dependent and adaptive non-IS-mediated ebgR mutations (P = 0.66 by likelihood ratio [G test] of contingency tables). This result sharply contrasts with the results of earlier studies that employed F′-borne targets for mutagenesis (4, 7, 19) and suggests that concerns about the generality of conclusions from F′-borne reporter genes are well justified.
TABLE 2.
Comparison of spectra of growth-dependent and adaptive non-IS-mediated mutationsa
| Category | Growth-dependent (early-arising) mutations | Adaptive (late-arising) mutations |
|---|---|---|
| A:T→G:C | 3 | 8 |
| G:C→A:T | 14 | 15 |
| G:C→T:A | 24 | 27 |
| G:C→C:G | 3 | 2 |
| A:T→C:G | 5 | 8 |
| A:T→T:A | 5 | 8 |
| −1 base frameshifts | 8 | 6 |
| +1 base frameshifts | 1 | 4 |
| Duplications | 2 | 2 |
| Deletions | 9 | 5 |
| Total | 74 | 85 |
In each of the two double mutants the two mutations were counted as independent events in this table.
The lacI gene is the most thoroughly studied gene in E. coli with respect to mutational spectra. Several studies have used the entire lacI gene, in which 59% of the mutations occur by frameshifts at one hot spot, as a target (14). If the spectra reported for lacI reflect general spontaneous mutation processes, then spontaneous mutations in other genes should exhibit spectra that do not differ significantly from the lacI spectra. Table 3 compares the spontaneous spectrum of lacI mutations (excluding the frameshift hot spot) with the spectrum of ebgR mutations (growth-dependent and adaptive mutations combined). The spectra are highly significantly different (P < 0.0001 by the likelihood ratio test). Transitions dominate the base substitutions in lacI, while transversions dominate in ebgR. Like ebgR, lacI specifies a repressor that controls expression of an adjacent β-galactosidase gene, and indeed the two genes share 25% homology at the amino acid level (13). The spectra of spontaneous mutations at lacI and ebgR differ significantly, and we do not know which, if either, spectrum is typical, but the difference between the spectra of these reasonably similar genes suggests that we should be cautious when generalizing about spontaneous mutational spectra on the basis of even extremely thorough and detailed studies of one gene.
TABLE 3.
Comparison of spectra of non-IS-mediated spontaneous mutations in ebgR and lacI
It is almost certainly not the case that there is a single mechanism for adaptive mutagenesis, any more than there is a single mechanism for DNA repair. At this time, the weight of the evidence indicates that (i) adaptive mutagenesis of F′-borne genes and chromosomal genes is dominated by different processes, and (ii) growth-dependent and adaptive mutagenesis of F′-borne genes are dominated by different processes. The finding that the spectra of non-IS-mediated growth-dependent and adaptive mutations do not differ fails to provide evidence for different processes acting at the chromosomal locus ebgR. In order to determine whether replicon location is a major factor, it will be necessary to compare the spectra of adaptive mutations at the same locus on F′ and on the chromosome.
ACKNOWLEDGMENTS
This study was supported by grant NP-932 from the American Cancer Society.
I am grateful to Jacqueline Toner for expert technical assistance and to George Kampo of the University of Rochester Core Nucleic Acid Facility for his expert advice and helpfulness.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 2.Foster P L. Adaptive mutation: the uses of adversity. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster P L. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster P L, Trimarchi J M. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall B G. Regulation of newly evolved enzymes. IV. Directed evolution of the ebg repressor. Genetics. 1978;90:673–691. doi: 10.1093/genetics/90.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall B G. Spontaneous point mutations that occur more often when they are advantageous than when they are neutral. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall B G. Spectrum of mutations that occur under selective and non-selective conditions in E. coli. Genetica. 1991;84:73–76. doi: 10.1007/BF00116545. [DOI] [PubMed] [Google Scholar]
- 8.Hall B G. Adaptive mutations in E. coli as a model for the multiple-mutational origins of tumors. Proc Natl Acad Sci USA. 1995;92:5669–5673. doi: 10.1073/pnas.92.12.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall B G. Genetics of selection-induced mutations. I. uvrA, uvrB, uvrC, and uvrD are selection-induced specific mutator loci. J Mol Evol. 1995;40:86–93. doi: 10.1007/BF00166599. [DOI] [PubMed] [Google Scholar]
- 10.Hall B G. On the specificity of adaptive mutations. Genetics. 1997;145:39–44. doi: 10.1093/genetics/145.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall B G. Adaptive mutagenesis at ebgR is regulated by PhoPQ. J Bacteriol. 1998;180:2862–2864. doi: 10.1128/jb.180.11.2862-2865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall B G. Adaptive mutagenesis: a process that generates almost exclusively beneficial mutations. Genetica. 1998;102/103:109–125. [PubMed] [Google Scholar]
- 13.Hall B G, Betts P W, Wootton J C. DNA sequence analysis of artificially evolved ebg enzyme and ebg repressor genes. Genetics. 1989;123:635–648. doi: 10.1093/genetics/123.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halliday J A, Glickman B W. Mechanisms of spontaneous mutation in DNA repair-proficient Escherichia coli. Mut Res. 1991;250:55–71. doi: 10.1016/0027-5107(91)90162-h. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura K, Torii Y, Matsuoka C, Yamamoto K. DNA sequence changes in mutations in the tonB gene on the chromosome of Escherichia coli K12: insertion elements dominate the mutational spectra. Jpn J Genet. 1995;70:35–46. doi: 10.1266/jjg.70.35. [DOI] [PubMed] [Google Scholar]
- 16.Luria S E, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Olasz F, Farkas T, Kiss J, Arini A, Arber W. Terminal inverted repeats of insertion sequence IS30 serve as targets for transposition. J Bacteriol. 1997;179:7551–7558. doi: 10.1128/jb.179.23.7551-7558.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg S M, Longerich S, Gee P, Harris R S. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 20.Schaaper R M, Danforth B N, Glickman B W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986;189:273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- 21.Schaaper R M, Dunn R L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci USA. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaaper R M, Dunn R L. Spontaneous mutations in the Escherichia coli lacI gene. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart F M, Gordon D M, Levin B R. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics. 1990;124:175–185. doi: 10.1093/genetics/124.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]