Abstract
The global pandemic of COVID‐19 began in December 2019 and is still continuing. The past 2 years have seen the emergence of several variants that were more vicious than each other. The emergence of Omicron (B.1.1.529) proved to be a huge epidemiological concern as the rate of infection of this particular strain was enormous. The strain was identified in South Africa on November 24, 2021 and was classified as a “Variant of Concern” on November 26, 2021. The Omicron variant possessed mutations in the key RBD region, the S region, thereby increasing the affinity of ACE2 for better transmission of the virus. Antibody resistance was found in this variant and it was able to reduce vaccine efficiency of vaccines. The need for a booster vaccine was brought forth due to the prevalence of the Omicron variant and, subsequently, this led to targeted research and development of variant‐specific vaccines and booster dosage. This review discusses broadly the genomic characters and features of Omicron along with its specific mutations, evolution, antibody resistance, and evasion, utilization of CRISPR‐Cas12a assay for Omicron detection, T‐cell immunity elicited by vaccines against Omicron, and strategies to decrease Omicron infection along with COVID‐19 and it also discusses on XE recombinant variant and on infectivity of BA.2 subvariant of Omicron.
Keywords: Omicron, recombinant variants, spike region, vaccine, variant of COVID‐19
1. INTRODUCTION
The emergence of the severe acute respiratory syndrome (SARS‐CoV‐2) or Coronavirus in the latter part of 2019, brought the whole world to a standstill plunging the world's economy and grouping the scientific community together to understand and investigate the virus' impact and to chalk out a way to understand its mechanism. The virus was quick to spread causing the WHO to declare it as a global pandemic and a concern for health. 1 In the last couple of months since the beginning of the pandemic, the SARS‐CoV‐2 virus has mutated continuously. Owing to its ability as an RNA virus and due to its shorter replication time, with high mutational changes and lower stability in its genome, it is able to mutate, spread rapidly, and adapt to newer environmental conditions thereby prompting continuous evolution in its genetic material. Therefore, it validates its resistance to vaccines and escapes the immunity of the host organism. 2 Due to these factors, new strains of SARS‐CoV‐2 have evolved having varying properties in its genome, with varying severity of infection, and evading immunity. 3 The mutated strains include Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Lambda (C.37), Mu (B.1621), Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1), and currently Omicron (B.1.1.529). All these variants are classified into three categories namely, Variant of Concern (VOC), Variant of Interest (VOI), and Variant under monitoring (VUM). 4 The Delta variant (B.617.2) was identified in the end of 2020 and by August of 2021 it went on to infect people from 163 different nations owing to its mutations. Therefore, the World Health Organization (WHO) redesignated the variant from VOI to VOC with a transmission rate of 40%–60%, which was dangerous in comparison to other variants 5 until the emergence of Omicron. Omicron was first reported to the WHO from South Africa on November 24, 2021, after the specimen collected from Botswana and South Africa 6 tested for an unusually high number of 32 mutations in the Spike (S) protein where the antibodies were targeted and the genomic sequence of South African strain revealed 45‐52 changes to amino acids and 26–32 changes to S‐Protein structure in contrast with earlier strains. 7 , 8 A few mutations in South African strains are N440K, T478K, and N501Y in the RBD region of Omicron that enhance human ACE2 bindin0067. The RBD mutations in conjunction with deletions at sites 69–70, 143–145, and 211 and insertion at between 214 and 215 significantly enhanced the strain's sensitivity against antibody neutralization activity 9 and a few other mutations present in the South African strain incudes, T95I, Y145D, A67V, L212I, G446S, G496S, T547K, N586K, L981F are prevalent with South African strain and a few mutations (common) are also shared by other variants such as N501Y shared with Alpha, Beta, Gamma and Mu, T478K that is shared with Delta variant. 10 In comparison, the Delta variant had only five mutations in its Spike protein/mutation on S protein receptor‐binding domain (RBD) 11 that including deletion at sites 156 and 157 in spike region (NTD domain), R158G, L452R, T478K, D614G, P681R, D950N, T19R, G142D and K417N (present in Delta plus variant). 12 Herein, mutation L452R or T478K contributes towards infectivity and transmissibility of Delta variant and the mutation P681R present in furin‐cleavage site is known to enhance viral entry into human cells and also contributes towards antibody resistance, the K417N mutation when combined with L452R mutation was seen to significantly evade vaccine‐induced immunity. 5 With the technical directive from the WHO's Technical Advisory Group on Virus Evolution (TAG‐VE) WHO categorized Omicron as a Variant of concern on November 26, 2021 issuing a warning to every country across the globe. 13 This article will brief on Omicron, its concerns, its physiology, preventive measure, possible way to detect the virus along with the possibility for an antigenic shift of the virus and the current trend of recombinant SARS‐CoV‐2 virus.
2. OMICRON‐VARIANT OF CONCERN
The variant Omicron exhibits an unusually high volume of mutations in the Spike region and key specific mutations of Omicron includes, N440K, G446S, G339D, E484A, A76V, Q493R, Q498R, G496S, T547K, Y505H, N679K, H655Y, N764K, N856K, D796Y, Q954H, S375F, L981F, N969K, S371L, L212I, and S373P which was revealed through the analysis of its genomic sequence. 14 This change is said to enhance Omicron's severity, infectivity, immune escape, higher binding affinity, and higher transmissibility, thus making Omicron a deadly variant. 15
The viral structure of the SARS‐CoV‐2 promotes four protein genes that include, M‐protein that binds to the host body's protein and structures the shape of the virus, N‐protein/Nucleocapsid protein modulates signaling, cell replication cycle and immune response of host towards SARS‐CoV‐2 infection, E‐protein/Envelope protein which portraits itself as an area for the production and maturation of the virus and the S protein/S‐Glycoprotein which divides itself into the region of the head (S1) and stem (S2) with a ratio of 3:2 where, the S1 region houses the N‐terminal domain (NTD), two C‐terminal domain (CTD), and receptor‐binding domain (RBD) that is crucial for new variants in conjunction with host receptor, Angiotensin‐converting enzyme 2 (ACE2). The efficiency and transmissibility of SARS‐CoV‐2 depends on the interaction of ACE2 receptor. 16 , 17 ACE2 acts as a mediator that facilitates the interaction between itself and SARS‐CoV‐2 and the other receptor being TMPRSS2 which acts as a coreceptor thereby regulating the infectivity level in living organisms. According to recent studies, genetic changes and vulnerability were found to play a crucial role in viral infections. 18
With the SARS‐CoV‐2 being an RNA virus, it is of no surprise regarding its capability to mutate and to evolve with greater virulence and to reform itself with hindrance towards the immune system. With the SARS‐CoV‐2 pandemic, we have seen the emergence of various variants with different mutability capacity. The origin of VOCs like Alpha, Beta, Gamma, Delta, and now Omicron along with various VOIs and VUMs showcases the virus' reinfection, increased resistance to the immune system and to vaccines. 19 Omicron was identified on November 24, 2021 in South Africa and the WHO quickly classified this new variant as VOC on November 26, 2021 due to its increased infection rate. Omicron was soon reported in several countries after the initial announcement. In countries like Hong Kong, Botswana, Belgium, South Africa, Canada, Australia, United States, United Kingdom, Denmark, and several other countries; the combination of both higher transmissibility and immune system evasion defined the Omicron's domination in comparison to other strains. This B.1.1.529 variant eventually branched out into two sublineages known as BA.1 and BA.2. 20 , 21 The receptor‐binding motif (RBM) of Omicron is reported to have an 11 times higher mutational rate in comparison to other variants.
The mutational changes are seen in the Spike protein, RBD region, receptor‐binding motif (RBM), and in the areas of S1 and S2. This dynamic change may have the ability to alter the interaction of host receptor ACE2 with RBD or the host's immune system and Omicron variant is shown to possess a stronger and higher Coulomb attraction force between ACE2, and Spike protein and it is shown to have increased electric charge in nucleocapsid protein thereby terming the Omicron as “Electric Virus.” 22 It was also indicated that Omicron had a close phylogenetic relation with the Alpha variant proving that Omicron was already in existence before its mass spreading. 23
As of 2022 Omicron is the prevalent strain globally and in particular, its subvariant BA.2 is a slowly increasing its dominance worldwide and it is dubbed as “Stealth Omicron” as this variant lacks deletion of 69–70 position in its Spike region that is a characteristic feature of variant alpha and BA.1, currently BA.2 have surpassed the infectivity rate of BA.1 prompting that transmissible rate of BA.2 is more than BA.1. 24
2.1. BA.2 subvariant of Omicron
Since January 2022, the infection of BA.2 have been steadily increasing and is being spread to multiple countries globally and it was reported that the reproduction rate of BA.2 is 1.40‐fold higher in comparison to BA.1 on average globally thereby becoming the domain subvariant of Omicron. 25 There has been considerable difference in the Spike region sequence between BA.1 and BA.2 subvariant of Omicron thus we can assume that infectivity rate, transmissibility, immune resistance, and neutralization properties can vary between these two subvariants; Yamasoba et al., have been investigating the molecular characteristics of BA.2 and have considerable findings related to dominance level of BA.2 subvariant.
Yamasoba et al., investigated the antisera resistance of BA.2 in comparison to BA.1, recent studies proved that BA.1 subvariant is extremely resistant to antisera provoked by vaccines, mRNA‐1273 and ChAdOx‐1, and BA.2 subvariant is completely resistant to monoclonal antibodies such as Sotrovimab, Imdevimab, and Casirivimab thus proving that both BA.1 and BA.2 are resistant to antibody therapeutics and antisera from immunization. Yamasoba investigated the immune resistance between BA.1 and BA.2 and have found that humoral immunity induced by BA.1 is less effective in comparison to BA.2. 25 The growth of BA.1 and BA.2 was compared in VeroE6/TMPRSS2 cell and was reported that BA.2 exhibited more replicative status in Nasal epithelial cells of humans and in Calu‐3 cells. 25
When compared for the fusogenicity difference between BA.1 and BA.2 it was found that BA.2 possess higher fusogenicity that directly depends on expression of TMPRSS2 upon target cell and it was also proved that TMPRSS2 expression on target cells have no effect on the infectivity levels of BA.1 and BA.2 thus Yamasoba et al., proved that BA.2 is more replicative and fusogenicity in comparison to BA.1. 25 Yamasoba et al., utilized hamster model to show the pathogenicity of BA.2 that is similar to Omicron variant B.1.1 and higher than BA.1, and they also showed that BA.2 spreads at a rapid rate in lung tissues such as alveolar tract, bronchioles, trachea, and epithelium tissues of lungs in comparison to BA.1. 25 Thus, from this investigation by Yamasoba et al., we can conclude and it is very evident that BA.2 might become a serious global problem in the future and steps needs to be taken to solve this arising subvariant. Table 1 depicts the information for COVID‐19 virus across different region globally along with their mutational presence.
Table 1.
A comprehensive view of arisen COVID‐19 virus across different region globally along with their mutational presence in different regions in their structure.
| Alpha (α) B.1.1.7 | United Kingdom | 2020, December | N501Y |
N501Y: Helps tight binding of ACE2. E484K: Evades Immune system Y144del: Limits binding affinity of antibodies. |
D1118H, T7161, S982A | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
| Beta (β) B.1.351 | South Africa | 2020, December | N501Y, K417N, E484K | K417N and E484K: Evades Immune system. | A701V | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
| Gamma (γ) B.1.1.128 (P.1) | Brazil | 2021, January | N501Y, K417T, E484K | E484K: Immune system Evasion. | V1176F, T10271 | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
| Delta (δ) B.1.617.2 | India | 2021, May | L452R, T478K |
L452R: Increases transmissibility. T478K‐ Shows increased affinity. |
D950N | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 ; Jhun et al. 30 ; Moghaddar et al. 31 |
| Kappa (κ) B.1.617.1 | India(Multiple countries) | 2021, April | E484Q, L452R | L452R: Increases transmissibility. | Q1071H | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
| Lambda (λ) C.47 | Peru | 2020, December | L452Q, F490S, T76I | L452Q, F490S, and T76I: Decreases sensitivity towards antibodies. | T859N | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 ; Moghaddar et al. 31 |
| Mu (μ) B.1.621 | Columbia | 2021, January | R346K, N501Y, E484K | N501Y and E484K: Evades immune system. | D950N | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 ; Xiong et al. 32 ; Rahimi et al. 33 |
| Eta (η) B.1.525 | Nigeria (Multiple countries) | 2021, March | E484K | E484K: Immune system Evasion. | F888L | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
| Iota (ι) B.1.526 | United States | 2020, November | E484K | E484K: Immune system Evasion. | A701V | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
| Omicron (ο) B.1.529 | South Africa | 2021, November | Y505H, G496S, N501Y, Q498R, G339D, Q493R, S371L, E484A, S373P, K417N, S375F, N440K, S477N, G446S, T478K | More than 30 mutations are present leading to high infectivity, immune evasion, higher binding properties, and S‐gene target failure. | L981F, N764K, Q954H, N856K, D796Y, N969K | Hadj Hassine 26 ; Cameroni et al. 27 ; Lippi et al. 28 ; Malik et al. 29 |
3. OMICRON DETECTION AND MUTATIONS
The Omicron is characterised with 37 amino acid changes in the Spike protein. Three deletions, one insertion, and three mutations have been observed in earlier variants, and 14 mutational areas have not been described earlier. The Spike is structured with three peptide chains along with RBD. The RBD is considered as the corresponding zone of the virus, where mutations are found in the RBD site of 333–527, and in Omicron 15 mutations were identified in this region. The mutations are known to interfere with protein–protein interaction complex thereby changing the dynamics of the virus. The mutational structure of Omicron has been of a significant concern due to its mutation in the Spike protein. The deletion of genomic positions H69 and H70 (also found in Alpha and Eta) facilitates the S‐assay of Taqpath polymerase chain reaction (PCR) tests to provide negative results thereby providing the virus with proxy prevalence also known as S‐gene target failure (SGFT). There are three amino acids “EPE” that are observed in position 214. This hotspot is also known as insertion hotspot and is known to induce structural dynamic changes to the Spike protein. 34 , 35
Omicron is said to possess increased transmissibility due to the mutation of H655Y, N679K, and P681H in the S1–S2 area thereby increasing spike cleavage. Due to the double mutation of Q498R and N501Y, it has led to an increase in the binding capability of ACE2. The mutation on E484A has made omicron to evade the immune system. The deletion of L3674, S3675, and G3676 in ORF1a (NSP6) promotes a possibility of innate immune system eluding, leading to degradation of immune cells' immunity. Its mutations consist of K856R, L2084I, S2083‐, A2710T, P3395H, L3674‐, T3255I, S3675‐, I3758V, and G3676‐. Mutations are present in regions P314L and I1566V at ORF1b and ORF9b and are known to provide an immune response when in contact with any virus. Mutation of its amino‐acids E27‐, N28‐, and A29‐are prone to result in the suppression of Interferon release; deletion at E31‐, S33‐, and R32‐. The mutations in the Nucleocapsid region of R203K, P13L, and G204R have exhibited increased RNA expression and viral loads. The E‐protein shows mutations only in the T9I region. D3G, A63T, and Q16E are mutations of the Matrix gene. 36 Figure 1 represents the spike protein structure of SARS‐CoV‐2 virus. In RBD the mutations of K417N, T478K, E484A, D614G, and N501Y are considered as important functions for the widespread infection and virulent activity.
Figure 1.
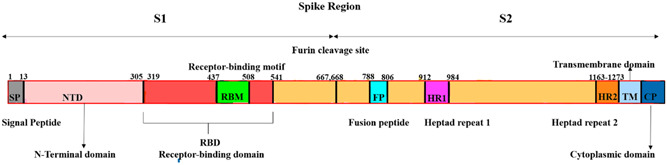
The Spike protein structure of SARS‐CoV‐2 virus. The Spike protein comprises the following structures: (1) Signal peptide: Guides virus to its targeted membrane; (2) N‐terminal domain: Helps in the binding interface of the virus; (3) Receptor‐binding domain/C‐terminal domain (CTD): Interacts with host's ACE2, extensively targeted for Covid‐19 vaccine development; (4) Fusion peptide: Helps in penetrating targeted cell membrane through either TMPRSS2 pathway or endosome‐cathepsin L pathway; (5) Heptad repeat 1&2: Forms the six‐helical package that is crucial for entry of S2 subunit into the host cell and also for viral fusion; (6) Transmembrane domain: Contributes towards membrane fusion and stabilizes trimeric structure; (7) Cytoplasmic domain: Anchors the Spike protein inside the viral membrane.
The mutations to the sites of K417N or K440N and S446K show slight increase towards the expression of RBD and are proven to impart resistance towards neutralizing monoclonal antibodies (mAbs). 37 , 38 The mutational change on region H655Y was reckoned to produce an increased level of infectivity and since H655Y is beside to the cleavage area of furin, there is a possibility of increased S protein cleavage aiding in transmission thereby leading towards the resistance of monoclonal antibody therapy. 39 Mutation in N679K is present nearby the cleavage area of furin and therefore it assists in demonstrating its fundamental character which might co‐relate with increased S‐protein cleavage and therefore increased transmission of Omicron. 40 The emergence of a new mutations in RBD include S371L, S373P, G339D, N440K, S375F, Q493K, G446S, G496S, S477N, Y505H, and Q498R. These could lead to increased capabilities to evade immune cells. 41 The prevalence of few Omicron strains that show distinctive properties than the conventional type raises an alarm as mutations on R346K were seen only in 8.3% of Omicron strains. This situation might affect the neutralization of monoclonal antibodies. With the prevalence of these mutations, further detailed studies are required to strategize an effective vaccine. 42 It is also worth noting the enhanced expression of ACE2 receptor with S protein. The combination of a mutation in the region of N201Y with Q498R and S477N causes enhanced binding of ACE2. The mutation on‐site D614G is associated with increased infectivity through the weakening of RBD‐down spike protein as shown in Figure 2 which may lead to an increase in infectivity. 43
Figure 2.
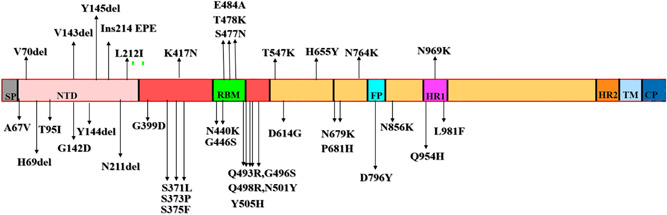
Amino acid mutations present on the Spike region of Omicron. Each domain regions are indicated in different colors.
Higher infection rates are a concern for SARS‐CoV‐2 and variants. The SARS‐CoV‐2 and its variants have shown to possess higher levels of infection rate in their Spike protein that binds with ACE2 receptor in comparison to its predecessors. The SARS‐CoV Spike protein region and the affinity of the ACE2 receptor with S‐protein in SARS‐CoV‐2 are around 10 nM that is 10 times higher than the binding capacity of SARS‐CoV. This hypothesis was proved with the emergence of variants with mutations in RBD region of the S‐protein. This evidence provides insights regarding higher infectivity of SARS‐CoV‐2. 44 With the emergence of variants, the Spike protein is able to bypass various parameters, such as temperature, as emerging variants are able to withstand the temperature resistance to promote higher binding of ACE2, with the facilitation of mutation in D614G and N501Y which can crucially have an impact on the interaction of ACE2‐Spike protein 45 and it was also reported that D614G mutation was concurrently present along with mutations C14408T, C3037T, and C241T. 46 , 47
Omicron has an increased positive electrostatic potential level in its RBD interaction with ACE2. With ACE2 possessing negative electrostatic potential on its surface, it is speculated that this interaction may lead to an increase in extension between ACE2 and RBD thereby progressing towards higher affinity. This may also contribute towards higher infectivity rate of Omicron. 48 Recent reports on binding free energy (BFE) that takes place between RBD region of the S‐Protein and ACE2 receptors directly correspond to the infectivity rate. To understand the SARS‐CoV‐2 infectivity, evolution changes to its genome, and its transmissibility tracking it is important to monitor its mutations in the RBD region along with BFE changes that will facilitate the interpretation regarding the infectivity of SARS‐CoV‐2 and its variants. A positive BFE change denotes a strong infectivity capacity of variants between its ACE2 receptors and S‐protein due to the generation of mutations in its region and a negative BFE change between S protein and ACE2 receptor denotes weak infectivity of a SARS‐CoV‐2 variant. This proves that Omicron emulates the strengthening infectivity pathway of natural selection. 21 Figure 3 reveals the information about variable number of mutations present in spike protein.
Figure 3.
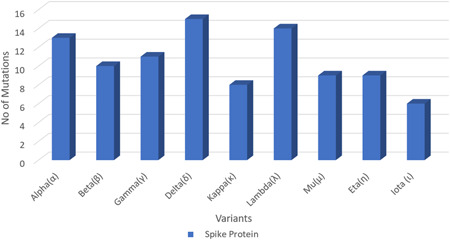
Shows the variable number of mutations present in Spike specific region in variants of SARS‐CoV‐2 virus
4. EVOLUTION AND IMMUNE EVASION OF OMICRON
Analysis of the RBD region provides evidence regarding the presence of two sub‐clades of Omicron. The sub‐clade 1 have mutations at 446 G, 417 K, and 446 G. The sub‐clade 2 has mutations at 446 S, 417 N, and 440 K. Analysis of Omicron and other variants suggest omicron to be evolved in parallel with variants and the possibility of divergence from other strains could have happened in mid of 2020. The phylogenetic analysis of the SARS‐CoV‐2 sequence has provided evidence on the evolution of Omicron from the strain that was found in Mexico (B.1.1.519). The other possible evolution is linked with an HIV‐infected person with an immunocompromised condition where possible mutations could have occurred. This possibility can also be supported due to the high prevalence of HIV in South Africa which could have given rise to Omicron. Other possible scenarios include natural selection towards evolution by altering the Spike region for mutation. The imbalance between low vaccination coverage and high infectivity provides a favorable situation for the virus to evolve. Nevertheless, the precise origin of Omicron is yet to be ascertained due to the lack of evolutionary evidence. 36
The Spike protein of Omicron is predominantly responsible for the occurrence of majority of the mutations. This is apparently due to its immune evasion property which therefore disables the neutralizing capability of the pre‐existent antibodies. However, the extent of this is yet to be ascertained. 49 The Spike protein and its regions have been the focus of attention in the past few years, primarily due to its capability to mutate thereby making themselves a target for vaccine development. One of the main challenges faced by the current vaccines are their diminishing efficiency against the SARS‐CoV‐2 virus.
Initial reports suggest that Omicron is capable of evading every clinically approved currently available antibody‐based remedy. This critically debars the humoral immunity induced either through immunization or by natural infection, thus indicating a probably altered structure in the antigen. This may be the cause of the increased transmission rate of the virus. This is further supported by mutations that occur in the RBD region compromising the neutralization activity. It is well known that mutations can potentially affect the binding of most antibody classes. 50 The two targeted regions for neutralization are in the RBD and NTD region. New mutations in G496S, S477N, Q493R, Y505H, and Q498R in conjunction with existing mutations continue to alter the antigenic activities thereby prompting virus evasion from antibodies shown in Table 2. 51 , 52
Table 2.
The table showing few specific key gene changes that occurred in Omicron
| Mutational gene | Changes in Spike region of Omicron (addition/deletion) | Reference |
|---|---|---|
| Y505 | Change in α‐helix region; H‐bond removal with ACE2 E37‐ | Pascarella et al. 48 ; Cao et al. 53 ; Planas et al. 54 |
| Q498R | Salt bridge formation with ACE2 D38‐ | Pascarella et al. 48 ; Cao et al. 53 ; Planas et al. 54 |
| G496S | H‐bond removal between ACE2 D38‐ | Pascarella et al. 48 ; Cao et al. 53 ; Planas et al. 54 |
| Q493R | Salt bridge formation with ACE2 E35‐ | Pascarella et al. 48 ; Cao et al. 53 ; Planas et al. 54 |
| E484A | Salt bridge formation between ACE2 K31‐ and E484A | Pascarella et al. 48 ; Cao et al. 53 ; Planas et al. 54 |
| K417N | Change in C‐terminal area of α‐helix region; Salt bridge removal with ACE2 D30‐ | Pascarella et al 48 ; Cao et al. 53 ; Planas et al. 54 |
| G339D | Possibility of H‐bond formation towards NAG protein | Pascarella et al. 48 ; Cao et al. 53 ; Planas et al. 54 |
New‐emerging variants carry the evolved mutations that are resistant to antibodies/vaccines. Mutations on Y449S and Y449H contribute to vaccine resistance. The important binding sites of antibodies are Y449, F456, L455, F486, E484, Y489, Q493, N487, Y505, and S494. Mutations in such sites may enable the virus for immune evasion. 55
A study performed by Lu et al., reveals that Omicron evades the neutralization of antibodies induced by CoronaVac or BNT162b2 and the presence of R346K mutation in one of the samples did not influence the neutralization response. It was also seen that mutation in G446S and Q493R sites coincide with modification of monoclonal antibodies. These findings are in agreement with the findings reported by Cao et al., who also demonstrated that Omicron evasion by monoclonal antibodies has also shown that single mutations in Omicron can evade the NAbs owing to various mutations in the epitome of the Spike region. Therefore, this has provided a possibility for antigenic shifting. 53 , 56 Such mutations have drastically reduced the efficiency of several major vaccines in the market. The vaccine efficiency of Pfizer was reportedly reduced from 86% to 43%; Johnson & Johnson's vaccine efficiency dropped from 86% to 13%. Moderna vaccine was down at 58% from 89%. These may be attributed to the effects of mutations, reinfection, and limitations in the neutralization of antibodies. 36 , 57 Figure 4 represents the map of emergence of VOCs, VUM, and VOI in different parts of the world.
Figure 4.
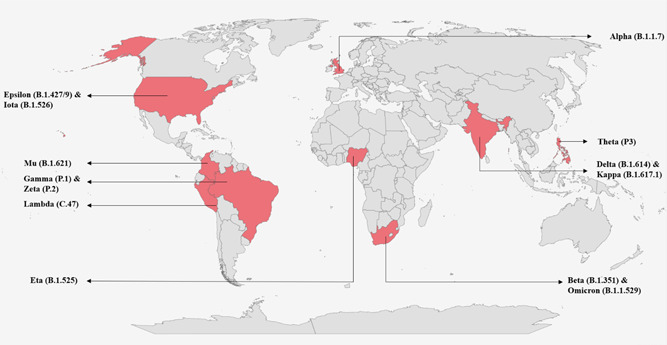
The map showing the emergence of VOCs, VUM, and VOI in different parts of the world. VOC, Variant of Concern; VOI, Variant of Interest; VUM, Variant under monitoring.
5. ANTIBODY EVASION AND RESISTANCE
The RBD identifies the ACE2 receptor and regulates the viral entry inside the host, therefore being a key target to stop viral infectivity. Consequently, RBD‐distinct neutralizing antibodies play a major role in the neutralization of a virus. This neutralization antibodies are divided into Classes 1, 2, 3, and 4 based on the binding affinity of RBD and ACE2. The variants Delta, Mu, and C.1.2 in particular are known to evade the neutralization antibodies (nAbs) belonging to Classes 2 and 3. The Beta variant predominantly evades from nAbs of Class 1 and 2. It is worth noting that these aforementioned variants do not interact with Class 4 antibodies. Interestingly, Omicron annihilated the neutralization of all four classes (Classes 1–4) of nAbs (exception S309). This may explain the immune evasive capability of Omicron. 58 , 59
Recognition of the Omicron RBD by Class 1 neutralizing antibodies was disrupted due to mutations in K417N and Q493R. A mutation in E484 results in the complete destruction of Class 2 nAbs. Mutation at G446S restricts Imdevimab's (a Class 3 antibody) binding affinity of nAbs to RBD. The resistance of Omicron towards Class 4 nAbs is mediated by S371L. Despite the presence of N440 and G339 mutation in its epitopes, the S309 maintains its stronger binding affinity towards Omicron's RBD. 60
Anti‐RBD nAbs towards SARS‐CoV‐2 have largely been identified from vaccines and infected personnel. Mutations in the S region of RBD may render the nAbs ineffective. 53 Several nAbs have been reported in the literature which is capable to neutralize SARS‐CoV‐2.
Eli Lilly Mab (LY‐CoV555) also recognized as Bamlanivimab, was subjected to interaction with Spike protein of Omicron with mutations. It was found that N501Y, K417N, E484A, and Q493R mutations reduced the efficiency of LY‐CoV555. Etesevimab (LY‐CoV016) has also shown a reduced affinity towards K417N, Y505H, and N501Y, thereby, being prone to Omicron mutations. 61 , 62 Regeneron mAbs also known as Casirivimab (REGN10933) portrays a critical role to neutralize the SARS‐CoV‐2 virus precisely. The G446K, E484A, and K417N mutations slightly weaken the complex but only mild changes were seen in its efficacy. Imdevimab (REGN10987) has similar mutations with Casirivimab thereby failing to neutralize Omicron. 61 , 62
Regdanvimab (CT‐P59) when in contact with Omicron shows mutations in the areas namely, Q498R, E484A, and Q493R. Therefore, this agent should be used with caution toward Omicron. 61 , 62 C135 antibodies from Rockefeller University show a critical mutation on S317L and R346K thus succumbing to Omicron infection. The antibody C144 have consequential reception towards Omicron due to change in E484A thus undermining its efficiency towards Omicron. The mAbs of Cilgavimab and Andintrevimab show mild resistance towards Omicron. Sotrovimab was least affected by mutations caused by Omicron. 54 , 61 , 62 , 63
The Pfizer‐BioNTech BNT162b2 vaccinated individuals have shown reduced neutralization against Omicron. Table 3 depicts the information about the list of therapeutic drugs used in various stages of development of SARS‐CoV‐2. This has led to an incomplete evasion from messenger RNA (mRNA) vaccine‐driven antibody neutralization. 27 It has also been reported that ChAdOx1 vaccinated individuals possessed no neutralization efficacy. Omicron reportedly promotes the decline in T‐cell mediated immunity. 36
Table 3.
SARS‐CoV‐2 therapeutic drugs in various stages of development
| Drug | Manufacturing Institution | Phase | Reference |
|---|---|---|---|
| Sotrovimab | GSK/Vir Biotech | Emergency (EUA) | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| Baricitinib (Olumiant) | Eli‐Lilly | Emergency (EUA) | Fiolet et al. 67 ; Raman et al. 65 ; Janik et al. 66 |
| LY‐CoV016 (Etesevimab) and LY‐CoV555 (Bamlanivimab) | Eli‐Lilly | Emergency (EUA) | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| REGN‐COV2 (Casirivimab and Imdevimab) | Regeneron/Sanofi | Emergency (EUA) | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| Opaganib | Redhill | Phase‐III | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al 66 |
| MK‐4482 | Merck | Phase‐III | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| GSK4182136 | GSK/Vir Biotech | Phase‐III | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| Aplidin (Plitidepsin) | Pharma Mar | Phase‐II | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| PF‐07321332 | Pfizer | Phase‐I | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
| Remdesivir | Gilead Sciences Inc. | Approved | Fiolet et al. 64 ; Raman et al. 65 ; Janik et al. 66 |
Abbreviation: EUA, emergency use authorization.
6. T‐CELL IMMUNITY AGAINST SARS‐COV‐2 OMICRON
Robust immune response in varying requirements are needed to recover patients infected with Omicron or other variants and SARS‐CoV‐2 specific B‐ and T‐cell immune response play a pivotal role in the pathogenesis of SARS‐CoV‐2 and its variants. 68 The cytotoxic (CD8) T cells are activated once they recognize the viral peptides unveiled by human leukocyte antigens (HLA) class I molecules present on the surface of antigen‐presenting cells (APC), and the helper T cells (CD4) are activated through the association between T‐cell receptors (TCR) and viral peptides unveiled by HLA Class II proteins and an important function of the CD4 cells is that it conveys the activation signal to B cells, which is crucial for antibody production 68 and the T‐cell response to the Omicron variant was found to have recognized ~80% antigenic peptides by T‐cell and identical to ancestral strain. 69 The coordination between CD8+ T cells, neutralizing antibodies acquired through infection/vaccination, and CD4+ T cells were seen to lower disease infectivity and severity, T‐cell cross‐reactivity was examined by Keeton et al., here the T‐cell response was investigated with participants who received vaccines of Ad26.COV2.S, BNT162b2 (either one or two doses), and from COVID‐19 recovered individuals and it is was seen that greater than 85% of vaccinees were able to generate a T‐cell response to vaccination after 22–32 days after the last vaccine dose, herein both the vaccination and infection were able to induce CD4+ T‐cell response but was significantly lower in comparison to the ancestral strain and the CD8 was detected in lower level, 70 it was also observed that no serious difference in cross‐reactive of CD4+ T, CD8+ T response towards variants Beta, Delta, and Omicron. 70
In another study by Paul Naaber et al., where T‐cell response against Omicron was studied after the booster dose, here participants were vaccinated with two doses of BNT162b2 vaccine and after a gap period of 9 months were vaccinated with a booster dose from BNT162b2, before the booster vaccination the Spike‐RBD immunoglobulin G (IgG) was seen to be in decline after second vaccination but 2 weeks after the booster dose the IgG levels were found to be higher than the previous level, however, 3 months after the booster dose the IgG level was seen to be lower in comparison to its initial levels. In this study, 71 Omicron‐specific CD4+ and CD8+ T‐cell responses and activation‐induced marker (AIM) memory T cells in CD4+ and CD8+ T cells were investigated after the second vaccination and after the third/booster vaccination. Before the administering the third dose the 84% of vaccinated participants were found to have CD4+ memory response and 58% of participants with CD8+ memory response after the booster dose the number was increased to 100% in CD4+ and 90% CD8+ and the Spike‐specific T cells were found to be higher after 2 weeks of booster dose in CD4+ in comparison with CD8+ levels and after 3 months of booster vaccination the Spike specific CD4+ T cells were still present in vaccinees. 71 Overall, this investigation showcases that vaccinated individuals after a second dose were seen to have stronger specific memory T‐cell responses against the SARS‐CoV‐2 virus, and this was further elevated after the third vaccination along with the long time presence of both CD4+ and CD8+ levels with a little lower response towards Omicron variant. 71
7. MITIGATION STRATEGIES TO LIMIT COVID‐19 VARIANTS AND OMICRON
7.1. Booster usage
The currently available vaccines are designed to interact with the Spike region of SARS‐CoV‐2 thereby inducing immunity. Mutations in this region may change the scenario for antibody evasion as is the case with the Covid‐19 variants. These emerging variants may bypass the resistance from the antibodies, threatening the action of COVID‐19 vaccination. 60 , 72 So, the topic of administering booster vaccines to vaccinated individuals was a subject of debate. The imbalance between fully vaccinated individuals who had been administered booster vaccines and individuals who are yet to receive their first dose of vaccine provides the virus with a landscape to evolve with higher infectivity, and transmission rate. These events could have led to the possible emergence of Omicron. Currently, a booster dose is being administered to individuals in nations with high resource capability. 72 , 73
The individuals administered with a booster dose showed higher neutralizing capacity against Omicron, paving the way for booster vaccination. A study report states that the homologous booster vaccine works well with individuals who were heterologous‐vaccinated, thereby improving efficacy. Further evidence has justified this claim. A randomized study named COVBOOST compared the cell‐mediated and humoral antibody responses against the Delta variant and wild‐type virus. Here, people vaccinated with Pfizer–BioNTech and Oxford‐AstraZeneca were administered with booster vaccines from Pfizer, Moderna, Janssen, Novax, and Valneva. The observed immunogenicity levels in the BioNTech vaccine were found to work better with the Moderna booster. The AstraZeneca vaccine was found to work well with Moderna, proving the high reactivity of the Moderna booster. 74 , 75 Another study showed a higher immune response (humoral and cell‐mediated) when individuals who received CoronaVac were boosted with AZD1222, proving the consideration for the choice of booster intake. 76 A study performed by Henning et al., assessed the neutralization activity of antibodies of the BNT162b2 vaccine along with a single booster dose of BNT162b2. Surprisingly, this kind of immunization has proved effective to improve the immune response against Omicron and stopping the Omicron infection. It is speculated that this is possible only because of vaccination, as vaccination leads to affinity maturation. 77 However, further studies are required to understand this phenomenon in detail.
Eddy Perez et al., experimented with administering two vaccine doses of mRNA vaccine/CoronaVac as it was previously shown that Omicron can evade the neutralizing antibodies. Another study group specifically reported the use of booster dose to enhance the neutralizing antibodies of CoronaVac vaccine, therefore recommending booster vaccine. But data from Eddy Perez experiments suggest that even post booster immunization (BNT162b2) the vaccines have a lower effectivity rate against Omicron infection. These findings may also provide implications for countries that had vaccinated their population with CoronaVac. 78
According to the weekly epidemiology report from WHO (dated, May 8, 2022) in case of severe COVID‐19 disease the vaccine efficiency after the administering the booster mRNA vaccine to Pfizer, Ad26.COV2.S and Sinovac‐CoronaVac dual vaccinated population stand at 70% post the booster dosage whereas, in the case of symptomatic infection the mRNA booster after initially administering shows estimated vaccine efficiency between an estimation of 50%–70% but with time the efficiency of booster vaccine reduced drastically to below ≤50% as seen with Sinovac‐Coronavac and AstraZeneca. 79
The safety and immunogenicity of the Pfizer/BioNTech mRNA booster vaccine were evaluated by Yohei Seki et al., on Japanese health workers, herein they noticed a decrease in the neutralization activity of antibodies against Omicron but was able to 95% effectiveness against ancestral strain, the booster mRNA vaccine administered to health workers showed a threefold increase in neutralizing antibodies against Omicron and it was also observed that booster vaccine was able to induce humoral immunity and cross‐reactivity against both strains of Omicron and Delta. 80
The effectiveness of the ChAdOx1‐S booster vaccine against Omicron in England was evaluated by Freja Kirsebom et al., in England, the booster vaccine widely utilized was BNT162b2, mRNA‐1273, and ChAdOx1‐S. Herein, the group has compared the booster vaccine efficiency between BNT162b2 and ChAdOx1‐S in England, vaccine efficiency after the initial booster dosage of BNT162b2 and ChAdOx1‐S stood at ≥50% and ≥60% and decreased to 30.6% and 37.2% after more than 15 weeks of dosage for the age group of 40–64 years, for aged 65 years and above the booster vaccine efficiency stood at 66.1% for ChAdOx1‐S and 68.5% BNT162b2 that was reduced to 44.5% in ChAdOx1‐S and 54.1% in BNT162b2 after 5–9 weeks of booster vaccination, due to limited study the vaccine efficiency after 10 weeks were limited but evidence speculate further decrement of vaccine efficiency, but this doesn't limit the efficiency of ChAdOx1‐S or BNT162b2 as both vaccines protect against SARS‐CoV‐2 virus and its variants. 81
But the question that arises here is the effectiveness of the booster vaccine? And is it feasible to invest in them? Countries vaccinating their population with booster vaccines have been experiencing increasing cases of COVID‐19 in recent times whereas South Africa with no booster vaccination and only 30% of the population fully vaccinated shows signs of a decline in COVID‐19 cases. Therefore, in‐depth studies are required to study the various facets of molecular genetics involved. 76
7.2. Development of variant‐specific vaccine
The reinfection rates in vaccinated individuals and the increased risk of contracting the Omicron variant have brought the attention of researchers towards vaccine efficiency. The emergence of Omicron has shown that the administration of a booster vaccine is the only preventive measure that could be executed against Omicron. 82
The importance given to the development of vaccines that are specific to new emerging variants is on the rise. Most vaccines have effective neutralization potential against new variants. However, several studies have reported certain vaccines to be found ineffective or less effective against WT SATS‐CoV‐2. 67 Some reports suggest that mRNA‐1273.213, a multifunctional vaccine developed by Moderna neutralizes mutations present in Omicron, Delta, and Beta. This vaccine's immunogenicity is yet to be ascertained and further in‐depth studies are needed. 15
7.3. CRISPR‐Cas12a‐based detection assay for Omicron
With enhanced mutations in its Spike region have provided variant Omicron with strengthened capabilities towards COVID‐19 virus vaccines, the deletion of spike protein 69–70 prompts the Reverse transcription and polymerase chain reaction (RT‐PCR) to reveal a negative result or known as spike gene target failure (SGFT) (Bal et al., 2021). The 69–70 deletions were earlier detected in Alpha variant and were used to distinguish this variant, however, with the emergence of sub‐lineage of Omicron namely, BA.1, BA.2, and BA.3 it is difficult to distinguish the variants with the deletions 83 and lineage BA.2 do not possess the deletion 69–70 in its spike region making the detection of variant gruelling. To solve this problem Liang Y et al., developed a CRISPR‐Cas12a‐based assay by combining with RT‐PCR that detects and distinguishes variants in any condition. 84
Liang et al. designed and developed two Cas12a‐crRNA complex specific to variant Omicron namely, crRNA‐S‐37X (an extra PAM sequence was added) to detect the mutation S371L, S375F, and S373P and crRNA‐S‐49X for the detection of G496S, Q493R, and Q498R. the designed system was evaluated by utilizing other variant for detection and the designed assay was able to distinguish and detect the required VOC. 84 The crRNA complex possesses 3‐4 mutations of Omicron that can validate, diagnose and distinguish Omicron thereby providing a simpler path towards tracking and detecting Omicron. 84 This Omicron‐specific assay can significantly detect Omicron thus helping in monitoring and detecting Omicron globally.
8. RECENT DEVELOPMENTS IN OMICRON
As Omicron is continuing to infect the global population on a large scale, the dominance of this variant can change the infection rate of the COVID‐19 pandemic with its broad range of mutations, infectivity, transmissibility, and its role in downplaying the functions of immune system. Currently, four lineages of Omicron namely, B.1.1.529, BA.1, BA.2, and BA.3 are spreading rapidly with B.1.1529 becoming the dominant strain infecting millions of people. 85
The WHO on January 21, 2022, published its 7th recommended briefings and actions to be undertaken against Omicron. In this latest report, the WHO specified that Omicron exhibits a lower mortality rate in comparison to other variants, but due to its high transmission rate it results in an increased hospitalization rate and subsequent increase in COVID‐19 cases worldwide. Other factors that cause increased hospitalizations are shorter intervals of infection and an increase in the rate of asymptomatic infections which was proved with data published from the Republic of Korea showing 2.22 days for serial infection with Omicron. 85 , 86 Due to the lower immunization rates, possibilities of reinfection with COVID‐19 is higher than expected. In addition, higher mutation rates have added further to the burden. Currently, the WHO is evaluating the efficiency of COVID‐19 vaccines and has set up the Technical Advisory Group on COVID‐19 Vaccine Composition (TAG‐CO‐VAC) to evaluate and mitigate emerging VOCs along with gauging vaccine efficacy for COVID‐19. 85 The WHO is continuing to maintain Omicron as a variant of concern and has implemented various measures to curb the spread of this virus. This statement comes after the UK Health Security Agency (UKHSA) reported on the emergence of more than 1000 cases of Omicron BA.2 lineage. It is also reported that BA.2 is shown to have a higher transmission rate in comparison to BA.1. In addition, the BA.2 lineage has shown an increased growth rate in most regions of England. But it was also reported that the current vaccines are still effective against BA.2 and booster dose was shown to elevate the immune response against BA.2. 87 Recently, reports on Antibody Neutralization of Omicron linages (with BNT162b2 vaccine), BA.1 and BA.2 were reported by Jingyou Yu et al. They have shown the similarities in neutralization of antibodies between BA.1 and BA.2. The study concluded that BA.1 shows a reduced (1.3–1.4‐fold) neutralization against BA.2 and the usage of a booster BNT162b2 vaccine shows consistent neutralizing antibody response towards BA.1 and BA.2. It is also suggested that surge of BA.2 infection is very likely related to increased transmission instead of strengthened immune evasion. 88
The NTD domain of SARS‐CoV‐2 and its variants stimulates the releases of various cytokines in humans that increase the severity of the disease. The presence of a high number of NTD mutations in Omicron shows no changes towards its regulation of cytokine activity. 89 Marina et al. discovered the usage of Olverembatinib and Ponatinib for the inhibition of NTD‐regulated cytokine activity, as they can inhibit the cytokine release of NTD thereby suppressing the actions of Omicron NTD. This has justified the therapeutic usage of Olverembatinib and Ponatinib towards the treatment of COVID‐19. 89
9. NEOCOV: A POSSIBLE ARRIVAL OF A NEW VARIANT OR AN ANTIGENIC SHIFT?
The Coronavirus family members have been known to cause three major outbreaks in the last two decades. These were SARS‐CoV, MERS‐CoV, and the current SARS‐CoV‐2. 32 , 90 The lineages of MERS‐CoV are NeoCoV, PDF‐2180‐CoV, HKU5‐CoV, and EriCoV‐HKU31 (hedgehog coronaviruses). The NeoCoV and PDF‐2180‐CoV have shown genome‐level similarity with MERS‐CoV. A recent study by Qing et al. has shown that NeoCoV and PDF‐2180‐CoV can interact with the ACE2 receptor of bats and use it as their functional receptor. 90 In addition, they found the use of S1‐CTD as the RBD for their specific interaction with ACE2 receptors. This raised major concern since NeoCoV and PDF‐2180‐CoV have the ability to cross the species barrier, therefore, infecting the human population. However, their current genomic sequence is not efficient to use the ACE2 receptor in humans. 32 In NeoCoV, incompatible residue presented in 337‐342 is responsible for this inefficiency and it was also revealed that N338 is a significant residue that restricts the usage of human ACE2 receptors. 32 It was hypothesized that an increase in hydrophobicity in the residue region of T510 of NeoCoV might develop a potency to bind with human ACE2 receptor. The humoral immunity stimulation due to immunization or past infection presumably is ineffective to protect humans from this infection because neither MERS‐CoV antibodies or SARS‐CoV‐2 vaccination cannot stop the infection due to its higher transmissibility rate. 32
Therefore, close monitoring of antigen shift of these viruses is required due to the fact of natural selection, evolution, and cross‐species transmission. Considering the mutational landscape of Omicron and other variants the possibility of these viruses' potentiality to infect humans through natural adaptation is very likely.
10. RECOMBINANT VARIANT XE?
Variant or could be said as a recombinant variant of both strains of Omicron BA.1 and BA.2 is currently making headlines globally, in recent days recombinant strains are emerging all over the world making it as a matter of concern and the possibility for the emergence of a recombinant strains occurs when a living organism is infected with two or more than two strains of SARS‐CoV‐2 virus thus leading to merging of its genetic material during its replication and leading to formation of a new strain within the host's body and this happened due to the prevalence of multiple variants in environment. 87
XE is a recombinant strain from Omicron's sublineages and was detected in the United Kingdom on January 19 2022 and was assessed to have a growth rate of 9.8% than of BA.2; however, this statement requires further investigation according to UK Health Security Agency (UKHSA) the current cases of XE as of March 22, 2022 is 763. 91 According to UKHSA and WHO, XE variant contains NSP1‐6 mutations derived from BA.1 and the rest of the genome contains BA.2 genome, currently more evidence is needed to study the variant's transmissibility, and vaccine effectiveness, and the strain's severity in humans. XE variant have spread to other countries such as Thailand, New Zealand, Israel, Cambodia, and recently India reported it's first case of XE variant on April 8, 2022, currently more investigation is necessary along with appropriate precaution by the people to eradicate the variants of COVID‐19. 91 , 92 Table 4 represent the information for all available COVID vaccines.
Table 4.
List of available COVID‐19 vaccines developed by various pharmaceutical enterprises.
| Vaccine | Manufacturing company | Country of Origin | Vaccines Type | Targeted region | Efficacy against SARS‐CoV‐2 (varies) | Maintenance temperature | Dose | Reference |
|---|---|---|---|---|---|---|---|---|
| AZD1222 ChAdOx1 nCoV‐19 | AstraZeneca, Oxford University, and Serum Institute | USA, UK, and Europe | Nonreplicating viral vector | Spike protein | 70%–90% |
No freezing required Unopened: +2°C to +8°C Usage temp (6 h): Up to +30°C |
0.5 ml/dose | Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| mRNA‐1273 | Moderna | USA | mRNA | Spike protein | 94% |
Frozen: −50°C to −15°C Unopened: +2°C to +8°C Unopened 24 h: +2°C to +25°C |
0.5 ml/dose 100 μg |
Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| BNT162b2 and BNT162b1 | Pfizer‐BioNTech | USA | mRNA | Spike protein | 94% |
Frozen/Unopened: 80°C to −60°C Before dilution: +2°C to +8°C |
0.3 ml/dose 30 μg |
Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| Ad26.COV2.S | Johnson & Johnson | USA | Non‐replicating viral vector | Spike protein | 66%–80% |
Light sensitive storage: +2°C to +8°C Unopened vial (24 h): +9°C to +25°C |
0.5 ml/dose | Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| Covaxin | Bharat Biotech | India | Inactivated viral load | Complete virus | 77.8% |
No freezing storage: +2°C to +8°C |
0.5 ml/dose 6 μg |
Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| CoronaVac | Sinovac | China | Inactivated viral load | Complete virus | 50%–84% |
No freezing light sensitive storage and usage: +2°C to +8°C |
0.5 ml/dose 3 μg |
Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| Ad5‐nCoV | CanSioBio | China | Nonreplicating viral vector | Spike protein | 67.2% |
No freezing storage and usage: +2°C to +8°C |
0.5 ml/dose | Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| Sputnik/Gam‐COVID‐Vac | Gamaleya | Russia | Nonreplicating viral vector | Spike protein | 70%–90% | Storage and usage: +2°C to +8°C | 0.5 ml/dose | Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| BBIBP‐CorV | Sinopharm | China | Inactivated viral load | Complete virus | 78% |
No freezing light sensitive storage and usage: +2°C to +8°C |
0.5 ml/dose 4 μg |
Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
| CVnCoV | CureVac | Germany | Inactivated viral load | Spike Protein |
47% (Phase‐III trails) |
Frozen: −60°C Unopened vial (3 months): +2°C to +8°C |
12 μg | Hadj Hassine 26 ; Kumar et al. 93 ; Forchette et al. 94 ; Fiolet et al. 64 |
11. CONCLUSION
The Omicron variant has downplayed all other arisen variants. It emerged in South Africa and Botswana. The variant was a result of multiple mutations in the Spike region larger than every other arisen variant. Omicron was declared as a VOC on November 26, 2021. Omicron was quick to spread at a rapid pace and outpowering the previous variants with its enhanced mutation and spearheading the immune evasion property has led to widespread reinfections. Omicron was proved to have a higher infectivity rate, better ability to evade the immune system, capable of neutralizing antibodies from vaccines, and having higher binding affinity of ACE2 provides it with high transmissibility. The broad mutation discovered in its spike region is the primary reason for the induction of these changes along with newer mutations. The identification of Omicron infection in vaccinated individuals or individuals with past infection history shows the immune evasion property and re‐infective nature of Omicron. 41
RT‐PCR which is commonly employed in the detection of SARS‐CoV‐2 was proved to be ineffective to detect Omicron due to its S‐genome target failure (SGFT) thereby bypassing this diagnostic test. However, a genome sequence analysis can detect Omicron. Therefore, the current rapid antigen test requires to be modified for the detection of Omicron.
The presence of mutations within the RBD complex, NTD region, RBM, Furin site, S1, and S2 regions have a consequential action in the interaction of ACE2 receptors making Omicron deadly. 95 The mutation of key areas such as H655Y, N679K, E484A, and P681H had increased the binding affinity of the ACE2‐RBD region. R203K, P13L, and G204R mutational areas generally elevate the RNA expression level. Due to a high number of mutations in the RBD region, there will be a possibility for receptor shift in Omicron and ACE2 may not function as a receptor for Omicron. 96 Omicron was capable to abolish the neutralization activities of all antibody Classes 1–4 with few critical mutations in Q493R, N440K, and G446S, and with this variant, the vaccine efficiency was drastically reduced. The presence of R346K mutation in Omicron proved fatal for antibody resistance to the vaccine. 97 Neutralizing antibodies and the Spike protein region are key to understanding the S region epitome and nAb activity which is key towards vaccination development. 98
The development of a vaccine to curb Omicron and its variants is proving to be effective along with the need for a booster vaccine was initiated due to the rapid prevalence of Omicron and currently, major countries provide booster vaccination. The prevalence of an unvaccinated population has further provided an impetus for the further evolution of the virus. The current booster vaccinations have proven to be very effective against Omicron while few vaccines have shown ineffectiveness against it. The arrival of various variants with a mutation in the Spike region prompted the scientific community to design variant‐specific vaccines based on the S region to combat future arising variants and this may require further analysis and in‐depth studies.
WHO recently published its weekly epidemiological update on the COVID‐19 pandemic and according to the report from February 28, 2022 to March 6, 2022 the global SARS‐CoV‐2 virus cases continue to decline by 5% and 8% in comparison to last week and only the Western Pacific region indicates the increase in COVID‐19 cases by 46% and the weekly COVID‐19 related death also increased to 29% and with a decrease in SARS‐CoV‐2 virus cases in global level, several countries have reduced relaxation in their respective regions but it is worth noting that Omicron variant is the dominating VOC around the globe. 99 The new weekly epidemiological update on the COVID‐19 pandemic until May 8, 2022 published on May 11, 2022 shows a decline in SARS‐CoV‐2 virus since March 2022 and there was a 12% decrease in SARS‐CoV‐2 cases globally with a 25% decrease in COVID‐19 related death globally with only American region and African region showing an increase in COVID‐19 cases 79 but WHO has cautioned countries regarding the declining trend of COVID‐19 that is to be interpreted cautiously.
Therefore, to control COVID‐19, driving up the vaccination process is the only solution to eradicate the virus. Given the current scenario with the rise of BA.2 subvariant and the prevalence of recombinant strains globally, the need for better precautions, community surveillance, and due diligence from every government is necessary to tackle the current prevailing situation and it is highly likely that there might be more variants on the way. Past experience and the cooperation between scientific communities and health bodies across the globe may help to understand the virus better and to develop therapeutics that may overcome the COVID‐19 pandemic.
AUTHOR CONTRIBUTIONS
Akash K: Conceptualize and written the whole manuscript. Deepak Kumar, Sachin K. Singh, and Gaurav Gupta: Participated in the design of study and improves grammar, Dinesh K. Chellappan and Kamal Dua: helps in review and editing, Rupak Nagraik Avinash Sharma: Design, Validate and supervised the whole manuscript.
CONFLICT OF INTEREST
The author declares no conflict of interest.
ACKNOWLEDGMENTS
The authors are thankful to the University to provide the appropriate facilities and data to complete this review.
K A, Sharma A, Kumar D, et al. Molecular aspects of Omicron, vaccine development, and recombinant strain XE: a review. J Med Virol. 2022;94:4628‐4643. 10.1002/jmv.27936
DATA AVAILABILITY STATEMENT
The data to support the findings for this study are available with the corresponding author upon request.
REFERENCES
- 1. Rastogi YR, Sharma A, Nagraik R, Aygün A, Şen F. The novel coronavirus 2019‐nCoV: its evolution and transmission into humans causing global COVID‐19 pandemic. Int J Environ Sci Technol. 2020;17(10):4381‐4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Islam MR, Hoque MN, Rahman MS, et al. Genome‐wide analysis of SARS‐CoV‐2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. 2020; 10(1):14004. Erratum in: Sci Rep. 2021 Oct 12;11(1):20568 10.1038/s41598-020-70812-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dyson L, Hill EM, Moore S, et al. Possible future waves of SARS‐CoV‐2 infection generated by variants of concern with a range of characteristics. Nat Commun. 2021;2(1):5730. 10.1038/s41467-021-25915-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization . Tracking SARS‐CoV‐2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/; 2022. [PubMed]
- 5. Bian L, Gao Q, Gao F, et al. Impact of the delta variant on vaccine efficacy and response strategies. Expert Rev Vaccines. 2021;20(10):1201‐1209. 10.1080/14760584.2021.1976153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choudhary OP, Dhawan M, Priyanka. Omicron variant (B.1.1.529) of SARS‐CoV‐2: threat assessment and plan of action. Int J Surg. 2022;97:106187. 10.1016/j.ijsu.2021.106187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fenwick C, Turelli P, Ni D, et al. SARS‐CoV‐2 Omicron potently neutralized by a novel antibody with unique Spike binding properties. bioRxiv. 2022. 10.1101/2022.03.18.484873 [DOI]
- 8. Cloete J, Kruger A, Masha M, et al. Paediatric hospitalisations due to COVID‐19 during the first SARS‐CoV‐2 Omicron (B.1.1.529) variant wave in South Africa: a multicentre observational study. Lancet Child Adolesc Health. 2022;6(5):294‐302. 10.1016/S2352-4642(22)00027-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS‐CoV‐2 Omicron variant in Southern Africa. Nature. 2022;603(7902):679‐686. 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS‐CoV‐2 Omicron variant in South Africa: a data linkage study. Lancet. 2022;399(10323):437‐446. 10.1016/S0140-6736(22)00017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Prevention and Control. Science brief: Omicron (B.1.1.529) variant; 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html [PubMed]
- 12. Baral P, Bhattarai N, Hossen ML, et al. Mutation‐induced changes in the receptor‐binding interface of the SARS‐CoV‐2 delta variant B.1.617.2 and implications for immune evasion. Biochem Biophys Res Commun. 2021;574:14‐19. 10.1016/j.bbrc.2021.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. World Health Organization . Update on Omicron. who.int/news/item/28-11-2021-update-on-omicron
- 14. Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS‐CoV‐2 variant: unique features and their impact on pre‐existing antibodies. J Autoimmun. 2022;126:102779. 10.1016/j.jaut.2021.102779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He X, Hong W, Pan X, Lu G, Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm. 2020;2(4):838‐845. 10.1002/mco2.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bakhshandeh B, Jahanafrooz Z, Abbasi A, et al. Mutations in SARS‐CoV‐2; consequences in structure, function, and pathogenicity of the virus. Microb Pathog. 2021;154:104831. 10.1016/j.micpath.2021.104831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhola S, Trisal J, Thakur V, et al. Neurological toll of COVID‐19. Neurol Sci. 2022;43:2171‐ 2186. 10.1007/s10072-022-05875-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torre‐Fuentes L, Matías‐Guiu J, Hernández‐Lorenzo L, et al. ACE2, TMPRSS2, and furin variants and SARS‐CoV‐2 infection in Madrid, Spain. J Med Virol. 2021;93(2):863‐869. 10.1002/jmv.26319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khan NA, Al‐Thani H, El‐Menyar A. The emergence of new SARS‐CoV‐2 variant (Omicron) and increasing calls for COVID‐19 vaccine boosters—the debate continues. Travel Med Infect Dis. 2021;45:102246. 10.1016/j.tmaid.2021.102246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petersen E, Ntoumi F, Hui DS, et al. Emergence of new SARS‐CoV‐2 variant of concern Omicron (B.1.1.529)—highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID‐19 response and control efforts. Int J Infect Dis. 2022;114:268‐272. 10.1016/j.ijid.2021.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ingraham NE, Ingbar DH. The Omicron variant of SARS‐CoV‐2: understanding the known and living with unknowns. Clin Transl Med. 2021;11(12):e685. 10.1002/ctm2.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pawłowski P. SARS‐CoV‐2 variant Omicron (B.1.1.529) is in a rising trend of mutations increasing the positive electric charge in crucial regions of the spike protein S. Acta Biochim Pol. 2021;69:263‐264. 10.18388/abp.2020_6072 [DOI] [PubMed] [Google Scholar]
- 23. Kandeel M, Mohamed MEM, Abd El‐Lateef HM, Venugopala KN, El‐Beltagi HS. Omicron variant genome evolution and phylogenetics. J Med Virol. 2021;1:6‐1632. 10.1002/jmv.27515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McLean G, Kamil J, Lee B, et al. The impact of evolving SARS‐CoV‐2 mutations and variants on COVID‐19 vaccines. mBio. 2022;13:e0297921. 10.1128/mbio.02979-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamasoba D, Kimura I, Nasser H. Virological characteristics of SARS‐CoV‐2 BA.2 variant. bioRxiv. 2022. 10.1101/2022.02.14.480335 [DOI]
- 26. Hadj Hassine I. Covid‐19 vaccines and variants of concern: a review. Rev Med Virol. 2021:e2313. 10.1002/rmv.2313 [DOI] [PMC free article] [PubMed]
- 27. Cameroni E, Bowen JE, Rosen LE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. Nature. 2021;602:664‐670. 10.1038/s41586-021-04386-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lippi G, Adeli K, Plebani M. Commercial immunoassays for detection of anti‐SARS‐CoV‐2 spike and RBD antibodies: urgent call for validation against new and highly mutated variants. Clin Chem Lab Med. 2021;60:338‐ 342. 10.1515/cclm-2021-1287 [DOI] [PubMed] [Google Scholar]
- 29. Malik JA, Ahmed S, Mir A, et al. The SARS‐CoV‐2 mutations versus vaccine effectiveness: new opportunities to new challenges. J Infect Public Health. 2022;15(2):228‐240. 10.1016/j.jiph.2021.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jhun H, Park HY, Hisham Y, Song CS, Kim S. SARS‐CoV‐2 Delta (B.1.617.2) variant: a unique T478K mutation in receptor binding motif (RBM) of spike gene. Immune Netw. 2021;21(5):e32. 10.4110/in.2021.21.e32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moghaddar M, Radman R, Macreadie I. Severity, pathogenicity and transmissibility of Delta and Lambda variants of SARS‐CoV‐2, toxicity of spike protein and possibilities for future prevention of COVID‐19. Microorganisms. 2021;9(10):2167. 10.3390/microorganisms9102167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiong Q, Cao L, Ma C, et al. Close relatives of MERS‐CoV in bats use ACE2 as their functional receptors. bioRxiv. 2022. 10.1101/2022.01.24.477490 [DOI] [PMC free article] [PubMed]
- 33. Rahimi F, Kamali N, Bezmin Abadi AT. The Mu strain: the last but not least circulating ‘variant of interest’ potentially affecting the COVID‐19 pandemic. Future Virol. 2021;17:5‐8. 10.2217/fvl-2021-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gerdol M, Dishnica K, Giorgetti A. Emergence of a recurrent insertion in the N‐terminal domain of the SARS‐CoV‐2 spike glycoprotein. Virus Res. 2022;310:198674. 10.1016/j.virusres.2022.198674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rath SL, Padhi AK, Mandal N. Scanning the RBD‐ACE2 molecular interactions in Omicron variant. BioRxiv. 2021. 10.1101/2021.12.12.472253 [DOI] [PMC free article] [PubMed]
- 36. Thakur V, Ratho RK. OMICRON (B.1.1.529): a new SARS‐CoV‐2 variant of concern mounting worldwide fear. J Med Virol. 2021;94:1821‐1824. 10.1002/jmv.27541 [DOI] [PubMed] [Google Scholar]
- 37. Kannan S, Shaik Syed Ali P, Sheeza A. Omicron (B.1.1.529)—variant of concern—molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci. 2021;25(24):8019‐8022. 10.26355/eurrev_202112_27653 [DOI] [PubMed] [Google Scholar]
- 38. Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant omicron in South Africa. J Med Virol. 2021;94:1728‐1733. 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 39. Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS‐CoV‐2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35(I13):109292. 10.1016/j.celrep.2021.109292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS‐CoV‐2 variants. Nat Rev Genet. 2021;22(12):757‐773. 10.1038/s41576-021-00408-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang X, Wu S, Wu B, et al. SARS‐CoV‐2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Sig Transduct Target Ther. 2021;6:430. 10.1038/s41392-021-00852-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS‐CoV‐2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28:490‐495. 10.1038/s41591-021-01678-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller NL, Clark T, Raman R, Sasisekharan R. Insights on the mutational landscape of the SARS‐CoV‐2 Omicron variant. bioRxiv [Preprint]. 2021. 10.1101/2021.12.06.471499 [DOI] [PMC free article] [PubMed]
- 44. Zahradník J, Marciano S, Shemesh M, et al. SARS‐CoV‐2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6:1188‐1198. 10.1038/s41564-021-00954-4 [DOI] [PubMed] [Google Scholar]
- 45. Gong SYu, Chatterjee D, Richard J. Contribution of single mutations to selected SARS‐CoV‐2 emerging variants Spike antigenicity. bioRxiv. 2021. 10.1101/2021.08.04.455140 [DOI] [PMC free article] [PubMed]
- 46. Winger A, Caspari T. The spike of concern—the novel variants of SARS‐CoV‐2. Viruses. 2021;13(6):1002. 10.3390/v13061002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Al‐Qahtani AA. Mutations in the genome of severe acute respiratory syndrome coronavirus 2: implications for COVID‐19 severity and progression. J Int Med Res. 2022;50(3):3000605221086433. 10.1177/03000605221086433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pascarella S, Ciccozzi M, Bianchi M, Benvenuto D, Cauda R, Cassone A. The electrostatic potential of the Omicron variant spike is higher than in delta and delta‐plus variants: a hint to higher transmissibility? J Med Virol. 2021. 10.1002/jmv.27528 [DOI] [PubMed]
- 49. Redd AD, Nardin A, Kared H. Minimal cross‐over between mutations associated with Omicron variant of SARS‐CoV‐2 and CD8+ T cell epitopes identified in COVID‐19 convalescent individuals. bioRxiv [Preprint]. 2021. 10.1101/2021.12.06.471446 [DOI] [PMC free article] [PubMed]
- 50. Ma W, Yang J, Fu H, et al. Genomic perspectives on the emerging SARS‐CoV‐2 Omicron variant. bioRxiv. 2022. 10.1101/2022.01.05.474231 [DOI] [PMC free article] [PubMed]
- 51. Cui Z, Liu P, Wang N, et al. Structural and functional characterizations of altered infectivity and immune evasion of SARS‐CoV‐2 Omicron variant. bioRxiv. 2021. 10.1101/2021.12.29.474402 [DOI] [PMC free article] [PubMed]
- 52. Lupala CS, Ye Y, Chen H, Su XD, Liu H. Mutations on RBD of SARS‐CoV‐2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun. 2022;590:34‐41. 10.1016/j.bbrc.2021.12.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature. 2021;602:657‐663. 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature. 2021;602:671‐675. 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 55. Wang R, Chen J, Wei GW. Mechanisms of SARS‐CoV‐2 evolution revealing vaccine‐resistant mutations in Europe and America. J Phys Chem Lett. 2021;12(49):11850‐11857. 10.1021/acs.jpclett.1c03380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu L, Mok BW, Chen LL, et al. Neutralization of SARS‐CoV‐2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021;73:ciab1041. 10.1093/cid/ciab1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cohn BA, Cirillo PM, Murphy CC, Krigbaum NY, Wallace AW. SARSCoV‐2 vaccine protection and deaths among US veterans during 2021. Science. 2021;375:0620. 10.1126/SCIENCE.ABM0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barnes CO, Jette CA, Abernathy ME, et al. SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682‐687. 10.1038/s41586-020-2852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yan R, Wang R, Ju B, et al. Structural basis for bivalent binding and inhibition of SARS‐CoV‐2 infection by human potent neutralizing antibodies. Cell Res. 2021;31:517‐525. 10.1038/s41422-021-00487-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ju B, Zheng Q, Guo H, et al. Molecular basis of broad neutralization against SARS‐CoV‐2 variants including Omicron by a human antibody. bioRxiv. 2022. 10.1101/2022.01.19.476892 [DOI]
- 61. Chen J, Gao K, Wang R, Wei GW. Revealing the threat of emerging SARS‐CoV‐2 mutations to antibody therapies. J Mol Biol. 2021;433(18):167155. 10.1016/j.jmb.2021.167155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen J, Wang R, Gilby NB, Wei GW. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. ArXiv [Preprint]. 2021. [DOI] [PMC free article] [PubMed]
- 63. McCallum M, Czudnochowski N, Rosen LE, et al. Structural basis of SARS‐CoV‐2 Omicron immune evasion and receptor engagement. Science. 2022;375:eabn8652‐eabn8868. 10.1126/science.abn8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fiolet T, Kherabi Y, MacDonald CJ, Ghosn J, Peiffer‐Smadja N. Comparing COVID‐19 vaccines for their characteristics, efficacy and effectiveness against SARS‐CoV‐2 and variants of concern: a narrative review. Clin Microbiol Infect. 2022;28(2):202‐221. 10.1016/j.cmi.2021.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Raman R, Patel KJ, Ranjan K. COVID‐19: unmasking emerging SARS‐CoV‐2 variants, vaccines and therapeutic strategies. Biomolecules. 2021;11(7):993. 10.3390/biom11070993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Janik E, Niemcewicz M, Podogrocki M, Saluk‐Bijak J, Bijak M. Existing drugs considered as promising in COVID‐19 therapy. Int J Mol Sci. 2021;22(11):5434. 10.3390/ijms22115434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. He C, Yang J, He X, et al. A bivalent recombinant vaccine targeting the S1 protein induces neutralizing antibodies against both SARS‐CoV‐2 variants and wild‐type of the virus. MedComm. 2021;2(3):430‐441. 10.1002/mco2.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nersisyan S, Zhiyanov A, Zakharova M, et al. Alterations in SARS‐CoV‐2 Omicron and Delta peptides presentation by HLA molecules. bioRxiv. 2022;27:e13354. 10.1101/2022.02.21.481175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Y, Wang X, Jin J, et al. T‐cell responses to SARS‐CoV‐2 Omicron spike epitopes with mutations after the third booster dose of an inactivated vaccine. J Med Virol. 2022;94:3998‐4004. 10.1002/jmv.27814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Keeton R, Tincho MB, Ngomti A, et al. T cell responses to SARS‐CoV‐2 spike cross‐recognize Omicron. Nature. 2022;603(7901):488‐492. 10.1038/s41586-022-04460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Naaber Paul, Tserel Liina, Kangro Kadri, et al. Protective antibodies and T cell responses to Omicron variant three months after the booster dose of BNT162b2 vaccine. medRxiv. 2022. 10.1101/2022.03.04.22271890 [DOI] [PMC free article] [PubMed]
- 72. Goldberg Y, Mandel M, Bar‐On YM, et al. Waning immunity after the BNT162b2 vaccine in Israel. N Engl J Med. 2021;385:e85. 10.1056/NEJMoa2114228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chagla Z, Pai M. COVID‐19 boosters in rich nations will delay vaccines for all. Nat Med. 2021;27:1659‐1660. 10.1038/s41591-021-01494-4 [DOI] [PubMed] [Google Scholar]
- 74. Munro APS, Janani L, Cornelius V, et al. Safety and immunogenicity of seven COVID‐19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov‐19 or BNT162b2 in the UK (COV‐BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet. 2021;398:2258‐2276. 10.1016/S0140-6736(21)02717-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yorsaeng R, Suntronwong N, Phowatthanasathian H, et al. Immunogenicity of a third dose viral‐vectored COVID‐19 vaccine after receiving two–dose inactivated vaccines in healthy adults. Vaccine. 2021;40:524‐530. 10.1016/j.vaccine.2021.11.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Singhal T. The emergence of Omicron: challenging times are here again! Indian J Pediatr. 2022;1–7:490‐496. 10.1007/s12098-022-04077-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gruell H, Vanshylla K, Tober‐Lau P, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS‐CoV‐2 Omicron variant. Nat Med. 2022;28:477‐480. 10.1038/s41591-021-01676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pérez‐Then E, Lucas C, Monteiro VS, et al. Neutralizing antibodies against the SARS‐CoV‐2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med. 2022;28:481‐485. 10.1038/s41591-022-01705-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. World Health Organization . Weekly epidemiological update on COVID‐19. Accessed May 11, 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---11-may-2022.
- 80. Seki Y, Yoshihara Y, Nojima K, et al. Safety and immunogenicity of Pfizer/BioNTech SARS‐CoV‐2 mRNA third booster vaccine against SARS‐CoV‐2 Omicron variant in Japanese healthcare workers. medRxiv. 2022. 10.1101/2022.01.20.22269587 [DOI]
- 81. Kirsebom F, Andrews N, Sachdeva R, Stowe J, Ramsay M, Bernal JL. Effectiveness of ChAdOx1‐S COVID‐19 Booster Vaccination against the Omicron and Delta variants in England. medRxiv. 2022. 10.1101/2022.04.29.22274483 [DOI] [PMC free article] [PubMed]
- 82. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654‐656. 10.1038/s41586-021-04387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bal A, Destras G, Gaymard A, et al. COVID‐Diagnosis HCL Study Group . Two‐step strategy for the identification of SARS‐CoV‐2 variant of concern 202012/01 and other variants with spike deletion H69‐V70, France, August to December 2020. Euro Surveill. 2021;26(3):2100008. 10.2807/1560-7917.ES.2021.26.3.2100008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liang Y, Lin H, Zou L, Deng X, Tang S. Rapid detection and tracking of Omicron variant of SARS‐CoV‐2 using CRISPR‐Cas12a‐based assay. Biosens Bioelectron. 2022;205:114098. 10.1016/j.bios.2022.114098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Parums DV. Editorial: The 2022 World Health Organization (WHO) priority recommendations and response to the Omicron variant (B.1.1.529) of SARS‐CoV‐2. Med Sci Monit. 2022;28:e936199. 10.12659/MSM.936199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hwang H, Lim JS, Song SA, et al. Transmission dynamics of the Delta variant of SARS‐CoV‐2 infections in South Korea. J Infect Dis. 2021;225:jiab586‐jiab799. 10.1093/infdis/jiab586 [DOI] [PubMed] [Google Scholar]
- 87. Mahase E. Omicron sub‐lineage BA.2 may have “substantial growth advantage,” UKHSA reports. BMJ. 2022;376:o263. 10.1136/bmj.o263 [DOI] [PubMed] [Google Scholar]
- 88. Yu J, Collier AY, Rowe M, et al. Comparable neutralization of the SARS‐CoV‐2 Omicron BA.1 and BA.2 Variants. medRxiv. 2022. 10.1101/2022.02.06.22270533 [DOI] [PMC free article] [PubMed]
- 89. Chan M, Holland EC, Gujral TS. Olverembatinib inhibits SARS‐CoV‐2‐Omicron variant‐mediated cytokine release. bioRxiv. 2022. 10.1101/2022.02.07.479443 [DOI] [PMC free article] [PubMed]
- 90. Lau SKP, Fan RYY, Luk HKH, et al. Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins. Emerg Microbes Infect. 2018;7(1):1‐11. 10.1038/s41426-018-0208-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.UK Health Security Agency—GOV.UK, SARS‐CoV‐2 variants of concern and variants under investigation in England: technical briefing 39. https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings
- 92. World Health Organization . Weekly epidemiological update on COVID‐19. Accessed April 5, 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---5-april-2022.
- 93. Kumar VM, Pandi‐Perumal SR, Trakht I, Thyagarajan SP. Strategy for COVID‐19 vaccination in India: the country with the second highest population and number of cases. NPJ Vaccines. 2021;6(1):60. 10.1038/s41541-021-00327-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Forchette L, Sebastian W, Liu T. A comprehensive review of COVID‐19 virology, vaccines, variants, and therapeutics. Curr Med Sci. 2021;41(6):1037‐1051. 10.1007/s11596-021-2395-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wu Cr, Yin Wc, Jiang Y, Xu HE. Structure genomics of SARS‐CoV‐2 and its Omicron variant: drug design templates for COVID‐19. Acta Pharmacol Sin. 2022;1‐3. 10.1038/s41401-021-00851-w [DOI] [PMC free article] [PubMed]
- 96. Saxena SK, Kumar S, Ansari S, et al. Characterization of the novel SARS‐CoV‐2 Omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol. 2021;94:1738‐1744. 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 97. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS‐CoV‐2. Nature. 2021;602:676‐681. 10.1038/s41586-021-04388-0 [DOI] [PubMed] [Google Scholar]
- 98. Metzger CMJA, Lienhard R, Seth‐Smith HMB, et al. PCR performance in the SARS‐CoV‐2 Omicron variant of concern? Swiss Med Wkly. 2021;151:w30120. 10.4414/smw.2021.w30120 [DOI] [PubMed] [Google Scholar]
- 99. World Health Organization . Weekly epidemiological update on COVID‐19. Accessed March 8, 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---8-march-2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data to support the findings for this study are available with the corresponding author upon request.


