Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has a worldwide distribution in humans and many other mammalian species. In late September 2021, 12 animals maintained by the Chicago Zoological Society's Brookfield Zoo were observed with variable clinical signs. The Delta variant of SARS‐CoV‐2 was detected in faeces and nasal swabs by qRT‐PCR, including the first detection in animals from the families Procyonidae and Viverridae. Test positivity rate was 12.5% for 35 animals tested. All animals had been vaccinated with at least one dose of a recombinant vaccine designed for animals and all recovered with variable supportive treatment. Sequence analysis showed that six zoo animal strains were closely correlated with 18 human SARS‐CoV‐2 strains, suggestive of potential human‐to‐animal transmission events. This report documents the expanding host range of COVID‐19 during the ongoing pandemic.
Keywords: carnivore, Procyonidae, SARS‐CoV‐2, severe acute respiratory syndrome coronavirus, Viverridae, zoo
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) originally described in December of 2019 in the Wuhan region of China is caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (Zhang & Holmes, 2020). SARS‐CoV‐2 is a single‐stranded, positive‐sense, enveloped RNA virus in the genus Betacoronavirus of family Coronaviridae. The SARS‐CoV‐2 genome encodes 16 non‐structural proteins (Nsp1‐16), four major structural protein (spike [S] glycoprotein, nucleocapsid [N] protein, membrane [M] protein, and envelope [E] protein), and some accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) (Forster et al., 2020; Mohamadian et al., 2021; Thakur et al., 2020). Two months after its initial identification, SARS‐CoV‐2 caused a global pandemic and at the close of 2021, it accounted for more than 5.1 million human mortalities and 280 million infections (World Health Organization, 2020). The continued evolution of SARS‐CoV‐2 in human populations resulted in the emergence of variants such as B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta) and B.1.1.529 (Omicron) (World Health Organization, 2020; Shiehzadegan et al., 2021). Initially identified in India in late 2020 (Yang & Jeffrey, 2021), the Delta variant has an increased transmissibility and has been reported in many countries (Shiehzadegan et al., 2021).
SARS‐CoV‐2 is believed to originate from bat coronaviruses and shares the highest sequence identity (96%) to a bat coronavirus strain RaTG13 (Prince et al., 2021; Zhang & Holmes, 2020; Zhou et al., 2020). It remains unclear about whether SARS‐CoV‐2 directly evolved from a bat coronavirus or through an intermediate host (Lean et al., 2021). Natural SARS‐CoV‐2 infection has been detected in various animal species including companion (Calvet et al., 2021; Ferasin et al., 2021; Garigliany et al., 2020; Jairak et al., 2021; Miro et al., 2021; Sit et al., 2020), captive (Bartlett et al., 2021; McAloose et al., 2020; Mitchell et al., 2021), farm (Fenollar et al., 2021; Oreshkova et al., 2020; Oude Munnink et al., 2021; Sharun et al., 2021), and wild animals (Chandler et al., 2021; Hale et al., 2021). SARS‐CoV‐2 was first detected in two Hong Kong dogs as a result of human‐to‐animal transmission events in February 2020 (Sit et al., 2020). Since then, SARS‐CoV‐2 infection in pet dogs and cats has been reported in different countries (Calvet et al., 2021; Garigliany et al., 2020; Jairak et al., 2021; Miro et al., 2021). Companion ferrets also were reported SARS‐CoV‐2 positive in households with human COVID‐19 patients (Racnik, et al., 2021). In addition to companion animals, SARS‐CoV‐2 in zoo‐managed animals was first reported in April 2020 at the Bronx Zoo where positive tigers (Panthera tigris jacksoni and Panthera tigris altaica) and lions (Panthera leo krugeri) showed mild respiratory signs from possible contact with asymptomatic, COVID‐positive zookeepers (McAloose et al., 2020; Wang et al., 2020). Other zoo‐maintained animals including cougar (Puma concolor), snow leopard (Panthera uncia), lynx (Lynx canadensis), gorilla (Gorilla gorilla), and Asian small‐clawed otter (Aonyx cinereus) have tested positive for SARS‐CoV‐2 RNA (U.S. Department of Agriculture, 2022). As of December 2021, SARS‐CoV‐2 was reported in animals maintained in zoos and aquariums in 20 states in the United States and in other countries including the Czech Republic, India, Singapore, Spain, Sweden, and South Africa (U.S. Department of Agriculture, 2022). All documented evidence showed that companion and zoo‐maintained animals contracted SARS‐CoV‐2 through reverse zoonotic events (Wang et al., 2020), which have been proposed as likely sources of infection in animals (Prince et al., 2021). In the case of farmed animals, SARS‐CoV‐2 was first detected in farmed minks exposed to COVID‐19‐positive workers (Oreshkova et al., 2020) and subsequently reported in numerous mink farms in 11 countries (Fenollar et al., 2021). Whole‐genome sequencing of outbreaks on 16 mink farms and farm workers revealed that both human‐to‐mink and mink‐to‐human transmissions could occur (Oude Munnink et al., 2021). A USDA serosurveillance study showed that 40% of 385 serum samples collected from wild white‐tailed deer in four different states were positive for SARS‐CoV‐2 antibody, suggestive of widespread SARS‐CoV‐2 exposure in deer populations (Chandler et al., 2021). A recent study revealed that 35.8% of 360 nasal swabs collected from white‐tailed deer in six different Ohio locations were positive for SARS‐CoV‐2 (Hale et al., 2021). Genomic analysis showed that deer were infected with three different SARS‐CoV‐2 lineages and it was hypothesized that human‐to‐deer as well as deer‐to‐deer transmission events were occurring (Hale et al., 2021). Spread of SARS‐CoV‐2 to and between wild animals complicates control efforts.
The specific objective was to report a SARS‐CoV‐2 outbreak that occurred during the COVID‐19 pandemic in a zoological institution in September of 2021. In this outbreak, among six different animal species infected, the virus was detected in three new species including fishing cat (Prionailurus viverrinus), binturong (Arctictis binturong), and coati (Nasua nasua). The epidemiology of this outbreak, clinical manifestations, viral shedding, and genome sequences are presented.
2. MATERIALS AND METHODS
2.1. Animal facility and care
Chicago Zoological Society's Brookfield Zoo (CZS) cares for over 3000 animals representing more than 500 species and subspecies. The Fragile Kingdom habitat is composed of indoor and outdoor spaces housing mammals, reptiles, and amphibians. Originally built as a single indoor facility for lions in 1931, it was divided into the current format of two U‐shaped spaces in the late 1980s.
Daily care for animals is provided by a team of personnel who provide food and water, sanitation, and behavioural enrichment. Animals had unique access to each space and no animals shared spaces with animals of a different species. Personal protective equipment (PPE) for animal care staff was instituted in April 2020 (approximately 18 months prior to the described outbreak). PPE included disposable nitrile or vinyl gloves and surgical masks. In addition, access to susceptible animals was restricted to full‐time staff, and human safety protocols were implemented such as daily self‐symptom checks and staying at home if sick.
2.2. Vaccination
In early September 2021, vaccination of animals with an experimental SARS‐CoV‐2 recombinant vaccine (1.0 ml SQ or IM, Zoetis International) was initiated. The vaccination schedule was designed to occur in phases based on proposed susceptibility (Damas et al., 2020) with felids and primates as the priority. Animals were vaccinated with a two‐dose series separated by at least 3 weeks but no more than 5 weeks apart.
Vaccination of all mammals in the affected habitat began on 6 September 2021 (n = 3) and animals were administered their first dose on 7 (n = 10), 8 (n = 14), 9 (n = 2), 10 (n = 1), and 16 (n = 1) September. The first animal to show clinical signs (Amur tiger, Panthera tigris altaica) was the last animal to start the vaccination series (the first dose was given on 16 September). Animals that remained negative and non‐clinical were vaccinated with their second dose on schedule. Animals that tested positive were subsequently revaccinated with the two‐dose vaccine series no sooner than 14 days after the last positive test (total three doses). All animals were fully vaccinated by 21 December 2021.
2.3. Sample collection
Samples were collected from animals either through trained voluntary nasal swabs, anesthetized examinations with nasal swabs, or passive faecal collection. Faecal samples were collected off the ground of their indoor exhibit space (concrete) during normal routine care of the animals and then spaces were disinfected each day. In addition, after the onset of the outbreak, swabs of air filters on inlet and outflow sides were collected. This testing was repeated at the conclusion of the outbreak prior to opening the building to the public.
All samples were processed similarly by placing the sample in a sterile individual collection tube with the animal identification number or location of the air filter and date on the label. Samples were stored at −20 or −80°C until shipment on ice packs to the University of Illinois Veterinary Diagnostic Laboratory.
2.4. Imaging
Computed tomography (CT) scans were accomplished with patients anesthetized and positioned in sternal recumbency. Images were acquired in the transverse plane using a multislice 16‐detector CT system (Toshiba Aquilion LB, Tustin, CA). The following technical factors were used for acquisition: 120 kVp, 100 mAs, and 16 detectors × 1.0 mm collimation. When used, intravenous contrast medium was administered via a cephalic venous catheter: iohexol 240 (Omnipaque, GE Healthcare, Princeton, NJ) at a dose of 600 mg I/kg. All studies were reconstructed into a high‐frequency bone algorithm, lung algorithm, and a soft tissue algorithm.
2.5. Epidemiologic investigation
Subsequent to the development of clinical signs and positive test results in carnivores, an epidemiologic investigation into possible human infections in staff working with these animals was conducted by the Cook County and Illinois Departments of Public Health (IDPH). The investigation focused on the time period 1 week prior to the onset of the first case through 2 weeks after the first clinical signs in animals were observed. All animal care staff that had direct contact with any animal or food preparation during this period were surveyed for clinical signs, contact with infected individuals, and tested for SARS‐CoV‐2 using either a rapid antigen test (QuickVue At Home OTC COVID‐19 Test, Quidel Corporation, 9975 Summers Ridge Road, San Diego, CA) or qRT‐PCR assay (IDPH testing site).
2.6. Real‐time RT‐PCR
RNA extraction and quantitative PCR was performed as previously described for animal samples (McAloose et al., 2020). RNA samples were extracted on the KingFisher Flex using the MagMAX Pathogen RNA/DNA Kit. Real‐time RT‐PCR was performed on Applied Biosystems™ 7500 Real‐Time PCR Systems using AgPath‐ID One‐Step RT‐PCR Kit and either CDC N1 or N2 primers and probes (both not typically applied for initial testing at the University of Illinois). The thermocycler conditions are with one cycle of 48°C for 10 min and 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 45 s.
2.7. Sequencing
Whole‐genome sequencing was performed at the National Veterinary Services Laboratories as previously described (Hale et al., 2021). Viral RNA was amplified by PCR38 and cDNA libraries were prepared using the Nextera XT DNA Sample Preparation Kit according to manufacturer instructions. Sequencing was performed using the 500‐cycle MiSeq Reagent Kit v2. Sequences were assembled using IRMA v.0.6.7 and DNAStar SeqMan NGen v.14.0.1. Additional sequencing was performed at the University of Illinois Veterinary Diagnostic Laboratory targeting the spike and ORF3a genes on MiSeq using a targeted amplification method with four pairs of primers. Sequences were deposited into GISAID (Elbe & Buckland‐Merrett, 2017). Reference SARS‐CoV‐2 sequences were downloaded from GISAID. Sequence alignment and construction of phylogenetic tree with maximum likelihood were performed using MEGA 7.0.26.
2.8. Wildlife pathology surveillance
Complete gross necropsies were performed on seven wild raccoons that died naturally or were humanely euthanized as part of disease surveillance efforts and were collected contemporaneously with zoo cases. Sections of brain, pituitary gland, adrenal gland, thyroid gland, eye, skin, skeletal muscle, peripheral nerve, tongue, salivary gland, trachea, oesophagus, lung, heart, diaphragm, stomach, pancreas, small intestine, cecum, colon, kidney, urinary bladder, liver, spleen, bone, bone marrow, lymph nodes, and either testis or ovary and uterus were collected and fixed in 10% neutral buffered formalin. Sections of lung, trachea, nasal turbinates, brain, heart, liver, spleen, and lymph nodes were saved at −80°C and sections of lung, trachea, and nasal turbinates were stored in RNAlater® (Invitrogen, Carlsbad, CA). Tissues were routinely processed for histopathology, embedded in paraffin, sections cut at 3–5 mm and stained with haematoxylin–eosin, and reviewed by a board‐certified veterinary pathologist.
2.9. Statistical analysis
Summary statistics were performed to include days positive, days negative, presence and prevalence of clinical signs, prevalence of clinical signs, and 7‐day positivity rate. Comparisons between days positive and presence of clinical signs and days positive by species were performed using negative binomial regression with the MASS package in R.
To quantify the viral dynamics of the infected animals, a within‐host model was used that was previously constructed to understand SARS‐CoV‐2 infection in humans (Ke, Martinez, et al., 2021; Ke, Zitmann, et al., 2021). A population non‐linear mixed effect modelling approach was used to fit the model to longitudinal viral loads of 10 animals where at least two samples were positive for SARS‐CoV‐2 (i.e. animals with less than two positive RT‐PCR results during the study period were excluded). Tables 1 and 2 provide parameter values. In the model, the total numbers of target cells (T), cells in the eclipse phase of infection (E) (i.e. infected cells not yet producing virus), productively infected cells (I), and viruses (V) were recorded. In addition, a prototypical innate response (e.g. type‐I interferon) was included. Immune mediators are produced from infected cells and bind to receptors on target cells stimulating an antiviral response that makes cells refractory to viral infection (R). Such cells are said to be refractory cells or cells in an antiviral state (Garcia‐Sastre & Biron, 2006). For simplicity and due to a lack of data, the specific immune mediators or their concentration were not explicitly considered. Instead, the quasi‐steady‐state assumption that the immune dynamics are fast and thus the concentration of immune mediators is proportional to the number of infected cells was used. The ordinary differential equations are as follows:
TABLE 1.
The fixed parameters in the model and their values
| Parameter | Description | Values |
|---|---|---|
| T 0 | Total number of (infection free) target cells | 8 × 107 cells |
| I 0 | Initial number of infected cells | 1 cell |
| c | Virus clearance rate | 10/day |
| k | 1/k is the eclipse period | 4/day |
Note: The initial values of variables are set to 0 except for the variables listed below.
TABLE 2.
Parameter values of the best fit of the extended target cell model to the full dataset
| ID | t 0 (days) | β (10−8 /day) | δ (/day) | π (/day) | Φ (10−6 /day) | ρ (/day) |
|---|---|---|---|---|---|---|
| 2475 | 5.7 | 318.5 | 1.1 | 0.5 | 475.24 | 0 |
| 4839 | 5.6 | 311.2 | 1.3 | 0.5 | 173.09 | 0 |
| 5541 | 5.4 | 249.1 | 1.3 | 0.5 | 23.94 | 0 |
| 7227 | 5.5 | 264 | 0.9 | 0.5 | 69.37 | 0 |
| 8046 | 5.5 | 276.4 | 0.4 | 0.6 | 11.23 | 0 |
| 9185 | 5.5 | 253.4 | 1 | 0.5 | 41.71 | 0 |
| 9191 | 5.5 | 103.3 | 0.9 | 0.3 | 33.06 | 0 |
| 9192 | 5.6 | 261.3 | 0.5 | 0.4 | 360.64 | 0 |
| 9311 | 5.4 | 218.2 | 0.8 | 0.6 | 9.81 | 0 |
| 9578 | 5.6 | 288 | 1.1 | 0.5 | 136.7 | 0 |
| Mean | 5.5 | 244.9 | 0.9 | 0.5 | 63.23 | 0 |
| SD | 0.02 | 0.32 | 0.36 | 0.24 | 1.36 | 0.01 |
Abbreviation: SD, standard deviation.
In this model, target cells are infected by virus with rate constant β. Cells in the eclipse phase become productively infected cells at per capita rate k. Productively infected cells die at per capita rate δ. The rate, π, is the product of the viral production rate per infected cell and the proportion of virus that is sampled. Φ is a constant describing the rate that innate signaling makes a target cell refractory, and ρ is the rate that refractory cells transition back to target cells. Viruses are cleared at per capita rate c.
3. RESULTS
3.1. SARS‐CoV‐2 detection and distribution
On 23 September 2021 (day 1), the first report of clinical signs consistent with SARS‐CoV‐2 was observed in a single Amur tiger. Four days later, this animal tested positive via faeces and voluntary nasal swab. Over the course of the next 60 days, an additional 11 animals representing six species from three families (Felidae, Viverridae, and Procyonidae) tested positive for SARS‐CoV‐2 RNA (Tables 3 and 4; Figure 1). During that 60‐day period, a total of 392 samples were submitted from 35 animals as part of surveillance efforts representing 17 species from three orders (Carnivora, Rodentia, and Pholidota) and eight families (Felidae, Canidae, Ursidae, Viverridae, Herpestidae, Sciuridae, Hystricidae, and Manidae). SARS‐CoV‐2 viral RNA was detected in 49 samples for a total sample positivity of 12.5%. Snow leopards had the highest viral load of all positive species estimated from RT‐qPCR Ct value (Figure 2). The 7‐day case positivity peaked in the first week (34%) and declined to 0% by day 53 (Figure 1). All positive animals were in the order Carnivora. The mean age of positive carnivores on the first day of the outbreak was 7.17 years (median: 5.93 years, range: 1.34–14.33 years), which was not significantly different from the mean age of negative carnivores, 8.47 years (median: 10.96, range: 1.23–18.71 years). Peak detection was observed on days 8 and 9, with 11 total detections each day (Figure 1). SARS‐CoV‐2 RNA was detected in individuals for a variable amount of time ranging from 2 to 20 days with a median of 6 days and mean of 8.75 days (Table 3). Six animals (55%) demonstrated at least one intermittent shedding period with a maximum of 9 days negative between positive tests. Three of those animals had a second intermittent period with a minimum of 4 days and maximum of 15 days between positive tests. Felidae had the highest number of total cases but had a lower prevalence (n = 7; 53%) than both Procyonidae (n = 2; 100%) and Viverridae (n = 3; 67%). After 24 November (2 months after initial index case), 14 carnivores were tested twice weekly through January 2022 with no SARS‐CoV‐2 viral detection using qRT‐PCR noted during this period.
TABLE 3.
Summary of demographic and clinical features from 12 carnivores detected with SARS‐CoV‐2 at Chicago Zoological Society's Brookfield Zoo in 2021
| Species | Sex | Age (years) | Number of days positive | Intermittent | Intermittent twice | Number of days between first intermittent period | Number of days between second intermittent period | Diarrhoea | Hyporexia | Lethargy | Coughing |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Binturong | Female | 8.63 | 6 | No | No | 0 | 1 | 1 | 0 | ||
| Binturong | Male | 10.31 | 5 | No | No | 0 | 0 | 0 | 0 | ||
| Fishing cat | Male | 5.32 | 10 | Yes | Yes | 8 | 4 | 1 | 1 | 1 | 0 |
| Lion | Male | 5.58 | 20 | Yes | Yes | 2 | 15 | 0 | 1 | 1 | 0 |
| Lion | Male | 5.58 | 14 | Yes | Yes | 2 | 4 | 0 | 1 | 1 | 0 |
| Snow leopard | Female | 6.27 | 5 | Yes | No | 2 | 0 | 0 | 0 | 0 | |
| Snow leopard | Male | 8.33 | 19 | Yes | No | 9 | 0 | 1 | 1 | 1 | |
| Snow leopard | Female | 1.34 | 11 | Yes | No | 2 | 1 | 1 | 0 | 0 | |
| Tiger | Female | 14.33 | 5 | No | No | 0 | 0 | 0 | 0 | ||
| Tiger | Female | 11.54 | 6 | No | No | 0 | 1 | 1 | 1 | ||
| White‐nosed coati | Male | 4.39 | 2 | No | No | 0 | 0 | 0 | 0 | ||
| White‐nosed coati | Female | 4.41 | 2 | No | No | 0 | 0 | 0 | 0 |
Note: Age was determined at the time of the outbreak.
TABLE 4.
Epidemiologic characteristics of the six carnivore families tested for SARS‐CoV‐2 at the Chicago Zoological Society's Brookfield Zoo in 2021
| Family | Positive | Negative | Prevalence | Median days positive |
|---|---|---|---|---|
| Felidae | 8 | 7 | 53% | 10.5 |
| Viverridae | 2 | 1 | 67% | 5.5 |
| Procyonidae | 2 | 0 | 100% | 2 |
| Herpestidae | 0 | 4 | 0% | NA |
| Ursidae | 0 | 2 | 0% | NA |
| Canidae | 0 | 1 | 0% | NA |
FIGURE 1.
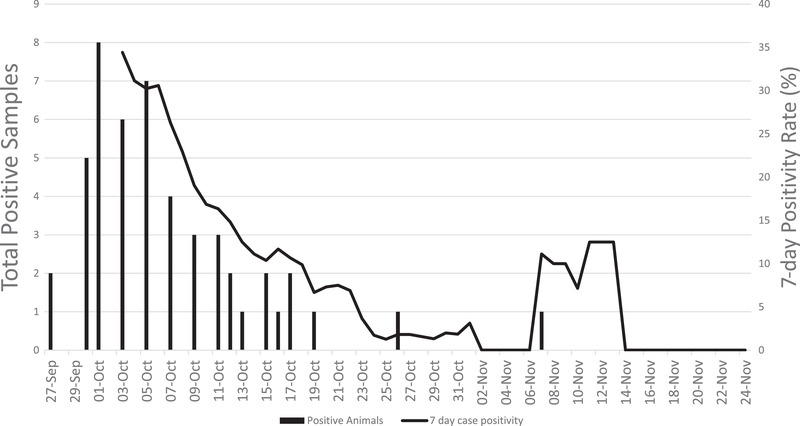
SARS‐CoV‐2 case positivity rate zoological animals. Daily (vertical bars; left axis) and 7‐day test positivity rate (line, right axis) in 12 carnivores with SARS‐CoV‐2 detected at the Chicago Zoological Society's Brookfield Zoo in 2021
FIGURE 2.
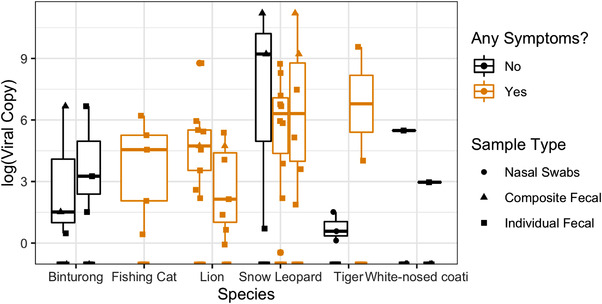
SARS‐CoV‐2 viral copies between species at a zoological institution. Viral copy number in 12 carnivores detected with SARS‐CoV‐2 with and without clinical signs at Chicago Zoological Society's Brookfield Zoo in 2021
3.2. Clinical signs of SARS‐CoV‐2
Only four classifications of clinical signs were observed, all in felids: hyporexia, lethargy, diarrhoea, and coughing (Table 3). Many felids were non‐clinical on days they tested positive; all clinical signs resolved by 6 October. The proportion of positive felids having clinical signs decreased from 40% on 30 September to 20% by 3 October; however, animals that had clinical signs at any point in their disease course had a significantly greater number of days positive (median: 18.5 days, range: 7–32, p < .0001) than animals that never showed clinical signs (median: 5.5 days, range: 1–8; Figure 3). Overall, hyporexia (n = 7; 58%) and lethargy (n = 6; 50%) were the most common clinical signs, followed by diarrhoea (n = 2; 17%) and coughing (n = 2; 17%).
FIGURE 3.
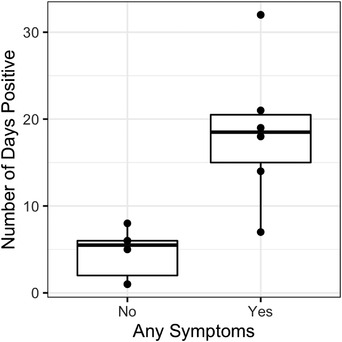
Clinical signs in SARS‐CoV‐2 positive and negative zoological animals. Number of days positive in 12 carnivores detected with SARS‐CoV‐2 at the Chicago Zoological Society's Brookfield Zoo in 2021
3.3. SARS‐CoV‐2 presence in air filtration system
SARS‐CoV‐2 RNA was detected in air handlers 3 and 4 from enclosure intake filter samples taken on 8 October. SARS‐CoV‐2 RNA was not detected in outflow filters from the enclosure samples. On 8 November, all air filters were changed using standard protocols. Three weeks later, air filter sampling was repeated in a similar manner and all results were negative.
3.4. Imaging comparison
A CT scan performed in December 2021 on a binturong, 2 months after testing positive for SARS‐CoV‐2, was compared to an earlier scan from the same individual from October 2020. Compared to the prior scan, the binturong developed bilateral, multifocal, patchy linear unstructured interstitial patterns within all lung lobes; the pulmonary infiltrates were predominantly peripheral in location in both the dependent and non‐dependent portions of the lungs (Figure 4).
FIGURE 4.
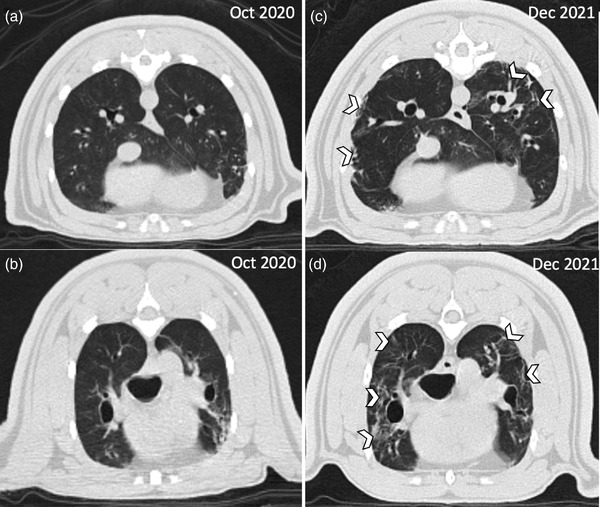
Imaging of a binturong prior to and after SARS‐CoV‐2 infection. Computed tomography of a female binturong (Arctictis binturong) 1 year before (a, b) and 2 months post (c, d) SARS‐CoV‐2 infection at the Chicago Zoological Society's Brookfield Zoo in 2021. Comparison images at similar locations at two time points; the white chevrons highlight areas of peripheral linear unstructured interstitial pattern.
3.5. Human SARS‐CoV‐2 infection
A single, fully vaccinated animal care staff employee tested positive by RT‐PCR concurrently with mild clinical signs (headache and cough) on 27 September 2021. This person had been off‐site for over a week until the morning of 23 September, worked with affected animals on 25 September and 26 September, and then off‐site again. The same animal care staff employee tested positive by a rapid SARS‐COV‐2 antigen test (LumiraDX SARS‐COV‐2 Rapid Result Antigen Test) on 29 September, and was sampled again on 1 October, testing positive by RT‐PCR by IDPH on 4 October 2021. The animal care staff employee remained off‐site per IDPH and CZS health and safety protocols, consistent with CDC recommendations at the time. Following confirmation of a positive animal (5 days after signs in first positive tiger), a total of 30 employees were tested: 18 from zoo veterinary services (VS) staff, eight animal care staff from the affected area, and four from zoo nutrition services. A second round of onsite employee testing took place 3 days later, where 13 VS staff were tested. All animal care staff were tested a second time by RT‐PCR at an IDPH site. All employees tested negative with either rapid test and/or RT‐PCR, except the first person tested.
3.6. Genomic sequencing of SARS‐CoV‐2 isolates
Six complete genome sequences of animal SARS‐CoV‐2 isolates (hCoV‐19/fishing_cat/USA/IL‐21‐029527‐002, hCoV‐19/binturong/USA/IL‐21‐029528‐001, hCoV‐19/tiger/USA/IL‐21‐028948‐002/2021, hCoV‐19/snow_leopard/USA/IL‐21‐029401‐002, hCoV‐19/snow_leopard/USA/IL‐21‐029401‐007, and hCoV‐19/lion/USA/IL‐21‐029401‐005) and one partial genome hCoV‐19/tiger/USA/IL‐21‐40753 (the same sample with hCoV‐19/tiger/USA/IL‐21‐028948‐002/2021) with complete spike and ORF3a genes were obtained. Sequence analysis showed the six animal SARS‐CoV‐2 strains were delta variants and closely related with each other (99.9%–100% nucleotide identity in the complete genome and only 1–4 nucleotide differences between them) (Figure 5). These strains shared the highest nucleotide identities 99.9% with 18 human SARS‐CoV‐2 clonal strains from 10 states circulating in the period from July to September including hCoV/human/USA/IN‐CDC‐STM‐000721646/2021. Compared to hCoV/human/USA/IN‐CDC‐STM‐000721646/2021, the tiger and snow leopard strains had only 2–3 nucleotide differences, and fishing cat, binturong, and lion strains had 4 nucleotide differences. In both S and E genes, there was one synonymous change each (C21595T and C26458T, respectively) between all six animal and human isolates, while four nonsynonymous changes with three isolates in the ORF1ab (fishing cat strain: C5850A [Pro to His] and G15380T [Ser to Ile], binturong strain: T5571C [Met to Thr]) and 1 in N gene (lion strain: G28349T [Gly to Val]).
FIGURE 5.
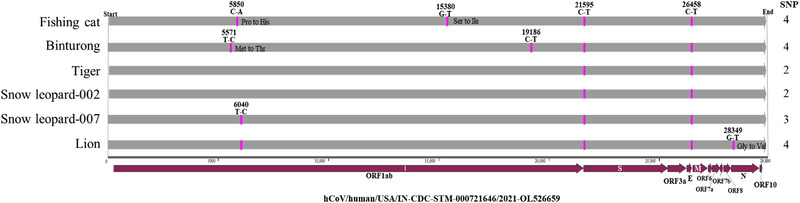
Complete genome comparison of six zoo SARS‐CoV‐2 strains from this study using the closest human strain (hCoV/human/USA/IN‐CDC‐STM‐000721646/2021, GenBank accession number: OL526659) as a reference. Two to four nucleotide differences observed in them.
These data strongly suggest that a single introduction contributed to the outbreak with potential human‐to‐animal transmission. Phylogenetic analysis of Delta variants detected in different hosts showed that six isolates from the present study formed a small cluster and closely related with five animal isolates (hCoV‐19/cat/USA/IN‐21‐032644‐001/2021, hCoV‐19/tiger/USA/NE‐21‐034299‐001/2021, hCoV‐19/dog/USA/OH‐OSU‐0508‐031718/2021, hCoV‐19/lion/USA/UT‐21‐031970‐001/2021, and hCoV‐19/tiger/USA/CA‐21‐032685‐002/2021) as well as those 18 human isolates in the complete genome (Figure 6). On the spike tree, the six Illinois zoo animal isolates are related with several animal isolates and the human isolates (Figure 7).
FIGURE 6.
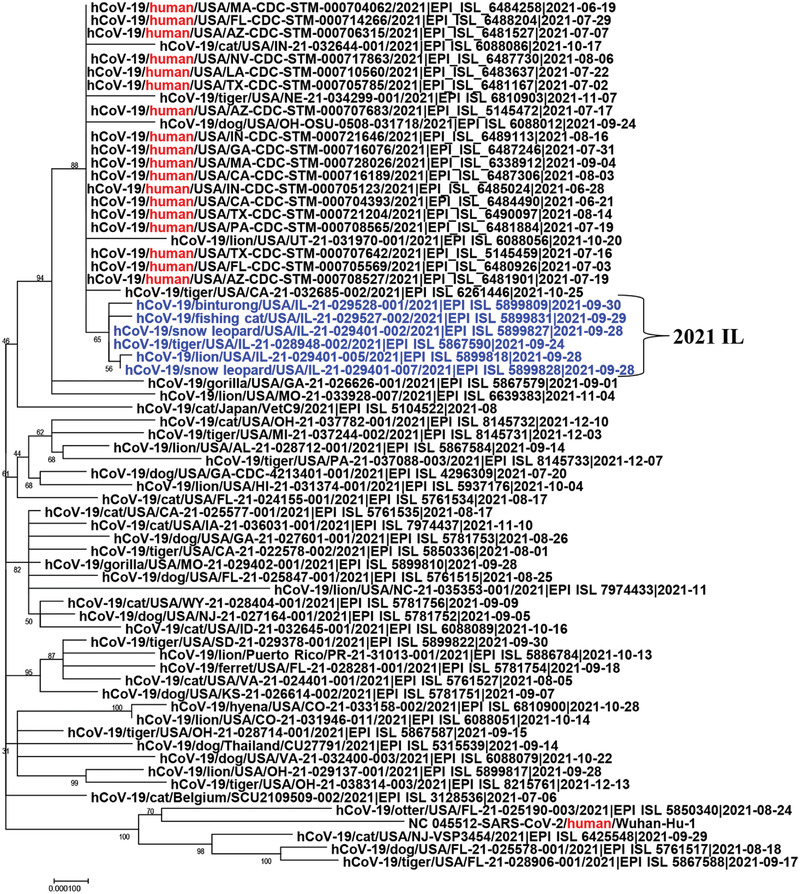
Phylogenetic tree analysis of complete genome of SARS‐CoV‐2 Delta variants including six zoo animal strains, 18 human strains from the United States, and the original Wuhan human strain (Wuhan‐Hu‐1)
FIGURE 7.

Phylogenetic tree analysis of spike gene of SARS‐CoV‐2 Delta variants including six zoo animal strains, 18 human strains from the United States, and the original Wuhan human strain (Wuhan‐Hu‐1)
3.7. Disease surveillance
Gross and histologic findings in wild raccoons were similar to findings in wild raccoons from this region and in the literature (Church et al., 2018). There were no lesions suggestive of SARS‐CoV‐2 pneumonia. Four raccoons had a bronchopneumonia with intralesional inclusions and viral antigen consistent with canine distemper virus infection. Lung was submitted for SARS‐CoV‐2 RT‐PCR and was negative in all cases. No lesions were noted in other sections of the respiratory tract or in the heart.
3.8. SARS‐CoV‐2 dynamics
We estimated that the three infected snow leopards had higher peak viral loads and longer periods of viral shedding than other animals. The clearance rates (i.e. the rate of viral decline after peak viral load) are similar across different animal groups (Figure 8), and their values are similar to those estimated from humans (Ke, Martinez, et al., 2021; Ke, Zitmann, et al., 2021). Overall, the viral dynamics in these animals are similar to those seen in humans (Ke, Martinez, et al., 2021; Wölfel et al., 2020).
FIGURE 8.
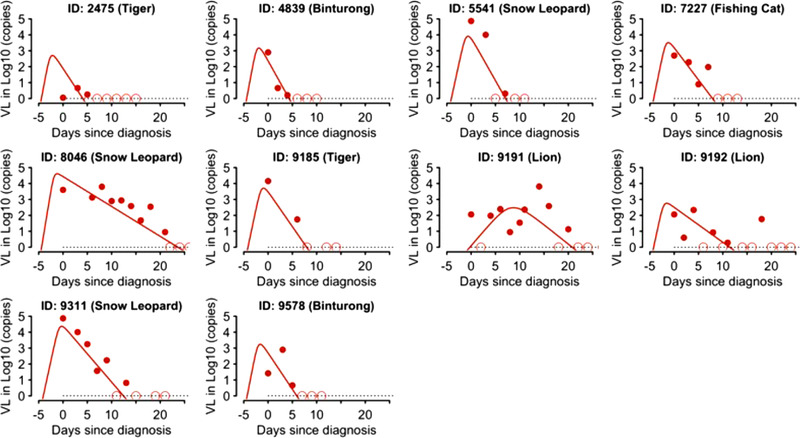
Individual host response in zoological animals with SARS‐CoV‐2. Fitting results of the viral dynamic model (lines) to data (dots) in 12 carnivores detected with SARS‐CoV‐2 at the Chicago Zoological Society's Brookfield Zoo in 2021. Open circles denote censored data (i.e. viral load below the limit of detection, black dashed lines)
4. DISCUSSION
Since the start of the SARS‐CoV‐2 pandemic, millions of human infections and deaths have been observed (World Health Organization, 2020). In addition to humans, SARS‐CoV‐2 has been detected in 17 different animal species of three different animal orders (i.e. Carnivora, Artiodactyla, and Primates) (do Vale et al., 2021) (Figure 9). To date, SARS‐CoV‐2 has been reported in 14 carnivore species of six families, including Felidae (cat, tiger, lion, snow leopard, cougar, lynx, and fishing cat), Viverridae (binturong), Hyaenidae (hyena), Canidae (dog), Mustelidae (mink, otter, and ferret), and Procyonidae (coati), whereas the virus has been reported in only two families of the order Artiodactyla (i.e. Cervidae and Hippopotamidae) and one family of the order Primate (i.e. Hominidae) (do Vale et al., 2021) (Figure 9; Hale et al., 2021; U.S. Department of Agriculture, 2022). The present study is the first report of SARS‐CoV‐2 in three new species: Prionailurus viverrinus (fishing cat), Arctictis binturong (binturong), and Nasua nasua (coati). These species represent three taxonomic families, two of which are new families (Viverridae and Procyonidae) to be reported with SARS‐CoV‐2.
FIGURE 9.
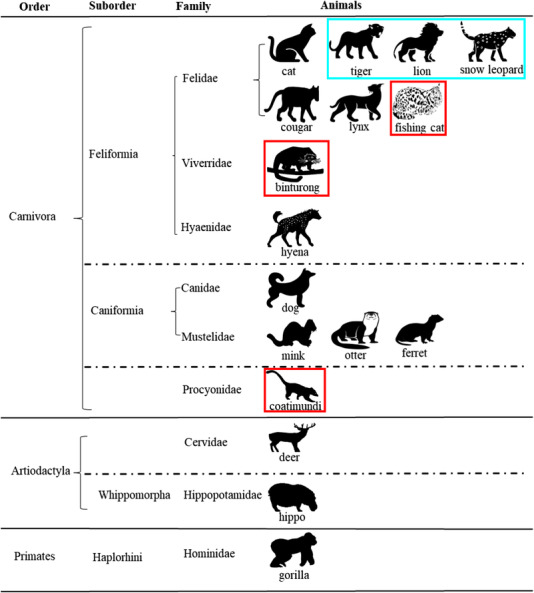
Current taxonomic diversity of wildlife detected with SARS‐CoV‐2. The diagram shows 17 animal species tested positive for SARS‐CoV‐2 within three animal orders Carnivora, Artiodactyla, and Primates. Information on order, suborder, and family of these animal species is listed. In the present study, three new animal species tested positive for SARS‐CoV‐2 and they are highlighted with a red colour frame; the other three animal species that tested positive are highlighted with an aqua colour frame.
Clinical and subclinical disease has been reported in humans (Tajbakhsh et al., 2021; Taleghani & Taghipour, 2020; Hu et al., 2021) and zoo‐maintained felids (McAloose et al., 2020; Mitchell et al., 2021). In the current report, clinical signs were rare and mild to moderate in all cases. Supportive care (anti‐nausea medications [n = 4], short‐term dexamethasone [n = 1]) was administered in four animals, but no treatments were used in the other eight positive animals. Animals with clinical signs shed virus longer than animals without clinical signs, which can be used to help guide appropriate duration of faecal monitoring during future outbreaks. A single female binturong was identified with SARS‐CoV‐2 with no clinical signs during the initial outbreak, but CT imaging showed evidence of chronic pulmonary disease 3 months after testing positive. Linear opacities within the lungs in humans are commonly observed with SARS‐CoV‐2 infections, but gradually resolve following a peak appearance of lesions at 10 days (Fields et al., 2021; Salehi et al., 2020). It is possible that the lesions observed on CT of the positive binturong were due to causes other than SARS‐CoV‐2 qRT‐PCR, but no other clinical features of pneumonia were observed in this animal. There is a paucity of imaging studies in zoo‐maintained species with SARS‐CoV‐2, but opportunities to image animals at various stages of infection may help to determine likelihood of acute or chronic diseases associated with SARS‐CoV‐2.
In the past 2 years, SARS‐CoV‐2 has continually evolved in humans and several variants have emerged. Since most animal SARS‐CoV‐2 cases were presumed to be due to human‐to‐animal transmission events (Ekstrand et al., 2021; Garigliany et al., 2020; Jo et al., 2020; McAloose et al., 2020; Miro et al., 2021; Prince et al., 2021; Racnik et al., 2021), SARS‐CoV‐2 strains detected in animals also are likely to change over time. Highlighting this reverse zoonosis, animal SARS‐CoV‐2 strains and variants have mirrored those within the human population. In 2020, non‐variant strains had been detected in tigers and lions at the Bronx Zoo (B lineage in lion and B.1 lineage in tigers) and in a tiger at a zoo in Tennessee (B.1.2 lineage) (Bartlett et al., 2021; Cushing et al., 2021). In contrast, in 2021 variants were detected in animals including the Alpha variant in domestic dogs and cats in the United Kingdom (Ferasin et al., 2021), tigers and lions at the Prague Zoo in the Czech Republic, and tigers at the Virginia Zoo (Mitchell et al., 2021) and the Delta variant in 14 animal species, including the current report (cat, dog, mink, gorilla, hyena, hippopotamus, ferret, tiger, lion, snow leopard, otter, binturong, fishing cat, and coatimundi) in Belgium, India, Japan, Netherlands, Poland, Singapore, South Africa, Thailand, and the United States (U.S. Department of Agriculture, 2022). In the United States, the Delta variant was detected in animals of multiple U.S. states (World Health Organization, 2020). Overall, these data suggest that the Delta variant has a higher transmissibility than other variants in animals, similar to transmission characteristics in humans. Since this outbreak occurred, the Omicron variant emerged and has shown increased transmissibility in humans, but no zoological cases have been confirmed.
In the present study, we report a SARS‐CoV‐2 outbreak in 12 zoo animals representing six animal species. Similar to previous studies, these zoo animals had variable shedding periods ranging from 2 to 29 days (Bartlett et al., 2021; Cushing et al., 2021; Fernandez‐Bellon et al., 2021). Many of the animals in the reported outbreak had no viral detection past 20 days, but viral RNA was detectable in some animals up to 53 days after the outbreak started. Faecal sampling is a non‐invasive detection method that has been used in humans (Zheng et al., 2020) and felids (Cushing et al., 2021). In humans, it has been observed that faecal shedding persists longer than oro/nasopharyngeal shedding (Wang et al., 2020; Xing et al., 2020; Zheng et al., 2020). In some human cases, faecal samples are positive even when oro/nasopharyngeal samples are not (Xing et al., 2020). Further studies have shown 100% agreement of faecal and oro/nasopharyngeal samples (Jeong et al., 2020). ACE2 receptors are positively expressed in enterocytes and thus it seems plausible that faecal detection is a sensitive method of diagnosis for systemic infections (Xiao et al., 2020). In humans, faecal samples peak 2–3 weeks after peak in oro/nasopharyngeal samples (Wang et al., 2020; Xing et al., 2020). If carnivores follow a similar pattern, then the original exposure in these cats occurred in early to mid‐September. A diagnostic advantage of qRT‐PCR is relative quantification, and CT (cycle threshold) values greater than 34 have been determined to be non‐contagious in humans (La Scola et al., 2020). If a similar pattern exists in carnivores, then many of the animals in this report had cleared the contagious stage of the infection by 8 October (∼14 days after first detection). A single lion had CT values below 30 during clinical disease, then >35 for several days, and finally <33 for several days through the 20 October. In humans, live virus has not been considered likely past day 8 based on CT values, but it seems that carnivores, and lions in particular, may be contagious for much longer (La Scola et al., 2020). Severity of infection has been linked to shedding intensity (CT) and duration in humans (Magleby et al., 2021; Zheng et al., 2020). Both lions, a female tiger, and a male snow leopard had the most significant clinical signs, but only the snow leopards were observed with statistically higher viral loads. Unfortunately, faecal surveillance was not performed prior to the outbreak, so the exact timing and intensity of the current outbreak is unknown, thus it is possible that the tiger and lions had higher viral loads prior to detection. This seems unlikely based on the timing of clinical signs but cannot be ruled out. Strategic surveillance in zoological institutions may be warranted to better characterize the epidemiology in susceptible animals.
Unlike other reported cases in zoos, all animals in the affected habitat had been vaccinated with only a single dose of a recombinant vaccine. It is possible that immune responses induced by the first dose might be not high enough to protect animals against SARS‐CoV‐2, but somewhat lessened the severity of clinical disease. Specifically, snow leopards have been observed with the greatest mortality across North American and European zoos (Allender, personal observation). In the outbreak discussed here, snow leopards had the highest viral load, but clinical signs were not severe enough to be life‐threatening. Following resolution of both clinical signs and qRT‐PCR detection, infected animals were vaccinated with a full two‐dose vaccine series, thus 12 individuals received three doses of the vaccine. Clinical signs during the outbreak were less severe and shorter in duration in both coati and binturong; it is unclear if this was due to differences in species susceptibility based on ACE2 receptor sequence (Ekstrand et al., 2021) or individual immune response due to either vaccine or exposure. In addition, several other species including sloth bears, meerkats, bat‐eared fox, and naked mole rats were near or adjacent to infected animals, but did not test positive for viral RNA nor show any clinical signs. The absence of detection in these species is important in understanding the epidemiology of this virus across taxa. It has been proposed that the greatest susceptibility is in primates, followed closely by carnivores and cetaceans based on ACE2 receptor similarity to humans (Martínez‐Hernández et al., 2020), thus it is not surprising carnivores were detected as SARS‐CoV‐2 positive during this outbreak, but somewhat interesting that carnivore members of the Ursidae family were not detected with the virus at any point. There are no primates in the affected building to help assess comparative susceptibility, but no primate at CZS has been detected with SARS‐CoV‐2 despite extensive sampling. All animals in the affected habitat were potentially exposed to virus because viral RNA was detected in two air handlers collecting circulating air from the building. Aerosolized SARS‐CoV‐2 has proven to be poorly recovered from hospitals, even when directly sampled in positive patient hospital rooms (Lane et al., 2021; Mallach et al., 2021). It is interesting to note that RNA was detected in the habitat only on the intake side of the filter and not on the outflow side, indicating that despite viral particles possibly making it to the air handler, it was not being recirculated. Air filters were not changed during the outbreak due to unknown human safety considerations, but resumed as normal in November 2021. In the 3 months following the outbreak, no animal tested positive despite twice weekly sampling. Human re‐infections have been documented as soon as 60 days after initial infection, but it is unknown if carnivores may get re‐infected or when the earliest possible time for reinfection would be. Additional surveillance is needed to identify intermittent or recurrent shedding of SARS‐CoV‐2 in carnivores.
The six animal SARS‐CoV‐2 strains in this study were closely related with 18 human strains (99.9% identity), with 4‐nucleotide differences at most. In fact, the sequence of the positive sample collected from a zoo animal care staff employee was genotyped as the Delta subvariant AY.25 lineage but not included in the analysis due to the low quality. Based on the timing of the keeper's work schedule, infection to or from the keeper seems unlikely. Overall, these data strongly suggest human‐to‐animal transmission events might have occurred in this outbreak. The public walkway of the enclosure building was open to the public at the start of the outbreak and thus the possibility that a member of the public transmitted the initial infection to a zoo animal cannot be dismissed. The only access that members of the public had to the first case (tiger) was outdoor viewing and transmission seems unlikely. It is possible that a member of the public transmitted the infection to an indoor animal and then animal‐to‐animal transmission events occurred throughout the building, but no genetic or epidemiologic evidence supports this because members of the public were not sampled at the zoo. The lack of regular testing of staff and animals prior to the outbreak precludes any definitive source determination.
In summary, our study is the first report of SARS‐CoV‐2 infection in three animal species (Prionailurus viverrinus, fishing cat; Arctictis binturong, binturong; and Nasua nasua, coati) as well as the first report in two families (Procyonidae and Viverridae) that occurred during an outbreak of the SARS‐CoV‐2 Delta variant among six species managed in a zoological institution in Illinois in 2021. Although these animals were partially immunized with COVID‐19 vaccine, it is difficult to evaluate the vaccine efficacy. Zoological institutions play key roles in conservation worldwide; and SARS‐CoV‐2 not only threatens individual health as identified in this report, but also negatively affects conservation efforts in the field (Fine et al., 2022). It is imperative to continue to use evidence‐based decision‐making to improve the welfare of zoo‐maintained and free‐living wildlife during the ongoing pandemic.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization: Matthew C. Allender, Michael J. Adkesson, and Leyi Wang. Methodology: Matthew C. Allender, Michael J. Adkesson, and Leyi Wang. Investigation: Matthew C. Allender, Michael J. Adkesson, Jennifer N. Langan, Katie W. Delk, Thomas Meehan, Copper Aitken‐Palmer, Michael M. McEntire, Mary L. Killian, Mia Torchetti, Shirley A. Morales, Connie Austin, Richard Fredrickson, Colleen Olmstead, Ruian Ke, Rebecca Smith, Eric T. Hostnik, Karen Terio, and Leyi Wang. Funding acquisition: Matthew C. Allender, Michael J. Adkesson, Richard Fredrickson, Karen Terio, and Leyi Wang. Project administration: Matthew C. Allender and Leyi Wang. Supervision: Matthew C. Allender, Richard Fredrickson, and Leyi Wang. Initial draft: Matthew C. Allender, Leyi Wang, Karen Terio, and Rebecca Smith. Review and revision: Matthew C. Allender, Michael J. Adkesson, Jennifer N. Langan, Katie W. Delk, Copper Aitken‐Palmer, Michael M. McEntire, Mary L. Killian, Mia Torchetti, Shirley A. Morales, Connie Austin, Richard Fredrickson, Ruian Ke, Rebecca Smith, Eric T. Hostnik, Karen Terio, and Leyi Wang.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required for this work because the information reported here involves standard laboratory analyses and routine care of zoo animals, which is governed by relevant federal and state laws that set standards of humane animal care and treatment, and monitor and achieve compliance with the U.S. Animal Welfare Act (7 USC §§ 2131–2159; 18 USC § 49) through inspections, education, and cooperative efforts.
ACKNOWLEDGEMENTS
The authors thank Bill Zeigler, Tim Sullivan, Sondi Dornhecker, and all animal care staff and veterinary technicians at the Chicago Zoological Society for their dedicated work in identifying, adapting, and implementing procedural changes during this outbreak. The findings and conclusions in this publication are those of the authors and should not be construed to represent any official U.S. Department of Agriculture or U.S. Government determination or policy. Funding for case investigation was provided by the Chicago Zoological Society, sequencing was financed by the University of Illinois Veterinary Diagnostic Laboratory, and support for molecular work performed at NVSL was provided by NVSL.
Allender, M. C. , Adkesson, M. J. , Langan, J. N. , Delk, K. W. , Meehan, T. , Aitken‐Palmer, C. , McEntire, M. M. , Killian, M. L. , Torchetti, M. , Morales, S. A. , Austin, C. , Fredrickson, R. , Olmstead, C. , Ke, R. , Smith, R. , Hostnik, E. T. , Terio, K. , & Wang, L. (2022). Multi‐species outbreak of SARS‐CoV‐2 Delta variant in a zoological institution, with the detection in two new families of carnivores. Transboundary and Emerging Diseases, 00, 1–16. 10.1111/tbed.14662
Contributor Information
Matthew C. Allender, Email: mcallend@illinois.edu.
Karen Terio, Email: kterio@illinois.edu.
Leyi Wang, Email: leyiwang@illinois.edu.
DATA AVAILABILITY STATEMENT
All patient identifiable data are available upon request. Data that specifically identify patients, their location, or disposition after the study concluded are unavailable due to safety and privacy concerns.
REFERENCES
- Bartlett, S. L. , Diel, D. G. , Wang, L. , Zec, S. , Laverack, M. , Martins, M. , Caserta, L. C. , Killian, M. L. , Terio, K. , Olmstead, C. , Delaney, M. A. , Stokol, T. , Ivancic, M. , Jenkins‐Moore, M. , Ingerman, K. , Teegan, T. , McCann, C. , Thomas, P. , McAloose, D. , … Calle, P. P. (2021). SARS‐CoV‐2 infection and longitudinal fecal screening in Malayan tigers (Panthera tigris jacksoni), Amur tigers (Panthera tigris altaica), and African lions (Panthera leo krugeri) at the Bronx Zoo, New York, USA. Journal of Zoo and Wildlife Medicine, 51, 733–744. 10.1638/2020-0171 [DOI] [PubMed] [Google Scholar]
- Calvet, G. A. , Pereira, S. A. , Ogrzewalska, M. , Pauvolid‐Correa, A. , Resende, P. C. , Tassinari, W. S. , Costa, A. P. , Keidel, L. O. , da Rocha, A. S. B. , da Silva, M. F. B. , Dos Santos, S. A. , Lima, A. B. M. , de Moraes, I. C. V. , Mendes Junior, A. A. V. , Souza, T. D. C. , Martins, E. B. , Ornellas, R. O. , Correa, M. L. , Antonio, I. , … Menezes, R. C. (2021). Investigation of SARS‐CoV‐2 infection in dogs and cats of humans diagnosed with COVID‐19 in Rio de Janeiro, Brazil. PLoS ONE, 16, e0250853. 10.1371/journal.pone.0250853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, J. C. , Bevins, S. N. , Ellis, J. W. , Linder, T. J. , Tell, R. M. , Jenkins‐Moore, M. , Root, J. J. , Lenoch, J. B. , Robbe‐Austerman, S. , DeLiberto, T. J. , Gidlewski, T. , Kim Torchetti, M. , & Shriner, S. A. (2021). SARS‐CoV‐2 exposure in wild white‐tailed deer (Odocoileus virginianus). Proceeding of the National Academy of Sciences of the United States of America, 118(47), e2114828118. 10.1073/pnas.2114828118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, M. E. , Terio, K. A. , & Keel, M. K. (2018). Procyonidae, viverridae, hyenidae, herpestidae, eupleridae and prionodontidae. In Terio K. A., McAloose D., & St. Leger J. (Eds.), Pathology of wildlife and zoo animals (pp. 305–322). Academic Press. 10.1016/B978-0-12-805306-5.00012-2 [DOI] [Google Scholar]
- Cushing, A. C. , Sawatzki, K. , Grome, H. N. , Puryear, W. B. , Kelly, N. , & Runstadler, J. (2021). Duration of antigen shedding and development of antibody titers in Malayan tigers (Panthera tigris jacksoni) naturally infected with SARS‐CoV‐2. Journal of Zoo and Wildlife Medicine, 52, 1224–1228. 10.1638/2021-0042 [DOI] [PubMed] [Google Scholar]
- Damas, J. , Hughes, G. M. , Keough, K. C. , Painter, C. A. , Persky, N. S. , Corbo, M. , Hiller, M. , Koepfli, K. P. , Pfenning, A. R. , Zhao, H. , Genereux, D. P. , Swofford, R. , Pollard, K. S. , Ryder, O. A. , Nweeia, M. T. , Lindblad‐Toh, K. , Teeling, E. C. , Karlsson, E. K. , & Lewin, H. A. (2020). Broad host range of SARS‐CoV‐2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proceedings of the National Academy of Sciences of the United States of America, 117(36), 22311–22322. 10.1073/pnas.2010146117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Vale, B. , Lopes, A. P. , Fontes, M. D. C. , Silvestre, M. , Cardoso, L. , & Coelho, A. C. (2021). Bats, pangolins, minks and other animals ‐ Villains or victims of SARS‐CoV‐2? Veterinary Research Communications, 45, 1–19. 10.1007/s11259-021-09787-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand, K. , Flanagan, A. J. , Lin, I. E. , Vejseli, B. , Cole, A. , Lally, A. P. , Morris, R. L. , & Morgan, K. N. (2021). Animal transmission of SARS‐CoV‐2 and the welfare of animals during the COVID‐19 pandemic. Animals, 11, 2044. 10.3390/ani11072044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe, S. , & Buckland‐Merrett, G. (2017). Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Challenges, 1, 33–46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenollar, F. , Mediannikov, O. , Maurin, M. , Devaux, C. , Colson, P. , Levasseur, A. , Fournier, P. E. , & Raoult, D. (2021). Mink, SARS‐CoV‐2, and the human‐animal interface. Frontiers in Microbiology, 12, 663815. 10.3389/fmicb.2021.663815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferasin, L. , Fritz, M. , Ferasin, H. , Becquart, P. , Corbet, S. , Ar Gouilh, M. , Legros, V. , & Leroy, E. M. (2021). Infection with SARS‐CoV‐2 variant B.1.1.7 detected in a group of dogs and cats with suspected myocarditis. Veterinary Record, 189(9), e944. 10.1002/vetr.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Bellon, H. , Rodon, J. , Fernandez‐Bastit, L. , Almagro, V. , Padilla‐Sole, P. , Lorca‐Oro, C. , Valle, R. , Roca, N. , Grazioli, S. , Trogu, T. , Bensaid, A. , Carrillo, J. , Izquierdo‐Useros, N. , Blanco, J. , Parera, M. , Noguera‐Julian, M. , Clotet, B. , Moreno, A. , Segales, J. , & Vergara‐Alert, J. (2021). Monitoring natural SARS‐CoV‐2 infection in lions (Panthera leo) at the Barcelona Zoo: Viral dynamics and host responses. Viruses, 13, 1683. 10.3390/v13091683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, B. K. K. , Demirjian, N. L. , Dadgar, H. , & Gholamrezanezhad, A. (2021). Imaging of COVID‐19: CT, MRI, and PET. Seminars in Nuclear Medicine, 51, 312–320. 10.1053/j.semnuclmed.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine, L. , Barnes, C. , Niedbalski, A. , & Deem, S. L. (2022). Staff perceptions of COVID‐19 impacts on wildlife conservation at a zoological institution. Zoo Biology, 41(3), 234–243. 10.1002/zoo.21669 [DOI] [PubMed] [Google Scholar]
- Forster, P. , Forster, L. , Renfrew, C. , & Forster, M. (2020). Phylogenetic network analysis of SARS‐CoV‐2 genomes. Proceedings of the National Academy of Sciences of the United States of America, 117, 9241–9243. 10.1073/pnas.2004999117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Sastre, A. , & Biron, C. A. (2006). Type 1 interferons and the virus‐host relationship: A lesson in détente. Science, 312, 879–882. 10.1126/science.1125676 [DOI] [PubMed] [Google Scholar]
- Garigliany, M. , Van Laere, A. S. , Clercx, C. , Giet, D. , Escriou, N. , Huon, C. , van der Werf, S. , Eloit, M. , & Desmecht, D. (2020). SARS‐CoV‐2 natural transmission from human to cat, Belgium, March 2020. Emerging Infectious Diseases, 26, 3069–3071. 10.3201/eid2612.202223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, V. L. , Dennis, P. M. , McBride, D. S. , Nolting, J. M. , Madden, C. , Huey, D. , Ehrlich, M. , Grieser, J. , Winston, J. , Lombardi, D. , Gibson, S. , Saif, L. , Killian, M. L. , Lantz, K. , Tell, R. , Torchetti, M. , Robbe‐Austerman, S. , Nelson, M. I. , Faith, S. A. , & Bowman, A. S. (2021). SARS‐CoV‐2 infection in free‐ranging white‐tailed deer. Nature, 602, 481–486. 10.1038/s41586-021-04353-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Guo, H. , Zhou, P. , & Shi, Z. L. (2021). Characteristics of SARS‐CoV‐2 and COVID‐19. Nature Reviews Microbiology, 19, 141–154. 10.1038/s41579-020-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairak, W. , Charoenkul, K. , Chamsai, E. , Udom, K. , Chaiyawong, S. , Bunpapong, N. , Boonyapisitsopa, S. , Tantilertcharoen, R. , Techakriengkrai, N. , Surachetpong, S. , Tangwangvivat, R. , Suwannakarn, K. , & Amonsin, A. (2021). First cases of SARS‐CoV‐2 infection in dogs and cats in Thailand. Transboundary and Emerging Diseases, 69, e979–e991. 10.1111/tbed.14383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H. W. , Kim, S.‐M. , Kim, H.‐S. , Kim, Y.‐I. , Hyoung Kim, J. , Cho, J. Y. , Kim, S.‐H. , Kang, H. , Kim, S.‐G. , Park, S.‐J. , Kim, E.‐H. , & Choi, Y. K. (2020). Viable SARS‐CoV‐2 in various specimens from COVID‐19 patients. Clinical Microbiology and Infection, 26, 1520–1524. 10.1016/j.cmi.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, W. K. , Oliveira‐Filho, E. F. , Rasche, A. , Greenwood, A. D. , Osterrieder, K. , & Drexler, J. F. (2020). Potential zoonotic sources of SARS‐CoV‐2 infections. Transboundary and Emerging Diseases, 68, 1824–1834. 10.1111/tbed.13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, R. , Martinez, P. P. , Smith, R. L. , Gibson, L. L. , Mirza, A. , Conte, M. , Gallagher, N. , Luo, C. H. , Jarrett, J. , Conte, A. , Liu, T. , Farjo, M. , Walden, K. K. O. , Rendon, G. , Fields, C. J. , Wang, L. , Fredrickson, R. , Edmonson, D. C. , Baughman, M. E. , … Brooke, C. B. (2021a). Daily sampling of early SARS‐CoV‐2 infection reveals substantial heterogeneity in infectiousness. medRxiv . 10.1101/2021.07.12.21260208 [DOI] [PMC free article] [PubMed]
- Ke, R. , Zitmann, C. , Ho, D. D. , Ribeiro, R. M. , & Perelson, A. S. (2021b). In vivo kinetics of SARS‐CoV‐2 infection and its relationship with a person's infectiousness. Proceedings of the National Academy of Sciences of the United States of America, 118, e2111477118. 10.1073/pnas.2111477118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola, B. , Le Bideau, M. , Andreani, J. , Hoang, V. T. , Grimaldier, C. , Colson, P. , Gautret, P. , & Raoult, D. (2020). Viral RNA load as determined by cell culture as a management tool for discharge of SARS‐CoV‐2 patients from infectious disease wards. European Journal of Clinical Microbiology and Infectious Diseases, 39, 1059–1061. 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, M. A. , Walawender, M. , Webster, A. S. , Brownsword, E. A. , Ingersoll, J. M. , Miller, C. , Waggoner, J. , Uyeki, T. M. , Lindsley, W. G. , & Kraft, C. S. (2021). Sampling for SARS‐CoV‐2 aerosols in hospital patient rooms. Viruses, 13, 2347. 10.3390/v13122347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lean, F. Z. X. , Nunez, A. , Spiro, S. , Priestnall, S. L. , Vreman, S. , Bailey, D. , James, J. , Wrigglesworth, E. , Suarez‐Bonnet, A. , Conceicao, C. , Thakur, N. , Byrne, A. M. P. , Ackroyd, S. , Delahay, R. J. , van der Poel, W. H. M. , Brown, I. H. , Fooks, A. R. , & Brookes, S. M. (2021). Differential susceptibility of SARS‐CoV‐2 in animals: Evidence of ACE2 host receptor distribution in companion animals, livestock, and wildlife by immunohistochemical charaterisation. Transboundary and Emerging Diseases, 69, 2275–2286. 10.1111/tbed.14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby, R. , Westblade, L. F. , Trzebucki, A. , Simon, M. S. , Rajan, M. , Park, J. , Goyal, P. , Safford, M. M. , & Satlin, M. J. (2021). Impact of severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clinical Infectious Diseases, 73, e4197–e4205. 10.1093/cid/ciaa851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallach, G. , Kasloff, S. B. , Kovesi, T. , Kumar, A. , Kulka, R. , Krishnan, J. , Robert, B. , McGuinty, M. , den Otter‐Moore, S. , Yazji, B. , & Cutts, T. (2021). Aerosol SARS‐CoV‐2 in hospitals and long‐term care homes during the COVID‐19 pandemic. PLoS ONE, 16, e0258151. 10.1371/journal.pone.0258151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Hernández, F. , Isaak‐Delgado, A. B. , Alfonso‐Toledo, J. A. , Munoz‐Garcia, C. I. , Villalobos, G. , Arechiga‐Ceballos, N. , & Rendon‐Franco, E. (2020). Assessing the SARS‐CoV‐2 threat to wildlife: Potential risk to a broad range of mammals. Perspectives in Ecology and Conservation, 18, 223–234. 10.1016/j.pecon.2020.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAloose, D. , Laverack, M. , Wang, L. , Killian, M. L. , Caserta, L. C. , Yuan, F. , Mitchell, P. K. , Queen, K. , Mauldin, M. R. , Cronk, B. D. , Bartlett, S. L. , Sykes, J. M. , Zec, S. , Stokol, T. , Ingerman, K. , Delaney, M. A. , Fredrickson, R. , Ivancic, M. , Jenkins‐Moore, M. , … Diel, D. G. (2020). From people to Panthera: Natural SARS‐CoV‐2 infection in tigers and lions at the Bronx Zoo. mBio, 11(5), e02220–20. 10.1128/mBio.02220-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miro, G. , Regidor‐Cerrillo, J. , Checa, R. , Diezma‐Diaz, C. , Montoya, A. , Garcia‐Cantalejo, J. , Botias, P. , Arroyo, J. , & Ortega‐Mora, L. M. (2021). SARS‐CoV‐2 infection in one cat and three dogs living in COVID‐19‐positive households in Madrid, Spain. Frontiers in Veterinary Science, 8, 779341. 10.3389/fvets.2021.779341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. K. , Martins, M. , Reily, T. , Caserta, L. C. , Anderson, R. R. , Cronk, B. D. , Murphy, J. , Goodrich, E. L. , & Diel, D. G. (2021). SARS‐CoV‐2 B.1.1.7 variant infection in Malayan tigers in Virginia, USA. Emerging Infectious Diseases, 27, 3171–3173. 10.3201/eid2712.211234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadian, M. , Chiti, H. , Shoghli, A. , Biglari, S. , Parsamanesh, N. , & Esmaeilzadeh, A. (2021). COVID‐19: Virology, biology and novel laboratory diagnosis. The Journal of Gene Medicine, 23, e3303. 10.1002/jgm.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova, N. , Molenaar, R. J. , Vreman, S. , Harders, F. , Oude Munnink, B. B. , Hakze‐van der Honing, R. W. , Gerhards, N. , Tolsma, P. , Bouwstra, R. , Sikkema, R. S. , Tacken, M. G. , de Rooij, M. M. , Weesendorp, E. , Engelsma, M. Y. , Bruschke, C. J. , Smit, L. A. , Koopmans, M. , van der Poel, W. H. , & Stegeman, A. (2020). SARS‐CoV‐2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance, 25, 2001005. 10.2807/1560-7917.ES.2020.25.23.2001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink, B. B. , Sikkema, R. S. , Nieuwenhuijse, D. F. , Molenaar, R. J. , Munger, E. , Molenkamp, R. , van der Spek, A. , Tolsma, P. , Rietveld, A. , Brouwer, M. , Bouwmeester‐Vincken, N. , Harders, F. , Hakze‐van der Honing, R. , Wegdam‐Blans, M. C. A. , Bouwstra, R. J. , GeurtsvanKessel, C. , van der Eijk, A. A. , Velkers, F. C. , Smit, L. A. M. , … Koopmans, M. P. G. (2021). Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science, 371, 172–177. 10.1126/science.abe5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, T. , Smith, S. L. , Radford, A. D. , Solomon, T. , Hughes, G. L. , & Patterson, E. I. (2021). SARS‐CoV‐2 infections in animals: Reservoirs for reverse zoonosis and models for study. Viruses, 13, 494. 10.3390/v13030494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racnik, J. , Kocevar, A. , Slavec, B. , Korva, M. , Rus, K. R. , Zakotnik, S. , Zorec, T. M. , Poljak, M. , Matko, M. , Rojs, O. Z. , & Zupanc, T. A. (2021). Transmission of SARS‐CoV‐2 from human to domestic ferret. Emerging Infectious Diseases, 27, 2450–2453. 10.3201/eid2709.210774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi, S. , Abedi, A. , Balakrishnan, S. , & Gholamrezanezhad, A. (2020). Coronavirus disease 2019 (COVID‐19): A systematic review of imaging findings in 919 patients. American Journal of Roentgenology, 215, 87–93. 10.2214/ajr.20.23034 [DOI] [PubMed] [Google Scholar]
- Sharun, K. , Tiwari, R. , Natesan, S. , & Dhama, K. (2021). SARS‐CoV‐2 infection in farmed minks, associated zoonotic concerns, and importance of the One Health approach during the ongoing COVID‐19 pandemic. The Veterinary Quarterly, 41, 50–60. 10.1080/01652176.2020.1867776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiehzadegan, S. , Alaghemand, N. , Fox, M. , & Venketaraman, V. (2021). Analysis of the delta variant B.1.617.2 COVID‐19. Clinics and Practice, 11, 778–784. 10.3390/clinpract11040093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit, T. H. C. , Brackman, C. J. , Ip, S. M. , Tam, K. W. S. , Law, P. Y. T. , To, E. M. W. , Yu, V. Y. T. , Sims, L. D. , Tsang, D. N. C. , Chu, D. K. W. , Perera, R. , Poon, L. L. M. , & Peiris, M. (2020). Infection of dogs with SARS‐CoV‐2. Nature, 586, 776–778. 10.1038/s41586-020-2334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh, A. , Gheibi Hayat, S. M. , Taghizadeh, H. , Akbari, A. , Inabadi, M. , Savardashtaki, A. , Johnston, T. P. , & Sahebkar, A. (2021). COVID‐19 and cardiac injury: Clinical manifestations, biomarkers, mechanisms, diagnosis, treatment, and follow up. Expert Review of Anti‐Infective Therapy, 19, 345–357. 10.1080/14787210.2020.1822737 [DOI] [PubMed] [Google Scholar]
- Taleghani, N. , & Taghipour, F. (2020). Diagnosis of COVID‐19 for controlling the pandemic: A review of the state‐of‐the‐art. Biosensors and Bioelectronics, 15(174), 112830. 10.1016/j.bios.2020.112830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, M. , Singh, A. , Joshi, B. D. , Ghosh, A. , Singh, S. K. , Singh, N. , Sharma, L. K. , & Chandra, K. (2020). Time‐lapse sentinel surveillance of SARS‐CoV‐2 spread in India. PLoS ONE, 15, e0241172. 10.1371/journal.pone.0241172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U. S. Department of Agriculture . (2022). Confirmed cases of SARS‐CoV‐2 in animals in the United States . https://www.aphis.usda.gov/aphis/dashboards/tableau/sars‐dashboard
- Wang, L. , Mitchell, P. K. , Calle, P. P. , Bartlett, S. L. , McAloose, D. , Killian, M. L. , Yuan, F. , Fang, Y. , Goodman, L. B. , Fredrickson, R. , Elvinger, F. , Terio, K. , Franzen, K. , Stuber, T. , Diel, D. G. , & Torchetti, M. K. (2020). Complete genome sequence of SARS‐CoV‐2 in a tiger from a U.S. zoological collection. Microbiology Resource Announcements, 9(22), e00468–20. 10.1128/mra.00468-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Muller, M. A. , Niemeyer, D. , Jones, T. C. , Vollmar, P. , Rother, C. , Hoelscher, M. , Bleicker, T. , Brunink, S. , Schneider, J. , Ehmann, R. , Zwirglmaier, K. , Drosten, C. , & Wendtner, C. (2020). Virological assessment of hospitalized patients with COVID‐2019. Nature, 581, 465–469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2020). Coronavirus disease (COVID‐19) dashboard 2020 . https://covid19.who.int/
- Xiao, F. , Tang, M. , Zheng, X. , Liu, Y. , Li, X. , & Shan, H. (2020). Evidence for gastrointestinal infection of SARS‐CoV‐2. Gastroenterology, 158, 1831–1833. 10.1053/j.gastro.2020.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y.‐H. , Ni, W. , Wu, Q. , Li, W.‐J. , Li, G.‐J. , Wang, W.‐D. , Tong, J.‐N. , Song, X.‐F. , Wong, G. W.‐K. , & Xing, Q.‐S. (2020). Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. Journal of Microbiology, Immunology and Infection, 53, 4747–480. 10.1016/j.jmii.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , & Jeffrey, S. (2021). COVID‐19 pandemic dynamics in India and impact of the SARS‐CoV‐2 Delta (B.1.617.2) variant. medRxiv . 10.1101/2021.06.21.21259268 [DOI]
- Zhang, Y. Z. , & Holmes, E. C. (2020). A genomic perspective on the origin and emergence of SARS‐CoV‐2. Cell, 181, 223–227. 10.1016/j.cell.2020.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S. , Fan, S. J. , Yu, E. , Feng, B. , Lou, B. , Zou, Q. , Xie, G. , Lin, S. , Wang, R. , Wang, Q. , Zhang, D. , Liu, Y. , Gong, R. , Ma, Z. , Lu, A. , Xiao, Y. , Gu, Y. , Zhang, J. , Yao, H. , … Liang, T. (2020). Viral load dynamics and disease severity in patients infected with SARS‐CoV‐2 in Zhejiang province, China, January‐March 2020: Retrospective cohort study. BMJ, 369, m1443. 10.1136/bmj.m1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C. L. , Chen, H. D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R. D. , Liu, M. Q. , Chen, Y. , Shen, X. R. , Wang, X. , … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All patient identifiable data are available upon request. Data that specifically identify patients, their location, or disposition after the study concluded are unavailable due to safety and privacy concerns.


