Abstract
The novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) caused the ongoing pandemic named COVID‐19 which causes a serious emergency on public health hazards of international concern. In the face of a critical medical emergency, repositioning of drugs is one of the most authentic options to design an adequate treatment for infected patients immediately. In this strategy, Remdesivir (Veklury), Hydroxychloroquine appears to be the drug of choice and garnered unprecedented attention as potential therapeutic agents against the pandemic realized worldwide due to SARS‐CoV‐2 infection. These are the breathtaking instances of possible repositioning of drugs, whose pharmacokinetics and optimal dosage are familiar. In this review, we provide an overview of these medications, their synthesis, and the possible mechanism of action against SARS‐CoV‐2.
Comprehensive overview on synthesis and mode of actions of antimalarial and antiviral drugs (Remdesivir, Chloroquine, and Hydroxychloroquine) have been furnished to contribute toward the drug repositioning for treatment of severe acute respiratory syndrome coronavirus 2 infection.
1. INTRODUCTION
The virus severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) appeared on December 2019 and spread swiftly to worldwide, posing an immense threat to the world economy and human health [1, 2, 3, 4, 5, 6]. According to the literature, the previous report of coronavirus triggered the severe acute respiratory syndrome coronavirus (SARS‐CoV) outburst, in addition, severe acute diarrhea syndrome coronavirus ravaged livestock creation by causing fatal diseases in pigs. The above‐mentioned epidemics were originated in China by a coronaviruses of bat origin [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17]. In 2019, Fan and workers retraced the initial case of SARS which was reported in November 2012 in Foshan city of Guangdong Province in China [18]. In particular, SARS‐CoV‐2, a novel betacoronavirus, (previously designated 2019‐nCoV) is the root of this global health crisis [19, 20, 21]. SARS‐CoV‐2 is found to contain a spherical morphology along with projection of spike on the surface. It was verified that SARS‐CoV‐2 has high identity of sequence with that of bat SARS‐like coronavirus and SARS‐CoV [22]. Remarkably, SARS‐CoV‐2 has higher transmissibility but decreased pathogenicity from one human to another human related with SARS‐CoV [23, 24, 25, 26, 27, 28, 29, 30]. As of April 14, 2022, over 500,186,525 cases have been confirmed, including 6,190,349 deaths in 235 countries according to the WHO [31]. The coronavirus pandemic (COVID‐19) has become the greatest global health crisis of recent time after the second world war. Together the World Bank and the World Health Organization (WHO) in 2019 estimated the impact of the pandemic at 2.2%–2.8% of global GDP with a fear of socio‐economic imbalance.
Such massive numbers of infected people and human casualties urge for an imperative demand of very efficient, accessible, and inexpensive medications to control and decline the pandemic. Thus, it is crucial to recognize vital antiviral molecules these could suppress the infection at the solitary phase and terminate its advancement over the globe. The health calamity has imposed the relocation of approved drugs in different therapeutic areas [32] and it has been proposed to be the most dynamic way to find a treatment for this virus [33, 34, 35]. Chloroquine (CQ) and Hydroxychloroquine (HCQ) have emerged as the most‐effective therapeutic option and labeled as potential “game‐changers” in popular press for current state of pandemic COVID‐19 [35]. Here in this review, the organization of SARS‐CoV‐2, its pathogenesis, synthesis, and chemotherapeutic role of Remdesivir, CQ or HCQ in COVID‐19 with the perspective of in silico approach of possible prevention of SARS‐CoV‐2 pathogenesis are discussed.
2. SARS‐COV‐2 AND PATHOGENESIS
2.1. The virus
SARS‐CoV‐2 associated to b‐group of the coronavirus family responsible for acute respiratory distress syndrome in human beings like other SARS viruses and transmitted through human contacts through respiratory droplets (Figure 1). Therefore, SARS‐CoV‐2 is extremely fatal to the patients with pre‐existed respiratory diseases. The COVID‐19 virus size, determined from electron micrograph was 70–90 nm [36].
FIGURE 1.
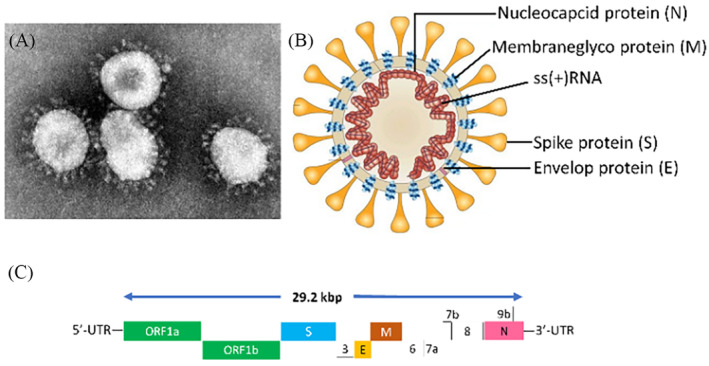
(A) Electron micrograph of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (image credit: Content source(s): CDC/Dr. Murphy F). This material comes with an identification number #4814 from the Centers for Disease Control and Prevention's Public Health Image Library. (B) Schematic presentation of SARS‐CoV‐2 showing spike (S) glycoprotein protein, membrane glyco (M) protein, envelope (E) protein, and nucleocapsid (N) protein (adapted from: Peiris et al.). (C) Large single‐stranded, RNA genome (positive‐sense) of SARS‐CoV‐2 [36]
2.2. Genome organization
The virus, SARS‐CoV‐2 corresponds to the family of coronavirus which consists a large single‐stranded, RNA genome of positive‐sense with molecular weight of around 29.2 kbp (Figure 1). Their genomes are typically made of a 3′‐poly‐A tail at the 3′‐end and a 5′‐methylguanosine cap at the 5′‐end. These genes are highly conserved. Genome analysis has shown that the genetic sequence of SARS‐CoVs consists of open reading frames (ORFs) in variable numbers (6–11). The translation of the two important polyproteins (pp1a and pp1b), which is necessary to encode 16 non‐structural proteins (NSP), is carried out by the first ORF (ORF1a/b) and other accessory and structural proteins are encoded by the remaining ORFs. The four other essential structural proteins, comprising spike (S) glycoprotein, membrane (M) protein, matrix, nucleocapsid (N) protein and small envelope (E) protein, and also several accessory proteins interfering with the host‐innate immune response is encoded by remaining viral genome [37]. Although, SARS‐CoV‐2 genes are subjected to undergo several mutations in different provinces, there are some similarities in the genomic structure with SARS‐CoV, especially the S‐glycoprotein genome is similar to SARS‐CoV along with the receptor‐binding domain (RBD) this feature became important for human‐to‐human viral infection innate immune response [38].
3. STRUCTURAL PROTEINS OF SARS‐COV‐2
3.1. Spike protein
The viral envelope consists of three protein components among which the Spike‐glycoprotein is of utmost importance. Recognition of host cells by the spike protein is the entry point of SARS‐CoV‐2 to the host cells. The S protein is a large transmembrane protein consisting of N‐exo, C‐endotrimeric assembly forming the impression of the spikes. The S protein consists of a small C terminal end of 71 residues and another large unit of 1162–1452 residues. Each monomeric unit, before glycosylation, has a molecular weight of 128–160 KDa. The S protein can be easily glycosylated via the modification of the N terminal end. The S protein has two protein subunits S1 and S2. The subunit S1 consists of the receptor‐binding site. The SARS‐CoV‐2 S protein provides an open binding site that allows better attachment and structural modification possibilities to the virus. The S proteins contain 30–50 cysteine residues whose positions are fixed with various types of coronaviruses, although their activity is not well known till now. The SARS‐CoV‐2 cooperate with the angiotensin‐converting enzyme 2 (ACE2) receptor as they consist of 75% similarly in the surface amino acids and 50% similarity with receptor binding motifs (Figure 2).
FIGURE 2.
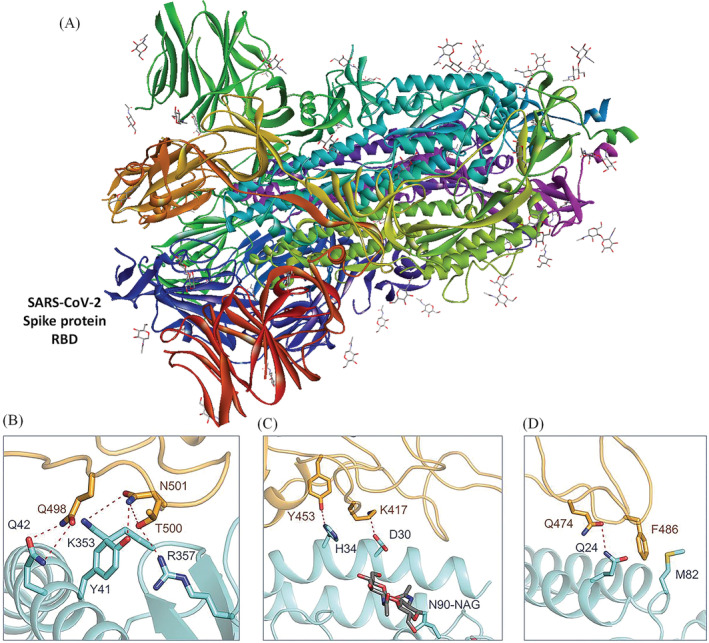
(A) The spike (S) glycoprotein protein of severe acute respiratory syndrome coronavirus 2 (SARS CoV‐2). The red colored residue indicated the receptor binding domain (PDB Id: 6VXX). (B–D) Interacting amino acids of receptor‐binding domain of SARS‐CoV‐2 and binding domain of angiotensin‐converting enzyme 2 receptor of the host cells at different angles (adapted from Yan et al. [38 ])
The previous reports suggest that there are 14 key sites for binding with SARS‐CoV binding sites with human angiotensin‐converting enzyme 2 as follows: Y491, G488, T487, T486, Y484 N479, Y475, N473, L472, Y442, Y440, Y436, R426, and T402. In SARS‐CoV‐2, eight positions are strictly conserved, and others are substituted conservatively. The modified parts are T487SARS‐CoVN501SARS‐CoV‐2, Y484SARS‐CoVQ498SARS‐CoV‐2, N479SARS‐CoVQ493SARS‐CoV‐2, L472SARS‐CoVF486SARS‐CoV‐2, Y442SARS‐CoVL455SARS‐CoV‐2, and R426SARS‐CoVN439SARS‐CoV‐2. The virus recognizes the host cells through the non‐covalent interaction between these sites, and subsequently, the envelope layer is dissolved and the viral genome is inserted in the cells [39].
3.2. Membrane protein
The most abundant protein present in the coronavirus is the membrane (M) protein. Before glycosylation, the molecular weight of M glycoprotein has been reported as 25–30 kDa (221–262 amino acid residue) in the SARS‐CoV virus. The M glycoprotein is multispanning and a small N terminal part is situated in the exterior of the virus. The other three transmembrane sequences and a large C‐terminal domain constitute the whole M protein. The M protein is involved in the development of the envelope and governs the shape of new virions. While new virions are created, the M protein binds with the S, E, N, and M proteins and creates the envelope for new virions [40].
3.3. Envelope protein
Envelope (E) protein is the most important protein in the SARS‐CoV‐2 and it is the very small protein of molecular weight 8.4–12 kDa (76–109 amino acid residue). The arrangements in the E proteins are very diverse across the different types of corona viruses. It is constructed of a short hydrophilic N‐terminus of 8–12 amino acids and a large hydrophobic region of 21–29 amino acid residues, 2–4 cysteine's and hydrophilic C‐terminal tail of 39–76 residues. The hydrophobic domain forms a hairpin‐like structure, facilitating the insertion of E proteins in the endoplasmic reticulum‐Golgi pocket. The E proteins take part in the trafficking of the new viral genome and other proteins to the endoplasmic reticulum‐Golgi pocket and bind with M protein to promote the assembly process of new viruses [41].
3.4. Nucleocapsid (N) protein
The N protein is a phosphor‐protein of a molecular weight of 43–50 kDa and the protein binds as beads‐on‐a‐string manner with the viral RNA genome. It consists of different domains in which three conserved domains are separated by two variable spacers. The domains are rich in arginine and lysine residues which favors the binding of N proteins with the viral RNA. The N and C terminal sites of N proteins are susceptible to bind with the RNA. The bound N protein‐RNA nucleocapsid is the active viral genome [41].
4. PATHOGENESIS AND HOST‐IMMUNE RESPONSES
The pathogenesis of SARS‐CoVs is almost similar to other SARS viruses where the spike protein plays a pivotal role in recognizing the ACE2 receptor of the host alveoli epithelial cells (Figure 3).
FIGURE 3.
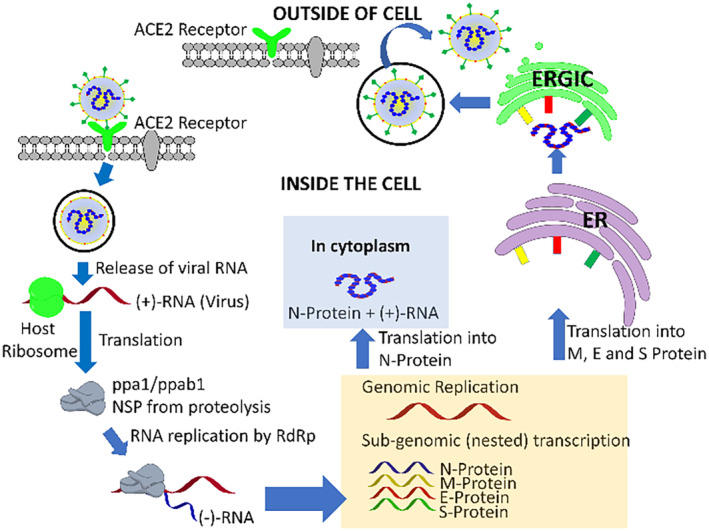
Pathogenesis of severe acute respiratory syndrome coronavirus 2 in host pneumonocyte cells [42, 43, 44, 45, 46, 47, 48, 49, 50]
After the receptor recognition and receptor mediated endocytosis, the viral genome encapsulated nucleocapsid protein released into host cellular cytoplasm media. Then occurs the translation of ORF1a and ORF1b genes for production of two polyproteins (PPs), pp1a and pp1b, which has the primary operation to modulate host ribosomes for the translation process of viral genome. Both pp1a and pp1b play a major role for the generation of the replication transcription complex. The polyproteins are activated by protease and produces 16 NSPs which carries out their specific functions including (i) NSP1 and NSP2 suppresses the host gene expression, (ii) NSP3, NSP5 forms of a multidomain complex (M protease), plays important character in replication, (iii) The role of transmembrane (TM) proteins is carried out by NSP4 and NSP6, (iv) The role of a primase is carried out by NSP7 and NSP8, (v) NSP9 acts as a RNA‐binding protein which has an important role during viral infection, (vi) The role of cofactor is played by NSP10, for the activation of the replicative enzyme, (vii) RNA‐dependent RNA polymerase activity is exhibited by NSP12, (viii) helicase activity exhibited by NSP13, exoribonuclease activity exhibited by NSP14, (ix) endoribonuclease activity pertains to NSP15, and (x) NSP16 has a crucial role in methyltransferase activity. The replication and transcription process of the viral genome is regulated by all NSPs and they perform a pivotal character in the processes. The structural proteins such as M proteins, E proteins, and S proteins are synthesized and for developing the structure of viral envelope, embedded inside the endoplasmic reticulum (ER)‐Golgi intermediate compartment (ERGIC) complex. Simultaneously the replicated (+)‐genome (RNA) gets bound to nucleocapsid (N) protein which initiates formation of the ribonucleoprotein (RNP) complex. Additionally, the new viral genome outer cover is formulated through structural S proteins, M proteins, and E proteins and eventually, the virus particle reveals from the compartment endoplasmic‐reticulum‐Golgi intermediate compartment (ERGIC) by forming a bud‐like structure through exocytosis. These mature virions come out of the cell by formation of vesicle and discharge the virus particles into the extracellular region by fusing with the plasma membrane [42, 43, 44, 45, 46, 47, 48, 49, 50].
The release of replicas of SARS‐CoV‐2 activates inflammatory mediators which stimulate macrophages and release specific cytokines (Interleukin‐1, Interleukin‐6, and TNF‐a). This causes vasodilatation and endothelial contraction leading to alveolar edema, hypoxemia and damages the lung cells causing acute distress respiratory syndrome [51].
4.1. Immuno‐ or chemo‐therapeutic option for SARS‐CoV‐2
Till now there is no efficient therapeutic option for SARS‐CoV‐2. However, symptomatic treatment strategies are recommended for clinical intervention. There are several antiviral agents that are found potentially important in the medication for SARS‐CoV‐2. Among them CQ and HCQ have shown immense potential to treat pneumonia associated with COVID‐19 in recent clinical studies [52, 53]. In China, the addition of CQ has been made to the list of trial drug molecules for the cure of COVID‐19 [54, 55]. Wang et al. recently described that two drugs, Remdesivir (GS‐5734), a nucleoside analog of the prodrug established by Gilead Sciences (USA) and CQ phosphate, is described to suppress the SARS‐CoV‐2 infection in vitro [56, 57]. A recent report exhibited that, the clinical situation of the first patient infected by SARS‐CoV‐2 in the United States has been upgraded by the treatment method involving Remdesivir [58] for which it gained to be emergency use authorization from the FDA on May 1, 2020 [59].
4.1.1. Interferon (Type‐I)
Type I Interferon's are the antiviral cytokines which involve inducing a large spectrum of proteins that can inhibit viral replication in targeted cells. Earlier studies revealed that IFN‐b was effective against SARS virus. Synergic antiviral effects of leukocytic IFN‐α with ribavirin and IFN‐β with ribavirin against SARS‐CoV has also been illustrated in vitro [60].
5. ANTIVIRAL AGENTS
The CQ (N 4‐[7‐Chloro‐4‐quinolinyl]‐N 1,N 1‐diethyl‐1,4‐pentanediamine) medication was significantly reduced due to infrequent consumption in clinical practice for treating malaria as a derivative of CQ was established to be much reduced (~40%) toxicity than CQ in animals by instigating a hydroxyl group (‐OH) onto CQ ring and it has been termed as HCQ sulfate [61]. The greater doses of HCQ (up to 600 mg/day) has been implemented for decades toward the cure of autoimmune diseases like rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) [62]. The mode of action of HCQ and CQ falls in to the category of a weak base and immunomodulator, as well as oral consumption of HCQ and CQ in human beings is very much reasonable. In animals, the two medicines share parallel tissue disposition patterns with large concentrations in the kidney, liver, lungs and spleen with a reaching level of 200–700 times superior compared to plasma [63]. It was stated that HCQ sulfate in optimal dosage (6–6.5 mg/kg per day) could produce serum levels of 1.4–1.5 μM in humans [64]. Consequently, SARS‐CoV‐2 infection can be prevented by the concentration of HCQ in the above tissues over a safe dosage. Clinical examinations revealed that the severely ill patients affected with SARS‐CoV‐2 are found to have cytokines in high concentration in the plasma, which suggests that cytokine storm was accompanying with disease intensity [65]. Apart from its antiviral action, HCQ is a benign and effective anti‐inflammatory drug which has been greatly used to treat autoimmune diseases and in reducing the significant production of pro‐inflammatory factors of cytokines. In this regard, Remdesivir, HCQ became the most effective, simplest and cheapest drugs to treat and prevent infectious global pandemic coronavirus disease 2019 (COVID‐19) caused by the SARS‐CoV‐2.
Remdesivir inhibited RNA dependent RNA polymerase (RdRp) during viral pathogenesis in host cells. Remdesivir was tested in vitro against SARS‐CoV‐2 and entered into phase 3 clinical trial [66]. Favipiravir also found to inhibit RdRp and a randomized trial has been started. Ribavirin tested in vitro against SARS‐CoV‐2, it generally inhibits the viral RNA synthesis and mRNA capping [67]. Randomized trial of Ribavirin has been initiated. Lopinavir and Ritonavir inhibit the 3C protease like 3Clpro. Both antiviral drugs have entered into the phase 3 clinical trials [68]. HIV protease inhibitors like Darunavir, Cobicistat, and ASC09F have successfully entered into the phase 3 clinical trial [69]. Arbidol (Umifenovir) which inhibit generally viral fusion in host cells, are also selected for phase 3 clinical trials [70].
5.1. The advantages of HCQ over other antimalarial drugs for the treatment of COVID‐19
Due to the swift spread of the novel SARS‐CoV‐2, the WHO acknowledged the epidemic of COVID‐19 as a pandemic on March 12, 2020 [71]. Thus, there is a crucial necessity for an efficacious medicine toward treatment of symptomatic patients as well as to reduce the extent of virus carriage in order to bound the spreading in the community. Several drugs such as CQ, nafamostat, nitazoxanide, penciclovir, ribavirin and two well‐accepted comprehensive antiviral drugs such as favipiravir (T‐705) and Remdesivir (GS5734) have been recommended as medications that could diminish viral spreading [56, 57]. Drugs, such as interferon, ribavirin, corticosteroids, and lopinavir‐ritonavir have also been employed in patients having MERS or SARS (Middle East Respiratory Syndrome) though the efficacy of some drugs remains contentious [72]. Regrettably, toward the cure of SARS‐CoV‐2 infection, until now, no drugs have approved by WHO. In this regard, among these drugs to cure the COVID‐19, replacement of old medications to utilize as antiviral treatment is a fascinating approach because of their well‐known knowledge on drug interactions, safety profile, posology, and side effects [32, 33, 34, 35]. Delightfully, numerous in‐vitro studies have revealed that SARS‐CoV‐2 transmission can be inhibited by both CQ and HCQ, through alkalinization of the intracellular phagolysosome, which was responsible for the uncoating and virion fusion and consequently the viral spread [54, 55, 56, 57, 73].
In precise, CQ is extensively used as immunomodulator with anti‐malarial activity [74, 75, 76, 77, 78]. Recently, an in vitro study revealed that, SARS‐CoV‐2 growth was found to be inhibited by CQ and these results have been reinforced by clinical trials on approximately 100 patients infected by SARS‐CoV‐2 [56, 57, 79, 80]. In addition, it is more toxic than HCQ [54, 55] and an overdose of CQ can be the reason of acute poisoning and death [81].
CQ can inhibit the viral cell signaling process by inhibiting phosphorylation. The immunomodulatory and antimalarial drug HCQ has seized greater awareness among the myriad therapeutics progressed as possible repurposing candidates in viral clearance in patients with SARS‐CoV‐2. Moreover, it was demonstrated that, HCQ has in vitro anti‐SARS‐CoV action in the former SARS outbreak [82] and this encourages that it may be a promising pharmacological agent to treat COVID‐19 infection. Very recently, several in vitro studies exposed that HCQ have better anti‐SARS‐CoV‐2 activity than CQ [54, 55]. Additionally, verification through EC50 values, which were consistently lesser for HCQ when compared to CQ, indicating the more potent antiviral activity of HCQ [73]. In another, results of some studies proposed that the viral loading in patients with COVID‐19 could be minimized by combined medication of HCQ with azithromycin (Table 1) [83, 84, 85, 86, 87]. In addition, a study on 30 patients with SARS‐CoV‐2 infection by Chen et al. [85]; when HCQ was associated as regular treatment, nasopharyngeal viral carriage on the 7th day seems to be unaltered. However, elucidating the results of this survey; providing associated antivirals, which might have assisted as confounders.
TABLE 1.
| Ref | Study type (no. of patients) | Duration | Primary outcomes | ICU patients (n/N) | Mortality (n/N) |
|---|---|---|---|---|---|
| Treatment | Control group (no. of patients) | Clinical outcomes | Adverse events (n/N) | ||
| 49 | Prospective open‐label, non‐randomized trial (n = 42) | 10 days | Viral load (nasopharyngeal swab): absence or presence of SARS‐CoV‐2 at Day 6 | 0/36 | 0/36 |
| HCQ (200 mg every 8 h) alone (n = 14) or with azithromycin (500 mg on Day 1, 250 mg on Days 2–5; n = 6) | Yes (n = 16) | NR (not reported) | NR | ||
| 49 | Prospective observational study (n = 80) | 10 days | Disease progression: need for oxygen or ICU admission | 3/80 | 1/80 |
| HCQ (200 mg every 8 h) and azithromycin (500 mg on Day 1, 250 mg on Days 2–5) | No | Viral load, hospital length of stay | 7/80 | ||
| 49 | Randomized Controlled Trial (RCT) (n = 30) | 7 days | Viral load (nasopharyngeal swab): presence of SARS‐CoV‐2 at Day 7 | 0/30 | 0/30 |
| HCQ (200 mg every 12 h) | Yes (n = 15) | NR | 4/15 | ||
| 49 | RCT (n = 62) | 5 days | Time to clinical recovery | 0/62 | 0/62 |
| HCQ (200 mg every 12 h) | Yes (n = 31) | Pulmonary recovery, adverse events | 2/31 | ||
| 49 | Prospective observational study (n = 11) | 10 days | Viral load (nasopharyngeal swab): presence of SARS‐CoV‐2 on Days 5–6 | 2/11 | 1/11 |
| HCQ (200 mg every 8 h) and azithromycin (500 mg on Day 1, 250 mg on Days 2–5) | No | NR | 1/11 |
Again, in another trial on 62 number of patients, Chen and co‐workers reported, the patients required short period of time for clinical recovery (cough, temperature) on administering HCQ for treatment than placebo [86].
The MOA of CQ and HCQ has not yet been entirely clarified. Although it was revealed from the previous studies, the coronavirus may get inhibited through a progressive step by the medication of CQ and HCQ. First, the change in pH at the cell membrane surface occurs by the intake of drugs and resulting in an inhibitory action on virus fusion to the cell membrane. Moreover, the replication of nucleic acid can be prevented by this drug. Nevertheless, glycosylation of viral proteins, virus assembly, virus release, new virus particle transport, and related processes to accomplish the antiviral action of this drug [88].
In the survey by Wang and co‐workers [56, 57], after a 48‐h incubation time with an EC50 value of 1.13 μM of CQ, appeared to have a repressive effect on SARS‐CoV‐2 infection. These findings are comparable with the study of Yao et al. [73], where EC50 value was reported to be 5.47 μM by an in vitro CQ study. In this study, the EC50 values were reduced on longer incubation times for both HCQ and CQ. This recommends that the drug's antiviral activity get influenced by incubation time. In addition, it was reported that HCQ and CQ can accumulate in cells [89]. Possibly, drug will have sufficient time to accumulate in higher intracellular concentrations on providing longer incubation time and resulting a good potency on antiviral activity [90].
Based on HCQ higher prophylactic activity as well as antiviral action along with acceptable safety profile in comparison with CQ [91], the US FDA (United States Food and Drug Administration) operated its emergency authority only the second time ever to approve the administration of a drug for an unapproved symptom [92]. HCQ has been approved to treat COVID‐19 in conditions when the clinical trials are inaccessible or infeasible [93].
Additionally, it has been reported in some of the studies that, mechanical ventilation risk in hospitalized patients with COVID‐19 does not get lowered with medication of HCQ either with or without azithromycin [87, 94, 95, 96]. Basically, azithromycin has been appeared with potent activity in vitro against Ebola and Zika viruses [97, 98, 99] and to restrict severe infections of respiratory tract when administered to viral infected patients [100].
According to a study, there were a total of 70 deaths out of 368 patients appraised with. The lowest death rate has been reported for patients who are not treated with HCQ when compared to HCQ and azithromycin associates [94].
India had revealed a stumble direction of SARS‐CoV‐2 transmission, with 43,039,023 cases diagnosed as of April 11, 2022 [95]. As no major side‐effects of HCQ were found in studies in India, the Indian Council of Medical Research under the Ministry of Health and Family Welfare, has endorsed chemoprophylaxis with HCQ as preventive treatment for COVID‐19 [96]. However, all drugs have the risks of adverse effects and there might be other risks when HCQ sulfate is used for treating COVID‐19 earlier. Moreover, risks related with these drugs include low blood sugar, particularly among people with anemia, diabetes and other blood problems, retina (layer of eye tissue) damage that can cause vision problems, serious heart rhythm problems, worsening of seizures, and other neurology (brain) problems. Also, problems can arise because of interactions between HCQ sulfate and other drugs which results in serious health issues. As mentioned above, the conflicting findings of HCQ administration were reported recently where the mortality rates are higher in comparison with the initial data available for the SARS‐CoV‐2 treatment [101].
Hence, it is advised to patients to inform their health care provider regarding the medications which are already administered for other purposes [93].
6. SYNTHESIS OF CQ AND HCQ
CQ and HCQ belong to a similar molecular family of substituted 4‐aminoquinolines. HCQ differs from CQ by the existence of a hydroxyl functionality (‐OH) at the terminal of the side chain. In particular, CQ is known as an amine acidotropic derivative of quinine which was prepared in Germany by Bayer in 1934 and appeared around 70 years ago as a potent surrogate for natural quinine [102]. Additionally, quinine is a compound originate in the bark of Cinchona trees which are found in Peru and was the earlier drug of choice to treat malaria [103]. During the past decades, CQ was the prescribed medicine for the prophylaxis and cure of malaria disease and also remain among the most recommended drug worldwide [104]. Very recently, it has been described as an antiviral drug with broad‐spectrum of potential [105, 106, 107].
HCQ is one among the oldest prescribed drugs still used in clinical practice for the treatment of malaria due to its relatively low‐cost and tolerance. HCQ sulfate is an orally administered drug which is another form of HCQ. These drugs have been acknowledged to be effectual in autoimmune diseases like RA and SLE as well as a pharmacologic agent to reduce heart rate [108, 109, 110]. In 1946, Alexander Surrey and Henry Hammer fabricated HCQ sulfate by introducing a hydroxyl group into one of the N‐ethyl groups of CQ phosphate which may dramatically decrease the latter's toxicity [111].
6.1. Synthesis of CQ
CQ (N 4‐[7‐Chloro‐4‐quinolinyl]‐N 1,N 1‐diethyl‐1,4‐pentanediamine) is synthesized by reacting 4,7‐dichloroquinoline with 4‐diethylamino1‐methylbutylamine at 180°C (Scheme 1) [112, 113].
SCHEME 1.
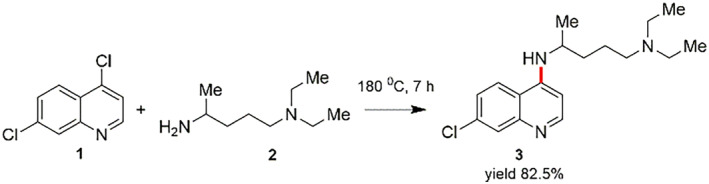
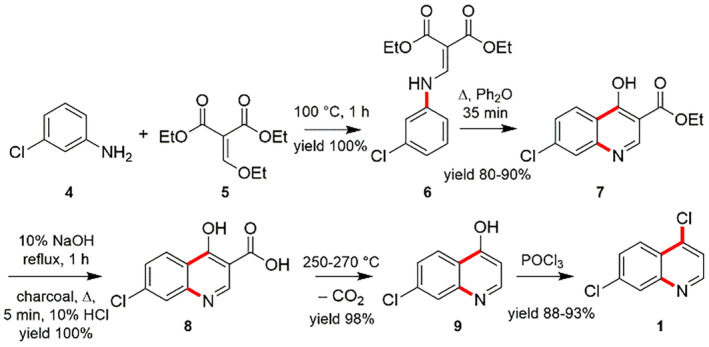
In order to appreciate the preparation of CQ, the essential 4,7‐dichloroquinoline (1) is assembled in diverse routes from 3‐chloroaniline (4). One of such protocol focused on the reaction between 3‐chloroaniline (4) and ethoxymethylenmalonic ester (5) to obtain the (3‐choroanilino)‐methylenemalonic ester (6), which afterwards experiences the hetero‐cyclization reaction at high‐temperature to afford the ethyl ester of 7‐chloro‐4‐hydroxyquinolin‐3‐carboxylic acid (7). After that, the hydrolysis of compound 7 using sodium hydroxide (NaOH) delivers the 7‐chloro‐4‐hydroxyquinoline‐3‐carboxylic acid (8), which on heating at 250–270°C gets decarboxylated to form 7‐chloro‐4‐hydroxyquinoline (9). The product 9 on treating in the presence of phosphorus oxychloride (POCl3) produces the 4,7‐dichloroquinoline (1) (Scheme 2) [114, 115].
SCHEME 2.
In another, 3‐chloroaniline (4) with diethyl oxaloacetate (10) were used for the preparation of 4,7‐dichloroquinoline (1). The reaction of 4 and 10 in participation of acetic acid and heating to 250°C resulted in the consistent enamine (11). It encounters hetero‐cyclization to realize the ethyl ester of 7‐chloro‐4‐hydroxyquinolin‐2‐carboxylic acid (7) accompanied with a slight amount of 5‐chloro‐4‐hydroxyquinolin‐2‐carboxylic acid (12), which is isolated using crystallization process from the mixture of main product in presence acetic acid. After that, alkaline hydrolysis of 7 results in 7‐chloro‐4‐hydroxyquinoline‐2‐carboxylic acid (8) and successive high temperature decarboxylation of the formed acid 8 gives 7‐chloro‐4‐hydroxyquinoline (9). Consequently, the reaction of 7‐chloro‐4‐hydroxyquinoline (9) with phosphorus oxychloride (POCl3) gives 4,7‐dichloroquinoline (1) (Scheme 3) [111].
SCHEME 3.
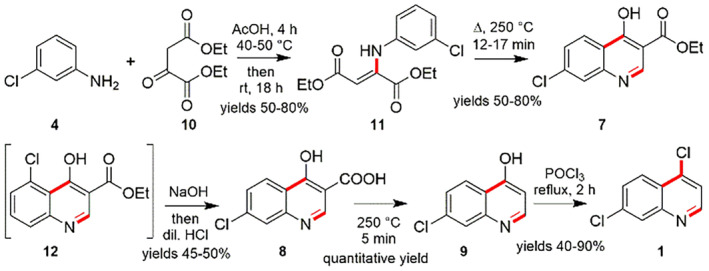
Synthesis of 4,7‐dichloroquineoline (1) using 3‐chloroaniline (4) and diethyl oxaloacetate (10) [111]
Additionally, the proposed alternatives for synthesizing 4,7‐dichloroquinoline (1) compose of the reaction between 3‐chloroaniline (4) and the ethyl ester of formylacetic acid (13) to prepare the corresponding enamine (14). Then, the enamine (14) experiences cyclization reactions upon heating to deliver the 7‐chloro‐4‐hydroxyquinoline (9) and reaction using phosphorus oxychloride dispatches the 4,7‐dichloroquinoline (1) (Scheme 4) [116].
SCHEME 4.
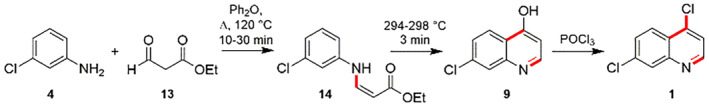
Synthesis of 4,7‐dichloroquineoline (1) using 3‐chloroaniline (4) and ethyl ester of formylacetic acid (13) [116]
The essential component for the synthesis of CQ is 4‐diethylamino‐1‐methylbutylamine (2), which was also made in various synthetic pathways. Alkylating ethyl acetoacetate (15) using 2‐diethylaminoethylchloride (16) delivers 2‐diethylaminoethylacetoacetic acid ester (17), which produces 1‐diethylamino‐4‐pentanone (18) followed by acidic hydrolysis using hydrochloric acid and rapid decarboxylation process. Reductive amination of 18 with H2 and NH3 in presence of Raney nickel gives 4‐diethylamino‐1‐methylbutylamine (2) (Scheme 5) [117].
SCHEME 5.
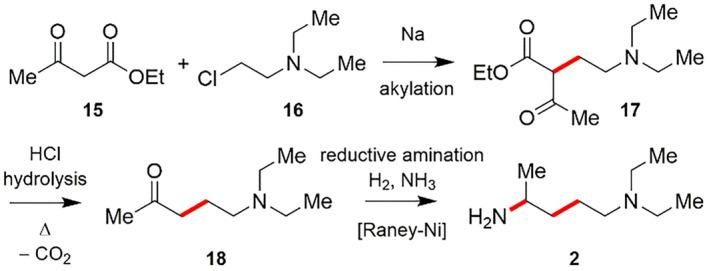
Synthesis of 4‐diethylamino‐1‐methylbutylamine (2) [117]
The compound 3‐acetylbutyrolactone (20) has been utilized as a starting component for the synthesis of 4‐diethylamino‐1‐methylbutylamine (2). The compound 3‐acetylbutyrolactone (20) is made by reacting ethyl acetoacetate (15) with ethylenoxide (19). Then, acidic hydrolysis using HBr of the ester group in 3‐acetylbutyrolactone (20) followed by subsequent decarboxylation process afforded 1‐bromo‐4‐pentanone (21). Next, reacting 21 with diethyl amine gives 1‐diethylamino‐4‐pentanone (18), followed by reductive amination with H2 and NH3 employing Raney nickel catalyst results 4‐diethyl‐1‐methylbutylamine (2) (Scheme 6) [118].
SCHEME 6.
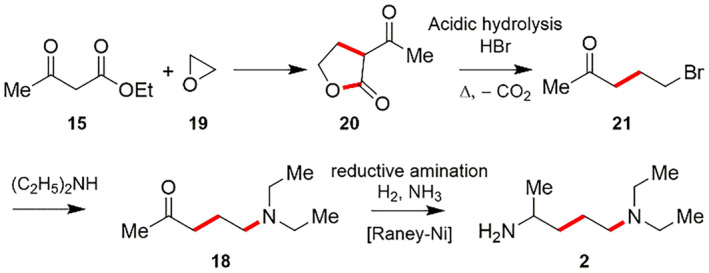
Synthesis of 4‐diethylamino‐1‐methylbutylamine (2) by using ethyl acetoacetate (15) with ethylenoxide (19) [118]
6.2. Synthesis of HCQ
HCQ (7‐chloro‐4‐[4‐[ethyl(2‐hydroxyethyl)amino]1‐methylbutylamino]quinoline) (22), is prepared by the Scheme similar to that of the construction of CQ molecule. The reaction between 1‐chloro‐4‐pentanone (23) and 2‐ethylaminoethanol (24) delivers the corresponding amino ketone 25. The reductive amination of 25 with H2 and NH3 using Raney nickel as a catalyst results in 4‐[ethyl(2‐hydroxyethyl)amino]‐1‐methylbutylamine (26). Next, the reaction of component 18 with 4,7‐dichloroquinoline (1) by the use of phenol and potassium iodide (KI) at the temperature ranging from 125 to 130°C for 18 h gives the desired HCQ (22) (Scheme 7) [119].
SCHEME 7.
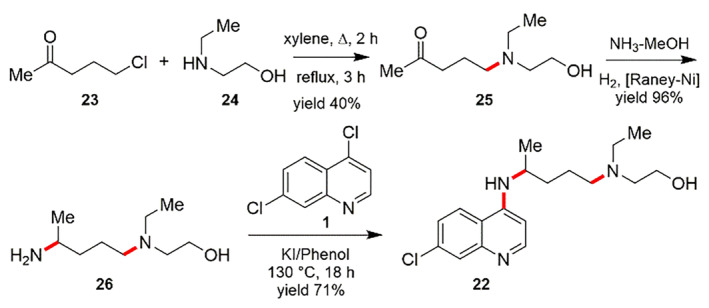
Synthesis of Hydroxychloroquine (22) [119]
The compounds 4,7‐dichloroquinoline (1) and 4‐[ethyl(2‐hydroxyethyl)amino]‐1‐methylbutylamine (26) in a molar ratio of 1:1.1 were positioned into a high‐pressure reactor. The internal pressure of the reactor is then regulated within the range of 5 to 20 bars and preferably 10 to 15 bars by nitrogen pressure. The compound 4,7‐dichloroquinoline is made to dissolve completely by stirring the reactor at 80°C for 30 min, accompanied by supplemental stirring at a temperature of 100 to 120°C for 4 to 6 h. This development allows the excellent production of HCQ with high yield by significantly decreasing the reaction temperature and time using pressure without making use of any catalyst and solvent [120].
7. SYNTHESIS OF REMDESIVIR (VEKLURY, GS‐5734)
Remdesivir has doubtlessly become a significant scaffold as most promising drug for COVID‐19 [58, 121]. The insistence of global pandemic of COVID‐19 has impelled the governments of India, the European Union, United States, Italy, Japan, etc. to accept the Remdesivir as a precise medication [122]. Recent clinical studies revealed it can efficiently abridge the retrieval period of patients and diminish the chance of death by 62% for critical hospitalized patients [123], and so it can significantly reduce the number of casualties and no of cases that benefits doctors and hospitals during the pandemic. Gilead Sciences, in 2020 conducted almost 2 million Remdesivir medication courses and they intend to do better by 2021 [124]. However, Remdesivir is prepared as a P‐stereogenic prodrug of a parent nucleoside (GS441524) by employing an amino acid ester‐ (27) and aryloxy‐substituted phosphoryl group (28) with the hydroxyl functionality of the nucleoside (30) [125, 126]. Mainly, the establishment of the phosphorus‐linked stereogeniccenter is the crucial stage for the synthesis of Remdesivir. The synthetic methods of Remdesivir (31) (first generation) are displayed in Scheme 8. By using stoichiometric amounts of N‐methyl imidazole along with GS441524 and P‐racemic phosphoryl chloride, the desired enantiopure Sp‐phosphoramidate is synthesized, followed by chiral preparative HPLC segregation of the two diastereoisomer products [125].
SCHEME 8.
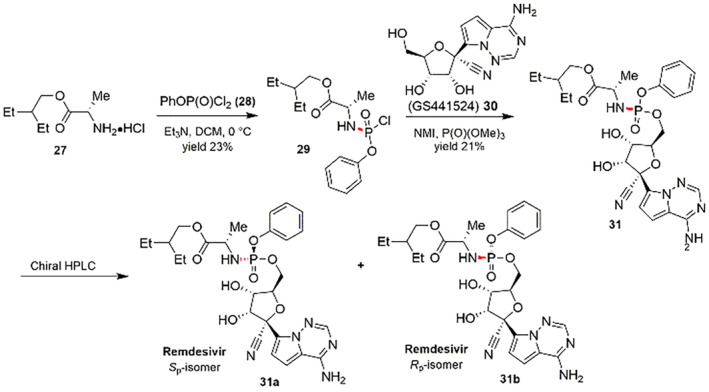
First generation synthesis of Remdesivir (31) [125]
The second generation of preparation procedure of Remdesivir is showcased in Scheme 9 which embraced another method including separation from the accompanying composed combination of two diastereoisomer intermediates followed by reaction of the enantiopure phosphorylating agent via nucleophilic substitutions [126].
SCHEME 9.
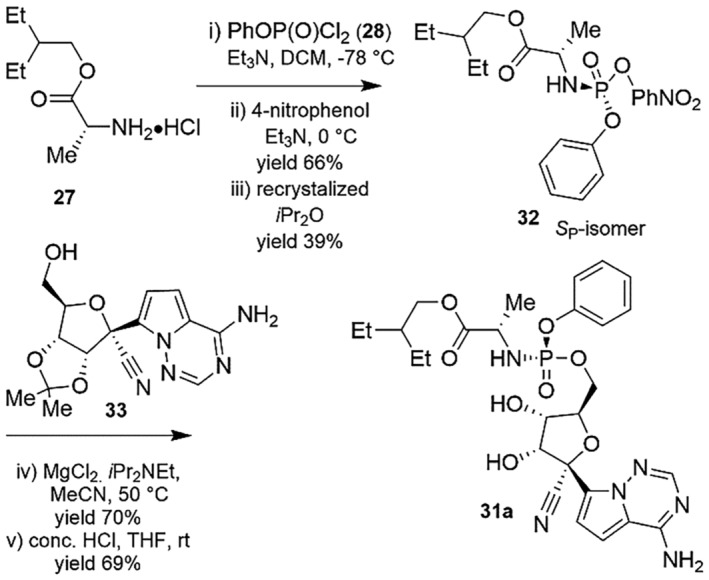
Second generation synthesis of Remdesivir (31) [126]
Recently, Wang and co‐workers showcased an asymmetric synthetic route of Remdesivir using a bicyclic imidazole‐catalyzed DyKAT, followed by reaction between protected nucleoside and P‐racemic phosphoryl chloride via coupling reaction (Scheme 10) [127].
SCHEME 10.
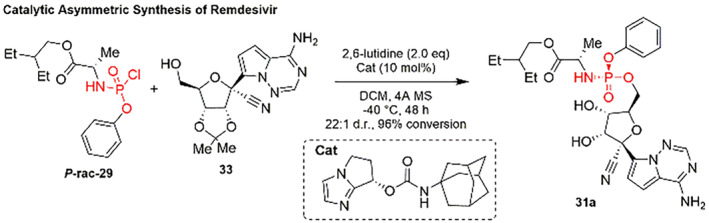
Catalytic asymmetric synthesis of Remdesivir (31) [127]
8. PERSPECTIVE OF CQ AND HCQ AS INHIBITORS OF SPIKE PROTEIN: IN SILICO APPROACH
In the last few decades, computational chemistry has emerged as a helpful aspect for the aid of understanding chemical phenomena. Molecular docking is a crucial aspect for visualization of the interaction between host proteins and foreign guest molecules, which gives us insights to understand the pharmacophore for a specific target protein. In our current approach, the specific target protein is the spike protein of the SARS‐CoV‐2 and the pharmacophores to be CQ and HCQ. As per the literature, it is evident that the spike protein is liable toward binding of the virus with the receptor ACE2 protein. The active RBD of SARS‐CoV‐2 was obtained from protein data bank (PDB ID: 6VW1) [39]. The structures of CQ and HCQ were optimized by DFT calculation using Gaussian09 software [128]. Prior to the molecular docking calculation, excess water molecules were deleted and polar hydrogen was added in the target protein (6VW1) followed by computation of Kollman charges. CQ and HCQ were docked in the RBD of spike protein of SARS‐CoV‐2 in Autodock 4 [129] docking software with a grid of 75 × 110 × 70 (x dimension × y dimension × z dimension) consisting with 598,959 total grid points, with a grid center of −35.612, 31.244, 7.128 (x, y, z coordinates). The docking calculation was done in Genetic algorithm with 10 runs, population size of 150 and 25,000,000 evaluations with rate of gene mutation 0.02 and rate of crossover 0.8. For visualization of docked pose and calculation of interactions, Discovery Studio 09 software was used. The docked pose (Figure 4) indicated us that CQ and HCQ has the potential ability to bind with the RBD of spike protein with binding constants of −4.63 and −3.85 Kcal/mol, inhibition constants (Ki) of 1.52 and 0.4 mM respectively. The docked poses were analyzed by BIOVIA Discovery Studio 9 molecular graphics program software [130]. The docked pose is stabilized by extensive hydrogen bonding between CQ/HCQ and the RBD.
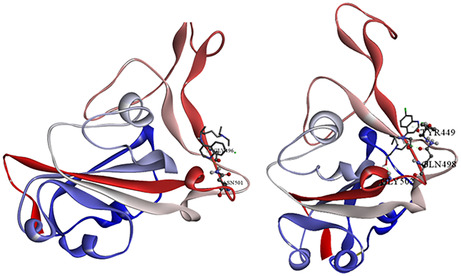
FIGURE 4.
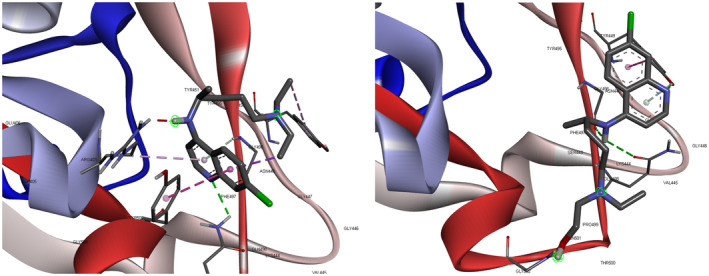
Docked pose of Chloroquine (A) and Hydroxychloroquine (B) on the receptor binding domain of the SARS‐CoV‐2 spike protein [130]
CQ is subjected to hydrogen bonding with ASN 501 of RBD and CQ hydrogen along with π‐donor hydrogen bonding between GLY 496 hydrogen and π‐orbitals of CQ. HCQ is associated with hydrogen bonding as TYS 449 (H) – HCQ (N1), GLY 502 (H) – HCQ (O), HCQ 29 (H) – GLY 496 (O), HCQ 29 (H) – GLN 498 (O), TYR 449 (H) – HCQ (O) (Table 2). The binding energy details, and the hydrogen bonding interactions are depicted in Table 3 and Figure 2. From the inhibition constant and binding energy, it is deductible that CQ is a better option than HCQ in terms of spike protein inhibition.
TABLE 2.
| Compound | Type of bond | Donor | Acceptor | Bond distance (Å) |
|---|---|---|---|---|
| Chloroquine | Hydrogen bond | ASN 501 (H) | CQ (N) | 2.61 |
| Chloroquine | Π‐donor Hydrogen bond | GLY 496 (H) | CQ (π‐orbitals) | 2.86 |
| Hydroxychloroquine | Hydrogen bond | TYR 449 (H) | HCQ (N) | 1.93 |
| Hydroxychloroquine | Hydrogen bond | GLY 502 (H) | HCQ (O) | 1.81 |
| Hydroxychloroquine | Hydrogen bond | HCQ 29 (H) | GLY 496 (O) | 2.30 |
| Hydroxychloroquine | Hydrogen bond | HCQ 29 (H) | GLN 498 (O) | 3.09 |
| Hydroxychloroquine | Π‐donor Hydrogen bond | TYR 449 (H) | HCQ (O) | 2.77 |
TABLE 3.
Binding energy of the docked poses of Chloroquine and Hydroxychloroquine on receptor binding domain of spike protein [128, 129]
| Compound | Binding energy (kcal/mol) | Intermolecular energy (kcal/mol) | Electrostatic energy (kcal/mol) | Torsional energy (kcal/mol) |
|---|---|---|---|---|
| Inhibition constant (mM) | Van der Waals + hydrogen bonding + desolvation energy (kcal/mol) | Total internal energy (kcal/mol) | Unbound energy (kcal/mol) | |
| Chloroquine | −4.63 | −7.02 | 0.1 | 2.39 |
| 0.404 | −7.11 | −1.5 | −1.5 | |
| Hydroxychloroquine | −3.85 | −6.83 | −0.05 | 2.78 |
| 1.52 | −6.78 | −0.71 | −0.71 |
9. SUMMARY
In conclusion, COVID‐19 has created a great risk to the safety and human health; hence, to administer the proliferation of the epidemic and faster control of the mortality is the prime concern. The successful option of antiviral therapy and vaccines such as Pfizer‐BioNTech, Moderna, CoronaVac, Covaxin, Sputnik V, Oxford‐AstraZeneca etc. are currently under evaluation and development. However, based on the current and previously published evidence, scientists all over the globe predict that Remdesivir as well as HCQ has prime efficacy to fight this widespread disease. In this perspective, we systematically summarize the synthesis, mechanism of action of Remdesivir, CQ, and HCQ as well as infection source, transmission route, pathogenesis, clinical characteristics, prevention, and treatment of SARS‐CoV‐2. This review is written in the hope of helping the public and deal with the novel coronavirus and providing the references for follow‐up research, therapy, and prohibition. Moreover, it may support readers to have the recent comprehension of this new infectious disease.
ACKNOWLEDGMENTS
Chandi C. Malakar appreciate Science and Engineering Research Board (SERB), New Delhi and NIT Manipur for financial support in the form of research grant (ECR/2016/000337 and CRG/2020/004509). Mithun Roy gratefully thank the Board of Research in Nuclear Science (BRNS), Mumbai (37(2)/14/18/2017‐BRNS) and the Science and Engineering Research Board, Government of India, New Delhi for financial support [ECR‐2016‐000839/CS]. Chandi C. Malakar and Mithun Roy greatly appreciate Technical Education Quality Improvement Program, Phase‐III (TEQIP‐III) for financial assistance as Minor Research Grant. Arup K. Kabi and Raghuram Gujjarappa grateful to Ministry of Education, New Delhi for fellowship support. Maynak Pal is thankful to BRNS for scholarship.
Kabi A. K., Pal M., Gujjarappa R., Malakar C. C., Roy M., J. Heterocycl. Chem. 2022, 1. 10.1002/jhet.4541
Arup K. Kabi and Maynak Pal contributed equally.
Funding information Ministry of Education, New Delhi; Board of Research in Nuclear Science (BRNS), Mumbai, Grant/Award Number: 37(2)/14/18/2017; NIT Manipur; Science and Engineering Research Board (SERB), Grant/Award Numbers: CRG/2020/004509, ECR/2016/000337; Science and Engineering Research Board (SERB), New Delhi, Grant/Award Number: ECR‐2016‐000839/CS
Contributor Information
Chandi C. Malakar, Email: chdeepm@gmail.com.
Mithun Roy, Email: mithunroy.iisc@gmail.com.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- 1. Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D. S. C., Du B., Li L., Zeng G., Yuen K.‐Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J.‐L., Liang Z., Peng Y., Wei L., Liu Y., Hu Y.‐H., Peng P., Wang J.‐M., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N., Engl N., J. Med. 2020, 382, 1708. [Google Scholar]
- 2. Lai C. C., Shih T. P., Ko W. C., Tang H. J., Hsueh P. R., Int. J. Antimicrob. Agents 2020, 55, 105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang L. S., Wang Y. R., Ye D. W., Liu Q. Q., Int. J. Antimicrob. Agents 2020, 55, 105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. El‐Aziz T. M. A., Stockand J. D., Infect. Genet. Evol. 2020, 83, 104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baildya N., Ghosh N. N., Chattopadhyay A. P., J. Mol. Struct. 2020, 1219, 128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. SanJuan‐Reyes S., Gómez‐Oliván L. M., Islas‐Flores H., Chemosphere 2020, 263, 127973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou P., Fan H., Lan T., Yang X. L., Shi W. F., Zhang W., Zhu Y., Zhang Y. W., Xie Q. M., Mani S., Zheng X. S., Li B., Li J. M., Guo H., Pei G. Q., An X. P., Chen J. W., Zhou L., Mai K. J., Wu Z. X., Li D., Anderson D. E., Zhang L. B., Li S. Y., Mi Z. Q., He T. T., Cong F., Guo P. J., Huang R., Luo Y., Liu X. L., Chen J., Huang Y., Sun Q., Zhang X. L. L., Wang Y. Y., Xing S. Z., Chen Y. S., Sun Y., Li J., Daszak P., Wang L. F., Shi Z. L., Tong Y. G., Ma J. Y., Nature 2018, 556, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drosten C., Günther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A. M., Berger A., Burguière A. M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J. C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H. D., Osterhaus A. D. M. E., Schmitz H., Doerr H. W., N. Engl. J. Med. 2003, 348, 1967. [DOI] [PubMed] [Google Scholar]
- 9. Guan Y., Zheng B. J., He Y. Q., Liu X. L., Zhuang Z. X., Cheung C. L., Luo S. W., Li P. H., Zhang L. J., Guan Y. J., Butt K. M., Wong K. L., Chan K. W., Lim W., Shortridge K. F., Yuen K. Y., Peiris J. S. M., Poon L. L. M., Science 2003, 302, 276. [DOI] [PubMed] [Google Scholar]
- 10. Lau S. K. P., Woo P. C. Y., Li K. S. M., Huang Y., Tsoi H. W., Wong B. H. L., Wong S. S. Y., Leung S. Y., Chan K. H., Yuen K. Y., Proc. Natl. Acad. Sci. USA 2005, 102, 14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J. H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B. T., Zhang S., Wang L. F., Science 2005, 310, 676. [DOI] [PubMed] [Google Scholar]
- 12. Ge X. Y., Li J. L., Yang X. L., Chmura A. A., Zhu G., Epstein J. H., Mazet J. K., Hu B., Zhang W., Peng C., Zhang Y. J., Luo C. M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S. Y., Wang L. F., Daszak P., Shi Z. L., Nature 2013, 503, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y., Guo H., Zhang H., Tu C., J. Virol. 2014, 88, 7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang X. L., Hu B., Wang B., Wang M. N., Zhang Q., Zhang W., Wu L. J., Ge X. Y., Zhang Y. Z., Daszak P., Wang L. F., Shi Z. L., J. Virol. 2016, 90, 3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu Z., Yang L., Ren X., Zhang J., Yang F., Zhang S., Jin Q., J. Infect. Dis 2016, 213, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang L., Fu S., Cao Y., Zhang H., Feng Y., Yang W., Nie K., Ma X., Liang G., Emerg. Microbes. Infect 2017, 6, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu B., Zeng L. P., Yang X. L., Ge X. Y., Zhang W., Li B., Xie J. Z., Shen X. R., Zhang Y. Z., Wang N., Luo D. S., Zheng X. S., Wang M. N., Daszak P., Wang L. F., Cui J., Shi Z. L., PLoS Pathog. 2017, 13, e1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan Y., Zhao K., Shi Z. L., Zhou P., Viruses 2019, 11, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao F., Tan W. A., N. Engl. J. Med. 2020, 382, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., Nat. Med. 2020, 26, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T., Emerg. Microbes. Infect. 2020, 9, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Zhou Y., Zhang V., Wang H., Zhao Q., Liu J., Antimicrob. Agents. Chemother. 2020, 64, e00483‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El‐hoshoudy A. N., J. Mol. Liq. 2020, 318, 113968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abo‐zeid Y., Ismail N. S. M., McLean G. R., Hamdy N. M., Eur. J. Pharm. Sci. 2020, 153, 105465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohanty S., Rashid M. H. A. I., Mridul M., Mohanty C., Swayamsiddha S., Metab. Syndr. Clin. Res. Rev. 2020, 14, 1027. [Google Scholar]
- 27. Rahmani H., Davoudi‐Monfared E., Nourian A., Khalili H., Hajizadeh N., Jalalabadi N. Z., Fazeli M. R., Ghazaeian M., Yekaninejad M. S., Int. Immunopharmacol. 2020, 88, 106903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Antunes A. E. C., Vinderola G., Xavier‐Santos D., Sivieri K., Food. Res. Int. 2020, 136, 109577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ismail S., Ahmad S., Azam S. S., J. Mol. Liq. 2020, 314, 113612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar M., Taki K., Gahlot R., Sharma A., Dhangar K., Sci. Total Environ. 2020, 734, 139278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. World Health Organization , https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed: April 14 2022).
- 32. Mullard A., Rev. Drug Discov. 2012, 11, 505. [DOI] [PubMed] [Google Scholar]
- 33. Colson P., Rolain J. M., Raoult D., Int. J. Antimicrob. Agents 2020, 55, 105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colson P., Rolain J. M., Lagier J. C., Brouqui P., Raoult D., Int. J. Antimicrob. Agents 2020, 55, 105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim A. H. J., Sparks J. A., Liew J. W., Putman M. S., Berenbaum F., Duarte‐García A., Graef E. R., Korsten P., Sattui S. E., Sirotich E., Ugarte‐Gil M. F., Webb K., Grainger R. A., Ann. Intern. Med. 2020, 172, 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.a) Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.‐D., Rappuoli R., Nat. Rev. Microbiol. 2003, 1, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]; b)Kim J. M., Chung Y. S., Jo H. J., Lee N. J., Kim M. S., Woo S. H., Park S., Kim J. W., Kim H. M., Han M. G., Osong. Public Health Res. Perspect. 2020, 11, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mousavizadeh L., Ghasemi S., J. Microbiol. Immunol. Infect. 2021, 54, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q., Science 2020, 367, 1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F., Nature 2020, 581, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wan Y., Shang J., Graham R., Baric R. S., Li F., J. Virol. 2020, 94, e00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siu Y. L., Teoh K. T., Lo J., Chan C. M., Kien F., Escriou N., Tsao S. W., Nicholls J. M., Altmeyer R., Peiris J. S. M., Bruzzone R., Nal B., J. Virol. 2008, 82, 11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alanagreh L., Alzoughool F., Atoum M., Pathogens 2020, 9, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stobart C. C., Sexton N. R., Munjal H., Lu X., Molland K. L., Tomar S., Mesecar A. D., Denison M. R., J. Virol. 2013, 87, 12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Qin C., Viruses 2019, 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu T., Chen C., Li H., Dou Y., Zhou M., Lu D., Zong Q., Li Y., Yang C., Zhong Z., Singh N., Hu H., Zhang R., Yang H., Su D., Protein Sci. 2017, 26, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peiris J. S. M., Guan Y., Yuen K. Y., Nat. Med. 2004, 10, S88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Narayanan K., Maeda A., Maeda J., Makino S., J. Virol. 2000, 74, 8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Wit E., Van Doremalen N., Falzarano D., Munster V. J., Nat. Rev. Microbiol. 2016, 14, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nieto‐Torres J. L., DeDiego M. L., Álvarez E., Jiménez‐Guardeño J. M., Regla‐Nava J. A., Llorente M., Kremer L., Shuo S., Enjuanes L., Virology 2011, 415, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Du L., He Y., Zhou Y., Liu S., Zheng B. J., Jiang S., Nat. Rev. Microbiol. 2009, 7, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singhal T. A., Indian J. Pediatr. 2020, 87, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gao J., Tian Z., Yang X., BioScience Trends 2020, 14, 72. [DOI] [PubMed] [Google Scholar]
- 53. Fantini J., Scala C. D., Chahinian H., Yahi N., Int. J. Antimicrob. Agents 2020, 55, 105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M., Cell Discov. 2020, 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marmor M. F., Kellner U., Lai T. Y. Y., Melles R. B., Mieler W. F., Ophthalmology 2016, 123, 1386. [DOI] [PubMed] [Google Scholar]
- 56. Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G., Cell Res. 2020, 30, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dong L., Hu S., Gao J., Drug Discov. Ther. 2020, 14, 58. [DOI] [PubMed] [Google Scholar]
- 58. Holshue M. L., DeBolt C., Lindquist S., Lofy K. H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S. I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M. A., Weldon W. C., Biggs H. M., Uyeki T. M., Pillai S. K., N. Engl. J. Med. 2020, 382, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Food and Drug Administration . 2020, https://www.fda.gov/media/137564/download (accessed: September 08 2020).
- 60. Morgenstern B., Michaelis M., Baer P. C., Doerr H. W., J. Cinatl. Biochem. Biophys. Res. Commun. 2005, 326, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McChesney E. W., Am. J. Med. 1983, 75, 11.6408923 [Google Scholar]
- 62. Lee S. J., Silverman E., Bargman J. M., Nat. Rev. Nephrol. 2011, 7, 718. [DOI] [PubMed] [Google Scholar]
- 63. Popert A. J., Rheumatology Rehabil 1976, 15, 235. [DOI] [PubMed] [Google Scholar]
- 64. Laaksonen A. L., Koskiahde V., Juva K., Scand. J. Rheumatol. 1974, 3, 103. [DOI] [PubMed] [Google Scholar]
- 65. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Lancet 2020, 395, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F. G., Horby P. W., Cao B., Wang C., Lancet 2020, 395, 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shetty R., Ghosh A., Honavar S. G., Khamar P., Sethu S., Indian J. Ophthalmol. 2020, 68, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu X., Wang X. J., J. Genet. Genomics. 2020, 47, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y. S., Singh K. P., Chaicumpa W., Hum. Vaccin. Immunother. 2020, 16, 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sanders J. M., Monogue M. L., Jodlowski T. Z., Cutrell J. B., J. Am. Med. Assoc. 2020, 323, 1824. [DOI] [PubMed] [Google Scholar]
- 71. WHO Director‐General's opening remarks at the media briefing on COVID‐19 . 2020, https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed: September 08 2020).
- 72. Zumla A., Chan J. F. W., Azhar E. I., Hui D. S. C., Yuen K. Y., Nat. Rev. Drug Discov. 2016, 5, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D., Clin. Infect. Dis. 2020, 71, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Romanelli F., Smith K. M., Hoven A. D., Curr. Pharm. Des. 2004, 10, 2643. [DOI] [PubMed] [Google Scholar]
- 75. Keyaerts E., Vijgen L., Maes P., Neyts J., Ranst M. V., Biochem. Biophys. Res. Commun. 2004, 323, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vincent M. J., Bergeron E., Benjannet S., Erickson B. R., Rollin P. E., Ksiazek T. G., Seidah N. G., Nichol S. T., Virol. J. 2005, 2, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ooi E. E., Chew J. S. W., Loh J. P., Chua R. C. S., Virol. J. 2006, 3, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li C., Zhu X., Ji X., Quanquin N., Deng Y. Q., Tian M., Aliyari R., Zuo X., Yuan L., Afridi S. K., Li X. F., Jung J. U., Nielsen‐Saines K., Qin F. X. F., Qin C. F., Xu Z., Cheng G., EBioMedicine 2017, 24, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huang J.. Efficacy of Chloroquine and Lopinavir/ Ritonavir in mild/general novel coronavirus (CoVID‐19) infections: a prospective, open‐label, multicenter randomized controlled clinical study. 2020, http://www.chictr.org.cn/showproj.aspx?proj=49263 (accessed: September 08 2020).
- 80. Li Y.. Ministry of Science and Technology of China: chloroquine phosphate is effective in the treatment of novel coronavirus pneumonia. 2020, http://news.ynet.com/2020/02/17/2388070t70.html (accessed: September 08 2020).
- 81. Weniger H., Bull. World Health. 1979, 79, 906. [Google Scholar]
- 82. Biot C., Daher W., Chavain N., Fandeur T., Khalife J., Dive D., De Clercq E., J. Med. Chem. 2006, 49, 2845. [DOI] [PubMed] [Google Scholar]
- 83. Gautret P., Lagier J. C., Parola P., Hoang V. T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V. E., Dupont H. T., Honoré S., Colson P., Chabrière E., Scola B. L., Rolain J. M., Brouqui P., Raoult D., Int. J. Antimicrob. Agents 2020, 56, 105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84. Gautret P., Lagier J. C., Parola P., Hoang V. T., Meddeb L., Sevestre J., Mailhe M., Doudier B., Aubry C., Amrane S., Seng P., Hocquart M., Eldin C., Finance J., Vieira V. E., Dupont H. T., Honoré S., Stein A., Million M., Colson P., Scola B. L., Veit V., Jacquier A., Deharo J. C., Drancourt M., Fournier P. E., Rolain J. M., Brouqui P., Raoult D., Travel Med. Infect. Dis. 2020, 34, 101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jun C., Danping L., Li L., Ping L., Qingnian X., Lu X., Yun L., Dan H., Shuli S., Dandan Z., Zhiping Q., Tao L., Yinzhong S., Hongzhou L., J. Zhejiang Univ. (Med. Sci.). 2020, 49, 215. [Google Scholar]
- 86. Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., Zhang Z., medRxiv 2020, 1, 1. 10.1101/2020.03.22.20040758 [DOI] [Google Scholar]
- 87. Molina J. M., Delaugerre C., Goff J. L., Mela‐Lima B., Ponscarme D., Goldwirt L., de Castro N., Med. Mal. Infect. 2020, 50, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fox R. I., Semin. Arthritis Rheum. 1993, 23, 82. [DOI] [PubMed] [Google Scholar]
- 89. Quintart J., Leroy‐Houyet M. A., Trouet A., Baudhuin P., J. Cell Biol. 1979, 82, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ying C., De Clercq E., Neyts J., J. Viral Hepat. 2000, 7, 79. [DOI] [PubMed] [Google Scholar]
- 91. Furst D. E., Lupus 1996, 5, 11.8646219 [Google Scholar]
- 92. Rome B. N., Avorn J., N. Engl. J. Med. 2020, 382, 2282. [DOI] [PubMed] [Google Scholar]
- 93. Fact Sheet For Health Care Providers Emergency Use Authorization (EUA) of Hydroxychloroquine Sulfate Supplied From The Strategic National Stockpile For Treatment of Covid‐19 In Certain Hospitalized Patients . 2020, https://www.fda.gov/media/136537/download (accessed: September 08 2020).
- 94. Magagnoli J., Narendran S., Pereira F., Cummings T., Hardin J. W., Sutton S. S., J. Ambati. Adv. Clin. Exp. Med. 2020, 1, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Our World in Data; Total and daily confirmed COVID‐19 cases, India . 2020, https://ourworldindata.org/grapher/covid-daily-vs-total-cases?tab=table&time=2025-08-02&country=~IND (accessed: April 11 2022).
- 96. National Taskforce for COVID‐19. Revised advisory on the use of Hydroxychloroquine (HCQ) as prophylaxis for SARS‐CoV‐2 infection . 2020, https://www.icmr.gov.in/pdf/covid/techdoc/V5_Revised_advisory_on_the_use_of_HCQ_SARS_CoV2_infection.pdf (accessed: September 08 2020).
- 97. Retallack H., Lullo E. D., Arias C., Knopp K. A., Laurie M. T., Sandoval‐Espinosa C., Leon W. R. M., Krencik R., Ullian E. M., Spatazz J., Pollen A. A., Mandel‐Brehm C., Nowakowski T. J., Kriegstein A. R., DeRisi J. L., Proc. Natl. Acad. Sci. USA. 2016, 113, 4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Madrid P. B., Panchal R. G., Warren T. K., Shurtleff A. C., Endsley A. N., Green C. E., Kolokoltsov A., Davey R., Manger I. D., Gilfillan L., Bavari S., Tanga M. J., ACS Infect. Dis. 2015, 1, 317. [DOI] [PubMed] [Google Scholar]
- 99. Bosseboeuf E., Aubry M., Nhan T., de Pina J. J., Rolain J. M., Raoult D., Musso D., J. Antivir. Antiretrovir. 2018, 10, 6. [Google Scholar]
- 100. Bacharier L. B., Guilbert T. W., Mauger D. T., Boehmer S., Beigelman A., Fitzpatrick A. M., Jackson D. J., Baxi S. N., Benson M., Burnham C. A. D., Cabana M., Castro M., Chmiel J. F., Covar R., Daines M., Gaffin J. M., Gentile D. A., Holguin F., Israel E., Kelly H. W., Lazarus S. C., LemanskeJr R. F., Ly N., Meade K., Morgan W., Moy J., Olin T., Peters S. P., Phipatanakul W., Pongracic J. A., Raissy H. H., Ross K., Sheehan W. J., Sorkness C., Szefler S. J., Teague W. G., Thyne S., Martinez F. D., J. Am. Med. Assoc. 2015, 314, 2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.a) Axfors C., Schmitt A. M., Janiaud P., Van't Hooft J., Abd‐Elsalam S., Abdo E. F., Abella B. S., Akram J., Amaravadi R. K., Angus D. C., Arabi Y. M., Azhar S., Baden L. R., Baker A. W., Belkhir L., Benfield T., Berrevoets M. A. H., Chen C. P., Chen T. C., Cheng S. H., Cheng C. Y., Chung W. S., Cohen Y. Z., Cowan L. N., Dalgard O., de Almeida E Val F. F., de Lacerda M. V. G., de Melo G. C., Derde L., Dubee V., Elfakir A., Gordon A. C., Hernandez‐Cardenas C. M., Hills T., Hoepelman A. I. M., Huang Y. W., Igau B., Jin R., Jurado‐Camacho F., Khan K. S., Kremsner P. G., Kreuels B., Kuo C. Y., Le T., Lin Y. C., Lin W. P., Lin T. H., Lyngbakken M. N., McArthur C., McVerry B. J., Meza‐Meneses P., Monteiro W. M., Morpeth S. C., Mourad A., Mulligan M. J., Murthy S., Naggie S., Narayanasamy S., Nichol A., Novack L. A., O'Brien S. M., Okeke N. L., Perez L., Perez‐Padilla R., Perrin L., Remigio‐Luna A., Rivera‐Martinez N. E., Rockhold F. W., Rodriguez‐Llamazares S., Rolfe R., Rosa R., Røsjø H., Sampaio V. S., Seto T. B., Shahzad M., Soliman S., Stout J. E., Thirion‐Romero I., Troxel A. B., Tseng T. Y., Turner N. A., Ulrich R. J., Walsh S. R., Webb S. A., Weehuizen J. M., Velinova M., Wong H. L., Wrenn R., Zampieri F. G., Zhong W., Moher D., Goodman S. N., Ioannidis J. P. A., Hemkens L. G., Nature Commun. 2021, 12, 2349. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bignardi P. R., Vengrusa C. S., Aquinoa B. M., Neto A. C., Pathog Global Health 2021, 115, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen Y., Li M.‐X., Lu G.‐D., Shen H.‐M., Zhou J., Int. J. Biol. Sci. 2021, 17, 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.a)Winzeler E. A., Nature 2008, 455, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Parhizgar A. R., Tahghighi A., Iran J. Med. Sci. 2017, 42, 115. [PMC free article] [PubMed] [Google Scholar]
- 103. Devaux C. A., Rolain J. M., Colson P., Raoult D., Int. J. Antimicrob. Agents. 2020, 55, 105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. White N. J., Pukrittayakamee S., Hien T. T., Faiz M. A., Mokuolu O. A., Dondorp A. M., Malaria. Lancet 2014, 383, 723. [DOI] [PubMed] [Google Scholar]
- 105. Savarino A., Trani L. D., Donatelli I., Cauda R., Cassone A., Lancet Infect. Dis. 2006, 6, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S., J. Crit. Care. 2020, 57, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Akpovwa H., Cell Biochem. Funct. 2016, 34, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hage M. P., Al‐Badri M. R., Azar S. T., Ther. Adv. Endocrinol. Metab. 2014, 5, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Poorvashree J., Suneela D., Drug Delivery Transl. Res. 2017, 7, 709. [DOI] [PubMed] [Google Scholar]
- 110. Capel R. A., Herring N., Kalla M., Yavari A., Mirams G. R., Douglas G., Bub G., Channon K., Paterson D. J., Terrar D. A., Burton R. A. B., Heart Rhythm 2015, 12, 2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Surrey A. R., Hammer H. F., J. Am. Chem. Soc. 1946, 68, 113. [DOI] [PubMed] [Google Scholar]
- 112. Andersag H., Breitner S., Jung H.. US2233970. 1941.
- 113. Drake N. L., Creech H. J., Garman J. A., Haywood S. T., Peck R. M., Van Hook J. O., Walton E., J. Am. Chem. Soc. 1946, 68, 1214.20990953 [Google Scholar]
- 114. Price C. C., Roberts R. M., J. Am. Chem. Soc. 1946, 68, 1204. [DOI] [PubMed] [Google Scholar]
- 115. Gerschickter C. F., Rice L.. US2947664. 1960.
- 116. Northey E. H., Dreisbach P. F.. US2478125. 1949.
- 117. Schuelemann W., Schoenhoefer F., Wingler A.. DE486079. 1924.
- 118. Elderfield R. C., Gensler W. J., Brody F., Head J. D., Dickerman S. C., Wiederhold L., Cramer C. B., Hageman H. A., Kreyse F. J., Griffing J. M., Kupchan S. M., Newman B., Maynard J. T., J. Am. Chem. Soc. 1946, 68, 1579.20994986 [Google Scholar]
- 119.a) Surrey A. R., Hammer H. F., J. Am. Chem. Soc 1950, 72, 1814. [Google Scholar]; b) Surrey A. R.. US2546658. 1951.
- 120. Min Y. S., Cho H. S., Mo K. W.. WO2010027150. 2010.
- 121. An Open Letter from Daniel O'Day, Chairman & CEO, Gilead Sciences, Daniel O'Day. 2020.
- 122. US Food and Drug Administration . Remdesivir EUA letter of authorization. 2020, https://www.fda.gov/media/137564/ (accessed: December 30 2021).
- 123. An Open Letter from Daniel O'Day, Chairman & CEO, Gilead Sciences. 2020, https://www.gilead.com/stories/articles/an-openletter-from-daniel-oday-june-29
- 124. Gilead Sciences Update on Veklury® (Remdesivir) Manufacturing Network. 2020, https://www.gilead.com/news-andpress/company-statements/gilead-sciences-update-on-vekluryremdesivir-manufacturing-network
- 125. Siegel D., Hui H. C., Doerffler E., Clarke M. O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K. M., Barauskas O., Feng J. Y., Xu Y., Lee G., Rheingold A. L., Ray A. S., Bannister R., Strickley R., Swaminathan S., Lee W. A., Bavari S., Cihlar T., Lo M. K., Warren T. K., Mackman R. L., J. Med. Chem. 2017, 60, 1648. [DOI] [PubMed] [Google Scholar]
- 126. Warren T. K., Jordan R., Lo M. K., Ray A. S., Mackman R. L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H. C., Larson N., Strickley R., Wells J., Stuthman K. S., Van Tongeren S. A., Garza N. L., Donnelly G., Shurtleff A. C., Retterer C. J., Gharaibeh D., Zamani R., Kenny T., Eaton B. P., Grimes E., Welch L. S., Gomba L., Wilhelmsen C. L., Nichols D. K., Nuss J. E., Nagle E. R., Kugelman J. R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M. O., Spiropoulou C. F., Lee W. A., Nichol S. T., Cihlar T., Bavari S., Nature 2016, 531, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang M., Zhang L., Huo X., Zhang Z., Yuan Q., Li P., Chen J., Zou Y., Wu Z., Zhang W., Angew. Chem., Int. Ed. 2020, 59, 20814. [DOI] [PubMed] [Google Scholar]
- 128. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Petersson G. A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B. G., Gomperts R., Mennucci B., Hratchian H. P., Ortiz J. V., Izmaylov A. F., Sonnenberg J. L., Williams‐Young D., Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V. G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Keith T., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Millam J. M., Klene M., Adamo C., Cammi R., Ochterski J. W., Martin R. L., Morokuma K., Farkas O., Foresman J. B., Fox D. J., Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT: 2016. https://gaussian.com/g09citation/ (accessed: September 08 2020). [Google Scholar]
- 129. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J., J. Comput. Chem. 2009, 30, 2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. BIOVIA, DassaultSystèmes, San Diego . DassaultSystèmes, 2015, https://www.3ds.com/products-services/biovia/resource-center/citations-and-references/ (accessed: September 08 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


