Abstract
Heterogeneity in symptoms associated with COVID‐19 in infected patients remains unclear. ACE2 and TMPRSS2 gene variants are considered possible risk factors for COVID‐19. In this study, a retrospective comparative genome analysis of the ACE2 and TMPRSS2 variants from 946 whole‐exome sequencing data was conducted. Allele frequencies of all variants were calculated and filtered to remove variants with allele frequencies lower than 0.003 and to prioritize functional coding variants. The majority of detected variants were intronic, only two ACE2 and three TMPRSS2 nonsynonymous variants were detected in the analyzed cohort. The main ACE2 variants that putatively have a protective or susceptibility effect on SARS‐CoV‐2 have not yet been determined in the Turkish population. The Turkish genetic makeup likely lacks any ACE2 variant that increases susceptibility to SARS‐CoV‐2 infection. TMPRSS2 rs75603675 and rs12329760 variants that were previously defined as common variants that have different allele frequencies among populations and may have a role in SARS‐CoV‐2 attachment to host cells were determined in the population. Overall, these data will contribute to the formation of a national variation database and may also contribute to further studies of ACE2 and TMPRSS2 in the Turkish population and differences in SARS‐CoV‐2 infection among other populations.
Keywords: ACE2, COVID‐19, SARS‐CoV‐2, TMPRSS2, variant
1. INTRODUCTION
After the pneumonia cases of unknown cause were reported to the World Health Organization (WHO) in Wuhan Province of China in December 2019, the factor causing the disease was identified as a novel coronavirus strain. Cases were spread worldwide and the WHO declared a pandemic on March 11, 2020. The new coronavirus strain was named SARS‐CoV‐2 as they share a remarkable genetic identity with the known SARS‐CoV and the disease was referred to as coronavirus disease 2019 (COVID‐19). 1 SARS‐CoV‐2, which has a much higher transmission rate than the known human coronavirus strains, damages the lung tissue, causing respiratory failure and leading to death. Individuals over 65 years old, smokers, and people with chronic diseases such as hypertension, diabetes, and kidney failure are more severely affected. Patients commonly show symptoms of dry cough, high fever, and shortness of breath, while some patients with abdominal pain, diarrhea, and headache are also reported. Some infected individuals, on the other hand, remain asymptomatic. 2 As of the end of March 2021, the number of cases reported as SARS‐CoV‐2 positive worldwide has exceeded 128 million, and over 2 million deaths were reported to the WHO. 3
Considering the cases worldwide, it was observed that SARS‐CoV‐2 was strangely and tragically selective. While only some infected people have been reported to be sick and most of the critical patients are elderly or people with chronic problems; some of those who die from the disease are individuals who do not have any chronic disease and are relatively young. A great variation in cases and mortality rates among countries were also detected. Along with factors including the number of tests performed, percentage of smokers, average age, and environmental factors, it is thought that genetic characteristics might also affect susceptibility to SARS‐CoV‐2 infection.
The entry of enveloped viruses into cells is initiated by the binding of its spike (S) proteins to cell surface receptors. Previous reports indicated that angiotensin‐converting enzyme 2 (ACE2) is one of the host receptors for the novel coronavirus, SARS‐CoV‐2. 1 , 4 ACE2 is a transmembrane protein encoded by the ACE2 (MIM# 300335) gene on the Xp22.2 chromosome and has a transcript composed of 3339 bp and 21 exons. It is responsible for the conversion of angiotensin I to angiotensin 1−9 and angiotensin II to vasodilator angiotensin 1−7 and has roles in renal and cardiovascular function. 5 In addition to cell surface receptors, another factor required for the entry of viruses into host cells is proteases. Proteases cleave and activate viral envelope glycoproteins and form domains catalyzing membrane melding, which is a process called priming. 6 Transmembrane protease serine 2 (TMPRSS2) is shown to be involved in priming SARS‐CoV‐2 by cleaving the S protein at the S1/S2 and S2 sites. 7 TMPRSS2 is encoded by TMPRSS2 (MIM# 602060) gene on chromosome 21q22.3, producing a 3250 bp‐long transcript with 14 exons according to the NCBI database.
Expression levels and variations in ACE2 and TMPRSS2 in different individuals may facilitate or slow down the entrance of the virus into host cells and this might explain the dramatic variability of SARS‐CoV‐2 infection through individuals and populations. Likewise, variations in expression quantitative trait loci (eQTL) regions, known to regulate the ACE2 gene expression, may lead to changes in protein synthesis hence the course of infection. In a recent study, ACE2 and eQTL variation data from worldwide populations in ChinaMap, 1000 Genomes Project, and gnomAD databases were examined. Even though there is no direct evidence supporting the presence of ACE2 variations causing resistance to coronavirus S‐protein binding among populations, the study suggested that the eQTL variants associated with higher ACE2 expression have much higher allele frequencies in East Asian populations that may have an effect on different sensitivity or response from different populations to COVID‐19 under similar conditions. 8 In the Italian population, where the disease caused more severe results compared to Asian and European countries, four TMPRSS2 variants were found to have significantly different allele frequencies. Furthermore, concerning the eQTL variants, population‐specific haplotypes were detected that are expected to upregulate TMPRSS2 gene expression. 9
In light of these works, we conducted a retrospective comparative genome analysis of the ACE2 and TMPRSS2 gene variants in the Turkish population.
2. MATERIAL AND METHODS
2.1. Data collection and analysis
To investigate the allele frequencies of all functional coding variants of ACE2 and TMPRSS2, variation data from 946 unique individuals were collected from a total of 10 centers and hospitals around Turkey. As these individuals were randomly selected from centers located in various cities over the whole geographical parts of Turkey, we believe that the data represent the population of the country. This study was approved by the institutional review board (approval no: YDU/2020/79‐1103). The name of the center, sequencing platform, panels, and bioinformatic pipelines used are listed in Supporting Information: Table 1. Allele frequencies of all ACE2 and TMPRSS2 variants were calculated and then filtered to remove variants with allele frequencies lower than 0.003. Individuals with unknown gender (fetus) and without sufficient variant information were removed from the analysis. Public databases including Database of single‐nucleotide polymorphism (dbSNP), genome aggregation database (gnomAD v2.1.1), and Ensembl were used to prioritize functional coding variants and to obtain global and population‐based allele frequencies for comparison. 10 , 11 , 12
2.2. In silico analysis
Crystal structures of ACE2‐Spike (PDB ID:6LZG) and TMPRSS2 (PDB ID: 7MEQ) were retrieved from the protein data bank. PyMol program (http://pymol.sourceforge.net) was used to visualize and generate in silico mutant proteins. 13
3. RESULTS
3.1. ACE2 gene variant analysis
A total of 2948 variants from 617 individuals were analyzed and 451 different variants were detected. Among the 451 variants, 9 of them were nonsynonymous. When the variants that have allele frequencies lower than 0.003 were removed, 70 variants remained and 2 of those were missense variants, one coding sequence synonymous variant, and the others were intronic variants. Details of the 70 variants, calculated allele frequencies in the Turkish population, and global and population‐based allele frequencies obtained from public databases are represented in Table 1.
Table 1.
The list of ACE2 gene variant analysis from the whole‐exome sequencing data (only the variants ≥0.003 are listed)
| dbSNP‐ID | c.DNA | Allele frequency | Coding consequence | Amino acid change | g1000 | GnomAD | GLOBAL | EUR |
|---|---|---|---|---|---|---|---|---|
| rs971249 | c.584‐71A>G | 0.581 | Intronic | 0.803 | 0.695 | 0.644 | 0.609 | |
| rs113691336, rs4646158 | c.1297 + 68_1297 + 69insCTTAT | 0.455 | Intronic | 0.834 | 0.729 | 0.629 | ||
| rs4646174 | c.1896 + 147G>C | 0.404 | Intronic | 0.683 | 0.621 | 0.683 | 0.649 | |
| rs2285666 | c.439 + 4G>A | 0.326 | Intronic, splice donor | 0.350 | 0.273 | 0.350 | 0.230 | |
| rs11340646, rs769765211, rs775397699 | c.1443‐97del | 0.128 | Intronic | 0.0016 | — | |||
| rs4646156 | c.1071‐605T>A | 0.078 | Intronic | 0.802 | 0.699 | 0.803 | 0.651 | |
| rs4646143 | c.900 + 1879A>G | 0.072 | Intronic | 0.826 | 0.727 | 0.828 | 0.650 | |
| rs397822493 | c.187‐1538dup | 0.070 | Intronic | 0.835 | 0.732 | |||
| rs111691073 | c.1997 + 520_1997 + 527del | 0.060 | Intronic | 0.971 | 0.940 | |||
| rs35803318 | c.2247G>A | 0.060 | Coding sequence variant; synonymous | 0.020 | 0.038 | 0.020 | 0.050 | |
| rs4646152 | c.1070 + 1320T>C | 0.058 | Intronic | 0.832 | 0.729 | 0.832 | 0.651 | |
| rs879922 | c.1542‐361G>C | 0.056 | Intronic | 0.682 | 0.619 | 0.682 | 0.646 | |
| rs4240157 | c.1897‐1015G>A | 0.056 | Intronic | 0.682 | 0.617 | 0.682 | 0.641 | |
| rs397686765, rs398087648, rs4646131, rs869127567 | c.345 + 524delT | 0.053 | Intronic | 0.803 | 0.701 | |||
| rs233575 | c.211‐625C>T | 0.051 | Intronic | 0.863 | 0.771 | 0.863 | 0.664 | |
| rs1514279 | c.802 + 101C>T | 0.048 | Intronic | 0.803 | 0.698 | 0.803 | 0.646 | |
| rs2158083 | c.584‐807G>A | 0.047 | Intronic | 0.808 | 0.703 | 0.808 | 0.648 | |
| rs2048683 | c.584‐920A>C | 0.046 | Intronic | 0.803 | 0.698 | 0.803 | 0.648 | |
| rs4646153 | c.1071‐1397G>A | 0.044 | Intronic | 0.831 | 0.730 | 0.832 | 0.649 | |
| rs2316904 | c.901‐1178G>A | 0.042 | Intronic | 0.623 | 0.827 | 0.828 | 0.650 | |
| rs146122606, rs57823828, rs754565978 | c.346‐1077_346‐1070dupCCTTCCTT | 0.041 | Intronic | NA | — | |||
| rs776459296, rs759499720, rs752472046 | c.584‐8dupA | 0.041 | Intronic | NA | — | — | ||
| rs4646124 | c.186 + 2053A>G | 0.037 | Intronic | 0.803 | 0.699 | 0.804 | 0.651 | |
| rs1978124 | c.186 + 786A>G | 0.035 | Intronic | 0.794 | 0.625 | 0.795 | 0.529 | |
| rs4646127 | c.187‐2327T>C | 0.035 | Intronic | 0.809 | 0.692 | 0.809 | 0.651 | |
| rs138373349, rs4646148 | c.901‐380_901‐379insTTAA | 0.035 | Intronic | NA | — | — | ||
| rs11394305 | c.901‐1367dup | 0.034 | Intronic | |||||
| rs2074192 | c.2115‐449G>A | 0.033 | Intronic | 0.363 | 0.424 | 0.364 | 0.426 | |
| rs200672831 | c.1443‐187G>C | 0.032 | Intronic | NA | — | — | ||
| rs2048684 | c.901‐702T>G | 0.032 | Intronic | 0.832 | 0.729 | 0.832 | 0.651 | |
| rs11374008 | c.1298‐936dup | 0.032 | Intronic | NA | — | — | ||
| rs34481900 | c.186 + 2745dup | 0.031 | Intronic | 0.809 | ||||
| rs1514280 | c.1897‐499T>C | 0.030 | Intronic | 0.802 | 0.719 | 0.802 | 0.655 | |
| rs233574 | c.2115‐268A>G | 0.029 | Intronic | 0.842 | 0.748 | 0.842 | 0.670 | |
| rs4646120 | c.186 + 1113C>T | 0.028 | Intronic | 0.735 | 0.567 | 0.735 | 0.527 | |
| rs4646142 | c.900 + 534C>G | 0.028 | Intronic | 0.357 | 0.235 | 0.359 | 0.238 | |
| rs199544436 | c.1443‐168G>C | 0.027 | Intronic | — | 0.0001 | — | ||
| rs2023802 | c.187‐1019C>T | 0.027 | Intronic | 0.804 | 0.702 | 0.804 | 0.651 | |
| rs4646147 | c.901‐1231A>T | 0.024 | Intronic | 0.828 | 0.724 | 0.828 | 0.651 | |
| rs2316903 | c.901‐1761C>A | 0.019 | Intronic | 0.827 | 0.724 | 0.828 | 0.651 | |
| rs2106809 | c.186 + 788T>C | 0.016 | Intronic | 0.316 | 0.191 | 0.316 | 0.247 | |
| rs41303171 | c.2158A>G | 0.016 | Coding sequence missense variant | p. Asn720Asp | 0.0045 | 0.016 | 0.023 | 0.018 |
| rs714205 | c.2114 + 472G>C | 0.015 | Intronic | 0.308 | 0.184 | 0.308 | 0.205 | |
| rs757066 | c.583 + 884G>A | 0.015 | Intronic | 0.856 | 0.750 | 0.856 | 0.649 | |
| rs892503408 | c.1443‐200C>T | 0.007 | Intronic | NA | — | — | ||
| rs1132186 | c.2309 + 6768T>G | 0.006 | Intronic | 0.688 | 0.629 | 0.688 | 0.649 | |
| rs73195521 | c.346‐143A>T | 0.006 | Intronic | 0.0013 | 0.00303 | 0.001 | 0.005 | |
| rs73195520 | c.439 + 24G>A | 0.006 | Intronic | 0.0013 | 0.00307 | 0.001 | 0.005 | |
| rs542683073 | c.440‐133G>A | 0.005 | Intronic | — | 0.00014 | — | ||
| ‐ | c.1297 + 70_1297 + 71insTATGA | 0.004 | NA | — | — | |||
| rs146598386 | c.187‐1124C>A | 0.004 | Intronic | 0.0026 | 0.01235 | 0.003 | 0.009 | |
| rs755489152 | c.2115‐274del | 0.004 | Intronic | NA | — | — | ||
| rs4830542 | c.2309 + 5541G>T | 0.004 | Intronic | 0.684 | 0.623 | 0.684 | 0.649 | |
| rs10551988 | c.2310‐701_2310‐696del | 0.004 | Intronic | No data | ||||
| rs780782488 | c.584‐19T>A | 0.004 | Intronic | — | 0.000223 | — | ||
| rs34161673 | c.697‐161del | 0.004 | Intronic | 0.0019 | 0.00638 | 0.002 | 0.007 | |
| rs41297301 | c.900 + 90C>A | 0.004 | Intronic | 0.0037 | 0.01453 | 0.004 | 0.012 | |
| rs4646188 | c.901‐1830T>C | 0.004 | Intronic | 0.0437 | 0.10405 | 0.044 | 0.131 | |
| rs1043432251 | c.901‐1890del | 0.004 | Intronic | NA | — | — | ||
| rs934301151 | c.901‐72C>T | 0.004 | Intronic | — | 0.00037 | 0.00034 | — | |
| — | c.*812C>A | 0.003 | NA | — | — | |||
| rs200260858 | c.1442 + 90_1442 + 91delCA | 0.003 | Intronic | 0.0074 | — | 0.007 | 0.004 | |
| — | c.186 + 73G>A | 0.003 | NA | — | — | |||
| — | c.186 + 74G>A | 0.003 | NA | — | — | |||
| — | c.186 + 75G>A | 0.003 | NA | — | — | |||
| — | c.186 + 79T>A | 0.003 | NA | — | — | |||
| rs187959864 | c.186 + 80C>A | 0.003 | Intronic | 0.0003 | 0.00009 | 0.000264 | 0.001 | |
| rs777042582 | c.2114 + 44CAA | 0.003 | Intronic | — | 0.00014 | 0.00013 | 0.00028 | |
| rs4646140 | c.802 + 24G>A | 0.003 | Intronic | 0.0601 | 0.0336 | 0.060 | 0.001 | |
| rs4646116 | c.77A>G | 0.003 | Coding sequence missense variant | p. Lys26Arg | 0.002 | 0.003 | 0.000 | 0.010 |
3.2. TMPRSS2 gene variant analysis
A total of 13 382 variants from 1072 individuals were analyzed and 490 different variants were detected. Among these variants, 9 were missense and 1 was deletion causing a frameshift. When the variants that have allele frequencies lower than 0.003 were removed, 192 variants remained. Three of those were missense variants, eight were coding sequence synonymous variants, seven were 3′UTR variants, and nine were upstream variants. Details of the 192 variants, calculated allele frequencies in the Turkish population, and global and population‐based allele frequencies obtained from public databases are represented in Table 2.
Table 2.
The list of TMPRSS2 gene variant analysis from the whole‐exome sequencing data (only the variants ≥0.003 are listed)
| dbSNP‐ID | c.DNA | Allele frequency | Coding consequence | Amino acid change | g1000 | GnomAD | GLOBAL | EUR |
|---|---|---|---|---|---|---|---|---|
| rs140530035 | c.795‐15_795‐14del, c.684‐15_684‐14del | 0.449 | Intronic | 0.81 | 0.90 | 0.813 | 0.977 | |
| rs17854725 | c.768T>C | 0.302 | Coding sequence; synonymous variant | 0.36 | 0.47 | 0.339 | 0.458 | |
| rs422471 | c.445 + 14G>A | 0.286 | Intronic | 0.55 | 0.61 | 0.555 | 0.698 | |
| rs386416 | c.326‐45C>G | 0.267 | Intronic | 0.55 | — | 0.555 | 0.700 | |
| rs464431 | c.1011‐52T>C | 0.242 | Intronic | 0.87 | 0.96 | 0.874 | 0.980 | |
| rs112132031 | c.1076‐44_1076‐43insCCCGAGGCCTTAG | 0.211 | Intronic | 0.83 | — | 0.830 | 0.979 | |
| rs75603675 | c.−57 + 99G>T, c.23G>T | 0.205 | Coding sequence; missense_variant | p. Gly8Val | 0.24 | 0.36 | 0.244 | 0.405 |
| rs462321 | c.1172‐115A>G | 0.161 | Intronic | 0.58 | 0.68 | 0.578 | 0.784 | |
| rs462326 | c.1172‐130C>G | 0.135 | Intronic | 0.57 | 0.68 | 0.573 | 0.784 | |
| rs12329760 | c.478G>A, c.589G>A | 0.129 | Coding sequence; missense_variant | p. Val197Met | 0.26 | 0.28 | 0.261 | 0.236 |
| rs2298659 | c.777C>T | 0.121 | Coding sequence; synonymous variant | 0.20 | 0.25 | 0.209 | 0.230 | |
| rs458280 | c.1011‐144A>C | 0.120 | Intronic | 0.88 | 0.96 | 0.879 | 0.980 | |
| rs455922 | c.1076‐164A>G | 0.112 | Intronic | 0.88 | 0.96 | 0.878 | 0.981 | |
| rs9975014 | c.683 + 93T>C | 0.106 | Intronic | 0.26 | 0.25 | 0.262 | 0.254 | |
| rs734056 | c.572 + 83G>T | 0.100 | Intronic | 0.28 | 0.37 | 0.285 | 0.489 | |
| rs458213 | c.1011‐54A>T | 0.099 | Intronic | 0.23 | 0.32 | 0.225 | 0.441 | |
| rs465576 | c.1076‐184G>T | 0.094 | Intronic | 0.83 | 0.93 | 0.834 | 0.979 | |
| rs3787950 | c.225A>G | 0.083 | Coding sequence; synonymous variant | 0.16 | 0.11 | 0.163 | 0.079 | |
| rs9974933 | c.683 + 122T>C | 0.066 | Intronic | 0.26 | 0.25 | 0.262 | 0.254 | |
| rs429442 | c.325 + 102G>A | 0.063 | Intronic | 0.28 | 0.25 | 0.280 | 0.232 | |
| rs7364083 | c.1011‐149C>T | 0.062 | Intronic | 0.64 | 0.62 | 0.639 | 0.538 | |
| rs2838042 | c.238 + 176A>G | 0.058 | Intronic | 0.23 | 0.23 | 0.233 | 0.243 | |
| rs455281 | c.1467 + 589C>A | 0.058 | Intronic | 0.74 | 0.89 | 0.736 | 0.967 | |
| rs28524972 | c.1076‐101G>C | 0.054 | Intronic | 0.29 | — | 0.287 | 0.303 | |
| rs2094881 | c.795‐288A>G, c.684‐288A>G | 0.053 | Intronic | 0.53 | 0.63 | 0.530 | 0.748 | |
| rs4816720 | c.445 + 2877G>A | 0.053 | Intronic | 0.82 | 0.92 | 0.820 | 0.977 | |
| rs9985159 | c.684‐137G>A | 0.053 | Intronic | 0.33 | 0.33 | 0.335 | 0.230 | |
| rs386638 | c.238 + 1236G>A | 0.052 | Intronic | 0.84 | 0.95 | 0.844 | 0.976 | |
| rs2298662 | c.727 + 389C>G | 0.050 | Intronic | 0.88 | 0.96 | 0.123 | 0.021 | |
| rs365724 | c.445 + 1099C>G | 0.050 | Intronic | 0.56 | 0.62 | 0.555 | 0.715 | |
| rs112132031, rs71951459 | c.1187‐43_1187‐42insCCGAGGCCTTAGT, c.1076‐44_1076‐43insCCCGAGGCCTTAG | 0.049 | Intronic | 0.83 | — | 0.830 | 0.979 | |
| rs456016 | c.1076‐279A>G | 0.049 | Intronic | 0.87 | 0.96 | 0.874 | 0.981 | |
| rs4818241 | c.445 + 2975T>C | 0.048 | Intronic | 0.87 | 0.96 | 0.872 | 0.978 | |
| rs415731 | c.126 + 983T>C | 0.043 | Intronic | 0.71 | 0.70 | 0.714 | 0.651 | |
| rs2298663 | c.727 + 317G>A | 0.043 | Intronic | 0.53 | 0.62 | 0.525 | 0.747 | |
| rs417443 | c.239‐1658T>C | 0.043 | Intronic | 0.86 | — | 0.865 | 0.978 | |
| rs457909 | c.1467 + 465C>T | 0.043 | Intronic | 0.99 | 0.99 | 0.994 | — | |
| rs2156300 | c.445 + 3565C>T | 0.042 | Intronic | 0.87 | 0.96 | 0.872 | 0.978 | |
| rs138365638; rs557282706; rs869112255 | c.684‐358_684‐357del, c.573‐358_573‐357del | 0.042 | Intronic | 0.87 | 0.96 | 0.874 | 0.979 | |
| rs2156301 | c.445 + 3372A>G | 0.041 | Intronic | 0.87 | 0.96 | 0.871 | 0.978 | |
| rs402303 | c.238 + 1132A>G | 0.039 | Intronic | 0.52 | 0.58 | 0.519 | 0.709 | |
| rs4818240 | c.445 + 3019A>G | 0.038 | Intronic | 0.82 | 0.92 | 0.821 | 0.977 | |
| rs3787947 | c.326‐153G>A | 0.037 | Intronic | 0.31 | 0.34 | 0.307 | 0.279 | |
| rs467375 | c.1075 + 168C>T | 0.036 | Intronic | 0.22 | 0.32 | 0.223 | 0.441 | |
| rs7277080 | c.−57 + 3608G>A | 0.036 | Intronic | 0.23 | 0.35 | 0.230 | 0.369 | |
| rs55964536 | c.728‐215G>A | 0.035 | Intronic | 0.24 | 0.35 | 0.242 | 0.483 | |
| rs435877 | c.−56‐2781C>G | 0.033 | Intronic | 0.81 | 0.83 | 0.812 | 0.852 | |
| rs2410430 | c.446‐3587T>C | 0.033 | Intronic | 0.87 | 0.96 | 0.869 | 0.978 | |
| rs402197 | c.445 + 651A>G | 0.033 | Intronic | 0.87 | 0.96 | 0.874 | 0.978 | |
| rs462448 | c.1172‐407A>G | 0.033 | Intronic | 0.88 | 0.97 | 0.879 | 0.981 | |
| rs8131648 | c.684‐587A>G | 0.033 | Intronic | 0.56 | 0.64 | 0.555 | 0.744 | |
| rs429524 | c.‐56‐1825C>G | 0.032 | Intronic | 0.86 | 0.87 | 0.860 | 0.853 | |
| rs8131649 | c.684‐590A>G | 0.032 | Intronic | 0.56 | 0.64 | 0.555 | 0.746 | |
| rs2104810 | c.795‐550C>T, c.684‐550C>T | 0.031 | Intronic | 0.53 | 0.63 | 0.532 | 0.745 | |
| rs3819138 | c.326‐54G>C | 0.030 | Intronic | 0.069 | — | 0.068 | 0.151 | |
| rs2410429 | c.446‐3519T>G | 0.029 | Intronic | 0.62 | 0.74 | 0.620 | 0.744 | |
| rs461194 | c.1467 + 362G>C | 0.029 | Intronic | 0.87 | 0.96 | 0.869 | 0.969 | |
| rs55896064 | c.1468‐118C>T | 0.029 | Intronic | 0.078 | 0.13 | 0.078 | 0.133 | |
| rs5844077 | insA | 0.028 | Upstream variant | 0.78 | 0.74 | 0.779 | 0.730 | |
| rs417888 | c.239‐1806T>C | 0.028 | Intronic | 0.62 | 0.62 | 0.625 | 0.515 | |
| rs73905370 | c.1468‐58T>A | 0.028 | Intronic | 0.078 | 0.13 | 0.078 | 0.133 | |
| rs456298 | c.*1318A>T | 0.027 | 3′UTR variant | 0.63 | 0.72 | 0.628 | 0.831 | |
| rs462471 | c.*1593T>C | 0.027 | 3′UTR variant | 0.63 | 0.74 | 0.630 | 0.831 | |
| rs35899679 | c.239‐1800G>T | 0.026 | Intronic | 0.24 | — | 0.238 | 0.463 | |
| rs381179 | c.445 + 2679A>G | 0.026 | Intronic | — | — | 0.001 | — | |
| rs392370 | c.238 + 2117T>G | 0.026 | Intronic | 0.30 | 0.26 | 0.302 | 0.240 | |
| rs462574 | c.*1340T>C | 0.026 | 3′UTR variant | 0.74 | 0.90 | 0.743 | 0.966 | |
| rs8126497 | c.‐57 + 284C>T | 0.026 | Intronic | 0.10 | 0.13 | 0.101 | 0.199 | |
| rs398061769 | 0.026 | Intronic | — | — | 0.555 | 0.715 | ||
| rs9974589 | c.1171 + 452T>G | 0.025 | Intronic | 0.60 | 0.60 | 0.604 | 0.536 | |
| rs383510 | c.445 + 1954A>G | 0.025 | Intronic | 0.60 | 0.57 | 0.604 | 0.515 | |
| rs415918 | c.445 + 445G>A | 0.025 | intronic | 0.56 | 0.62 | 0.558 | 0.713 | |
| rs61735794 | c.1155G>A, c.1266G>A | 0.025 | Coding sequence; synonymous variant | 0.009 | 0.02 | 0.009 | 0.030 | |
| rs2298661 | c.728‐219G>T | 0.023 | Intronic | 0.27 | 0.26 | 0.268 | 0.230 | |
| rs365025 | c.445 + 1254C>G | 0.023 | Intronic | 0.56 | 0.62 | 0.559 | 0.715 | |
| rs378616 | c.‐57 + 2466G>T | 0.023 | Intronic | 0.71 | 0.69 | 0.714 | 0.717 | |
| rs8134203 | c.684‐695G>A | 0.023 | Intronic | 0.54 | 0.63 | 0.536 | 0.744 | |
| rs375408 | c.445 + 717C>T | 0.023 | Intronic | 0.88 | 0.96 | 0.877 | 0.979 | |
| rs456142 | c.*1573A>G | 0.023 | 3′UTR variant | 0.63 | 0.73 | 0.630 | 0.831 | |
| rs11701576 | c.56‐146T>C | 0.022 | Intronic | 0.16 | 0.099 | 0.162 | 0.106 | |
| rs2070788 | c.1282 + 587C>T, c.1171 + 587C>T | 0.021 | Intronic | 0.60 | 0.59 | 0.603 | 0.536 | |
| rs460976 | 0.021 | Intronic | 0.87 | 0.96 | 0.872 | 0.968 | ||
| rs4290734 | c.446‐554T>C | 0.020 | Intronic | 0.24 | 0.34 | 0.245 | 0.487 | |
| rs4818239 | c.683 + 1024A>G | 0.020 | Intronic | 0.30 | 0.39 | 0.302 | 0.502 | |
| rs8134216 | c.684‐711G>A | 0.020 | Intronic | 0.54 | 0.63 | 0.536 | 0.745 | |
| rs9974995 | c.683 + 188G>A | 0.020 | Intronic | 0.26 | 0.25 | 0.261 | 0.253 | |
| rs455045 | c.126 + 1158G>A | 0.020 | Intronic | 0.63 | 0.56 | 0.627 | 0.493 | |
| rs2070787 | c.1282 + 446A>C, c.1171 + 446A>C | 0.020 | Intronic | 0.29 | — | 0.293 | 0.308 | |
| rs34769294 | c.238 + 2137dup | 0.020 | Intronic | 0.24 | — | 0.245 | 0.238 | |
| rs61170417; rs67617179 | c.1172‐773_1172‐772del | 0.020 | Intronic | 0.27 | 0.26 | 0.266 | 0.308 | |
| rs35041537 | c.239‐1849G>A | 0.019 | Intronic | 0.24 | 0.34 | 0.240 | 0.463 | |
| rs4283504 | 0.019 | Upstream variant | 0.87 | 0.89 | 0.874 | 0.887 | ||
| rs7279603 | c.1172‐759A>G | 0.019 | Intronic | 0.33 | 0.32 | 0.334 | 0.310 | |
| rs2298664 | c.325 + 253C>G | 0.018 | Intronic | 0.32 | — | 0.322 | 0.278 | |
| rs2298660 | c.728‐210G>A | 0.017 | Intronic | 0.26 | 0.27 | 0.259 | 0.201 | |
| rs56097233 | c.445 + 1040_445 + 1041del | 0.017 | Intronic | 0.56 | 0.61 | 0.555 | 0.715 | |
| rs62217531 | c.445 + 2420G>A | 0.017 | Intronic | 0.30 | 0.39 | 0.298 | 0.478 | |
| rs430915 | c.238 + 1540T>C | 0.017 | Intronic | 0.62 | 0.62 | 0.623 | 0.514 | |
| rs7275220 | c.238 + 959C>T | 0.016 | Intronic | 0.53 | 0.66 | 0.529 | 0.750 | |
| rs139374762; rs75929377 | c.557‐671_557‐666delTGTCTG | 0.016 | Intronic | 0.25 | 0.34 | 0.253 | 0.487 | |
| rs34624090 | c.1075 + 291dup | 0.015 | Intronic | 0.22 | 0.32 | 0.224 | 0.442 | |
| rs2070793 | c.1282 + 998T>C, c.1171 + 998T>C | 0.015 | Intronic | 0.33 | 0.32 | 0.332 | 0.309 | |
| rs57474639 | c.1468‐188G>A | 0.015 | Intronic | 0.08 | 0.14 | 0.085 | 0.133 | |
| rs8129713 | c.‐57 + 3410A>G | 0.014 | Intronic | 0.11 | 0.13 | 0.109 | 0.199 | |
| rs386818798 | c.1011‐54_1011‐52delACTinsTCC | 0.014 | Intronic | NA | — | — | ||
| rs2070790 | c.1282 + 888C>G, c.1171 + 888C>G | 0.013 | Intronic | 0.29 | 0.29 | 0.292 | 0.307 | |
| rs2070792 | c.1282 + 965C>T, c.1171 + 965C>T | 0.013 | Intronic | 0.33 | 0.32 | 0.332 | 0.309 | |
| rs10154090 | c.239‐2203A>T | 0.013 | Intronic | 0.30 | 0.33 | 0.298 | 0.273 | |
| rs11702475 | c.556 + 2753G>A, c.445 + 2753G>A | 0.013 | Intronic | 0.26 | 0.35 | 0.259 | 0.491 | |
| rs2298857 | c.445 + 2340C>T | 0.013 | Intronic | 0.30 | 0.26 | 0.299 | 0.236 | |
| rs915823 | c.573‐245T>G | 0.013 | Intronic | 0.16 | 0.22 | 0.161 | 0.204 | |
| rs9976780 | c.446‐2706G>A | 0.013 | Intronic | 0.56 | 0.61 | 0.561 | 0.723 | |
| rs928871 | c.239‐1011G>A | 0.012 | Intronic | 0.37 | — | 0.372 | 0.279 | |
| rs9636988 | c.683 + 1054A>G | 0.012 | Intronic | 0.26 | 0.25 | 0.261 | 0.257 | |
| rs34783969 | c.446‐2109T>A | 0.012 | Intronic | 0.26 | 0.35 | 0.257 | 0.488 | |
| rs11088551 | 0.011 | Upstream variant | 0.27 | 0.36 | 0.246 | 0.411 | ||
| rs375760 | c.445 + 635C>A | 0.011 | Intronic | 0.22 | 0.20 | 0.223 | 0.232 | |
| rs4303795 | 0.011 | Upstream variant | 0.25 | 0.39 | 0.246 | 0.411 | ||
| rs66575656 | c.727 + 569G>A | 0.011 | Intronic | 0.25 | 0.24 | 0.245 | 0.257 | |
| rs4303794 | 0.010 | Upstream variant | 0.25 | 0.36 | 0.246 | 0.411 | ||
| rs6517669 | c.239‐1416T>C | 0.010 | Intronic | 0.37 | 0.39 | 0.369 | 0.278 | |
| rs7364088 | c.1011‐222C>T | 0.010 | Intronic | 0.30 | 0.30 | 0.304 | 0.263 | |
| rs364289 | c.445 + 1456C>T | 0.010 | Intronic | 0.30 | 0.26 | 0.308 | 0.224 | |
| rs8128074 | 0.010 | Upstream variant | 0.87 | 0.89 | 0.874 | 0.886 | ||
| rs9305744 | c.1076‐318C>T | 0.010 | Intronic | 0.31 | 0.31 | 0.313 | 0.233 | |
| rs9977234 | c.446‐3035C>A | 0.010 | Intronic | 0.20 | 0.19 | 0.207 | 0.232 | |
| rs117696554 | c.437‐92C>T, c.326‐92C>T | 0.009 | Intronic | 0.008 | 0.019 | 0.008 | 0.028 | |
| rs2257202 | c.445 + 2606A>G | 0.009 | Intronic | 0.22 | 0.19 | 0.217 | 0.233 | |
| rs28548447 | c.1011‐330C>T | 0.009 | Intronic | 0.26 | 0.26 | 0.264 | 0.303 | |
| rs34983238 | c.126 + 1049T>G | 0.009 | Intronic | 0.058 | 0.08 | 0.058 | 0.117 | |
| rs61735792 | c.300C>T, c.189C>T | 0.009 | Coding sequence variant; synonymous | 0.005 | 0.009 | 0.005 | 0.017 | |
| rs73230068 | c.1010 + 85C>G | 0.009 | Intronic | 0.013 | 0.026 | 0.013 | 0.039 | |
| rs10668560, rs150454800 | c.445 + 3305_445 + 3312del | 0.009 | Intronic | NA | — | — | ||
| rs34561135 | c.683 + 92C>T | 0.009 | Intronic | 0.017 | 0.046 | 0.017 | 0.053 | |
| rs3787946 | c.727 + 769C>G | 0.009 | Intronic | 0.28 | 0.28 | 0.285 | 0.231 | |
| rs61299115 | 0.009 | Upstream variant | 0.25 | 0.36 | 0.246 | 0.411 | ||
| rs9305745 | c.238 + 2209G>A | 0.009 | Intronic | 0.29 | 0.33 | 0.292 | 0.273 | |
| rs391099 | c.239‐2259A>G | 0.008 | Intronic | 0.30 | 0.27 | 0.304 | 0.240 | |
| rs56695953 | c.126 + 311C>T | 0.008 | Intronic | 0.12 | 0.13 | 0.108 | 0.200 | |
| rs9983252 | c.445 + 2999G>C | 0.008 | Intronic | 0.31 | 0.34 | 0.313 | 0.253 | |
| rs401371 | c.238 + 1471C>G | 0.008 | Intronic | 0.20 | 0.17 | 0.196 | 0.210 | |
| rs56066678 | c.445 + 3842G>A | 0.008 | Intronic | 0.29 | 0.26 | 0.293 | 0.236 | |
| rs743542 | c.1425 + 151C>T | 0.008 | Intronic | 0.15 | 0.10 | 0.149 | 0.063 | |
| rs918360768 | c.239‐480T>C | 0.008 | Intronic | NA | — | — | ||
| rs145283231 | c.838 + 1237del. c.727 + 1237del | 0.007 | Intronic | — | 0.25 | — | — | |
| rs2070786 | c.1282 + 372A>G. c.1171 + 372A>G | 0.007 | Intronic | 0.30 | 0.29 | 0.298 | 0.308 | |
| rs9983330 | c.683 + 846T>C | 0.007 | Intronic | 0.26 | 0.28 | 0.261 | 0.235 | |
| rs112467088 | c.55 + 474T>A, c.‐57 + 605T>A | 0.007 | Intronic | 0.19 | 0.28 | 0.186 | 0.281 | |
| rs11911394 | c.350‐755A>G | 0.007 | Intronic | 0.37 | 0.39 | 0.369 | 0.279 | |
| rs61735793 | c.224C>T | 0.007 | Coding sequence; missense_variant | p. Thr75Ile | 0.003 | 0.009 | 0.003 | 0.008 |
| rs62217525 | c.*221G>A | 0.007 | 3′UTR variant | 0.02 | 0.035 | 0.021 | 0.055 | |
| rs144192191 | c.839‐422_839‐419dup, c.728‐422_728‐419dup | 0.007 | Intronic | 0.28 | 0.22 | 0.283 | 0.263 | |
| rs62217527 | c.727 + 285G>A | 0.007 | Intronic | 0.046 | 0.078 | 0.046 | 0.117 | |
| rs73230088 | c.55 + 273G>T | 0.007 | Intronic | 0.08 | 0.13 | 0.080 | 0.157 | |
| rs12481984 | 0.006 | Upstream variant | 0.24 | 0.36 | 0.240 | 0.406 | ||
| rs2070789 | c.1282 + 771G>A, c.1171 + 771G>A | 0.006 | Intronic | 0.32 | 0.32 | 0.315 | 0.229 | |
| rs28360562 | c.55 + 1751T>G | 0.006 | Intronic | 0.06 | 0.08 | 0.057 | 0.117 | |
| rs34205539 | c.‐56‐1430dup | 0.006 | Intronic | 0.06 | 0.08 | 0.057 | 0.117 | |
| rs55704664 | c.55 + 1266G>A | 0.006 | Intronic | 0.10 | 0.13 | 0.057 | 0.117 | |
| rs2838043 | c.‐56‐1104G>A | 0.006 | Intronic | 0.11 | 0.13 | 0.109 | 0.200 | |
| rs395584 | c.‐57 + 3561A>G | 0.006 | Intronic | 0.22 | 0.11 | 0.224 | 0.019 | |
| rs55760462 | c.127‐1701A>G | 0.006 | Intronic | 0.16 | 0.24 | 0.165 | 0.239 | |
| rs61735789 | c.540C>T | 0.006 | Coding sequence variant; synonymous | 0.004 | 0.010 | 0.004 | 0.013 | |
| rs7283324 | c.1172‐364G>A | 0.006 | Intronic | 0.31 | 0.31 | 0.312 | 0.224 | |
| rs73372166 | c.1467 + 623C>T | 0.006 | Intronic | 0.13 | 0.18 | 0.133 | 0.136 | |
| rs2838039 | c.445 + 3777A>G | 0.005 | Intronic | 0.31 | 0.34 | 0.308 | 0.254 | |
| rs75756279 | c.838 + 47G>A | 0.005 | Intronic | 0.004 | 0.004 | 0.004 | 0.008 | |
| rs73372163 | c.1467 + 669C>T | 0.005 | Intronic | 0.13 | 0.18 | 0.132 | 0.136 | |
| rs73905371 | c.1467 + 674G>C | 0.005 | Intronic | 0.08 | 0.14 | 0.085 | 0.133 | |
| rs3761373 | c.56‐406G>A | 0.005 | Intronic | 0.16 | 0.10 | 0.163 | 0.106 | |
| rs460751 | 0.005 | Intronic | 0.83 | 0.92 | 0.826 | 0.965 | ||
| rs111220497 | c.838 + 1292C>T, c.727 + 1292C>T | 0.004 | Intronic | — | 0.31 | — | — | |
| rs1003030 | c.126 + 440T>C | 0.004 | Intronic | 0.16 | 0.10 | 0.163 | 0.106 | |
| rs111220481 | c.838 + 1319C>G, c.727 + 1319C>G | 0.004 | Intronic | — | 0.28 | — | — | |
| rs143680939 | c.*1583del | 0.004 | 3′UTR variant | 0.08 | — | 0.082 | 0.133 | |
| rs201627185 | c.557‐2706delG | 0.004 | Intronic | NA | — | — | — | |
| rs2187238 | c.‐56‐2635A>G | 0.004 | Intronic | 0.11 | 0.14 | 0.112 | 0.199 | |
| rs2838040 | c.238 + 1591T>C | 0.004 | Intronic | 0.33 | 0.36 | 0.331 | 0.275 | |
| rs28707508 | 0.004 | Upstream variant | 0.23 | 0.34 | 0.230 | 0.384 | ||
| rs34256269 | c.126 + 1170C>T | 0.004 | Intronic | 0.08 | 0.13 | 0.079 | 0.159 | |
| rs61728255 | c.727 + 1468T>C | 0.004 | Intronic | 0.88 | 0.92 | 0.880 | 0.980 | |
| rs141788162 | c.759C>T | 0.003 | Coding sequence variant; synonymous | 0.002 | 0.004 | 0.002 | 0.003 | |
| rs199824558 | c.210C>T | 0.003 | Coding sequence variant; synonymous | 0.001 | 0.0002 | 0.001 | — | |
| rs422761 | c.‐56‐877C>T | 0.003 | Intronic | 0.22 | 0.11 | 0.225 | 0.019 | |
| rs61459778 | c.1468‐343G>C | 0.003 | Intronic | 0.14 | 0.19 | 0.136 | 0.136 | |
| rs777860329 | 0.003 | Intronic | NA | — | — | — | ||
| rs113506821 | c.795‐200G>A, c.684‐200G>A | 0.003 | Intronic | 0.02 | 0.05 | 0.022 | 0.050 | |
| rs35871560 | c.445 + 2741del | 0.003 | Intronic | 0.54 | — | 0.536 | 0.680 | |
| rs56136037 | c.445 + 185C>A | 0.003 | Intronic | 0.014 | 0.04 | 0.014 | 0.046 | |
| rs74749793 | c.55 + 4225C>A | 0.003 | Intronic | 0.16 | 0.098 | 0.158 | 0.103 | |
| rs75200570 | c.‐57 + 1396A>G | 0.003 | Intronic | 0.06 | 0.03 | 0.056 | 0.011 | |
| rs76000363 | c.*1592C>T | 0.003 | 3′UTR variant | 0.08 | 0.14 | 0.082 | 0.133 |
3.3. In silico findings and functional predictions
The crystal structure of ACE2 (PDB ID: 6LZG) revealed that N‐linked glycan molecules are attached to Asn53, Asn90, and Asn322. 14 Asn90 is a conserved amino acid in a number of bats in which coronaviruses cannot infect through ACE2. Glycosylation of this amino acid regulates the Spike−ACE2 interaction in bats. 15 Glycosylation of Asn90 and its subsequent branching is suggested to decrease the ACE2−Spike binding affinity through steric effects. 16 Lys26 of ACE2 generates critical polar and salt bridge interactions with sugar moiety and nearby amino acids, Glu22 and Asn90 (Figure 1A). To analyze the effect of Lys26Arg mutation we generated in silico mutant on the ACE2−Spike structure using the crystal structure having PDB ID of 6LZG. 14 Since Arg has a larger side chain than Lys, the side chain of Arg cannot fit in the same space. The sterically most favorable orientation of in silico mutation showed that Arg side chain cannot generate polar interactions as in the case of Lys amino acid. Instead, Arg may generate a salt bridge with Asp30 (Figure 1B). Asp30 forms a salt bridge with Lys417 of Spike in the crystal structure (Figure 1). In Lys26Arg mutation, Arg can stabilize the Asp30−Lys417 interaction which may result in higher infectivity of the SARS‐CoV‐2.
Figure 1.
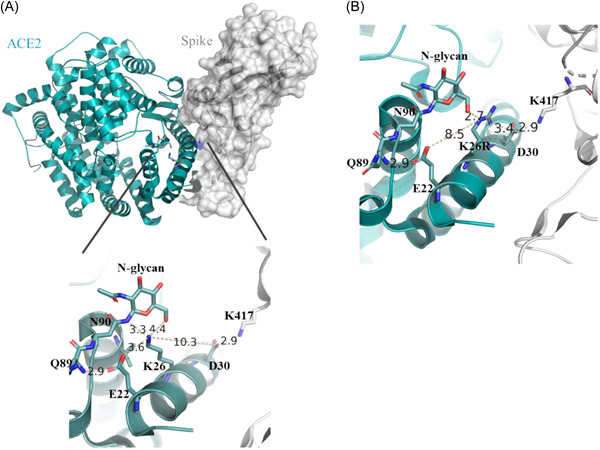
Structure of ACE2‐Spike (6LZG). (A) ACE2 is shown as a cyan cartoon and Spike is shown on a white surface. At the zoom‐in structure, Lys26 interacting amino acids are shown. (B) In silico generated Lys26Arg ACE2 mutation and its proposed interaction scheme is shown.
The TMPRRS2 has three regions: cytoplasmic, transmembrane, and extracellular. Val160 is found in the extracellular region of the protein. 17 TMPRSS2 structure (PDB ID: 7MEQ) shows that Val160 is located on a beta‐strand structure and surrounded by hydrophobic residues Leu225, Val171, Leu158, and Val149 (Figure 2A). Mutation of Val160 to a less hydrophobic residue may disturb this interaction network. To analyze the effect of Val160Met on TMPRSS2, we mutated valine to methionine in silico. Molecular analysis of Val160Met shows that (i) hydrophobic network cannot be maintained and (ii) there is a steric clash between Met and nearby amino acids for example with Tyr222 which suggests that the mutation may destabilize the protein (Figure 2B). In addition to this analysis, we used DUET server to predict the effect of mutation on the TMPRSS2. 18 DUET uses two previously developed approaches for predictions: knowledge‐based and graph‐based signature methods. All type of calculations in DUET predicts that Val160Met mutation destabilizes the protein (Table 3).
Figure 2.
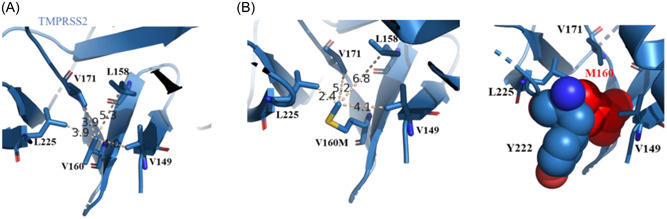
Structure of TMPRSS2 (7MEQ). (A) TMPRSS2 is shown as a blue cartoon representation. Val160 and its interacting amino acids are shown. (B) In silico generated Val160Met TMPRSS2 mutation is shown. To show a steric clash between mutant Met160 and a nearby Tyr222, amino acids are shown in red and blue spheres (proportional to their van der Waals radius) (at right).
Table 3.
In silico analysis of TMPRSS2 Val160Met. ∆∆G < 0 indicates destabilization
| DUET Results (kcal/mol) | |
|---|---|
| mCSM (∆∆G) | −0.847 |
| SDM (∆∆G) | −2.39 |
| DUET (∆∆G) | −1.251 |
4. DISCUSSION
Many studies have demonstrated that the symptoms of COVID‐19 vary greatly among patients. Understanding the reason underlying this heterogeneity in risk of progression to a severe form has been a challenge since the start of the pandemic. There are many known factors that can potentially affect the severity of COVID‐19 infection including greater age, presence of co‐morbidities, smoking, and air pollution. 19 , 20 , 21 In addition to these clinical and environmental factors, genetic variability can also account for the susceptibility to SARS‐CoV‐2 infection and the different clinical presentations observed in COVID‐19 patients. 22 ACE2 and TMPRSS2 are transmembrane surface proteins that play critical roles in viral attachment and host cell entry for SARS‐CoV and SARS‐CoV‐2. SARS‐CoV‐2 binds to ACE2 through the receptor‐binding domain in spike proteins, which are then cleaved by TMPRSS2 to allow fusion with the host cell membrane. 7 , 23 Therefore, polymorphisms in genes encoding these proteins can affect the binding affinity of the viral spike protein to host cells as well as membrane fusion efficiency, modulating the host susceptibility to SARS‐CoV‐2. In this context, we investigated the genetic variability of ACE2 and TMPRSS2 in the Turkish population to show the existence of any enrichment of missense or indel variants in coding regions that may potentially affect the binding dynamics of the virus to host cells and also wanted to compare our results with previous epidemiological studies in different populations.
For both ACE2 and TMPRSS2, majority of variants detected in the Turkish population were intronic. Only 2/70 of ACE2 variants (c.2158A>G;p.Asn720Asp; NM_021804.2 (rs41303171) and c.77A>G;p.Lys26Arg; NM_021804.2 (rs4646116)) (Table 1) and 3/192 of TMPRSS2 variants (c.23G>T;p.Gly8Val; NM_001135099.1 (rs75603675), c.589G>A;p.Val197Met; NM_001135099.1 (rs12329760) and c.224C>T;p.Thr75Ile; NM_005656.3 (rs61735793)) (Table 2) that have allele frequencies above 0.003 were identified as coding variant missense variations.
The most frequent ACE2 variant was identified as rs971249 variant with an allele frequency of 0.581, followed by rs113691336 which has an allele frequency of 0.455 and the third most frequent ACE2 variant was found to be rs4646174 with an allele frequency of 0.404 in the Turkish population. All frequent variants that have allele frequencies above 0.06 were intronic.
Considering the missense variants that potentially affect protein structure or function, ACE2 rs41303171 has a detected allele frequency of 0.016 in the Turkish population. Previous in silico structural analyses have demonstrated that this variation causes ACE2 protein to have a higher binding affinity to TMPRSS2 and may facilitate entry of the virus to the host cells. 24 The global allele frequency of this variant is 0.023, 0.018 in European populations, and 0.001 in the Southern Asian population according to the dbSNP database. The variation was previously mentioned by different groups from Italy, India, and Iran. It was found to be frequent in the study where ACE2 variants in a cohort of SARS‐CoV‐2‐positive Italian patients were investigated. 25 Likely, the variant was reported as a common missense change (AF 0.011) together with rs4646116 and c.631G>A; p.(Gly211Arg) variants in a study conducted with whole‐exome data of 6930 Italian control individuals, which are predicted to affect protein structure and stabilization. 26 c.1051C>G;p.(Leu351Val) and c.1166C>A;p.(Pro389His) were the rare variants detected in this cohort predicted to interfere with the internalization process but were not present in our studied group. In the same study, WES data of 131 patients and 258 controls were compared. The allelic variability in the control group was detected to be statistically significant even though no single variant was significantly enriched between the two groups. c.1166C>A;p.(Pro389His) was one of the missense variants, along with c.1174A>C;p.(Lys392Gln), c.1178C>G;p.(Thr393Ser), and c.1312C>G;p.(Gln438Glu) that was listed as variants leading to an increase in interaction affinity between TMPRSS2 and SARS‐CoV‐2 S protein, where c.1409G>T;p.(Arg470Ile) and c.1247A>G;p.(Tyr416Cys) were found to cause a decrease in a recent study that performed molecular docking analyses. 27 None of these variants were present in the Turkish population included in the present study. In a comprehensive retrospective study, c.1166C>A;p.(Pro389His) was stated to be present only in the Latino/Admixed American population, with an allele frequency of 0.015%. 28
Intronic c.439+4G>A (rs2285666) and c.1888G>C;p.(Asp630His) variants were also detected in the Italian COVID‐19 patient cohort. 25 p.(Asp630His) variant is not present in our study group. However, rs2285666 is the fourth common variant detected in the Turkish population with an allele frequency of 0.326. Allele frequency of the variant in EUR‐TSI (Italy) is 0.186, lower than other populations reported in dbSNP. Additionally, in another study, this variant was found to be the most frequent ACE2 variant detected among clinical exome data of 103 individuals from India. 29 In the same study, the rs4646116 variant was detected in one individual. The variant was shown to potentially affect the binding affinity of SARS‐CoV‐2 spike protein to ACE2 receptor and is not frequent in the Turkish population (0.003) whereas it is not detected in the Italian population according to dbSNP variation data. 30 Consistent with the previous analysis, 31 our in silico model predicts that rs4646116 variation (p. Lys26Arg ACE2) may facilitate the SARS‐CoV‐2 infection via stronger Spike−ACE2 interaction (Figure 1). It was shown to be not associated with COVID‐19 clinical outcomes in Iranian patients. 32 Synonymous exonic c.2247G>A;p.(Val749=) (rs35803318) variant was detected in groups of COVID‐19 patients with different clinical symptoms (mild, severe, and death) in the Iranian population, according to the same study. This variation is relatively common in the Turkish population (AF 0.06) compared to Global (AF 0.02).
p.Arg708Trp, p.Arg710Cys, p.Arg710His, and p.Arg716Cys ACE2 variants that are located in the dimeric interface of ACE2 with TMPRSS2 were found to be present in European, Eastern, Asian, and Latino/Admixed American populations but not present in our Turkish study population in the present study. 28
In a more recent study, in a total of 1378 whole‐exome sequences of individuals from the Middle Eastern populations (Iran, Qatar, and Kuwait), the prevalence of the rs41303171 was noted to be highest among Europeans (2.5%), Iranians (0.6%) when compared to Kuwaitis (0.3%), Qataris (0.2%), and other global populations (0.4%) and minör allele frequency of this variant significantly correlated with the case fatality rates (p < 0.0003) in the corresponding countries as of December 2020. 33 In the same study, they also propose that the rs41303171 variant may enhance TMPRSS2 activation and subsequent viral entry.
Cao et al. investigated allele frequency distributions of 1700 ACE2 variants among different populations. Uneven distribution of some variants between populations was observed in this study. For example, ACE2 rs4646127 intronic variant was shown to be associated with higher expression levels in East‐Asian populations with an allele frequency of 0.993 according to dbSNP data. This variant was also detected in the studied Turkish population with an allele frequency of 0.035.
The most frequent TMPRSS2 variant detected in the Turkish population was the intronic rs140530035 variant with an allele frequency of 0.449. The second most frequent variant is a coding sequence synonymous variant, rs17854725. It has an allele frequency of 0.302. This variant was reported to be rare in the Latin American population and is frequent in the Eastern Asian populations according to the databases. The third most frequent variant in the studied population is the intronic rs422471 variant with the calculated allele frequency of 0.286.
TMPRSS2 rs75603675 and rs12329760 were the missense variants detected in the Turkish population with allele frequencies of 0.205 and 0.129 respectively. Both were within the 10 most frequent variants detected in the studied population. In a recent study, these variants were referred to as variants whose allele frequencies vary by ancestry and geography, differing between East Asians and other populations. 34 Importantly, rs12329760 was predicted to be deleterious by SIFT, PolyPhen‐2, and PROVEAN which suggest altered protein function. Our in silico analyses suggest that the rs12329760 variant (p.Val160Met TMPRSS2) may disrupt the hydrophobic interaction core of TMPRSS2 and destabilize the protein (Figure 2, Table 3). It is in a highly conserved exonic splicing enhancer region of the gene and is strongly associated with TMPRSS2‐ERG fusion translocation in prostate cancer due to the increased risk of exon skipping. 35 Rs75603675, on the other hand, was considered deleterious only by PolyPhen‐2 software. Both could potentially affect the function of TMPRSS2 in facilitating SARS‐CoV‐2 cell entry and therefore may possess a protective role. 36 It was noted in a study that the rs12329760‐T variant allele may have altered the highly conserved scavenger receptor cysteine‐rich (SRCR) domain of TMPRSS2 and also decreased protein stability thus impairing the processing of the spike protein of the SARS‐CoV‐2 A2a subtype. 37 , 38 This may result in the protection of East Asians from the SARS‐CoV‐2 A2a subtype as the variant has a higher allele frequency in that region compared to others and also the Turkish population.
Rs12329760 was reported in 4.85% of individuals studied in India as well. 29 Rs383510, rs2298662, and rs2070788 are three variants, that are known to increase susceptibility to Influenza A (H7N9) and may also affect COVID‐19 infectivity was reported to have low allele frequencies in the Indian population as well as in the Turkish population in our study. 29
A very recent study, which analyzed the association between the rs12329760 and COVID‐19 severity in 2244 critically ill patients with COVID‐19 from the UK intensive care units has shown that the T allele of rs12329760 is associated with a reduced likelihood of developing severe COVID‐19. Results of this study further identified TMPRSS2 protein as a promising drug target, with a potential role for camostat mesylate, which is a drug approved for the treatment of postoperative reflux esophagitis and chronic pancreatitis, in COVID‐19 treatment. 39
In another study among Italian COVID‐19 patients, the rare rs114363287; p.Gly111Arg TMPRSS2 variant was detected with a higher frequency compared to other populations. This variant is missing in our cohort. On the other hand, rs75603675 and rs12329760 which are among frequent TMPRSS2 variants in the general Turkish population were detected in lower frequencies in the COVID‐19 patients, which supports the possible protective role of these two variants against COVID‐19. 40
The other TMPRSS2 missense variant detected in the Turkish population was rs61735793 with an allele frequency of 0.007. The variant has low allele frequencies in all reported populations in the dbSNP database. No studies are associating this variant with COVID‐19 susceptibility or disease severity in any population.
Irham et. al. investigated TMPRSS2 variants affecting expression among populations from different continents. They identified four variants: rs464397, rs469390, rs2070788, and rs383510 that influence TMPRSS2 protein expression in the lungs. 41 Rs464397 and rs469390 variants were not detected in the studied cohort of the Turkish population, whereas rs2070788 and rs383510 were detected with frequencies of 0.021 and 0.025 respectively. These frequencies are lower than other studied populations.
Considering the large population size of Turkey, the sample size may be a limitation in our study. Additionally, we conducted this analysis on the general population. A study with a larger sample size that will include COVID‐19 infected and control groups can be designed for further analysis of alleles affecting susceptibility and disease severity.
5. CONCLUSION
Overall, our data suggests enrichment of the rs4646116 ACE2 functional allele in the Turkish population, which was demonstrated to potentially enhance the binding of the SARS‐CoV‐2 to the receptor by in silico modelling. The two TMPRSS2 missense variants, rs12329760 and rs75603675, that were detected in the Turkish population and have differential frequency distributions in dbSNP may have a role in population‐specific outcomes in COVID‐19 severity. To conclude, new SARS‐CoV‐2 variants and their potentially different transmission abilities, as well as ACE2 and TMPRSS2 gene variants should be considered while developing therapeutics for COVID‐19 disease.
AUTHOR CONTRIBUTIONS
Conceived and designed the analysis: Gulten Tuncel, Mahmut Cerkez Ergoren, Sehime Gulsun Temel. Collected the data: Nilgun Duman, Atil Bisgin, Sevcan Tug Bozdogan, Sebnem Ozemri Sag, Aslihan Kiraz, Burhan Balta, Murat Erdogan, Bulent Uyanik, Sezin Canbek, Pinar Ata, Bilgen Bilge Geckinli, Esra Arslan Ates, Ceren Alavanda, Sevda Yesim Ozdemir, Ozlem Sezer, Gulay Oner Ozgon, Hakan Gurkan, Kubra Guler, Ibrahim Boga, Niyazi Kaya, Adem Alemdar, Murat Sayan, Munis Dundar, Sehime Gulsun Temel. Contributed data or analysis tools: Nilgun Duman, Gulten Tuncel, Atil Bisgin, Sevcan Tug Bozdogan, Sebnem Ozemri Sag, Seref Gul, Aslihan Kiraz, Burhan Balta, Murat Erdogan, Bulent Uyanik, Sezin Canbek, Pinar Ata, Bilgen Bilge Geckinli, Esra Arslan Ates, Ceren Alavanda, Sevda Yesim Ozdemir, Ozlem Sezer, Gulay Oner Ozgon, Hakan Gurkan, Kubra Guler, Ibrahim Boga, Niyazi Kaya, Adem Alemdar, Murat Sayan, Munis Dundar, Mahmut Cerkez Ergoren, Sehime Gulsun Temel. Performed analysis: Nilgun Duman, Gulten Tuncel, Atil Bisgin, Sevcan Tug Bozdogan, Sebnem Ozemri Sag, Seref Gul, Aslihan Kiraz, Burhan Balta, Murat Erdogan, Bulent Uyanik, Sezin Canbek, Pinar Ata, Bilgen Bilge Geckinli, Esra Arslan Ates, Ceren Alavanda, Sevda Yesim Ozdemir, Ozlem Sezer, Gulay Oner Ozgon, Hakan Gurkan, Kubra Guler, Ibrahim Boga, Niyazi Kaya, Adem Alemdar, Murat Sayan, Munis Dundar, Mahmut Cerkez Ergoren, Sehime Gulsun Temel. Wrote the paper: Gulten Tuncel, Seref Gul, Mahmut Cerkez Ergoren, Sehime Gulsun Temel. Read and revised the paper: Nilgun Duman, Gulten Tuncel, Atil Bisgin, Sevcan Tug Bozdogan, Sebnem Ozemri Sag, Seref Gul, Aslihan Kiraz, Burhan Balta, Murat Erdogan, Bulent Uyanik, Sezin Canbek, Pinar Ata, Bilgen Bilge Geckinli, Esra Arslan Ates, Ceren Alavanda, Sevda Yesim Ozdemir, Ozlem Sezer, Gulay Oner Ozgon, Hakan Gurkan, Kubra Guler, Ibrahim Boga, Niyazi Kaya, Adem Alemdar, Murat Sayan, Munis Dundar, Mahmut Cerkez Ergoren, Sehime Gulsun Temel.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
All procedures performed in this study were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards (approval number: YDU/2020/78‐1055).
Supporting information
Supplementary information.
Supplementary information.
Duman N, Tuncel G, Bisgin A, et al. Analysis of ACE2 and TMPRSS2 coding variants as a risk factor for SARS‐CoV‐2 from 946 whole‐exome sequencing data in the Turkish population. J Med Virol. 2022;94:5225‐5243. 10.1002/jmv.27976
Nilgun Duman, Gulten Tuncel, Mahmut Cerkez Ergoren, and Sehime Gulsun Temel contributed equally to this study and should be considered joint first/last authors.
Contributor Information
Mahmut Cerkez Ergoren, Email: mahmutcerkez.ergoren@neu.edu.tr.
Sehime Gulsun Temel, Email: sehime@uludag.edu.tr.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grant MC, Geoghegan L, Arbyn M, et al. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS‐CoV‐2; COVID‐19): a systematic review and meta‐analysis of 148 studies from 9 countries. PLoS One. 2020;15:0234765. 10.1371/journal.pone.0234765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO . WHO Coronavirus (COVID‐19) Dashboard. World Health Organization; 2021. https://covid19.who.int/ [Google Scholar]
- 4. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuba K, Imai Y, Penninger JM. Multiple functions of angiotensin‐converting enzyme 2 and its relevance in cardiovascular diseases. Circ J. 2013;77(2):301‐308. 10.1253/circj.CJ-12-1544 [DOI] [PubMed] [Google Scholar]
- 6. Earnest JT, Hantak MP, Park J‐E, Gallagher T. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin‐enriched membrane microdomains. J Virol. 2015;89(11):6093‐6104. 10.1128/jvi.00543-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao Y, Li L, Feng Z, et al. Comparative genetic analysis of the novel coronavirus (2019‐nCoV/SARS‐CoV‐2) receptor ACE2 in different populations. Cell Discov. 2020;6(1):1‐4. 10.1038/s41421-020-0147-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID‐19 severity in Italy. Aging. 2020;12(11):10087‐10098. 10.18632/aging.103415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434‐443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28(1):352‐355. 10.1093/nar/28.1.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682‐D688. 10.1093/nar/gkz966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeLano WL (2008). The PyMOL Molecular Graphics System. https://pymol.org/2/
- 14. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell. 2020;181(4):894‐904e9. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W, Zhang C, Sui J, et al. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634‐1643. 10.1038/sj.emboj.7600640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demogines A, Farzan M, Sawyer SL. Evidence for ACE2‐utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J Virol. 2012;86(11):6350‐6353. 10.1128/JVI.00311-12/SUPPL_FILE/TABLES1-S2_FIGS1.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. David A, Khanna T, Beykou M, Hanna G, Sternberg MJE. Structure, function and variants analysis of the androgen‐regulated TMPRSS2, a drug target candidate for COVID‐19 infection. bioRxiv. 2020. 10.1101/2020.05.26.116608 [DOI]
- 18. Pires DEV, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42(W1):W314‐W319. 10.1093/NAR/GKU411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu W, Tao ZW, Wang L, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J. 2020;133(9):1032‐1038. 10.1097/CM9.0000000000000775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williamson EJ, Walker AJ, Bhaskaran K, et al. Factors associated with COVID‐19‐related death using OpenSAFELY. Nature. 2020;584(7821):430‐436. 10.1038/s41586-020-2521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Tang M, Guo F, et al. Associations between air pollution and COVID‐19 epidemic during quarantine period in China. Environ Pollut. 2021;268(Pt A):115897. 10.1016/j.envpol.2020.115897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anastassopoulou C, Gkizarioti Z, Patrinos GP, Tsakris A. Human genetic factors associated with susceptibility to SARS‐CoV‐2 infection and COVID‐19 disease severity. Hum Genomics. 2020;14(1):40. 10.1186/s40246-020-00290-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippi G, Lavie CJ, Henry BM, Sanchis‐Gomar F. Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in coronavirus disease 2019 (COVID‐19)? Clin Chem Lab Med. 2020;58(9):1415‐1422. 10.1515/cclm-2020-0727 [DOI] [PubMed] [Google Scholar]
- 24. Mohammad A, Marafie SK, Alshawaf E, Abu‐Farha M, Abubaker J, Al‐Mulla F. Structural analysis of ACE2 variant N720D demonstrates a higher binding affinity to TMPRSS2. Life Sci. 2020;259:118219. 10.1016/j.lfs.2020.118219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Novelli A, Biancolella M, Borgiani P, et al. Analysis of ACE2 genetic variants in 131 Italian SARS‐CoV‐2‐positive patients. Hum Genomics. 2020;14(1):29. 10.1186/s40246-020-00279-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benetti E, Tita R, Spiga O, et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID‐19 in the Italian population. Eur J Hum Genet. 2020;28(11):1602‐1614. 10.1038/s41431-020-0691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nouira F, Hamdi M, Redissi A, et al. In silico screening of TMPRSS2 SNPs that affect its binding with SARS‐CoV2 spike protein and directly involved in the interaction affinity changes. bioRxiv. 2021. 10.1101/2021.09.29.462283 [DOI]
- 28. Hou Y, Zhao J, Martin W, et al. New insights into genetic susceptibility of COVID‐19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):216. 10.1186/s12916-020-01673-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iyer GR, Samajder S, Zubeda S, et al. Infectivity and progression of COVID‐19 based on selected host candidate gene variants. Front Genet. 2020;11:11. 10.3389/fgene.2020.00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bosso M, Thanaraj TA, Abu‐Farha M, Alanbaei M, Abubaker J, Al‐Mulla F. The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID‐19. Mol Ther—Methods Clin Dev. 2020;18:321‐327. 10.1016/j.omtm.2020.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stawiski EW, Diwanji D, Suryamohan K, et al. Human ACE2 receptor polymorphisms predict SARS‐CoV‐2 susceptibility.bioRxiv.2021; 10.1038/s42003-021-02030-3 [DOI] [PMC free article] [PubMed]
- 32. Ardeshirdavani A, Zakeri P, Mehrtash A, et al. Clinical population genetic analysis of variants in the SARS‐CoV‐2 receptor ACE2. medRxiv. 2020. 10.1101/2020.05.27.20115071; [DOI]
- 33. Al‐Mulla F, Mohammad A, Al Madhoun A, et al. ACE2 and FURIN variants are potential predictors of SARS‐CoV‐2 outcome: a time to implement precision medicine against COVID‐19. Heliyon. 2021;7(2):2405. 10.1016/J.HELIYON.2021.E06133; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS‐CoV‐2 infection. Mayo Clin Proc. 2020;95(9):1989‐1999. 10.1016/j.mayocp.2020.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhanushali A, Rao P, Raman V, et al. Status of TMPRSS2–ERG fusion in prostate cancer patients from India: correlation with clinico‐pathological details and TMPRSS2 Met160Val polymorphism. Prostate Int. 2018;6(4):145‐150. 10.1016/j.prnil.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paniri A, Hosseini MM, Akhavan‐Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS‐CoV‐2 among different populations. J Biomol Struct Dyn. 2020;39:1‐18. 10.1080/07391102.2020.1767690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhattacharyya C, Das C, Ghosh A, et al. Global spread of SARS‐CoV‐2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv. 2020. 10.1101/2020.05.04.075911 [DOI]
- 38. Sharma S, Singh I, Haider S, Zubbair Malik M, Ponnusamy K, Rai E. ACE2 homo‐dimerization, human genomic variants and interaction of host proteins explain high population specific differences in outcomes of COVID19. bioRxiv. 2020. 10.1101/2020.04.24.050534; [DOI]
- 39. David A, Parkinson N, Peacock TP, et al. A common TMPRSS2 variant has a protective effect against severe COVID‐19. Curr Res Transl Med. 2022;70(2):2452. 10.1016/J.RETRAM.2022.103333; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Latini A, Agolini E, Novelli A, et al. COVID‐19 and genetic variants of protein involved in the SARS‐CoV‐2 entry into the host cells. Genes. 2020;11(9):1‐8. 10.3390/genes11091010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Irham LM, Chou WH, Calkins MJ, Adikusuma W, Hsieh SL, Chang WC. Genetic variants that influence SARS‐CoV‐2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem Biophys Res Commun. 2020;529(2):263‐269. 10.1016/j.bbrc.2020.05.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


