Abstract
Thirty-two plasmid insertion mutants were independently isolated from two strains of Xanthomonas campestris pv. campestris in Taiwan. Of the 32 mutants, 14 (44%), 8 (25%), and 4 (12%) mutants resulted from separate insertions of an IS3 family member, IS476, and two new insertion sequences (IS), IS1478 and IS1479. While IS1478 does not have significant sequence homology with any IS elements in the EMBL/GenBank/DDBJ database, IS1479 demonstrated 73% sequence homology with IS1051 in X. campestris pv. dieffenbachiae, 62% homology with IS52 in Pseudomonas syringae pv. glycinea, and 60% homology with IS5 in Escherichia coli. Based on the predicted transposase sequences as well as the terminal nucleotide sequences, IS1478 by itself constitutes a new subfamily of the widespread IS5 family, whereas IS1479, along with IS1051, IS52, and IS5, belongs to the IS5 subfamily of the IS5 family. All but one of the IS476 insertions had duplications of 4 bp at the target sites without sequence preference and were randomly distributed. An IS476 insertion carried a duplication of 952 bp at the target site. A model for generating these long direct repeats is proposed. Insertions of IS1478 and IS1479, on the other hand, were not random, and IS1478 and IS1479 each showed conservation of PyPuNTTA and PyTAPu sequences (Py is a pyrimidine, Pu is a purine, and N is any nucleotide) for duplications at the target sites. The results of Southern blot hybridization analysis indicated that multiple copies of IS476, IS1478, and IS1479 are present in the genomes of all seven X. campestris pv. campestris strains tested and several X. campestris pathovars.
Insertion sequences (IS) are mobile DNA elements capable of mediating various types of DNA rearrangements such as transposition, deletion, inversion, and cointegration. They are usually 0.8 to 2.5 kb long and encode a transposase protein. Most IS carry 10- to 40-bp inverted repeats (IR) at their ends and generate direct repeats of short target sequences upon insertion (7). To date, more than 500 IS elements have been isolated from both eubacteria and archaea. Except for those highly similar variants from the same or related hosts, IS elements are considered heterogeneous at the nucleotide sequence level. Many can be grouped into families on the basis of conservation of motifs in their presumptive transposase amino acid sequences and their terminal nucleotide sequences. The IS3 and IS5 families are the two largest families (14).
Members of the IS3 family contain two overlapping open reading frames (ORFs), and the transposase protein is generated by programmed ribosomal frameshifting between the two ORFs. The transposase C-terminal region contains the characteristic DD(35)E motif, i.e., conservation of the acidic amino acid triad with several additional residues and 35 residues between the last two conserved acidic residues, D and E. Most members carry the dinucleotide TG at the 5′ ends (14). It has been suggested that the acidic amino acid triad interacts with the terminal 2 or 3 bp of the element to correctly position the IS ends in the catalytic site during transposition (10, 14). The IS3 family can be divided into the subgroups IS407, IS2, IS3, IS51, and IS150 on the basis of alignment of the transposase sequences (14). The IS5 family, on the other hand, is relatively heterogeneous. Either the members contain two ORFs and produce transposase by frameshifting, like the IS3 family members, or they contain a long ORF which covers most of the length of one strand and encodes transposase. In any case, their transposases carry another type of DDE motif, i.e., a spacing of 71 to 76 residues between the first two conserved acidic residues and a spacing of 40 to 67 residues between the last two acidic residues, in addition to conservation of D(1)GY in the region containing the second acidic residue and conservation of R(3)E(6)K in the region containing the third acidic residue (14, 18). All members carry the nucleotide G at the 5′ end. The IS5 family can be divided into the subgroups IS5, IS427, IS903, IS1031, ISH1, and ISL2 on the basis of alignment of the transposase sequences, particularly the residues near the three conserved acidic residues (14).
Xanthomonas campestris is a plant-pathogenic bacterium, consisting of more than 125 pathovars based on host specificity (26). Kearney et al. (11) isolated a mutant strain of X. campestris pv. vesicatoria from a pepper field and demonstrated that the mutant had an insertion of IS476 in the avr locus, leading to virulence on an otherwise resistant pepper cultivar. This implies that the bacteria may extend the host range to include plants previously resistant through transposition. In order to investigate the role that IS elements play in the diversity of this species, first we systemically analyzed IS elements from an important pathovar, X. campestris pv. campestris, the agent causing crucifer black rot. A plasmid system developed by Gay et al. (8) was used to isolate IS elements in bacteria. In this system, the broad-host-range plasmid pUCD800 carrying the sucrose-sensitive Bacillus subtilis sacB gene with its cis-regulatory sequence, sacR was transferred into bacteria, and plasmid mutants with an insertion of chromosomal IS element in the sacRB gene were identified from survivors on agar plates containing 5% sucrose. We report here the successful isolation of 32 plasmid insertion mutants of X. campestris pv. campestris by using this system. Twenty-six (81%) were characterized and had insertions of IS476 and two newly identified IS elements, IS1478 and IS1479. Their insertion specificity and distribution in related bacteria are presented.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Oligonucleotides were obtained from the Regional Instruments Center at National Chung Hsing University and Gibco BRL. Bacteria were grown in Luria broth (LB) (19) at 37°C (for Escherichia coli) or 28°C (for other bacteria). To select for sucrose-sensitive mutants, a modified TYG medium (4) was used, in which 5% glucose was substituted for 5% sucrose. Kanamycin or ampicillin at 50 μg per ml was added to the medium to avoid segregation of the plasmids.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or sourcea |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−hsdR17 recA1 supE44 endA1 gyrA96 relA1 Δ(argF-lacZYA)U169 thi-1 φ80d lacZΔM15 λ− | BRL |
| E. coli HB101 | F−hsdR20 ara-14 leuB6 proA2 recA13 lacY1 galK2 rpsL20 xyl-5 thi-1 mtl-1 supE44 λ | 2 |
| E. coli RR1 | F−hsdR20 ara-14 leuB6 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44 λ | 1 |
| X. campestris pv. campestris 11 (Xc11) | Apr, Taiwanese strain | Y.-H. Tseng |
| X. campestris pv. campestris 11A | Apr, a spontaneous avirulent mutant of Xc11 | Y.-H. Tseng |
| X. campestris pv. campestris 17 | Apr, Taiwanese strain | Y.-H. Tseng |
| X. campestris pv. campestris 2 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. campestris 6 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. campestris 85 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. campestris 88 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. begoniae | ATCC 49082 | ATCC |
| X. campestris pv. citri 60 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. dieffenbachiae 65 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. glycinea 69 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. mangiferaeindicae 38 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. phaseoli 73 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. vesicatoria 64 | Taiwanese strain | K.-C. Tzeng |
| X. campestris pv. oryzae 1 | Taiwanese strain | K.-C. Tzeng |
| Agrobacterium tumefaciens LBA4404 | Strr | Clontech |
| Erwinia carotovora subsp. carotovora ZL4 | Taiwanese strain | K.-C. Tzeng |
| Pseudomonas solanacearum RD4 | Taiwanese strain | K.-C. Tzeng |
| Rhizobium leguminosarum 128C53 | str-29, harboring a 200-kb, Kmr plasmid pVW3JI | CCRC |
| Plasmids | ||
| pUCD800 | Kmr; carrying sucrose-sensitive sacRB | 8 |
| pRK2013 | Kmr; mobilization helper plasmid | 5 |
| pUC18 | Apr; α-lacZ/MCSb or cloning vector | 28 |
BRL, Bethesda Research Laboratories; ATCC, American Type Culture Collection; CCRC, Culture Collection and Research Center, Food Industry Research and Development Institute, Hsinchu, Taiwan.
α-lacZ/MCS, DNA encoding the α-fragment of β-galactosidase (the lacZ gene product) with a multiple cloning site.
Triparental mating.
Plasmid pUCD800 was transferred into X. campestris pv. campestris 11 (Xc11) and 17 (Xc17) by conjugation with pUCD800-containing E. coli DH5α (donor), promoted by pRK2013-containing E. coli HB101 (helper). Mating was performed on solid medium as outlined by Simon (23) with slight modifications. Briefly, about 108 mid-log-phase cells each of the donor and helper cells were mixed with 109 mid-log-phase cells of Xc11 or Xc17 on a membrane disk on a nonselective LB plate. The plate was incubated at 28°C for 24 h, and bacterial cells on the disk were resuspended in LB and plated on medium containing ampicillin and kanamycin in order to select the transconjugants.
Genome and plasmid DNA extraction.
The alkaline lysis method described by Sambrook et al. (19) was used for plasmid extraction. For extraction of the total cellular DNA, bacterial cells were gently lysed by sodium dodecyl sulfate (SDS)-proteinase K treatment followed by the phenol-chloroform extraction and ethanol precipitation methods described by Scordilis et al. (22). Since Xanthomonas cells secrete exopolysaccharides which will coprecipitate with the DNA, Xanthomonas cells were washed with buffer consisting of 10 mM Tris-HCl, pH 7.6, and 1 M NaCl prior to DNA extraction.
DNA manipulation and Southern hybridization.
Restriction enzyme digestions were performed according to the instructions provided by the suppliers. Cloning, PCR, plasmid transformation, and random primer labeling were performed by the methods described by Sambrook et al. (19). Southern hybridization was performed with Hybond N membranes (Amersham), using the protocols provided by the manufacturer. After hybridization with the labeled probe, the nylon membrane was washed under high-stringency conditions, namely, two 30-min washes with 2× SSC–0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature, followed by one 30-min wash with 0.5× SSC–0.1% SDS at room temperature and one 30-min wash with 0.1× SSC–0.1% SDS at 50°C.
DNA sequencing.
Double-strand DNA sequencing was performed on both strands of the plasmid DNA template by the dideoxy-chain termination method (20) using Sequenase version 2.0 DNA sequencing kit and 35S-dATP (Amersham). When a long sequence reading was preferred, the DNA fragment was cloned into plasmid pUC18 before sequencing. For resolving G-C compressions, dITP was used according to the instructions provided by Amersham.
Nucleotide sequence accession number.
The nucleotide sequences of IS476B, IS1478A, IS1479A, and IS1479B have been deposited in GenBank under accession nos. U62552, U59749, U56973, and U56974, respectively.
RESULTS AND DISCUSSION
Isolation and grouping of 32 independent plasmid insertion mutants.
Plasmid pUCD800 was successfully transferred into two strains of X. campestris pv. campestris, Xc11 and Xc17, by conjugation promoted by triparental mating with pRK2013. Four independent subcultures of pUCD800-containing Xc17 and three independent subcultures of pUCD800-containing Xc11 were collected, and dilution plating was performed on agar plates containing 5% sucrose. For each subculture, one plate with about 40 survivors was used to screen for plasmid insertion mutants. Since plasmid DNA extracted from Xanthomonas cells is generally refractory to enzymatic digestion, plasmid DNA of each survivor was extracted, transformed into E. coli DH5α, and extracted again from the transformant for restriction mapping with BamHI and PstI. The restriction mapping results showed that about one-third of the survivors were plasmid insertion mutants, all of which had insertions in the 2.6-kb BamHI-PstI sacRB fragment. One mutant (Xc17-47) had an additional 1.4-kb insertion in the plasmid other than that in the 2.6-kb BamHI-PstI sacRB region. These mutant plasmid DNAs were further mapped with DraI, EcoRI, EcoRV, HindIII, KpnI, and SmaI, and their restriction patterns were compared. For each subculture, one insertion mutant of the same plasmid restriction pattern was collected. In this way, we collected a total of 32 independent plasmid insertion mutants. The restriction patterns of the 32 mutant plasmids were compared with that of pUCD800 (8, 24), and information about the insertion fragments, such as their locations within sacRB, their sizes, as well as restriction enzyme sites carried by the insertion fragments, was obtained and is shown in Table 2. Twenty-seven of the 32 mutants could be divided into four groups, group I to IV, with 13, 8, 4, and 2 members, respectively, on the basis of their restriction enzyme sites and size of insertion.
TABLE 2.
Characteristics of the 32 independent plasmid insertion mutants
| Mutant | Isolated from: | Size (kb) of insertion | No. of restriction enzyme sitesa in the insertion
|
Group | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B | D | EI | EV | H | K | P | S | ||||
| Xc11-12 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc11-8 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc11-6 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc11-10 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc11-14 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc11-17 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc17-2 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc17-48 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc17-6 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc17-47 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| 1.4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| Xc17-49 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc17-51 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc17-3 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | I |
| Xc11-1 | Xc11 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc11-2 | Xc11 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc11-13 | Xc11 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc17-13 | Xc17 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc17-15 | Xc17 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc17-25 | Xc17 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc17-14 | Xc17 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc17-7 | Xc17 | 1.5 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | II |
| Xc11-25 | Xc11 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | III |
| Xc17-27 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | III |
| Xc17-5 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | III |
| Xc17-4 | Xc17 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | III |
| Xc11-7 | Xc11 | 1.3 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | IV |
| Xc17-35 | Xc17 | 1.3 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | IV |
| Xc11-5 | Xc11 | 0.8 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| Xc17-17 | Xc17 | 0.8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Xc11-15 | Xc11 | 1.2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Xc11-11 | Xc11 | 2.1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | |
| Xc17-1 | Xc17 | 7.0 | 1 | 0 | 1 | 2 | 0 | 0 | 5 | 2 | |
Restriction enzyme sites abbreviations: B, BamHI; D, DraI; EI, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; P, PstI; S, SmaI.
The 13 group I mutants and mutant Xc11-11 had insertions of IS476 in their plasmids.
Two mutants from each of the group I, II, and III mutants were randomly picked for complete sequence determination of the insertion fragments. The mutants selected included mutants Xc11-12 and Xc17-3 from the group I mutants, Xc11-2 and Xc11-13 from the group II mutants, and Xc17-4 and Xc17-5 from the group III mutants (Table 2). Based on the location of the insertion in the 2.6-kb BamHI-PstI sacRB DNA fragment and the nucleotide sequence of the sacRB fragment (24), for each insertion fragment to be sequenced, one pair of 16-mer oligonucleotides identical to the sacRB sequence close to the insertion site was synthesized and used as primers to sequence from the neighboring sacRB region into the insertion fragment. Oligonucleotide primers identical to the end sequences of the readings were again synthesized, and sequencings were performed to read further inward into the insertion fragments, and so forth. The complete sequences of the six insertion fragments could thus be obtained.
The sequencing results indicated that both group I insertion fragments were 1,226 bp long and flanked by 4-bp direct repeats of the target sequences. The sequence of the insertion fragment in mutant Xc11-12 was identical to that of the IS476 sequence (accession no. M28557; 12, 16), whereas the sequence in mutant Xc17-3 had differences from that of the IS476 sequence at three positions, i.e., G466C, C467G, and C1051G substitutions. We named the former IS476A, and we named the latter IS476B. IS476A (IS476) was originally isolated in X. campestris pv. vesicatoria and was characterized as an IS407 group member of the widespread IS3 family (14, 16). IS476B, carrying the three base substitutions and thus changes of A118R and H312D in its presumptive transposase sequence, belongs to the IS407 group as well.
A 1.2-kb BspMI fragment comprising 97% of IS476A sequence was excised from the Xc17-3 mutant plasmid and used as a probe for Southern hybridization with the 32 mutant plasmids. The result showed that the 13 group I and Xc11-11 mutant plasmids hybridized with the probe (data not shown). Note that the Xc11-11 insertion fragment was 0.9 kb longer than the 13 group I insertion fragments and carried one additional EcoRI site and one additional DraI site (Table 2). Two 15-mer primers identical to the IS476A sequence from bp 102 to 88 and from bp 1096 to 1110 were synthesized, and the insertion junction sequences in the 13 group I and the Xc11-11 mutant plasmids were determined by sequencing outward into sacRB with the two primers. The results showed that all 13 group I insertion fragments had about 100 bp of the IS476A(B) (either IS476A or IS476B) terminal sequences and were flanked by duplications of 4-bp target sequences. The Xc11-11 insertion fragment had about 100 bp of the terminal IS476A(B) sequences but was not flanked by a 4-bp target site duplication (Fig. 1). We concluded that the 13 group I mutants and mutant Xc11-11 all had insertions of IS476 variants in their plasmids. The facts that X. campestris pv. campestris has at least two IS476 variants and the sequence of one is identical to that of an IS476 copy derived from X. campestris pv. vesicatoria suggest that horizontal transfer occurred between the two bacteria in recent years.
FIG. 1.
Insertion junction sequences in the group I, II, III, and Xc11-11 mutant plasmids. Only about 30 bp of the sacRB sequences flanking the insertions is listed. For the group I and the Xc11-11 mutant plasmids, arrows indicate insertions of IS476 variants and their orientations. For the group II and group III mutant plasmids, arrows indicate insertions of IS1478 and IS1479 variants separately and their orientations. Numbers to the left of the arrows indicate the sacRB coordinates of the bases 5′ to the insertions. For the Xc11-11 mutant plasmid, the number to the right of the arrow indicates the sacRB coordinate of the base 3′ to the insertion. Duplicated target sequences are indicated by capital letters. Bases in the flanking sacRB sequences that are conserved within the same group of mutant plasmids are in bold type.
The 13 group I mutants carried 4-bp target site duplications at the insertion junctions without sequence preference, whereas mutant Xc11-11 carried a duplication of 952 bp at the insertion junction.
The insertion junction sequences in the 13 group I mutants were aligned. As shown in Fig. 1, there was no sequence conservation or preference in either the 4-bp target sequences or the sacRB sequences upstream and downstream from the insertion sites. The 13 insertions were randomly distributed in sacRB in both orientations at about equal frequencies. In addition to IS476, seven IS407 group members of the IS3 family were reported, i.e., IS407 in Pseudomonas cepacia, IS511 in Caulobacter crescentus, IS1222 in Enterobacter agglomerans, IS1400 in Yersinia enterocolitica, ISD1 in Desulfovibrio vulgaris, ISRm6 in Rhizobium meliloti, and ISR1 in Rhizobium lupini (6, 14). Their 4-bp target duplications were GGAT (for IS407) (27), GCGG (for IS511) (15), TTTC (for IS1400) (3), AAGG and AATT (for ISD1) (6), CCCA (for ISRm6) (29), and TGCC (for ISR1) (17). (Target duplication for IS1222 was not detected.) It seemed that these six IS407 group members had at least two successive identical bases within the 4-bp target sequences. IS476 is distinct from them in that it did not have this or any type of specificity in its 4-bp target sequence. That the IS476 insertion detected in X. campestris pv. vesicatoria had the target duplication of GATG further supports our observation (11).
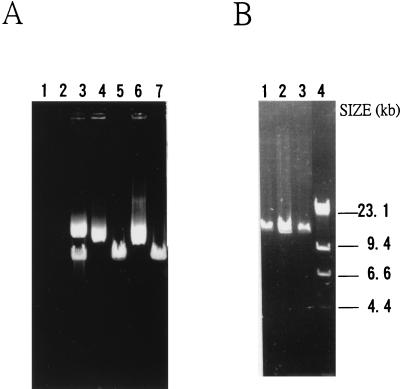
The Xc11-11 mutant plasmid carrying an insertion of IS476 without obvious target sequence duplication was further examined through detailed restriction mapping and sequencing with primers identical to several regions in sacRB. The result indicated that mutant Xc11-11 actually had a duplication of 952 bp (from sacBR coordinates 513 to 1464) at the insertion site. The 952-bp region contains one DraI site and one EcoRI site, resulting in an extra DraI site and an extra EcoRI site in the Xc11-11 insertion fragment compared with the mapping results of the group I mutant plasmids (Table 2 and Fig. 1). To further study this, plasmid pUCD800 DNAs were extracted from Xc11 and Xc17 and E. coli DH5α, HB101 (both recA), and RR1 (recA+) backgrounds and analyzed by agarose gel electrophoresis. As shown in Fig. 2A, plasmid preparations from Xc17 and E. coli RR1 contained only the large species, while preparations from E. coli DH5α and HB101 contained only the small species. Interestingly, both species were present in about equal amounts in the Xc11 preparation. The DNA preparations from the three E. coli backgrounds were digested with EcoRI, which cut pUCD800 once and analyzed by agarose gel electrophoresis. As shown in Fig. 2B, all three preparations demonstrated DNA fragments of the size of pUCD800 (14.5 kb), indicating that the small species was the monomer and the large species was the dimer (or multimer). A two-step mechanism for generation of Xc11-11 mutant plasmid is thus proposed. Insertion of IS476 occurred first in the pUCD800 dimer molecule in Xc11 at coordinate 1464 in one sacRB copy, and an IS-mediated adjacent deletion occurred later from one end of IS476 to coordinate 512 in the other sacRB copy.
FIG. 2.
(A) Agarose gel electrophoresis of pUCD800 DNAs isolated from Xc11, Xc17, E. coli DH5α, E. coli RR1, and E. coli HB101 (lanes 3 to 7). Endogenous plasmid preparations of Xc11 (lane 1) and Xc17 (lane 2) were electrophoresed simultaneously. (B) Agarose gel electrophoresis of EcoRI-digested pUCD800 DNAs isolated from E. coli DH5α, E. coli RR1, and E. coli HB101 (lanes 1 to 3). The HindIII digests of λ DNA (lane 4) were used as size markers, and the sizes of the resulting fragments are indicated to the right of the gel.
The eight group II mutants had insertions of IS1478, which itself constitutes a new subgroup of the IS5 family.
The two insertion fragments in the two randomly picked group II mutants, Xc11-2 and Xc11-13, displayed identical 1,506-bp nucleotide sequence. Both carry perfect inverted repeats of 20 bp at their ends and were flanked by 6-bp direct target repeats. A Blast search in the EMBL/GenBank/DDBJ database did not reveal significant nucleotide sequence similarity with any known IS element. This newly identified insertion sequence was named IS1478A, as we recently found a chromosomal copy carrying at least one base substitution in the sequence, which was named IS1478B (unpublished data).
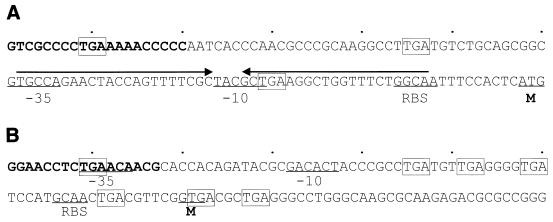
IS1478A contains a long ORF, from ATG at bp 118 to TGA at bp 1485, presumably encoding a transposase protein of 455 amino acids. A ς70-like promoter (bp 61 to 89) and a ribosome binding site (RBS) (bp 104 to 108) with an IR (bp 62 to 107) inbetween were found upstream of the ORF. Only in the case of transcription from upstream into the IS could the mRNA form a stable stem-loop structure (ΔG = −29.2 kcal/mol [25]). Located within the stem structure, the RBS could then be obscured, perturbing translation of the transposase. This would be a means of preventing transposition of an IS from external activation, as postulated for IS10 and IS150 (13, 21). Figure 3A shows the first 120-bp IS1478A sequence with these sequence elements indicated.
FIG. 3.
The first 120-bp nucleotide sequences of IS1478A (A) and IS1479A (B). The left terminal IR sequences are in bold type. Predicted RBS, the −35 and −10 regions of ς70-like promoters, and the methionine (M) start codons for translation into the cognate transposase proteins are indicated. Imperfect IRs are indicated by arrows over the sequence, and TGA stop codons are boxed.
The predicted IS1478A transposase sequence, which is shown in Fig. 4A, carries the DDE motif characteristic of the IS5 family, such as conservation of D(1)GY in the region containing the second conserved acidic residue, conservation of R(3)E(6)K in the region containing the third acidic residue, and 36 residues between the second and third conserved acidic residues (14, 18). The 5′ terminal G nucleotide characteristic of the IS5 family members is conserved in IS1478A (14). Therefore, IS1478A belongs to the IS5 family. A BLAST search with nonredundant protein database revealed only low level of resemblance to the IS5 transposase. IS1478A is distinct from all other IS5 family members in its terminal 3-bp sequence (5′-GTC-3′), size of direct target repeats (6 bp), and spacing between the first two conserved acidic residues in its transposase DDE motif (192 residues) and cannot be grouped with any of the five existing subgroups (14). A new IS1478 subgroup of the IS5 family is thus proposed. In fact, IS1478A is distinct from all reported DDE-type IS elements in that the spacing between the first two conserved acidic residues (192 residues) in the transposase DDE motif is about triple the sizes of the others (51 to 85 residues) (14).
FIG. 4.
(A) Alignment of the transposase amino acid sequences of IS1478A, IS5, and IS1479A. Amino acids are indicated in the single-letter code. Shaded amino acids are identical or homologous amino acids in two of the three sequences. Shaded and boxed amino acids are conserved in the three sequences. Amino acids conserved in the D(1)GY and R(3)E(6)K signatures for the IS5 family members (18) are indicated by plus signs above the sequence. Consensus sequence motifs for the IS5 subgroup (14) are indicated under the three sequences, in which capital letters indicate conservation and lowercase letters indicate predominance of that particular amino acid. The three conserved acidic residues of the DDE motif are shown as large bold letters. Note that the third conserved acidic residue, E, in IS5 (and IS1479A) is shown at different positions according to the two reports (14, 18). Homologous amino acids are grouped as follows: I, L, V, and M; F, Y, and W; H, K, and R; E and D; N and Q; G and A; S and T; C; and P (18). Gaps introduced in the sequences to maximize the alignment are indicated by the dashes. (B) Alignment of the left and right IR (IRL and IRR, respectively) sequences of IS1478A, IS5, and IS1479A. Nucleotides identical in two of the three elements are shaded. Nucleotides conserved in the three elements are shaded and boxed. Gaps introduced in the sequences to maximize the alignment are indicated by the dashes.
Transposase sequences and the terminal IR sequences from IS5 and IS1478A were aligned, and the results are shown in Fig. 4A and B, respectively. Those sequences from IS1479A, which belongs to the IS5 subgroup of the IS5 family (see below), were aligned for comparison. The alignments clearly showed that limited but significant homologies exits among the transposase sequences and the terminal IR sequences of the three IS5 family members.
Southern hybridization with the 32 mutant plasmids was performed with the PCR-amplified IS1478A DNA fragment as a probe. The result indicated that only the eight group II mutant plasmids hybridized to the probe (data not shown). Two 15-mer primers identical to the IS1478A sequence from bp 82 to 68 and from bp 1420 to 1434 were synthesized, and insertion junction sequences in the eight group II mutant plasmids were determined by sequencing outward into sacRB with the two primers. As shown in Fig. 1, all eight group II insertion fragments had about 80 bp of the IS1478A terminal sequences and were flanked by duplications of 6-bp target sequences, indicating that all of the eight group II mutants had insertions of IS1478A or its variants in their plasmids.
The four group III mutants had insertions of IS1479, which belongs to the IS5 subgroup of the IS5 family.
Nucleotide sequencing with the two randomly picked group III mutants, Xc17-4 and Xc17-5, indicated that the two insertion fragments were 1,154 bp long and flanked by 4-bp directed target repeats (Fig. 1). The two sequences differ at six positions (G610, G643, C661, A677, G679, and C943 for the Xc17-4 insertion fragment; A610, A643, T661, G677, A679, and G943 for the Xc17-5 insertion fragment), but have the same 17-bp imperfect terminal IRs. Both sequences carry an ORF starting with GTG at bp 80 and ending with TAA at bp 1039, presumably encoding a transposase protein of 319 amino acids. A putative RBS and a ς70-like promoter were found upstream of the ORF. Several TGA stop codons were found in the 5′ noncoding regions of the two sequences, which might be a means of premature terminating the readthrough transcript from the external promoter due to tight coupling of transcription and translation in prokaryotic organisms (13). Figure 3B shows the first 120-bp sequence with these sequence elements indicated. A Blast search of the nucleotide sequences in the EMBL/GenBank/DDBJ database indicated that both sequences have 73% sequence homology with IS1051 in X. campestris pv. dieffenbachiae (accession no. X70380), 62% homology with IS52 in Pseudomonas syringae pv. glycinea (accession no. M14366), and 60% homology with IS5 in E. coli (accession no. J01735). These two 1,154-bp insertion fragments were named IS1479A and IS1479B. IS1479A and IS1479B differ in one amino acid residue (T200 for IS1479A and A200 for IS1479B) in their presumptive transposase sequences, and like IS1051, IS52, and IS5, carry the DDE motif of the IS5 subgroup of the IS5 family (14), which is shown in Fig. 4A. IS1479A(B), therefore, belongs to the IS5 subgroup of the IS5 family. The presumptive transposase sequences and the terminal IR sequences of IS1478A, IS1479A, and IS5 were aligned, and the results are shown in Fig. 4A and B, respectively.
Southern hybridization with the 32 mutant plasmids was performed with PCR-amplified IS1479A DNA fragment as a probe. The result indicated that only the four group III mutant plasmids hybridized to the probe (data not shown). Two 15-mer primers identical to the IS1479A sequence from bp 76 to 62 and bp 1081 to 1095 were synthesized, and insertion junction sequences in the other two group II mutant plasmids were determined by sequencing outward into sacRB with the two primers. As shown in Fig. 1, insertion fragments in these two group III mutant plasmids had about 70 bp of the IS1479A(B) terminal sequences and were flanked by duplications of 4-bp target sequences. Thus, the four group III mutants all had insertions of IS1479 variants in their plasmids.
Insertions of IS1478 and IS1479 had preferred orientations and target site specificities.
The junction sequences of the eight IS1478 insertions and four IS1479 insertions, shown in Fig. 1, were examined in more detail. It was found that the eight IS1478 insertions demonstrated conservation of PyPuNTTA (Py is a pyrimidine, Pu is a purine, and N is any nucleotide) in their 6-bp target sequences. Insertions of IS1478 showed a strong preference in one orientation and were not random, as three insertions occurred at sacRB coordinate 1695 and two occurred at coordinate 1587. When ca. 30-bp sacRB sequences up- and downstream from the five IS1478 insertion sites were aligned, a consensus sequence AAN16AN10C 8 bp upstream from the insertion sites was found. Similar phenomena were observed for the four IS1479 insertions. These included conservation of sequence PyTAPu for the 4-bp target duplications, two insertions detected at sacBR coordinate 1467, and consensus sequences AN8AAN2C 15 bp upstream and TGN5G 22 bp downstream from the insertion sites. It is likely that an A 20 bp upstream or a sequence AN(10 or 12)C upstream from the insertion site is important for selection of target sites by these two IS5 family members. Recently, Hu and Derbyshire (9) examined 63 insertion sites of an IS5 family member, IS903, and found preferences for insertions at four regions in a 55-kb plasmid, and several sites occurred more than once. When one preferred region was cloned in a plasmid and the insertion sites were examined, they observed strong preference of insertions of IS903 in one orientation and conservation of 5-bp sequences on both sides of the target sequences. Although we examined only eight IS1478 and four IS1479 insertion sites, our results are fairly consistent with theirs.
Presence of IS476, IS1478, and IS1479 in strains of X. campestris pv. campestris and in related bacteria.
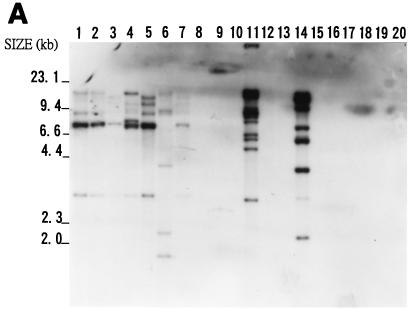
It was suspected that IS476, IS1478, and IS1479 might be widespread in nature. Twenty bacteria were chosen for examination of the presence of the three elements. These bacteria included Agrobacterium tumefaciens, Erwinia carotovora subsp. carotovora, E. coli, Pseudomonas solanacearum, Rhizobium leguminosarum, X. campestris pv. begoniae, X. campestris pv. citri, X. campestris pv. dieffenbachiae, X. campestris pv. glycinea, X. campestris pv. mangiferaeindicae, X. campestris pv. oryzae, X. campestris pv. phaseoli, X. campestris pv. vesicatoria, and six strains of X. campestris pv. campestris, including Xc11 and Xc17. A spontaneous avirulent mutant of Xc11 (Xc11A) was also included. Southern blot hybridization of EcoRI-digested genomic DNAs of these bacteria was performed with either the 1.2-kb BspMI IS476A DNA fragment or the PCR-amplified IS1478A or IS1479A DNA fragment as a probe. Figure 5 shows the hybridization results, which indicate that all seven strains of X. campestris pv. campestris, X. campestris pv. vesicatoria, and X. campestris pv. begoniae had multiple copies of the three elements in their genomes. In addition, X. campestris pv. citri, X. campestris pv. glycinea, X. campestris pv. oryzae, and X. campestris pv. phaseoli had multiple copies of either IS1478, IS1479, or both in their genomes. Of the 20 bacteria tested, only X. campestris pv. mangiferaeindicae, X. campestris pv. dieffenbachiae, and the five non-X. campestris bacteria did not contain any of the three elements. Thus, the three insertion elements are widespread only in X. campestris pathovars. It is interesting that six of the seven X. campestris pv. campestris strains tested and X. campestris pv. begoniae carried more than 20 copies of IS1478 in their genomes (Fig. 5B).
FIG. 5.
Southern blot hybridization analyses of total DNAs from 20 bacteria. Samples of about 3 μg of EcoRI-digested genomic DNAs were analyzed on an agarose gel, transferred to a nylon membrane, and hybridized with the IS476A probe (A), IS1478A probe (B), or IS1479A probe (C). DNAs from X. campestris pv. campestris 11A (lane 1), X. campestris pv. campestris 11 (lane 2), X. campestris pv. campestris 17 (lane 3), X. campestris pv. campestris 2 (lane 4), X. campestris pv. campestris 6 (lane 5), X. campestris pv. campestris 85 (lane 6), X. campestris pv. campestris 88 (lane 7), X. campestris pv. mangiferaeindicae 38 (lane 8), X. campestris pv. glycinea 69 (lane 9), X. campestris pv. dieffenbachiae 65 (lane 10), X. campestris pv. vesicatoria 64 (lane 11), X. campestris pv. phaseoli 73 (lane 12), X. campestris pv. citri (lane 13), X. campestris pv. begoniae (lane 14), X. campestris pv. oryzae 1 (lane 15), Agrobacterium tumefaciens LBA4404 (lane 16), Erwinia carotovora subsp. carotovora ZL4 (lane 17), Pseudomonas solanacearum RD4 (lane 18), Rhizobium leguminosarum 128C53 (lane 19), and E. coli DH5α (lane 20) were analyzed. The sizes of HindIII-digested λ DNA fragments are indicated to the left of the gels.
There are other six plasmid insertion mutants, including the two group IV mutants, that did not show homology with either IS476, IS1478, or IS1479. Insertions in these mutants ranged from 0.8 to 7.1 kb in size (Table 2). They accounted for 19% of the total insertion mutants isolated and were not analyzed in this study.
ACKNOWLEDGMENTS
We thank Y.-H. Tseng for helpful suggestions, C. I. Kado for providing pUCD800, and Y.-H. Tseng and K.-C. Tzeng for providing the bacterial strains.
This work was supported by research grants NCS-81-0211-B-005-555 and NSC-83-0211-B-005-043 from the National Science Council of the Republic of China.
REFERENCES
- 1.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 2.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 3.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J-H, Hsu W-B, Hwang J-L. Two amino acid residues of transposase contributing to differential transposability of IS1 elements in Escherichia coli. J Bacteriol. 1998;180:5279–5283. doi: 10.1128/jb.180.19.5279-5283.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu R, Voordouw G. ISD1, an insertion element from the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough: structure, transposition, and distribution. Appl Environ Microbiol. 1998;64:53–61. doi: 10.1128/aem.64.1.53-61.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 8.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu W-Y, Derbyshire K M. Target choice and orientation preference of the insertion sequence IS903. J Bacteriol. 1998;180:3039–3048. doi: 10.1128/jb.180.12.3039-3048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearney B, Ronald P C, Dahlbeck D, Staskawicz B J. Molecular basis for evasion of plant host defence in bacterial spot disease of pepper. Nature. 1988;332:541–543. [Google Scholar]
- 12.Kearney B, Staskawicz B J. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J Bacteriol. 1990;172:143–148. doi: 10.1128/jb.172.1.143-148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleckner N. Regulation of transposition in bacteria. Annu Rev Cell Biol. 1990;6:297–327. doi: 10.1146/annurev.cb.06.110190.001501. [DOI] [PubMed] [Google Scholar]
- 14.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta N, Mullin D A, Tarleton J, Ely B, Newton A. Identification, distribution, and sequence analysis of new insertion elements in Caulobacter crescentus. J Bacteriol. 1990;172:236–242. doi: 10.1128/jb.172.1.236-242.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prere M-F, Chandler M, Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Priefer U B, Kalinowski J, Ruger B, Heumann W, Puhler A. ISR1, a transposable DNA sequence resident in Rhizobium class IV strains, shows structural characteristics of classical insertion elements. Plasmid. 1989;21:120–128. doi: 10.1016/0147-619x(89)90055-3. [DOI] [PubMed] [Google Scholar]
- 18.Rezsohazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz E, Kroger M, Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988;16:6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scordilis G E, Ree H, Lessie T G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987;169:8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196:413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 24.Steinmetz M, Le Coq D, Aymerich S, Gonzy-Treboul G, Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985;200:220–228. doi: 10.1007/BF00425427. [DOI] [PubMed] [Google Scholar]
- 25.Tinoco I, Jr, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nature. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 26.Van den Mooter M, Swings J, Gossele F, Kersters K, De Ley J. The taxonomy of Xanthomonas Dowson 1939. In: Civerolo E L, Collmer A, Davis R E, Gillaspie A G, editors. Plant pathogenic bacteria. Boston, Mass: Martinus Nijhoff Publishers; 1987. pp. 795–796. [Google Scholar]
- 27.Wood M S, Byrne A, Lessie T G. IS406 and IS407, two gene-activating insertion sequences from Pseudomonas cepacia. Gene. 1991;105:101–105. doi: 10.1016/0378-1119(91)90519-h. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 29.Zekri S, Toro N. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm6, a small transposable element that belongs to the IS3 family. Gene. 1996;175:43–48. doi: 10.1016/0378-1119(96)00118-7. [DOI] [PubMed] [Google Scholar]