Abstract
Since the onset of the coronavirus disease 2019 (COVID‐19) pandemic, preventive social paradigms and vaccine development have undergone serious renovations, which drastically reduced the viral spread and increased collective immunity. Although the technological advancements in diagnostic systems for severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) detection are groundbreaking, the lack of sensitive, robust, and consumer‐end point‐of‐care (POC) devices with smartphone connectivity are conspicuously felt. Despite its revolutionary impact on biotechnology and molecular diagnostics, the reverse transcription polymerase chain reaction technique as the gold standard in COVID‐19 diagnosis is not suitable for rapid testing. Today's POC tests are dominated by the lateral flow assay technique, with inadequate sensitivity and lack of internet connectivity. Herein, the biosensing advancements in Internet of Things (IoT)‐integrated electroanalytical tools as superior POC devices for SARS‐CoV‐2 detection will be demonstrated. Meanwhile, the impeding factors pivotal for the successful deployment of such novel bioanalytical devices, including the incongruous standards, redundant guidelines, and the limitations of IoT modules will be discussed.
Keywords: coronavirus disease 2019, biosurveillance, electrochemical biosensors, rapid test, Internet of Things
One of the first steps to combat a highly transmittable infectious disease is to detect it locally, before it spreads globally. The recent electrochemical biosensors are promising electroanalytical tools, pivotal for developing a biosurveillance ecosystem for rapid, point‐of‐care, and Internet of Things‐integrated detection of severe acute respiratory syndrome coronavirus‐2, and possibly, other infectious pathogens.

1. Introduction
One of the first steps to combat a highly transmittable infectious disease is to detect it in a cost‐effective and timely fashion locally before it spreads globally. Since the beginning of the coronavirus disease 2019 (COVID‐19) pandemic, few widely accepted techniques and tools have been approved and utilized to diagnose its causative virus, severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2).[ 1 ] Lateral flow assays (LFA) are still a dominant point of care (POC) tool for biosensing SARS‐CoV‐2 antigens as listed on the food and drug administration (FDA) website.[ 2 ] Moreover, the reverse‐transcriptase polymerase chain reaction (RT‐PCR) technique has become the only gold standard method to detect SARS‐CoV‐2 genetic material detection, widely required by physicians and authorities such as airports to approve its diagnosis.[ 3 ]
On one side, social norms and rules have undergone serious reconsideration to raise people's awareness about preventive manners. Social preventive paradigms, namely mandatory masking, social distancing, strict quarantine rules, lockdowns, and guidelines, have become the “new norm” of societies in such a short period.[ 4 ] Moreover, governments, industries, pharmaceuticals, and universities, cooperatively surge novel and highly effective vaccine development methods in such a compressed time.[ 5 ]
However, such renovations have not been administered to rapid biosurveillance systems for SARS‐CoV‐2. The molecular mechanism and the design of most commercial LFAs are not much different from dozens of years ago. The same goes for the RT‐PCR technique, and despite its groundbreaking advancements in genetic, biotechnology, and molecular diagnostics, it is not suitable for rapid diagnosis.[ 6 ] Moreover, the gold standard method for approving COVID‐19‐related lung infection is computed tomography (CT) scan.[ 7 ] Though it is a fast and non‐invasive imaging method, a CT scan cannot be performed repeatedly to monitor the degree of lung infection due to X‐ray exposure complications. Also, the equipment cost and the trained staff required for a CT scan make them un‐deliverable and impractical in countries and areas with low healthcare resources.[ 8 ]
One overlooked factor in COVID‐19 diagnosis is the sampling method, still dominated by nasopharyngeal swab sampling. Though it is a non‐invasive sampling, it still needs training and careful consideration as it may cause significant errors due to patient discomfort and reactions such as coughing and gagging, resulting in early swab removal from the nasopharyngeal area.[ 9 ] Altogether, the monopolies of diagnostic methods from sampling (nasopharyngeal), imaging (CT‐scan), and molecular detection (LFA, RT‐PCR) need to be reconsidered and improved by viable and comparable alternatives if we want to surge the way to combat not only COVID‐19, but also other emerging infectious diseases.
The fundamentals of SARS‐CoV‐2 infection pathways, its diagnosis and eradication have been reviewed by several research groups. Kevadiya et al. investigated the principle and fundamentals of gold standard methods to detect SARS‐CoV‐2 antigen and antibody, including RT‐PCR and RT loop‐mediated isothermal amplification (RT‐LAMP).[ 10 ] Udugama et al. reviewed the COVID‐19 patient and sample workflow during the outbreak.[ 1 ] They investigated SARS‐CoV‐2 morphology, PCR detection method, developed emerging technologies for diagnostic assays, developmental phases of diagnostic tests, and the role of smartphones in medical diagnostics. Pokhrel et al. analyzed the mortality rate of common infectious diseases as a function of the transmission rate.[ 9 ] Moreover, they showed the importance of early testing in reducing the mortality rate of COVID‐19 patients. Also, they categorized diagnostic kits for SARS‐CoV‐2 nucleic acid and antibody detection. Kotru et al. introduced the basics of modern potentiostats used in biosensor development.[ 11 ] They also investigated emerging electrochemical biosensors for detecting various biomarkers in COVID‐19 patients, including neutrophils, platelets, interleukins, C‐reactive protein (CRP), interferons, breath pH, and lymphocytes.
Mahshid et al. reviewed advancements in nucleic acid‐based electrochemical biosensors and antibody‐based immunosensors for analyzing viruses similar to SARS‐CoV‐2 such as Influenza.[ 12 ] Kaushik et al. provided a comprehensive review paper about COVID‐19, elaborating on different aspects of this outbreak, including the early attempts to understand its spread, its structure, the passage of infection through ACE 2 receptors, preventive and treatment strategies, the roadmap of vaccine development, clinical setup and molecular bioassays, nanosystem assisted photo‐degradation of the virus, nano‐enabled biosensing strategies for SARS, MERS, and SARS‐CoV‐2 detection, and the importance of bioinformatics and artificial intelligence to optimize therapeutics.[ 13 ] Serrano‐Aroca et al. focused on carbon‐based nanomaterials used as antiviral agents with broad antimicrobial properties, including graphene, carbon nanotubes, fullerene, graphene oxide, and carbon quantum dots.[ 14 ] Similarly, Prakash et al. reviewed antiviral photocatalysts based on titanium oxide nanoparticles as potential practical tools for the inactivation of SARS‐CoV‐2.[ 15 ] Jain et al. and Mujawar et al. reviewed point‐of‐care‐testing (POCT) for infectious diseases, including malaria, influenza A, HIV, Ebola, Zika, SARS, MERS, and SARS‐CoV‐2.[ 16 ] They also explained the role of the Internet of Things (IoT) and the Internet of Medical Things (IoMT). They provided their viewpoint on integrating nano‐sensors, AI, and IoMT for intelligent healthcare management. Tiwari et al. provided an opinion article to project the importance of nano‐enabled protective gear, gloves, masks, sheets, disinfectants, and filtration units to facilitate quarantine plans.[ 17 ] Gage et al. reviewed the new variants of SARS‐COV‐2, their severe effects, and nano‐enabled tools for virus eradication through nanoparticle‐based air filters and purifiers.[ 18 ] Herein, the newly developed and commercial SARS‐CoV‐2 electrochemical biosensors are introduced. Moreover, the limitations that impede the real‐world application of such devices will be thoroughly discussed; from the biosensing aspect, standards, guidelines, POC testing criteria, and the necessity of IoT integration for smart and rapid biosurveillance of infectious pathogens.
2. POC Devices Criteria
Given that few countries are equipped with the latest advances in healthcare, global criteria are required to be deployable in developing countries with less capital allocation in healthcare. A set of criteria was introduced by the World Health Organization Special Programme for Research and Training in Tropical Diseases (world health organization (WHO)/TDR) in 2003, namely ASSURED (affordable, sensitive, specific, user‐friendly, rapid, equipment‐free, and delivered) to address the ideal rapid diagnostic tests for sexually transmitted infections and tropical infectious diseases.[ 19 ] Meanwhile, fundraisers such as Bill and Melinda Gates foundation along with the innovative paper‐based diagnostic platforms introduced by Whitesides research team have implemented unique POC testing systems, particularly in low‐income countries.[ 20 ]
2.1. REASSURED Criteria
In a provident publication in early 2019, Chen's and Peeling's research groups reconsidered the ASSURED criteria, highlighting the advances in digital technology and mobile health (m‐health), and introduced REASSURED as the new criteria for ideal POC testing (Figure 1 ).[ 21 ] Real‐time connectivity is the first new criterion, meaning all the diagnostic tests’ parameters should be accessible via some sort of connectivity, such as smartphones, data communication‐enabled readers (radio‐frequency identification (RFID), Bluetooth, and Wi‐Fi), or cloud/fog storage platforms. Also, the connectivity should not only be limited to the raw data collection from POC systems since data analysis is required to provide useful and quantitative information. Analyzing huge raw data sets requires advanced data analysis techniques powered by artificial intelligence (AI) and machine‐learning algorithms to produce information, feedback, and predictive results. In fact, the paradigm of such real‐time connectivity is not a new concept, already known as the IoT introduced by Ashton et al. in 1999.[ 22 ] The next introduced criterion of REASSURED is the ease of sample collection and environmental friendliness.[ 21 ] Ideally, specimen collection should be non‐invasive without the requirement of advanced training. Moreover, if we envisage deploying ideal POC testing systems globally, we cannot deny their impact on the environment, either in their production or disposal. If neglected, the accumulation of millions or even billions of assays and devices can cause environmental catastrophes. Hence, the ideal POC device needs to be sustainable and environmentally friendly, either to be recycled in a cost‐effective fashion or to be disposed safely. For instance, natural‐based polymers such as cellulose, chitin, and their (nano)derivatives can be of great choices as substrates for biosensors, owing to their inherent characteristics, including abundance, affordability, biodegradability, biocompatibility, flexibility, and capillary wicking property.[ 23 ]
Figure 1.
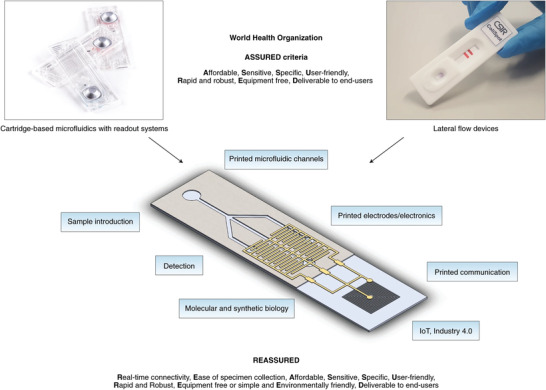
Integration of standard lateral flow diagnostic tests with printed electronics and IoT modules would result in ideal diagnostic tests known as REASSURED diagnostics to fulfill the sustainable development goals of global biosurveillance. Adapted with permission.[ 21 ] Copyright 2019, Springer Nature.
2.2. Implementation of IoT in Bioanalytical Tools
One of the commonly used definitions of IoT is introduced by the US National Intelligence Council “The “IoT” is the general idea of things, especially everyday objects, that are readable, recognizable, locatable, addressable, and controllable via the Internet, whether via RFID, wireless LAN, wide‐area network, or other means.”[ 24 ] IoT has intertwined with our daily lives, from navigation systems, weather forecast software, smartwatches, smart wristbands, smart fridges, and smart home devices, are all connected mainly via the internet (Wi‐Fi, cellular networks) or Bluetooth to our smartphones, tablets, and computer devices.[ 25 ] Such connectivity is not limited to consumer services, but it has also been deployed in many industries such as production monitoring, food and agriculture, environmental sensing, and healthcare.[ 23 ]
Though IoT has revolutionized the above‐mentioned industries, some shortcomings exist in implementing IoT in (bio)analytical chemistry, medical diagnostics, and POC devices. Mayer et al. reviewed the developments and challenges in Internet of Analytical Things (IoAT), highlighting the lack of centralized, access‐free databases for bioanalytical data produced from medical diagnosis and pathobiology laboratories or POC devices.[ 22 ] Needless to say, many analytical instruments and POC tools are equipped with a connectivity module, but data collection, transmission, storage, and analysis steps are decentralized. In other words, intra‐connectivity between user and device is achieved without inter‐connectivity across different users and devices.[ 26 ] Such limitation in inter‐connectivity has raised serious concerns regarding the management of the COVID‐19 pandemic. For instance, airlines do not have any access to the passengers’ COVID‐19‐related history and passengers are only required to bring the latest RT‐PCR test result, commonly within 72 h before their flight.[ 27 ] This approach puts a question mark on the authenticity of such results that could be easily manipulated, resulting in global transmission of the virus. Perhaps, a centralized IoT‐based platform for biometrics results—not only for fingerprints and criminal records but for medical history and bioanalytical test results—is required to assess and monitor individual's health by authorities, especially in the time of crisis such as global pandemics, though it might raise serious concerns about people's privacy. Moreover, many new infrastructures, namely cloud/fog/edge computing, AI and algorithm training along with unified protocols, need to be established to reach such a goal.[ 28 ] Substantial progress has been made to integrate electrochemical analyzers with communication modules, evidently in successful commercial products of PalmSens instruments.[ 29 ] The modern electrochemical analyzers are much smaller with interesting capabilities of battery‐powered, handheld, wireless communication via Bluetooth or USB connection. Besides, various smartphone‐based potentiostats have been developed with novel applications in flexible electronics, wearables, and molecular diagnostics. The typical modules used in such integrated systems are based on RFID communication, namely near field communication (NFC), Bluetooth, and Wi‐Fi.[ 24 , 29 , 30 ]
NFC is referred to communication in short distances (less than 10 cm) with the frequency of 13.56 MHz.[ 24 ] The main component of an NFC‐enabled device is loops of a coiled antenna to power the electronics via inductive coupling between itself and a reader, such as most modern smartphones. Features such as battery‐less energy harvesting, and wireless data communication, make NFC devices attractive for efficient and straightforward IoT modules for portable, flexible, and wearable electrochemical devices.[ 31 ] However, the produced power may not be sufficient for advanced potentiates/galvanostats/impedance devices, and they suffer from short communication distances as well. Bluetooth data transfer is another common RFID communication with more range than NFC, extending to tens of meters.[ 24 ] Modern Bluetooth technology consumes significantly lower powers, also known as Bluetooth low energy (BLE). However, it still needs an external power source to operate. Wi‐Fi or IEEE 802.11 data communication protocol uses a higher frequency range (2.4 GHz).[ 32 ] Though Wi‐Fi offers broader transmission with significantly rapid data transfer, its power consumption is more than NFC or BLE modules. Despite the notable progress in the integration of IoT modules with biosensors, the remaining challenges need to be addressed:
-
•
Efficient power consumption is required for data communication and storage, either with built‐in storage units or smartphones’ memory.
-
•
Specialized file storage and synchronization services based on fog and cloud computing should be dedicated to the diagnostics data sets. Standard security protocols customized for data transfer and data storage from decentralized users (people) and centralized organizations (diagnostic laboratories, hospitals, airports) should be provided to avoid data breach and manipulation.
A detailed IoT model has been proposed by Hosseinifard et al. regarding data management, cloud and fog computing, machine learning, and artificial intelligence in a pandemic.[ 33 ] It was suggested that smartphones could be ideal IoT gateways due to their inherent characteristics such as wide availability (94% of the world's population), powerful integrated processors, multiple RFID‐based communication (NFC, BLE, Wi‐Fi, and cellular networks), built‐in data storage (memory) and data management tools.[ 33 , 34 ]
2.3. Standardization and Commercialization
We witnessed an unprecedented acceleration in therapeutics and vaccination approval for COVID‐19 since organizations such as the FDA and the WHO have facilitated all the paths required for their mass production and deployment.[ 5e ] Meanwhile, such success and development have been rarely seen in COVID‐19 diagnostics, particularly rapid in vitro diagnostics (IVD) tools. A contributing factor in the delay of rapid POC systems could be the divergence of standards and guidelines in IVD regulations.[ 35 ] As mentioned earlier, an ideal POC system encompasses several components, including materials, biochemical compounds, electronics, data communication and storage, analytical performance, disposal, and waste management. It appears the guidelines and standards for such POC systems are still incongruous and cannot be deployed easily on novel POC devices (Table S1, Supporting Information).[ 36 ] Meanwhile, the analytical performance of the developed device must be evaluated carefully including limit of detection (LOD), linear range, sensitivity, cross‐reactivity, microbial interference, exogenous/endogenous interference, and other limitations. Therefore, the divergence between these standards and guidelines can impede the development of ideal POC devices in a crisis. Thus, a new paradigm of converging standards and guidelines is required for the massive deployment of POC systems.
3. Novel Advancements in Electroanalytical Tools for SARS‐CoV‐2 Detection
Tremendous multidisciplinary efforts involving chemistry, biology, nanoscience, biotechnology, and m‐health resulted in advanced nano‐bio‐diagnostic tools to diagnose viral infections.[ 1 , 8 , 9 , 10 , 37 ] Herein, we focus on the recent advanced electrochemical methods to detect SARS‐CoV‐2 spike proteins and its genetic materials, categorized in Table S2, Supporting Information based on the electrode material, mechanism of sensing, and analytical performance.
3.1. Electrochemical Immunosensors
Simply put, the concept of immunosensors is based on a transducer's response (commonly optical and electrochemical) due to the binding affinity between the biorecognition element (i.e., antibody) and the target. Immunosensor fabrication starts with modifying an electrode surface to produce proper surface chemistry to immobilize the antibody. This can be achieved through various approaches such as chemisorption of thiolated antibodies on a gold electrode's surface and carbodiimide crosslinker chemistry to create an amide bond between carboxylated carbon electrodes, amine‐terminated antibodies, and streptavidin‐avidin affinity binding.[ 38 ] The antibodies act as a lock for specific keys (antigens); thus, by adding the target antigen to the modified electrode, a complex of antigen‐antibody can be formed. Voltametric and impedimetric techniques are common electrochemical methods used to monitor the complex formation.[ 39 ] Without the presence of a target, the redox reagent can be oxidized or reduced at the working electrode, thus creating a significant current analyzed by a potentiostat. Alternately, target‐receptor binding causes a spatial hindrance for the redox molecules to migrate from the bulk solution to the electrode surface, thus decreasing the amount of produced current.[ 39 ] Enzyme‐linked electrochemical biosensors are another type of immunosensors commonly used in magnetic‐assisted electrochemical assays.[ 40 ] In enzyme‐linked methods, detection antibodies and capture antibodies are immobilized on the electrode surface and magnetic beads (MBs), respectively. Moreover, MBs’ surface is modified with an enzyme, commonly horseradish peroxidase (HRP). Introducing the target antigen to the magnetic bead solution forms a complex with the capture antibody. Then, a sandwich hybrid is formed after incubating the magnetic‐bead‐antigen complex with the immobilized detection antibodies on the electrode surface. Next, substrates (commonly 3,3′,5,5′‐ tetramethylbenzidine) are added to the sandwich complex, consequently oxidized by the HRP enzyme. The number of electrons generated by substrate oxidation can be monitored by amperometric techniques, commonly chronoamperometry.[ 41 ] No binding happens in the absence of antigen; thus, the non‐binding MBs capture antibodies would be washed away and no enzymatic reaction can occur.
Herein, some recent SARS‐CoV‐2 electrochemical immunosensors will be introduced. For instance, Zhao et al. developed a supersandwich immunosensor for highly sensitive SARS‐CoV‐2 viral RNA detection.[ 42 ] This immunosensor comprises three main parts. A) Magnetic nanocomposites of AuNP@Fe3O4 conjugated with capture probes (CPs). B) Host–guest recognition elements of immobilized calixarene molecules on rGO for enrichment of toluidine blue (TB) electrochemical mediators, AuNP for sensitivity enhancement, label probes (LP), and auxiliary probes (AP). C) A screen‐printed carbon electrode. In the presence of the viral RNA, a highly specific bioconjugate between the viral RNA and the nanocomposites could be formed, which is further analyzed by a smartphone‐based differential pulse voltammetry (DPV) analyzer. Another report demonstrated a microfluidic magneto immunosensor for COVID‐19 NP measurement.[ 43 ] Labeled (HRP) magnetic nanobeads (MNBs) were utilized for signal amplification and magnetic enrichment of the captured biomarker. This approach led to highly sensitive chronoamperometric detection of NP (10 pg mL−1) in small serum volumes (<50 µL). Gao's team developed a smartphone‐based multiplexed immunosensor, SARS‐CoV‐2 RapidPlex, for four COVID‐19 related biomarkers, including S1‐IgM, S1‐IgG, CRP, and antigen nucleocapsid protein (NP) in blood and saliva (Figure 2 ).[ 44 ] Laser‐engraved graphene was used as the sensing electrode since it offers superior properties, including high surface area, fast charge mobility, and well‐established modification techniques. Graphene was modified with a pyrene derivative, 1‐pyrenebutyric acid (PBA), to carboxylate its surface for receptor binding since it does not disrupt the graphene sheets’ conjugation. Moreover, they integrated the biosensors with a custom PCB‐based wireless potentiostat for chronoamperometric measurements of the biomarkers. A sandwich‐type immunosensor was reported by Arduini et al. targeting either S or N spike proteins in saliva.[ 45 ] In the presence of the virus, its S or N surface proteins could be sandwiched between monoclonal antibody conjugated MBs and polyclonal antibody (PAb) conjugated alkaline phosphatase (AP) enzyme, producing 1‐naphtol as an electroactive enzymatic byproduct detected by DPV method (Figure 3 ).
Figure 2.
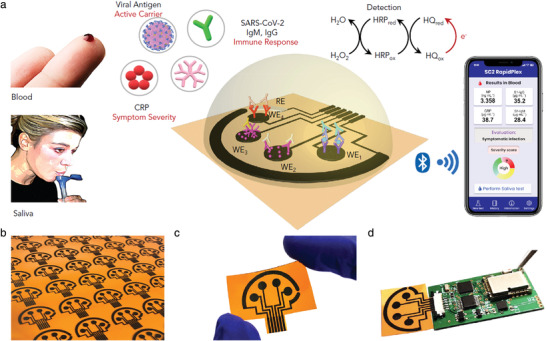
Multiplex, wireless, and graphene‐based biosensors for SARS‐CoV‐2 antigen and antibody detection (RapidPlex). a) Schematic illustration of salivary and blood viral antigen multisensing using enzyme labeled antibodies as biorecognition element. The produced current from substrate oxidation is transferred by a BLE module to a smartphone with a customized app. b) Rapid and cheap ($0.05) mass production of laser‐engraved flexible sensor arrays, c) disposable and flexible sensors, and d) rapidplex system integration with a PCB with built‐in potentiostat, signal processing, and Bluetooth communication. Adapted with permission.[ 44 ] Copyright 2020, Elsevier.
Figure 3.
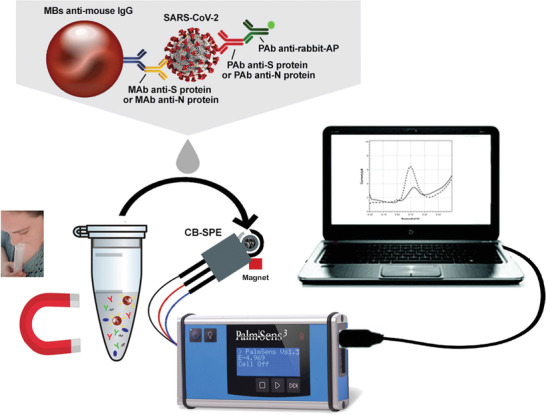
Magnetic‐amplified SARS‐CoV‐2 biosensor. Enzyme‐linked antibody conjugation with magnetic beads can bind to the target with excellent sensitivity owing to magnetic bead's high surface area and facile magnetic preconcentration on a screen‐printed support. Adapted with permission.[ 45 ] Copyright 2021, Elsevier.
3.2. Artificial Bioreceptors
Although antibody‐based biorecognition elements offer high selectivity and sensitivity, their production cost and batch‐to‐batch structural difference might hinder their application in commercial IVD tools.[ 46 ] On the contrary, artificially‐produced bioreceptors, namely aptamers and molecularly imprinted polymers (MIP), can be produced cost‐effectively with a consistent structural form.[ 46 ] Particularly, high quality research has been conducted for aptamer development for SARS‐CoV‐2 detection. Aptamers are short single‐stranded DNA or RNA molecules (oligonucleotide) or short peptides produced in vitro by a process known as systematic evolution of ligands by exponential enrichment (SELEX) to bind to a target ligand specifically.[ 47 ] Electrochemical aptamer‐based (EAB) biosensors have received tremendous attention due to the fast and cost‐effective production of customized aptamers with higher stability than in vivo‐produced antibodies.[ 48 ] Each end of an aptamer strand can be modified with a desired functional group for two primary purposes in oligonucleotide EAB sensors: a) chemical immobilization on an electrode by modifying the 5′ end with thiol or amine groups and b) covalent attachment of a redox molecule such as (methylene blue, MB) on the 3′ end.[ 49 ] The common mechanisms of EAB biosensing are based on a) spatial hindrance due to the formation of aptamer‐target complex or b) aptamer conformational change in the presence of its target.[ 48 ] In case of spatial hindrance, the aptamer‐target complex would impede the redox molecules to migrate from the bulk solution to the electrode layer; thus, a decrease in the produced current is anticipated. In case of the induced conformational change, the aptamer's end (either 3′ or 5′) would get closer or further to/from the electrode, therefore causing a shift in the redox activity and the produced current.[ 48 ] For example, Zhang et al. designed a highly sensitive aptasensor based on dimeric DNA aptamers (DSA1N5) to detect salivary spike proteins. The binding affinity of this dimeric aptamer against variants of SARS‐CoV‐2 was investigated. Interestingly, it has a semi‐selective affinity to the common variants with different dissociation constants of 120, 290, and 480 pm against wildtype, Alpha, and Delta variants, respectively. The aptamer was designed based on multivalent ligation of two or more monomers to achieve picomolar sensitivity. This approach resulted in enhanced binding with spike proteins since each virus carries around 30 trimeric spike proteins on its surface with an average spacing distance of 13–15 nm between each other. The developed aptamer's length was more than 15 nm. Thus, it can surround and bind the spikes more significantly than monomeric aptamers. Moreover, a gold SPE‐based electrochemical biosensor was fabricated to test spike proteins in saliva samples rapidly. The synthesized aptamer was thiolated at the 3′ end to facilitate chemisorption on the gold working electrode. Also, tris(2‐carboxyethyl)phosphine was mixed with the thiolated aptamer to reduce all the S–S bound to S–H, which improves the chemisorption efficiency on the gold's surface. Then, the electrode was incubated with thiolated polyethylene glycol (Mw = 6000 Da) to reduce the non‐specific absorption of other biomolecules. Ferro/ferricyanide redox was utilized to study the electrochemical charge transfer on the modified gold electrode. In the presence of spike proteins, the complex between aptamers and spikes passivates the gold's surface, hindering the charge transfer of the redox‐active molecules. Electrochemical impedance spectroscopy technique was utilized to measure the charge transfer resistance of the redox reagents from the bulk solution to the electrode's surface, resulting in highly specific detection of viral proteins with 80.5% sensitivity. In another report, Alafeef et al. designed four antisense oligonucleotides (ssDNA) aptamers targeting N gene at four segments simultaneously.[ 50 ] The biosensor consisted of a paper substrate coated with graphene and then aptamer‐capped AuNPs. The aptamers were designed in the Soligo software with some constraints and considerations such as folding temperature and ionic strength to predict the binding energy (≤−8 kcal mol−1 cutoff) of RNA−DNA hybridization. Also, the biosensor was integrated with a costumed Arduino‐based circuit board equipped with IoT modules such as Bluetooth and Wi‐Fi to provide a rapid and portable POC system. There is a common problem in electrochemical biosensors known as biofouling due to accumulations of molecules and particles on the sensing electrode. Li et al. addressed the biofouling issue by designing an electrochemical biosensor with enhanced antifouling property. Bovin serum albumin (BSA), a relatively cheap and abundant protein for electrode blocking, was used to reduce the non‐specific adsorption of biomaterials on the electrode surface (Figure 4 ).[ 51 ] The key point of this paper is that the BSA was covalently bounded to polymerized aniline nanowire (PANI‐NW) to enhance the electrode blocking, reduce the BSA leaking, and alleviate BSA blocking effect on electrode conductivity. Moreover, a commercial peptide aptamer against COVID‐19 immunoglobin G (IgG) was immobilized on BSA using 4‐(N‐Maleimidomethyl) cyclohexane‐1‐carboxylic acid 3‐sulfoN‐hydroxy‐succinimide ester sodium salt (sulfo‐SMCC) to covalently bond the amine groups of BSA and the aptamer's Cys‐terminal thiols. Overall, this biosensor showed significant antifouling properties without diminishing the electron transfer of redox‐active molecules at the electrode surface. Chaibun et al. demonstrated an amplification method for ultrasensitive S and N genes of SARS‐CoV‐2 biosensing (Figure 5 ).[ 52 ] The CP consisted of streptavidin‐MNB incubated with biotin labeled aptamers. The reporter probes (RP) were prepared as follows: First, silica nanoparticles (SiNPs) were synthesized using a modified sol–gel (Stöber) process. Then redox‐active dyes MB and acridine orange (AO) were bounded to the SiNPs via polyelectrolyte‐assisted layer by layer assembly through activated surface functional groups to produce SiMB and SiAO redox probes. Then, the redox probes were incubated with biotin‐reporter primers to provide the Si‐RP probe. Isothermal rolling circle amplification (RCA) method was used to greatly enhance the sensitivity of the biosensor. The detection procedure included a one‐step hybridization method by mixing the prepared CP‐MNB and RP‐Si with the target (or RCA amplicons) to produce a sandwich hybrid. DPV method was utilized to measure the current induced by the hybridization between the redox‐labeled reporter probe and the magnetic CP. Moreover, a portable Palmsens potentiostat was used to facilitate POC biosensing. Another type of artificial bioreceptors is MIP. They can (electro)chemically attach or physically entrapped on an electrode's surface.[ 53 ] These polymers have micron to nanometer‐sized cavities which only fit their target. Therefore, the target analyte can saturate the MIP's cavities; thus, redox molecules cannot penetrate through the cavities to reach the electrode's surface. As a result, charge transfer and the produced current would decrease, which can be monitored by voltametric methods.[ 53 ] Raziq et al. developed a MIP‐based biosensor to detect nucleocapsid protein (ncovNP), the protection layer of SARS‐CoV‐2 RNA.[ 54 ] Phenylenediamine monomers were polymerized on a thin film of gold electrode and further functionalized with 4‐aminothiophenol (4‐ATP) followed by ncovNPs immobilization and the final washing step to provide the molecular cavities. Moreover, a lysis step is necessary to extract ncovNPs from the infected samples followed by a DPV scan in the presence of ferro/ferricyanide redox agent. Clearly, the charge transfer is hindered when ncovNPs are trapped in the cavities, causing a significant decrease in the DPV peak currents.
Figure 4.
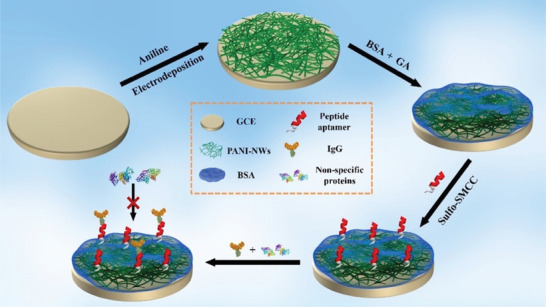
Peptide‐based SARS‐CoV‐2 aptasensor with enhanced anifouling capability. Polyaniline nanowires‐bovine serum albumin cross‐links significantly reduces non‐specific absorption of proteins and other biomolecules. Adapted with permission.[ 51 ] Copyright 2021, American Chemical Society.
Figure 5.
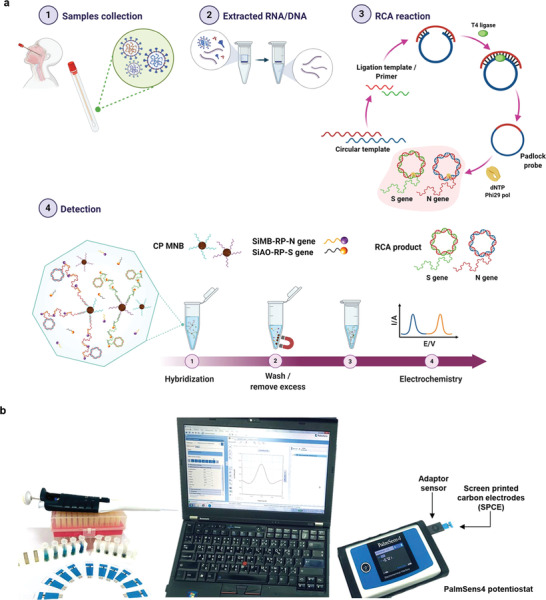
Workflow of SARS‐CoV‐2 S and N genes detection. a) Extracted genes are isothermally amplified by RCA technique, then the amplicons are hybridized with redox‐tagged aptamers, which are subsequently detected by voltametric techniques. b) The picture of hand‐held PalmSens4 potentiostat with Bluetooth connectivity for data transfer to a personal computer. Adapted with permission.[ 52 ] Copyright 2021, Springer Nature.
3.3. CRISPR‐Based Molecular Circuits
Besides its astonishing genome editing ability, the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR‐associated proteins (Cas) systems have paved a new era of biosensing applications due to their isothermal signal amplification and high base resolution.[ 55 ] The discovery origin of CRISPR goes back to 1984 when microbiologists stumbled upon short extrachromosomal DNA of the bacteria Escherichia coli and Salmonella typhimurium, which was named repetitive extragenic palindromic (REP) genes.[ 56 ] Later, it was found REP gene is responsible for producing membrane proteins as a defense mechanism against invading viral bacteriophages.[ 57 ] In 2002, for the first time, the term CRISPR was introduced to define the repetitive nucleotide sequences that were only found in bacteria/archaea but not in Eukaryotes or viruses.[ 58 ] A characteristic of the CRISPRs, not seen in any other class of repetitive DNA, is that the repeats of the CRISPRs are interspaced by similarly sized nonrepetitive DNA. In 2005, it was suggested that the spacer elements in these genes are the traces of past invasions by viruses, and they offer cell immunity against bacteriophage invasion by coding an anti‐sense RNA.[ 59 ] Nowadays, CRISPR is usually understood as a molecular machine, or simply as a molecular scissor, consisting of two main parts: an effector called CRISPR‐associated effector (Cas) and a single guide RNA (sgRNA).[ 60 ] The effector's responsibility is to locate and bind to a specific part of the target's DNA/RNA, known as a protospacer adjacent motif (PAM), and then the sgRNA breaks the double helix of that DNA/RNA.[ 61 ] It is possible to use such specific cleavage activity of the CRISPR/Cas system to sense genomic materials of viral infections.[ 62 ]
For instance, Dai et al. demonstrated a CRISPR‐based biochemical circuit as a highly sensitive SARS‐CoV‐2 biosensor.[ 63 ] Inspired by the conventional microcontrollers, they innovated a biochemical circuit with the potential capability of an electrical circuit but programmed for genetic analysis (Figure 6 ). Comparable to a microcontroller, this platform can identify, convert, translate, amplify, and transduce biological signals. Target's specific genome sequence identification, which is the first step of this biochemical circuit, was initiated by arrays of CRISPR mutant nickase (Cas9 D10A). Two different sgRNA were used to guide the Cas9 complex to cut two distant regions of the target's genome. The cut product was then amplified using primer exchange reaction (PER) to produce signaling concatemers.
Figure 6.
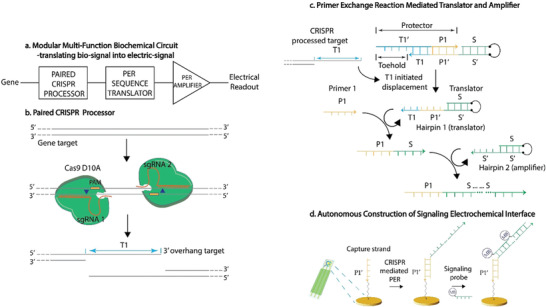
CRISPR‐based biochemical circuit combined with electrochemical biosensing. a) A heterogeneous biochemical circuit composed of paired CRISPR processor, amplification using primer exchange reaction (PER), and genetic data processor and translator into electrical signal. b) Two offset sgRNAs guide a pair of CRISPR (Cas9 D10A) to detect two PAM regions of the target, which subsequently cleaves the gene and cut it into a 3′‐overhang strand. c) Translation and amplification by PER technique. Hairpin 1 functions as a translator, only operating with the presence of the overhang target. Hairpin 2 functions as an amplifier and catalyzes concatemer formation. d) A nucleic acid‐based capture strand is immobilized on the gold electrode to bind with the produced concatemer. A redox‐tagged signal probe forms complementary binding to the concatemer and produces electrochemical signal detected by SWV technique. Adapted with permission.[ 63 ] Copyright 2020, John Wiley and Sons.
PER is a novel and powerful DNA synthesizing technique that produces arbitrary ssDNA in an autonomous and programmable fashion.[ 64 ] The PER cascade begins with a specific DNA primer and is then extended by a catalytic DNA hairpin and a user‐specified primer. The catalytic DNA hairpin is a substrate for amplifying the primer, consisting of a stem, loop, and an exposed region.[ 64 ] The primer bonds to the hairpin's exposed region, and then a polymerase enzyme starts the displacement elongation to produce an elongated strand. The polymerase would stop synthesis before the loop region at a given stop codon. Next, the displaced strand releases from the hairpin via a three‐branched migration process.[ 63 ] Consequently, the hairpin's exposed region is free again to be used for another synthesis cycle. In order to quantify the amount of the amplified concatemers for PER amplification, an SPE‐based electrochemical interface was utilized.[ 63 ] The SPE consisted of a gold working electrode, modified with a thiolated ssDNA CP. The nucleotide sequences of the capture's probe are only complementary to the overhang of PER amplified concatemers; thus, only in the presence of the target's genome can the concatemers hybridize with the CP via base pairs hydrogen binding. Then, an MB tagged oligomer, known as a signal probe, was cast on top of the hybridized concatemer to make a secondary hybrid with the other end of the concatemer. The excess probes and regents were washed away so that only hybridized CP/concatemer/MB‐tagged signaling probe would remain on top of the gold working electrode. Finally, square wave voltammetry (SWV) technique was applied to the electrode to measure the resulting current from the hybridized MB tag. Only in the presence of PER amplified concatemers MB tagged signal probe can form a hybrid. Otherwise, the signal probe would be removed in the final washing step, and without MB, no electron transfer could happen between the bulk solution, CP, and gold surface. Therefore, the more concatemer is present, the more signaling probe can be hybridized, resulting in higher current intensity from SWV technique.
3.4. Cellulosic‐Based Biosensors
Given the low‐cost, biodegradability, abundance, wicking properties, and well‐developed functionalization techniques of cellulosic paper, they are viable candidates for developing cost‐effective biosensors. Compared to a common LFA, electrochemical paper‐based sensors can be designed without the specific requirement of different antibodies. Moreover, in situ diagnosis of COVID‐19 is of significant importance to fasten the data collection without distinct sample collection, pretreatment, and analysis. For instance, Yakoh et al. fabricated a label‐free, 3D‐origami shaped, paper‐based electrochemical immunosensor (ePAD) to detect SARS‐CoV‐2 antibodies (IgG and IgM) (Figure 7 ).[ 23b ] After immobilizing spike protein RBD on graphene oxide electrodes, ferro/ferric redox indicator was added to the test zone. In the presence of antibodies, the conversion of the redox indicator was interrupted, resulting in a decreased current response monitored by SWV technique. Eisa et al. innovated an immunosensor to detect COVID‐19 antibodies in situ in nasopharyngeal samples via a cotton‐tipped biosensor.[ 65 ] The biosensor's electrode consisted of carbon nanofiber (CNF) SPE, suitable for functionalization with a large surface area. Diazonium salt of aminobenzoic acid was electrografted on the CNF's surface by a two‐step CV, completely flattened the cathodic and anodic peak as the carboxyphenyl (CP) layer retarded the electron transfer process due to the presence of aromatic layer and electrical repulsion from the negatively charged carboxyl groups. The CP functionalized electrode was further modified by EDC/NHS coupling the NH2 residue of COVID‐19 N protein, shielding the negative charge of carboxylic group and enabling electron transfer. The whole biosensor was put into a cotton tip to absorb the nasopharyngeal samples readily via capillary wicking. Ferro/ferricyanide redox indicator and a portable potentiostat were used to probe the electron transfer via SWV technique.
Figure 7.
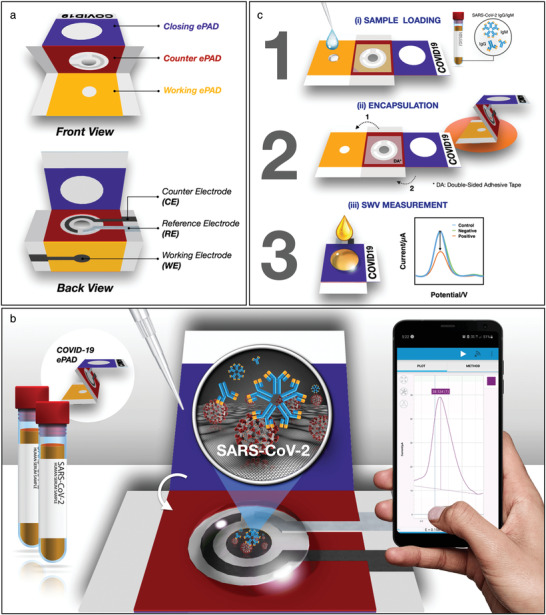
3D‐origami‐paper‐based SARS‐CoV‐2 biosensor. A) Device components including three foldable paper‐based electrodes. B) Detection of antibodies against SARS‐CoV‐2 using RBD proteins as the biorecognition element and SWV monitoring of the electrode's response upon target binding. C) The results can be transferred wirelessly to a smartphone. Adapted with permission.[ 24b ] Copyright 2021, Elsevier.
3.5. Reagent‐Free Biosensors
Developing reagent‐free methods for direct viral infections can be a viable solution for cost‐effective and rapid analysis of infectious diseases. Yousefi et al. developed a fast (5 min) direct method to detect viral particles of SARS‐CoV‐2 in unprocessed saliva (Figure 8 ).[ 66 ] Negatively charged, amine terminated DNA linkers were immobilized on a gold electrode and they were further conjugated with ferrocene as the redox reagent and cAb against S1 protein. When a positive potential was applied to the electrode, viral particles moved toward the sensing layer, causing an increase in hydrodynamic drag force and a faradaic response. This sensing platform can potentially be used for easy‐to‐use, rapid, and reagent‐free detection of COVID‐19 or other viral targets. A recognition element‐free electrochemical sensor was demonstrated by Hashemi et al. based on the emerged patterns (fingerprints) from DPV analysis at different voltages.[ 67 ] The sensor comprised gold nano stars (AU NS) decorated at a GO/8‐ hydroxyquinoline (8H) electrode, which could differentiate between different glycoproteins owing to the differential interaction of different glycoproteins to 8H.
Figure 8.
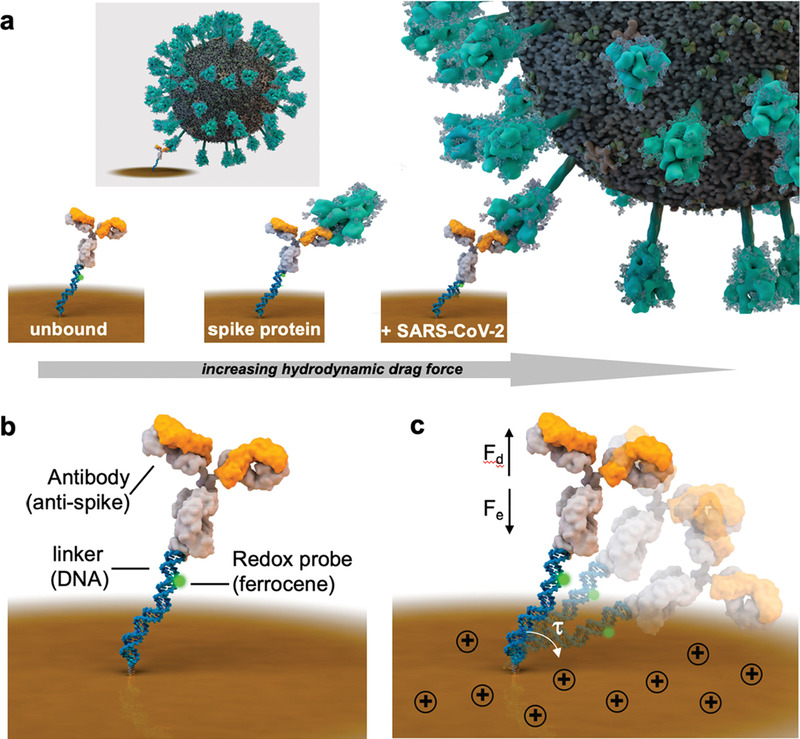
Reagent‐free SARS‐CoV‐2 biosensor. a) Receptor binding with viral particles increases the hydrodynamic diameter and subsequently influences the required time for the redox tag to contact the electrode's surface. b) Biosensor architecture based on immobilized DNA linker with a redox tag, conjugated with antibody against spike proteins and c) negatively charged bioreceptor swings across the electrode when a positive potential is applied, which cause a shift in the hydrodynamic drag force detected by chronoamperometric methods. Adapted with permission.[ 66 ] Copyright 2021, American Chemical Society.
3.6. Field‐Effect Transistor‐Based Biosensors
In short, field‐effect transistors (FET) are a branch of transistors that pass through current—from its conducting gate proportional to the amount of electric field applied between the semiconducting source and gate.[ 68 ] FET‐based biosensors incorporate a biorecognition element on the gate, in which the amount of due to target‐receptor binding can be quantified by the change in the surface charge distribution and the current it passes through accordingly.[ 69 ] There are few reports on SARS‐CoV‐2 detection using FET‐based biosensors.[ 70 ] For instance, Seo et al. developed a FET‐based biosensor in which SiO2/Si substrate utilized as the source and drain (Figure 9 ). The gate encompassed antibodies against spike proteins, immobilized on a graphene‐based gate using pyrenebutyric acid N‐hydroxysuccinimide ester linker.
Figure 9.
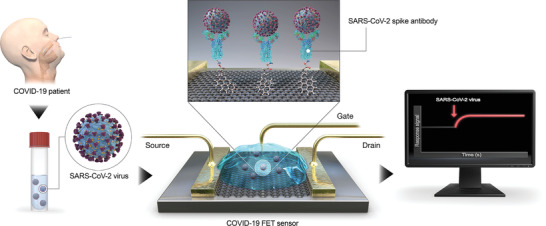
FET‐based SARS‐CoV‐2 biosensor. Anti‐spike antibodies are immobilized on the gate of a graphene‐based transistor. Target‐receptor binding causes a change in the graphene's surface charge distribution, which consequently changes the current passing through the drain. Adapted with permission.[ 72b ] Copyright 2020, American Chemical Society.
3.7. Commercial Products
Most FDA‐approved in vitro diagnostic devices are dominated by optical and RT‐PCR techniques. However, two electrochemical‐based medical devices, namely Sampinute[ 71 ] and ePlex,[ 72 ] offer alternative approaches for SARS‐CoV‐2 diagnosis, depicted in Figure 10 . Commercial electrochemical biosensors for SARS‐CoV‐2 detection. A) ePlex: digital microfluidics fluid handling integrated with RT‐PCR amplification and detection via nucleic acid hybridization,[ 43 ] B) Sampinute: magnetic force‐assisted electrochemical sandwich immunoassay.[ 42 ] Sampinute analyzer is a magnetic force‐assisted electrochemical sandwich immunoassay for the qualitative detection of RBD spike proteins of SARS‐CoV‐2 within 1 h (LOD = 30 TCID50 mL−1). The analyzer is equipped with a cartridge, encompasses magnetic nanoparticles (MNPs) and an electrode, both coated with antibodies against RBD, hence in case of the virus presence, it would be sandwiched between the MNPs and the electrode surface. Developed by GenMark, ePlex SARS‐Cov‐2 assay consists of digital microfluidics for fluid handling/mixing, followed by an automated RT‐PCR amplification, and finally electrochemical detection. The detection mechanism is based on DNA hybridization of the target's DNA and capture's probe, determined by voltammetry. Antibody analysis is also of great importance as it provides valuable information on diagnostics, post‐infection immunity, vaccine immunity, and health decisions. For instance, Roche Diagnostics has developed rapid (18 min), high‐throughput, and automated electrochemiluminescence immunoassay for total anti‐SARS‐CoV‐2 antibodies in the range of 0.4–250 U mL−1.[ 73 ] Moreover, a handful of LFAs are listed on the FDA website for qualitative antibody detection, which are cheaper and can be available for POC use, particularly the fingerstick whole blood tests.[ 74 ]
Figure 10.
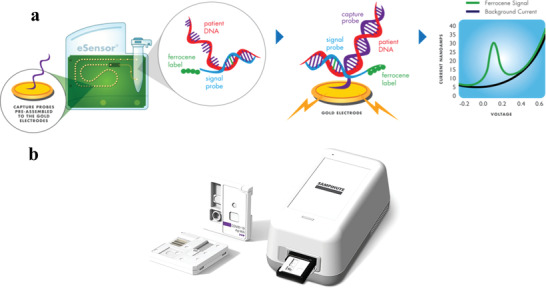
Commercial electrochemical biosensors for SARS‐CoV‐2 detection. a) ePlex: Digital microfluidics fluid handling integrated with RT‐PCR amplification and detection via nucleic acid hybridization. The target DNA forms complementary binding to the ferrocene‐tagged signal probe then it can be transferred by digital microfluidics to a gold electrode with a DNA capture probe. The target‐receptor binding can be analyzed by voltametric methods. Adapted with permission.[ 72 ] Copyright 2022, GenMark Diagnostics. b) Sampinute: a magnetic force‐assisted electrochemical sandwich immunoassay for qualitative detection of RBD spike proteins. Antibody conjugated magnetic nanobeads are utilized to target the RBD region of spike proteins, which subsequently forms a sandwich complex with the detection probe on the working electrode. Adapted with permission.[ 71 ] Copyright 2022, Celltrion Group.
4. Challenges and Perspectives in Biosurveillance of COVID‐19
Currently, the major bottlenecks of global‐scale COVID‐19 screening are:
-
1.
Insufficient self‐administered POC diagnostic tools
-
•
Using accessible samples including blood, respiratory mucosa, saliva, or exhaled breath
-
•
-
2.
Disproportionate standards (Table S1, Supporting Information)
-
•
Food and Drug Administration (FDA), World Health Organization (WHO), International organization for standardization (ISO), and international electrotechnical commission technical committee (IEC TC)
-
•
-
3.
Poor IoT implementation in POC tests
-
•
Current lateral flow assays are not equipped with an IoT‐based platform (e.g., smartphones)
-
•
Future POC electroanalytical devices need to be integrated with IoT modules and smartphones
-
•
-
4.
Inadequate testing coordination
-
•
By expanding the availability of testing sites (e.g., office space, in‐car‐testing, or self‐testing) patients and susceptible samples can be monitored while minimizing exposure.
-
•
-
5.
Lack of accessible cloud storage systems
-
•
Dedicated to the data of travel history, medical history, physical symptoms, medical imaging, and molecular diagnostics
-
•
-
6.
Insufficient smart telemedicine systems
-
•
To deconvolute and analyze the diagnostic big data, gathered from cloud storage systems
-
•
Integrated IoT‐based electrochemical biosensors are powerful POCT tools that can address the issues mentioned above for rapid and sensitive SARS‐CoV‐2 detection on a global scale. In Figure 11 the workflow of electrochemical biosurveillance systems is shown. Sampling is the first step in acquiring biofluids for molecular analysis. Although nasopharyngeal swab is a standard sampling method, other biofluids such as urine, saliva, and sweat can be extracted non‐invasively. The sample can be introduced into a microfluidic‐based electrochemical biosensor to detect the target using various bioreceptors, including mono/PAb, enzyme‐linked antibody, nucleic acid, aptamer, molecularly imprinted polymers, and CRISPR‐based molecular circuit. An RFID‐based IoT module (NFC, Bluetooth, and Wi‐Fi) can transfer the sensor's response to a reader such as a smartphone to locally store the data. The stored data from great numbers of smartphones across the world (as a decentralized IoT gateway) can be transferred to a cloud‐based data storage platform via the internet (Wi‐Fi, 4G, and 5G), followed by (big) data analysis methods, machine learning algorithms, and artificial intelligence to filter, purify, transcode, and deconvolute countless raw datasets. The output of such data analytics can be accessed by healthcare professionals, authorities, and governments for remote monitoring and real‐time decision‐making.
Figure 11.
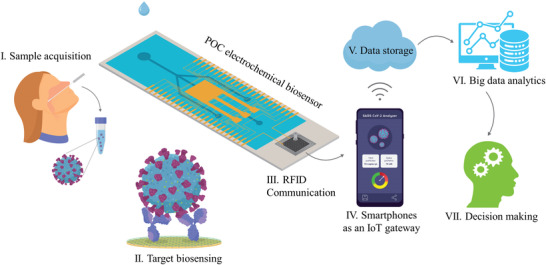
Electrochemical biosurveilance systems for real‐time monitoring of COVID‐19. IoT integrated electrochemical biosensors enable remote monitoring of patients and a cloud‐based service sets can allow patients and physicians to communicate 24/7 using smartphones, tablets, or computers.
5. Conclusion
This perspective strives to answer several nurturing questions regarding the potential to utilize electroanalytical tools for POC rapid SARS‐CoV‐2 detection. We aim to highlight impeding factors pivotal for the successful deployment of such novel bioanalytical devices and encourage further electrochemical technology developments to fight against this deadly and prevalent disease. Looking in the future, we have identified several important areas of exploration for electrochemical diagnostic, we hope to answer:
-
1.
What biosensor is the most promising, and what technological advancements are needed to utilize these devices for POC rapid SARS‐CoV‐2 detection? Any biorecognition element (artificial or antibody‐based) coupled with an amplification method (PCR, isothermal, and magnetic) has shown high potential for COVID‐19 diagnosis. However, these methods often require multistep processes for sample analysis; thus, they are more difficult and time‐consuming. Meanwhile, reagent‐free electrochemical detection of unprocessed saliva would be an ideal rapid test, though it might suffer from poor selectivity.
-
2.
Would the combined electrochemical recognition and amplification be feasible in terms of cost? Magnetic enrichment is one of the cost‐effective methods, but it is not as effective as molecular amplification methods. However, molecular amplification requires highly purified and expensive biochemicals (e.g., PCR, RCA).
-
3.
Is the highly sensitive assay possible without the amplification? Yes, the CRISPR‐based biosensing does not necessarily imply an amplification step by offering a novel mechanism of biosensing, and according to the reports, it is much more sensitive, selective, and faster than classical ELISA or fluorometric monitoring of amplicons. In fact, there are CRISPR‐based commercial products that have been approved by the FDA, though they are based on optical detection.[ 2a ]
-
4.
What is the status of smart biosurveillance technology for SARS‐CoV‐2 detection? Data communication between smart nano‐bio‐electro‐analytical devices needs to be improved by the advances in IoT, machine learning, and m‐health technologies. There is still no homogenous database and communication systems to provide real‐time data on the performed diagnostic tests and results along with exposure risk, clinical, or POC test result history, and travel history.
-
5.
How does electrochemical sensing address the problem of new variance of SARS‐CoV‐2? How e‐sensor technology competes with RT‐PCR in this respect? Since variants have slightly different nucleotide sequence in their RNA and slightly different amino acids in their spike proteins, it is not feasible to distinguish them by the common electrochemical biosensors. The variants are still detected by highly sensitive genetic sequencing tools and RT‐PCR assays. However, it may be possible to discriminate against SARS‐CoV‐2 variants using electrochemical sensor arrays (chemical nose/tongue) using different aptamers at each working electrode.[ 75 ]
Compared to the current RT‐PCR test, the introduced electrochemical biosensors have the potential to meet the RESSSURED criteria, resulting in efficient, rapid, and smart biosurveillance of COVID‐19. We envisage the integration of IoT modules with electrochemical biosensors allows patients to be efficiently screened and protects patients, clinicians, and the community from direct exposure. A cloud‐based service dedicated to diagnostic data sets can allow patients and physicians to communicate 24/7 using smartphones, tablets, or computers. Doctors and health care providers can effortlessly obtain detailed exposure and travel histories, physical symptoms (e.g., body temperature, coughing, and breathing rate), molecular diagnostic data (e.g., quantitative results of viral proteins, nucleic acids, and antibodies), and imaging data (e.g., CT scan) using the diagnostic cloud‐based service, acquired from IoT‐integrated electroanalytical devices.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
This work was carried out with financial support of Canada Foundation for Innovation CFI‐JELF (Project No. 36874) and the New Brunswick Innovation Foundation NBIF and New Brunswick Health Research Fund NBHRF (Project No. COV‐0000000063).
Biographies
Sina Ardalan is a Ph.D. student in the Chemistry Department of the University of New Brunswick under the supervision of Dr. Anna Ignaszak. He is the recipient of the Dr. William S. Lewis Doctoral Fellowship. He obtained his B.Sc. from the Sharif University of Technology and his M.Sc. from Chemistry and Chemical Engineering Research Center of Iran. His current research focuses on the design and fabrication of electrochemical biosensors for rapid detection of infectious diseases.

Anna Ignaszak is an associate professor at the University of New Brunswick (Canada), before that she was a junior professor (W1) at the Friedrich‐Schiller University in Jena (Germany). She obtained a Ph.D. in her native Poland in 2006 and moved to Canada to take on a position as a research associate at The University of British Columbia, and at the National Research Council of Canada. She has a diverse background in materials for electrochemical sensors and electrochemical energy applications. Her group at UNB currently develops electrochemical sensing platforms for early detection of Covid‐19, Lyme bacteria, and cancer biomarkers.

Ardalan S., Ignaszak A., Innovations and Challenges in Electroanalytical Tools for Rapid Biosurveillance of SARS‐CoV‐2. Adv. Mater. Technol. 2022, 2200208. 10.1002/admt.202200208
References
- 1. Udugama B., Kadhiresan P., Kozlowski H. N., Malekjahani A., Osborne M., Li V. Y. C., Chen H., Mubareka S., Gubbay J. B., Chan W. C. W., ACS Nano 2020, 14, 3822. [DOI] [PubMed] [Google Scholar]
- 2.a) In Vitro Diagnostics EUAs—Molecular Diagnostic Tests for SARS‐CoV‐2, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2 (accessed: June 2022);; b) In Vitro Diagnostics EUAs—Antigen Diagnostic Tests for SARS‐CoV‐2, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-antigen-diagnostic-tests-sars-cov-2 (accessed: June 2022).
- 3. Bustin S. A., Nolan T., Int. J. Mol. Sci. 2020, 21, 3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Vos J., Transp. Res. Interdiscip. Perspect. 2020, 5, 100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Liu C., Zhou Q., Li Y., Garner L. V., Watkins S. P., Carter L. J., Smoot J., Gregg A. C., Daniels A. D., Jervey S., Albaiu D., ACS Cent. Sci. 2020, 6, 315; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Marc G. P., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W., Hammitt L. L., Türeci O., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C., Group C. C. T., N. Engl. J. Med. 2020, 383, 2603; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Baden L. R., El Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., et al,, N. Engl. J. Med. 2021, 384, 403;33378609 [Google Scholar]; d) Le T. Thanh, Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S., Nat. Rev. Drug Discovery 2020, 19, 305; [DOI] [PubMed] [Google Scholar]; e) Lurie N., Saville M., Hatchett R., Halton J., N. Engl. J. Med. 2020, 382, 1969; [DOI] [PubMed] [Google Scholar]; f) Ye T., Zhong Z., García‐Sastre A., Schotsaert M., De Geest B. G., Angew. Chem., Int. Ed. 2020, 59, 18885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Merkoçi A., Li C.‐z., Lechuga L. M., Ozcan A., Biosens. Bioelectron. 2021, 178, 113046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L., Radiology 2020, 296, E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng W., Newbigging A. M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D. B., Cui M., Tao J., Tyrrell D. L., Zhang X. E., Zhang H., Le X. C., Anal. Chem. 2020, 92, 10196. [DOI] [PubMed] [Google Scholar]
- 9. Pokhrel P., Hu C., Mao H., ACS Sens. 2020, 5, 2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kevadiya B. D., Machhi J., Herskovitz J., Oleynikov M. D., Blomberg W. R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A. M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C. B., Reid S. P. M., Punyadeera C., Chang L., Gendelman H. E., Nat. Mater. 2021, 20, 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kotru S., Klimuntowski M., Ridha H., Uddin Z., Askhar A. A., Singh G., Howlader M. M. R., TrAC, Trends Anal. Chem. 2021, 136, 116198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mahshid S. S., Flynn S. E., Mahshid S., Biosens. Bioelectron. 2021, 176, 112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaushik A. K., Dhau J. S., Gohel H., Mishra Y. K., Kateb B., Kim N.‐Y., Goswami D. Y., ACS Appl. Bio Mater. 2020, 3, 7306. [DOI] [PubMed] [Google Scholar]
- 14. Serrano‐Aroca Á., Takayama K., Tuñón‐Molina A., Seyran M., Hassan S. S., Pal Choudhury P., Uversky V. N., Lundstrom K., Adadi P., Palù G., Aljabali A. A. A., Chauhan G., Kandimalla R., Tambuwala M. M., Lal A., Abd El‐Aziz T. M., Sherchan S., Barh D., Redwan E. M., Bazan N. G., Mishra Y. K., Uhal B. D., Brufsky A., ACS Nano 2021, 15, 8069. [DOI] [PubMed] [Google Scholar]
- 15. Prakash J., Cho J., Mishra Y. K., Micro Nano Eng. 2022, 14, 100100. [Google Scholar]
- 16.a) Mujawar M. A., Gohel H., Bhardwaj S. K., Srinivasan S., Hickman N., Kaushik A., Mater. Today Chem. 2020, 17, 100306; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jain S., Nehra M., Kumar R., Dilbaghi N., Hu T., Kumar S., Kaushik A., Li C.‐z., Biosens. Bioelectron. 2021, 179, 113074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garg P., Attri P., Sharma R., Chauhan M., Chaudhary G. R., Front. Nanotechnol. 2022, 4, 898411. [Google Scholar]
- 18. Gage A., Brunson K., Morris K., Wallen S. L., Dhau J., Gohel H., Kaushik A., Front. Nanotechnol. 2021, 3, 700888. [Google Scholar]
- 19. Kettler H., White K., Hawkes S. J., Scientific Working Group on Leishmaniasis, Meeting & UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases, Geneva, Switzerland 2004.
- 20.a) $30 Million Research Effort to Develop New Tests for Deadly Infectious Diseases, https://www.gatesfoundation.org/ideas/media-center/press-releases/2003/05/new-tests-for-deadly-infectious-diseases (accessed: June 2022); [PubMed]; b) Martinez A. W., Phillips S. T., Whitesides G. M., Carrilho E., Anal. Chem. 2010, 82, 3. [DOI] [PubMed] [Google Scholar]
- 21. Land K. J., Boeras D. I., Chen X.‐S., Ramsay A. R., Peeling R. W., Nat. Microbiol. 2019, 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer M., Baeumner A. J., Chem. Rev. 2019, 119, 7996. [DOI] [PubMed] [Google Scholar]
- 23.a) Golmohammadi H., Morales‐Narváez E., Naghdi T., Merkoçi A., Chem. Mater. 2017, 29, 5426; [Google Scholar]; b) Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S., Biosens. Bioelectron. 2021, 176, 112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costa F., Genovesi S., Borgese M., Michel A., Dicandia F. A., Manara G., Sensors 2021, 21, 3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gubbi J., Buyya R., Marusic S., Palaniswami M., Future Gener. Comput. Syst. 2013, 29, 1645. [Google Scholar]
- 26. Chamola V., Hassija V., Gupta V., Guizani M., IEEE Access 2020, 8, 90225. [Google Scholar]
- 27.Requirement for Proof of Negative COVID‐19 Test or Documentation of Recovery from COVID‐19, https://www.cdc.gov/coronavirus/2019-ncov/travelers/testing-international-air-travelers.html (accessed: January 2022).
- 28. Morales‐Narváez E., Dincer C., Biosens. Bioelectron. 2020, 163, 112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lukas H., Xu C., Yu Y., Gao W., ACS Nano 2020, 14, 16180. [DOI] [PubMed] [Google Scholar]
- 30. Sadighbayan D., Ghafar‐Zadeh E., IEEE Sens. J. 2021, 21, 10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aqeel ur R., Abbasi A. Z., Islam N., Shaikh Z. A., Comput. Stand. Interfaces 2014, 36, 263. [Google Scholar]
- 32. Talla V., Pellerano S., Xu H., Ravi A., Palaskas Y., in 2015 IEEE Int. Conf. RFID (RFID), IEEE, Piscataway: 2015, pp. 47–54. [Google Scholar]
- 33. Hosseinifard M., Naghdi T., Morales‐Narváez E., Golmohammadi H., Front. Bioeng. Biotechnol. 2021, 9, 637203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ting D. S. W., Carin L., Dzau V., Wong T. Y., Nat. Med. 2020, 26, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reyes D. R., van Heeren H., Guha S., Herbertson L., Tzannis A. P., Ducrée J., Bissig H., Becker H., Lab Chip 2021, 21, 9. [DOI] [PubMed] [Google Scholar]
- 36.a) Misra S., Huddy J., Hanna G., Oliver N., in Medical Biosensors for Point of Care (POC) Applications, (Ed: Narayan R. J.), Woodhead Publishing, Sawston: 2017, pp. 27–44; [Google Scholar]; b) Mazzocchi R. A., ACS Sens. 2016, 1, 1167. [Google Scholar]
- 37.a) Narita F., Wang Z., Kurita H., Li Z., Shi Y., Jia Y., Soutis C., Adv. Mater. 2020, 33, 2005448; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sharafeldin M., Davis J. J., Anal. Chem. 2021, 93, 184; [DOI] [PubMed] [Google Scholar]; c) Cesewski E., Johnson B. N., Biosens. Bioelectron. 2020, 159, 112214; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yüce M., Filiztekin E., Özkaya K. G., Biosens. Bioelectron. 2021, 172, 112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J., Park M., Biosensors 2021, 11, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leva‐Bueno J., Peyman S. A., Millner P. A., Med. Microbiol. Immunol. 2020, 209, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rocha‐Santos T. A. P., TrAC, Trends Anal. Chem. 2014, 62, 28. [Google Scholar]
- 41. Nguyen H. H., Lee S. H., Lee U. J., Fermin C. D., Kim M., Materials 2019, 12, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao H., Liu F., Xie W., Zhou T. C., OuYang J., Jin L., Li H., Zhao C. Y., Zhang L., Wei J., Zhang Y. P., Li C. P., Sens. Actuators, B 2021, 327, 128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J., Lillehoj P. B., ACS Sens. 2021, 6, 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Torrente‐Rodriguez R. M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H. B., Gao W., Matter 2020, 3, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F., Biosens. Bioelectron. 2021, 171, 112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singhal C., Bruno J. G., Kaushal A., Sharma T. K., ACS Appl. Bio Mater. 2021, 4, 3962. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y., Lai B. S., Juhas M., Molecules 2019, 24, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dai Y., Liu C. C., Angew. Chem., Int. Ed. 2019, 58, 12355. [DOI] [PubMed] [Google Scholar]
- 49. De Girolamo A., McKeague M., Pascale M., Cortese M., DeRosa M. C., in Aptamers for Analytical Applications, Wiley, Weinheim: 2018, pp. 85–126. [Google Scholar]
- 50. Alafeef M., Dighe K., Moitra P., Pan D., ACS Nano 2020, 14, 17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Y., Han R., Chen M., Zhang L., Wang G., Luo X., Anal. Chem. 2021, 93, 4326. [DOI] [PubMed] [Google Scholar]
- 52. Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A. P., Vongpunsawad S., Poovorawan Y., Lee S. Y., Lertanantawong B., Nat. Commun. 2021, 12, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramanavičius S., Morkvėnaitė‐Vilkončienė I., Samukaitė‐Bubnienė U., Ratautaitė V., Plikusienė I., Viter R., Ramanavičius A., Sensors 2022, 22, 1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raziq A., Kidakova A., Boroznjak R., Reut J., Opik A., Syritski V., Biosens. Bioelectron. 2021, 178, 113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bruch R., Urban G. A., Dincer C., Trends Biotechnol. 2019, 37, 791. [DOI] [PubMed] [Google Scholar]
- 56. Stern M. J., Ames G. F.‐L., Smith N. H., Clare Robinson E., Higgins C. F., Cell 1984, 37, 1015. [DOI] [PubMed] [Google Scholar]
- 57. Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A., J. Bacteriol. 1987, 169, 5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jansen R., van Embden J. D. A., Gaastra W., Schouls L. M., Mol. Microbiol. 2002, 43, 1565. [DOI] [PubMed] [Google Scholar]
- 59. Mojica F. J. M., Díez‐Villaseñor C. s., García‐Martínez J., Soria E., J. Mol. Evol. 2005, 60, 174. [DOI] [PubMed] [Google Scholar]
- 60. Garneau J. E., Dupuis M.‐È., Villion M., Romero D. A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A. H., Moineau S., Nature 2010, 468, 67. [DOI] [PubMed] [Google Scholar]
- 61. Wiedenheft B., Sternberg S. H., Doudna J. A., Nature 2012, 482, 331. [DOI] [PubMed] [Google Scholar]
- 62. Zhang Y., Wu Y., Wu Y., Chang Y., Liu M., TrAC, Trends Anal. Chem. 2021, 137, 116210. [Google Scholar]
- 63. Dai Y., Xu W., Somoza R. A., Welter J. F., Caplan A. I., Liu C. C., Angew. Chem., Int. Ed. 2020, 59, 20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kishi J. Y., Schaus T. E., Gopalkrishnan N., Xuan F., Yin P., Nat. Chem. 2018, 10, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eissa S., Zourob M., Anal. Chem. 2021, 93, 1826. [DOI] [PubMed] [Google Scholar]
- 66. Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J. B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., McGeer A., Christie N., Gray‐Owen S., Cochrane A., Rini J. M., Sargent E. H., Kelley S. O., J. Am. Chem. Soc. 2021, 143, 1722. [DOI] [PubMed] [Google Scholar]
- 67. Hashemi S. A., Golab Behbahan N. G., Bahrani S., Mousavi S. M., Gholami A., Ramakrishna S., Firoozsani M., Moghadami M., Lankarani K. B., Omidifar N., Biosens. Bioelectron. 2021, 171, 112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bergveld P., IEEE Trans. Biomed. Eng. 1970, BME‐17, 70. [DOI] [PubMed] [Google Scholar]
- 69. Panahi A., Sadighbayan D., Forouhi S., Ghafar‐Zadeh E., Biosensors 2021, 11, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.a) Li Y., Peng Z., Holl N. J., Hassan M. R., Pappas J. M., Wei C., Izadi O. H., Wang Y., Dong X., Wang C., Huang Y. W., Kim D., Wu C., ACS Omega 2021, 6, 6643; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Seo G., Lee G., Kim M. J., Baek S. H., Choi M., Ku K. B., Lee C. S., Jun S., Park D., Kim H. G., Kim S. J., Lee J. O., Kim B. T., Park E. C., Kim S. I., ACS Nano 2020, 14, 5135; [DOI] [PubMed] [Google Scholar]; c) Fathi‐Hafshejani P., Azam N., Wang L., Kuroda M. A., Hamilton M. C., Hasim S., Mahjouri‐Samani M., ACS Nano 2021, 15, 11461. [DOI] [PubMed] [Google Scholar]
- 71.Sampinute | Celltrion, https://www.celltrion.com/en-us/kit/sampinute (accessed: June 2022).
- 72.ePlex System | GenMark Diagnostics, https://www.genmarkdx.com/education/technology/ (accessed: June 2022).
- 73. Resman Rus K., Korva M., Knap N., Avsic Zupanc T., Poljak M., J. Clin. Virol. 2021, 139, 104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.In Vitro Diagnostics EUAs—Serology and Other Adaptive Immune Response Tests for SARS‐CoV‐2, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-serology-and-other-adaptive-immune-response-tests-sars-cov-2 (accessed: June 2022).
- 75. Geng Y., Peveler W. J., Rotello V. M., Angew. Chem., Int. Ed. 2019, 58, 5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


