Abstract
Immunological memory is the basis of protective immunity provided by vaccines and previous infections. Immunological memory can develop from multiple branches of the adaptive immune system, including CD4 T cells, CD8 T cells, B cells, and long‐lasting antibody responses. Extraordinary progress has been made in understanding memory to SARS‐CoV‐2 infection and COVID‐19 vaccines, addressing development; quantitative and qualitative features of different cellular and anatomical compartments; and durability of each cellular component and antibodies. Given the sophistication of the measurements; the size of the human studies; the use of longitudinal samples and cross‐sectional studies; and head‐to‐head comparisons between infection and vaccines or between multiple vaccines, the understanding of immune memory for 1 year to SARS‐CoV‐2 infection and vaccines already supersedes that of any other acute infectious disease. This knowledge may help inform public policies regarding COVID‐19 and COVID‐19 vaccines, as well as the scientific development of future vaccines against SARS‐CoV‐2 and other diseases.
Keywords: adenoviral vectors, coronavirus, hybrid immunity, memory B cells, memory T cells, mRNA vaccines, natural immunity, protein vaccine
1. INTRODUCTION
Protective immunity provided by vaccines is predicated on the existence of immunological memory: the capacity of the adaptive immune system to not only recognize a novel pathogen but to also remember it. Only in the past few decades have the cellular and molecular sources of immunological memory been defined, and much remains to be determined. The three main branches of the adaptive immune system are B cells (the source of antibodies, “Abs”), CD4 T cells, and CD8 T cells. Immune memory is encoded in four main compartments of adaptive immunity: memory CD8 T cells, memory CD4 T cells, memory B cells (BMem), and circulating Abs 1 (Figure 1). There is evidence of roles for B cells (including Abs), CD4 T cells, and CD8 T cells in protective immunity to SARS‐CoV‐2, and thus, it is important to study immune memory to SARS‐CoV‐2 and COVID‐19 vaccines to understand protective immunity against COVID‐19. Because of the size and scope of immunological studies of SARS‐CoV‐2 in humans, the large number of first‐time infections, the large number of first‐time vaccinations, and the diversity of COVID‐19 vaccines developed in a short period of time, there are now more data on human antigen‐specific immune responses to SARS‐CoV‐2 than any other acute pathogen. As a result, immune memory to SARS‐CoV‐2 is now a benchmark in human immunology for understanding antigen‐specific T cell and B cell memory.
Figure 1.
Components of immune memory. Virus‐specific CD4 T cells, CD8 T cells, Abs, and BMem cells constitute the four major components of immune memory to a viral infection
Immune memory to SARS‐CoV‐2 can be generated by infection (classically referred to as “natural immunity”), vaccination, or hybrid immunity. Hybrid immunity is the combination of infection‐induced immunity and vaccine‐induced immunity. 2 Each of these causes of immune memory is discussed in each section of this review. Overall, immune memory from prior infection, vaccination, or hybrid immunity each have distinctive characteristics. Previous infection can generate robust immune memory, 3 , 4 including memory CD8 T cell, CD4 T cell, BMem, durable Abs, and local immune memory (Figure 2). Epidemiological data on protective immunity in previously infected individuals are consistent with the immune memory measurements. Multiple large studies observe that prior infection provides approximately 80%–95% protection against symptomatic COVID‐19 reinfections for 8+ months, for SARS‐CoV‐2 ancestral strain and the Alpha through Delta VOCs, 5 , 6 , 7 , 8 , 9 , 10 , 11 and significant protection against disease with Omicron. 12 , 13 , 14
Figure 2.
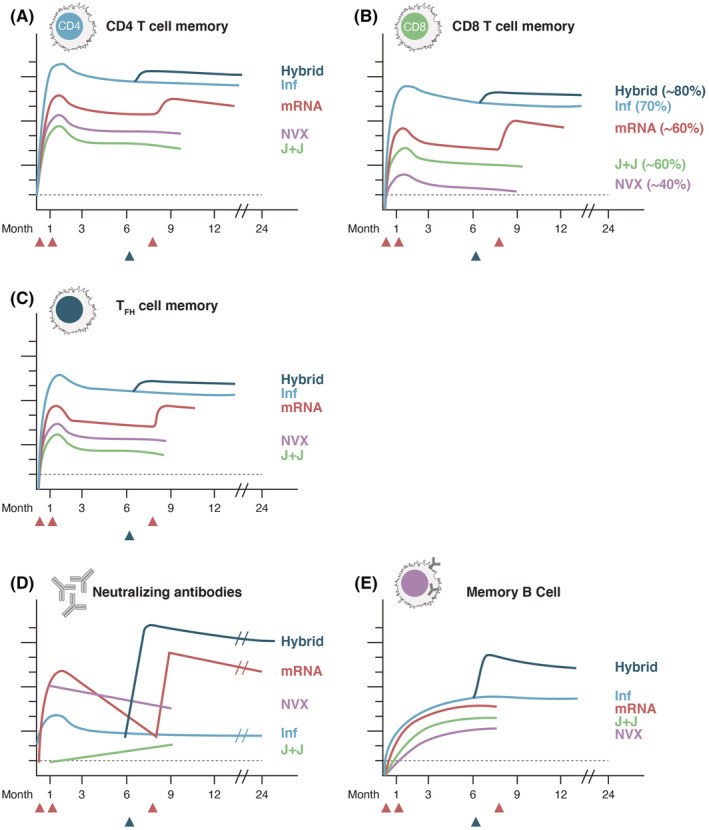
Kinetics of immune memory to SARS‐CoV‐2 infection and COVID‐19 vaccines. Schematics of immune memory components against SARS‐CoV‐2. (A) Memory CD8 T cells, (B) memory CD4 T cells, (C) memory TFH cells, (D) neutralizing antibodies, and (E) BMem cells. For T cell memory, with vaccines memory is to spike, and with infection, memory is to the entire virus. For B cell memory, spike‐specific is shown in all cases. "Inf" = SARS‐CoV‐2 infected. "Hybrid" = Hybrid immunity, infected and then vaccinated. "mRNA" = Moderna mRNA‐1273 or Pfizer/BioNTech BNT162b2, a 3 dose regimen. "NVX" = Novavax NVX‐CoV2373, given as the 2‐dose regimen in the main clinical trials. "J&J" = Janssen Ad26.COV2.S, given as the 1‐dose approved by EUAs. Lines are color coded by vaccine. CD8 T cell % indicates the % of individuals with detectable CD8 T cell memory at 3–6 months. Scales are non‐quantitative, but the antibody scale approximates log10 and the cellular scales approximate log2. For hybrid immunity, in this schematic, the vaccination occurs at approximately 6 months, indicated by the blue triangle. For mRNA vaccines, in this schematic, the 1st two dose are given at d1 and d21‐28, with the 3rd dose ("booster") given at approximately 8 months, indicated by the red arrows
COVID‐19 vaccines, focusing for the moment on 2‐dose mRNA vaccines (Moderna mRNA‐1273, Pfizer/BioNTech BNT162b2), clearly provide high levels of protective immunity against SARS‐CoV‐2 ancestral strain and the Alpha through Delta VOCs. 9 , 15 , 16 However, the high levels of vaccine immunity against detectable SARS‐CoV‐2 infections wanes over a period of months. 17 , 18 Immunity provided by 2‐dose ChAdOx1 (AstraZeneza/Oxford ChAdOx1‐nCoV‐19 AZD1222 “ChAdOx1”) is somewhat lower and wanes faster than the mRNA vaccines. 19 With hybrid immunity, neutralizing Ab (nAb) titers and breadth of recognition of SARS‐CoV‐2 variants are dramatically higher in previously infected individuals receiving at least one dose of a COVID‐19 RNA vaccine 2 (Figure 2). Hybrid immunity from vaccination plus subsequent infection (breakthrough infection) also results in similarly robust immune responses. 20 , 21 Multiple epidemiological studies have now validated those immunological findings, by observing that hybrid immunity results in more robust protection against COVID‐19 than either previous infection immunity or vaccine‐induced immunity. 9 , 11 These types of antigen exposure(s)—infection‐induced memory, vaccine‐induced memory, and hybrid immunity—each have distinct characteristics of immune memory (Figure 2), which are key to the observed protective immunity in each case. High circulating nAb titers can clearly provide protective immunity. However, such high nAb titers are not present in many cases, particularly after SARS‐CoV‐2 infection, and there is substantial evidence for roles of T cells, BMem, and local tissue immunity in protection from COVID‐19 22 , 23 (Goldblatt et al., this volume). 24 For example, previous infection provides substantial immunity with significantly lower circulating nAb titers than after BNT162b2 or ChAdOx1 vaccination. 19 Local tissue immunity contributes to protective immunity after infection (Figure 3). Different adaptive immunity mechanisms may be involved in protective immunity to differing degrees with immune memory generated by previous infection, vaccination, or hybrid immunity. Thus, each is discussed in this review for CD8 T cells, CD4 T cells, BMem, and Abs.
Figure 3.
Components of local tissue immunity. Human immune responses are most often measured in blood, but immune responses at local sites of infection and/or portals of entry are important and may not be directly reflected by blood measurements
2. CD8 T CELL MEMORY TO SARS‐CoV‐2
In general, CD8 T cells are important in the control and clearance of viral infections. 25 In particular, several lines of evidence suggest that CD8 T cells are a relevant and valuable component of the overall adaptive immune response to SARS‐CoV‐2. One line of evidence is derived from studies in acute SARS‐CoV‐2 infection, which observed that early CD8 T cell responses were significantly associated with milder disease. 26 An inverse association between CD8 T cell response magnitude and disease severity was also reported in 4 of 5 additional independent studies. 4 , 27 , 28 , 29 , 30 Additional evidence for a role of CD8 T cells comes from studies in non‐human primates. McMahan et al. 31 directly showed that depletion of CD8 T cells in COVID convalescent animals affected immunity against SARS‐CoV‐2 re‐challenge. An important role for CD8 T cell responses was also reported in an antibody‐independent COVID‐19 vaccine study. 32 See companion article by Goldblatt et al. for a review of T cells in protection 24 and ref. 33.
Multiple techniques are commonly utilized to measure antigen‐specific T cell responses, with antigen specificity ensured by the use of SARS‐CoV‐2 derived peptides or defined epitopes, utilized as pools or isolated epitopes. The techniques utilized included activation induced marker (AIM), ICS (intracellular cytokine staining), ELISPOT, and tetramer staining assays and their strengths and weaknesses are reviewed elsewhere. 34 , 35
2.1. CD8 T cell memory to SARS‐CoV‐2 infection
SARS‐CoV‐2‐specific CD8 T cells are detectable in approximately 70% of COVID‐19 cases 1 month after infection. 3 , 4 The frequency of responders then declines to approximately 50% by 8 months post‐infection. 3 , 4 The Dan et al. study included SARS‐CoV‐2‐specific CD8 T cell measurements from 169 COVID‐19 case subjects and the Cohen et al. study included 114 subjects, making them the two largest studies of CD8 T cell memory to an acute viral infection examining a 6+ month period. The estimated SARS‐CoV‐2‐specific memory CD8 T cell kinetics from the Dan et al. study was t1/2 = 125–190 days, while the Cohen et al. t1/2 was 196 days. That is strong concordance between the two studies, given that the studies utilized different CD8 T cell assays (AIM and ICS) and the calculations were based predominantly on cross‐sectional sampling. By comparison, a rigorous study using in vivo deuterium labeling found yellow fever virus (YFV) vaccine CD8 T cell responses to have an initial t1/2 of 123 days. 36 Given that the YFV vaccine is highly effective, elicits robust CD8 T cells as a live attenuated viral vaccine, and deuterium labeling determined a subsequent t1/2 of 460 days, 36 the observation of similar initial t1/2for memory CD8 T cells after a SARS‐CoV‐2 infection indicates the generation of long‐lasting memory SARS‐CoV‐2‐specific CD8 T cells 3 , 4 (Figure 2). Memory CD8 T cells to SARS‐CoV were detected 17 years post‐infection. 37
SARS‐CoV‐2‐specific CD8 T cells generated in response to SARS‐CoV‐2 infection predominantly express IFNγ and granzyme B (GzB), 4 , 38 , 39 with some expression of TNF and IL‐2. 4 Eight months post‐infection, effector memory (TEM) and CD45RA+ effector memory (TEMRA) phenotype memory CD8 T cells predominate, with a smaller fraction of central memory (TCM) phenotype cells. 3 , 4 Other studies have corroborated these central findings, with fewer COVID‐19 cases, shorter study periods, or inferred CD8 T cells from PBMC ELISPOT assays. 40 , 41 Bystander CD8 T cell activation can occur during COVID‐19, 42 and some AIM markers can represent bystander activation of CD8 T cells and must be used carefully, depending on the experiment context, to avoid miscalculation of antigen‐specific CD8 T cells. SARS‐CoV‐2‐specific memory CD8 T cells have also been identified with MHCI tetramers/multimers. 27 , 43 , 44 , 45 , 46 , 47 There has been some confusion about the expression of PD‐1 on CD8 T cells in COVID‐19. PD‐1 is expressed on virtually all activated CD8 T cells. PD‐1 is also a marker of exhausted CD8 cells in some contexts. Expression of PD‐1 by itself does not indicate an exhausted T cell. While CD8 T cells have been observed to express PD‐1 after COVID‐19, and increased activation/exhaustion markers were noted in mild as compared to hospitalization‐level or severe disease 48 , 49 ; subsequent studies using tetramers reported that at later time points the memory PD‐1‐expressing CD8 T cells are not exhausted, and appear to be highly functional. 46
Notably, SARS‐CoV‐2‐specific memory CD8 T cell responses are undetectable in approximately 30% of COVID‐19 cases, 3 , 4 , 38 even when testing the full SARS‐CoV‐2 ORFeome of epitopes. 3 It is unknown whether those subjects have CD8 T cell responses to untested epitopes, CD8 T cell responses just below the technical limit of detection of the assays used, or whether those subjects truly did not develop CD8 T cell response to SARS‐CoV‐2 infection.
Lower SARS‐CoV‐2‐specific CD8 T cell responses have been observed in hospitalized COVID‐19 cases in multiple studies, 4 , 26 , 29 including memory, consistent with a weak CD8 T cell response predisposing for more severe COVID‐19, particularly in older adults possessing fewer naive CD8 T cells. 26 This contrasts with CD8 T cell responses to YFV vaccine, which positively correlate with viral load. This also contrasts with positive correlations in SARS‐CoV‐2 infections between spike Ab titers, BMem cell frequencies, and disease severity, indicating that the humoral immune response and memory development correlates with acute SARS‐CoV‐2 viral loads (see BMem cell and Antibody sections). One interpretation of these data is that acute T cell responses are important for the control and clearance of SARS‐CoV‐2 infection. 22
Many SARS‐CoV‐2 antigens are recognized by human CD8 T cell responses in SARS‐CoV‐2‐infected individuals, 38 with an estimated median of 17 epitopes per individual. 50 Recognized class I epitopes are distributed throughout the SARS‐CoV‐2 ORFeome, but structural proteins (spike, nucleocapsid, and M) are relatively immunodominant. 38 , 50 CD8 T cell epitopes are in general well conserved between variants. 51 , 52 , 53 SARS‐CoV‐2 CD8 T cell specificities have been recently reviewed elsewhere. 54 , 55
The frequencies of circulating memory CD8 T cells after SARS‐CoV‐2 infection are lower than the frequencies reported for influenza in humans, 56 though influenza memory reflects the accumulation of many exposures throughout life. The frequencies of circulating SARS‐CoV‐2 memory CD8 T cells can be considered relatively low in comparison with frequencies generated to viruses in animal models associated with high levels of CD8 T cell‐mediated protective immunity. However, the frequency of memory CD8 T cell required for control of an infection is generally highly dependent on the speed of the clinical disease. Most animal models of CD8 T cell‐mediated protection require CD8 T cell immediate effector functions (e.g., cell killing at the site of infection within hours), or viral clearance within a few days. However, once activated CD8 T cell proliferate rapidly and migrate to infected tissues (sites of inflammation), with the cells potentially expanding in number 10‐fold every 24 hours. Thus, within 72 hours of activation memory, CD8 T cell can increase in number close to 1000‐fold. Importantly, progression of hospitalization with COVID‐19 is relatively slow, often occurring 10 days post‐infection or later. 57 , 58 Thus, a role for CD8 T cell in prevention of severe COVID‐19 may require quite low levels of circulating memory CD8 T cells at the time of infection. In contrast, since CD8 T cells must exert their functions directly on infected cells, a role for CD8 T cells in prevention of SARS‐CoV‐2 transmission (half of which occurs within the first 5 days), before reported symptoms 59 , 60 , 61 is a high bar and likely requires a large number of tissue‐resident memory (TRM) CD8 T cells in the URT (Figure 3), which nevertheless can be achieved by immunization. 32
TRM are an important category of T cells for protective immunity. They are underreported in the human immunology literature because it is much more difficult to obtain human tissue samples and isolate T cells from diverse tissues in comparison with blood. It is also much more difficult to identify antigen‐specific T cells from such samples, due to limited cell numbers recovered and different cellular phenotypic characteristics. 61 In the context of SARS‐CoV‐2, immune memory in tissues is particularly relevant in the nasal passages, oral cavity, throat, and lungs (Figure 3). SARS‐CoV‐2‐specific CD8 T cell TRM have been identified in humans. 62 , 63 Lung tissue was examined from four subjects up to 6+ months after unremarkable (non‐hospitalized) cases of COVID‐19. Lymphoid tissues such as blood and lymph nodes (LNs) were examined for comparison. While SARS‐CoV‐2‐specific CD8 T cell was identified in blood from only 2 out of 4 subjects (consistent with the detection rate in much larger studies 3 , 4 ), SARS‐CoV‐2‐specific CD8 T cell was identified in lung LNs from 4 out of 4 subjects, and SARS‐CoV‐2‐specific CD8 T cell was identified in lung tissue from 3 or 4 out of 4 subjects. 62 Additionally, substantial fractions of the CD8 T cells in both lungs and lung LNs were TRM cells. Notably, in two studies SARS‐CoV‐2‐specific CD8 T cell frequencies in lung tissue or bronchoalveolar lavage (BAL) were substantially higher than blood. 62 , 63 SARS‐CoV‐2 is also present in gut tissues. While intestinal tissue was not available, intestinal LNs were examined, and SARS‐CoV‐2‐specific CD8 T cell and CD8 T cell TRM were found in all three samples tested, suggesting that substantial CD8 T cell TRM may be present in intestinal tissue of previously infected individuals. 62 Thus, blood samples may significantly underreport SARS‐CoV‐2‐specific CD8 T cell memory in previously infected individuals. SARS‐CoV‐2‐specific TRM from throat, nasal passages, or the oral cavity after COVID‐19 are less well studied, 64 representing a major knowledge gap for immune memory.
Development of pediatric T cell memory is of particular interest in the context of the well‐known lower disease susceptibility of children to severe COVID‐19, and in the context of the many differences in adult versus pediatric immune reactivity. 65 Conflicting data have been reported regarding pediatric T cell responses to COVID‐19. In one study, significantly stronger acute and memory T cell responses were reported. 66 In another study, significantly lower acute and memory CD8 T cell responses were reported. 67 Of potential interest is also the observation that T cell memory in children may have differences in immunodominance, perhaps as a result of different exposure to common cold coronaviruses. 66 , 67 A small fraction of pediatric SARS‐CoV‐2 infections later develop a serious hyperinflammatory condition, multisystem inflammatory syndrome in children (MIS‐C). Conflicting data have been reported regarding the relative strength of T cell memory in MISC versus non‐MISC cases. 68 , 69 , 70 , 71 , 72 , 73
Long COVID (aka post‐acute sequelae of SARS‐CoV‐2 infection [PASC]) is an important collection of conditions, with unclear etiology. 74 Given that SARS‐CoV‐2 viral RNA and protein have been detected 90+ days post‐infection in gut biopsies from unremarkable COVID‐19 cases (not hospitalized, and not long COVID), it is plausible that significant SARS‐CoV‐2 viral persistence occurs in some individuals in some tissues. This could be a source of inflammation and symptoms for at least some cases of long COVID. It is unclear why CD8 T cell would not clear SARS‐CoV‐2 infection from intestines within a few weeks [Goldblatt et al]. 24 Efficacy of CD8 T cell clearance of virus in humans remains an important knowledge gap. Notably, SARS‐CoV‐2‐specific CD8 T cell responses are undetectable in approximately 30% of COVID‐19 cases, 3 , 4 , 38 even when testing the full SARS‐CoV‐2 ORFeome of epitopes. 3 It is unknown whether those subjects have CD8 T cell responses to untested epitopes, CD8 T cell responses just below the technical limit of detection of the assays used, or whether those subjects truly have no CD8 T cell response to SARS‐CoV‐2 infection. It is also possible that some subjects in which SARS‐CoV‐2‐specific CD8 T cell responses are undetectable in the periphery, might nevertheless have TRM antigen‐specific cells residing in the lung or upper respiratory tract tissues. 62 , 75 Separately, CXCR6+ CD8 T cells in lungs have been associated with extended periods of COVID‐19 inflammation and may have a role in long COVID. 63
2.2. CD8 T cell memory to vaccination
Spike‐specific CD8 T cell responses are detected in approximately 70%–90% of individuals weeks after receiving 2‐dose mRNA COVID‐19 vaccines, 76 , 77 , 78 , 79 and memory CD8 T cells are detectable in approximately 41%–65% of individuals at 6 months after the 2nd dose (7 months from 1st dose). 76 , 77 , 78 , 80 A low dose (25 μg) of mRNA‐1273 was found to generate memory CD8 T cells at similar frequencies as previous infection, comparing 6 months after the 2nd dose to 6 months after infection, indicating similar spike‐specific CD8 T cell responses between mRNA vaccination and infection. A separate study also found similar spike‐specific CD8 T cell responses at earlier times. 81 Among individuals with detectable CD8 T cell memory to mRNA vaccines months after immunization, the magnitude of the memory is generally observed to be low, 76 , 77 , 79 , 80 , 82 both in comparison with spike‐specific CD4 T cell memory, and memory to influenza. 56
There was initial confusion about whether both BNT162b2 and mRNA‐1273 mRNA COVID‐19 vaccines generated CD8 T cell responses. 43 , 83 , 84 More recently, multiple groups have observed similar CD8 T cell responses to both vaccines, 77 including in head‐to‐head comparisons. 77 , 79 , 82 Methodological differences measuring antigen‐specific T cells can result in different findings, as studies not detecting memory CD8 T cells usually utilized a less than optimal short stimulation ICS protocol, or less sensitive ELISPOT formats. The mRNA vaccine‐elicited memory CD8 T cells detected predominantly have a TEM surface phenotype, consistently express IFNγ, and have proliferative capacity. 44 , 79
The first 6 months are likely to be the period with the fastest decline in T cell memory. 36 The observation of approximately twofold declines at 6 months in spike‐specific CD8 T cell memory from peak cytokine‐positive CD8 T cell frequencies is encouraging evidence that CD8 T cell memory to mRNA vaccines is long‐lived and may last many years 76 , 78 , 79 (Figure 2). Memory CD4 T cells exhibit similar kinetics, as discussed in the CD4 T cell section below.
The adenoviral vector vaccines ChAdOx1 and Ad26.COV2.S elicit spike‐specific CD8 T cell responses in 51%–64% of individuals in immunogenicity clinical trials, 85 though the response rate drops to 24%–36% in individuals >65 years old. 85 Stable CD8 T cell memory to Ad26.COV2.S is observed to 8 months. 86 Some comparisons between mRNA vaccines and adenoviral vector vaccines are available for CD8 T cell memory. Similar (within approximately twofold) spike‐specific CD8 T cell responses to mRNA vaccines and adenoviral vector vaccines are observed approximately 1 month after immunization, including the 1‐dose Ad26.COV2.S, 77 , 79 , 82 , 87 2‐dose Ad26.COV2.S, 87 or 2‐dose ChAdOx1. 88 , 89 , 90 Two studies that assessed CD8 T cell memory at 5+ months determined that mRNA‐1273 elicited larger spike‐specific CD8 T cell memory that Ad26.COV2.S. 77 , 79 Two others studies reported the opposite, 91 , 92 with a clinical trial reporting 45/57 Ad26.COV2.S subjects and only 20/116 BNT162b2 or mRNA‐1273 subjects positive for CD8 T cell memory. 92 In PBMC IFNγ ELISPOT assays, T cell memory 2–4 months after ChAdOx1 and BNT162b2 appears to be equivalent, though CD8 and CD4 T cells were not distinguished 93 , 94 (bulk PBMC IFNγ ELISPOT assay signal comes from a mixture of CD8 T cells and CD4 T cells. 43 ). A similar observation was made for Ad.COV2.S. 95 In sum, CD8 T cell memory to mRNA and adenoviral vector COVID‐19 vaccines appears to be similar in magnitude and % responders (Figure 2), but conclusions vary depending on the study. Additional head‐to‐head studies or memory are warranted, including examination of CD8 T cell functionality.
Memory CD8 T cells elicited by mRNA vaccines recognize diverse spike epitopes. 50 , 51 , 77 Memory CD8 T cells largely have conserved recognition of variants, including Omicron. 77 , 81 , 87 , 96 , 97 , 98 Mix & match adenoviral vector + mRNA vaccine approaches may increase CD8 T cell responses 88 , 89 , 95 and thereby may alter CD8 T cell memory. Perplexingly, lower CD8 T cell responses were reported to vaccine extended dose intervals, with the caveat that minimal CD8 T cells were measurable in any group. 99
2.3. CD8 T cell memory in hybrid immunity
Modest differences have been observed between vaccination only and hybrid immunity for circulating spike‐specific CD8 T cells in most studies, 99 as well as no difference based on symptomatic or asymptomatic infection 100 (Figure 2). In one study, no difference in memory IFNγ+ spike‐specific CD8 T cells was observed between vaccination only and hybrid immunity after 6 months. 80 Multiple studies observed increased T cell responses in hybrid immunity compared to infection or vaccination alone without distinguishing between CD4 and CD8 T cells. 81 , 99 CD8 T cell repertoire diversity is maintained after multiple exposures. 101
3. CD4 T CELL MEMORY TO SARS‐CoV‐2
Memory CD4 T cells are important in the control and clearance of viral infections, both directly and by the effects exerted in the support and amplification of antibody responses. Several different subsets of CD4 T cells can differentiate in antigen‐specific responses to infections. This heterogeneity is manifested at the level of different memory subsets, each associated with distinctive patterns of cytokine secretion, transcription factors, and differentiation profiles (TH1, TH2, TH17, TFH, and others 102 ). This heterogeneity is further amplified by a diverse array of functional roles. TFH cells play a key role in orchestrating the development and maturation of antibody responses, 103 , 104 while Th1 and cytotoxic CD4 T cells (CD4‐CTL) can exert direct antiviral functions. 105 , 106 , 107 Multiple lines of evidence suggest that CD4 T cell is a relevant and valuable component of the overall adaptive immune response to SARS‐CoV‐2. 22 , 33 , 108 Protective effects of CD4 T cells against COVID‐19 are fully reviewed in the companion article [Goldblatt et al., this volume]. 24
3.1. CD4 T cell memory to SARS‐CoV‐2 infection
Dan et al. found that antibody, CD4 T cell, CD8 T cell, and B cell memory responses were durable over 8 months after infection, with 95% of the subjects still retaining multiple measurable memory responses, including memory CD4 T cells. 3 Notably, memory CD4 T cells are detectable in 93% of COVID‐19 cases 1 month after infection 3 and still 92% at >6 months post‐infection. 3 Multiple studies have reported similar memory CD4 T cell findings, 4 , 39 , 40 , 109 with a notable large longitudinal study. 4 The estimated t1/2 of memory SARS‐CoV‐2‐specific CD4 T cells is 94–207 days during the first 8 months, 3 , 4 with the t1/2 likely increasing substantially over time, based on a study determining a t1/2 of 377 days for memory SARS‐CoV‐2‐specific CD4 T cells at 6–15 months post‐infection 110 (Figure 2), as well as similar data on CD8 T cell memory against a different virus. 36 These SARS‐CoV‐2 infection data are consistent with T cell memory to SARS‐CoV being detected 17 years post‐infection. 37 , 111 , 112 , 113
SARS‐CoV‐2‐specific memory CD4 T cells after SARS‐CoV‐2 infection predominantly are TH1, TFH, and CD4‐CTL cells. 3 , 4 , 110 Eight months post‐infection, the memory CD4 T cells predominantly have central memory (TCM) and effector memory (TEM) surface phenotypes. 3 , 4 Virus‐specific TH2 cells and TH17 cells are generally not detectable. 4 , 26 , 38 Regarding the TH1 memory cells, they are a stably maintained population, 4 predominantly expressing CD40L and IFNγ, 4 with significant expression of TNF and some expression of IL‐2. 4 , 29 , 56 , 100 , 110 , 114 The CD4‐CTL cells express CD40L and granzyme B (GzB). 4 , 79 , 115 , 116 CD4‐CTL cells are of interest because SARS‐CoV‐2‐infected epithelial cells upregulate class II expression, 117 and CD8 T cell responses appear to be low in many individuals, leaving open the possibly that memory CD4‐CTL may compensate. 117 , 118 Memory TFH cells are generated after SARS‐CoV‐2 infection and are stably maintained after a brief decline 3 , 110 , 119 (Figure 2). The memory TFH cells are highly functional, as they greatly enhance nAb responses to COVID‐19 vaccines. 2 , 120 , 121 Some memory TFH cells express CCR6, which is associated with lung homing. 3 , 119 Other memory TFH cells express CXCR3, which is associated with rapid anamnestic antibody responses, 104 , 122 while the CXCR3neg memory TFH population is associated with higher quality germinal center and antibody responses. 123 , 124 Germinal centers are discussed in the B cell memory section.
In terms of anatomical location of memory T cells, memory CD4 T cells are detectable in the bone marrow, spleen, lung, and multiple lymph nodes (LNs) for 6+ months after infection. 62 Memory TFH cells are observed in LNs. 62 CD4 TRM cells were present at substantial frequencies in lungs 62 and BAL 63 (Figure 3). Less is known regarding SARS‐CoV‐2‐specific CD4 TRM in the URT and oral cavity.
Many SARS‐CoV‐2 antigens are recognized by human memory CD4 T cells in previously infected individuals, 38 with an estimated median of 19 epitopes per individual. 50 Recognized class II epitopes are distributed throughout the SARS‐CoV‐2 ORFeome, but structural proteins (spike, nucleocapsid, and M) are relatively immunodominant. 4 , 38 , 50 , 125 CD4 T cell epitopes are in general well conserved between variants. 51 , 52 , 53 SARS‐CoV‐2 CD4 T cell specificities have been recently reviewed elsewhere. 54 , 55
Human antiviral immune memory may be influenced by variables such as viral factors related to the infection event such as viral dose and tissue distribution, and host factors such as age, sex, and general health of the host. 126 A distinctive feature of SARS‐CoV‐2 infection and COVID‐19 disease is the wide range of clinical outcomes, ranging from fully asymptomatic infection to severe disease and death. The heterogeneity in clinical outcomes associated with SARS‐CoV‐2 infection and COVID‐19 disease is paralleled by large variations in heterogeneity at the level of CD4 T cell responses. 3 , 26 Specifically, SARS‐CoV‐2 T cell responses are influenced by older age and the size of the pool of naive T cells. 26 No significant difference in CD4 T cell memory is observed between males and females. 3 , 4 Pre‐existing comorbidities in COVID patients have been reported to affect magnitude and helper T cell subset composition. 127 T cell responses of a homogenous group of healthy young males were still widely heterogenous, 128 suggesting that heterogeneity of responses is driven by variables other than predisposing conditions, age, and sex. As noted above, conflicting data have been reported regarding pediatric T cell responses to COVID‐19. In one study, stronger acute and memory CD4 T cell responses were reported in one study, 66 but significantly lower acute and memory CD4 T cell responses were reported in another study. 67 Conflicting data have been reported regarding CD4 T cell memory in MISC versus non‐MISC cases. 68 , 69 , 70 , 71 , 72 , 73
Asymptomatic SARS‐CoV‐2 infections may reflect shorter infections with lower overall viral loads in tissues, which could be expected to have lower T cell memory as a result of lower antigen exposure. Comparison between asymptomatic and mild symptomatic COVID‐19 cases revealed slightly lower T cell memory among asymptomatic COVID‐19 cases. 39 , 129 The relation between COVID‐19 severity and quality of memory CD4 T cells is still an open topic of debate and investigation. 115 , 125 , 130 , 131 In the context of long COVID, 74 alterations of T cell responses lasting several months post‐infection have been reported, 63 as have increased total T cell accumulation in BAL. 132
3.2. Crossreactive memory T cells
Memory CD4 T cells able to recognize SARS‐CoV‐2 have been demonstrated in unexposed subjects, with the clearest evidence comping from blood samples obtained during pre‐pandemic times. 37 , 38 , 115 , 133 , 134 It was hypothesized that these cells may predominantly be memory T cell to previous common cold coronavirus (CCC) infections. 54 , 135 Indeed, at least in some cases, these memory T cells cross‐recognize SARS‐CoV‐2 and CCCs. 28 , 46 , 125 , 136 , 137 , 138 , 139 Detailed studies suggest that cross‐recognition across distant viral species can occur, but rather infrequently, 140 , 141 and is observed for SARS‐CoV‐2 sequences. 129 , 142 In general, SARS‐CoV‐2 crossreactive memory T cells have been most often described in the case of CD4 T cells, and less often for CD8 T cells. 38 In terms of antigen specificity, the sequences associated with crossreactive memory are often derived from non‐structural antigens encoded in the Orf1ab, which correlates with the higher degree of conservation of across the genome of CCCs and other coronaviruses. 54
It has been debated to what extent this pre‐existing crossreactive T cell memory is functional and biologically relevant. 135 , 143 The T cells associated with this pre‐existing immunity display classical memory markers, 136 and were detected by a variety of assays. However, this crossreactive recognition can be of low affinity, particularly in the case of more distant unrelated viruses. 142 Additionally, SARS‐CoV‐2 infection is associated with development of T cell response that is largely focus on novel epitopes. 50 Nevertheless, it has now been demonstrated that the crossreactive memory T cells are biologically functional. Pre‐existing crossreactive memory T cells exert a positive influence on COVID‐19 vaccination outcomes. 76 , 144 , 145 This is consistent with two reports that persons with CCC infections within recent years preferentially had less severe COVID‐19 outcomes, 146 , 147 while a different study found no association. 148 Healthcare workers were observed to have high levels of SARS‐CoV‐2 crossreactive memory T cells, and CCC‐specific T cells. 149 It was further shown that the presence of these crossreactive T cells was linked to favorable outcomes in a large healthcare worker cohort during the first wave of the pandemic, with crossreactive memory CD4 T cells possibly providing protection resulting in abortive SARS‐CoV‐2 infection. 150 Evidence of protection was also observed in a household contacts study. 151 Overall, crossreactive memory CD4 T cells recognizing SARS‐CoV‐2 existed in approximately 50% of individuals pre‐pandemic, those crossreactive CD4 T cells have functional properties in vivo, and they have been associated with some degree of protection from COVID‐19.
3.3. CD4 T cell memory to vaccination
COVID‐19 vaccines can elicit robust CD4 T cell memory. Spike‐specific CD4 T cell responses are detected in close to 100% of individuals weeks after receiving 2‐dose mRNA COVID‐19 vaccines, 76 , 77 , 78 , 79 , 80 and memory CD4 T cells are detectable in approximately 100% of individuals at 6 months after the 2nd dose (7 months from 1st dose). 76 , 77 , 78 , 80 mRNA‐1273 generated spike‐specific memory CD4 T cell frequencies higher than seen in previously infected individuals, 79 while BNT162b2 generate spike‐specific memory CD4 T cell frequencies similar to infection. 79 , 80 A dose of mRNA‐1273 similar to that of BNT162b2 generated spike‐specific memory CD4 T cells at frequencies comparable to previous infection, 76 indicating that differences between memory CD4 T cells after the two mRNA vaccines most likely predominantly relate to the different doses of the two vaccines. Reductions in memory CD4 T cell frequencies over 6 months were modest, and half‐lives of memory CD4 T cells after mRNA COVID‐19 vaccines appear to be at least as long as after infection 76 , 79 , 80 (Figure 2). Memory CD4 T cells are generated after a single dose of mRNA vaccine or Ad26.COV2.S that maintained at least several months. 79 , 86 , 95 , 99 In the context of vaccine interval extension protocols, IL‐2+ memory CD4 T cells were increases in vaccinees who waited longer before the 2nd mRNA vaccine dose. 99
After vaccination, memory TH1 cells are stably maintained, 77 , 78 , 79 , 80 , 97 and express CD40L, IFNγ, TNF, and IL‐2. 78 , 79 Memory TFH cells represent approximately 25% of CD4 T cell memory after mRNA immunization, 79 and the abundance of the cTFH cells is associated with the magnitude of the nAb response. 76 , 79 , 120 , 121 , 152 Vaccine‐elicited memory TFH cells in blood are stably maintained with minimal decline over 6+ months, 79 though the cTFH may change phenotype during the first months. 80 , 110 , 153 Active germinal center TFH (GC‐TFH) cells are found in LNs for at least 6 months and appear to be critical for maintaining germinal centers and development of nAbs after vaccination. 152 , 153 Durable TFH cell memory in blood was observed for mRNA vaccines, Ad26.COV2.S, and NVX‐CoV2373. 79 Memory CD4‐CTL cells are also generated in response to several COVID‐19 vaccines and are stably maintained for at least 6 months. 79 Overall, each major subset of memory CD4 T cells is maintained for at least 6 months after vaccination with BNT162b2, mRNA‐1273, Ad26.COV2.S, and NVX‐CoV2373, with kinetics that indicate the memory CD4 T cells will be substantially maintained for years (Figure 2).
After ChAdOx1‐nCoV‐19 immunization, polyfunctional TH1 memory CD4 T cells are induced. 154 , 155 Similar (within approximately twofold) spike‐specific CD4 T cell responses to mRNA vaccines and ChAdOx1‐nCoV‐19 are observed approximately 1 month after 2 doses. 88 Immunization with 2 doses of the inactivated SARS‐CoV‐2 alum and imidazoquinolin adjuvanted vaccine BBV152 (Covaxin) generates memory CD4 T cell responses comparable to that seen in infected individuals, stably maintained for over 6 months. 156 Limited T cell memory data are available for several other vaccines, including Coronavac, Sinopharm, and Sputnik.
Memory CD4 T cells elicited by mRNA vaccines recognize diverse spike epitopes. 50 , 51 , 77 Recognition of variants by memory CD4 T cells is maintained in mRNA, Ad26.COV2.S, and NVX‐CoV2373 vaccinees. 77 , 81 , 96 , 97
3.4. CD4 T cell memory in hybrid immunity
Modest differences have been observed between vaccination only and hybrid immunity for circulating spike‐specific memory CD4 T cells in most studies, 99 , 114 , 120 , 157 as well as no difference based on symptomatic or asymptomatic infection. 100 In one study, no difference in memory spike‐specific CD4 T cells was observed between vaccination only and hybrid immunity after 6 months 80 (Figure 2). Multiple studies observed increased T cell responses in hybrid immunity compared to infection or vaccination alone using IFNγ ELISPOTs that do not distinguish between CD4 and CD8 T cells, 81 , 99 suggesting functional changes may occur. Indeed, a distinct population of spike‐specific IFNγ+ IL‐10+ TH1 memory cells is observed in hybrid immunity but not after vaccination alone, demonstrating a function of imprinting on the memory TH1 cells by infection. 114 There is dramatic enhancement of antibody and B cell responses in persons with hybrid immunity, demonstrating a strong functional role for memory TFH cells in hybrid immunity, discussed elsewhere.
An additional important aspect of hybrid immunity is the location of the T cell memory. Intramuscular vaccination is expected to generated almost exclusively circulating T cell memory. In contrast, SARS‐CoV‐2 infection generates both circulating T cell memory and TRM (Figure 3). Thus, hybrid immunity is expected to result in both circulating and TRM, but it is unclear if the vaccines enhance TRM already present from infection. Lastly, if the order is vax+infection, it is unknown whether the TRM are qualitatively or quantitative different than what occurs after infection alone.
4. B CELL MEMORY TO SARS‐CoV‐2
Human BMem cells can be exceptionally long‐lived, with smallpox vaccine BMem lasting >50 years, 158 and BMem cells generated from infections during the 1918 pandemic lasting at least 90 years. 159 BMem cells are re‐activated upon an infection and are the source of classic anamnestic antibody responses. BMem cells serve two purposes. The first is a cellular source for the anamnestic antibody response. BMem cells can plausibly reactive and generate an anamnestic antibody response within 3–5 days. 160 The second important value of BMem cells is to serve as a library of predictions by the immune system of possible future viral variants. 2 , 161 The COVID‐19 pandemic has dramatically demonstrated the importance of BMem cell diversity in the recognition of a pathogen and variants, also highlighting the brilliance of the immune system at predicting viral mutations, embedding those predictions in the BMem cell repertoire. BMem cells likely play a role in protective immunity against SARS‐CoV‐2 infection by both of the mechanisms above, and protection by BMem cells is reviewed in the accompanying article [Goldblatt et al.]. 24
4.1. B cell memory to SARS‐CoV‐2 infection
Detectable BMem cells develop within two weeks of symptom onset after SARS‐CoV‐2 infection. 3 , 4 Strikingly, BMem cell frequencies continuously increase over the course of 3–6 months post‐infection. 3 , 4 , 162 Spike‐, RBD‐, and nucleocapsid‐specific BMem cells all exhibit this increase, in a cohort of 160 individuals. 3 SARS‐CoV‐2‐specific BMem cell frequencies stabilize approximately 4 months post‐infection 3 and are maintained for at least 15 months 162 , 163 (Figure 2). These spike‐ and RBD‐binding BMem cell frequency increases are associated with substantial somatic hypermutation (SHM) for 6 months, 162 , 164 , 165 continuing for at least 12 months. 162 The BMem cell antibody mutations accumulated over 6–12 months demonstrated increased affinity maturation and increased neutralization potency, particularly against variants. 162 , 165 These patterns are all indicative of long‐lasting germinal centers after SARS‐CoV‐2 infection; an exception, however, is fatal COVID‐19, in which profound disruption of germinal centers can be observed in autopsies. 166 , 167 The high quality of the BMem cells after SARS‐CoV‐2 infection is also evidenced by the anamnestic nAb responses to variants after a subsequent vaccination or infection, as discussed in the “Antibody durability” sections below.
While IgM+ BMem cells initially comprise approximately 1/3rd of SARS‐CoV‐2‐specific BMem cells, IgM+ cells decline rapidly and are mostly undetectable after 5 months. 3 , 4 IgA+ BMem cells are uncommon, comprising only approximately 5% of spike or RBD‐specific BMem cells on average, 3 , 4 , 165 but the IgA+ BMem cells are stably maintained over 8 months post‐infection, in contrast to the IgM+ BMem cells. 3 BMem cells can have diverse phenotypes. After SARS‐CoV‐2 infection, activated SARS‐CoV‐2‐specific BMem cell frequencies are initially high, but decline over the course of 7 months, with a reciprocal increase in resting BMem cells. 164
COVID‐19 severity does impact the magnitude of the BMem cell response. Patients with hospitalization‐level COVID‐19 develop higher RBD‐specific BMem cell frequencies compared to individuals with mild COVID‐19, 3 , 164 similar to what is observed for antibody titers. 168 Asymptomatic cases develop similar Spike‐specific BMem cell frequencies compared to symptomatic but non‐hospitalized COVID‐19 cases [Crotty manuscript in prep].
The detailed study of SARS‐CoV‐2‐specific BMem cells in response to infection, over periods of 6–12 months, in multiple large independent cohorts, with a range of disease severities, and intensive BCR sequencing, amounts to the most detailed understanding of the development of B cell memory to any acute infection. In a small data set from two YFV vaccine (a live viral vaccine) recipients, increases in BMem cell frequencies were observed for 6 months, increases in affinity maturation were observed for over 6 months, and declining frequencies of IgM+ or activated BMem cells were observed over 6+ months. 169 All of those features are commonalities shared with BMem cell responses to SARS‐CoV‐2 infection. Ebola infection BMem cell responses also have some commonalities, though the severity of Ebola disease and longevity of high viral loads may alter that response. 170 Overall, the BMem cell response to SARS‐CoV‐2 infection is quite impressive, with substantial RBD‐ and spike‐specific BMem cell generation; and with only exposure to a single viral strain, the BMem cell compartment develops over several months to contain BMem cells with high neutralization potency and BMem cells capable of recognizing and neutralizing a range of variants.
Tissue‐resident BMem cells (BRM) can exist in some cases. Pathogen‐specific tissue BRM have been observed in lungs of mouse models, 171 and BRM phenotype BMem cells are found in multiple human tissues. 172 BRM have now been demonstrated in humans after SARS‐CoV‐2 infection 62 (Figure 3). By studying organ donors with a previous history of unremarkable COVID‐19 (non‐hospitalized), it was shown that spike/RBD‐specific BRM were observed in the lungs of all subjects. 62 Notably, the frequencies of spike/RBD‐specific BMem cells in lungs were significantly higher than in spleen, indicating enrichment of local memory in lungs after SARS‐CoV‐2‐infection. Spike/RBD‐specific BMem cells were also found in bone marrow, lung LNs, and gut LNs. 62 RBD‐specific BRM have also been observed in BAL. 63 Local reactivation of BRM in lungs could result in faster anamnestic antibody responses than from circulating BMem cells, based on data from a mouse influenza model. 171 URT tissues were not available for BMem cell analysis, and this remains a knowledge gap, given that the URT is the primary site of SARS‐CoV‐2 replication and transmission.
Regarding long COVID, there is no SARS‐CoV‐2‐specific BMem cell study reported for long COVID. As noted in the memory T cell sections, it is plausible that at least some long COVID cases are due to persistent infection in some tissue sites. As such, some of the extended SHM of BMem cells may be due to new SARS‐CoV‐2 antigen generation after the acute phase of the disease was resolved. These are significant remaining knowledge gaps.
4.2. B cell memory to vaccination
BMem cells are generated in response to COVID‐19 vaccines. Similar frequencies of RBD‐binding IgG+ BMem cells are generated after 2‐dose RNA vaccines or SARS‐CoV‐2 infection 80 (Figure 2). SHM levels are also substantial, and comparable between 2‐dose RNA vaccines and SARS‐CoV‐2 infection at 5 months. 80 , 173 A substantial fraction of RBD‐binding IgG+ BMem cells from 2‐dose RNA‐vaccinated individuals also bind to VOC RBDs. 77 , 80 Thus, 2‐dose RNA vaccines generate substantial affinity matured BMem cells. Nevertheless, the affinity maturation after a standard 2‐dose RNA vaccine regimen is qualitatively poorer than that after SARS‐CoV‐2 infection. Substantial improvements in nAb breadth were observed months after infection but not after RNA vaccination (e.g., 69% of nAbs from previously infected subjects had improved potency, but on 19% of nAbs did from 2‐dose RNA vaccinees). 173 These qualitative differences may be related to the narrow time between dose 1 and dose 2 of the RNA vaccines. The priming period can be important for the quality of a B cell response. Extending the priming period can result in better nAb responses to HIV. 174 , 175 , 176 , 177 , 178 Extending the dose interval between RNA vaccine immunizations from 3 to 10 weeks significantly improves nAb titers and nAb breadth, 99 most likely by impacting affinity maturation.
Spike and RBD IgG+ BMem cell frequencies increase between 3 and 6 months after immunization with mRNA vaccines, 79 , 80 , 173 an adenoviral vector vaccine, 79 or a recombinant protein vaccine. 79 1‐dose Ad26.COV2.S vaccine elicits significantly lower spike and RBD IgG+ BMem cell frequencies than the mRNA vaccines 79 (Figure 2). NVX‐CoV2373 also elicits lower spike and RBD IgG+ BMem cell frequencies than the mRNA COVID‐19 vaccines 79 (Figure 2). Additionally, BMem cell frequencies are somewhat higher after mRNA‐1273 compared to BNT162b2 vaccination. 79 Less is known regarding BMem cells after ChAdOx1 immunizations.
Germinal centers appear to be central to the immune responses to COVID‐19 vaccines. Most nAb responses, class‐switched BMem cells, and durable antibody responses to viral infections and vaccines are dependent on germinal centers. 104 For COVID‐19 vaccines, nAb responses are substantially reduced in many immunocompromised individuals, such as kidney transplant recipients. Direct examination of germinal centers in healthy subjects compared to kidney transplant recipients revealed dramatically weaker germinal center responses in kidney transplant recipients after mRNA vaccination. 152 The smaller germinal center responses may be due to weaker GC‐TFH cell responses, as GC‐TFH cell frequencies were severely reduced and associated with poor nAb titers. 152 Germinal centers are observed to continue in draining LNs of healthy vaccinated individuals for at least 6 months after BNT162b2 immunization. 153 , 179 , 180 This is associated with presence of vaccine mRNA in germinal centers for at least a month, as well as detectable spike protein in the germinal centers. 167 Since the vast majority of the BMem cell response to COVID‐19 vaccines is class‐switched and contains SHMs, the data indicate that the vast majority of the BMem cell response, and nAb response, to COVID‐19 vaccines is TFH‐dependent and germinal center‐dependent.
4.3. B cell memory in hybrid immunity
Circulating spike and RBD IgG+ BMem cell frequencies increase substantially in hybrid immunity, 80 , 162 , 181 but become similar to 2‐dose mRNA vaccination after 6 months 80 (Figure 2). In hybrid immunity, the RBD‐binding BMem cells have substantially more SHM and affinity maturation than after vaccination alone. 80 , 162 , 181 Functionally, this is observed most clearly with the significantly higher potency and variant breadth of nAbs from BMem cells in people with hybrid immunity compared to vaccination alone or infection alone. 162 , 181 The robustness and quality of these responses are likely driven by memory TFH cells and BMem cells, and can occur after infection + vax or vax + infection (“breakthrough”), discussed in the ”Antibody durability” section below.
5. ANTIBODY DURABILITY TO SARS‐CoV‐2 INFECTION OR COVID‐19 VACCINES
Abs are key components of protective immunity against SARS‐CoV‐2. Thus, durability of Abs is a major topic of interest for protective immunity against SARS‐CoV‐2 for previously infected, vaccinated, or person with hybrid immunity. Acute Ab responses are primarily generated by B cells differentiating into short‐lived plasma cells (short‐lived BPC). These short‐lived BPC only live for a few days. IgG protein has a long half‐life of 21–28 days in the blood, and thus, a large short‐lived BPC response can result in detectable antibody titers in blood for months. Long‐lived BPC can survive for many years producing large quantities of Abs daily. Long‐lived BPC are typically the product of germinal center B (BGC) cells.
5.1. Antibody durability to infection
The vast majority of SARS‐CoV‐2‐infected individuals seroconvert and develop nAbs (91%–99%). 182 , 183 , 184 While nAb titers decline during the first few months post‐infection, nAb titers stabilize between 4 and 6 months post‐infection, with little evidence of decline thereafter. After the initial decay phase (dominated by short‐lived BPC), the estimate SARS‐CoV‐2 nAb t1/2 is 254 days. 4 This may further stabilize over time (Figure 2). NAbs titers are detectable in approximately 80%–90% of SARS‐CoV‐2‐infected individuals at 6 months and 12 months post‐infection. 3 , 4 Nevertheless, SARS‐CoV‐2 nAb titers in previously infected individuals are relatively low, resulting in enhanced interest in understanding all of the other compartments of immune memory nAb titers in previously infected individuals are relatively low. The led to concern that low circulating nAb titers would be insufficient for protection, and increased interest in defining other branches of potential immunity to SARS‐CoV‐2, such as T cells. Nevertheless, immune memory overall in previously infected individuals was robust, 3 leading to a conclusion that natural immunity was likely sufficient to prevent reinfections of significant clinical concern in the majority of people for years. 3 , 185
SARS‐CoV‐2 spike‐ and RBD‐binding IgG titers exhibit similar kinetics to that of nAbs, 3 , 4 , 186 , 187 , 188 , 189 though not identical, depending on the study, likely due to affinity differences between the assays. A multi‐phasic decay kinetic is observed, with a t1/2 of >700 days by 6–9 months post‐infection. 187 , 188 Long‐lived BPC are found in bone marrow 7–8 months after infection. 189 SARS‐CoV‐2‐specific IgM responses are not durable, consistent with IgM responses being short‐lived for most antigen exposures. SARS‐CoV‐2‐specific serum IgA responses are relatively low but are durable at low levels in most individuals, 3 , 4 , 190 with SARS‐CoV‐2‐specific IgA long‐live BPC detected in approximately 50% of individuals. 189
Long‐term antibody titers are lower in asymptomatic cases at 6–16 months post‐infection, 191 , 192 with some individuals being seronegative, though some amount of this difference is due to false‐positive PCR tests with high C t values. 192
Antibody titers against other HCoVs are also relatively stable over time. 4 This is consistent with human immunology findings for multiple acute viral infections and the live attenuated YFV, measles, and smallpox vaccines. 158 , 193 , 194
Nucleocapsid antibody assays have been found to not be trustworthy indicators of previous infection at timepoints >6 months post‐infection. This may be due to a faster decay kinetics of nucleocapsid Abs, 195 or a high background signal from crossreactive nucleocapsid Abs against other HCoVs 187 , 196 which may be more problematic in certain assays formats, making it more challenging to definitively distinguish SARS‐CoV‐2 nucleocapsid IgG. Thus, RBD IgG is more widely used as a serodiagnostic marker, though it cannot distinguish infection from vaccination.
Local immunity is important, and Abs are a key factor of local immunity, as they are the only component of adaptive immunity capable of providing sterilizing immunity (Figure 3). Circulating IgG is transudated into most mucosal tissues, and circulating IgG can provide protective immunity at mucosal surfaces. The most dramatic example of this is the human papillomavirus (HPV) vaccine, which provides 99% protective immunity in the vaginal tract, even though the vaccine is a conventional intramuscular immunization and elicits circulating IgG. For SARS‐CoV‐2 previously infected individuals, the titers of circulating IgG correlate with saliva IgG, 56 , 190 , 197 and the correlation was sustained over a period of 9 months. 56 Correlation between circulating IgA and saliva IgA in previously infected individuals was also substantial over a 9‐month period. 56 A more rapid decay of IgG and IgA titers was observed in saliva compared to blood, possibly indicating local production of Abs in salivary tissue for a limited number of months post‐infection. 56 Few studies have examined nasal passage Abs, but spike IgG is detectable 6 months after infection in the majority of individuals. 100 Overall, the evidence suggests that nAbs at nasal, oral, and lung tissues are important for protective immunity against SARS‐CoV‐2 and are likely present proportionally to serum IgG titers after infection.
5.2. Antibody durability to vaccination
Two doses of an COVID‐19 mRNA vaccine are incredibly successful at eliciting high titers of nAbs. However, the biggest shortcoming of the mRNA COVID‐19 vaccines has been that the nAb titers decline continuously over a period of months. Vaccination with the mRNA‐1273 mRNA vaccine generates peak RBD IgG and SARS‐CoV‐2 nAb titers twofold higher than the BNT162b2 mRNA vaccine, on average 92 , 198 ; as a result, Ab durability analyses are confounded in studies that mix vaccinees receiving the two vaccines. In one large study of 2600 recipients of the 2‐dose BNT162b2 vaccine in Israel, RBD IgG continuously declined over 7 months from peak after the 2nd dose, with a 16‐fold reduction in RBD IgG from peak 199 (Figure 2). In that study, a vaccinated cohort and a previous‐infected cohort were directly compared; the RBD IgG titers declined extensively in the vaccinated individuals but were largely stable in the previously infected individuals. 199 NAb titers and RBD IgG similarly declined fivefold and 10‐fold to the mRNA‐1273 vaccine from peak over 6 months after the 2nd dose, ending with low but detectable levels of nAbs in 100% of subjects. 200 , 201 , 202 Ninefold to 10‐fold nAb declines were also observed for a low dose (25 μg) of the mRNA‐1273 mRNA COVID vaccine (instead of 100 μg), 76 comparable to the BNT162b2 dose (30 μg), indicating that the durability of the Ab responses to 2‐dose mRNA vaccines is consistent, and the kinetics are not determined by the vaccine dose, though the absolute magnitude of the Ab response is higher with a higher vaccine dose.
Long‐lived plasma cells specific for SARS‐CoV‐2 spike are detectable in a majority of individuals 6 months after 2‐dose mRNA vaccination 179 ; however, given that nAb and RBD IgG titers continue to decline for at least 8 months after 2‐dose RNA vaccination, these long‐lived BPCs apparently represent low frequencies, or do not have the durability observed for BPCs generated to other antigen exposures.
Due to the precipitous drop in nAb titers over 6–8 months after two doses, and the emergence of VOCs Delta and Omicron, 3‐dose mRNA vaccine regimens have been implemented as the norm in many countries (i.e., 2‐dose regimen plus a “booster” at approximately 6 months) (Figure 3). A critical question about 3‐dose mRNA regimens is whether they induce more durable Ab responses than 2‐dose regimens. Given that the 2‐dose mRNA vaccines were immunogenic and elicited substantial memory CD8 T cells, memory CD4 T cells, BMem cells, and at least a few long‐lived PCs, it was reasonable to predict that 3‐dose mRNA vaccine regimens would induce substantially more durable Abs than the 2‐dose regimen. Results from previously infected individuals cleared demonstrated that the human immune system is capable of making durable Ab responses to SARS‐CoV‐2, and hybrid immunity also demonstrated that the human immune system is clearly capable of making high nAb titers to SARS‐CoV‐2. Additionally, many vaccines are three dose regimens, with durable Abs only being developed after the third dose. Teleologically, one can consider that this is because the immune system performs a cost‐benefit analysis of durable memory to each antigen exposure. Durable Ab responses for 10 years or more have a high caloric resource cost commitment, whereas durable BMem cells or T cells have significantly lower caloric costs. As such, it frequently takes multiple antigen exposures to trigger significant durable Ab responses. Nevertheless, it was unclear if mRNA vaccines were capable of triggering durable Ab responses.
Acute Ab responses to a 3rd dose of mRNA vaccine were strong, with peak nAb titers above that of 2‐dose immunization. 203 , 204 Two studies found much more durable nAb titers at 4 or 6 months after a 3rd dose of mRNA vaccine compared to 2‐doses. 202 , 205 NAb titers against Ac SARS‐CoV‐2 only declined 1.6‐fold for the BNT162b2 vaccine at 4 months and 2.3‐fold for the mRNA‐1273 vaccine at 6 months after a 3rd dose. 202 , 205 Those findings indicate robust long‐lived Ab production after 3 doses (Figure 2). However, not all results agree. In the study of mRNA‐1273 vaccinees, there was a substantial discordance between the durability of nAbs against Ac SARS‐CoV‐2 or Omicron, with nAbs against Ac SARS‐CoV‐2 only declining 2.3‐fold after 6 months, but nAbs against Omicron declining 6.3‐fold. 202 In a third study, of an Israeli population receiving the BNT162b2 vaccine, Ac SARS‐CoV‐2 nAbs declined 5.5‐fold over approximately 4 months after 3 doses. 206 Thus, conclusions about durability of Abs after 3‐dose mRNA vaccination remain uncertain.
Adenoviral vector COVID‐19 vaccines ChAdOx1 (2‐dose) and Ad26.COV2.S (1‐dose) initially elicit substantially lower Ab responses than mRNA vaccines. Spike or RBD IgG titers after 1‐dose Ad26.COV2.S are approximately 70‐fold to 355‐fold lower than 2‐dose mRNA vaccines. 79 , 82 , 207 NAb titers are approximately 10‐ to 70‐fold lower 30 to 60 days after Ad26.COV2.S compared to mRNA vaccines. 79 , 82 , 92 NAb titers increase some over time after Ad26.COV2.S in some individuals, but this is variable 79 , 82 , 86 , 208 (Figure 2). NAb titers are at least stably maintained in most individuals who receive Ad26.COV2.S vaccination, such that 6‐8 months after Ad26.COV2.S vaccination nAb titers are detectable in almost all individuals, albeit at low levels, 79 , 86 though not all studies agree, with some cohorts reporting many negative individuals (table 40 ref. 208 ) and one study reporting higher levels. 91 ChAdOx1 vaccine (2‐dose) generates early nAb titers that are also substantially lower than mRNA vaccine generated nAb titers. nAb titers are 8.3‐fold lower after ChAdOx1 compared to BNT162b2 one month after immunization. 93 Spike IgG titers after 2‐dose ChAdOx1 decline with a t1/2 comparable to that of 2‐dose BNT162b2, 19 with no evidence of greater durability. However, in one study, the difference between ChAdOx1 and BNT162b2 nAb titers was only approximately twofold at 3 months after the 2nd dose. 94 There are limited data for 2‐dose ChAdOx1 at longer time points with head‐to‐head comparisons, but when comparing ChAdOx1/ChAdOx1 to ChAdOx1/ BNT162b2, spike Ab titers at 6 months were sixfold lower in the ChAdOx1/ChAdOx1 group.
Protein‐based COVID‐19 vaccines are in two categories, recombinant spike vaccines, such as NVX‐CoV2373, and inactivated virus vaccines such as BBV152 and Sinovac. Minimal data on Ab durability are available for inactivated vaccines. NVX‐CoV2373 generates significant nAb titers after two doses. 209 , 210 NAb titers 6 months after the 2nd dose are approximately equivalent between individuals receiving NVX‐CoV2373, mRNA‐1273, or BNT162b2 vaccines (Figure 2), suggesting that high nAb titers may wane after 2‐dose NVX‐CoV2373. 79
Mix & match vaccine strategies could potentially elicit more durable Ab responses. Limited data are available on immune memory after mix & match vaccination. 92 , 93 , 94 Given that peak nAb titers are higher with 2‐dose mRNA or recombinant protein vaccination compared to adenoviral vaccines, but that adenoviral vaccines might elicit more stable Ab responses at 6 months, it is plausible that mix & match approaches may combine the best of both and result in higher level durable nAb titers. This may also be relevant for boosters. 94
Regarding local Abs in the respiratory tract and oral mucosa, the mRNA vaccines do elicit some circulating IgA. 100 , 173 IgA and IgG are detectable in saliva and nasal swabs in a fraction of vaccinated individuals, but both the IgA and IgG decline substantially over the course of 6 months after 2‐dose mRNA vaccination, mirroring the declines in circulating IgG and IgA. 100 , 211 , 212 In immunized non‐human primates, RBD IgG and IgA in bronchoalveolar lavage and nasal swabs directly correlated with circulating IgG and IgA levels. 213 Exploring development of intranasal vaccines is of substantial interest for potentially improving mucosal immune memory. 214
5.3. Antibody durability in hybrid immunity
The most prominent characteristic of hybrid immunity is the impressive improvement in nAb titers and the breadth of neutralization of SARS‐CoV‐2 variants. In some individuals, SARS‐CoV‐2 nAb titers increase 100‐fold after a single mRNA vaccination. Equally impressive, the nAbs are not only able to neutralize every known SARS‐CoV‐2 variant, including Omicron, they are also able to neutralize a different viral species, SARS‐CoV. BMem cells and memory CD4 T cells are at the root of these impressive outcomes. While circulating nAb titers are frequently low in previously infected individuals, without much evidence of breadth, some BMem cells from those same individuals encode Abs with impressive potency and breadth. 162 , 165 , 215 , 216 Those BMem cells are then recalled after vaccination to generate an anamnestic Ab response, now composed of Abs capable of neutralizing breadth against VOCs such as Omicron, and even neutralization of SARS‐CoV, 20 , 80 , 157 , 162 , 217 , 218 irrespective of original COVID‐19 severity. 219
Hybrid immunity can also occur in the reverse order—vaccination and then infection—with similarly high titer and broad nAb responses, irrespective of whether the infection was Alpha, Delta, or Omicron, and irrespective of disease severity. 20 , 21 , 212 , 220 , 221 , 222 These responses are again derived from BMem cells, in this case BMem cells generated in response to vaccination. 173 , 223
Ab durability at 6 months is robust in a majority of individuals with hybrid immunity, as measured by nAb titers (Figure 2). NAb titers were stable in a majority of hybrid immunity individuals, declined less than twofold over 6 months. 20 , 80 Of note, RBD‐binding titers exhibited larger declines, for unclear reasons. 80 , 100 After 6 months, people with hybrid immunity maintained fivefold to 17‐fold higher nAb titers against ancestral SARS‐CoV‐2, Beta, or Delta compared to individuals who were 2‐dose mRNA vaccinated 20 , 80 ; compared to individuals who were previously infected alone, people with hybrid immunity maintained 10‐ to 51‐fold higher nAb titers against ancestral SARS‐CoV‐2, Beta, Delta, or Omicron. 20
Higher nasal RBD IgG and IgA are found in individuals with hybrid immunity (either inf+vax or vax+inf) when sampled up to 10 months after vaccination. 100 , 212
6. INTERRELATIONSHIPS BETWEEN IMMUNE MEMORY COMPARTMENTS
Studies of SARS‐CoV‐2 memory are the first time that large datasets have been collected of multiple antigen‐specific memory cell compartments over a period of 6+ months after an acute infection. This provides key opportunities to understand relationships between different aspects of immune memory. It was observed that each compartment of immune memory after infection exhibit distinct kinetics over time, and different quantitative relationships to the other compartments of immune memory. 3 Some of the relationships changed dramatically over time. 3 Perhaps most importantly from a practical perspective, serum RBD IgG titers were not quantitatively predictive of the other components of immune memory, notably memory T cells. 3 Nevertheless, other relationships were observed. 3 TFH cells, BMem cells, and circulating Ab titers are functionally associated. 104 However, cTFH cell frequencies after infection were not quantitatively predictive of germinal centers, 153 or nAb titers, 110 suggesting more complex relationships between circulating T cell memory, germinal centers, and nAbs.
For mRNA vaccines, relationships between nAbs and memory CD4 T cells are clear, and early TFH cell responses do correlate with subsequent nAb titers. 79 However, at any given memory timepoint, no clear association is observed between serum Ab titers and memory CD4 T cell and memory CD8 T cell frequencies. 79 CD4 T cells provide help for CD8 T cell differentiation and memory CD8 T cells in multiple contexts. 224 Nevertheless, memory CD4 T cell frequencies and memory CD8 T cell frequencies do not show a strong relationship in mRNA‐vaccinated individuals. 79 Overall, interrelationships between immune memory compartments exist, but much remains to be learned.
7. IMMUNE MEMORY IN SPECIAL POPULATIONS
7.1. Immune memory in the immunocompromised or suppressed
Immune responses to COVID‐19 vaccines in immunocompromised or immunosuppressed individuals vary depending on the specific immunocompromised or immunosuppressed condition. B cell depleted individuals (i.e., anti‐CD20 mAb treatment) have defective Ab, BMem cell, and TFH cell responses to mRNA COVID‐19 vaccines, but their TH1 and CD8 T cell responses are normal or elevated. 225 Thus, it is expected that immune memory will be substantially defect in those individuals for durable Abs, BMem cells, and memory TFH cells; however, memory TH1 cells and CD8 T cells may or may not be compromised. Solid organ transplant patients frequently have reduced responses to COVID‐19 vaccines because of their immunosuppressive drug therapies. Immune memory in such individuals is not well understood, but based on the severity of the germinal center, TFH cell, TH1 cell, and CD8 T cell response defects in kidney transplant individuals responding to COVID‐19 mRNA vaccines, 152 it is likely that there are severe immune memory defects in those patients. Certain categories of cancer patients on similar immunosuppressive drug regimens are likely to also have immune memory defects. Fingolimod, the S1P receptor antagonist, appears to cause an almost complete block of Ab and T cell responses to COVID‐19 vaccines, 226 and would be expected to result in severe immune memory defects to COVID‐19 vaccines. More information is needed about immune memory to COVID‐19 vaccines in a diverse range of immunocompromised or immunosuppressed individuals, given that the efficacy of the vaccines is predicated on immune memory.
7.2. Boosters in persons with hybrid immunity
Many papers show substantial immunological and epidemiological evidence that hybrid immunity is the most robust immunity against COVID‐19. 2 , 9 This includes vax+vax+inf, 20 , 212 , 227 inf+vax, and inf+vax+vax. These individuals have the best neutralizing Ab breadth—able to recognize every single known variant, include Omicron, and even able to recognize another species of virus (SARS) 20 , 157 , 228 —and they also have substantially better local immunity in the nose and mouth, 62 , 212 which is not generated by intramuscular vaccination. They also have more durable Ab responses, based on the available data. 3 , 4 , 100 , 186 , 200 , 229 , 230 However, many governments have booster vaccination requirements within 90 days of an infection. For people who had breakthrough infections with Delta or Omicron after being double vaccinated, this is most likely to be far sooner than needed, and may be counterproductive. It is plausible that such individuals may have such good immune memory that they do not need a booster for years. The quality of the Ab response needs time to develop. The immune system has done an amazing job making Ab responses and memory B cells against SARS‐CoV‐2 that are educated guesses about potential future variants. 2 , 20 , 162 , 165 , 215 , 229 , 231 , 232 That is important for immunity against this virus, but takes time in germinal centers, 233 and it is likely disrupted by a new immunization. Hence, immunizations too close together are shortsighted and result in poorer quality immunity. We also know that memory B cell frequencies increase for almost 6 months after infection, 3 , 215 , 229 or after vaccination. 162 , 229 We know that germinal centers can persist for more than 6 months after SARS‐CoV‐2 infection. 62 We know that germinal centers can persist and be productive for more than 6 months after two doses of COVID‐19 mRNA vaccines. 180 , 234 , 235 In addition, we know that the quality of neutralizing Abs can improve over 3–6 months, 86 , 162 , 173 , 215 , 236 reflective of outcomes of these long processes of developing higher quality immune memory. Boosters too close together may disrupt those processes of generating broader protection against future variants.
8. CONCLUDING REMARKS
A remarkable amount has been learned about immune memory after SARS‐CoV‐2 infection. A remarkable amount has also been learned about a multitude of COVID‐19 vaccines, and hybrid immunity. Increasing our understanding of the deterministic relationships between early immune responses and immune memory outcomes remain a major knowledge gap for further research. There is much to be learned about local immune memory in mucosal tissues such as nasal passages, oral cavity, the URT broadly, the intestinal tract, and lungs. Investigation of the relationships between local immune memory and systemic immune memory is of particular significance. Longer term durability of each compartment of immune memory after SARS‐CoV‐2 infection or COVID‐19 vaccination of course remains to be empirically determined. However, the wealth of scientific literature already accumulated regarding immune memory provides strong predictions regarding the durability of T cell memory, B cell memory, and long‐lasting antibody responses that can be extrapolated for several years, if not decades, and may provide determining factors of sustained protection against disease. Lastly, clearly this knowledge and experience can also be leveraged toward vaccines against other diseases that affect humanity now, and prevent future plagues.
CONFLICT OF INTEREST
SC has consulted for GSK, JP Morgan, Citi, Morgan Stanley, Avalia NZ, Nutcracker Therapeutics, University of California, California State Universities, United Airlines, and Roche. A.S. is a consultant for Gritstone Bio, Flow Pharma, ImmunoScape, Moderna, AstraZeneca, Avalia, Fortress, Repertoire, Gilead, Gerson Lehrman Group, RiverVest, MedaCorp, and Guggenheim.
ACKNOWLEDGEMENTS
Thanks to Zeli Zhang and Parham Ramezani‐Rad for helpful input. Thanks to graphical assistance by Christina Corbaci. This work was funded by the NIH NIAID under award AI142742 (Cooperative Centers for Human Immunology, CCHI) and CHAVD UM1 AI144462, and was additionally supported in part by La Jolla Institute for Immunology Institutional Funds and an anonymous donor.
Sette A, Crotty S. Immunological memory to SARS‐CoV‐2 infection and COVID‐19 vaccines. Immunol Rev. 2022;00:1‐20. doi: 10.1111/imr.13089
*This article introduces a series of reviews covering SARS‐CoV‐2 Vaccines appearing in Volume 310 of Immunological Reviews.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Schoenberger SP, Crotty S. Immunological memory. In: Paul WE, ed. Immunological Memory. 6th ed. Lippincott Wiliams & Wilkins; 2010. [Google Scholar]
- 2. Crotty S. Hybrid immunity. Science. 2021;372(6549):1392‐1393. doi: 10.1126/science.abj2258 [DOI] [Google Scholar]
- 3. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science. 2021;371(6529):eabf4063. doi: 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen KW, Linderman SL, Moodie Z, et al. Longitudinal analysis shows durable and broad immune memory after SARS‐CoV‐2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. 2021;2(7):100354. doi: 10.1016/j.xcrm.2021.100354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lumley SF, O’Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS‐CoV‐2 infection in health care workers. N Engl J Med. 2020;384(6):533‐540. doi: 10.1056/nejmoa2034545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abu‐Raddad LJ, Chemaitelly H, Coyle P, et al. SARS‐CoV‐2 antibody‐positivity protects against reinfection for at least seven months with 95% efficacy. EClinicalMedicine. 2021;35:100861. doi: 10.1016/j.eclinm.2021.100861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall VJ, Foulkes S, Charlett A, et al. SARS‐CoV‐2 infection rates of antibody‐positive compared with antibody‐negative health‐care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. 2021;397(10283):1459‐1469. doi: 10.1016/s0140-6736(21)00675-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leidi A, Koegler F, Dumont R, et al. Risk of reinfection after seroconversion to SARS‐CoV‐2: a population‐based propensity‐score matched cohort study. Clin Infect Dis. 2021;74:622‐629. doi: 10.1093/cid/ciab495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pouwels KB, Pritchard E, Matthews PC, et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS‐CoV‐2 infections in the UK. Nat Med. 2021;27:2127‐2135. doi: 10.1038/s41591-021-01548-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altarawneh HN, Chemaitelly H, Hasan MR, et al. Protection against the omicron variant from previous SARS‐CoV‐2 infection. N Engl J Med. 2022;386:1288‐1290. doi: 10.1056/nejmc2200133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. León TM, Dorabawila V, Nelson L, et al. COVID‐19 cases and hospitalizations by COVID‐19 vaccination status and previous COVID‐19 diagnosis — California and New York, May–November 2021. MMWR Morb Mortal Wkly Rep. 2022;71(4):125‐131. doi: 10.15585/mmwr.mm7104e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS‐CoV‐2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet Lond Engl. 2022;399:1303‐1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes among patients infected with Omicron (B.1.1.529) SARS‐CoV‐2 variant in southern California. medRxiv. 2022;2022.01.11.22269045. doi: 10.1101/2022.01.11.22269045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Madhi SA, Kwatra G, Myers JE, et al. Population immunity and Covid‐19 severity with omicron variant in South Africa. N Engl J Med. 2022;386:1314‐1326. doi: 10.1056/nejmoa2119658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. New Engl J Med. 2020;383(27):2603‐2615. doi: 10.1056/nejmoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahly HME, Baden LR, Essink B, et al. Efficacy of the mRNA‐1273 SARS‐CoV‐2 vaccine at completion of blinded phase. N Engl J Med. 2021;385(19):1774‐1785. doi: 10.1056/nejmoa2113017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tartof SY, Slezak JM, Fischer H, et al. Effectiveness of mRNA BNT162b2 COVID‐19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021;398(10309):1407‐1416. doi: 10.1016/s0140-6736(21)02183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldberg Y, Mandel M, Bar‐On YM, et al. Waning immunity after the BNT162b2 vaccine in Israel. N Engl J Med. 2021;385(24):NEJMoa2114228. doi: 10.1056/nejmoa2114228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei J, Pouwels KB, Stoesser N, et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat Med. 2022;28:1072‐1082. doi: 10.1038/s41591-022-01721-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walls AC, Sprouse KR, Bowen JE, et al. SARS‐CoV‐2 breakthrough infections elicit potent, broad and durable neutralizing antibody responses. Cell. 2022;185(5):872‐880.e3. doi: 10.1016/j.cell.2022.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wratil PR, Stern M, Priller A, et al. Three exposures to the spike protein of SARS‐CoV‐2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat Med. 2022;28(3):496‐503. doi: 10.1038/s41591-022-01715-4 [DOI] [PubMed] [Google Scholar]
- 22. Sette A, Crotty S. Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell. 2021;184(4):861‐880. doi: 10.1016/j.cell.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadarangani M, Marchant A, Kollmann TR. Immunological mechanisms of vaccine‐induced protection against COVID‐19 in humans. Nat Rev Immunol. 2021;21(8):475‐484. doi: 10.1038/s41577-021-00578-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldblatt D, Alter G, Crotty S, Plotkin S. Correlates of protection against SARS‐CoV‐2 infection and COVID‐19 disease. Immunological Reviews. 2022. (This volume). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15(12):1104‐1115. doi: 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moderbacher CR, Ramirez SI, Dan JM, et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell. 2020;183(4):996‐1012.e19. doi: 10.1016/j.cell.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng Y, Felce SL, Dong D, et al. An immunodominant NP105–113‐B*07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID‐19 disease. Nat Immunol. 2022;23(1):50‐61. doi: 10.1038/s41590-021-01084-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sekine T, Perez‐Potti A, Rivera‐Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell. 2020;183(1):158‐168.e14. doi: 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS‐CoV‐2 in UK convalescent individuals following COVID‐19. Nat Immunol. 2020;21(11):1336‐1345. doi: 10.1038/s41590-020-0782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thieme CJ, Anft M, Paniskaki K, et al. Robust T cell response toward spike, membrane, and nucleocapsid SARS‐CoV‐2 proteins is not associated with recovery in critical COVID‐19 patients. Cell Rep Med. 2020;1(6):100092. doi: 10.1016/j.xcrm.2020.100092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS‐CoV‐2 in rhesus macaques. Nature. 2020;590:630‐634. doi: 10.1038/s41586-020-03041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishii H, Nomura T, Yamamoto H, et al. Neutralizing antibody‐independent SARS‐CoV‐2 control correlated with intranasal vaccine‐induced CD8+ T‐cell responses. Cell Rep Med. 2022;3(2):100520. doi: 10.1016/j.xcrm.2022.100520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kedzierska K, Thomas PG. Count on us: T cells in SARS‐CoV‐2 infection and vaccination. Cell Rep Med. 2022;3(3):100562‐100562. doi: 10.1016/j.xcrm.2022.100562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sidney J, Peters B, Sette A. Epitope prediction and identification‐ adaptive T cell responses in humans. Semin Immunol. 2020;50:101418. doi: 10.1016/j.smim.2020.101418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reiss S, Baxter AE, Cirelli KM, et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen‐specific CD4 T cells. PLoS One. 2017;12(10):e0186998. doi: 10.1371/journal.pone.0186998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akondy RS, Fitch M, Edupuganti S, et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552(7685):362‐367. doi: 10.1038/nature24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bert NL, Tan AT, Kunasegaran K, et al. SARS‐CoV‐2‐specific T cell immunity in cases of COVID‐19 and SARS, and uninfected controls. Nature. 2020;584(7821):457‐462. doi: 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 38. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020;181(7):1489‐1501.e15. doi: 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuo J, Dowell AC, Pearce H, et al. Robust SARS‐CoV‐2‐specific T‐cell immunity is maintained at 6 months following primary infection. Nat Immunol. 2021;22(5):620‐626. doi: 10.1038/s41590-021-00902-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodda LB, Netland J, Shehata L, et al. Functional SARS‐CoV‐2‐specific immune memory persists after mild COVID‐19. Cell. 2020;184:169‐183.e17. doi: 10.1016/j.cell.2020.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bilich T, Nelde A, Heitmann JS, et al. T cell and antibody kinetics delineate SARS‐CoV‐2 peptides mediating long‐term immune responses in COVID‐19 convalescent individuals. Sci Transl Med. 2021;13(590):eabf7517. doi: 10.1126/scitranslmed.abf7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bergamaschi L, Mescia F, Turner L, et al. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID‐19 from mild disease. Immunity. 2021;54(6):1257‐1275.e8. doi: 10.1016/j.immuni.2021.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sahin U, Muik A, Vogler I, et al. BNT162b2 vaccine induces neutralizing antibodies and poly‐specific T cells in humans. Nature. 2021;595:572‐577. doi: 10.1038/s41586-021-03653-6 [DOI] [PubMed] [Google Scholar]
- 44. Oberhardt V, Luxenburger H, Kemming J, et al. Rapid and stable mobilization of CD8+ T cells by SARS‐CoV‐2 mRNA vaccine. Nature. 2021;597(7875):268‐273. doi: 10.1038/s41586-021-03841-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gangaev A, Ketelaars SLC, Isaeva OI, et al. Identification and characterization of a SARS‐CoV‐2 specific CD8+ T cell response with immunodominant features. Nat Commun. 2021;12(1):2593. doi: 10.1038/s41467-021-22811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rha MS, Jeong HW, Ko JH, et al. PD‐1‐expressing SARS‐CoV‐2‐specific CD8+ T cells are not exhausted, but functional in patients with COVID‐19. Immunity. 2020;54(1):44‐52.e3. doi: 10.1016/j.immuni.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nguyen THO, Rowntree LC, Petersen J, et al. CD8+ T cells specific for an immunodominant SARS‐CoV‐2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54(5):1066‐1082.e5. doi: 10.1016/j.immuni.2021.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Files JK, Boppana S, Perez MD, et al. Sustained cellular immune dysregulation in individuals recovering from SARS‐CoV‐2 infection. J Clin Invest. 2020;131(1):e140491. doi: 10.1172/jci140491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kusnadi A, Ramírez‐Suástegui C, Fajardo V, et al. Severely ill COVID‐19 patients display impaired exhaustion features in SARS‐CoV‐2‐reactive CD8+ T cells. Sci Immunol. 2021;6(55):eabe4782. doi: 10.1126/sciimmunol.abe4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tarke A, Sidney J, Kidd CK, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS‐CoV‐2 epitopes in COVID‐19 cases. Cell Rep Med. 2021;2(2):100204. doi: 10.1016/j.xcrm.2021.100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tarke A, Sidney J, Methot N, et al. Impact of SARS‐CoV‐2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2(7):100355. doi: 10.1016/j.xcrm.2021.100355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woldemeskel BA, Garliss CC, Blankson JN. SARS‐CoV‐2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS‐CoV‐2 variants and HCoV‐NL63. J Clin Invest. 2021;131(10):e149335. doi: 10.1172/jci149335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Redd AD, Nardin A, Kared H, et al. CD8+ T‐cell responses in COVID‐19 convalescent individuals target conserved epitopes from multiple prominent SARS‐CoV‐2 circulating variants. Open Forum Infect Dis. 2021;8(7):ofab143. doi: 10.1093/ofid/ofab143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grifoni A, Sidney J, Vita R, et al. SARS‐CoV‐2 human T cell epitopes: adaptive immune response against COVID‐19. Cell Host Microbe. 2021;29(7):1076‐1092. doi: 10.1016/j.chom.2021.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quadeer AA, Ahmed SF, McKay MR. Landscape of epitopes targeted by T cells in 852 individuals recovered from COVID‐19: Meta‐analysis, immunoprevalence, and web platform. Cell Rep Med. 2021;2(6):100312. doi: 10.1016/j.xcrm.2021.100312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Law JC, Girard M, Chao GYC, et al. Persistence of T cell and antibody responses to SARS‐CoV‐2 up to 9 months after symptom onset. J Immunol. 2022;208(2):429‐443. doi: 10.4049/jimmunol.2100727 [DOI] [PubMed] [Google Scholar]
- 57. Garg S, Patel K, Pham H, et al. Clinical trends among U.S. adults hospitalized with COVID‐19, March to December 2020. Ann Intern Med. 2021;174(10):1409‐1419. doi: 10.7326/m21-1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID‐19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577‐582. doi: 10.7326/m20-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Killingley B, Mann A, Kalinova M, et al. Safety, tolerability and viral kinetics during SARS‐CoV‐2 human challenge in young adults. Nat Med. 2022;28:1031‐1041. [DOI] [PubMed] [Google Scholar]
- 60. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26(5):672‐675. doi: 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 61. Steinert EM, Schenkel JM, Fraser KA, et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 2015;161(4):737‐749. doi: 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Poon MML, Rybkina K, Kato Y, et al. SARS‐CoV‐2 infection generates tissue‐localized immunological memory in humans. Sci Immunol. 2021;6(65):eabl9105. doi: 10.1126/sciimmunol.abl9105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheon I, Li C, Son YM, et al. Immune signatures underlying post‐acute COVID‐19 lung sequelae. Sci Immunol. 2021;6:eabk1741. doi: 10.1126/sciimmunol.abk1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roukens AHE, Pothast CR, König M, et al. Prolonged activation of nasal immune cell populations and development of tissue‐resident SARS‐CoV‐2‐specific CD8+ T cell responses following COVID‐19. Nat Immunol. 2022;23(1):23‐32. doi: 10.1038/s41590-021-01095-w [DOI] [PubMed] [Google Scholar]
- 65. Chou J, Thomas PG, Randolph AG. Immunology of SARS‐CoV‐2 infection in children. Nat Immunol. 2022;23(2):177‐185. doi: 10.1038/s41590-021-01123-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dowell AC, Butler MS, Jinks E, et al. Children develop robust and sustained cross‐reactive spike‐specific immune responses to SARS‐CoV‐2 infection. Nat Immunol. 2022;23(1):40‐49. doi: 10.1038/s41590-021-01089-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cohen CA, Li APY, Hachim A, et al. SARS‐CoV‐2 specific T cell responses are lower in children and increase with age and time after infection. Nat Commun. 2021;12(1):4678. doi: 10.1038/s41467-021-24938-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Consiglio CR, Cotugno N, Sardh F, et al. The immunology of multisystem inflammatory syndrome in children with COVID‐19. Cell. 2020;183(4):968‐981.e7. doi: 10.1016/j.cell.2020.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ramaswamy A, Brodsky NN, Sumida TS, et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS‐CoV‐2‐associated multisystem inflammatory syndrome in children. Immunity. 2021;54(5):1083‐1095.e7. doi: 10.1016/j.immuni.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hsieh L, Grifoni A, Sidney J, et al. Characterization of SARS‐CoV‐2 and common cold coronavirus‐specific T cell responses in MIS‐C and Kawasaki disease children. Eur J Immunol. 2021;52:123‐137. doi: 10.1002/eji.202149556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Singh V, Obregon‐Perko V, Lapp SA, et al. Limited induction of SARS‐CoV‐2–specific T cell responses in children with multisystem inflammatory syndrome compared with COVID‐19. JCI Insight. 2022;7(4):e155145. doi: 10.1172/jci.insight.155145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Conway SR, Lazarski CA, Field NE, et al. SARS‐CoV‐2‐specific T cell responses are stronger in children with multisystem inflammatory syndrome compared to children with uncomplicated SARS‐CoV‐2 infection. Front Immunol. 2022;12:793197. doi: 10.3389/fimmu.2021.793197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sacco K, Castagnoli R, Vakkilainen S, et al. Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID‐19. Nat Med. 2022;28:1050‐1062. doi: 10.1038/s41591-022-01724-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Merad M, Blish CA, Sallusto F, Iwasaki A. The immunology and immunopathology of COVID‐19. Science. 2022;375(6585):1122‐1127. doi: 10.1126/science.abm8108 [DOI] [PubMed] [Google Scholar]
- 75. Niessl J, Sekine T, Lange J, et al. Identification of resident memory CD8+ T cells with functional specificity for SARS‐CoV‐2 in unexposed oropharyngeal lymphoid tissue. Sci Immunol. 2021;6(64):eabk0894. doi: 10.1126/sciimmunol.abk0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mateus J, Dan JM, Zhang Z, et al. Low‐dose mRNA‐1273 COVID‐19 vaccine generates durable memory enhanced by cross‐reactive T cells. Science. 2021;374(6566):eabj9853. doi: 10.1126/science.abj9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tarke A, Coelho CH, Zhang Z, et al. SARS‐CoV‐2 vaccination induces immunological T cell memory able to cross‐recognize variants from Alpha to Omicron. Cell. 2022;185(5):847‐859.e11. doi: 10.1016/j.cell.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guerrera G, Picozza M, D’Orso S, et al. BNT162b2 vaccination induces durable SARS‐CoV‐2 specific T cells with a stem cell memory phenotype. Sci Immunol. 2021;6:eabl5344. doi: 10.1126/sciimmunol.abl5344 [DOI] [PubMed] [Google Scholar]
- 79. Zhang Z, Mateus J, Coelho CH, et al. Humoral and cellular immune memory to four COVID‐19 vaccines. bioRxiv. 2022;2022.03.18.484953. doi: 10.1101/2022.03.18.484953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goel RR, Painter MM, Apostolidis SA, et al. mRNA vaccines induce durable immune memory to SARS‐CoV‐2 and variants of concern. Science. 2021;374(6572):abm0829. doi: 10.1126/science.abm0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Naranbhai V, Nathan A, Kaseke C, et al. T‐cell reactivity to the SARS‐CoV‐2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185:1041‐1051.e6. doi: 10.1016/j.cell.2022.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Naranbhai V, Garcia‐Beltran WF, Chang CC, et al. Comparative immunogenicity and effectiveness of mRNA‐1273, BNT162b2 and Ad26.COV2.S COVID‐19 vaccines. J Infect Dis. 2021;225(7):1141‐1150. doi: 10.1093/infdis/jiab593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS‐CoV‐2 — preliminary report. N Engl J Med. 2020;383(20):1920‐1931. doi: 10.1056/nejmoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Corbett KS, Flynn B, Foulds KE, et al. Evaluation of the mRNA‐1273 Vaccine against SARS‐CoV‐2 in nonhuman primates. N Engl J Med. 2020;383(16):1544‐1555. doi: 10.1056/nejmoa2024671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sadoff J, Gars ML, Shukarev G, et al. Interim results of a phase 1–2a trial of Ad26.COV2.S Covid‐19 vaccine. N Engl J Med. 2021;384:1824‐1835. doi: 10.1056/nejmoa2034201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Barouch DH, Stephenson KE, Sadoff J, et al. Durable humoral and cellular immune responses 8 months after Ad26.COV2.S vaccination. N Engl J Med. 2021;385:951‐953. doi: 10.1056/nejmc2108829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Keeton R, Tincho MB, Ngomti A, et al. T cell responses to SARS‐CoV‐2 spike cross‐recognize Omicron. Nature. 2022;603:488‐492. doi: 10.1038/s41586-022-04460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schmidt T, Klemis V, Schub D, et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV‐19/mRNA vaccination. Nat Med. 2021;27(9):1530‐1535. doi: 10.1038/s41591-021-01464-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pozzetto B, Legros V, Djebali S, et al. Immunogenicity and efficacy of heterologous ChAdOx1–BNT162b2 vaccination. Nature. 2021;600(7890):701‐706. doi: 10.1038/s41586-021-04120-y [DOI] [PubMed] [Google Scholar]
- 90. Schmidt T, Klemis V, Schub D, et al. Cellular immunity predominates over humoral immunity after homologous and heterologous mRNA and vector‐based COVID‐19 vaccine regimens in solid organ transplant recipients. Am J Transplant. 2021;21(12):3990‐4002. doi: 10.1111/ajt.16818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Collier ARY, Yu J, McMahan K, et al. Differential kinetics of immune responses elicited by Covid‐19 vaccines. N Engl J Med. 2021;385(21):2010‐2012. doi: 10.1056/nejmc2115596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Atmar RL, Lyke KE, Deming ME, et al. Homologous and heterologous Covid‐19 booster vaccinations. New Engl J Med. 2022;386(11):1046‐1057. doi: 10.1056/nejmoa2116414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu X, Shaw RH, Stuart ASV, et al. Safety and immunogenicity of heterologous versus homologous prime‐boost schedules with an adenoviral vectored and mRNA COVID‐19 vaccine (Com‐COV): a single‐blind, randomised, non‐inferiority trial. Lancet. 2021;398(10303):856‐869. doi: 10.1016/s0140-6736(21)01694-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Munro APS, Janani L, Cornelius V, et al. Safety and immunogenicity of seven COVID‐19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov‐19 or BNT162b2 in the UK (COV‐BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet. 2021;398(10318):2258‐2276. doi: 10.1016/s0140-6736(21)02717-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Khoo NKH, Lim JME, Gill US, et al. Differential immunogenicity of homologous versus heterologous boost in Ad26.COV2.S vaccine recipients. Med (N Y). 2022;3(2):104‐118.e4. doi: 10.1016/j.medj.2021.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gao Y, Cai C, Grifoni A, et al. Ancestral SARS‐CoV‐2‐specific T cells cross‐recognize the Omicron variant. Nat Med. 2022;28:472‐476. doi: 10.1038/s41591-022-01700-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu J, Chandrashekar A, Sellers D, et al. Vaccines elicit highly conserved cellular immunity to SARS‐CoV‐2 Omicron. Nature. 2022;603(7901):493‐496. doi: 10.1038/s41586-022-04465-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Geers D, Shamier MC, Bogers S, et al. SARS‐CoV‐2 variants of concern partially escape humoral but not T cell responses in COVID‐19 convalescent donors and vaccine recipients. Sci Immunol. 2021;6(59):eabj1750. doi: 10.1126/sciimmunol.abj1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Payne RP, Longet S, Austin JA, et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184(23):5699‐5714.e11. doi: 10.1016/j.cell.2021.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sajadi MM, Myers A, Logue J, et al. Mucosal and systemic responses to SARS‐CoV‐2 vaccination in infection naïve and experienced individuals. bioRxiv. 2021;2021.12.13.472159. doi: 10.1101/2021.12.13.472159 [DOI] [Google Scholar]
- 101. Minervina AA, Pogorelyy MV, Kirk AM, et al. SARS‐CoV‐2 antigen exposure history shapes phenotypes and specificity of memory CD8+ T cells. Nat Immunol. 2022;23:781‐790. doi: 10.1038/s41590-022-01184-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Weaver C, Murphy K. Janeway’s Immunobiology. W. W. Norton; 2016. https://wwnorton.com/books/9780815345053/about‐the‐book. Accessed October 31, 2020. [Google Scholar]
- 103. Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185‐189. doi: 10.1038/nri3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50(5):1132‐1148. doi: 10.1016/j.immuni.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wilkinson TM, Li CKF, Chui CSC, et al. Preexisting influenza‐specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274‐280. doi: 10.1038/nm.2612 [DOI] [PubMed] [Google Scholar]
- 106. Weiskopf D, Bangs DJ, Sidney J, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci USA. 2015;112(31):E4256‐E4263. doi: 10.1073/pnas.1505956112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CM. Cytotoxic CD4 T cells‐friend or foe during viral infection? Front Immunol. 2017;8(2–463):19. doi: 10.3389/fimmu.2017.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Vardhana S, Baldo L, MoriceII WG, Wherry EJ. Understanding T‐cell responses to COVID‐19 is essential for informing public health strategies. Sci Immunol. 2022:eabo1303. doi: 10.1126/sciimmunol.abo1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Breton G, Mendoza P, Hägglöf T, et al. Persistent cellular immunity to SARS‐CoV‐2 infection. J Exp Med. 2021;218(4):e20202515. doi: 10.1084/jem.20202515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wragg KM, Lee WS, Koutsakos M, et al. Establishment and recall of SARS‐CoV‐2 spike epitope‐specific CD4+ T cell memory. Nat Immunol. 2022;23:768‐780. doi: 10.1038/s41590-022-01175-5 [DOI] [PubMed] [Google Scholar]
- 111. Ng OW, Chia A, Tan AT, et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post‐infection. Vaccine. 2016;34(17):2008‐2014. doi: 10.1016/j.vaccine.2016.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Li CK, Wu H, Yan H, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181(8):5490‐5500. doi: 10.4049/jimmunol.181.8.5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yang LT, Peng H, Zhu ZL, et al. Long‐lived effector/central memory T‐cell responses to severe acute respiratory syndrome coronavirus (SARS‐CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120(2):171‐178. doi: 10.1016/j.clim.2006.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rodda LB, Morawski PA, Pruner KB, et al. Imprinted SARS‐CoV‐2‐specific memory lymphocytes define hybrid immunity. Cell. 2022;185:1588‐1601.e14. doi: 10.1016/j.cell.2022.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Meckiff BJ, Ramírez‐Suástegui C, Fajardo V, et al. Imbalance of regulatory and cytotoxic SARS‐CoV‐2‐reactive CD4+ T cells in COVID‐19. Cell. 2020;183(5):1340‐1353.e16. doi: 10.1016/j.cell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kaneko N, Boucau J, Kuo HH, et al. Expansion of cytotoxic CD4+ T cells in the lungs in severe COVID‐19. SSRN Electron J. 2021. doi: 10.2139/ssrn.3813278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kaneko N, Boucau J, Kuo HH, et al. Expansion of Cytotoxic CD4+ T cells in the lungs in severe COVID‐19. medRxiv. 2021;2021.03.23.21253885. doi: 10.1101/2021.03.23.21253885 [DOI] [Google Scholar]
- 118. Dong T, Liu G, Felce S, et al. Memory cytotoxic SARS‐CoV‐2 spike protein‐specific CD4+ T cells associate with viral control. Int Arch Allergy Immunol. 2022;183:350‐359. doi: 10.21203/rs.3.rs-1317569/v1 34794147 [DOI] [Google Scholar]
- 119. Juno JA, Tan HX, Lee WS, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID‐19. Nat Med. 2020;26:1428‐1434. doi: 10.1038/s41591-020-0995-0 [DOI] [PubMed] [Google Scholar]
- 120. Painter MM, Mathew D, Goel RR, et al. Rapid induction of antigen‐specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS‐CoV‐2 mRNA vaccination. Immunity. 2021;54(9):2133‐2142.e3. doi: 10.1016/j.immuni.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tauzin A, Nayrac M, Benlarbi M, et al. A single dose of the SARS‐CoV‐2 vaccine BNT162b2 elicits Fc‐mediated antibody effector functions and T‐cell responses. Cell Host Microbe. 2021;29(7):1137‐1150.e6. doi: 10.1016/j.chom.2021.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bentebibel SE, Khurana S, Schmitt N, et al. ICOS(+)PD‐1(+)CXCR3(+) T follicular helper cells contribute to the generation of high‐avidity antibodies following influenza vaccination. Sci Rep. 2016;6:26494. doi: 10.1038/srep26494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Locci M, Havenar‐Daughton C, Landais E, et al. Human circulating PD‐1+CXCR3−CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758‐769. doi: 10.1016/j.immuni.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Crotty S. Do memory CD4 T cells keep their cell‐type programming: plasticity versus fate commitment? Complexities of interpretation due to the heterogeneity of memory CD4 T cells, including T follicular helper cells. Cold Spring Harb Perspect Biol. 2018;10(3):a032102. doi: 10.1101/cshperspect.a032102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Nelde A, Bilich T, Heitmann JS, et al. SARS‐CoV‐2‐derived peptides define heterologous and COVID‐19‐induced T cell recognition. Nat Immunol. 2021;22(1):74‐85. doi: 10.1038/s41590-020-00808-x [DOI] [PubMed] [Google Scholar]
- 126. Poon MML, Byington E, Meng W, et al. Heterogeneity of human anti‐viral immunity shaped by virus, tissue, age, and sex. Cell Rep. 2021;37(9):110071‐110071. doi: 10.1016/j.celrep.2021.110071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sattler A, Angermair S, Stockmann H, et al. SARS‐CoV‐2 specific T‐cell responses and correlations with COVID‐19 patient predisposition. J Clin Invest. 2020;130(12):6477‐6489. doi: 10.1172/jci140965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bert NL, Chia WN, Wan WY, et al. Widely heterogeneous humoral and cellular immunity after mild SARS‐CoV‐2 infection in a homogeneous population of healthy young men: Heterogenous immunity to SARS‐CoV‐2. Emerg Microbes Infect. 2021;10(1):2141‐2150. doi: 10.1080/22221751.2021.1999777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bert NL, Clapham HE, Tan AT, et al. Highly functional virus‐specific cellular immune response in asymptomatic SARS‐CoV‐2 infection. J Exp Med. 2021;218(5):e20202617. doi: 10.1084/jem.20202617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Boyton RJ, Altmann DM. The immunology of asymptomatic SARS‐CoV‐2 infection: what are the key questions? Nat Rev Immunol. 2021;21(12):1‐7. doi: 10.1038/s41577-021-00631-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Reynolds CJ, Swadling L, Gibbons JM, et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS‐CoV‐2 infection. Sci Immunol. 2020;54(5):eabf3698. doi: 10.1126/sciimmunol.abf3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Vijayakumar B, Boustani K, Ogger PP, et al. Immuno‐proteomic profiling reveals aberrant immune cell regulation in the airways of individuals with ongoing post‐COVID‐19 respiratory disease. Immunity. 2022;55(3):542‐556.e5. doi: 10.1016/j.immuni.2022.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Braun J, Loyal L, Frentsch M, et al. SARS‐CoV‐2‐reactive T cells in healthy donors and patients with COVID‐19. Nature. 2020;587(7833):270‐274. doi: 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- 134. Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype and kinetics of SARS‐CoV‐2‐specific T cells in COVID‐19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sette A, Crotty S. Pre‐existing immunity to SARS‐CoV‐2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457‐458. doi: 10.1038/s41577-020-0389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Mateus J, Grifoni A, Tarke A, et al. Selective and cross‐reactive SARS‐CoV‐2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89‐94. doi: 10.1126/science.abd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Schulien I, Kemming J, Oberhardt V, et al. Characterization of pre‐existing and induced SARS‐CoV‐2‐specific CD8+ T cells. Nat Med. 2020;27:78‐85. doi: 10.1038/s41591-020-01143-2 [DOI] [PubMed] [Google Scholar]
- 138. Shomuradova AS, Vagida MS, Sheetikov SA, et al. SARS‐CoV‐2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53(6):1245‐1257.e5. doi: 10.1016/j.immuni.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Prakash S, Srivastava R, Coulon PG, et al. Genome‐wide B cell, CD4+, and CD8+ T cell epitopes that are highly conserved between human and animal coronaviruses, identified from SARS‐CoV‐2 as targets for preemptive pan‐coronavirus vaccines. J Immunol. 2021;206(11):ji2001438. doi: 10.4049/jimmunol.2001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ishizuka J, Grebe K, Shenderov E, et al. Quantitating T cell cross‐reactivity for unrelated peptide antigens. J Immunol. 2009;183(7):4337‐4345. doi: 10.4049/jimmunol.0901607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Grifoni A, Voic H, Dhanda SK, et al. T Cell responses induced by attenuated flavivirus vaccination are specific and show limited cross‐reactivity with other flavivirus species. J Virol. 2020;94(10):e00089‐20. doi: 10.1128/jvi.00089-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Bacher P, Rosati E, Esser D, et al. Low‐avidity CD4+ T cell responses to SARS‐CoV‐2 in unexposed individuals and humans with severe COVID‐19. Immunity. 2020;53(6):1258‐1271.e5. doi: 10.1016/j.immuni.2020.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lipsitch M, Grad YH, Sette A, Crotty S. Cross‐reactive memory T cells and herd immunity to SARS‐CoV‐2. Nat Rev Immunol. 2020;20(11):709‐713. doi: 10.1038/s41577-020-00460-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Loyal L, Braun J, Henze L, et al. Cross‐reactive CD4 + T cells enhance SARS‐CoV‐2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Meyer‐Arndt L, Schwarz T, Loyal L, et al. Cutting edge: serum but not mucosal antibody responses are associated with pre‐existing SARS‐CoV‐2 spike cross‐reactive CD4 + T cells following BNT162b2 vaccination in the elderly. J Immunol. 2022;208(5):1001‐1005. doi: 10.4049/jimmunol.2100990 [DOI] [PubMed] [Google Scholar]
- 146. Sagar M, Reifler K, Rossi M, et al. Recent endemic coronavirus infection is associated with less severe COVID‐19. J Clin Invest. 2020;131(1):e143380. doi: 10.1172/jci143380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Aran D, Beachler DC, Lanes S, Overhage JM. Prior presumed coronavirus infection reduces COVID‐19 risk: a cohort study. J Infect. 2020;81(6):923‐930. doi: 10.1016/j.jinf.2020.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Gombar S, Bergquist T, Pejaver V, et al. SARS‐CoV‐2 infection and COVID‐19 severity in individuals with prior seasonal coronavirus infection. Diagn Microbiol Infect Dis. 2021;100(2):115338. doi: 10.1016/j.diagmicrobio.2021.115338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Antunes RS, Pallikkuth S, Williams E, et al. Differential T cell reactivity to endemic coronaviruses and SARS‐CoV‐2 in community and health care workers. J Infect Dis. 2021;224(1):jiab176. doi: 10.1093/infdis/jiab176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Swadling L, Diniz MO, Schmidt NM, et al. Pre‐existing polymerase‐specific T cells expand in abortive seronegative SARS‐CoV‐2. Nature. 2021;601(7891):110‐117. doi: 10.1038/s41586-021-04186-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kundu R, Narean JS, Wang L, et al. Cross‐reactive memory T cells associate with protection against SARS‐CoV‐2 infection in COVID‐19 contacts. Nat Commun. 2022;13(1):80. doi: 10.1038/s41467-021-27674-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lederer K, Bettini E, Parvathaneni K, et al. Germinal center responses to SARS‐CoV‐2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022;185(6):1008‐1024.e15. doi: 10.1016/j.cell.2022.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Mudd PA, Minervina AA, Pogorelyy MV, et al. SARS‐CoV‐2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185(4):603‐613.e15. doi: 10.1016/j.cell.2021.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ewer KJ, Barrett JR, Belij‐Rammerstorfer S, et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV‐19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat Med. 2021;27(2):270‐278. doi: 10.1038/s41591-020-01194-5 [DOI] [PubMed] [Google Scholar]
- 155. Folegatti PM, Ewer KJ, Aley PK, et al. Safety and immunogenicity of the ChAdOx1 nCoV‐19 vaccine against SARS‐CoV‐2: a preliminary report of a phase 1/2, single‐blind, randomised controlled trial. Lancet. 2020;396(10249):467‐478. doi: 10.1016/s0140-6736(20)31604-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Vikkurthi R, Ansari A, Pai AR, et al. Inactivated virus vaccine BBV152/Covaxin elicits robust cellular immune memory to SARS‐CoV‐2 and variants of concern. medRxiv. 2021;2021.11.14.21266294. doi: 10.1101/2021.11.14.21266294 [DOI] [Google Scholar]
- 157. Stamatatos L, Czartoski J, Wan YH, et al. mRNA vaccination boosts cross‐variant neutralizing antibodies elicited by SARS‐CoV‐2 infection. Science. 2021;372(6549):1413‐1418. doi: 10.1126/science.abg9175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting Edge: Long‐Term B Cell Memory in Humans after Smallpox Vaccination. J Immunol. 2003;171(10):4969‐4973. doi: 10.4049/jimmunol.171.10.4969 [DOI] [PubMed] [Google Scholar]
- 159. Yu X, Tsibane T, McGraw PA, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455(7212):532‐536. doi: 10.1038/nature07231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Li GM, Chiu C, Wrammert J, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross‐reactive memory B cells. Proc Natl Acad Sci USA. 2012;109(23):9047‐9052. doi: 10.1073/pnas.1118979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long‐lived plasma cells, possess antigen specificities for viral escape mutants. J Exp Med. 2011;208(13):2599‐2606. doi: 10.1084/jem.20110740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Wang Z, Muecksch F, Schaefer‐Babajew D, et al. Naturally enhanced neutralizing breadth against SARS‐CoV‐2 one year after infection. Nature. 2021;595(7867):426‐431. doi: 10.1038/s41586-021-03696-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Marcotte H, Piralla A, Zuo F, et al. Immunity to SARS‐CoV‐2 up to 15 months after infection. Iscience. 2022;25(2):103743. doi: 10.1016/j.isci.2022.103743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Sokal A, Chappert P, Barba‐Spaeth G, et al. Maturation and persistence of the anti‐SARS‐CoV‐2 memory B cell response. Cell. 2021;184(5):1201‐1213.e14. doi: 10.1016/j.cell.2021.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Sakharkar M, Rappazzo CG, Wieland‐Alter WF, et al. Prolonged evolution of the human B cell response to SARS‐CoV‐2 infection. Sci Immunol. 2021;6(56):eabg6916. doi: 10.1126/sciimmunol.abg6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kaneko N, Kuo HH, Boucau J, et al. Loss of Bcl‐6‐expressing T follicular helper cells and germinal centers in COVID‐19. Cell. 2020;183(1):143‐157.e13. doi: 10.1016/j.cell.2020.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Röltgen K, Nielsen SCA, Silva O, et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS‐CoV‐2 infection and vaccination. Cell. 2022;185(6):1025‐1040.e14. doi: 10.1016/j.cell.2022.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Piccoli L, Park YJ, Tortorici MA, et al. Mapping neutralizing and immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell. 2020;183(4):1024‐1042.e21. doi: 10.1016/j.cell.2020.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Wec AZ, Haslwanter D, Abdiche YN, et al. Longitudinal dynamics of the human B cell response to the yellow fever 17D vaccine. Proc Natl Acad Sci USA. 2020;117(12):6675‐6685. doi: 10.1073/pnas.1921388117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Davis CW, Jackson KJL, McElroy AK, et al. Longitudinal analysis of the human B cell response to Ebola virus infection. Cell. 2019;177(6):1566‐1582.e17. doi: 10.1016/j.cell.2019.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Allie SR, Bradley JE, Mudunuru U, et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol. 2019;20(1):97‐108. doi: 10.1038/s41590-018-0260-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Weisel NM, Weisel FJ, Farber DL, et al. Comprehensive analyses of B‐cell compartments across the human body reveal novel subsets and a gut‐resident memory phenotype. Blood. 2020;136(24):2774‐2785. doi: 10.1182/blood.2019002782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Cho A, Muecksch F, Schaefer‐Babajew D, et al. Anti‐SARS‐CoV‐2 receptor‐binding domain antibody evolution after mRNA vaccination. Nature. 2021;600(7889):517‐522. doi: 10.1038/s41586-021-04060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Pauthner M, Havenar‐Daughton C, Sok D, et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46(6):1073‐1088.e6. doi: 10.1016/j.immuni.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Havenar‐Daughton C, Lee JH, Crotty S. Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol Rev. 2017;275(1):49‐61. doi: 10.1111/imr.12512 [DOI] [PubMed] [Google Scholar]
- 176. Havenar‐Daughton C, Carnathan DG, Peña AT de la, et al. Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 2016;17(9):2195‐2209. doi: 10.1016/j.celrep.2016.10.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Cirelli KM, Carnathan DG, Nogal B, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177(5):1153‐1171.e28. doi: 10.1016/j.cell.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lee JH, Sutton H, Cottrell CA, et al. Long‐lasting germinal center responses to a priming immunization with continuous proliferation and somatic mutation. bioRxiv. 2021;2021.12.20.473537. doi: 10.1101/2021.12.20.473537 [DOI] [Google Scholar]
- 179. Kim W, Zhou JQ, Horvath SC, et al. Germinal centre‐driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141‐145. doi: 10.1038/s41586-022-04527-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Turner JS, O’Halloran JA, Kalaidina E, et al. SARS‐CoV‐2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596(7870):109‐113. doi: 10.1038/s41586-021-03738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Sokal A, Barba‐Spaeth G, Fernández I, et al. mRNA vaccination of naive and COVID‐19‐recovered individuals elicits potent memory B cells that recognize SARS‐CoV‐2 variants. Immunity. 2021;54:2893‐2907.e5. doi: 10.1016/j.immuni.2021.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS‐CoV‐2 in Iceland. N Engl J Med. 2020;383(18):1724‐1734. doi: 10.1056/nejmoa2026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science. 2020;370(6521):1227‐1230. doi: 10.1126/science.abd7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Qaseem A, Yost J, Etxeandia‐Ikobaltzeta I, et al. What is the antibody response and role in conferring natural immunity after SARS‐CoV‐2 infection? Rapid, living practice points from the american college of physicians (Version 2). Ann Intern Med. 2022;175:556‐565. doi: 10.7326/m21-3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Mandavilli A. Immunity to the coronavirus may last years, new data hint. The New York Times. 2020. https://www.nytimes.com/2020/11/17/health/coronavirus‐immunity.html. Accessed January 1, 2022. [Google Scholar]
- 186. Li C, Yu D, Wu X, et al. Twelve‐month specific IgG response to SARS‐CoV‐2 receptor‐binding domain among COVID‐19 convalescent plasma donors in Wuhan. Nat Commun. 2021;12(1):4144. doi: 10.1038/s41467-021-24230-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Gallais F, Gantner P, Bruel T, et al. Evolution of antibody responses up to 13 months after SARS‐CoV‐2 infection and risk of reinfection. EBioMedicine. 2021;71:103561. doi: 10.1016/j.ebiom.2021.103561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kannenberg J, Trawinski H, Henschler R, Buhmann R, Hönemann M, Jassoy C. Antibody course and memory B‐cell response in the first year after SARS‐CoV‐2 infection. J Infect Dis. 2022;jiac034. doi: 10.1093/infdis/jiac034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Turner JS, Kim W, Kalaidina E, et al. SARS‐CoV‐2 infection induces long‐lived bone marrow plasma cells in humans. Nature. 2021;595(7867):421‐425. doi: 10.1038/s41586-021-03647-4 [DOI] [PubMed] [Google Scholar]
- 190. Isho B, Abe KT, Zuo M, et al. Persistence of serum and saliva antibody responses to SARS‐CoV‐2 spike antigens in COVID‐19 patients. Sci Immunol. 2020;5(52):eabe5511. doi: 10.1126/sciimmunol.abe5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Yang Y, Yang M, Peng Y, et al. Longitudinal analysis of antibody dynamics in COVID‐19 convalescents reveals neutralizing responses up to 16 months after infection. Nat Microbiol. 2022;7(3):423‐433. doi: 10.1038/s41564-021-01051-2 [DOI] [PubMed] [Google Scholar]
- 192. Wei J, Matthews PC, Stoesser N, et al. Anti‐spike antibody response to natural SARS‐CoV‐2 infection in the general population. Nat Commun. 2021;12(1):6250. doi: 10.1038/s41467-021-26479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Crotty S, Ahmed R. Immunological memory in humans. Semin Immunol. 2004;16(3):197‐203. doi: 10.1016/j.smim.2004.02.008 [DOI] [PubMed] [Google Scholar]
- 194. Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357(19):1903‐1915. doi: 10.1056/nejmoa066092 [DOI] [PubMed] [Google Scholar]
- 195. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science. 2021;372:815‐821. doi: 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short‐lasting. Nat Med. 2020;26(11):1691‐1693. doi: 10.1038/s41591-020-1083-1 [DOI] [PubMed] [Google Scholar]
- 197. Dobaño C, Alonso S, Vidal M, et al. Multiplex antibody analysis of IgM, IgA and IgG to SARS‐CoV‐2 in saliva and serum from infected children and their close contacts. Front Immunol. 2022;13:751705. doi: 10.3389/fimmu.2022.751705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Steensels D, Pierlet N, Penders J, Mesotten D, Heylen L. Comparison of SARS‐CoV‐2 antibody response following vaccination with BNT162b2 and mRNA‐1273. JAMA. 2021;326(15):1533‐1535. doi: 10.1001/jama.2021.15125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Israel A, Shenhar Y, Green I, et al. Large‐scale study of antibody titer decay following BNT162b2 mRNA vaccine or SARS‐CoV‐2 infection. Vaccines (Basel). 2021;10(1):64. doi: 10.3390/vaccines10010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Doria‐Rose N, Suthar MS, Makowski M, et al. Antibody persistence through 6 months after the second dose of mRNA‐1273 vaccine for Covid‐19. N Engl J Med. 2021;384:2259‐2261. doi: 10.1056/nejmc2103916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Pegu A, O’Connell SE, Schmidt SD, et al. Durability of mRNA‐1273 vaccine–induced antibodies against SARS‐CoV‐2 variants. Science. 2021;373(6561):eabj4176. doi: 10.1126/science.abj4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Pajon R, Doria‐Rose NA, Shen X, et al. SARS‐CoV‐2 omicron variant neutralization after mRNA‐1273 booster vaccination. N Engl J Med. 2022;386:1088‐1091. doi: 10.1056/nejmc2119912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Choi A, Koch M, Wu K, et al. Safety and immunogenicity of SARS‐CoV‐2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med. 2021;27(11):2025‐2031. doi: 10.1038/s41591-021-01527-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Falsey AR, Frenck RW, Walsh EE, et al. SARS‐CoV‐2 neutralization with BNT162b2 vaccine dose 3. N Engl J Med. 2021;385(17):NEJMc2113468. doi: 10.1056/nejmc2113468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Xia H, Zou J, Kurhade C, et al. Neutralization of omicron SARS‐CoV‐2 by 2 or 3 doses of BNT162b2 vaccine. bioRxiv. 2022;2022.01.21.476344. doi: 10.1101/2022.01.21.476344 [DOI] [Google Scholar]
- 206. Regev‐Yochay G, Gonen T, Gilboa M, et al. 4th dose COVID mRNA vaccines’ immunogenicity & efficacy against omicron VOC. medRxiv. 2022;2022.02.15.22270948. doi: 10.1101/2022.02.15.22270948 [DOI] [Google Scholar]
- 207. Self WH, Tenforde MW, Rhoads JP, et al. Comparative effectiveness of Moderna, Pfizer‐BioNTech, and Janssen (Johnson & Johnson) vaccines in preventing COVID‐19 hospitalizations among adults without immunocompromising conditions — United States, March–August 2021. MMWR Morb Mortal Wkly Rep. 2021;70(38):1337‐1343. doi: 10.15585/mmwr.mm7038e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Marks P. FDA Ad26.COV2.S COVID‐19 vaccine emergency use authorization amendment review memorandum. 2021. https://www.fda.gov/media/153439/download. Accessed December 1, 2021.
- 209. Shen X, Tang H, Pajon R, et al. Neutralization of SARS‐CoV‐2 variants B.1.429 and B.1.351. New Engl J Med. 2021;384:2352‐2354. doi: 10.1056/nejmc2103740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Keech C, Albert G, Cho I, et al. Phase 1–2 Trial of a SARS‐CoV‐2 Recombinant Spike Protein Nanoparticle Vaccine. New Engl J Med. 2020;383(24):2320‐2332. doi: 10.1056/nejmoa2026920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Sheikh‐Mohamed S, Isho B, Chao GYC, et al. Systemic and mucosal IgA responses are variably induced in response to SARS‐CoV‐2 mRNA vaccination and are associated with protection against subsequent infection. medRxiv. 2022;2021.08.01.21261297. doi: 10.1101/2021.08.01.21261297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Collier AY, Brown CM, Mcmahan K, et al. Immune responses in fully vaccinated individuals following breakthrough infection with the SARS‐CoV‐2 delta variant in Provincetown, Massachusetts. medRxiv. 2021:2021.10.18.21265113. doi: 10.1101/2021.10.18.21265113 [DOI] [Google Scholar]
- 213. Corbett KS, Nason MC, Flach B, et al. Immune correlates of protection by mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. Science. 2021;373(6561):eabj0299. doi: 10.1126/science.abj0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Lund FE, Randall TD. Scent of a vaccine. Science. 2021;373(6553):397‐399. doi: 10.1126/science.abg9857 [DOI] [PubMed] [Google Scholar]
- 215. Gaebler C, Wang Z, Lorenzi JCC, et al. Evolution of antibody immunity to SARS‐CoV‐2. Nature. 2021;591(7851):639‐644. doi: 10.1038/s41586-021-03207-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Muecksch F, Wang Z, Cho A, et al. Increased potency and breadth of SARS‐CoV‐2 neutralizing antibodies after a third mRNA vaccine dose. bioRxiv. 2022;2022.02.14.480394. doi: 10.1101/2022.02.14.480394 [DOI] [Google Scholar]
- 217. Andreano E, Paciello I, Piccini G, et al. Hybrid immunity improves B cells and antibodies against SARS‐CoV‐2 variants. Nature. 2021;600(7889):530‐535. doi: 10.1038/s41586-021-04117-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature. 2021;602:671‐675. doi: 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 219. Saadat S, Tehrani ZR, Logue J, et al. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS‐CoV‐2. Jama. 2021;325(14):1467‐1469. doi: 10.1001/jama.2021.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Richardson SI, Madzorera VS, Spencer H, et al. SARS‐CoV‐2 Omicron triggers cross‐reactive neutralization and Fc effector functions in previously vaccinated, but not unvaccinated individuals. medRxiv. 2022;2022.02.10.22270789. doi: 10.1101/2022.02.10.22270789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Gaebler C, DaSilva J, Bednarski E, et al. SARS‐CoV‐2 neutralization after mRNA vaccination and variant breakthrough infection. medRxiv. 2022;2022.02.09.22270692. doi: 10.1101/2022.02.09.22270692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Suryawanshi RK, Chen IP, Ma T, et al. Limited cross‐variant immunity after infection with the SARS‐CoV‐2 omicron variant without vaccination. medRxiv. 2022;2022.01.13.22269243. doi: 10.1101/2022.01.13.22269243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Kotaki R, Adachi Y, Moriyama S, et al. SARS‐CoV‐2 Omicron‐neutralizing memory B‐cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci Immunol. 2022;7:eabn8590. doi: 10.1126/sciimmunol.abn8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Janssen EM, Lemmens EE, Wolfe T, Christen U, van Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421(6925):852‐856. doi: 10.1038/nature01441 [DOI] [PubMed] [Google Scholar]
- 225. Apostolidis SA, Kakara M, Painter MM, et al. Cellular and humoral immune responses following SARS‐CoV‐2 mRNA vaccination in patients with multiple sclerosis on anti‐CD20 therapy. Nat Med. 2021;27(11):1990‐2001. doi: 10.1038/s41591-021-01507-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Tortorella C, Aiello A, Gasperini C, et al. Humoral‐ and T‐cell–specific immune responses to SARS‐CoV‐2 mRNA vaccination in patients with MS using different disease‐modifying therapies. Neurology. 2022;98(5):e541‐e554. doi: 10.1212/wnl.0000000000013108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Bates TA, McBride SK, Leier HC, et al. Vaccination before or after SARS‐CoV‐2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol. 2022;7:eabn8014. doi: 10.1126/sciimmunol.abn8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654‐656. doi: 10.1038/s41586-021-04387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Goel RR, Apostolidis SA, Painter MM, et al. Distinct antibody and memory B cell responses in SARS‐CoV‐2 naïve and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6(58):eabi6950. doi: 10.1126/sciimmunol.abi6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Rodda LB, Morawski PA, Pruner KB, et al. Imprinted SARS‐CoV‐2‐specific memory lymphocytes define hybrid immunity. medRxiv. 2022;2022.01.12.22269192. doi: 10.1101/2022.01.12.22269192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Brouillette M. Your immune system evolves to fight coronavirus variants – scientific American. Scientific American. 2021. https://www.scientificamerican.com/article/your‐immune‐system‐evolves‐to‐fight‐coronavirus‐variants/. Accessed December 1, 2021. [Google Scholar]
- 232. Tarke A, Coelho CH, Zhang Z, et al. SARS‐CoV‐2 vaccination induces immunological memory able to cross‐recognize variants from Alpha to Omicron. bioRxiv. 2021;2021.12.28.474333. doi: 10.1101/2021.12.28.474333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Laidlaw BJ, Ellebedy AH. The germinal centre B cell response to SARS‐CoV‐2. Nat Rev Immunol. 2021;22:7‐18. doi: 10.1038/s41577-021-00657-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Kim W, Zhou JQ, Sturtz AJ, et al. Germinal centre‐driven maturation of B cell response to SARS‐CoV‐2 vaccination. bioRxiv. 2021;2021.10.31.466651. doi: 10.1101/2021.10.31.466651 [DOI] [Google Scholar]
- 235. Lederer K, Parvathaneni K, Painter MM, et al. Germinal center responses to SARS‐CoV‐2 mRNA vaccines in healthy and immunocompromised individuals. medRxiv. 2021;2021.09.16.21263686. doi: 10.1101/2021.09.16.21263686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Muecksch F, Weisblum Y, Barnes CO, et al. Affinity maturation of SARS‐CoV‐2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853‐1868.e7. doi: 10.1016/j.immuni.2021.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


