Abstract
A novel coronavirus causing the severe and fatal respiratory syndrome was identified in China, is now producing outbreaks in more than 200 countries around the world, and became pandemic by the time. In this article, a modified version of the well‐known mathematical epidemic model susceptible‐infected‐recovered (SIR) is used to analyze the epidemic's course of COVID‐19 in eight different countries of the South Asian Association for Regional Cooperation (SAARC). To achieve this goal, the parameters of the SIR model are identified by using publicly available data for the corresponding countries: Afghanistan, Bangladesh, Bhutan, India, the Maldives, Nepal, Pakistan, and Sri Lanka. Based on the prediction model, we estimated the epidemic trend of COVID‐19 outbreak in SAARC countries for 20, 90, and 180 days, respectively. A short‐mid‐long term prediction model has been designed to understand the early dynamics of the COVID‐19 epidemic in the southeast Asian region. The maximum and minimum basic reproduction numbers (R 0 = 1.33 and 1.07) for SAARC countries are predicted to be in Pakistan and Bhutan. We equate simulation results with real data in the SAARC countries on the COVID‐19 outbreak, and predicted different scenarios using the modified SIR prediction model. Our results should provide policymakers with a method for evaluating the impacts of possible interventions, including lockdown and social distancing, as well as testing and contact tracking.
Keywords: COVID‐19, epidemics, modified susceptible‐infected‐recovered, prediction model, SAARC, SARS‐CoV‐2
An short‐mid‐long term prediction model has been designed to understand the early dynamics of the COVID‐19 epidemic in the southeast Asian region.
Based on the proposed prediction model we estimated the epidemic trend of COVID‐19 outbreak in SAARC countries for 20 days (short term), 90 days (mid‐term), and 180 days (long term), respectively.
Modified susceptible‐infected‐recovered model is used to calculate the transmission rate and recovery rate of each country in the SAARC regions.
1. INTRODUCTION
A novel coronavirus (SARS‐CoV‐2), called COVID‐19, caused an outbreak in the city of Wuhan, Hubei Province, China that is linked to the Huanan Seafood Wholesale Market 1 , 2 in late December 2019. Till now, more than 200 countries around the world have been infected by a novel coronavirus. As of May 30, 2020, according to the World Health Organization (WHO), globally 5,817,385 confirmed cases have reported with a death count of 362,705. 3 Among them, the south‐east Asia region has confirmed 4.2% of cases while the case fatality ratio is 2.86%. The very first COVID‐19 patient was treated for being coronavirus positive in Wuhan, China, at the beginning of December 2019. In the south‐east Asia region, the very first coronavirus infected patient was detected in Pakistan on February 26, 2020. 4 On January 30, 2020, WHO officially declared this outbreak of COVID‐19 as global pandemic. 5 To mitigate the spread of COVID‐19, the affected countries of the world have taken various measures, including citywide lockdown, social distancing, traffic halt, community management, and information on health education knowledge. More importantly, the outbreak of COVID‐19 possessed a massive threat to global health and economics all over the world. One of the significant feature of novel coronavirus unlike other infectious diseases like severe acute respiratory syndrome (SARS), and Middle East respiratory syndrome (MERS), it causes asymptomatic infections (symptoms are very mild). 6 , 7 , 8 Responses to COVID‐19 in South Asian Association for Regional Cooperation (SAARC) nations in 2020 were reported and examined with available data in a recent study by Malik et al. 9 For the months of April 2020 to December 2020, the authors utilized the exponential growth approach to calculate the reproductive number R t. Different epidemiological model parameters were also used to establish a correlation with the COVID‐19 transmission pattern. According to this study, immunization and accurate advice on COVID‐19 prevention measures may assist to reduce the risk of infection. Similar work has been proposed by Malek and Hoque 10 in the domain of COVID‐19 epidemic trend analysis. The COVID‐19 trend in South Asian countries was explored by the authors using the SEIATR model, an upgraded version of the SEIR system. However, their recommended results only anticipated the COVID cases until June 2020 and only for four nations including India, Pakistan, Bangladesh, and Afghanistan. In another study, Tiwari et al. 11 provided India‐centric mathematical modeling that focused on the repercussions of lockdown in another prediction of COVID‐19 disease spread. By studying nationwide lockdown in India, the authors used a SEIRD model with five compartments. Overall, the article argued that a global countrywide lockdown is an effective control policy for COVID‐19 spread. For analyzing the COVID‐19 spread across India, the authors in Reference 12 have presented a regression analysis model using the dataset available at Kaggle. However, the developed models is only capable of forecasting the next 6 days of COVID‐19 spread data and among the six utilized model, sixth‐degree polynomial regression outperformed others utilized models. In another study, the authors in Reference 13 used ML‐guided piecewise linear regression to predict COVID‐19 instances across India. The trend of COVID‐19 instances climbed linearly, then exponentially, according to the findings. However, only the six separate states of India were considered for the next 50 days in the projection of cases. In order to estimate the spread of COVID‐19 cases in India, the authors in Reference 14 suggested a deep LSTM architecture and attained an accuracy of 97.59% utilizing WHO data for India. Moreover, an extensive review of the available worldwide datasets for COVID‐19 has been reviewed thoroughly. The authors in Reference 15 provide a time‐dependent based susceptible‐infected‐recovered (SIR) model that is capable of detecting transmission and recovery rates using data provided by China. This research also proposed two distinct ways to social separation in order to minimize the effective reproduction number. This study only emphasized eight different world counters in five separate projection categories, including the United States, the United Kingdom, France, Iran, Spain, Italy, Germany, and the Republic of Korea. In these circumstances, the rate of transmission among a large number of people can increase within no time. According to the latest World Health Organization survey, only 87.9% of COVID‐19 patients have a fever, and 67.7% have dry cough. 16 Therefore, this is highly crucial to estimate the intensity of the COVID‐19 epidemic and predict the time course, peak time, total duration, and so on. In recent times, the authors in Reference 17 have investigated and projected COVID‐19 instances using three distinct models (SIR, SEIQR, and ML), concentrating on three different nations (Australia, the United Kingdom, and the United States). The research found that the Prophet Algorithm fared better in the UK and the United States than in Australia among the ML models. However, to the best of our knowledge, no previous research has been conducted to predict COVID‐19 instances focused on countries in the South Asian region. Therefore, our study focuses on the COVID‐19 case prediction using a modified SIR (M‐SIR) model in the countries of the SAARC. SAARC is considered as an intergovernmental organization and geopolitical union of states in South Asia. Its member states are Afghanistan, Bangladesh, Bhutan, India, the Maldives, Nepal, Pakistan, and Sri Lanka. Our aim is to develop a prediction model for the SAARC countries to understand the epidemiological trend of novel coronavirus outbreak in these countries. Here we explore a modified version based on the SIR epidemic model to predict the short term (20 days), mid‐term (90 days), and long term (180 days) evaluation of COVID‐19 situation in these countries of SAARC regions.
2. METHODOLOGY
The research methodology is divided into several phases. First, appropriate data were collected from four different data sources including Johns Hopkins University (JHU), COVID‐19 Dataset available at Kaggle, COVID‐19 Open Research Dataset, and population by country 2020 dataset. The data were then evaluated and preprocessed to eliminate any duplicate or missing values. This section also includes the working principle of M‐SIR model, and then discussed the proposed algorithm of this research.
2.1. Dataset analysis
For this exploration, we have used datasets from various sources for our analysis and building the model. We have used four different sources of the dataset including, COVID‐19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (January–May 2020), 18 COVID‐19 Dataset (January–May 2020), 19 COVID‐19 Open Research Dataset (CORD‐19), 20 and population by country 2020 dataset. 21 Table 1 provides insight on each dataset and their respective data files with their column description.
TABLE 1.
Tabular representation of different data sources of COVID‐19
| Dataset | Description | Columns |
|---|---|---|
| Novel Coronavirus 2019 Dataset | ||
| COVID19_line_list_data.csv | This file is an aggregated version of the Novel Coronavirus dataset collected by Johns Hopkins University | Id, case_in_country, reporting date, summary, location, country, gender, age, symptom_onset, If_onset_approximate, hosp_visit_date, exposure_start, exposure_end, visiting Wuhan, from Wuhan, death, recovered, symptom, source |
| covid_19_data.csv | Daily level information on the number of COVID‐19 affected cases across the globe | Sno, observation date, province/state, country/region, last update, confirmed, deaths, recovered |
| time_series_covid_19_confirmed.csv | Time series data on the number of confirmed cases | Province/state, country/region, lat, long, 1/22/20…0.5/6/20 |
| time_series_covid_19_deaths.csv | Time series data on the number of death cases | Province/state, country/region, lat, long, 1/22/20…0.5/6/20 |
| time_series_covid_19_recovered.csv | Time series data on the number of recovered cases | Province/state, country/region, lat, long, 1/22/20…0.5/6/20 |
| COVID‐19 (nCoV‐19) Coronavirus Spread Dataset | ||
| covid_19_clean_complete.csv | The file contains the cumulative count of confirmed, death and recovered cases of COVID‐19 from different countries from January 2020 | Province/state, country/region, lat, long, date, confirmed, deaths, recovered |
| COVID‐19 Open Research Dataset | ||
| metadata.csv | This dataset contains metadata for 59k articles on COVID‐19 | Cord_uid, sha, source_x, title, doi, pmcid, license, abstract, publish_time, authors, journal |
| Population by country 2020 dataset | ||
| population_by_country_2020.csv | This dataset contains the information from 235 countries along with their population till 2020 | Country (or dependency), population (2020), yearly change, net change, density (P/km2), land area (km2), migrants (net), fert. rate, med. age, urban pop %, world share |
We also developed a custom dataset to develop and evaluate our model. Table 2 is developed based on collected data from different sources.
TABLE 2.
Columns description of custom build COVID‐19 dataset
| Field name | Field data description |
|---|---|
| Sno. | Serial number |
| Province/state | Province or state of the observation |
| Country/region | Country of observation |
| Date | Date and time of the observation in MM/DD/YYYY HH:MM: SS |
| Confirmed | Number of confirmed cases |
| Deaths | Number of deaths |
| Recovered | Number of recovered cases |
| Active | Number of active cases |
| 1/22/20 | First reporting date |
| 5/18/2020 | Latest reporting date |
2.2. M‐SIR model
Predictive mathematical disease models are important for understanding the trajectory of the outbreak and for preparing successful response strategies. One commonly used model is the human‐to‐human transmission SIR model, which defines people's flow through three mutually exclusive stages of infection: susceptible (S), infected (I), and recovered (R). Most epidemic models are based on dividing the population into a small number of sections. Each person is identical in terms of their status with the considered disease. The SIR model based on three sections: susceptible (S) is the class for those who are susceptible to infection. This can include passive immune systems as soon as they lose their immunity. In the infected (I) class, the parasite level within the host is large enough, and there is a possibility of spreading the infection to other susceptible people. The recovered (R) class includes all infected and recovered individuals. This epidemiological model captures the dynamics of acute infections that, after recovery, confer lifelong immunity. In general, the overall size of the population is considered constant N = S + I + R. The two cases should be examined and characterized by the inclusion or exclusion of demographic factors. Let us assume in the SIR model; there is a natural host lifespan of 1/μ years. Then the rate at which individuals in an epidemiological compartment suffer from natural mortality is given by μ. It is important to emphasize that this factor is independent of the disease and should not reflect the pathogenicity of the infectious agent. Diachronically, it can be expressed in Equation (1).
| (1) |
So, the SIR model can be defined using Equation (2),
| (2) |
Here, the initial conditions S(0) > 0, I(0) ≥ 0, and R(0) ≥ 0. It is important to enter the expression of basic reproduction number R 0 for this model. The basic reproduction number R 0 is the parameter that estimates whether a disease has spread to the population or not. If the estimated R 0 < 1, we can assume that the disease will die out, and if R 0 = 1, the disease remains in the system and is stable. But if R 0 > 1, the disease will spread and cause an outbreak. The higher the value of R 0, the more difficult it is to control.
2.3. Proposed model and algorithm
In this exploration, we have used an SIR epidemic model. In a general SIR model, transmission rate (β) and recovery rate (γ) are considered as two time‐invariant variables. Moreover, several research studies have shown that a SIR model works much better in presenting the information contained in the confirmed case data than an SEIR model. 22 Therefore we have developed a model presented in Figure 1 that can dynamically adjust the crucial parameters while working on time‐varying data, which is also treated as an M‐SIR model. 23 However, in a basic SIR model, the reproduction number () is a simple division of transmission and recovery rates, as shown in Equation (3).
| (3) |
FIGURE 1.

Modified SIR (M‐SIR) model with the time‐varying transmission. It can dynamically adjust the crucial parameters while working on time‐varying data, which is also treated as an M‐SIR model.
For building the model, we modified the primary reproduction number, with respect to time (t). Equation (4) represents the changes that happened depending on time (t).
| (4) |
We also considered detectable () and nondetectable () infected persons for building our model effectively, as shown in Equation (5).
| (5) |
In general, detectable persons contain a lower transmission rate than nondetectable persons. Therefore we calculated the transmission rate (β) and recovery rate (γ) for each country of the SAARC region. For instance, Bangladesh has 165 million people, 24 and the first confirmed case reported in the country was on March 7, 2020. As of May 30, 2020, a total of 42,844 COVID‐19 infected people were detected in the country. 25 Depending on the M‐SIR model, Bangladesh contains a transmission rate of 0.63 with a recovery rate of 0.49. The initial reproduction number for the country is 1.27. Based on available data sources of every SAARC country, we have calculated these parameters for the prediction model, which is shown in Table 3. Algorithm 1 represents the working procedure of our proposed (M‐SIR) prediction model.
TABLE 3.
Transmission rate, recovery rate, and reproduction number of SAARC countries
| Countries | Transmission rate (β) | Recovery rate (γ) | Reproduction number (R t) |
|---|---|---|---|
| Afghanistan | 0.5997691 | 0.4889147 | 1.22673566575 |
| Bangladesh | 0.6286615 | 0.4932285 | 1.2745847006 |
| Bhutan | 0.5334682 | 0.4982385 | 1.07070850607 |
| India | 0.5060977 | 0.3955793 | 1.27938367857 |
| Maldives | 0.5774223 | 0.4920300 | 1.17355100299 |
| Nepal | 0.5406592 | 0.4954823 | 1.09117762632 |
| Pakistan | 0.5254487 | 0.3947732 | 1.33101411139 |
| Sri Lanka | 0.5595021 | 0.4933888 | 1.13399838018 |
Algorithm 1. M‐SIR prediction model.
Evaluate the transmission rate , recovery rate , within the dataset time period, T
Train multiple regression or ridge regression
Estimate number of an infected person after the dataset time period, T
Estimate number of a recovered person after the dataset time period, T
while dataset time period, T ≤ respective time, t ≤ estimated time for prediction do
Estimate predicted transmission rate and recovery rate
Predict the number of affected persons, the number of recovered persons, the number of active cases
end while
The transmission rate β and the recovery rate γ are two time‐invariant variables in the conventional SIR model. β denotes that each person has on average β contacts with randomly selected others per unit of time. The recovery rate γ, on the other hand, suggests that diseased individuals recover or die at a constant average rate γ. As the time‐varying feature of β and γ is ignored in the traditional SIR model, therefore, this assumption is too simplistic to accurately and effectively anticipate the disease's progression. 15 For this reason, to make the model robust, time‐dependent SIR (M‐SIR) model is proposed, in which both the transmission rate β and the recovery rate γ are functions of time t. Therefore, this type of M‐SIR model is far more effective at predicting disease spread, control, and future trends.
3. RESULTS
We performed our simulations and tabulated the predicted results (short term, midterm, and long term) with all countries from the SAARC region. We also explored 3D parameter space using gradient descent to minimize the error. However, a lag parameter is also used in this experiment to reduce the gap of first confirmed cases of SAARC countries' pandemic situation. The M‐SIR model was calculated using MATLAB R2020a software, and Tableau data analysis visualization tools were utilized to present the estimated data in a more dynamic manner. Table 3 represents the transmission rate (β), recovery rate (γ), and the reproduction numbers (R t) of all countries in the SAARC regions. Bangladesh contains the highest transmission rate of (β = ∼0.63) in the SAARC region, whereas India shows a much lower transmission rate of only ∼0.51 compared with other countries. Also, data analysis visualization tools have been employed (Figure 2) to understand the current (as of May 30, 2020) scenario of the countries (Afghanistan, Bangladesh, Bhutan, India, Maldives, Nepal, Pakistan, and Sri Lanka) in terms of their confirmed and death cases ratio.
FIGURE 2.
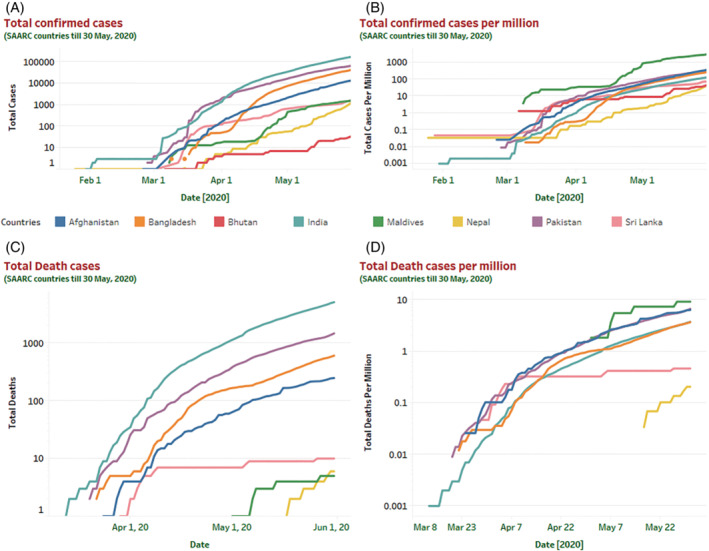
Graphical representation of countrywide COVID‐19 total confirmed and death cases along with the total number of confirmed and death cases per million till May 30, 2020. This graphical representation provides a comprehensive overview of the total number of confirmed and death cases in SAARC regions. Countrywise COVID‐19 confirmed and death cases along with the number of cases per million illustrated in (A–D) till May 30, 2020. Based on the illustration in (A) and (C), India has confirmed the highest number of infected cases (173,763) along with the most number of death cases (5164) in the region.
Surprisingly the slow growth of infections in the South‐Asian region could be a result of a lower number of testing and testing strategy. 26 Initially, the testing was limited to specific individuals who have come from high‐risk countries. Even their immediate contacts were also ignored primarily. The initial growth rate of COVID‐19 infection in the SAARC region is comparatively lower than countries like the US, France, Germany, Spain, China, Italy, and so on. However, based on the current scenario in the SAARC region (as of May 30, 2020), India confirmed the highest number of COVID‐19 reported cases. A short term model prediction for the next 20 days (till June 19, 2020) is illustrated in Figure 3 for all the countries of SAARC regions. Similarly, we also predicted the epidemic curve of SAARC regions for the midterm (90 days) and long term (180 days) COVID‐19 cases. After predicting the 20 days of the epidemic, we noticed that India, Bangladesh, Sri Lanka, and Pakistan are increasing gradually by minimizing the active cases, and their recovery rate is also rising over time. However, in this short period, the confirmed cases of these countries are also increasing till June 19, 2020. A comprehensive statistical analysis for the long‐term prediction model has been developed for each country of the SAARC region. To highlight the results, this research considered the different descriptive statistical measures including mean, median, standard error, standard deviation, range, min value, max value, and different quartile values. Based on the prediction, Afghanistan, Bangladesh, India, and Pakistan are being considered the most severely affected region due to the adverse effect of COVID‐19 (Table 4).
FIGURE 3.
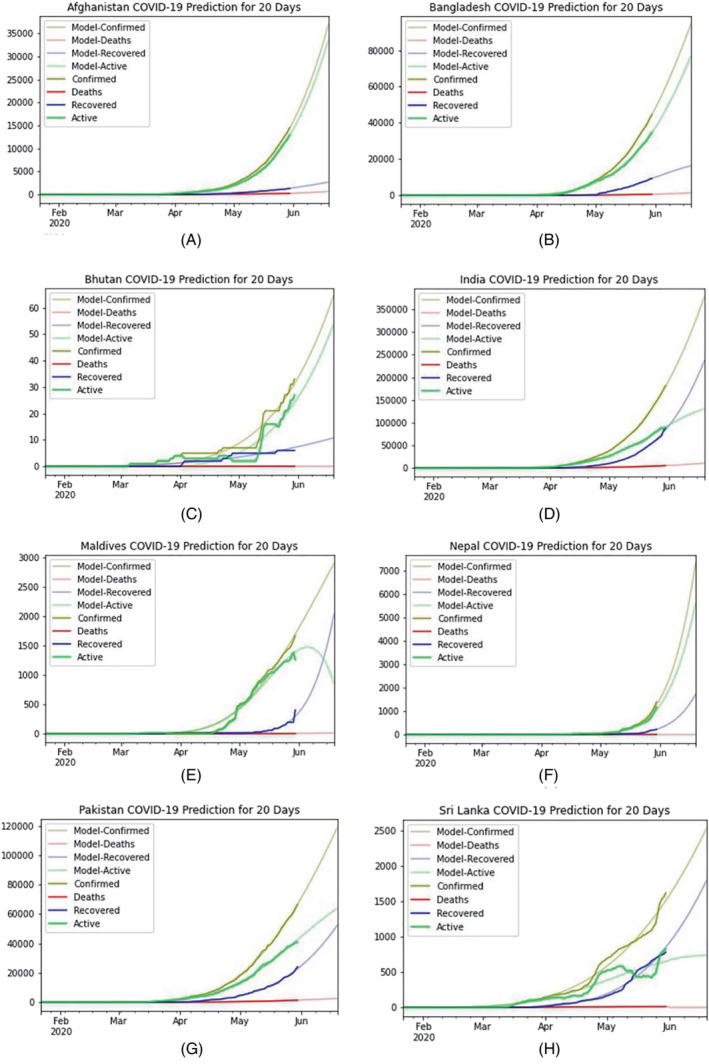
Graphical representation of a short‐term prediction curve till June 19, 2020, for all the countries of SAARC regions. This short‐term prediction model for the next 20 days provides a depiction of all SAARC countries' evaluation of COVID‐19 till mid‐June. All the countries prediction curves are denoted from (A–H), respectively, where (A) Afghanistan, (B) Bangladesh, (C) Bhutan, (D) India, (E) Maldives, (F) Nepal, (G) Pakistan, (H) Sri Lanka predicted the epidemiological curve for next 20 days successfully.
TABLE 4.
Statistical summary of 180 days prediction model for the most affected countries in the SAARC region
| Countries | Model‐confirmed | Model‐deaths | Model‐recovered | Model‐active | |
|---|---|---|---|---|---|
| Mean | AFG | 190,412.40 | 4643.50 | 11,126.18 | 174,642.719 |
| BAN | 155,231.18 | 2218.13 | 23,731.48 | 129,281.56 | |
| IND | 211,200.72 | 3017.89 | 27,867.53 | 180,315.3 | |
| PAK | 528,468.79 | 11,653.97 | 68,238.28 | 448,576.53 | |
| Standard error | AFG | 11,206.57 | 273.28 | 509.94 | 10,430.84 |
| BAN | 4659.14 | 66.57 | 456.77 | 4168.88 | |
| IND | 47,359.55 | 1479.88 | 48,017.68 | 2700.96 | |
| PAK | 25,914.54 | 571.47 | 1998.50 | 23,473.099 | |
| Standard deviation | AFG | 150,351.97 | 3666.56 | 6841.61 | 139,944.42 |
| BAN | 62,509.03 | 893.20 | 6128.31 | 55,931.46 | |
| IND | 635,395.08 | 19,854.72 | 644,224.84 | 36,237.26 | |
| PAK | 347,680.15 | 7667.16 | 26,812.71 | 314,924.67 | |
| Range | AFG | 485,970.31 | 11,851.13 | 21,504.85 | 452,614.31 |
| BAN | 200,118.47 | 2859.53 | 22,442.97 | 174,815.96 | |
| IND | 2,004,354.33 | 62,631.73 | 2,004,242.36 | 94,846.56 | |
| PAK | 1,119,327.13 | 24,683.79 | 86,381.49 | 1,008,261.84 | |
| Minimum | AFG | 7898.49 | 192.61 | 924.35 | 6781.52 |
| BAN | 26,187.94 | 374.20 | 5558.10 | 20,255.63 | |
| IND | 110,541.12 | 3454.17 | 44,567.18 | 0 | |
| PAK | 46,959.35 | 1035.56 | 13,124.96 | 32,798.82 | |
| Maximum | AFG | 493,868.81 | 12,043.75 | 22,429.20 | 459,395.84 |
| BAN | 226,306.41 | 3233.74 | 28,001.08 | 195,071.59 | |
| IND | 2,114,895.46 | 66,085.91 | 2,048,809.55 | 94,846.56 | |
| PAK | 1,166,286.48 | 25,719.35 | 99,506.45 | 1,041,060.66 | |
| First quartile (25%) | AFG | 51,363.72 | 1232.90 | 4631.15 | 44,692.82 |
| BAN | 104,429.75 | 1492.22 | 21,777.67 | 81,159.87 | |
| IND | 494,619.83 | 15,455.8 | 427,771.65 | 0 | |
| PAK | 202,699.9 | 4470.01 | 46,338.15 | 151,891.8 | |
| Median (50%) | AFG | 156,590.5 | 3818.70 | 10,947.42 | 141,824.4 |
| BAN | 174,802.19 | 2497.78 | 26,986.22 | 145,318.2 | |
| IND | 1,095,738.02 | 34,239.44 | 1,061,498.57 | 0 | |
| PAK | 487,562.5 | 10,751.9 | 76,114.45 | 400,696.1 | |
| Third quartile (75%) | AFG | 314,807 | 7724.63 | 17,425.18 | 291,608.2 |
| BAN | 211,200.72 | 3017.89 | 27,867.53 | 180,315.3 | |
| IND | 1,679,552.22 | 52,482.38 | 1,621,471.80 | 59,168.73 | |
| PAK | 835,283.1 | 18,419.96 | 92,331.4 | 724,531.8 |
Note: Count: 180.
4. DISCUSSION
The US, Italy, Spain, Germany, China are the top most affected countries in the world due to the pandemic of COVID‐19. Though the novel coronavirus originated and started to spread from China but, countries from the South‐Asian region are also infected faster than other countries globally. The overall population density of the countries in the SAARC regions is also too high. According to Table 5, we can observe the various parameters (population, land area, density, and world share) and their impact on analyzing the spread of novel coronavirus in these regions.
TABLE 5.
Population, total area, and density of SAARC countries with their individual world share 21
| Countries | Population | Land area (km2) | Density (P/km2) | World share % |
|---|---|---|---|---|
| Afghanistan | 38,742,911 | 652,860 | 60 | 0.50 |
| Bangladesh | 164,354,176 | 130,170 | 1265 | 2.11 |
| Bhutan | 769,867 | 38,117 | 20 | 0.01 |
| India | 1,377,233,523 | 2,973,190 | 464 | 17.70 |
| Maldives | 538,558 | 300 | 1802 | 0.01 |
| Nepal | 29,027,347 | 143,350 | 203 | 0.37 |
| Pakistan | 219,992,900 | 770,880 | 287 | 2.83 |
| Sri Lanka | 21,395,196 | 62,710 | 341 | 0.27 |
We further extended our prediction model for the next 90 days and 180 days, respectively. Besides, the total number of the population of a country is also parameterized because the total population of a country cannot be infected. Therefore, if we consider the total population as infected, then the probable number of infected persons remains unknown. Based on the prediction model, we tabulated the model predicted results of all SAARC countries for short term, midterm, and long term prediction, respectively (Table 6).
TABLE 6.
Predicted data based on M‐SIR model for the countries of SAARC region
| Countries | Short term prediction (20 days) | Mid‐term prediction (90 days) | Long term prediction (180 days) | Remarks | |
|---|---|---|---|---|---|
| Afghanistan | Confirmed | 35,286 | 297,264 | 1,092,714 |
|
| Deaths | 652 | 5498 | 3521 | ||
| Recovered | 2555 | 8521 | 13,416 | ||
| Active | 32,078 | 283,245 | 1,075,775 | ||
| Bangladesh | Confirmed | 100,037 | 451,840 | 867,700 |
|
| Deaths | 1346 | 6080 | 11,676 | ||
| Recovered | 17,724 | 29,418 | 30,476 | ||
| Active | 80,966 | 416,341 | 825,547 | ||
| Bhutan | Confirmed | 61 | 372 | 1683 |
|
| Deaths | 0 | 0 | 0 | ||
| Recovered | 11 | 32 | 79 | ||
| Active | 50 | 340 | 1603 | ||
| India | Confirmed | 378,657 | 1,717,831 | 3,709,965 |
|
| Deaths | 10,648 | 48,307 | 104,329 | ||
| Recovered | 217,681 | 1,269,646 | 2,609,589 | ||
| Active | 150,327 | 399,877 | 996,047 | ||
| Maldives | Confirmed | 2873 | 5970 | 7081 |
|
| Deaths | 14 | 0 | 0 | ||
| Recovered | 1139 | 5970 | 7081 | ||
| Active | 1719 | 0 | 0 | ||
| Nepal | Confirmed | 6295 | 185,528 | 1,089,821 |
|
| Deaths | 0 | 0 | 2955 | ||
| Recovered | 1783 | 99,292 | 658,568 | ||
| Active | 4512 | 86,236 | 428,297 | ||
| Pakistan | Confirmed | 118,775 | 332,528 | 497,800 |
|
| Deaths | 2534 | 7096 | 10,623 | ||
| Recovered | 49,434 | 254,510 | 487,176 | ||
| Active | 66,806 | 70,921 | 588,234 | ||
| Sri Lanka | Confirmed | 2496 | 8832 | 23,151 |
|
| Deaths | 0 | 40 | 106 | ||
| Recovered | 1840 | 8791 | 23,045 | ||
| Active | 655 | 0 | 0 | ||
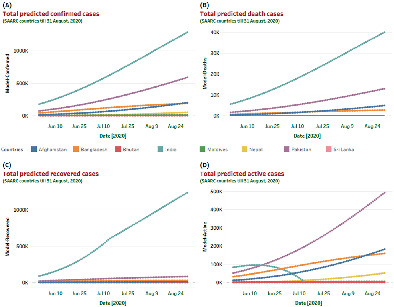
A midterm (90 days) prediction model is further assessed until August 31, 2020, and the nature of the epidemic curve has been depicted in Figure 4. For all the countries of the SAARC regions, we have predicted the total number of confirmed, death, recovery, and active cases.
FIGURE 4.
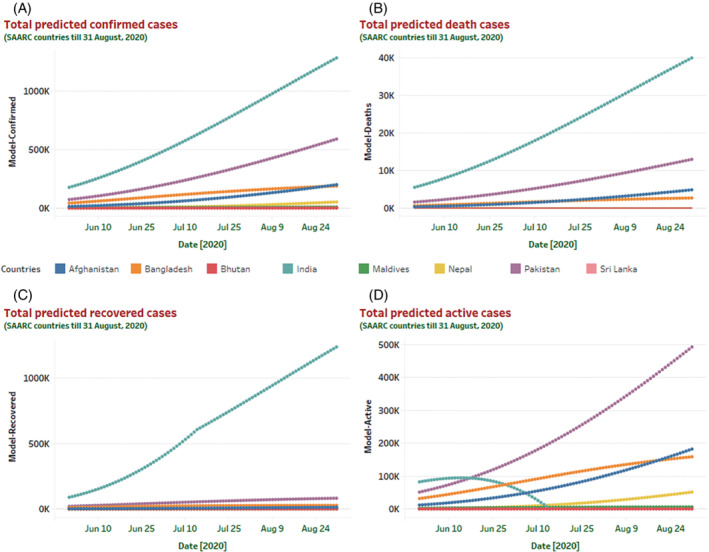
Visualization of a mid‐term prediction curve till August 31, 2020, for all the countries of SAARC regions. Based on the M‐SIR prediction model, prediction statistics for the upcoming 90 days for all the countries in the SAARC regions has depicted here. Different predicted case scenarios (confirmed, death, recovered, and active) till August 31, 2020, help to understand the epidemiological nature of COVID‐19 in the south Asian region. The predicted model indicates that in the next 90 days prediction curve will increase sharply for India (in terms of confirmed and death cases). Moreover, India will also show a substantial increase in its recovery rate. However, the number of active cases will increase in Pakistan till August 31, 2020.
Lastly, a long term prediction model for the next 180 days (till November 30, 2020) situation has been presented in Figure 5. According to the prediction model, active cases will fall to zero at the end of June in Sri Lanka, and at the beginning of August in India (Figure 5). However, Bangladesh, Maldives, and Pakistan will take more few months to reduce its active cases. On the other hand, countries like Afghanistan, Nepal, and Bhutan will show a steep increase in their active cases.
FIGURE 5.
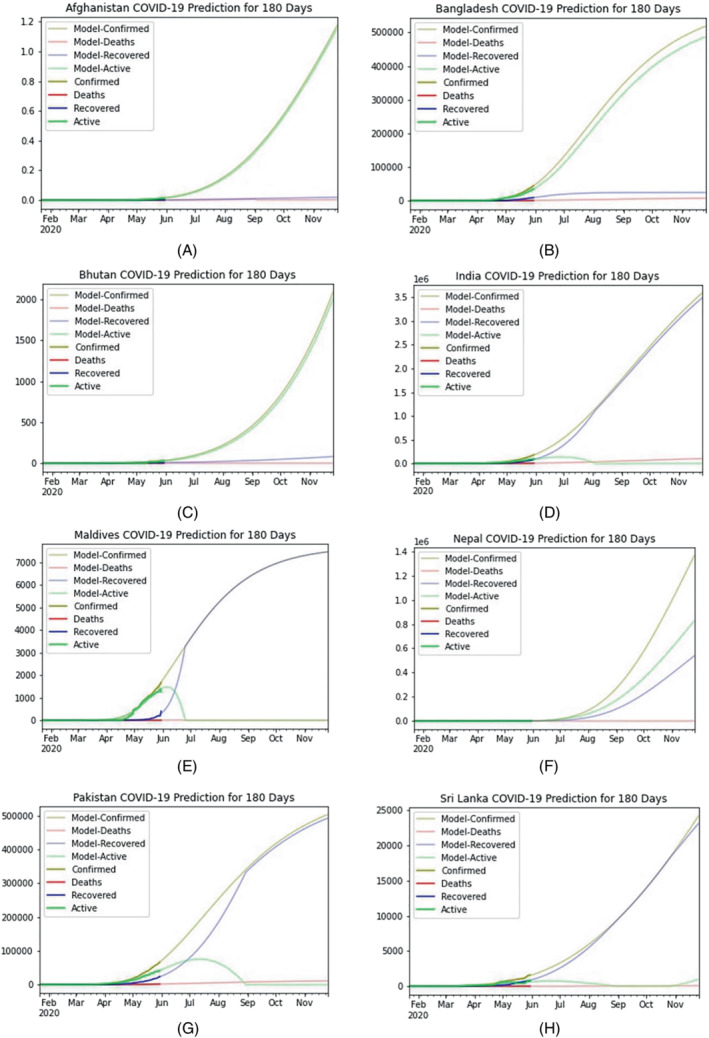
Graphical representation of a long‐term prediction curve till November 30, 2020, for all the countries of SAARC regions. This long‐term prediction model for the next 180 days provides a depiction of all SAARC countries' evaluation of COVID‐19 till mid‐November. All the countries prediction curves are denoted here from (A–H), respectively, where (A) Afghanistan, (B) Bangladesh, (C) Bhutan, (D) India, (E) Maldives, (F) Nepal, (G) Pakistan, (H) Sri Lanka predicted the epidemiological curve for next 180 days successfully.
Since the classic forms of SIR are deterministic, an improved version based on parameter optimization is suggested to improve the prediction. Moreover, all the forecasts showed in the article for the countries of the SAARC region without considering the conditions of quarantine and social distancing. Apart from that, analysis of the data based on available COVD‐19 cases till May 30, 2020. Therefore, the current data of the COVID‐19 case trend is radically different from what this article predicted. With such challenges, the future direction of this research can be extended by employing other epidemiology models (i.e., SEIQR) along with some machine learning (i.e., regression analysis) architecture. Also, consideration of social distancing and quarantine along with other observations including policy actions, human behavior, and restrictions that have the potential to improve forecast accuracy are encouraged for future studies.
5. CONCLUSION
The COVID‐19 epidemic has brought unprecedented health concerns for the community all around the globe. This research predicted the epidemic trend of COVID‐19 in SAARC countries on the basis of short‐term, midterm, and long‐term situations. We explored the transmission pattern of a new coronavirus using the M‐SIR compartmental epidemic model, a modified version of the SIR pandemic model. This study also calculated the transmission rate, recovery rate, and reproduction number for eight South Asian countries. In addition, nine different statistical metrics have been analyzed, and it has been determined that Afghanistan, Bangladesh, India, and Pakistan will continue to be the most affected countries in the SAARC area through November 2020. To the best of our knowledge, this study is the very first COVID‐19 prediction model which focused on the countries of SAARC regions. This epidemic modeling can be a helpful tool for estimating and predicting the scale and time course of COVID‐19, evaluation of the effectiveness of public health interventions, and information on public health policies in SAARC countries. In the future machine learning tools can be further used to identify and optimize the time profile for the confinement.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/eng2.12550.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Samrat Kumar Dey: Conceptualization (equal); investigation (equal); methodology (equal); software (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Md. Mahbubur Rahman: Conceptualization (equal); formal analysis (supporting); methodology (equal); project administration (lead); supervision (lead); writing – original draft (supporting); writing – review and editing (lead). Kabid Hassan Shibly: Conceptualization (equal); data curation (equal); investigation (equal); methodology (equal); software (supporting); writing – original draft (supporting); writing – review and editing (supporting). Umme Raihan Siddiqi: Investigation (supporting); project administration (supporting); supervision (supporting); writing – original draft (supporting); writing – review and editing (supporting). Arpita Howlader: Conceptualization (supporting); formal analysis (supporting); validation (supporting); writing – original draft (supporting).
Dey SK, Rahman MM, Shibly KH, Siddiqi UR, Howlader A. Epidemic trend analysis of SARS‐CoV‐2 in South Asian Association for Regional Cooperation countries using modified susceptible‐infected‐recovered predictive model. Engineering Reports. 2022;e12550. doi: 10.1002/eng2.12550
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
REFERENCES
- 1. Bogoch II, Watts A, Thomas‐Bachli A, Huber C, Kraemer MUG, Khan K. Potential for global spread of a novel coronavirus from China. J Travel Med. 2020;27(2):taaa011. doi: 10.1093/jtm/taaa011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019‐nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689‐697. doi: 10.1016/S0140-6736(20)30260-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization . Coronavirus disease (COVID‐19) situation report – 131; 2020. Accessed May 5, 2021; 2020. https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200530‐covid‐19‐sitrep‐131.pdf?sfvrsn=d31ba4b3_2
- 4. Abid K, Bari YA, Younas M, Tahir Javaid S, Imran A. Progress of COVID‐19 epidemic in Pakistan. Asia Pac J Public Health. 2020;32(4):154‐156. doi: 10.1177/1010539520927259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dey SK, Rahman MM, Siddiqi UR, Howlader A. Analyzing the epidemiological outbreak of COVID‐19: a visual exploratory data analysis approach. J Med Virol. 2020;92(6):632‐638. doi: 10.1002/jmv.25743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. New Engl J Med. 2020;382(18):1708‐1720. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahase E. Coronavirus: COVID‐19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ. 2020;368:m641. doi: 10.1136/bmj.m641 [DOI] [PubMed] [Google Scholar]
- 9. Malik YS, Obli Rajendran V, Ma I, et al. Responses to COVID‐19 in South Asian Association for Regional Cooperation (SAARC) countries in 2020, a data analysis during a world of crises. Chaos Solitons Fract. 2021;152:111311. doi: 10.1016/j.chaos.2021.111311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malek A, Hoque A. Trends of 2019‐nCoV in south Asian countries and prediction of the epidemic peaks. Virus Res. 2021;292:198230. doi: 10.1016/j.virusres.2020.198230 [DOI] [PubMed] [Google Scholar]
- 11. Tiwari V, Deyal N, Bisht NS. Mathematical modeling based study and prediction of COVID‐19 epidemic dissemination under the impact of lockdown in India. Front Phys. 2020;8:586899. doi: 10.3389/fphy.2020.586899 [DOI] [Google Scholar]
- 12. Yadav RS. Data analysis of COVID‐2019 epidemic using machine learning methods: a case study of India. Int J Inf Tecnol. 2020;12(4):1321‐1330. doi: 10.1007/s41870-020-00484-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Senapati A, Nag A, Mondal A, Maji S. A novel framework for COVID‐19 case prediction through piecewise regression in India. Int J Inf Technol. 2021;13(1):41‐48. doi: 10.1007/s41870-020-00552-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shastri S, Singh K, Kumar S, Kour P, Mansotra V. Deep‐LSTM ensemble framework to forecast COVID‐19: an insight to the global pandemic. Int J Inf Technol. 2021;13:1291‐1301. doi: 10.1007/s41870-020-00571-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y‐C, Lu P‐E, Chang C‐S, Liu T‐H. A time‐dependent SIR model for COVID‐19 with undetectable infected persons. IEEE Trans Netw Sci Eng. 2020;7(4):3279‐3294. doi: 10.1109/TNSE.2020.3024723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. WHO‐China Joint Mission . Report of the WHO‐China joint mission on coronavirus disease 2019 (COVID‐19); Accessed May 5, 2021. https://www.who.int/docs/default‐source/coronaviruse/who‐china‐joint‐mission‐on‐covid‐19‐final‐report.pdf
- 17. Rahimi I, Gandomi AH, Asteris PG, Chen F. Analysis and prediction of COVID‐19 using SIR, SEIQR, and machine learning models: Australia, Italy, and UK cases. Information. 2021;12(3):109. doi: 10.3390/info12030109 [DOI] [Google Scholar]
- 18. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20(5):533‐534. doi: 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Devakumar KP. COVID‐19 dataset; 2020. Accessed May 5, 2021. https://kaggle.com/imdevskp/corona‐virus‐report
- 20. Allen Institute for AI . COVID‐19 Open Research Dataset Challenge (CORD‐19); 2020. Accessed May 5, 2021. https://kaggle.com/allen‐institute‐for‐ai/CORD‐19‐research‐challenge
- 21. Prabhu TN. Population by country; 2020. Accessed May 5, 2021. https://kaggle.com/tanuprabhu/population‐by‐country‐2020
- 22. Roda WC, Varughese MB, Han D, Li MY. Why is it difficult to accurately predict the COVID‐19 epidemic? Infect Dis Modell. 2020;5:271‐281. doi: 10.1016/j.idm.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nesteruk I. Statistics‐based predictions of coronavirus epidemic spreading in mainland China. Innov Biosyst Bioeng. 2020;4(1):13‐18. doi: 10.20535/ibb.2020.4.1.195074 [DOI] [Google Scholar]
- 24. Uddin M, Ashrafi SAA, Azad AK, et al. Improving coverage of civil registration and vital statistics, Bangladesh. Bull World Health Organ. 2019;97(9):637‐641. doi: 10.2471/BLT.18.219162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dey SK, Rahman MM, Siddiqi UR, Howlader A. Exploring epidemiological behavior of novel coronavirus (COVID‐19) outbreak in Bangladesh. SN Compr Clin Med. 2020;2(10):1724‐1732. doi: 10.1007/s42399-020-00477-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gupta R, Pal SK. Trend analysis and forecasting of COVID‐19 outbreak in India. medRxiv:2020.03.26.20044511, 2020. 10.1101/2020.03.26.20044511 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.


