Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is one of seven coronaviruses known to infect humans. Different from other concerned coronavirus and influenza viruses, SARS‐CoV‐2 has a higher basic reproduction number and thus transmits more efficiently among hosts. Testing animals for SARS‐CoV‐2 may help decipher virus reservoirs, transmission and pathogenesis. Here, we report the first detection of SARS‐CoV‐2 in three snow leopards (Panthera uncia) in a zoo in Kentucky in 2020, the first year of the pandemic. Sequence analysis revealed that snow leopard SARS‐CoV‐2 strains were non‐variant B.1.2 lineage and closely correlated with human strains. One snow leopard shed SARS‐CoV‐2 in faeces up to 4 weeks. Based on clinical signs and viral shedding periods and levels in the three snow leopards, animal‐to‐animal transmission events could not be excluded. Further testing of SARS‐CoV‐2 in animals is needed.
Keywords: detection, Panthera uncia, SARS‐CoV‐2, snow leopard, viral shedding
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) was first described in December 2019 in Wuhan, China, and subsequently was confirmed to be caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (Zhou et al., 2020). Since 2020, SARS‐CoV‐2 has caused a global pandemic and resulted in millions of infections and deaths worldwide. In addition to humans, SARS‐CoV‐2 has also been detected in a variety of animal species including pets, farmed animals, and captive and free‐ranging wildlife.
In the case of captive animals, tigers (Panthera tigris) and lions (Panthera leo) were first detected SARS‐CoV‐2 positive at the Bronx Zoo in April of 2020 and sequence analysis of SARS‐CoV‐2 strains from animals and animal keepers revealed human‐to‐tiger transmission (McAloose et al., 2020; Wang et al., 2020). Following that, SARS‐CoV‐2 was detected in many other zoos in the United States and other countries, such as Spain and India (Fernandez‐Bellon et al., 2021; Karikalan et al., 2021; USDA, 2020). As of January 2022, a total of 14 captive animal species have tested positive for SARS‐CoV‐2 infection (Elbe & Buckland‐Merrett, 2017; USDA, 2020). The present study reports the first detection of SARS‐CoV‐2 in snow leopards in a zoo in the United States in December of 2020.
2. MATERIALS AND METHODS
2.1. Specimens
Three snow leopards (S1, S2 and S3 were 9, 3 and 5 years old, respectively) at the Louisville Zoo in Louisville, Kentucky, USA, were housed individually in adjacent pens but sharing the same air space and exhibit. Faeces from these three snow leopards were collected, initially in response to respiratory signs, and then daily thereafter for up to 4 weeks and shipped to the University of Illinois Veterinary Diagnostic Laboratory.
2.2. SARS‐CoV‐2 real‐time RT‐PCR
Real‐time RT‐PCR was performed using the N1 and N2 primers and probes as described previously (Lu et al., 2020) and AgPath‐ID One‐Step RT‐PCR Kit (ThermoFisher). Real‐time RT‐PCR was performed on ABI7500 with 1 cycle of 48°C for 10 min and 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 45 s.
2.3. SARS‐CoV‐2 sequencing and analysis
Sequencing of spike and ORF3a region of SARS‐CoV‐2 was conducted on MiSeq for the amplicons generated using five sets of primers (Table 1). The complete genome was obtained on MiSeq at the National Veterinary Services Laboratories as previously described (Hale et al., 2021). Sequences were deposited into GISAID (Elbe & Buckland‐Merrett, 2017). Sequence alignment and construction of maximum likelihood tree were performed using MEGA 7.0.26 (Kumar et al., 2016).
TABLE 1.
Five sets of primers used for amplification of spike and ORF3a genes
| Gene | Primer | Sequence (5′ to 3′) |
|---|---|---|
| Spike | Covid19‐S1‐F | CCAATTCAGTTGTCTTCCTATTC |
| Covid19‐S1515‐R | TTAGAATTCCAAGCTATAACGCA | |
| Covid19‐S‐1177‐F | gacACTAATGTCTATGCAGATTCAT a | |
| Covid19‐S‐2104‐R | CTGCACCAAGTGACATAGTGTAG | |
| Covid19‐S2168‐F | AACAACTCATATGAGTGTGACATACC | |
| Covid19‐S3207‐R | TGAAGTCTGCCTGTGATCAACCT | |
| Covid19‐S3019‐F | CACAGCAAGTGCACTTGGAA | |
| Covid19‐S4145‐R | GCTTGTATCGGTATCGTTGCAGT | |
| ORF3a | ORF3a‐F | CCAGTTGCTGTAGTTGTCTCAAG |
| ORF3a‐R | AGCGCAGTAAGGATGGCTAGT |
Lowercase nucleotides are non‐specific sequence used to increase GC contents of the primers.
3. RESULTS
On 22–23 November 2020, wheezing was reported from one (S1) of three snow leopards housed at the Louisville Zoo. A second snow leopard (S2) exhibited a dry hacking cough in the following days, while a third cat (S3) presented with similar minor and infrequent respiratory symptoms the following week. Faecal samples from these three animals were collected on 4 December 2020 and sent to the University of Illinois Veterinary Diagnostic Laboratory for SARS‐CoV‐2 PCR testing following approval by the state veterinarian. All three samples were positive for SARS‐CoV‐2 by the 2019‐nCoV real‐time RT‐PCR (N1 Ct values 34.00, 29.04 and 28.13 of S1, S2 and S3, respectively). The presumptive positive results of the three snow leopards were confirmed by the US Department of Agriculture National Veterinary Services Laboratories (Ames, IA, USA).
One sample of S2 was sequenced by both whole genome sequencing (KY‐20‐035685‐003, GISAID accession number: EPI_ISL_2928447) and spike as well as ORF3a sequencing (KY‐20‐43641, GISAID accession number: EPI_ISL_8144424) and another sample of S3 was sequenced by spike and ORF3a sequencing (KY‐20‐43642, GISAID accession number: EPI_ISL_8365447). Due to the relatively low viral load, the complete genome sequence was not obtained for KY‐20‐035685‐003 containing a total of six gaps with one gap located in the spike gene. KY‐20‐035685‐003 was classified as B.1.2 by the software Pangolin v.3.1.17 at GISAID (O'Toole et al., 2021). Both spike and ORF3a gene sequences of two samples (KY‐20‐43641 and KY‐20‐43642) were successfully obtained, and blast analysis showed that KY‐20‐43641 and KY‐20‐43642 shared 100% identities with human strains circulating in different US states during October to December 2020. A phylogenetic tree shows the snow leopard viruses clustered with several human strains collected during the time period (Figure 1). One week prior to observing symptoms in the first snow leopard, a zookeeper who cared for these animals tested positive for SARS‐CoV‐2. Unfortunately, the sample collected from the infected keeper was discarded and lost to follow up before any sequencing and comparison could be performed. Nevertheless, these data support that a human‐to‐animal transmission event occurred.
FIGURE 1.

Phylogenetic tree analysis of spike gene of SARS‐CoV‐2 strains of animals as well as human including two snow leopard strains KY‐20‐43641 and KY‐20‐43642 from our study marked with red colour. Alpha, Beta, Gamma, Delta, Epsilon and Lambda variant strains were included in the analysis
The snow leopards were quarantined off‐exhibit, and faeces were collected daily for non‐invasive monitoring of viral shedding by real‐time RT‐PCR between 10 December 2020 to 10 January 2021. S3 shed for 11 consecutive days and then tested negative for 9 days, subsequently testing positive again for 2 more days. In contrast, the other two snow leopards (S1 and S2) had intermittent shedding with positive PCR results for 2 days and 7 days, respectively (Table 2 and Figure 2). The results (Ct values) by sample for each of the two PCR targets, 2019‐nCoV N1 and N2, were comparable.
TABLE 2.
SARS‐CoV‐2 shedding in three snow leopards S1, S2 and S3 from 10 December 2020 to 10 January 2021, as measured by N1 and N2 real‐time RT‐PCR
| N1 | N2 | ||||||
|---|---|---|---|---|---|---|---|
| Month | Date | S1 | S2 | S3 | S1 | S2 | S3 |
| 2020 December | 10 | – | 40.00 | – | – | 37.47 | – |
| 11 | 40.00 | 40.00 | 32.98 | 40.00 | 40.00 | 35.00 | |
| 12 | 37.58 | 36.52 | 25.19 | 40.00 | 37.01 | 26.08 | |
| 13 | 40.00 | 37.56 | 29.11 | 40.00 | 40.00 | 31.41 | |
| 14 | 40.00 | 40.00 | 32.43 | 40.00 | 40.00 | 33.34 | |
| 15 | 40.00 | 40.00 | 30.31 | 40.00 | 38.03 | 30.13 | |
| 16 | 40.00 | 36.26 | 34.64 | 40.00 | 38.01 | 35.97 | |
| 17 | – | 40.00 | 34.93 | – | 40.00 | 36.12 | |
| 18 | 40.00 | 40.00 | 31.89 | 40.00 | 40.00 | 32.41 | |
| 19 | 40.00 | 40.00 | 35.05 | 40.00 | 40.00 | 35.79 | |
| 20 | 40.00 | 37.45 | 37.62 | 40.00 | 40.00 | 35.97 | |
| 21 | 40.00 | 40.00 | 35.02 | 40.00 | 40.00 | 34.86 | |
| 22 | 36.37 | 37.59 | 40.00 | 38.38 | 36.35 | 40.00 | |
| 23 | – | 40.00 | 40.00 | – | 40.00 | 40.00 | |
| 24 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 25 | 40.00 | 40.00 | – | 40.00 | 40.00 | – | |
| 26 | 40.00 | 40.00 | – | 40.00 | 40.00 | – | |
| 27 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 28 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 29 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 30 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 31 | – | 40.00 | 37.73 | – | 40.00 | 39.00 | |
| 2021 January | 1 | 40.00 | 40.00 | 37.51 | 40.00 | 40.00 | 40.00 |
| 2 | – | 40.00 | 40.00 | – | 40.00 | 40.00 | |
| 3 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 4 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 5 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 6 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 7 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 8 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 9 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | 40.00 | |
| 10 | – | – | 40.00 | – | – | 40.00 | |
Note: Ct values of N1 and N2 real‐time RT‐PCR are provided. Ct values of the samples with negative results were set to 40.00
–: no samples tested.
FIGURE 2.
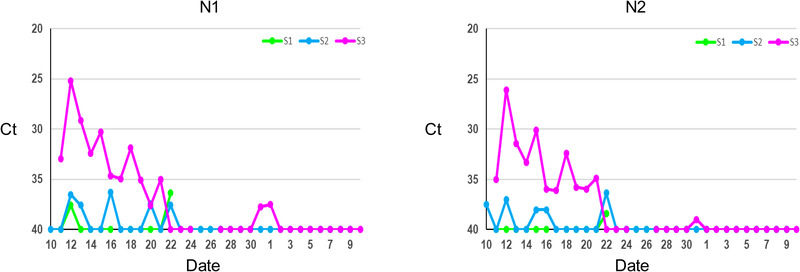
SARS‐CoV‐2 shedding in three snow leopards S1, S2 and S3 from 10 December 2020 to 10 January 2021, as measured by N1 and N2 real‐time RT‐PCR. Ct values of samples with negative results were set to 40
4. DISCUSSION
The SARS‐CoV‐2 pandemic has lasted for 2 years and is ongoing. Great efforts have been undertaken in attempts to understand viral host origin and range, transmission, pathogenesis and therapeutics. It is hypothesized that SARS‐CoV‐2 is a bat‐origin coronavirus based on the evidence that it shared the highest genome identity with a bat coronavirus RaTG13 strain (Zhou et al., 2020). The spillover mechanism of SARS‐CoV‐2 remains unknown. Several lines of evidence of both experimental and natural SARS‐CoV‐2 infection demonstrated that SARS‐CoV‐2 had a wide spectrum of susceptible hosts. Regarding natural infection, SARS‐CoV‐2 has been detected in at least 17 animal species within three orders: Carnivora, Artiodactyla and Primates. Except for two species in the Artiodactyla order (deer and hippopotamus) and one in the Primates order (gorilla), the remaining 14 animal species are within six families of Carnivora (Felidae, Viverridae, Hyaenidae, Canidae, Mustelidae and Procyonidae). The Felidae family has the highest number of species (7) with SARS‐CoV‐2 detections.
In the present study, we report the first detection of SARS‐CoV‐2 in snow leopards in December of 2020 (USDA, 2020). Since then, SARS‐CoV‐2 infections and mortalities have been detected in multiple snow leopards in zoos across four other states including California, Nebraska, South Dakota and Illinois (USDA). Introductions of SARS‐CoV‐2 to zoo animals have been due to human‐to‐animal transmission events; and thus, strains detected in zoo animals change with the progression of the pandemic. Therefore, it is not surprising to see that at different periods of the pandemic both non‐variant strains (e.g. in zoos of Kentucky and California) and variant strains (e.g. Delta in zoos of Nebraska, South Dakota, and Illinois) could infect snow leopards (Elbe & Buckland‐Merrett, 2017). In 2021, several unvaccinated snow leopards did not survive after infection with the Delta variant (NVSL personal communication). As the Delta lineage predominated in humans, the US animal detections between August 2021 and January 2022 were also predominated by Delta; however, it is unclear why this variant resulted in higher disease severity and outcomes specifically in snow leopards. A previous study indicated that low genetic diversity in snow leopards affects their adaptability to the environment and populations (Aruge et al., 2019). Whether susceptibly of snow leopards to SARS‐CoV‐2 is correlated with their genetic diversities needs future investigation.
There are only a few previous studies about SARS‐CoV‐2 shedding in large cats (Bartlett et al., 2021; Fernandez‐Bellon et al., 2021). Similar to observations in tigers and lions at the Bronx Zoo (Bartlett et al., 2021), our data showed that snow leopards had variable shedding periods by faecal analysis. It is hard to conclude that longer shedding periods are observed in younger animals (S2 and S3) than the older animal (S1) due to the limited number of animals in the present study. These three snow leopards were housed adjacent to one another and share exhibit and holding space at different times. Based upon PCR results reflecting virus shed and duration, it is possible that S3 may have been the index case of SARS‐CoV‐2 infection since the other two snow leopards (S1 and S2) shed virus for a shorter period and at lower levels. However, this hypothesis is not supported by the timeline of clinical signs in the three animals as S3 was the last cat to present clinically. It is also possible that these three snow leopards were infected at the same time through fomites such as contaminated feed or enrichment items.
In summary, we report detection of SARS‐CoV‐2 in three snow leopards in a zoo in Kentucky in 2020. Genomic characterization of the virus identified the B.1.2 lineage with no mutations compared to the identified human strains across the spike and ORF3a regions. Virus shedding was variable with one animal shedding up to 4 weeks after the initial detection and other two animals shedding for 2 and 7 days. This report highlights the importance of biosecurity measures for disease prevention at zoological and other conservation facilities. It is important to better understand viral transmission and pathogenesis as SARS‐CoV‐2 not only has significant zoonotic impacts, but also the potential to infect many different animal species.
The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or US Government determination or policy.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
No ethical approval was required as this study solely contains diagnostic samples collected from a zoo.
ACKNOWLEDGEMENTS
Follow‐up testing and sequencing were funded in part by the Food and Drug Administration Veterinary Laboratory Investigation and Response Network (FOA PAR‐17‐141) under grant 1U18FD006673‐01.
Wang, L. , Gyimesi, Z. S. , Killian, M. L. , Torchetti, M. , Olmstead, C. , Fredrickson, R. , & Terio, K. A. (2022). Detection of SARS‐CoV‐2 clade B.1.2 in three snow leopards. Transboundary and Emerging Diseases, 00, 1–6. 10.1111/tbed.14625
Contributor Information
Leyi Wang, Email: leyiwang@illinois.edu.
Karen A. Terio, Email: kterio@illinois.edu.
DATA AVAILABILITY STATEMENT
All the sequence data analysed in this study are publicly available at GISAID (https://www.gisaid.org/).
REFERENCES
- Aruge, S. , Batool, H. , Khan, F. M. , Fakhar, I. A. , & Janjua, S. (2019). A pilot study‐genetic diversity and population structure of snow leopards of Gilgit‐Baltistan, Pakistan, using molecular techniques. PeerJ, 7, e7672. 10.7717/peerj.7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, S. L. , Diel, D. G. , Wang, L. , Zec, S. , Laverack, M. , Martins, M. , Caserta, L. C. , Killian, M. L. , Terio, K. , Olmstead, C. , Delaney, M. A. , Stokol, T. , Ivancic, M. , Jenkins‐Moore, M. , Ingerman, K. , Teegan, T. , McCann, C. , Thomas, P. , McAloose, D. , … Calle, P. P. (2021). Sars‐Cov‐2 infection and longitudinal fecal screening in Malayan Tigers (Panthera Tigris Jacksoni), Amur Tigers (Panthera Tigris Altaica), and African Lions (Panthera Leo Krugeri) at the Bronx Zoo, New York, USA. Journal of Zoo and Wildlife Medicine, 51, 733–744. [DOI] [PubMed] [Google Scholar]
- Elbe, S. , & Buckland‐Merrett, G. (2017). Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Challenges, 1, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Bellon, H. , Rodon, J. , Fernandez‐Bastit, L. , Almagro, V. , Padilla‐Sole, P. , Lorca‐Oro, C. , Valle, R. , Roca, N. , Grazioli, S. , Trogu, T. , Bensaid, A. , Carrillo, J. , Izquierdo‐Useros, N. , Blanco, J. , Parera, M. , Noguera‐Julian, M. , Clotet, B. , Moreno, A. , Segales, J. , & Vergara‐Alert, J. (2021). Monitoring natural SARS‐CoV‐2 infection in lions (Panthera leo) at the Barcelona Zoo: Viral dynamics and host responses. Viruses, 13, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, V. L. , Dennis, P. M. , McBride, D. S. , Nolting, J. M. , Madden, C. , Huey, D. , Ehrlich, M. , Grieser, J. , Winston, J. , Lombardi, D. , Gibson, S. , Saif, L. , Killian, M. L. , Lantz, K. , Tell, R. , Torchetti, M. , Robbe‐Austerman, S. , Nelson, M. I. , Faith, S. A. , & Bowman, A. S. (2021). SARS‐CoV‐2 infection in free‐ranging white‐tailed deer. Nature, 602, 481–486. 10.1038/s41586-021-04353-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikalan, M. , Chander, V. , Mahajan, S. , Deol, P. , Agrawal, R. K. , Nandi, S. , Rai, S. K. , Mathur, A. , Pawde, A. , Singh, K. P. , & Sharma, G. K. (2021). Natural infection of Delta mutant of SARS‐CoV‐2 in Asiatic lions of India. Transboundary and Emerging Diseases, Advance online publication. [DOI] [PMC free article] [PubMed]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X. , Wang, L. , Sakthivel, S. K. , Whitaker, B. , Murray, J. , Kamili, S. , Lynch, B. , Malapati, L. , Burke, S. A. , Harcourt, J. , Tamin, A. , Thornburg, N. J. , Villanueva, J. M. , & Lindstrom, S. (2020). US CDC real‐time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases, 26, 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAloose, D. , Laverack, M. , Wang, L. , Killian, M. L. , Caserta, L. C. , Yuan, F. , Mitchell, P. K. , Queen, K. , Mauldin, M. R. , Cronk, B. D. , Bartlett, S. L. , Sykes, J. M. , Zec, S. , Stokol, T. , Ingerman, K. , Delaney, M. A. , Fredrickson, R. , Ivancic, M. , Jenkins‐Moore, M. , … Diel, D. G. (2020). From people to panthera: Natural SARS‐CoV‐2 infection in tigers and lions at the Bronx zoo. mBio, 11, e02220–20. 10.1128/mBio.02220-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, A. , Scher, E. , Underwood, A. , Jackson, B. , Hill, V. , McCrone, J. T. , Colquhoun, R. , Ruis, C. , Abu‐Dahab, K. , Taylor, B. , Yeats, C. , du Plessis, L. , Maloney, D. , Medd, N. , Attwood, S. W. , Aanensen, D. M. , Holmes, E. C. , Pybus, O. G. , & Rambaut, A. (2021). Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evolution, 7, veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA . (2022). Confirmed cases of SARS‐CoV‐2 in animals in the United States. Available at: https://www.aphis.usda.gov/aphis/dashboards/tableau/sars‐dashboard (accessed Jan 15 2022)
- USDA . (2020). Confirmation of COVID‐19 in a snow leopard at a Kentucky Zoo. Available at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder‐info/sa_by_date/sa‐2020/sa‐12/ky‐snow‐leopard‐covid
- Wang, L. , Mitchell, P. K. , Calle, P. P. , Bartlett, S. L. , McAloose, D. , Killian, M. L. , Yuan, F. , Fang, Y. , Goodman, L. B. , Fredrickson, R. , Elvinger, F. , Terio, K. , Franzen, K. , Stuber, T. , Diel, D. G. , & Torchetti, M. K. (2020). Complete genome sequence of SARS‐CoV‐2 in a tiger from a U.S. zoological collection. Microbiology Resource Announcements, 9, e00468–20. 10.1128/MRA.0046820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C. L. , Chen, H. D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R. D. , Liu, M. Q. , Chen, Y. , Shen, X. R. , … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the sequence data analysed in this study are publicly available at GISAID (https://www.gisaid.org/).


