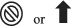Abstract
Since its emersion, coronavirus disease 2019 (COVID‐19) has been a significant global dilemma. Several mutations in the severe acute respiratory virus (SARS‐Co‐2) genome has given rise to different variants with various levels of transmissibility, severity and mortality. Up until November 2021, the variants of concern declared by the World Health Organization were Alpha, Beta, Delta and Gamma. Since then, a novel variant named Omicron (B.1.1.529) has been developed. BA.1, BA.1.1, BA.2 and BA.3 are four known subvariants of Omicron. The Omicron variant involves new mutations in its spike protein, most of which are in its receptor binding site, and increase its transmissibility and decrease its antibody and vaccine response. Understanding the virology and mutations of Omicron is necessary for developing diagnostic and therapeutic methods. Moreover, important issues, such as the risk of re‐infection, the response to different kinds of vaccines, the need for a booster vaccine dose and the increased risk of Omicron infection in pediatrics, need to be addressed. In this article, we provide an overview of the biological and immunopathological properties of Omicron and its subvariants, its clinical signs and symptoms, Omicron and pediatrics, vaccines against Omicron, re‐infection with Omicron, diagnostic approaches and specific challenges of Omicron in the successful control and management of the rapid global spread of this variant.
Keywords: biology, coronavirus disease 2019 (COVID‐19), genetic, Omicron, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), virology
Several mutations in SARS‐CoV‐2, especially deletion mutations in the spike (S) protein, have led to the development of a new variant of concern, named Omicron. Omicron has higher transmissibility and infectivity but milder symptoms compared with other SARS‐CoV‐2 variants. Currently, five subvariants of Omicron have been identified, including BA.1 and BA.1.1, BA.2, BA.3, BA.4 and BA.5. Laboratory diagnosis of Omicron includes genetic testing using reverse transcriptase PCR and next‐generation sequencing, serology testing for neutralizing IgM/IgG and imaging studies.
List of abbreviations
- ACE2
angiotensin‐converting enzyme 2
- COVID‐19
coronavirus disease 2019
- CRS
cytokine release syndrome
- IL‐6
interleukin‐6
- IVIG
intravenous immunoglobulin
- NAAT
nucleic acid amplification testing
- NGS
next‐generation sequencing
- RBD
receptor binding domain
- RDT
rapid diagnostic test
- RNA
ribonucleic acid
- RT‐PCR
reverse transcriptase‐polymerase chain reaction
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- VOC
variants of concern
- WHO
World Health Organization
1. INTRODUCTION
Two years ago, a novel coronavirus disease 2019 (COVID‐19) started in Wuhan, China. Caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), it has created a problematic and dangerous scenario worldwide. 1 , 2 , 3 As this pathogen swiftly spread around the globe, COVID‐19 was declared a global pandemic and a public health emergency of worldwide importance in early 2020. COVID‐19 infection manifestations can range from asymptomatic (no symptoms) to acute pneumonia and even multiple organ failure, and the severity of the disease varies from mild to deadly. 4 , 5 COVID‐19 has infected and killed over 406 and 5.8 million individuals, respectively, in the last two years. Based on the available data, 52.7% of the global population has received at least one dose of a SARS‐CoV‐2 vaccine to help eliminate this infection (COVID‐19 Map, Johns Hopkins Coronavirus Resource Center, https://coronavirus.jhu.edu/map.html, accessed 31 January 2022).
SARS‐CoV‐2 contains the capacity to undergo mutations and antigenic change over time. This incredibly infectious epidemic is being fought by scientists, academics and medical professionals, but the announcement of the new coronavirus variety has caused a shock recently. 6 Based on the Centers for Disease Control and Prevention, variation between SARS‐CoV‐2 subtypes contributes to disease progression, increased transmissibility, decreased treatment efficacy and other worrying aspects (‘What you need to know about variants’, https://www.cdc.gov/coronavirus/2019-ncov/variants/about‐ variants.html?s_cid = 11720:covid variant:sem.ga:p:RG:GM:gen: PTN:FY22, accessed 7 February 2022). Particular strains of SARS‐CoV‐2, known as variants of concern (VOC), are more infectious and are more likely to re‐infect previously vaccinated or infected individuals. The VOCs of SARS‐CoV‐2 include the following: the Alpha (B.1.1.7) variation which was first detected in the UK, the Beta variation (B.1.351) which was found in South Africa, the Gamma (P.1/B.1.1.28) variation which was first seen in Brazil, and the Delta (B.1.617.2) variant which was found in India. 7 The features of Delta and Omicron have been listed in Table 1.
TABLE 1.
Comparison of Delta and Omicron variants of concern
| Variant features | Delta | Omicron | Reference |
|---|---|---|---|
| Lineage | B.1.617.2 | B.1.1.529 | 8 |
| Origin | India | South Africa | 8 , 9 |
| Subvariants | AY.4.2 | BA.1/BA.1.1/BA.2/BA.3 | 10 |
| Total mutations | More than 13 | More than 50 | 11 , 12 |
| Spike mutations | 9 | 36 | 13 |
| Transmissibility |

|

|
12 , 14 , 15 |
| Infectiousness |

|

|
15 , 16 , 17 |
| Re‐infection |
?/
|

|
17 , 18 |
| Mortality |

|

|
19 |
| Pediatric infection |

|

|
16 , 20 |
| Vaccine efficacy |

|
?/
|
21 , 22 , 23 , 24 |
On 24 November 2021, a novel coronavirus variant, Omicron (B.1.1.529) was detected in a patient’s test sample in South Africa. The number of mutations in this strain was greater than in any prior strain of the virus, nearly 50, of which more than 26 are in its spike protein region. On 26 November, this novel variant was announced as a variant of concern (VOC) by the World Health Organization (WHO). Soon after this announcement, Omicron was reported in many other countries. Omicron is the most altered of the VOC variants, leading to expanded transmissibility. 25 There are several mutations in this new “Omicron variant” that might significantly impact its behavior. It may resist vaccine‐induced immunity and pose an expanded chance of re‐infection. Moreover, alterations in the spike protein cause the virus to show unique traits, such as increased transmissibility and severity, which can lead to more severe infection as well as higher morbidity and mortality rates. 26 The rapid spread of Omicron is an important matter of concern since the infection rate has been reported to be 7 times higher after only 2 weeks. 27
The increased pathogenicity of the Omicron variety has made it a public health concern. Notably, Omicron appeared when humanity was on the verge of achieving worldwide immunity with global vaccination against SARS‐CoV‐2. 14 The emergence of recent SARS‐CoV‐2 variations, such as Omicron, serves as a reminder that this disease pandemic is still far from over. Individuals must obtain the vaccine as soon as possible and keep following existing measures to reduce virus transmission, including physical separation, mask use, frequent handwashing and ventilation of interior splaces. 28 In this literature review, we aim to review the virological, biological and diagnostic implications of Omicron and discuss the specific challenges confronted in fighting this novel variant of SARS‐CoV‐2.
1.1. Immunopathogenesis of Omicron
SARS‐CoV‐2 contaminates many respiratory system cell types by attaching to the ACE2 receptor, including vascular endothelial cells, airway and alveolar epithelial cells, and macrophages. 29 Innate immune cells, mainly macrophages and neutrophils, identify the pathogen‐associated molecular patterns through their Toll‐like receptors and activate the host immune system. Interleukin‐6 (IL‐6), IFN‐γ and monocyte chemoattractant protein‐1 are secreted by innate and adaptive immune cells. 30 These cytokines and chemokines help monocytes, macrophages and neutrophils shift the infection. These cells release cytotoxic chemicals to eradicate the virus. Immune homeostasis may be altered owing to an imbalance in the immune system’s ability to respond to a viral infection. 31 , 32 An essential part of innate immunity is the production of inflammatory mediators and the recruitment of other immune cells into the lungs by alveolar macrophages and neutrophils. SARS‐CoV‐2 infection, on the other hand, is fought by CD4+ T helper cells, cytotoxic CD8+ T cells and B cells. CD8+ T cells kill virus‐infected cells and B cells emit neutralizing antibodies accompanied by T cell involvement in this stage. 33 Cytokine release syndrome (CRS) is related to the increased production of pro‐inflammatory cytokines such as TNF, IL‐1, ‐6, ‐12, ‐18, ‐33, and interferon‐I and ‐II, along with chemokines such as CCL2, 3, 5 and CXCL8, 9, 10. 2 , 34
According to investigations, tissue damage in COVID‐19 appears to be caused chiefly by immune system disturbances and not direct viral damage. The host’s immunological response to COVID‐19 appears to be crucial in the development of clinical symptoms and the pathogenesis of this disease. There is mounting evidence that the immunological characteristics of SARS‐COV‐2 patients are inextricably connected to the disease progression. Leukocytosis, lymphopenia and a high ratio of neutrophils to lymphocytes are the first changes in blood cells. 35 SARS‐CoV‐2 has been revealed to disrupt normal immune responses in severe COVID‐19 patients by draining the immune procedure and producing uncontrolled inflammatory reactions. Severe COVID‐19 manifests itself through raised cytokines, organ failure, lymphocyte hyperactivity and malfunction, lymphopenia, and granulocyte and monocyte irregularities. Studies have also demonstrated that the IL‐6 signaling pathway has a particular role in CRS pathogenesis. 36
There are two primary methods through which SARS‐CoV‐2 is transmitted, including direct viral spread and through droplets/airborne from infected patients. 2 In the pathophysiology of COVID‐19, the immune system plays a critical function, and an uncontrolled innate immune response and problems with adaptive immunity are two ways in which SARS‐CoV‐2 can induce overall tissue damage. 37 There are various pathways behind immunological dysregulation in COVID‐19 patients. Most people have leukocytosis, lymphopenia and a high ratio of neutrophils to lymphocytes, which are the first signs of immunological shifts. These processes may primarily damage lymphocytes, especially T cells, make macrophages more active in the body and finally cause a cytokine storm. 37 After the presentation of the viral antigen to the T cells in the lymph node, the T cells can take on a variety of tasks. They can either act as killing cells to destroy cells contaminated with the virus or operate as helper T cells to assist B cells in producing antibodies. Immune neutralization of the SARS‐CoV‐2 infection requires a cytotoxic T cell response, which depends on an individual’s genetics and does not need antibodies. When infected with SARS‐CoV‐2, CD8 + T lymphocytes secrete pore‐forming proteases and induce programmed cell death in infected cells. An effective cellular response is associated with fast and efficient protection against SARS‐CoV‐2. Moreover, the cellular reaction is long lasting and evokes immunoprotective memory. 38 Concerning the T cell reaction in Omicron variant, investigations indicated that T cell responses to Omicron are retained in most infected and vaccinated people. Shortly after the booster immunization, T cell responsiveness to Omicron is improved. 39 In 20% of individuals, T cell reactivity to the Omicron spike is lowered by more than 50%. 40 The majority of Omicron epitopes are sequence conserved and preserve affinity to HLA‐I. 41
In some cases, the immune response against SARS‐CoV‐2 may be dysregulated, leading to a severe hyperinflammatory condition, CRS. Acute respiratory distress syndrome and end organ failure are caused by CRS and are the most important cause of death in severe COVID‐19 cases. 42 Acute respiratory distress syndrome is a potentially life‐threatening disorder in which the respiratory system fails. It induces endothelial and alveolar epithelial destruction in the lungs and presents as difficulty in breathing, hypoxemia and the development of pulmonary edema. 43
Proinflammatory cytokines are overproduced in SARS‐CoV‐2, leading to lung pathogenesis and respiratory distress, resulting from the responses to the virus. There is an excessive expansion of monocytes, macrophages and neutrophils near the site of infection because of the presence of proinflammatory cytokines. Cytotoxic substances like reactive oxygen species, which are constructed by these cells, cause cell death and tissue damage. 44 Understanding the immunopathogenesis of COVID‐19 has helped in developing efficacious immunological therapies for COVID‐19; however, the exact mechanisms of the immunopathogenesis of Omicron are unclear. 45
Previous research uncovered that the Omicron variation might be 10 times more transmissible than the wild type. 46 The capacity of Omicron to escape the vaccination may be two times greater than that of the Delta version, according to 132 3D structures of antibody receptor binding domains (RBDs). 47 Omicron possesses a novel mode of entry into cells, making it more effective than Delta at entering cells and less efficient at causing lung illness. It is capable of causing upper respiratory infection but not lung disease, which makes Omicron’s activity more similar to that of prior human coronaviruses clinically. 48 There is still the possibility of severe illness; therefore, it is not identical to them, but it functions pretty similarly. Omicron virus replication is diminished in cells overexpressing TMPRSS2 but boosted in cells that only allow endosomal entrance. Additionally, by inhibiting cell‐surface fusion mediated by TMPRSS2 and endosomal fusion mediated by cathepsin, Omicron can enter via either pathway but prefers endosomal fusion over cell‐surface fusion. 49 , 50 The capability of Omicron to infect cells via both routes significantly expands the number of cell types that it can infect. Finally, research indicates that Omicron may utilize ACE2 receptors from a wider variety of host species than previous variations, particularly mice and domestic fowl. This raises the chance that SARS‐CoV‐2 might establish a long‐term reservoir in a new animal host, facilitating the virus’s spread in future human epidemics. However, while the full efficacy of vaccinations vs. Omicron has not been proved, it may give some protection and decrease the spread of COVID‐19. 50
1.2. Clinical signs and symptoms of Omicron
COVID‐19 prevention and control efforts depend heavily on SARS‐CoV‐2 diagnosis. SARS‐Cov‐2 diagnosis is based chiefly on epidemiological data, clinical symptoms and adjuvant technologies like nucleic acid detection and immunological testing. Omicron clinical signs and symptoms are divided into the most common, the less common and the severe. Fever, cough, tiredness and a loss of taste or smell are the most common complaints of patients. It is not uncommon for the Omicron to cause other symptoms, such as pharyngitis, headaches, skin rashes, discolored fingers and toes, red or itchy eyes, stomach cramps and diarrhea. Furthermore, it can cause severe symptoms such as chest discomfort, shortness of breath, difficulty moving, trouble breathing and disorientation. In case of respiratory involvement, pulmonary characteristics, such as cough, scratchy throat, shortness of breath and pneumonia, may also be observed. 51 Studies have reported that, compared with the Delta variant, Omicron is associated with a 50–70% reduced risk of hospitalization. 52
1.3. Laboratory diagnosis of Omicron
COVID‐19 assays, both PCR and rapid test, can detect all Omicron subvariants as COVID‐19, but additional testing is required to differentiate the subvariants from one another and other COVID‐19 variations. The use of swift and accurate diagnostic technologies to detect the virus and then pick appropriate and effective therapies is significant for epidemic control. Reverse transcriptase‐polymerase chain reaction (RT‐PCR), the gold‐standard test for laboratory diagnosis of SARS‐CoV‐2, remains the best method for determining if a patient has Omicron. The S gene has been most frequently affected by Omicron mutations. SARS‐CoV‐2 may also be identified by searching for the S genes that encode the spike glycoprotein. The E, Rd, Rp and N genes are the primary focus of the RT‐PCR diagnostic kits that have been approved for usage. 53 Using the most commonly used RT‐PCR kits makes it possible to identify the RT‐PCR test result as either positive or negative. The S gene mutation may or may not cause a positive result, depending on the test. Patients may be recommended to get their DNA sequenced to discover the new mutation. 54
Laboratory diagnosis of Omicron can be divided into two parts: first molecular diagnosis and then immunological diagnosis. Molecular diagnosis has two main sections: real‐time PCR and next‐generation sequencing (NGS). Immunological diagnosis entails looking for antigens and antibodies. Viral RNA may be amplified via molecular assays, allowing for the detection of viral infection. The term “nucleic acid amplification testing” also describes these tests (NAATs). The first step is to collect a sample from the nose or mouth of a potentially infected individual, where the virus could be present. A molecular diagnostic technique can identify millions of copies of viral genomic material if SARS‐CoV‐2 is present in the sample. To conduct molecular testing, secretions of nasopharyngeal surfaces, which are most likely to carry the virus, must be collected. Most tests now available or in development for COVID‐19 use nasopharyngeal or oral specimens to make testing easy. If someone is infected, viral RNA will be detected. Several molecular diagnostics are available, and some produce findings faster than traditional PCR‐based techniques. Rapid molecular diagnostics, such as LAMP, are specific but exceedingly sensitive since they intensify genetic material in the patient sample. All of these approaches follow the same basic procedure: identify DNA or RNA pathogens and amplify a region of the pathogen’s genome discovered. Then, if any amplified genetic material is present in the sample, provide an output measurement showing its amount. In BA.1, there is a small deletion mutation in the form of deletion 69–70, which changes the structure of the spike protein and allows it to be hidden from the PCR test. Thus the test result is doubtful for Omicron. 55 Other specialized tests can identify the strain, but in the BA.2 sub‐variant, this omission mutation does not exist, and despite being Omicron, the test result will be erroneous with other strains, especially the Delta variant in routine PCRs. 56 Type BA.2 is known in the USA as the Stealth Omicron because it spreads faster and carries mutations that make it difficult to quickly differentiate this subtype from the Delta type by routine PCR tests.
NGS sequencing methods have quickly established themselves as the technique of choice for various virological applications, including the discovery of new viruses. This method is critical in determining the origin of SARS‐CoV‐2. Most coronavirus and SARS‐CoV‐2 genomes found using NGS are currently publicly available for academics to use in their investigations on the origins of SARS‐CoV‐2. 57 SARS‐CoV‐2 has benefited from the use of both second‐ and third‐generation NGS technology and a variety of proprietary library preparation techniques created by various companies. When using NGS‐based techniques, whole‐length viral genomes may be rebuilt, even for viruses that have not been previously defined, which is one of the most critical advantages of NGS. 58 , 59 NGS can also be useful to detect probable novel variations of SARS‐CoV‐2 and Omicron subvariants. Therefore, sequencing of further specimens is necessary.
Antigen‐detecting diagnostic testing identifies viral proteins in upper respiratory samples or saliva to screen for SARS‐CoV‐2 infection. The use of antigen‐detecting rapid diagnostic tests (Ag‐RDTs), rather than NAATs, may be a more practical method of screening for SARS‐CoV‐2. Ag‐RDTs work best in those with a high viral load early in their illness and would be most accurate in areas where the SARS‐CoV‐2 prevalence is less than 5%. Ag‐RDTs have a low positive predictive value when there is a low transmission. Hence, NAATs are recommended for first‐line testing or the confirmation of Ag‐RDT positive results in these situations. Ag‐RDTs are less accurate than NAAT, especially in asymptomatic individuals. However, this can be mitigated by carefully selecting cohorts for testing. The WHO recommends Ag‐RDTs that meet 80% sensitivity and 97% specificity minimum performance parameters (‘Antigen‐detection in the diagnosis of SARS‐CoV‐2 infection’, https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov2infection-using-rapid-immunoassays, accessed 5 February 2022). 60
Sequencing is now the most frequently used approach for identifying, classifying and tracking SARS‐CoV‐2 variations. 61 Compared with other genotyping procedures like PCR, sequencing is slow and needs specialist mechanisms and interpretation. Although PCR techniques target preselected mutations, they are less expensive and more convenient than sequencing for SARS‐CoV‐2 genotyping, producing more rapid results. Because variations propagate throughout populations over time, these techniques may allow for an improved search of variants, particularly in resource‐limited contexts. 62 , 63 Genome sequencing is the gold standard for tracking the virus’s genesis and evolution, but it is expensive, time‐consuming and challenging to get. Using single‐nucleotide discriminating molecular beacons, multiplex quantitative RT‐PCR assays for SARS‐CoV‐2 have been created that identify all VOCs and additional variants of interest in the viral genome, for a total of nine mutations. Several significant mutations in the spike protein‐coding sequence have been identified using quantitative RT‐PCR experiments. Using readily accessible five‐color thermal cyclers, these tests may be quickly and inexpensively put into practice to monitor the spread of these variations. 64
SARS‐CoV‐2 antigens emanating from a spike glycoprotein or nucleocapsid will elicit one of three reactions: a robust, weak or difficult‐to‐detect reaction. Individuals with lesser antibody responses to infection had better viral clearance rates and prognoses, whereas patients with robust responses had more severe clinical outcomes. Therefore, the human antibody response to viral infection can be an independent prognostic indicator. Likewise, antibodies cannot shed viral particles from the body soon after release since a patient might even have extended virus shedding after seroconversion. 65
Serum and other body fluids can be tested for antibodies using serological tests. Immune reactions to microbes and foreign or one’s own proteins contain antibodies, which may be discovered by serological testing. A lack of commercially available reagents for serological tests mean that they are not routinely used to detect coronavirus infections. 53 When a virus enters the body and releases viral antigens into circulation, the human immune system produces IgA, IgM and IgG antibodies that are more effective and long‐lasting than the antigenic proteins. As a result, scientists are switching their attention to serological antibodies as a diagnostic option rather than focusing on viral antigens to create faster, more straightforward and more sensitive serological tests. 66 To identify SARS–CoV‐2, many studies have employed S or N protein‐specific antibodies. It was demonstrated that testing for virus‐specific antibodies was more accurate than testing for total immunoglobulin. 67
The intensity of a patient’s symptoms can be reflected in serological detection results, but RT‐PCR results can only provide a limited view. Compared with mild‐to‐moderate COVID‐19 patients, severe COVID‐19 patients had considerably higher levels of IgM and IgG antibodies. Most serological test kits presently utilize particular IgM and IgG for detection. Changes in a virus’s genome can change its proteins, making an antigen or serology test less effective. 68
1.4. Structure, mutations and biology of Omicron
SARS‐CoV‐2, a novel Beta coronavirus, is an enveloped virus with a single‐stranded RNA genome and a helical symmetry nucleocapsid. The host cell receptor for SARS‐CoV‐2 is angiotensin‐converting enzyme 2 (ACE2). 69 , 70 SARS‐CoV‐2 contains three main proteins. The envelope (E) protein is a tiny protein complex that helps to construct pentameric ion channels in the body. The M protein is the principal structural protein. The head S1 and stem S2 subunits make up the spike (S) protein. The S protein promotes the binding of the virus to ACE2 receptors on host cell surfaces. 3 It is usual for the S1 heterotrimer’s N terminus to attach to carbohydrate molecules on the cell surface. However, the C terminal domain only targets angiotensin II and aminopeptidase N. 71 The primary target tissue for SARS‐CoV‐2 is the respiratory epithelium. As the virus replicates and releases from the lungs, it causes non‐specific symptoms, including headache, muscle aches, fever and respiratory problems. 72 The location of ACE2 receptors in various organs may help clarify where infections occur. For instance, ACE2 receptors are also expressed by the intestinal epithelium and blood vessel endothelial cells, reflecting gastrointestinal complaints and cardiovascular issues. 73 , 74
It is important to remember that structural proteins, including those found in the spike, envelope, membrane and nucleocapsid, play an essential part in the virulence, pathogenicity and virion assembly processes. 75 The S protein binds to cell surface receptors, cleaving the receptor‐attached S protein by a host cell protease, which causes the host cell and virus membranes to merge. Virus RNA uses host facilities to induce specialized RNA polymerase and viral genomes and proteins. The viral M protein combines into the endoplasmic reticulum and promotes the creation of viral particles, which are subsequently created, exocytosed and released as new virions. 76 The spike of SARS‐CoV‐2 is a critical predictor of viral infectivity and mediates cell entrance via interactions with ACE2 and TMPRSS2 at the cathepsins in endosomes or the plasma membrane. 77
Since the beginning of 2020, there have been three significant COVID‐19 outbreaks in South Africa. The Beta and Delta variations, respectively, are responsible for two of them. Within roughly 100 days of the epidemic, the rate of infections linked to the Beta variation grew to 50% of all daily infections. The infection rate for the Delta version, on the other hand, increased to 80% within the same time frame. The Delta variety is more capable of spreading than the Beta variant. 16 In less than a month, Omicron had spread to 90% of the population in South Africa. Doubling times for Beta, Delta and Omicron types were estimated to be 1.7, 1.5 and 1.2 days. 78 Based on studies, Omicron is more efficiently transmitted than Beta or Delta. In addition, a recent study from South Africa suggests that Omicron may enhance the chance of SARS‐CoV‐2 re‐infection. 79
Various sequences studies have indicated that just a few mutations successfully induce a more severe illness with high disease transmission and pathogenicity. 79 COVID‐19 spread through the population was associated with several gene mutations in SARS‐CoV‐2, similar to other RNA viruses. According to phylogenetic analysis, the Omicron variation is intimately bonded to the Gamma (P.1) variant. 80 Compared with the previous variants, at least 50 mutations have been found in the Omicron genome, 32 impacting the spike protein. 81 Beta contained six and Delta had 10 variant‐specific mutations, while Omicron has 26. Omicron has 23 of the 30 signature mutations, and none of these mutations has ever been found in any other variant. Altogether, 46 distinct mutations in the S, ORF1a and ORF1b genes of the Omicron variant were identified, with over 50% frequency. 82
As mentioned earlier, numerous non‐synonymous mutations in the Omicron variants have been discovered through genomic sequence analysis. Most of these mutations are in spike proteins and are correlated to disease severity, transmission and immune evasion. Furthermore, more than 60 insertions, substitutions and deletions have been discovered in Omicron, creating the variant with the most mutation area of all SARSCoV‐2 variants reported to date. 83 , 84 The RBD of Omicron contains numerous threatening mutations. One of Omicron’s pathogenic properties appears to be its ability to connect to ACE2, which is a particular source of concern when virus mutations improve their capacity to connect to cell surface receptors. 84 Two mutations in the Omicron may significantly boost the S protein’s affinity to ACE2. Moreover, three mutations located near the cleavage site of Omicron have a potential chance of improving transmissibility, whereas other mutations may lead to immune system evasion. 85 Changing a hydrophilic amino acid to a hydrophobic amino acid in the most recent SARS‐CoV‐2 variant would affect the hACE2 and RBD connection. Omicron has an approximately 6–10 times extended mutation rate in the receptor‐binding motif compared with other VOC variants. 86 As a result, the virulence aspects of the Omicron variation may be superior to those of the other variants. It is crucial to mention that many of the “concerns” with the mutations stated above are theoretical.
1.5. Subvariants of omicron
The WHO classifies Omicron into four different substrains: BA.1, BA.1.1, BA.2 and BA.3 (Table 2). BA.1 and BA.2 differ significantly in their mutations in the most critical places that are often associated with higher transmissibility. When compared with Omicron subvariants, BA.2 is 1.5 times more transmittable. On the other hand, in comparison with BA.1, BA.2 has a significant growth advantage. When identifying the BA.2 variation, there are five distinct mutations on the spike (RBD) that are frequently related to increased transmissibility. 90 According to studies, the BA.2 subvariant infects individuals at a 33% higher rate than BA.1. Researchers at Statens Serum Institute have demonstrated that if you have been exposed to Omicron BA.2 in your family, you have a 39% chance of contracting the infection within 7 days. In contrast, if you have been exposed to BA.1, your risk is only 29%. 46 , 87 , 91 According to a Danish study, BA.2 or Omicron spreads about 1.5 times faster than Omicron itself, and researchers did not find any evidence that the disease was more severe than BA.1, but they said that it could cause a wave of infection. 46 Furthermore, laboratory results indicate an increase in the pathogenicity of BA.2 compared with BA.1. The new SARS‐CoV‐2 subvariants emerged concurrently from the same source with an equal probability of global spread. However, it is unclear why just BA.1 dominated the global expansion. This is most likely due to spike protein variations, which are critical for viral propagation and host cell entrance. 92
TABLE 2.
Characteristics of Omicron subvariants
According to the UK Health Security Agency, there have been over 1000 instances of BA.2. Compared with the original Omicron form (BA.1), BA.2 has an expanded growth rate in all regions of England. Moreover, there is more evidence that people with BA.2 were more likely to spread the disease to their families. It has been shown that 13.4% of BA.2‐infected individuals spread the virus, while this is only 10.3% in BA.1‐infected patients. The agency also stated that early research revealed no indication of BA.2 vaccination efficacy against symptomatic illness compared with BA.1. At least 25 weeks after two doses, vaccination efficacy against symptomatic infection was reported to be 9% for BA.1 and 13% for BA.2, which climbed to 63% for BA.1 and 70% for BA.2 2 weeks after a third booster dose. 93
Moreover, two novel subvariants of the BA.2 have emerged, recognized as BA.2.12 and BA.2.12.1. Already, these two sublineages of BA.2 encompass an estimated 80.6% of all COVID‐19 infections in New York. Even if the original BA.2 Omicron subvariant looks to be 23–27% more transmissible, the new BA.2.12.1 subvariant appears to be a significant time rush for the virus. The COVID‐19 vaccinations are as effective against BA.2.12 and BA.2.12.1 as BA.2. 94
New cases of SARS CoV‐2 Omicron strain subvariants are now being tracked by the WHO to determine whether they are more infectious or destructive. The WHO have announced that BA.4 and BA.5 have been added to its list of Omicron variations to track. 95 This will shed light on their immunological resilience. BA.1 and BA.2 are the subvariants that are presently prominent worldwide. BA.1.1 and BA.3 are already under the watchful eye of the WHO. Scientists are still studying the novel Omicron variety. As of right now, BA.5, found in South Africa, is spreading at an 84% faster rate than BA.2. 96 The BA.4 strain found in South Africa is now growing 63% faster than the BA.2 strain. In the Delta form, the L452R harmful mutation is present in both BA.4 and BA.5, 97 (‘What do we know about Omicron BA.4 and BA.5’, Medriva, https://medriva.com/what-do-we-know-about-Omicron-ba-4-and-ba-5/).
1.6. Transmission of Omicron
The Omicron variation spread over the world in days and the number of cases rose rapidly. There is not enough information available on infection rates to determine how contagious the newly discovered, highly altered Omicron strain is. Studies have shown that the Gamma variation had similar infection rates to the wild type. In contrast, the Beta variant had fewer infections, and Delta was approximately two‐fold more efficient in disease spread than the wild type. The Omicron variation has an infection rate four times greater than the wild type, while the Delta variant had an infection rate that was twice as high. 47 Various circumstances can influence the Omicron variant’s increased transmission capacity. The most important one is that over 30 mutations changed the S protein of SARS‐CoV‐2 in Omicron. 98 Furthermore, the rate of re‐infection is significantly increased in the Omicron variation. It has been reported that, in South Africa, the re‐infection rate of formerly COVID‐19‐infected individuals is significantly higher with Omicron. 99 This is attributed to the mutations of the spike protein in Omicron’s structure. According to further computational analysis, Omicron has been shown to have a greater affinity for ACE2 than Delta. Q498R, S371L and T478K are mutations in the RBD of Omicron variants’ S protein accountable to this enhanced affinity to ACE2. 100 Also, Omicron is not as good at causing cell fusion inside cells as Delta, and this is because of the imperfect cell‐to‐cell transmission capability of Omicron. Fusion cells are prevalent in tissues taken from the lungs after a destructive infection. To support the findings of the decreased cell entrance, an infection experiment utilizing lung cells used a live Omicron variation which showed that Omicron’s entrance into the target cell was significantly lower than that of the Delta variant. 101
An early clinical investigation in South Africa found that the Omicron variation might evade the immune system and spread more quickly, both of which could have serious repercussions. 99 , 102 Investigators gathered evidence of Omicron immune escape potential, discovering that alterations at the T478, N440K, and N501Y locations increase the infectivity of OMICRON ten times more than the original SARS‐CoV‐2 variant and two times more than Delta variant, respectively. 103 When neutralized with pseudo‐type Omicron virus, antibody titers were 36 times lower for convalescent serum from patients infected with early strains and 39 times lower for convalescent serum from patients infected with the Delta strain, indicating natural immune escape by Omicron. 104
Coronavirus vaccinations do not protect against the Omicron form, which can elude the immune system’s defenses. Thorough preventative measures, including immunization, will be critical for eradicating the Omicron strain. The mutation rate and consequences on viral dynamics between individual hosts alter the host’s response to transmission and infection. All of these elements affect the genesis of viral variations and the expansion of pandemics. 105
To neutralize SARS‐CoV2, the human immune system produces a wide range of antibody categories: class 1, class 2 and class 3. Each of these classes targets a distinct part of the spike protein. The spike proteins that include the full‐length trimer, the S1 subunit, the NTD and the RBD are the principal targets of immune responses to Omicron. The absence of one of the three target genes responsible for spike protein production is called S gene dropout or S gene target failure, and it can be detected utilizing the RT‐PCR technique. 106
1.7. Severity of Omicron
Omicron‐infected individuals have been demonstrated to have a broad spectrum of disease, from moderate disease to organ failure and death. The severity of COVID‐19 is determined by the need for hospitalization, the length of hospital stay, the use of ventilators, the time necessary to recover and mortality. 107 The most prevailing type of infection is asymptomatic. An increase in hospitalizations is expected as the Omicron variety spreads rapidly. People who have not been immunized, the elderly and those with preexisting medical conditions are at a higher risk of developing a severe infection. 108
A study in South Africa was designed to compare the severity of BA.1 and BA.2 using logistic regression models. This study revealed that the hospitalization rate and disease severity did not differ between the two groups and concluded that the clinical settings of BA.1 and BA.2 are similar, 109 despite the possible growth advantage of BA.2. 93 The previous immunization rate against SARS‐CoV‐2, economic status, healthcare system quality and the existence of comorbidities are among the most important determinants of the Omicron outcome. 110 The severity of Omicron will have to be investigated in more depth in a much larger group of people before a conclusive illustration can be produced. 5 , 111
1.8. Vaccine immune protection against omicron
Numerous vaccines against SARS‐CoV‐2 have been produced and given for therapeutic and preventive purposes (Figure 1). Current vaccinations continue to be effective against severe disease and mortality, including the Delta variant, and boosters provide additional protection. 113 Vaccinated individuals are highly protected against hospitalization owing to Delta or Alpha. 114 Although the Omicron version diminishes the efficacy of Pfizer‐BioNTech’s SARS‐CoV‐2 vaccine, the vaccination can still minimize the risk of hospitalization. 27 Compared with the Wuhan variant’s genome, six mutations in Omicron’s genome have expanded transmissibility and vaccination resistance. 115
FIGURE 1.
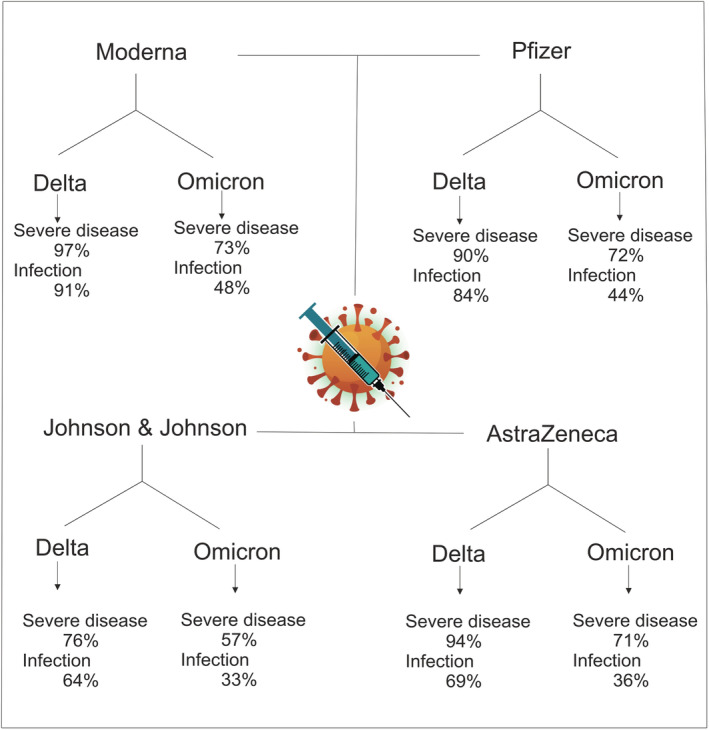
Vaccine protection in Delta and omicron variants. Created by Esmaeilzadeh et al. 112 (‘COVID‐19 vaccine efficacy summary’, Institute for Health Metrics and Evaluation, https://www.healthdata.org/covid/covid‐19‐vaccine‐efficacy‐summary, accessed 21 February 2022)
Low levels of neutralizing antibodies may protect against severe COVID‐19 infections. Since the surface features of the Omicron’s spike protein are targeted by T cells, which generally arise after immunization, T cells may be less affected than antibody responses by the Omicron mutations. BioNTech and Pfizer stated that two doses of the vaccine should still protect against severe illness. 116 , 117 On the other hand, according to computer modeling, Omicron appears to escape immunity consulted by T cells. Most patients evaluated who had received two doses of an mRNA vaccine or had recovered from COVID‐19 infection demonstrated that the poly mutant spike was resistant to neutralizing antibodies. 118
Omicron may bypass immunity induced by vaccinations or past infections and significantly reduce the efficacy of neutralizing antibodies, making current vaccines less effective against the Omicron. 119 According to research conducted by Discovery Ltd in South Africa, Omicron diminished vaccination efficacy against infection to 33%, down from 80% for Delta. Moreover, the effectiveness of the Pfizer‐BioNTech vaccine against severe disease and hospitalization has reduced from 93% to 70%. Individuals infected with the Delta variation have a 40% relative risk of getting the Omicron form. In contrast, those infected with the Beta variant had a 60% likelihood of re‐infection with Omicron at the start of 2020. 120 Since the Omicron infection has been recorded in people administered with the Johnson & Johnson, Pfizer–BioNTech and Oxford–AstraZeneca vaccines in South Africa, it can be concluded that Omicron infection cannot be prevented using the standard two‐dose mRNA vaccine. 121 , 122 However, the Pfizer vaccine was reported to improve SARS‐CoV‐2 neutralization. 116
Approximately 120 days after vaccination, the immune system’s response to SARS‐CoV‐2 declines. As a result, irrespective of VOC, a booster dose can prevent the transmission of this virus. Omicron may also be diminished by applying heterogeneous vaccinations; however, this assumption needs additional exploration. 123 Booster dosages have been advised to compensate for immunological evasion. Pfizer and BioNTech commented that booster dosage generates virus neutralization equivalent to the protection conferred by two doses against the prototype virus. 124 , 125
The frequency of mutations on the spike protein of Omicron variation predicts that the efficiency of antibodies generated by immunizations will be reduced. 126 Despite contradictory reports on the efficiency of the current vaccinations against the four SARS‐CoV‐2 mutations that preceded Omicron, their efficacy is acceptable. Vaccines have so far assisted in reducing the numbers of severe COVID‐19 infection, hospitalization and death, and the transmission of earlier mutations. The fact that vaccination effectiveness is dependent on T cell immune responses rather than antibodies might explain this hopeful outcome. 16 While CD4+ T cells do not prevent infection, they are essential for developing protective antibody responses and the maturation of CD8+ T cells. As a result, given the capacity of pathogenic variants to evade neutralization, the establishment and maintenance of strong SARS‐CoV‐2‐specific T cell responses may contribute to long‐term vaccination effectiveness against severe disease. Owing to the inability of HCoV‐specific antibodies and cellular responses to produce sterilizing immunity, there is a fear that protective immunity against SARS‐CoV‐2 will be similarly temporary. The information available at the moment paints a slightly contradictory picture. It is possible to be re‐infected with SARS‐CoV‐2; however, prior infection provides approximately 87% protection for 6 months with a stable profile for at least 10 months. 127
The sequencing of the seed strain should be modified during clinical trials according to the Omicron variation for the SARS‐CoV‐2 vaccine design and mixed vaccination to avoid immunological escape. 127 Broad vaccination in conjunction with highly efficient oral anti‐COVID‐19 medicines targeting conservative areas, such as RdRp, will significantly contribute to the epidemic’s abolition. 128
The immune protection of vaccines is being investigated in clinical trials. NCT05249829 is evaluating the immunogenicity of the mRNA‐1273.529 (Moderna) vaccine against B.1.1.529. A phase II study (NCT05238441) is designed to investigate the immune protection of SCTV01E against different SARS‐CoV‐2 variants. Two phase III trials, NCT05230953 and NCT05231005, are designed to evaluate the immunity given by the fourth dose of Moderna and Pfizer vaccines against Omicron.
1.9. Re‐infection with Omicron
Natural infection with the Alpha, Beta and Delta forms of the SARS‐CoV‐2 results in considerable protection against re‐infection. On the other hand, as discussed earlier, the Omicron carries several mutations that might facilitate immune evasion. This is associated with a higher potential for re‐infection and poor early estimates of vaccination efficacy against symptomatic infection. 129 Since SARS‐CoV‐2 re‐infection has been linked with negative S gene target failure data, most likely owing to random PCR target failure induced by the lower virus loads during re‐infection, scientists utilized genotype data to examine the influence of Omicron on re‐infection rates. Omicron was related to a 5.40‐fold increased risk of re‐infection compared with Delta after controlling vaccination status, age, sex, ethnicity, asymptomatic status, area and specimen data. To put this in context, the UK “SIREN” research on COVID infection in healthcare professionals in the pre‐Omicron period found that past infection provided 85% protection against a second COVID infection over 6 months. According to the present study’s anticipated re‐infection risk, this protection against an Omicron infection has dropped to 19%. 130 The previous infection was assessed to be 90.2% successful in avoiding re‐infection against the Alpha version, 85.7% effective against the Beta variation, 92.0% effective against the Delta variant and 56.0% effective against the Omicron form. 117 To conclude, current data have demonstrated that, owing to the mutations in the viral genome, immunity from previous COVID‐19 infection is less effective against re‐infection with Omicron. However, further studies are still required to evaluate the exact re‐infection probability and the risk of re‐infection associated with each variant.
1.10. Omicron and pediatrics
Despite the lower number of pediatric infections with previous forms of COVID‐19, Omicron has been shown to infect young individuals more often. During the Omicron outbreak, the number of new COVID‐19 hospitalizations in children exceeded an all‐time high. 131 Parents might take solace from knowing that Omicron does not appear to be associated with more severe disease in children than previous variations. Children, on average, continue to be at a lower risk of severe disease from COVID‐19 than adults. 132 On the other hand, children with particular health problems still have an elevated chance of severe disease. 20 The main theories suggested for the higher infection rate of Omicron in pediatrics are the late vaccination of children, the existence of naive B and T cells without prior immunity to Omicron antigens and a lack of cross‐immunity between prior immune responses to the Omicron spike protein. 133
Countries with substantial exposure to the Omicron form are reporting record numbers of child coronavirus hospitalizations, putting further strain on healthcare systems, even though the great majority of cases are mild, according to health specialists. Data from Europe and the USA suggest that there have been more child admissions recently than at any other time throughout the pandemic. However, severe cases are uncommon in the lowest age groups, and Omicron infections appear comparable with those of other typical respiratory illnesses. 132 It is crucial to keep an eye out for any signs of COVID‐19 infection as the disease progresses. Typically, manifestations of Omicron in children continue to be comparable with those produced by earlier variations. Among the symptoms of COVID‐19 in children are fever (a minimum temperature of 100°F), chills, cough, breathing difficulties, diarrhea, shortness of breath, fatigue, vomiting, etc. 134
According to research, pediatric SARS‐CoV‐2 infections and hospitalizations are on the rise in South Africa and the USA. One study found that the risk of hospitalization with Omicron variant among unvaccinated children under the age of 5 was one‐third that of the Delta variant, while the risk of an Emergency Department visit was less than one‐fifth, both of which were statistically significant. 132 Similar hospitalization and emrgency visit patterns were detected in children aged 5–11 and 12–17. 135 This is especially alarming considering that in the USA, children under the age of 5 are ineligible for COVID‐19 immunization, and children aged 5–11 do not qualify for boosters. 131 These findings imply that, although the numbers of pediatric SARS‐CoV‐2 infections and hospitalizations are increasing, the outcomes are milder following the introduction of the Omicron variation compared with the preceding dominant Delta variant period. One Swedish study confirmed previous findings that the SARS‐CoV‐2 virus is linked to neurological results in children. 136
Based on research, although children have the same symptoms and quantities of virus in their systems, their involvement with SARS‐CoV‐2 is more likely to induce antibodies against the virus than adults. In comparison with adults, children appear to have a more potent first immune response to COVID‐19 and can eliminate the infection quickly. 137 However, since antibodies are expected to play a role in preventing re‐infection, the findings raise concerns about how effectively protected infants are against subsequent diseases. Researchers collected nasal and throat swabs to determine patients’ viral RNA levels and blood samples to determine if they had IgG antibodies against the Omicron. They discovered that, while children and adults had comparable virus loads, only 37% of children developed antibodies, compared with 76% of adults. 138 Adults produce a more extensive range of antibodies than children, including a more significant number of virus‐blocking antibodies. Children may produce fewer antibodies than adults owing to a potent innate immune response. This is the initial defense against infections, and it is non‐specific. Additionally, children may be more adept at reacting to illnesses that enter the body by the throat or nose. This subject implies that the virus is cleared swiftly by the body and does not persist to induce the adaptive response that creates antibodies.
1.11. Challenges and alternative approaches
The new Omicron variant has shown partial resistance to natural immune antibodies, monoclonal antibodies and vaccine‐induced antibodies, and is also associated with new challenges compared with previous variants, including higher transmissibility, infectious rate and immune evasion. These challenges necessitate the development of novel technology‐based methods to tackle and neutralize the virus. In the case of diagnosis, nanofiber swabs are an example of more rapid tests that can be used to detect SARS‐CoV‐2. 139
Nanosystems can be used as carriers of the drugs for efficient delivery of the drugs to a specific organ. To achieve this aim, drugs are nano‐encapsulated (using different nanocarriers such as nanosomes, nanoemulsions, etc.). Nanosystems can be used as a therapeutic option for the successful delivery of drugs for Omicron. 140 Nano‐neutraceuticals, in the shape of nanosystem‐based masks, gloves and disinfectants, are another example of using nanotechnology for overcoming the higher transmissibility of Omicron. 141 Nanotechnology can also be used to improve therapeutic methods and develop vaccines with a higher efficacy (lipid nanoparticles). Nanosystems can be used for the delivery of vaccines to the target site of the virus using smart nanocarriers. Moreover, bioinformatics and artificial intelligence can be used to improve the computer simulation and evaluate the efficacy of novel drugs against SARS‐CoV‐2. 142 According to the several mutations of the Omicron variant, novel antiviral drugs should be developed for successfully tackling the virus. Paxlovid and Mulnopirvair are two examples of these specific drugs that have recently shown potent efficacy against Omicron. 143
In this context, specialists have studied the opto‐electro‐magnetic nanosystem using biosensing to diagnose the SARS‐CoV‐2 virus. These efficient‐miniaturized biosensors may be managed with a smartphone and advocate for early‐stage COVID‐19 infection diagnosis in clinical settings. 144 Researchers have created stimuli‐responsive nanotechnology that can capture aerosols of viral size and eliminate viruses upon external stimulation, such as nano‐enable photo‐sensitive virus destruction, to avoid the spread of SARS‐CoV‐2 virus infection. In terms of studying innovative therapeutic compounds with more efficacy and fewer and more tolerable adverse effects, the participation of biotech‐pharma companies seems to be of great importance. Although the SARS‐CoV‐2 virus is novel, it has shown variant variation, making therapy optimization difficult. However, biotechnology specialists examine all facets of bioinformatics to create and develop a successful cure based on innovative antiviral drugs, antibodies, CRISPR‐Cas and vaccines. 145 , 146
Orienting or boosting immunity through diet is another method for managing COVID‐19 infection. For instance, nutraceuticals have served as inhibitors to disrupt the interaction between the COVID‐19 virus and ACE2. Investigating features of nanomedicine might be a potential strategy for developing innovative therapeutics to treat COVID‐19 infection and strengthen the immune system and organs impacted by the SARS‐CoV‐2 virus. 147
Because of the multiple mutations outlined in Omicron, this variety has developed more elevated immune evasion than the previous ones, meaning that it is partially resistant to monoclonal antibodies, vaccination neutralizing antibodies and those constructed from natural immunity. Current results also imply that natural and vaccine‐induced immunity would will act differently in the case of Omicron infection. To successfully treat Omicron patients, the first phase is to diagnose Omicron patients correctly. As an important topic, the most reliable diagnostic techniques efficient in detecting Omicron variants related to S‐protein include PCR and antigen COVID‐19 testing. However, intense efforts are necessary for Omicron‐specific fast testing kits because these tests are time‐consuming and costly. 148
The literature contains proposals that can be adjusted to the variety and may be commercially available shortly. The use of nanoparticles to separate RNA or DNA from biological materials through a magnetic field can be an alternative means of immediate diagnosis. These nano‐enable SARSCoV‐2 sensors effectively identified a low level of specific viral concentration. In addition to the intelligent nanosystem, various efficient biomarkers, including antibodies, CIRSR/Cas, and segment‐specific DNA/RNA, have been studied to detect SARS‐CoV‐2 in actual samples. 149 Regardless, before they can be recommended for clinical and point‐of‐care testing, these nanotechnology‐supported biosensing devices must undergo comprehensive validation and population‐based confirmation. In addition, these nanobiosensors have not been evaluated against the Omicron, which must be the top priority. 150 Modern SARS‐CoV‐2 biosensors target the spike protein; therefore, they should theoretically function. Because Omicron includes more mutations at the spike protein than other variations, a reasonably designed and high‐throughput validation of existing diagnostic tools must be performed to diagnose a large population and establish whether COVID‐19 infection is due to Omicron. 151
2. CONCLUSION
The Omicron variant has thus far received less awareness, and further research is urgently needed to understand its threat correctly. Previous experiences with other variants, especially Delta strains, have shown that we can only better understand the transmission potential, vaccination effectiveness and severity of infection through continued compliance. Undoubtedly, it is not by chance that Omicron has developed in a country where vaccination rates are low. As this new variant emerges, it becomes even more critical to provide universal access to vaccination because allowing the virus to circulate freely in non‐vaccinated populations threatens them with severe COVID‐19 cases and deaths. Moreover, this will provide the virus with more opportunities to undergo mutations that can boost viral transmissibility and infectivity or conduct new harmful waves around the world. Vaccination and other preventative measures will continue to be crucial in halting the spread of COVID‐19 and preventing further outbreaks of severe illness and death.
3. FUTURE PERSPECTIVES
According to the research, Omicron infection is distinct from the COVID‐19 infection induced by other variations. Even if it is less severe than Delta, it has substantial health consequences. Furthermore, Omicron can re‐infect the COVID‐19‐infected population or those who have received both vaccine doses. 152 A thorough investigation of how Omicron may escape vaccine‐induced immune protection and how it could reduce immunity to infection is urgently needed, notwithstanding the possibility that the variant can evade immunity resulting from previous infections. 95 Nevertheless, comprehensive vaccination is suggested to reduce hospitalization and fatality rates; however, it is uncertain how successfully the antibodies generated by current vaccinations can neutralize Omicron variants, and this should be explored thoroughly. With early access to anti‐inflammatory, antiviral and immunotherapies, like MAbs, to treat Omicron infection, hospitalizations and deaths in high‐risk infected and exposed patients could be cut by 70%. 139
Pfizer and Moderna came up with the idea of a fourth booster dose, which could be followed by other doses. The health agencies agree with this claim because they want to develop timely and effective treatments. The appearance of the Omicron variation, with its high‐level transmission and quick expansion worldwide and its significant danger to several authorized COVID‐19 vaccines and therapies, demonstrated that new COVID19 variants are incredibly likely to reappear sooner or later. 99 As a result, biopharmaceutical firms may conclude an unmet need for the creation and development of innovative treatments and techniques that can forecast the evolutionary path of SARS‐CoV‐2 and its severity in the event of the appearance of a deadlier coronavirus in the future. Many studies must be conducted in corporate and academic settings to prevent such contagious infections.
CONFLICT OF INTEREST
The authors declare having no financial or scientific conflicts of interest.
AUTHOR CONTRIBUTION
M.B. contributed to data gathering, writing the primary manuscript, and designing tables and Figures. R.E. contributed to revising the manuscript and editing figures and Tables. A.E. contributed to the hypothesis, correspondence, and editing manuscript before submission.
ACKNOWLEDGEMENTS
The authors would like to express deep appreciation for healthcare workers in the field of COVID‐19 and wish all scholars be safe. We wish that proper control of the pandemic will be available by immediate global vaccination.
Bazargan M, Elahi R, Esmaeilzadeh A. OMICRON: Virology, immunopathogenesis, and laboratory diagnosis. J Gene Med. 2022;24(7):e3435. doi: 10.1002/jgm.3435
Funding information
The authors have received no funding for preparing the manuscript.
REFERENCES
- 1. Lvov DK, Alkhovsky SV. Source of the COVID‐19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS‐CoV, SARS‐CoV‐2 (subgenus Sarbecovirus), and MERS‐CoV (subgenus Merbecovirus). Vopr Virusol. 2020;65(2):62‐70. doi: 10.36233/0507-4088-2020-65-2-62-70 [DOI] [PubMed] [Google Scholar]
- 2. Elahi R, Karami P, Heidary AH, Esmaeilzadeh A. An updated overview of recent advances, challenges, and clinical considerations of IL‐6 signaling blockade in severe coronavirus disease 2019 (COVID‐19). Int Immunopharmacol. 2022;105:108536. doi: 10.1016/j.intimp.2022.108536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohamadian M, Chiti H, Shoghli A, Biglari S, Parsamanesh N, Esmaeilzadeh A. COVID‐19: Virology, biology and novel laboratory diagnosis. J Gene Med. 2021;23(2):e3303. doi: 10.1002/jgm.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ingraham NE, Ingbar DH. The omicron variant of SARS‐CoV‐2: Understanding the known and living with unknowns. Clin Transl Med. 2021;11(12):e685. doi: 10.1002/ctm2.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meo S, Meo A, Al‐Jassir F, Klonoff D. Omicron SARS‐CoV‐2 new variant: global prevalence and biological and clinical characteristics. Eur Rev Med Pharmacol Sci. 2021;25(24):8012‐8018. doi: 10.26355/eurrev_202112_27652 [DOI] [PubMed] [Google Scholar]
- 6. Callaway E. Heavily mutated coronavirus variant puts scientists on alert. Nature. 2021;600(7887):21. doi: 10.1038/d41586-021-03552-w [DOI] [PubMed] [Google Scholar]
- 7. Walensky RP, Walke HT, Fauci AS. SARS‐CoV‐2 variants of concern in the United States—challenges and opportunities. Jama. 2021;325(11):1037‐1038. doi: 10.1001/jama.2021.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duong D. Alpha, Beta, Delta, Gamma: What's important to know about SARS‐CoV‐2 variants of concern? Can Med Assoc. 2021;193(27):E1059‐E1060. doi: 10.1503/cmaj.1095949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS‐CoV‐2 variants. Nat Rev Genet. 2021;22(12):757‐773. doi: 10.1038/s41576-021-00408-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rajah MM, Hubert M, Bishop E, et al. SARS‐CoV‐2 Alpha, Beta, and Delta variants display enhanced Spike‐mediated syncytia formation. EMBO J. 2021;40(24):e108944. doi: 10.15252/embj.2021108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alizon S, Haim‐Boukobza S, Foulongne V, et al. Rapid spread of the SARS‐CoV‐2 Delta variant in some French regions, June 2021. Eurosurveillance. 2021;26(28):2100573. doi: 10.2807/1560-7917.ES.2021.26.28.2100573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khateeb J, Li Y, Zhang H. Emerging SARS‐CoV‐2 variants of concern and potential intervention approaches. Crit Care. 2021;25(1):1‐8. doi: 10.1186/s13054-021-03662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS‐CoV‐2 variants of concern: a retrospective cohort study comparing B. 1.1. 7 (Alpha), B. 1.315 (Beta), and B. 1.617. 2 (Delta). Clin Infect Dis. 2021:ciab721. doi: 10.1093/cid/ciab721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gu H, Krishnan P, Ng DY, et al. Probable transmission of SARS‐CoV‐2 omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg Infect Dis. 2022;28(2):460‐462. doi: 10.3201/eid2802.212422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li B, Deng A, Li K, et al. Viral infection and transmission in a large, well‐traced outbreak caused by the SARS‐CoV‐2 Delta variant. Nat Commun. 2022;13(1):1‐9. doi: 10.1038/s41467-022-28089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karim SSA, Karim QA. Omicron SARS‐CoV‐2 variant: a new chapter in the COVID‐19 pandemic. Lancet. 2021;398(10317):2126‐2128. doi: 10.1016/S0140-6736(21)02758-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altarawneh HN, Chemaitelly H, Hasan MR, et al. Protection against the Omicron Variant from Previous SARS‐CoV‐2 Infection. N Engl J Med. 2022;386(13):1288‐1290. doi: 10.1056/NEJMc2200133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chemaitelly H, Bertollini R, Abu‐Raddad LJ. Efficacy of Natural Immunity against SARS‐CoV‐2 Reinfection with the Beta Variant. N Engl J Med. 2021;385(27):2585‐2586. doi: 10.1056/NEJMc2110300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies MA, Kassanjee R, Rosseau P, et al. Outcomes of laboratory‐confirmed SARS‐CoV‐2 infection in the Omicron‐driven fourth wave compared with previous waves in the Western Cape Province, South Africa. medRxiv [Preprint]. 2022. doi: 10.1101/2022.01.12.22269148. Update in: Trop Med Int Health. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L‐L, Chua GT, Lu L, et al. Omicron variant susceptibility to neutralizing antibodies induced in children by natural SARS‐CoV‐2 infection or COVID‐19 vaccine. Emerg Microbes Infect. 2022;11(just‐accepted):1‐17. doi: 10.1080/22221751.2022.2035195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corbett KS, Gagne M, Wagner DA, et al. Protection against SARS‐CoV‐2 Beta variant in mRNA‐1273 vaccine–boosted nonhuman primates. Science. 2021;374(6573):1343‐1353. doi: 10.1126/science.abl8912 [DOI] [PubMed] [Google Scholar]
- 22. Wuertz KM, Barkei EK, Chen W‐H, et al. A SARS‐CoV‐2 spike ferritin nanoparticle vaccine protects hamsters against Alpha and Beta virus variant challenge. NPJ Vacc. 2021;6(1):1‐11. doi: 10.1038/s41541-021-00392-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carreño JM, Alshammary H, Tcheou J, et al. Activity of convalescent and vaccine serum against SARS‐CoV‐2 Omicron. Nature. 2022;602(7898):682‐688. doi: 10.1038/s41586-022-04399-5 [DOI] [PubMed] [Google Scholar]
- 24. Chemaitelly H, Tang P, Hasan MR, et al. Waning of BNT162b2 vaccine protection against SARS‐CoV‐2 infection in Qatar. N Engl J Med. 2021;385(24):e83. doi: 10.1056/NEJMoa2114114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torjesen I. Covid‐19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. British Medical Journal Publishing Group; 2021. doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 26. Rahmani S, Rezaei N. Omicron (B. 1.1. 529) variant: Development, dissemination, and dominance. J Med Virol. 2021;94(5):1787‐1788. doi: 10.1002/jmv.27563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Torjesen I. Covid restrictions tighten as omicron cases double every two to three days. British Medical Journal Publishing Group; 2021. [DOI] [PubMed] [Google Scholar]
- 28. Altarawneh H, Chemaitelly H, Tang P, et al. Protection afforded by prior infection against SARS‐CoV‐2 reinfection with the Omicron variant. medRxiv. 2022. [Google Scholar]
- 29. Shi Y, Wang Y, Shao C, et al. COVID‐19 infection: the perspectives on immune responses. Nature Publishing Group; 2020:1451‐1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mortaz E, Tabarsi P, Varahram M, Folkerts G, Adcock IM. The immune response and immunopathology of COVID‐19. Front Immunol. 2020;11:2037. doi: 10.3389/fimmu.2020.02037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schultze JL, Aschenbrenner AC. COVID‐19 and the human innate immune system. Cell. 2021;184(7):1671‐1692. doi: 10.1016/j.cell.2021.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valizadeh H, Abdolmohammadi‐vahid S, Danshina S, et al. Nano‐curcumin therapy, a promising method in modulating inflammatory cytokines in COVID‐19 patients. Int Immunopharmacol. 2020;89(Pt B):107088. doi: 10.1016/j.intimp.2020.107088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75(7):1564‐1581. doi: 10.1111/all.14364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hojyo S, Uchida M, Tanaka K, et al. How COVID‐19 induces cytokine storm with high mortality. Inflamm Regen. 2020;40(1):1‐7. doi: 10.1186/s41232-020-00146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Esmaeilzadeh A, Elahi R. Immunobiology and immunotherapy of COVID‐19: A clinically updated overview. J Cell Physiol. 2021;236(4):2519‐2543. doi: 10.1002/jcp.30076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu J, Li S, Liu J, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS‐CoV‐2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Esmaeilzadeh A, Jafari D, Tahmasebi S, Elahi R, Khosh E. Immune‐Based Therapy for COVID‐19. Coronavirus Disease‐COVID‐19. Springer; 2021:449‐468. [DOI] [PubMed] [Google Scholar]
- 38. Foix A, López D, Díez‐Fuertes F, McConnell MJ, Martín‐Galiano AJ. Predicted impact of the viral mutational landscape on the cytotoxic response against SARS‐CoV‐2. PLoS Comput Biol. 2022;18(2):e1009726. doi: 10.1371/journal.pcbi.1009726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen X. Boosting immunity to Omicron. Nat Med. 2022;28(3):445‐446. doi: 10.1038/s41591-022-01727-0 [DOI] [PubMed] [Google Scholar]
- 40. Kodera S, Rashed EA, Hirata A. Estimation of real‐world vaccination effectiveness of mRNA COVID‐19 vaccines against Delta and omicron variants in Japan. Vaccine. 2022;10(3):430. doi: 10.3390/vaccines10030430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nersisyan S, Zhiyanov A, Zakharova M, et al. Alterations in SARS‐CoV‐2 Omicron and Delta peptides presentation by HLA molecules. PeerJ. 2022;10:e13354. doi: 10.7717/peerj.13354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Esmaeilzadeh A, Rostami S, Yeganeh PM, Tahmasebi S, Ahmadi M. Recent advances in antibody‐based immunotherapy strategies for COVID‐19. J Cell Biochem. 2021;122(10):1389‐1412. doi: 10.1002/jcb.30017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Batah SS, Fabro AT. Pulmonary pathology of ARDS in COVID‐19: a pathological review for clinicians. Respir Med. 2021;176:106239. doi: 10.1016/j.rmed.2020.106239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033‐1034. doi: 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boechat JL, Chora I, Morais A, Delgado L. The immune response to SARS‐CoV‐2 and COVID‐19 immunopathology–current perspectives. Pulmonology. 2021;27(5):423‐437. doi: 10.1016/j.pulmoe.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lyngse FP, Kirkeby CT, Denwood M, et al. Transmission of SARS‐CoV‐2 Omicron VOC subvariants BA. 1 and BA. 2: evidence from Danish Households. MedRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mohsin M, Mahmud S. Omicron SARS‐CoV‐2 Variant of Concern: A Review on its Transmissibility, Immune Evasion, Reinfection, and Severity. Medicine (Baltimore). 2022;101(19):e29165. doi: 10.1097/MD.0000000000029165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fall A, Eldesouki RE, Sachithanandham J, et al. A Quick Displacement of the SARS‐CoV‐2 variant Delta with Omicron: Unprecedented Spike in COVID‐19 Cases Associated with Fewer Admissions and Comparable Upper Respiratory Viral Loads. medRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meng B, Abdullahi A, Ferreira IA, et al. Altered TMPRSS2 usage by SARS‐CoV‐2 Omicron impacts tropism and fusogenicity. Nature. 2022;603(7902):706‐714. doi: 10.1038/s41586-022-04474-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peacock TP, Brown JC, Zhou J, et al. The SARS‐CoV‐2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. BioRxiv. 2022. [Google Scholar]
- 51. Maisa A, Spaccaferri G, Fournier L, et al. First cases of Omicron in France are exhibiting mild symptoms, November 2021–January 2022. Infect Dis Now. 2022;52(3):160‐164. doi: 10.1016/j.idnow.2022.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mahase E. Covid‐19: Hospital admission 50–70% less likely with omicron than delta, but transmission a major concern. British Medical Journal Publishing Group; 2021. [DOI] [PubMed] [Google Scholar]
- 53. Tahmasebi S, Khosh E, Esmaeilzadeh A. The outlook for diagnostic purposes of the 2019‐novel coronavirus disease. J Cell Physiol. 2020;235(12):9211‐9229. doi: 10.1002/jcp.29804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ettaboina SK, Nakkala K, Laddha K. A Mini Review on SARS‐COVID‐19‐2 Omicron Variant (B. 1.1. 529). Sci Med J. 2021;3(4):399‐406. doi: 10.28991/SciMedJ-2021-0304-10 [DOI] [Google Scholar]
- 55. Kumar S, Karuppanan K, Subramaniam G. Omicron (BA. 1) and sub‐variants (BA. 1, BA. 2 and BA. 3) of SARS‐CoV‐2 spike infectivity and pathogenicity: A comparative sequence and structural‐based computational assessment. bioRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fonager J, Bennedbæk M, Bager P, et al. Molecular epidemiology of the SARS‐CoV‐2 variant Omicron BA. 2 sub‐lineage in Denmark, 29 November 2021 to 2 January 2022. Eurosurveillance. 2022;27(10):2200181. doi: 10.2807/1560-7917.ES.2022.27.10.2200181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen X, Kang Y, Luo J, et al. Next‐generation sequencing reveals the progression of COVID‐19. Front Cell Infect Microbiol. 2021;11:142. doi: 10.3389/fcimb.2021.632490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Allam M, Ismail A, Khumalo ZT, et al. Genome Sequencing of a Severe Acute Respiratory Syndrome Coronavirus 2 Isolate Obtained from a South African Patient with Coronavirus Disease 2019. Microbiol Res Announce. 2020;9(27):e00572‐e00520. doi: 10.1128/MRA.00572-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sahajpal NS, Mondal AK, Njau A, et al. High‐Throughput Next‐Generation Sequencing Respiratory Viral Panel: A Diagnostic and Epidemiologic Tool for SARS‐CoV‐2 and Other Viruses. Viruses. 2021;13(10):2063. doi: 10.3390/v13102063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mak GC, Cheng PK, Lau SS, et al. Evaluation of rapid antigen test for detection of SARS‐CoV‐2 virus. J Clin Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Izquierdo‐Lara R, Elsinga G, Heijnen L, et al. Monitoring SARS‐CoV‐2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg Infect Dis. 2021;27(5):1405‐1415. doi: 10.3201/eid2705.204410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fontanet A, Autran B, Lina B, Kieny MP, Karim SSA, Sridhar D. SARS‐CoV‐2 variants and ending the COVID‐19 pandemic. Lancet. 2021;397(10278):952‐954. doi: 10.1016/S0140-6736(21)00370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dikdan RJ, Marras SA, Field AP, et al. A Multiplex PCR Assay for Identifying All Major SARS‐CoV‐2 Variants. medRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vogels CB, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS‐COV‐2 qRT‐PCR assays. MedRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tan W, Lu Y, Zhang J, et al. Viral kinetics and antibody responses in patients with COVID‐19. MedRxiv. 2020. [Google Scholar]
- 66. Espejo AP, Akgun Y, al Mana AF, et al. Review of current advances in serologic testing for COVID‐19. Am J Clin Pathol. 2020;154(3):293‐304. doi: 10.1093/ajcp/aqaa112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li D, Li J. Immunologic testing for SARS‐CoV‐2 infection from the antigen perspective. J Clin Microbiol. 2020;59(5):e02160‐e02120. doi: 10.1128/JCM.02160-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ward S, Lindsley A, Courter J, Assa'ad A. Clinical testing for COVID‐19. J Allergy Clin Immunol. 2020;146(1):23‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu C‐r, Yin W‐c, Jiang Y, Xu HE. Structure genomics of SARS‐CoV‐2 and its Omicron variant: drug design templates for COVID‐19. Acta Pharmacol Sin. 2022;1‐13. doi: 10.1038/s41401-021-00851-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Trus I, Udenze D, Berube N, et al. CpG‐recoding in Zika virus genome causes host‐age‐dependent attenuation of infection with protection against lethal heterologous challenge in mice. Front Immunol. 2020;10:3077. doi: 10.3389/fimmu.2019.03077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cevik M, Bamford C, Ho A. COVID‐19 pandemic—a focused review for clinicians. Clin Microbiol Infect. 2020;26(7):842‐847. doi: 10.1016/j.cmi.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vanessa M, Hyesoo K, Patricia P, et al. Inhibition of SARS‐CoV‐2 Infections in Engineered Human Tissues Using Clinical‐Grade Soluble Human ACE2. Cell. 2020;181(4):905‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet. 2020;395(10234):1417‐1418. doi: 10.1016/S0140-6736(20)30937-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Weiss S, Leibowitz J. Advances in virus research. Adv Virus Res. 2011;81:85‐164. doi: 10.1016/B978-0-12-385885-6.00009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kannan S, Ali PSS, Sheeza A. Evolving biothreat of variant SARS‐CoV‐2‐molecular properties, virulence and epidemiology. Eur Rev Med Pharmacol Sci. 2021;25(12):4405‐4412. doi: 10.26355/eurrev_202106_26151 [DOI] [PubMed] [Google Scholar]
- 77. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Abbott S, Hellewell J, Thompson RN, et al. Estimating the time‐varying reproduction number of SARS‐CoV‐2 using national and subnational case counts [version 2; peer review: 1 approved with reservations]. Wellcome Open Res. 2020;5:112. doi: 10.12688/wellcomeopenres.16006.2 [DOI] [Google Scholar]
- 79. Walls AC, Park Y‐J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Spratt AN, Kannan SR, Woods LT, et al. Evolution, correlation, structural impact and dynamics of emerging SARS‐CoV‐2 variants. Comput Struct Biotechnol J. 2021;19:3799‐3809. doi: 10.1016/j.csbj.2021.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li B, Luo X, McAndrews KM, Kalluri R. Mutations in the spike RBD of SARS‐CoV‐2 omicron variant may increase infectivity without dramatically altering the efficacy of current multi‐dosage vaccinations. bioRxiv. 2021. [Google Scholar]
- 82. Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS‐CoV‐2 variant: Unique features and their impact on pre‐existing antibodies. J Autoimmun. 2022;126:102779. doi: 10.1016/j.jaut.2021.102779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shang J, Ye G, Shi K. et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581:221‐224. doi: 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Venkatakrishnan A, Anand P, Lenehan PJ et al. Omicron variant of SARS‐CoV‐2 harbors a unique insertion mutation of putative viral or human genomic origin. 2021.
- 85. Bayarri‐Olmos R, Jarlhelt I, Johnsen LB, et al. Functional Effects of Receptor‐Binding Domain Mutations of SARS‐CoV‐2 B. 1.351 and P. 1 Variants. Front Immunol. 2021;12:4145. doi: 10.3389/fimmu.2021.757197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang L, Cheng G. Sequence analysis of the Emerging Sars‐CoV‐2 Variant Omicron in South Africa. J Med Virol. 2021;94(4):1728‐1733. doi: 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 87. World Health Organization . Enhancing response to Omicron SARS‐CoV‐2 variant: Technical brief and priority actions for Member States. World Health Organization Headquarters, Geneva, Switzerland Update. 2022;(6).
- 88. Garcia‐Beltran WF, St. Denis KJ, Hoelzemer A, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell. 2022;185(3):457‐466.e4. doi: 10.1016/j.cell.2021.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Callaway E. Beyond Omicron: what's next for COVID's viral evolution. Nature Publishing Group; 2021, 600, 7888, 204, 207. doi: 10.1038/d41586-021-03619-8. [DOI] [PubMed] [Google Scholar]
- 90. Chen J, Wei G‐W. Omicron BA. 2 (B. 1.1. 529.2): high potential to becoming the next dominating variant. arXiv preprint arXiv:220205031. 2022.
- 91. Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA. 3 and its importance. J Med Virol. 2022;94(5):1808‐1810. doi: 10.1002/jmv.27601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mahase E. Omicron sub‐lineage BA. 2 may have “substantial growth advantage,” UKHSA reports. British Medical Journal Publishing Group; 2022. [DOI] [PubMed] [Google Scholar]
- 94. Chen J, Qiu Y, Wang R, Wei G‐W. Persistent Laplacian projected Omicron BA. 4 and BA. 5 to become new dominating variants arXiv preprint arXiv:220500532. 2022. [DOI] [PMC free article] [PubMed]
- 95. Cao Y, Yisimayi A, Jian F, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022. doi: 10.1038/s41586-022-04980-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tegally H, Moir M, Everatt J, et al. Continued Emergence and Evolution of Omicron in South Africa: New BA. 4 and BA. 5 lineages. medRxiv. 2022. [Google Scholar]
- 97. Yamasoba D, Kosugi Y, Kimura I, et al. Sensitivity of novel SARS‐CoV‐2 Omicron subvariants, BA. 2.11, BA. 2.12. 1, BA. 4 and BA. 5 to therapeutic monoclonal antibodies. bioRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lyngse FP, Mortensen LH, Denwood MJ, et al. SARS‐CoV‐2 Omicron VOC Transmission in Danish Households. medRxiv. 2021. [Google Scholar]
- 99. Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS‐CoV‐2 reinfection associated with emergence of the Omicron variant in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and Delta variant of SARS‐CoV‐2: a comparative computational study of spike protein. J Med Virol. 2022;94(4):1641‐1649. doi:10.1002/jmv.27526 [DOI] [PubMed] [Google Scholar]
- 101. Meng B, Ferreira I, Abdullahi A, et al. SARS‐CoV‐2 Omicron spike mediated immune escape, infectivity and cell‐cell fusion. BioRxiv. 2021. [Google Scholar]
- 102. Vaughan A. Omicron emerges. Elsevier; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen J, Wang R, Gilby NB, Wei GW. Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. 2022;ArXiv [Preprint]. 2021. arXiv:2112.01318v1. Update in: J Chem Inf Model. 2022;62(2):412‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang L, Li Q, Liang Z, et al. The significant immune escape of pseudotyped SARS‐CoV‐2 Variant Omicron. Emerg Microbes Infect. 2022;11(1):1‐5. doi: 10.1080/22221751.2021.2017757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bal A, Destras G, Gaymard A, et al. Two‐step strategy for the identification of SARS‐CoV‐2 variant of concern 202012/01 and other variants with spike deletion H69–V70, France, August to December 2020. Eurosurveillance. 2021;26(3):2100008. doi: 10.2807/1560-7917.ES.2021.26.3.2100008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Brits E, Adepoju P. Omicron potential under close scrutiny. Nature. 2021;10:197‐199. [Google Scholar]
- 107. Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes among patients infected with Omicron (B. 1.1. 529) SARS‐CoV‐2 variant in southern California. medRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ghaebi M, Tahmasebi S, Jozghorbani M, et al. Risk factors for adverse outcomes of COVID‐19 patients: Possible basis for diverse responses to the novel coronavirus SARS‐CoV‐2. Life Sci. 2021;277:119503. doi: 10.1016/j.lfs.2021.119503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wolter N, Jassat W, von Gottberg A, Cohen C. Clinical severity of Omicron sub‐lineage BA. 2 compared to BA. 1 in South Africa. medRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gao SJ, Guo H, Luo G. Omicron variant (B. 1.1. 529) of SARS‐CoV‐2, a global urgent public health alert! Wiley Online Library; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nealon J, Cowling BJ. Omicron severity: milder but not mild. Lancet. 2022;399(10323):412‐413. doi: 10.1016/S0140-6736(22)00056-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tseng HF, Ackerson BK, Luo Y, et al. Effectiveness of mRNA‐1273 against SARS‐CoV‐2 omicron and delta variants. medRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Callaway E, Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600(7888):197‐199. doi: 10.1038/d41586-021-03614-z [DOI] [PubMed] [Google Scholar]
- 114. Ren S‐Y, Wang W‐B, Gao R‐D, Zhou A‐M. Omicron variant (B. 1.1. 529) of SARS‐CoV‐2: Mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. 2022;10(1):1‐11. doi: 10.12998/wjcc.v10.i1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gowrisankar A, Priyanka T, Banerjee S. Omicron: a mysterious variant of concern. Eur Phys J Plus. 2022;137(1):1‐8. doi: 10.1140/epjp/s13360-021-02321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Basile K, Rockett RJ, McPhie K, et al. Improved neutralization of the SARS‐CoV‐2 Omicron variant after Pfizer‐BioNTech BNT162b2 COVID‐19 vaccine boosting. bioRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Khan NA, Al‐Thani H, El‐Menyar A. The emergence of new SARS‐CoV‐2 variant (Omicron) and increasing calls for COVID‐19 vaccine boosters‐The debate continues. Travel Med Infect Dis. 2022;45:102246. doi: 10.1016/j.tmaid.2021.102246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Graham F. Daily briefing: Omicron coronavirus variant puts scientists on alert. Nature. 2021. doi: 10.1038/d41586-021-03564-6 [DOI] [PubMed] [Google Scholar]
- 119. Kupferschmidt K, Vogel G. How bad is Omicron? Some clues are emerging. Science. 2021;374(6573):1304‐1305. doi: 10.1126/science.acx9782 [DOI] [PubMed] [Google Scholar]
- 120. Callaway E. Omicron likely to weaken COVID vaccine protection. Nature. 2021;600(7889):367‐368. [DOI] [PubMed] [Google Scholar]
- 121. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654‐656. doi: 10.1038/s41586-021-04387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kuhlmann C, Mayer CK, Claassen M et al. Breakthrough infections with SARS‐CoV‐2 Omicron variant despite booster dose of mRNA vaccine. 2021. Available at SSRN 3981711. [DOI] [PMC free article] [PubMed]
- 123. Ferré VM, Peiffer‐Smadja N, Visseaux B, Descamps D, Ghosn J, Charpentier C. Omicron SARS‐CoV‐2 variant: What we know and what we don't. Anaesth Crit Care Pain Med. 2022;41(1):100998. doi: 10.1016/j.accpm.2021.100998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhao X, Li D, Ruan W, et al. Reduced sera neutralization to Omicron SARS‐CoV‐2 by both inactivated and protein subunit vaccines and the convalescents. bioRxiv. 2021. [Google Scholar]
- 125. Cele S, Jackson L, Khoury DS, et al. SARS‐CoV‐2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. MedRxiv. 2021. doi: 10.1101/2021.12.08.21267417 [DOI] [Google Scholar]
- 126. Burki TK. Omicron variant and booster COVID‐19 vaccines. Lancet Respir Med. 2022;10(2):e17. doi: 10.1016/S2213-2600(21)00559-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hall VJ, Foulkes S, Charlett A, et al. SARS‐CoV‐2 infection rates of antibody‐positive compared with antibody‐negative health‐care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. 2021;397(10283):1459‐1469. doi: 10.1016/S0140-6736(21)00675-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Fan H, Lou F, Fan J, Li M, Tong Y. The emergence of powerful oral anti‐COVID‐19 drugs in the post‐vaccine era. Lancet Microbe. 2022;3(2):e91. doi: 10.1016/S2666-5247(21)00278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cameroni E, Bowen JE, Rosen LE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. Nature. 2021:1‐9. doi: 10.1038/d41586-021-03825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ferguson N, Ghani A, Cori A, Hogan A, Hinsley W, Volz E. Report 49: Growth, population distribution and immune escape of Omicron in England. Imperial College London (16‐12‐2021). 2021. doi: 10.25561/93038 [DOI]
- 131. Cloete J, Kruger A, Masha M, et al. Paediatric hospitalisations due to COVID‐19 during the first SARS‐CoV‐2 omicron (B. 1.1. 529) variant wave in South Africa: a multicentre observational study. Lancet Child Adolesc Health. 2022;6(5):294‐302. doi: 10.1016/S2352-4642(22)00027-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang L, Berger NA, Kaelber DC, Davis PB, Volkow ND, Xu R. COVID infection severity in children under 5 years old before and after Omicron emergence in the US. medRxiv. 2022. doi: 10.1101/2022.01.12.22269179 [DOI] [Google Scholar]
- 133. Tang J, Randolph AG, Novak T, et al. Systemic and Lower Respiratory Tract Immunity to SARS‐CoV‐2 Omicron and Variants in Pediatric Severe COVID‐19 and Mis‐C. Vaccine. 2022;10(2):270. doi: 10.3390/vaccines10020270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cloete J, Kruger A, Masha M, et al. Rapid rise in paediatric COVID‐19 hospitalisations during the early stages of the Omicron wave, Tshwane District, South Africa. medRxiv. 2021. [Google Scholar]
- 135. Wang L, Berger NA, Kaelber DC, Davis PB, Volkow ND, Xu R. Comparison of outcomes from COVID infection in pediatric and adult patients before and after the emergence of Omicron. medRxiv. 2022. doi: 10.1101/2021.12.30.21268495 [DOI] [Google Scholar]
- 136. Ludvigsson JF. Convulsions in children with COVID‐19 during the Omicron wave. Acta Paediatr. 2022;111(5):1023‐1026. doi: 10.1111/apa.16276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tang J, Novak T, Hecker J, et al. Cross‐reactive immunity against the SARS‐CoV‐2 Omicron variant is low in pediatric patients with prior COVID‐19 or MIS‐C. Nat Commun. 2022;13(1):1‐10. doi: 10.1038/s41467-022-30649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Toh ZQ, Anderson J, Mazarakis N, et al. Comparison of seroconversion in children and adults with mild COVID‐19. JAMA Netw Open. 2022;5(3):e221313. doi: 10.1001/jamanetworkopen.2022.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mostafavi E, Dubey AK, Teodori L, Ramakrishna S, Kaushik A. SARS‐CoV‐2 Omicron variant: A next phase of the COVID‐19 pandemic and a call to arms for system sciences and precision medicine. MedComm. 2022;3(1):e119. doi: 10.1002/mco2.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kaushik A. Manipulative magnetic nanomedicine: the future of COVID‐19 pandemic/endemic therapy. Expert Opin Drug Deliv. 2021;18(5):531‐534. doi: 10.1080/17425247.2021.1860938 [DOI] [PubMed] [Google Scholar]
- 141. Dubey AK, Chaudhry SK, Singh HB, Gupta VK, Kaushik A. Perspectives on nano‐nutraceuticals to manage pre and post COVID‐19 infections. Biotechnol Rep (Amst). 2022;33:e00712. doi: 10.1016/j.btre.2022.e00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tiwari S, Juneja S, Ghosal A, et al. Antibacterial and antiviral high‐performance nano‐systems to mitigate new SARS‐CoV‐2 variants of concerns. Curr Opinion Biomed Eng. 2022;21:100363. doi: 10.1016/j.cobme.2021.100363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mohapatra RK, Tiwari R, Sarangi AK, Islam MR, Chakraborty C, Dhama K. Omicron (B. 1.1. 529) variant of SARS‐CoV‐2: Concerns, challenges, and recent updates. J Med Virol. 2022;94(6):2336‐2342. doi: 10.1002/jmv.27633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kaushik AK, Dhau JS, Gohel H, et al. Electrochemical SARS‐CoV‐2 sensing at point‐of‐care and artificial intelligence for intelligent COVID‐19 management. ACS Appl Bio Mater. 2020;3(11):7306‐7325. doi: 10.1021/acsabm.0c01004 [DOI] [PubMed] [Google Scholar]
- 145. Mujawar MA, Gohel H, Bhardwaj SK, Srinivasan S, Hickman N, Kaushik A. Nano‐enabled biosensing systems for intelligent healthcare: towards COVID‐19 management. Mater Today Chem. 2020;17:100306. doi: 10.1016/j.mtchem.2020.100306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Paliwal P, Sargolzaei S, Bhardwaj SK, Bhardwaj V, Dixit C, Kaushik A. Grand challenges in bio‐nanotechnology to manage the COVID‐19 pandemic. Front Nanotechnol. 2020;2:571284. doi: 10.3389/fnano.2020.571284 [DOI] [Google Scholar]
- 147. Weiss C, Carriere M, Fusco L, et al. Toward nanotechnology‐enabled approaches against the COVID‐19 pandemic. ACS Nano. 2020;14(6):6383‐6406. doi: 10.1021/acsnano.0c03697 [DOI] [PubMed] [Google Scholar]
- 148. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature. 2022;602(7898):657‐663. doi: 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Phan QA, Truong LB, Medina‐Cruz D, Dincer C, Mostafavi E. CRISPR/Cas‐powered nanobiosensors for diagnostics. Biosens Bioelectron. 2022;197:113732. doi: 10.1016/j.bios.2021.113732 [DOI] [PubMed] [Google Scholar]
- 150. Saravanan M, Mostafavi E, Vincent S, et al. Nanotechnology‐based approaches for emerging and re‐emerging viruses: special emphasis on COVID‐19. Microb Pathog. 2021;156:104908. doi: 10.1016/j.micpath.2021.104908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Vernet‐Crua A, Medina‐Cruz D, Mostafavi E, et al. Nanobiosensors for theranostic applications. Handbook on Nanobiomaterials for Therapeutics and Diagnostic Applications. Elsevier; 2021:511‐543. [Google Scholar]
- 152. Ai J, Zhang H, Zhang Y, Lin K, et al. Omicron variant showed lower neutralizing sensitivity than other SARS‐CoV‐2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. 2022;11(1):337‐343. doi: 10.1080/22221751.2021.2022440 [DOI] [PMC free article] [PubMed] [Google Scholar]