Summary
Infection with SARS‐CoV‐2, the etiology of the ongoing COVID‐19 pandemic, has resulted in over 450 million cases with more than 6 million deaths worldwide, causing global disruptions since early 2020. Memory B cells and durable antibody protection from long‐lived plasma cells (LLPC) are the mainstay of most effective vaccines. However, ending the pandemic has been hampered by the lack of long‐lived immunity after infection or vaccination. Although immunizations offer protection from severe disease and hospitalization, breakthrough infections still occur, most likely due to new mutant viruses and the overall decline of neutralizing antibodies after 6 months. Here, we review the current knowledge of B cells, from extrafollicular to memory populations, with a focus on distinct plasma cell subsets, such as early‐minted blood antibody‐secreting cells and the bone marrow LLPC, and how these humoral compartments contribute to protection after SARS‐CoV‐2 infection and immunization.
Keywords: antibody secretion, antibody‐secreting cell, COVID‐19, long‐lived plasma cell, SARS‐CoV‐2
1. INTRODUCTION
The SARS‐CoV‐2 pandemic arose in China at the end of 2019 and has caused over 450 million infections and more than 6 million deaths worldwide. Initially, the virus caused a wide range of clinical manifestations from asymptomatic and mild to severe and critical, leading to death in ~1.5% of infected individuals. Elderly patients were initially at risk of severe disease together with those who had co‐morbid conditions, such as diabetes and obesity. Pneumonia and respiratory failure often led to hospitalization; however, this infection caused gastrointestinal and endothelial injury leading to systemic illness affecting multiple organ systems that included the brain and many others. 1 With mild illness even after recovery, post‐acute sequelae of SARS‐CoV‐2 infection (PASC) has also been reported without clear underlying pathophysiologic mechanisms. 2 At first, sequelae were thought to occur only after severe infection, but now PASC has been commonly reported after both mild and severe disease at frequencies as high as 10‐30% making it even more puzzling. Therefore, understanding immune mediators of protection from infection and severe disease as well as the immune mechanisms of the sequelae are critical to overcoming this pandemic.
Viral neutralizing antibodies (nAbs) secreted by LLPC provide durable protection after infection. Prior to COVID‐19, the best‐known pandemic was the 1918 H1N1 influenza virus, which offered life‐long serologic protection after primary infection. 3 However, reinfections could occur from new re‐assorted influenza viral mutants and not necessarily from the previously circulating strains. But, in COVID‐19, unlike influenza virus infections, antibody responses after SARS‐CoV‐2 infection whether it be mild or severe appear to persist for only 18‐20 months. 4 , 5 Thus, antibody protection after SARS‐CoV‐2 infection may not necessarily be long lasting and a cause of breakthrough infections. Additionally, similar to influenza viruses, the evolution of new viral variants of SARS‐CoV‐2 for which there is little cross‐protection may be another cause of repeat coronavirus infections with the recent Delta 6 and Omicron 7 mutants despite history of previous infection. 8 , 9 , 10
In the United States and then globally, vaccines to SARS‐CoV‐2 were introduced within a year after the start of the pandemic which was an incredible scientific achievement. These vaccines provided robust protection especially with high titers of nAbs and afforded safeguards for severe disease. However, the primary vaccine series were effective only short‐term and exhibited waning efficacy within months. 11 , 12 , 13 , 14 Thus, the CDC guidance now recommends a booster dose 6 months after the initial primary two‐dose immunization. Despite shielding from hospitalizations, waning vaccine titers were not necessarily effective against new viral variants, causing many breakthrough infections (BTI) even though most were mild. In all, following emerging viral mutants, the understanding of the mechanisms of durable humoral protection from infection and vaccination is vital in the fight against this pandemic.
2. B CELLS AND LONG‐LIVED PLASMA CELLS IN VIRAL INFECTION
During a canonical respiratory viral immune response, naive B cells encounter viral antigens, become activated, and differentiate into antibody‐secreting cells (ASC) from extrafollicular (EF) or germinal center (GC) B cells. Some naive B cells enter the GC to engage with the antigen and T follicular helper cells (Tfh) to undergo rounds of expansion, somatic hypermutation (SHM), and antigen‐specific positive selection. Ultimately, successful GC‐derived clones differentiate into high‐affinity ASC and memory B cells (MBC) and are thought to become long‐lived. The decision of GC B cells to remain in the GC, exit as MBC, or further differentiate into ASC have been studied in mouse models 15 but are not well described in human studies 16 (Figure 1).
FIGURE 1.
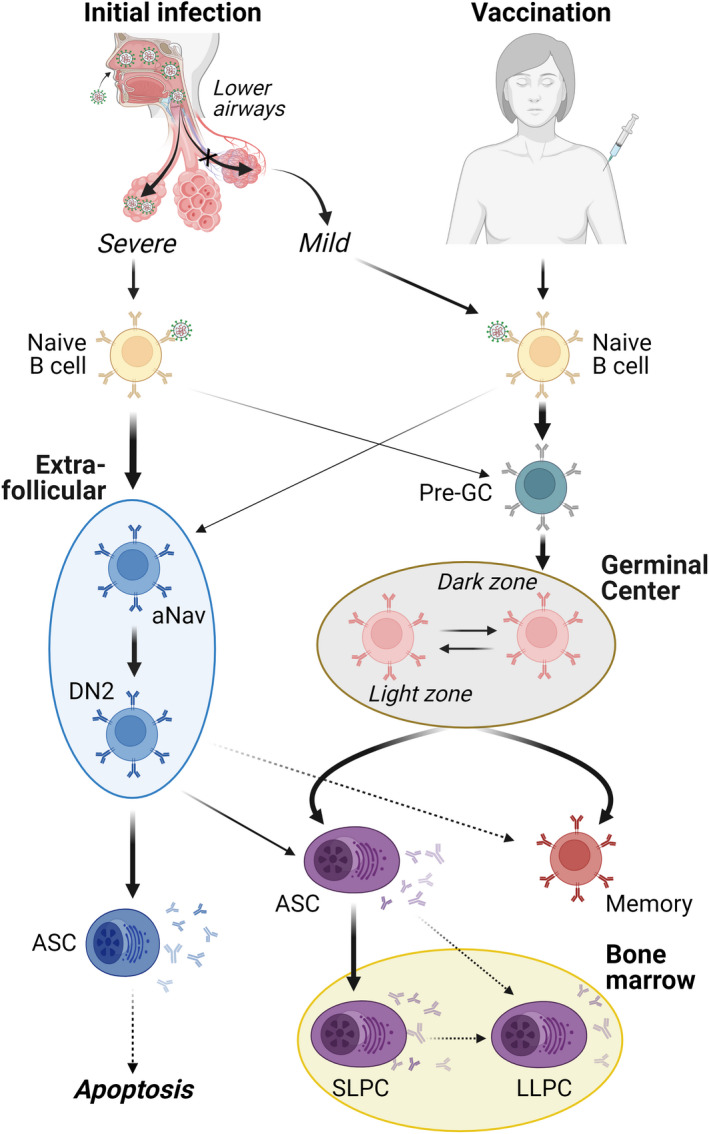
B cell response development in COVID‐19. Primary infection with SARS‐CoV‐2 results in a spectrum of disease severity with differing impacts on humoral response development. (Right) Mild COVID‐19 or vaccination results in a GC‐focused response, allowing normal accumulation of somatic hypermutation, affinity maturation, memory formation, and plasma cell development. The extent of LLPC development in GC‐focused COVID‐19 responses remains a critical open question with important implications in response longevity. (Left) Severe/critical COVID‐19 results in an extrafollicular (EF)‐biased response with the rapid development of low‐mutation effector B cells (DN2) and plasmablasts. While the neutralizing capability of these populations has been confirmed, the impact of EF‐biased responses on memory formation, plasma cell development, and bone marrow engraftment is less clear. Heavy arrows—dominant pathway; Light arrows—secondary pathway; Dotted arrows—unconfirmed pathway. GC, germinal center; aNav, activated naive B cells; DN2, double negative (i.e., IgD−CD27− B cells that also lack expression of CXCR5 and are involved in the EF response that is outside the GC but can still have T cell help); ASC, antibody‐secreting cell; SLPC, short‐lived plasma cell; LLPC, long‐lived plasma cell
At steady state, healthy humans have a low ASC frequency in the circulation (i.e., <1% of the total B cells) 17 , 18 , 19 but during acute viral infections, ASC rapidly burst into the bloodstream with a rise in protective pathogen‐specific antibody levels. 18 , 20 , 21 Typical ASC responses after acute infection range from 2 to 10% of total B cells; but in some specific infections such as with Hantavirus and Dengue viruses, ASC may account for up to 70‐80% of all circulating B cells. 17 , 18 , 22 , 23 , 24 Whether the magnitude of the responses reflect primary versus secondary exposures or result from the different types of viruses is not entirely clear.
Typical serum titer responses reveal an early GC‐independent phase with the appearance of low‐affinity primarily IgM, 21 , 25 followed by high‐affinity, class‐switched, pathogen‐specific durable IgG and IgA. After the initial robust rise in Ab titers, the decay kinetics of the antigen‐specific levels are twofold as shown in non‐human primate models. The first is a rapid fall‐off due to apoptosis of short‐lived ASC and then a slower decline or “memory” Ab after months likely from LLPC generation and maintenance. 26 The main source of serum “memory” Abs arises from circulating GC‐derived MBC which differentiate into ASC to mature into tissue‐resident LLPC, both of which are extremely rare and produce highly diverse and affinity‐matured Abs. 27 , 28 , 29 , 30 In mice, LLPC have been identified in the spleen, the gut, and the bone marrow (BM), and are found weeks following the initial induction. 31
The mechanisms of how human LLPC are generated and maintained are not entirely clear. However, it is known to include GC and MBC responses and the migration of ASC to long‐lived tissue sites such as the BM niches. In humans, LLPC were found in the BM CD19−CD38hiCD138+ compartment from natural viral infections that occurred over 40 years ago. However, exposures to repeat viral infections and vaccination were localized in other BM compartments such as CD19+CD38hiCD138+ subsets and the LLPC subsets. 32 After the initial burst into the blood, most early‐minted ASC or plasmablasts undergo apoptosis triggering the rapid primary decline of Ab titers. Only some ASC eventually enter long‐lived tissue sites such as the BM to submit to further development and maturation through factors provided by the specialized microniche. 33 These cells are likely responsible for the second slower Ab decay. Histology shows that LLPC have unique morphology from nascent ASC such as increased cytoplasm/nucleus ratio and higher number of mitochondria. Although LLPC are derived from early‐minted ASC, they are transcriptionally and epigenetically different illustrating ongoing maturation in the BM sites. 34 These special molecular and epigenetic pathways enhance longevity, minimize energetic needs, and upregulate programs to acquire resistance to apoptosis in order to maintain antibody secretion for a lifetime. 34 In this review, we will investigate whether ASC after SARS‐CoV‐2 infection and new COVID‐19 mRNA vaccines follow the canonical B cell and LLPC maturation programs or if these humoral responses are fundamentally altered.
3. ASC RESPONSE IN SARS‐COV‐2 INFECTION
3.1. Primary infection: virus‐specific antibodies are highly diverse, peak early, and decline
The majority of primary SARS‐CoV‐2 infections elicits a robust systemic viral‐specific Ab response initially within 1‐2 months, 35 , 36 , 37 although the Ab magnitude among infected individuals is heterogenous with peak levels varying over 200‐fold. 37 , 38 By and large, Ab levels were reduced by 5‐fold to 10‐fold compared to the peak at 5 months 35 , 36 , 37 , 39 with some studies showing that they remain detectable for 5‐12 months, 37 , 39 , 40 , 41 , 42 , 43 , 44 , 45 13‐14 months, 46 , 47 and some suggesting 18‐20 months, 4 , 5 in the absence of vaccination and reinfection. However, the pandemic started only 2 years ago, and so longer durability data are just not available.
After an early peak within 2‐5 weeks, Abs decline in a fashion that varied by isotype, viral antigen‐specificity, and age. 37 , 48 , 49 While IgM and IgA often wane rapidly and become undetectable after 2‐3 months, 50 , 51 IgG decays at a slower rate. Additionally, different viral antigens such as nucleocapsid (N), receptor‐binding domain (RBD), and spike (S) also give rise to variable kinetics. For example, the serum N‐Ab decay more rapidly compared to RBD‐ or S‐Ab. The estimated average half‐life in most infections of S‐specific IgG, IgM, or IgA1 is 14‐33, 8, or 6 weeks, respectively. 37 , 52 On average, the fastest waning Abs were N‐specific IgG with two‐third the levels at 4‐9 months and undetectable levels in 33% of the patients. By 1 year, almost all patients had no measurable N‐specific IgG. 53 , 54 , 55 , 56 , 57 S‐specific IgG decays slowest, waning to less than one‐third of the peak levels at 8‐10 months. However, nearly all patients (90‐97%) have detectable S‐IgG titers at 12‐13 months. 53 , 54 , 55 Finally, not all SARS‐CoV‐2‐infected patients developed demonstrable serum Abs, with some studies reporting 5% to 33% of PCR‐positive patients particularly in young adults who did not seroconvert. 58 , 59 , 60
Antibodies that functionally neutralize correlate with total virus‐specific Abs and RBD‐specific Abs. 61 Both total virus‐specific and nAbs usually peaked between 3 and 5 weeks after infection, but also rapidly decayed with an average half‐life of 8‐13 weeks. 37 , 41 , 50 , 52 , 62 , 63 , 64 , 65 However, it appears that in mild to moderate infections, nAbs could last for at least 5‐7 months. 14 , 39 , 66 , 67 , 68 , 69 , 70 Both total viral‐specific Abs and nAbs rapidly wane initially, but then declined at a much slower rate to remain relatively stable with time. 37 , 39 , 51 , 71 , 72 , 73 In all, infection‐induced serum S‐ and RBD‐specific IgG were positively correlated with nAbs, and these antibodies peaked within a few months and initially wane rapidly and then with a slower decay over the first year. 37 , 52 , 53 , 56 , 74 , 75 , 76 Whether this slower decay will ultimately plateau as seen in other infections to provide LLPC and life‐long protection remains at large.
Increased Ab responses were associated with older age, male sex, and hospitalization. 38 However, disease severity seemed to have the greatest effect on the magnitude of infection‐induced Abs. 38 , 73 , 77 , 78 In general, severe infections were associated with both a more rapidly rise and a higher peak in both binding and neutralizing Abs. 40 , 51 , 79 , 80 These Abs rocketed rapidly within days of symptom onset 40 , 81 especially in hospitalized or critically ill patients compared to mild (outpatients or asymptomatic) subjects. 40 , 50 , 51 , 54 , 56 , 73 , 79 Moreover, unlike conventional responses, the majority of these responses did not generate an early IgM response followed by the conventional class‐switched IgG and IgA. 50 , 77 Instead, a class‐switched IgG with neutralization was detected early in these critically ill patients. Later monoclonal antibody studies showed low or germline mutation frequencies found in severe infections, implicating unique nonconventional B cell origins. 81 , 82 , 83
3.2. Germinal centers are disrupted in severe COVID‐19 infection
Unlike typical viral infections, early studies showed that in severe SARS‐CoV‐2 infections, the GC are impaired 84 , 85 and are associated with large plasmablast expansions and enhanced Ab levels compared to mild disease 37 , 40 , 77 , 86 (Figure 1). The decreased numbers of Tfh in the draining lymph nodes (LN) and spleen provided evidence that functional GC fail to form during critical illness. 84 , 85 Furthermore, in these severely ill patients, a robust EF B cell response dominates with higher ASC expansion and correlated with nAb levels. 81 , 86 Corroborating this model, multiple potent nAbs were isolated from severe patients exhibiting only few mutations suggesting that EF responses can give rise to effective nAbs. 83 , 87 Interestingly enough, both mild and severe COVID‐19 infections showed evidence of class‐switched MBC with higher mutation frequencies 41 , 78 , 88 , 89 and strong Tfh cell responses. 90 , 91 , 92 Thus, a strong EF response may not always occur at the exclusion of GC B cell responses. However, the collapsed GC in the critically ill patients give rise to a massive early EF ASC response, causing the rapid rise in Ab titers.
3.3. SARS‐CoV‐2‐specific ASC responses
The rapid and transient expansion in the circulation of ASC is generally a hallmark of early B cell responses during acute viral infections. 21 Initial infections with SARS‐CoV‐2 give rise to an early Ab peak within the 2nd week post‐induction that wanes substantially and rapidly over time (declining by 5‐fold to 10‐fold within 3‐4 months or to <7% of the peak at 5‐6 months) (Figure 2). 19 , 35 , 36 , 37 , 51 , 71 , 72 , 73 , 77 , 93 , 94 , 95 This fast decay most probably reflects apoptosis of many circulating short‐lived IgG and IgA ASC, known to appear within a few days after initial antigen exposure. 20 , 22 , 23 , 24 , 32 , 96 , 97 In severe infections, circulating ASC defined as CD19+CD27hiCD38hi, which included CD138+ subsets were expanded although their frequency was not associated with virus‐specific IgM. 81 , 86 A similar pattern was seen in Dengue infections, where higher ASC expansions were associated with more severe illness. 86 , 98 , 99 Hence, the rapid antibody decay is a manifestation of apoptosis of the nascent blood ASC.
FIGURE 2.
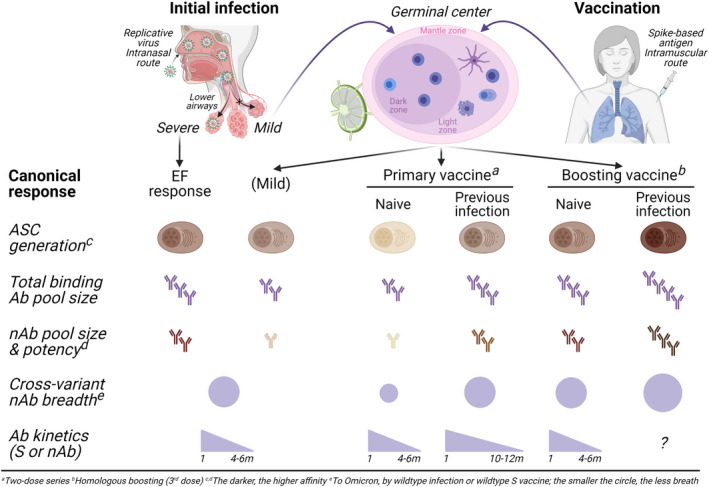
ASC kinetics and Ab effector functions during responses to infection with and vaccination against SARS‐CoV‐2. Initial infection induces ASC that produce virus‐specific, low‐affinity serum Abs. In general, mild infection, priming vaccination, or tertiary vaccination generates a GC response, by which the derived MBC undergo continued clonal evolution over 6‐12 mo, leading to the production of more potent and broader nAbs. The frequency of ASC generally correlates with the magnitude of the serum Ab levels (total binding Ab pool size). Dose 1 vaccine induces a robust GC response resulting in the generation of virus‐specific ASC (and MBC) including in infection‐naive subjects and which is substantially enhanced either by Dose 2 (in infection‐naive subjects) or in previously infected (recovered) subjects—and further enhanced by boosters (in infection‐naive subjects). The highest total binding Ab production is observed in recovered, tertiary vaccinees. Dose 1 ignites potent nAbs (in about half the subjects) that are enhanced by Dose 2 and further enhanced by booters—against the wildtype but less potent against variants (decreasing cross‐variant nAb potency). S‐specific and nAbs wane over 4‐6 mo following infection, although total binding Abs could be detected 18‐20 mo post‐infection. The nAb waning period of time in COVID‐19‐naive vaccinees also are usually 4‐6 mo; it may last longer in previously infected subjects (i.e., 10‐12 mo). Ab, antibody; nAb, neutralizing antibody; ASC, antibody‐secreting cell; EF, extrafollicular; S, spike
Early ASC may serve as a biomarker of disease severity, 40 , 81 which at the same time, raises concerns about a potential pathogenic role of ASC. 86 , 98 , 99 One study showed that the expansion of ASC in the circulation in hospitalized patients with COVID‐19 infection decreased 28‐day mortality although the differences were small, suggesting ASC might actually also serve as a marker of disease resolution. 100 Whether ASC expansions are pathogenic or bystander effects from certain proinflammatory cytokines supporting ASC survival, such as IL‐6 and TNF‐α, which are coincidently elevated in severe COVID‐19, 81 , 86 , 101 , 102 , 103 is not entirely clear.
A meaningful ASC response depends not only on quantity but also on quality, such as nAb and different isotypes. Different isotypes IgM, IgA, and IgG were notable in the serum and/or mucosal sites 1‐2 weeks post‐symptom onset. 40 , 77 , 104 In COVID‐19, although IgA is normally responsive at mucosal sites, virus‐specific IgA ASC were also expanded in the circulation. 95 , 105 Additionally, SARS‐CoV‐2 neutralization was correlated more closely with IgA than IgM or IgG in the first weeks after symptom onset. 95 Despite this result, the IgA responses were not associated with disease severity and serum IgA concentrations decreased by 1 month. However, mucosal neutralizing IgA remained detectable in the saliva for more than 3 months, suggesting locally differentiated IgA ASC may have a longer half‐life than systemic IgA ASC and confer protection from reinfection. 95 In all, IgA ASC can be found in the blood and mucosal sites during an acute infection. However, it is not clear if mucosal IgA ASC differentiate locally or systemically and then migrate to the mucosal sites in acute illness.
Another study showed that RBD‐specific ASC are released into the blood transiently during acute COVID‐19 with high IgM and low IgG ASC frequencies. 106 However, these results may have been skewed with antigen‐labeled flow cytometry which only select for ASC that retain surface BCR expression. From B cell to ASC differentiation, surface Ig receptors are often downregulated. 32 , 97 Interestingly, only IgM ASC preferentially express surface BCR compared to IgG ASC. 82 Hence, antigen‐specific surface flow cytometry of ASC may neglect the majority of blood ASC in this infection.
3.4. Memory B cell evolution and cross‐variant reactive antibodies in COVID‐19 infection
Understanding MBC specificity and kinetics is key to predicting durability of protection from reinfection. After infection, it is well‐established that a strong MBC response is elicited. While most Ab response metrics decrease within 4‐6 months, the frequency of circulating MBC remain relatively stable for 6‐9 months after infection (including mild and asymptomatic), 36 , 42 , 52 , 107 , 108 , 109 and may even increase before plateauing during convalescence. 37 , 41 , 52 , 78
It appears that even after viral clearance, the MBC response continues to mature. Perhaps more importantly, infection‐induced MBC continue to accumulate somatic mutations over 12 months comparable to those acquired in other acute viral infections. 85 , 110 This maturation results in the emergence of unique clones and the production of memory Abs with increased affinity. 36 , 107 , 111 Although class‐switched MBC evolved in both mild and severe COVID‐19, 41 , 78 , 88 such affinity maturation might not be the same. For example, B cell repertoires in severe patients are enriched for clonally expanded and unmutated ASC and MBC clones, consistent with EF‐dominant responses, 81 whereas in mild illness, they are characterized by clonally diverse and mutated MBC. 112 Evolution of MBC after infection was observed over 12 months together with persistence of GC after infection intimated antigen persistence. 36 , 42 , 113 Interestingly enough, some asymptomatic individuals 4 months after the onset of COVID‐19 infection showed persistence of SARS‐CoV‐2 nucleic acids in the intestinal biopsies, demonstrating antigenic persistence. 36 With each new emerging mutant, whether MBC in the LN continue to rapidly evolve to generate higher affinity clones that could provide a stronger and more cross‐reactive protection will require further study.
3.5. Lack of bona fide LLPC in response to COVID‐19 infection
High‐affinity “memory” nAbs in the serum are the effector molecules of long‐term protection. While ASC provide robust Ab response during the acute infection, tissue‐resident LLPC in the BM are the cellular origins of such persistent “memory” Abs. LLPC secrete Ab continuously in the absence of antigen. 114 After mild SARS‐CoV‐2 infections, plasma cells specific for SARS‐CoV‐2 have been identified in the BM 7‐11 months after infection. 45 However, BM niche is known to contain both LLPC and other shorter‐lived subsets 32 (Table 1), and this study 45 did not demonstrate whether these viral‐specific ASC were residents of the BM LLPC subset 32 (i.e., PopD; Table 1). Furthermore, the serologic data after acute infection 37 , 39 , 51 , 71 , 72 , 73 , 77 may not be consistent with the presence of LLPC, and thus, whether this infection generates bone fide LLPC still remains unknown (Figure 1).
TABLE 1.
Phenotype of blood and bone marrow ASC subsets
| Blood ASC subsets | Pop2 | Pop3 | Pop5 |
|---|---|---|---|
| CD19 | + | + | − |
| CD138 | − | + | + |
| CD38 | ++ | ++ | ++ |
| Bone marrow ASC subsets | PopA (SLPC a ) | PopB (Intermediate) | PopD (LLPC b ) |
|---|---|---|---|
| CD19 | + | + | − |
| CD138 | − | + | + |
| CD38 | ++ | ++ | + |
Short‐lived plasma cell.
Long‐lived plasma cell.
3.6. Transcriptional profiles of ASC in COVID‐19 infection
ASC single cell profiling from COVID‐19‐infected patients is often sorted from total peripheral blood mononuclear cell (PBMC) samples. 115 , 116 Despite acute and recovered time points and known expansions, these cells are relatively rare in the blood. Therefore, single‐cell studies using PBMC can at best enumerate the ASC, B cell, and other lymphocytes but have major limitations in understanding the transcriptional profiles of ASC due to the small number of ASC recovered from PBMC isolations. Using PBMCs, one study explored the transcriptional profile of ASC from COVID‐19 during acute infection from those who shed virus <7 days versus <14 days and healthy adults. COVID‐19 had higher percentage of ASC with significantly reduced naive BC frequencies as compared to healthy controls. As expected, they could only see higher level of B cell activation‐related genes and ASC differentiation were upregulated in the COVID‐19 patients. 115 Another PBMC study showed that ASC from a severe cohort had interferon responsive genes such as FOS, IFI6, and MX1, 117 suggesting the potential of EF B cell origins found in autoimmunity and recently described severe COVID‐19. 81 , 118 However, the ASC numbers analyzed were small. Qi et al. 119 re‐analyzed data from three published PBMC single‐cell datasets from mild and severe COVID‐19 and showed that metabolic genes regulating oxidative phosphorylation were expressed at highest level in ASC of severe COVID‐19. Although interesting, the progressive upregulation this pathway had been previously appreciated in B cell to ASC differentiation. 120 , 121
The novel single‐cell technologies have proven to be extremely powerful in deeply characterizing the transcriptional profiles and the VDJ sequences of plasma cells. However, the rare frequencies of ASC despite their large expansions together with the propensity for apoptosis are the major technical limitations of further enriching this population for single‐cell studies. Hence, using total PBMC isolations to study ASC on a single‐cell level has many limitations. To properly analyze the heterogeneity of ASC subsets and their possible role in severe and mild COVID‐19 infection, strategies for better enrichment will be needed to provide insights into the ASC metabolic, homing, survival, and maturation pathways to become a LLPC.
3.7. Neutralizing versus non‐neutralizing antibodies in COVID‐19 infection
Neutralization is thought to be the main mechanism of immune protection to most infections, including SARS‐CoV‐2. This mechanism is achieved by blocking the engagement of the SARS‐CoV‐2 S protein to its cognate receptor ACE2. As expected, many nAbs target the RBD. 122 , 123 During severe COVID‐19 illness, patients have higher levels of Abs and exhibit an oligoclonal ASC expansion. 86 Although higher nAb titers are seen in severe disease, 124 , 125 the potency of neutralization is actually associated with survival and favorable clinical outcomes. 126 , 127 Amanat et al. 128 showed that mRNA vaccines can elicit more potent antiviral polyclonal responses than those seen with infection, but vaccines can actually induce a majority of non‐nAbs. While the benefits of nAb are known, the exact role played by non‐nAbs is still under intense investigation.
Before vaccines were available, passive transfer with convalescent plasma was approved for clinical use. Concerns about potential risk of antibody‐dependent enhancement (ADE) of infection in SARS‐CoV‐2 with non‐nAbs were raised. 77 , 129 , 130 , 131 , 132 , 133 , 134 These concerns were based on evidence of that virus‐specific Abs can promote cellular infection through Fc receptors. This phenomenon has been seen with several other endemic coronaviruses (eCoV), 135 including feline infectious peritonitis virus, 136 SARS‐CoV‐1, 137 , 138 , 139 , 140 , 141 , 142 and MERS. 143 Additionally, in SARS‐CoV‐1, Fc‐mediated Ab function can skew macrophage activation to a more inflammatory state in the lung leading to tissue injury. 144 Furthermore, Abs against SARS‐CoV‐2 could facilitate viral entry into myeloid cells through Fc receptors in vitro. 145 , 146 Although studies have shown viral genetic and protein content inside macrophages, 147 , 148 , 149 , 150 there is still debate whether this cell type is permissive to productive SARS‐CoV‐2 viral replication. 151 To our knowledge, there is no evidence of clinically significant ADE with SARS‐CoV‐2 infection or vaccination.
In vivo animal models of SARS‐CoV‐2 infection have revealed that Fc‐mediated Ab function improves disease outcomes and reduces viral replication. Consistent results have been seen in mice, 145 , 152 , 153 , 154 , 155 hamsters, 154 and macaques. 145 , 156 , 157 In humans, Fc patterns differentially correlate with disease outcomes. Patients with clinically more severe COVID‐19 disease exhibited a more proinflammatory pattern of Ig Fc glycosylation than those with mild disease. 158 , 159 On the contrary, Fc‐mediated antiviral functions of non‐nAbs have also been observed in vitro, including antibody‐dependent complement deposition (ADCD), 156 antibody‐dependent cellular phagocytosis (ADCP), 152 , 156 and antibody‐dependent cellular cytotoxicity (ADCC). 160 , 161 ADCP was associated with lower inflammation and clinically milder COVID‐19 than ADCD. 162 Interestingly, adults after mRNA vaccination have a distinct pattern not seen with infection. 159 , 163 This finding demonstrates how different immunity to vaccination and infection can be. In another study, Zohar et al. 164 showed that in severe SARS‐CoV‐2 infection, Fcγ receptor binding and Fc effector activity were compromised and associated with COVID‐19 non‐survivors.
Another potential protective mechanism of non‐nAb is through the soluble Fcγ‐binding protein (Fcγbp) located on mucosal surfaces. Fcγbp is a large molecular weight mucin‐like secretory Fc receptor protein secreted by human goblet cells in the large and small intestine. Virus‐Ab complexes can engage the soluble Fcγbp attached to mucin and facilitate viral clearance. 165 Fcγbp may be one potentially protective non‐nAb functions in COVID‐19, and there are likely other innate‐like functions of non‐nAb. In sum, although non‐neutralizing, these Abs can cause pro‐ or anti‐inflammatory based on different Fc functionalities.
4. ASC RESPONSES TO SARS‐COV‐2 VACCINATION
4.1. Primary vaccine series: nAbs are robust and predictive of vaccine efficacy (VE) but wane
The currently available COVID‐19 vaccines use SARS‐CoV‐2S antigen and are developed from two distinct platforms: mRNA‐based and adenovirus‐based vector vaccines. 166 These vaccines exhibit high initial efficacy at preventing infections (91‐95%) as well as hospitalization and severe disease (97%). 11 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 , 175 In addition to individual protection provided to vaccine recipients, massive vaccination could reduce community transmission, 176 although the absence of residual mucosal IgA with systematic vaccines may have hindered this potential benefit 177 , 178 (Figure 3).
FIGURE 3.
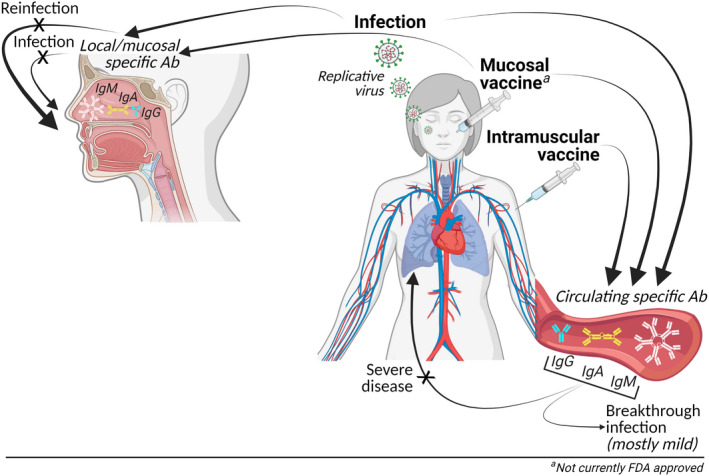
Mucosal and systemic antiviral responses after SARS‐CoV‐2 infection and vaccination. Mucosal exposure to viral antigen (by natural infection or by intranasal immunization) leads to in situ as well as systemic activation of virus‐specific adaptive immune cells. With intramuscular immunization, mucosal exposure to antigen is not present, therefore, only generating systemic but not mucosal immune responses. With mucosal antigen exposure, there is generation of tissue‐resident memory lymphocytes and ASC that locally prevent infection upon subsequent virus exposures. Without mucosal responses but in the presence of systemic antiviral responses, there is protection against severe disease but less so against the early infection at the mucosal entry site. Ab, antibody; BTI, breakthrough infection
Similar to SARS‐CoV‐2 infection, mRNA vaccination induces early and robust production of S‐specific IgM, IgA, and IgG in the circulation 179 , 180 , 181 , 182 , 183 (Figures 2 and 3). The GC disruptions present in severe COVID‐19 patients 81 , 84 are not observed after mRNA vaccination and active SARS‐CoV‐2‐specific GC responses can detected for several months. 183 However, with vaccination, the expansion of ASC is often less robust compared to acute infections (i.e., average of 2‐6% and mostly <20% of the total circulating B cells). 18 , 22 , 23 , 24 In contrast with infection that exposes the infected patient to epitopes across the entire viral proteome, vaccines only include S epitopes. 166 Therefore, as expected and unlike infection, vaccination incites a largely homogeneous S‐specific response among vaccinees. 184 , 185 , 186
After receiving the first dose of mRNA vaccine, about only half of the recipients produce nAbs, which, to most of the vaccinees, increase after the second dose. 187 , 188 , 189 In comparison with the two‐dose mRNA vaccination strategy, the single‐dose adenovirus vaccine used in the United States elicits lower S‐specific Abs. 175 , 190 , 191 However, it sufficiently primes the immune system and provokes a durable humoral and cellular immunity lasting up to 8 months. 192 As with infection, serum binding S‐specific IgG elicited by vaccination (both mRNA‐based and adenovirus‐vectored) positively corelate with nAbs 40 , 74 , 75 , 76 , 193 , 194 , 195 and are associated with VE. 74 , 193 , 194 , 195 , 196 Thus, like infection, nAbs have been identified as a surrogate marker/predictor of VE.
Consistent with epidemiological data of VE, S‐specific binding and neutralizing Abs induced by vaccination exhibit a time‐dependent reduction. 12 , 13 , 74 , 174 , 197 , 198 Moreover, most ASC undergo apoptosis rapidly after their peaks in peripheral blood (i.e., 5‐7 days post‐induction), resulting in a sharp fall of total Abs. 20 , 23 , 24 , 32 , 114 Ab waning often occurs within 4‐6 months, yet it starts to become evident at 3‐10 weeks after the second dose. 199 Ab decrease is more profound in immunosuppressed patients 13 , 200 and exhibits a more intense decline in the older individuals. 13 , 179 , 201 , 202 , 203 Nevertheless, Abs (including nAbs) may still be detected 6‐8 months post‐vaccine. 192 , 204
4.2. Breakthrough infections with waning Abs and emerging immune‐escape variants
Vaccine breakthrough infection (BTI) refers to individuals who get infected 2 weeks or more after the initial vaccination series. Despite the initial high VE against infection, BTI cases of COVID‐19 have become increasingly common—first by the Beta variant, 205 , 206 then quickly followed by the Delta variant, 13 , 174 , 197 , 207 , 208 , 209 , 210 , 211 , 212 , 213 , 214 , 215 which emerged in the late spring–summer of 2021, 6 , 8 , 216 , 217 and currently with the dominating Omicron variant. 7
While there is a significant reduction in VE (54‐85%) with most variants, numerous observational studies suggest that VE remains substantial (90‐100%) against hospitalization and severe infections. 8 , 12 , 206 , 218 , 219 , 220 , 221 Despite high viral loads and persistent viral RNA shedding, BTI are mostly mild or asymptomatic 209 , 222 and are associated with substantially lower risk of developing long COVID symptoms than infections in unvaccinated individuals. 223 The facts that most BTI are associated with lower disease severity 8 , 209 , 222 , 224 suggest that nAbs elicited by wildtype antigens remain protective against severe infection to SARS‐CoV‐2 variants. Nonetheless, such protection of severe disease in BTI may be attributed to vaccine‐induced S‐specific T cell responses, as variants can evade Abs but not the T cell immunity. 225 , 226 , 227 , 228 , 229 , 230 Of note, in addition to nAb evasion and waning immunity, the lack of protective mucosal IgA mucosal in the setting of mRNA‐based (i.e., intramuscular) vaccination 177 , 178 might also be a contributing factor in BTI (Figure 2).
The antigenic variants that emerge and become the predominant strain are mostly those that escape pre‐existing immunity. Compared with the wildtype, the Alpha, Beta, Gamma, and Delta variants exhibit a several‐fold drop in vaccination‐induced nAbs 231 , 232 , 233 (Table 2) that further decreases over time. 53 Moreover, VE against variants is predicted to lose more than half of its power at 12 months, 231 which may explain the BTI and reinfection with variants are increasingly occurring. 70 , 206 , 234 , 235
TABLE 2.
Protection induced by homologous vaccine boosting or wildtype virus infection against evolving SARS‐CoV‐2 virus
| Variant of concern | Wildtype | Alpha | Beta | Gamma | Delta | Omicron |
|---|---|---|---|---|---|---|
| Sequence mutation a | ||||||
| Genome | − | 25‐26 | 22‐23 | 25‐26 | 24‐36 | 56‐60 |
| S | − | 10‐11 | 10‐11 | 12‐13 | 10‐12 | 31‐37 |
| RBD | − | 1‐2 | 3‐4 | 3 | 1‐3 | 15‐16 |
| RBM | − | 1‐2 | 2 | 2 | 1‐3 | 8‐10 |
| nAb potency b | +++++ | ++++ | ++ | +++ | +++ | + |
| Vaccine‐ or infection‐induced protection b | +++++ | ++++ | ++ | +++ | +++ | + |
Note: Natural infection with wildtype SARS‐CoV‐2 virus or receipt of a homologous vaccine booster after completion of the two‐dose wildtype S‐based vaccine series induces nAb potency (and hence, protection) of the highest level against the wildtype virus, which decreases gradually against variants of concern. In general, the most or the least reduced protection is observed in the variant bearing the most numerous or the least numerous numbers of mutations, respectively, in RBD/RBM sequence (i.e., most epitopic changes or more conserved epitopes, respectively). It is the generated MBC that increase in the number and continue to clonally evolve for at least 6‐12 mo after (mild) infection or upon boosting that give rise to Abs with higher potency and broader breadth in neutralizing activities against the evolving virus. See texts for detail. Sequence mutation is retrieved from https://covdb.stanford.edu/page/mutation‐viewer/.
Abbreviations: nAb, neutralizing antibody; RBD, receptor‐binding domain; RBM, receptor‐binding motif; S, spike.
Varies by sublineages.
By homologous boosting (3rd dose) or wildtype virus infection; the more (+), the stronger.
All this was further complicated by the emergence of Omicron in the fall of 2021. Once identified, this highly transmissible Omicron variant spread rapidly worldwide and by mid‐winter it accounted for nearly 100% of new US infections. 7 Compared to the ancestral strain, Omicron has 56‐60 mutations throughout its genome (Table 2). Of these mutations, 31‐37 are in S with 15‐16 of those in RBD. 236 , 237 While RBD accounts for only 15% of S, it is the target of over 90% serum nAbs. 123 Importantly, 8‐10 of the 15‐16 RBD mutations are present in receptor‐binding motif (RBM), which directly interacts with ACE2 receptor (and most monoclonal nAbs). 236 , 237 For comparison, Delta and Gamma have 10‐12 and 12‐13S mutations—with 1‐3 and 3 localized in RBD, respectively. 237 Consequently, Omicron‐specific nAbs are low or undetectable in individuals that had previously had infection to other strains or have been vaccinated with wildtype S. This makes Omicron very effective at evading immune responses and VE drops to 57% with this variant. 210 , 238 , 239 , 240 Omicron also escapes 85% of existing monoclonal nAbs. 241 Vaccination, with or without a booster, provides better protection against Delta than Omicron, 238 , 240 but BTI with Delta are associated with higher disease severity than with Omicron. 221
4.3. Booster vaccines increase nAbs and reduce infection with nAb‐escaping variants
The rapid waning of VE (and correlatively, nAbs) and the everchanging SARS‐CoV‐2 have made necessary the implementation of an additional vaccine dose. This is usually a third dose called a booster for mRNA‐based vaccinations (or a second dose after the one‐dose adenovirus vector vaccine regimens). This approach of a COVID‐19 booster vaccination (administered 5‐9 months after the two‐dose regimens) has shown to reduce the infection risk, severe illness, and deaths, 238 , 240 , 242 including older individuals 243 , 244 , 245 and immunosuppressed patients. 246 It also more rapidly decreases the viral RNA loads in patients with BTI and temporarily restores the declining immunity previously evoked by two‐dose vaccination. 210 , 211 Regardless of the type of initial and vaccine technologies used, boosters substantially raise the levels of binding Abs (by 5‐fold to 55‐fold) and of cross‐variant nAbs (by 4‐fold to 73‐fold) against multiple strains, including Beta, Gamma, Delta, and Omicron. 231 , 238 , 247 , 248 , 249 , 250 Importantly, the highest nAb production against Omicron is observed in BTI with Delta (i.e., infection on two‐dose vaccination) or two‐dose vaccinated convalescent individuals, 251 , 252 , 253 emphasizing the superior neutralization potency of hybrid immunity against immune‐evading variants. Boosters can also potentially decrease the BTI infectiousness risk (i.e., disease transmission by variants). 210 , 239 Even though boosters contain wildtype S, they can restore waning immunity and expand its breadth, possibly prolonging protection against reinfection with either the ancestral strain or variants.
4.4. Memory B cell responses in COVID‐19 vaccination
Like infection, primary vaccination against SARS‐CoV‐2 also provokes a strong MBC recall response and with booster vaccines eliciting expansion of MBC that rapidly enhance production of cross‐variant nAbs. 183 , 247 Similarly, the frequency of circulating MBC remains relatively stable for 6‐9 months post‐vaccine. 181 In contrast to infection where class‐switched MBC continuously evolve over time, 36 , 42 , 113 the evolution of vaccine‐generated primary MBC is either little or no change in the blood or secondary lymphoid organs weeks after the second dose. 111 , 113 , 254
4.5. Lack of bona fide LLPC in response to COVID‐19 vaccination
To be long‐term effective, a vaccine must generate LLPC, which deliver durable recall protection through constitutively secreting circulating “memory” Abs as a rapid primary response. In reality, not all vaccines generate and maintain LLPC. For example, while tetanus, smallpox, or MMR vaccines offer long‐lasting protection (i.e., long‐lived vaccines: LLV), pneumococcal 23‐valent (PSV23), hepatitis B, or influenza vaccine confers short‐lived efficacy (i.e., short‐lived vaccines; SLV). 27 , 255 , 256 , 257 , 258 Although the mechanisms for LLV and the generation and maintenance of LLPC remain poorly understood, 34 , 114 infections with a whole, replicative virus often induce long‐lasting response even though viral Ag component‐based vaccines usually lead to short‐lived immunity. 114
Concerns have been raised that COVID‐19 vaccines more likely belong to the SLV group. 259 , 260 , 261 The acquired humoral immunity rapidly waning within 4‐6 months after completing two‐dose and post‐boosters (i.e., VE decreases to 66% and 78% within 4 months 262 ) (Figure 2) is inconsistent with LLPC being generated and maintained. Indeed, mRNA vaccination, instead of consistently provoking a primary LLPC response, may just trigger a secondary recall. 113 , 260 , 263 The nature of such recall response might be that of immunity conferred by pre‐existing cross‐reactive MBC and cross‐reactive memory T cells, 263 which were previously elicited from prior vaccination 113 or previous eCoV infection), which may be mostly non‐neutralizing and non‐protective against the newest virus.
In a single study, mRNA vaccines are reported to induce persistent GC reactions that last for months, 183 where blood circulating ASC peak around 3‐4 weeks and decline until becoming virtually absent at 7 weeks. The presence of genuine LLPC is again not entirely evident. 16 , 32 , 103 Surprisingly, S‐specific GC B cells and ASC residing in LN are detected for up to 6 months post‐vaccination. 254 At this time point, when Ag‐specific MBC are formed and display levels of mutation similar to the GC‐derived clones, highly mutated S‐specific ASC are present in the BM. 254 If persistent GC reactions and possibly reactivation of pre‐formed MBC are ongoing for up to 6 months (which is a very long time for typical GC activities), 254 they would keep seeding the BM with newly generated ASC. Hence, it is crucial to consider not only the presence of those newly generated ASC but also the actual timing of their arrival in the BM. Additionally, identification of ASC within the BM compartment does not necessarily mean they are LLPC due to the heterogeneity of the plasma cells in this site. 32 Consequently, new ASC arrivals in the BM may not have sufficient time to mature. Thus, BM samples collected within months post‐infection or post‐vaccination may or may not become LLPC. Currently, there are no sequential studies to assess the actual timing of newly generated ASC and their arrival in the BM. Moreover, there are no immune correlates of durability which can only be conclusively determined by a tincture of time. Thus, like natural infection, definitive evidence of bona fide LLPC 16 , 32 , 103 in response to vaccination against SARS‐CoV‐2 is currently lacking. This potential absence of vaccine‐induced LLPC might indeed be one of the reasons for reinfections and BTI within months. 68
4.6. Transcriptional profiles of ASC after vaccination
The exact mechanism of the efficiency of mRNA‐based vaccines against SARS‐CoV‐2 remains largely unclear. Recent studies have tried to resolve this query by characterizing the transcriptional profile of ASC post‐vaccination. Studies have shown that the first dose generated polyclonal non‐neutralizing IgA‐dominant ASC response with some S2‐specific plasmablasts with low SHM, whereas the second dose provided neutralizing B cell responses to S1 with RBD. 264 , 265 A mass cytometry‐based study identified expansion of metabolically active, class‐switched plasmablasts expressing CD71, CD98, and cytochrome C between day 0 and 28 post‐vaccine. 266 Similarly, Amanat et al. 128 reported that some of the isolated S2‐specific mAbs had cross‐reactivity toward human coronaviruses, suggestive of recall responses which were initially induced by seasonal beta‐coronavirus exposure. To support this model, some of the cross‐reactive mAbs showed extensive SHM. Ultimately, NTD and RBD antibodies co‐dominated the response induced by SARS‐CoV‐2 mRNA vaccination, indicating alternative targets for vaccine. 128
Although abovementioned studies have provided the initial characterization of ASC response after vaccination, they are mostly qualitative using PBMC, 267 , 268 thus resulting in the same limitations as previously described in single‐cell analysis of ASC after infection. Furthermore, these studies are devoid of transcriptional comparisons of ASC between SARS‐CoV‐2‐infected and vaccinated individuals but enumerate ASC subsets within total PBMC populations. For example, the CITE‐seq‐based study showed an enrichment of plasmablasts in COVID‐19‐infected patients but not after vaccination. 268 This may have resulted apoptosis of ASC using frozen PBMC. From the limited number of plasmablasts, these studies showed that COVID‐19 patients were enriched in oxidative phosphorylation, type I and type II IFN responses, fatty acid metabolism, and mTORC1 signaling genes as compared to healthy donors. Additionally, plasmablasts of mRNA vaccinated and healthy donors were transcriptionally overall similar except for TNF‐NFkB pathway activation. A similar observation was made where volunteers were vaccinated with inactive COVID‐19 vaccine. 267 Although interesting, limited numbers of ASC analyzed would require further validation with enriched ASC populations.
5. HYBRID IMMUNITY IN SARS‐COV‐2 INFECTION AND VACCINATION
5.1. Local vs systemic protection: Abs at the viral entry site
While infection incites a specific mucosal response dominated by potent neutralizing IgA early on at the site of viral entry, 95 intramuscular vaccines only lead to the production of circulating but not residual mucosal IgA 177 , 178 (Figure 3). However, intramuscular vaccines can induce virus‐specific IgG in the upper respiratory mucosal sites. 269 With intranasal (adenovirus‐vectored) vaccines, both circulating and mucosal IgA are accelerated to protect against infection and transmission in animals, 270 similar to mucosal immunity after infection. Altogether, as protection is not a single outcome with conclusive correlates, it would be important to understand the durability of both mucosal and systemic humoral responses.
5.2. Protection conferred by vaccination versus infection: equivalent, superior, or inferior?
To control reinfection at both population and individual levels, it is essential to understand whether protection elicited by vaccination might be more durable than by infection. 271 Initially, high levels of protection (>90‐95%) against reinfection were observed equally after vaccination 11 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 and infection, 69 , 235 in part due to similar early decay rate of nAbs after infection and vaccination (approximately 60 days). 74 , 179 Also, similar protective Ab response was also observed between individuals after two vaccine doses and those who had a previous COVID‐19 infection after only one vaccine dose. 272 Thus, protection elicited by vaccination and infection originally appeared to be relatively equivalent.
While infections with the 1918 influenza pandemic virus elicited life‐long protection, 3 after influenza vaccines, humoral immunity rapidly declines within 6 months. 273 For COVID‐19, protective immunity conferred by vaccines was most sufficient within 2 months, although it may last 4‐6 months. 11 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 Immunity after infection appeared to last 4‐9 months. 36 , 69 , 186 , 235 However, observational studies suggest prior infections, especially those caused by Delta, drove greater protection against reinfection and severe disease than did full vaccination at 3‐8 months. 14 , 70 , 274 , 275 The reinfection risk among the survivors from initial infection drops remarkably by 80‐95% over 6‐9 months and even at 12 months. 54 , 67 , 69 , 235 , 276 , 277 , 278 , 279 , 280 Even if patients were reinfected, they had lower incidence of severe disease. Thus, protection elicited by infection may be superior.
On the contrary, other studies suggest vaccines may provide better protection. For example, cross‐neutralization occurred sporadically in the sera among previously infected patients but if previously infected individuals are vaccinated, nearly all developed cross‐neutralizing titers against multiple variants. 281 Immunization of non‐infected patients also elicited cross‐neutralization but at lower rates. Yet, another study showed that there was higher fold reduction of neutralization titers to new spike variants in patients with history of COVID‐19 infection vs vaccine recipients. 282 Additionally, and in favor of vaccination, an epidemiologic study of COVID‐19‐hospitalized infections showed 5.5 times higher rates among previously infected patients compared to fully vaccinated adults within 90‐179 days after infection or vaccination. 283 Prior to the circulation of Delta, COVID‐19 infections were higher among persons who survived previous infection, suggesting that vaccination appeared superior; however, when Delta became the predominant circulating strain, case rates were higher among persons who were vaccinated compared to those who survived previous infection, demonstrating immunity from infection was indeed superior. 221 Whether this is true also for Omicron will require further study.
The differences between infection versus vaccine‐induced protective durability may be influenced by several immunological factors. The different aspects may drive MBC evolution and selection to distinctive Abs. For example, infection‐induced MBC appears to undergo greater affinity maturation than those induced by vaccination, possibly generating more robust and durable immunity. 36 , 42 , 107 , 111 , 113 Antigen persistence in infection is weeks while for mRNA vaccines, it is days. 36 The route of Ag delivery probably plays a role with mucosal routes in infection and intramuscular with the current mRNA vaccines. Infections with its sundry of proteins in the intact virus compared to the adynamic pre‐fusion S in the vaccines likely also manifests different immune responses. 284 In sum, while infection‐induced immunity may be generally superior to vaccine‐elicited one, 114 virtues of immunity provided by vaccination in addition to protection from infection can be appreciated.
5.3. Hybrid immunity confers better protection than vaccine or infection alone
Hybrid immunity, which is induced by prior infection in combination with vaccination, may drive stronger and longer‐lasting protection against reinfection and severe disease compared to either immunity from infection or vaccination alone during 3‐8 months from induction. During the Delta‐virus surge in the summer 2021, previously infected persons who received the vaccine were protected against reinfection and severe disease better than adults who received just two doses of the vaccine. 14 , 70 , 274 , 275 , 285 , 286 Although all immunity wanes, vaccination after infection induced a rapid nAb titer which had higher cross‐variant neutralizing activity compared to healthy adults after just two vaccine doses 42 , 107 , 187 , 188 , 189 , 197 , 272 , 281 , 287 , 288 , 289 , 290 (Figure 2). Also, BTI significantly enhanced Ab responses to elevate IgA production (possibly owning to the intranasal route of Ag exposure) and broaden cross‐nAb potency against variants. 291 Importantly, hybrid immunity appears the most protective against Omicron, which is the most mutated and most immune evasive variant to date. 5 , 251 , 252 , 253 , 292 , 293 Overall, hybrid immunity appears to offer an immune response that is more robust, more durable, and with the best cross‐variant neutralization than immunity from vaccine or infection alone.
6. THE ASC RESPONSE IN COVID‐19: PREDICTING LONG‐TERM ANTIBODIES
6.1. Protection is not a single outcome but correlated with nAbs that wane
The long‐term control of the COVID‐19 pandemic depends on understanding durability of protection, which is based on memory induced by infection and/or vaccination. Protection against symptomatic reinfection and severe illness is normally assessed epidemiologically since a single immune outcome is not available. Immune protection is likely attributed by multiple aspects of memory responses involving dynamic interplays of viral replication and pathogenesis with key humoral and cellular components that include Abs, B cells, and T cells (and secreted products). 195 , 225 , 229 , 294 , 295 The current lack of standardized or consensus quantitative Ab (particularly nAb) assays across studies further complicates this assessment. 296
Although Ab responsiveness represents only a partial picture of the overall immune responses, the magnitude of serum nAbs in most viral infections and vaccination is highly predictive of protection against reinfection. 74 , 126 , 297 Immunologically, this effect is based on nAb functions which are to block the entry of virus into its target cells through binding viral surface Ag epitopes. Indeed, the success of vaccines to date has relied on nAbs. For SARS‐CoV‐2, passive transfer of monoclonal nAbs, which mostly recognize viral RBD, 298 , 299 , 300 offers protection against infection and severe disease in outpatients 301 and macaques. 229 nAbs can contribute to over 68% vaccination‐induced protection. 195 Moreover, in severe disease, fatal outcomes are associated with the delayed nAb kinetics. 302 Overall, nAbs are positively correlated with protection against symptomatic (not asymptomatic) reinfection and severe disease. 126 Thus, while MBC have recently been proposed to serve as an indicator of protection beyond declining nAbs, nAbs may be a surrogate measure of protection in COVID‐19. 303 , 304
6.2. Predicting long‐term Abs against SARS‐CoV‐2: lessons from endemic coronaviruses
The maintenance of long‐term protection in COVID‐19 can only be conclusively defined with the passage of time. However, as SARS‐CoV‐2 and other coronaviruses, including SARS‐CoV‐1, MERS‐CoV, and eCoV, are related phylogenetically and antigenically, the natural history and immune durability of coronaviruses may provide insights to predict potential outcome for SARS‐CoV‐2. eCoV circulate worldwide and elicit SARS‐CoV‐1‐specific MBC (and memory T cells) in many adults. 263 , 305 , 306 , 307 , 308 Durability of the response to eCoV varies significantly and is also strain‐dependent. 309 Most infections with eCoV, such as OC43, NL63, 229E, or HKU1, as well as SARS‐CoV‐1 and MERS‐CoV, led to Ab responses that last for only several months, 310 , 311 although some wane within 12‐18 months. 306 , 307 , 308 Thus, they were thought to be short‐lived. However, one report showed they persist for up to 3 years while another suggested longer durability but they were just modeling studies. 68 , 312 , 313 SARS‐CoV‐1 nAbs appear 5‐10 days post‐symptom onset 314 but may wane even more rapidly than total Ag‐specific Abs, raising questions whether there is humoral durability after infection with any coronavirus. 312 During initial SARS‐CoV‐1 outbreaks, nAbs were detected for 16‐24 months. 306 , 307 , 308 , 312 , 315 Using linear mixed models, Ab levels associated with protection against reinfection last 1.5‐2 years. 74 , 316 In sum, it is not clear that after infection life‐long protection is maintained with coronaviruses.
6.3. Original antigenic sin and how pre‐existing immunity affects SARS‐CoV‐2 antibodies
After 2 years into the pandemic and the widespread administration of vaccines, the immune landscape of COVID‐19 is ever changing with a variety of MBC responses among individuals with vaccine‐induced, infection‐induced, or hybrid immunity. Questions arise whether MBC are always helpful or can be potentially harmful to new emerging variants. Original antigenic sin (OAS) refers to an immunological phenomenon when the recall immunity generated by a previous strain dominates over the primary response to the new virus, resulting in potentiating disease severity. 317 Dominance of pre‐existing Abs that cross‐react but do not likely neutralize the novel virus actually interferes with effective responses to the new infection. The best example of OAS is perhaps the 1918 influenza pandemic, which explains the increased morbidity of young adults. Recent studies suggest that these deaths may have resulted from past MBC responses, causing a rapid EF ASC response to the old but similar virus and delayed new naive B cell responses to the new viral subtype to mediate effective viral clearance. OAS is also known by various terms, such as immunological imprinting, Ag imprinting, Ag seniority, negative interference, 318 and recently, back‐boosting. 319 For SARS‐CoV‐2, it is not clear if OAS will be problematic as new emerging variants arise; thus, close attention will be required.
eCoV, such as 229E, NL63, OC42, and HKU1, are among the most common causes of respiratory infections worldwide. 263 , 305 , 306 , 307 , 308 The pre‐existing eCoV‐specific MBC may be cross‐activated upon SARS‐CoV‐2 exposure, which might influence the subsequent response to SARS‐CoV‐2 infection 263 —and probably vaccination. 113 While antigenic imprinting appear to be common and associated with disease severity in COVID‐19, their overall protective impact has to date been largely neutral. In general, no protective correlation is observed, 320 , 321 , 322 , 323 , 324 , 325 which is likely due to the inability of the raising eCoV‐specific Abs to normally neutralize the new virus. 35 , 323 However, cross‐reactivity induced by recent eCOV infection could be relevant clinically as it can lessen disease manifestations 322 , 323 or facilitate faster recovery in COVID‐19. 326 Notably, children develop robust and stable cross‐reactive Abs beyond 12 months, which may be linked to their often milder or asymptomatic COVID‐19. 327 Of note, recent in vitro studies using human FcγR‐expressing cells suggest that these cross‐reactive Abs may be worse than non‐protective since they can induce ADE of infection with SARS‐CoV‐2 virus in these cells. 325 , 328 Whether this in vitro phenomenon is relevant to patient disease is not known. In sum, infection with SARS‐CoV‐2 enhances pre‐existing, eCoV‐specific Abs that are cross‐reactive but mostly non‐neutralizing against the new virus, unveiling in COVID‐19 an OAS response that is often poorly protective and potentially harmful. In all, the current serum assays to study OAS are severely limited since serum cannot distinguish newly generated Abs arising from newly minted ASC from Abs secreted by previously established plasma cells in BM and spleen.
7. AUTOREACTIVE FEATURES OF ASC AFTER SARS‐COV‐2 INFECTION
7.1. Extrafollicular B cells in COVID‐19 infection
An increasingly important component in the investigation of primary humoral immunity has revolved around the identification of non‐canonical B cell activation pathways that initiate outside of traditional GC structures 329 (Figure 1). First described in mouse modeling of infectious disease, 330 the EF response pathway was initially described as an expedited pathway to the generation of short‐lived ASC responsible for the earliest waves of Ag‐directed Ab production. However, over the past decade, this model has developed nuance, with evidence that EF pathway effectors can undergo SHM and contribute in a limited manner through both the generation of EF‐derived memory and LLPC generation. 331 These findings implicate the need to view the EF pathway as a potential integral component in all phases of immunity, not just in acute response. As such, understanding of primary B cell and ASC development requires careful evaluation of both GC‐ and EF‐derived pathway activation.
While the balance of these pathways has been relatively limited in primary viral infection prior to the COVID‐19 pandemic, extensive work in autoimmune disorders such as systemic lupus erythematosus (SLE) has revealed that disease severity in these patients is directly correlated with the extent of EF response bias. These responses are easily recognizable through the emergence of two t‐bet‐driven effector B cells—CD11c + IgD + CD27‐activated naive (aN) cells and CD11c+IgD−CD27− double negative 2 (DN2) cells. 118 Both EF populations can be identified as expanded circulating components in patients with active/flaring autoimmune disease and have been directly linked by repertoire analysis to the expanded ASC populations that are widely identified in SLE as pathologic components of disease. 332 Importantly, ASC resulting from EF‐dominated B cell responses in those disease systems have undergone low levels of SHM, low levels of negative selection, and have been directly linked to the emergence of self‐reactivity within the humoral compartment. 332 Though both pathways can contribute to the formation of long‐term memory and persisting humoral immunity, their relative dominance in an ongoing immune response has important implications.
While extensive mouse studies had established EF response activation as an important component in both primary infections and autoimmune models, its relevance to human infectious response had remained less clear due to the difficulty in establishing primacy in severe primary infection. Studying naive‐derived responses to previously circulating seasonal viral infections in humans were often challenging to interpret due to unknown infection history and background memory B cells. However, the emergence of SARS‐CoV‐2 provided a unique opportunity to study a single primary “natural” viral infection in the global human population. Early in the pandemic, a lack of effective immunomodulatory therapies allowed scientists to observe the ‘natural’ response courses in infected individuals, studies which would not be possible today. Employing emerging technologies including high‐dimensional flow cytometry, 333 single‐cell RNA sequencing, 334 VDJ repertoire analysis, 110 and advanced serological screening methods, 40 several groups, including our own, took advantage of these unique circumstance to characterize the natural development of B cell responses across a spectrum of primary viral disease severity.
7.2. Autoreactive features of COVID‐19‐induced ASC
Despite early speculation that disease severity might correlate with a failure of B cell development and antibody production, these concerns proved unfounded with early reporting of nAb titers across a spectrum of disease severity in the acute and recovery phase of COVID‐19. 335 These serologically based studies were rapidly bolstered by cellular analyses identifying ASC expansion as a critical feature of patients with severe disease. 333 Importantly, dimensionality reduction and clustering analysis contained within that work revealed some indications of EF response intermediates in patients with highly expanded ASC, although a lack of markers dedicated to B cell classification made positive identification difficult. This robust ASC expansion was reminiscent of previous work in dengue where severe viral infection resulted in rapid ASC responses, 18 suggesting that the observed responses to COVID‐19 may not be entirely unique. To further probe these responses, our group made use of directed B cell panels tuned to the identification of EF activation pathways to investigate emerging B cell responses across highly characterized patient groups with both mild/moderate and severe/critical COVID‐19. Surprisingly, the responses identified were highly similar to activation profiles seen in patients with chronic autoimmune disorders 81 , whereas mild/moderate patients displayed relatively modest activation of the EF pathway, expanding transitional B cell populations and unswitched memory compartments. Analysis of severe disease revealed significant increases in aN, DN2, and ASC compartments, similar to the B cell compositions identified in SLE patients with high‐activity. 81
In addition, serum collected from these patients revealed an important hallmark of reduced peripheral tolerance with increased circulation of antibodies derived from B cells expressing IGHV4‐34 as a component of the BCR. This is significant because in germline configuration, these antibodies contain an inherent capacity for self‐targeting. 336 In healthy individuals, while IGHV4‐34+ clones can be readily identified in the naive B cell compartment, they are either negatively selected due to self‐reactivity, or “redeemed” through SHM that eliminates the self‐reactive potential of these clones. 337 Loss of this peripheral tolerance enforcement had been previously identified in flaring SLE and linked directly to the emergence of new autoreactivity. 332 This finding combined with extraordinarily low SHM frequencies identified by our group and others 110 within the ASC compartment were strongly suggestive that the course of severe infection may reflect some of the biology previously characterized in the context of autoimmune disease. This possibility of emerging autoreactivity was supported by several early reports of self‐targeted antibodies against phospholipids, 338 nuclear antigens, and immune components such as type 1 interferons. 339
However, despite the indications that some ASC targeting may be self‐directed, patients displaying strong activation of the EF pathway nonetheless displayed higher levels of nAbs at early time points during acute infection whereas mild/moderate disease had more memory‐oriented B cell composition. 81 Indeed, direct testing of individual ASC clonotypes emerging from this low‐selection environment displays high specificity to the virus with more than 65% confirmed as SARS‐CoV‐2 specific. 82 However, despite this specificity, these cells are also prone to self‐reactivity with clonotypes capable of binding nuclear antigens, naive B cells, and even glomerular basement membrane, a target often associated with pathology of the kidney and lung. Interestingly, these features appeared independently controlled. Individual clonotypes could display viral binding alone, self‐reactivity alone, or even both. 82 Thus, these findings are consistent with a general reduction in negative selection thresholds and suggest that documented emergence of autoreactivity in these patients is more likely a function of altered tolerance than the result of molecular mimicry or non‐specific clonal activation.
In patients with mild illness, a lack of these low‐mutation ASC clonotypes together with lower levels of identified autoreactivity suggests that these features of ASC selection are highly responsive to the local developmental microenvironment. In this model, the EF response pathway could be envisioned as an emergency response mechanism. Under highly inflammatory conditions (reflecting severe viral illness), the slow process of GC‐based B cell selection would be suppressed or even suspended 84 in favor of EF activation for the purpose of rapid antibody production and infection control. Previous work in mice suggests that even in these EF responses positive selection is likely to guide clonotype inclusion, thereby ensuring that the overall ASC mobilization will be generally viral specific. However, autoreactive clonotypes that have escaped central tolerance would also have the opportunity to respond under these circumstances, ultimately resulting in a mix of self‐reactive and viral‐reactive ASC pools. These mixed antibody responses, while actively and effectively participating in viral clearance, may also contribute to the overall inflammatory environment through innate activation and self‐targeting to create a feed‐forward loop of EF response bias (Figure 4). Ultimately, this bias may result in mounting tissue damage. Perhaps more interesting is how engagement of multiple antigens due to poor negative selection might combine to drive low‐affinity clones toward response inclusion, although extensive molecular and cellular study would be required to confirm this phenomenon.
FIGURE 4.
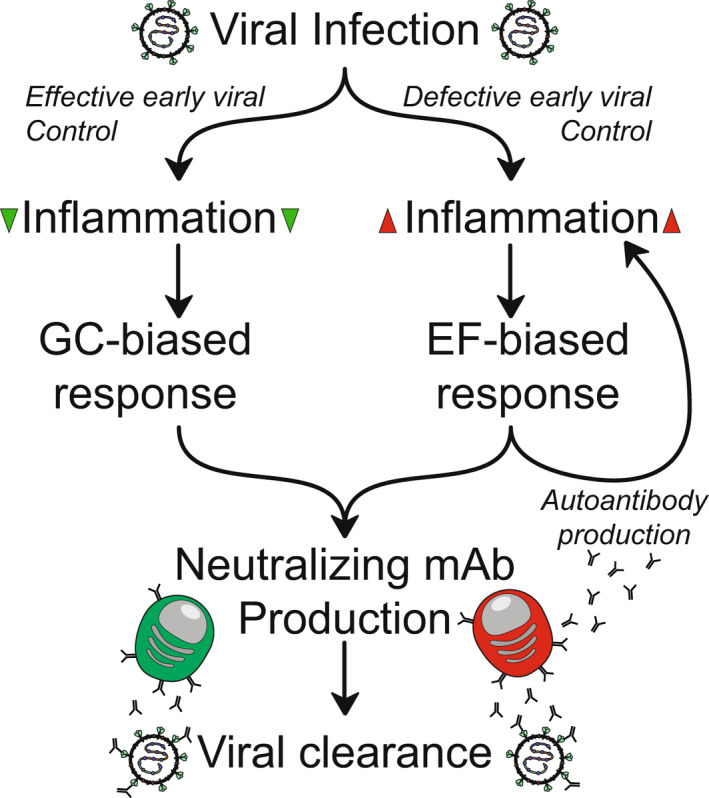
Model of autoantibody feedback. B cell activation pathway bias in COVID‐19 is dictated by early viral control. (Right) High‐inflammation microenvironments due to poor viral control drive EF‐biased responses that, while rapidly generating neutralizing Abs, can result in autoantibody production through relaxed tolerance enforcement. These autoantibodies may contribute to inflammation and tissue damage, potentially reenforcing EF‐biased response
7.3. Post‐acute sequelae of SARS‐CoV‐2 infection (PASC) and the role of auto‐Ab responses
Long COVID‐19 syndrome (LCS), COVID‐19 long hauler, post‐acute COVID‐19, long‐haul COVID‐19, or chronic COVID‐19 is all terms referring to post‐acute sequelae of SARS‐CoV‐2 infection (PASC). The incidence and prevalence of PASC are difficult to determine given that these non‐specific symptoms overlap with other clinical conditions. 8 , 340 , 341 However, PASC is becoming one of the most important healthcare problems of our time. 341 Since SARS‐CoV‐2 infection can elicit auto‐Ab responses, especially in those critically ill, 82 whether the autoimmune responses during the acute phase of infection persist to contribute to the pathogenic mechanisms in PASC is not known.
Targets of these auto‐Abs have been documented to include self‐Ag seen commonly in autoimmune conditions 342 , 343 , 344 , 345 and some have with molecular homology with SARS‐CoV‐2. 346 Some identified targets included phospholipids, 347 , 348 , 349 cytokines, 343 , 345 and type 1 interferons. 343 , 344 , 350 The disruption of these targeted self‐molecules could potentially explain procoagulant states, immune dysregulation, and the weakened antiviral responses, respectively, conditions commonly observed in COVID‐19. 351 Some researchers 352 have even hypothesized that anti‐idiotype auto‐Abs against SARS‐CoV‐2‐specific Abs could structurally resemble the SARS‐CoV‐2 S epitopes with the potential to cause cellular dysfunction by engaging its cognate receptor ACE2. Notably, anti‐ACE2 auto‐Abs have been reported in COVID‐19. 346 These proposed anti‐idiotype Abs would also be able to induce ADCC if the appropriate Fc functionality is present. It is still unknown whether the generation of auto‐Abs during acute infection correlates with PASC, but evidence is starting to emerge that patients with PASC harbor auto‐Abs for longer than the acute infection process, 342 , 344 , 347 and overall immune perturbations last longer than the acute period. 353 Interestingly, SARS‐CoV‐2 mRNA vaccination does not appear to trigger auto‐Ab responses. 354
Forecasting who will eventually develop PASC could be helpful in anticipating complications and possibly directing treatment. Prediction models of self‐reported symptoms and immune parameters have been suggested. 355 One showed particular IgM and IgG3 subclass signatures 356 and another one utilized a complex multi‐omics analysis to show that during acute illness, auto‐Abs and Th1‐like responses, along with type 2 diabetes, SARS‐CoV‐2 viremia, and Epstein–Barr virus viremia may anticipate PASC. 344 Interestingly, the study also showed a signature of atypical memory B cells which were likely the previously described T‐bet driven DN2 in SLE and severe COVID‐19. 81 , 118 Ultimately, a better understanding the longevity of autoreactive ASC after EF‐biased responses after acute COVID‐19 infection may provide insight into one immune mechanism of PASC.
8. CONCLUSIONS
After 2 years into the COVID‐19 pandemic, we are still witnessing an ongoing “arms race” between an everchanging SARS‐CoV‐2 virus and an evolving immunity induced by infection or vaccination. Much progress has been made in understanding the cellular origins of such responses yet many questions remain unanswered about the durability of long‐term protection. A better understanding of the balance between the EF and GC responses and the phenotype of ASC for the generation and maintenance of LLPC after infection and vaccination are still needed. The role of antigenic imprinting with each new emerging mutant will also be essential to develop a nimble vaccine strategy together with viral surveillance. Scientifically, the pandemic has proven itself to be an unprecedented opportunity to understand the immune response to primary viral infections. With the deep immunological insights of B cell and plasma cells, we will be prepared for the next one which is not a matter of if but when.
AUTHOR CONTRIBUTIONS
DCN, PAL, MCW, ASS, KEF, IS, and FEL wrote the manuscript.
FUNDING INFORMATION
This work was supported by National Institutes of Health grants: UL TR000424 (Emory Library IT), U54‐CA260563‐01 Emory SeroNet (IS, FEL), U19‐AI110483 Emory Autoimmunity Center of Excellence (IS), P01‐A1078907 (IS, FEL), P01‐AI125180‐01 (IS, FEL), R37‐AI049660 (IS), 2U19AI110483‐06 (IS), 1R01AI12125 (FEL), 1U01AI141993 (FEL), T32‐HL116271‐07 (PAL), and Bill & Melinda Gates Foundation Grant INV‐002351 (FEL).
CONFLICT OF INTEREST
Competing interests: FEL is the founder of Micro‐Bplex, Inc. FEL serves on the scientific advisory board of Be Biopharma, is a recipient of grants from the BMGF and Genentech, Inc. FEL has also served as a consultant for Astra Zeneca. IS has consulted for GSK, Pfizer, Kayverna, Johnson & Johnson, Celgene, Bristol Myer Squibb, and Visterra. The other authors declare no conflicts of interest.
Nguyen DC, Lamothe PA, Woodruff MC, et al. COVID‐19 and plasma cells: Is there long‐lived protection? Immunol Rev. 2022;309:40‐63. doi: 10.1111/imr.13115
*This article is part of a series of reviews covering SARS‐CoV‐2 Immunity appearing in Volume 309 of Immunological Reviews.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Jain U. Effect of COVID‐19 on the organs. Cureus. 2020;12(8):e9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Groff D, Sun A, Ssentongo AE, et al. Short‐term and Long‐term rates of Postacute sequelae of SARS‐CoV‐2 infection: a systematic review. JAMA Netw Open. 2021;4(10):e2128568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu X, Tsibane T, McGraw PA, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455(7212):532‐536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alejo JL, Mitchell J, Chang A, et al. Prevalence and durability of SARS‐CoV‐2 antibodies among unvaccinated US adults by history of COVID‐19. JAMA. 2022;327(11):1085‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mobaraki PD, Wang C, Floridi A, Floridi A, Zaidi AK. Long‐Term Persistence of IgG Antibodies in recovered COVID‐19 individuals at 18 months and the impact of two‐dose BNT162b2 (Pfizer‐BioNTech) mRNA vaccination on the antibody response. 2022. https://www.medrxiv.org/content/10.1101/2022.01.18.22269349v1. Accessed February 27, 2022. [DOI] [PMC free article] [PubMed]
- 6. Del Rio C, Malani PN, Omer SB. Confronting the delta variant of SARS‐CoV‐2, summer 2021. JAMA. 2021;326(11):1001‐1002. [DOI] [PubMed] [Google Scholar]
- 7. Del Rio C, Omer SB, Malani PN. Winter of omicron‐the evolving COVID‐19 pandemic. JAMA. 2022;327(4):319‐320. [DOI] [PubMed] [Google Scholar]
- 8. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid‐19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bates TA, Leier HC, Lyski ZL, et al. Neutralization of SARS‐CoV‐2 variants by convalescent and BNT162b2 vaccinated serum. Nat Commun. 2021;12(1):5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen RE, Zhang X, Case JB, et al. Resistance of SARS‐CoV‐2 variants to neutralization by monoclonal and serum‐derived polyclonal antibodies. Nat Med. 2021;27(4):717‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas SJ, Moreira ED Jr, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine through 6 months. N Engl J Med. 2021;385(19):1761‐1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tartof SY, Slezak JM, Fischer H, et al. Effectiveness of mRNA BNT162b2 COVID‐19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021;398(10309):1407‐1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levin EG, Lustig Y, Cohen C, et al. Waning immune humoral response to BNT162b2 Covid‐19 vaccine over 6 months. N Engl J Med. 2021;385(24):e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goldberg Y, Mandel M, Bar‐On Y, et al. Protection and waning of natural and hybrid COVID‐19 immunity. 2021. https://www.medrxiv.org/content/10.1101/2021.12.04.21267114v1. Accessed January 07, 2022. [DOI] [PMC free article] [PubMed]
- 15. Zuccarino‐Catania GV, Sadanand S, Weisel FJ, et al. CD80 and PD‐L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15(7):631‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanz I, Wei C, Jenks SA, et al. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Front Immunol. 2019;10:2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garcia M, Iglesias A, Landoni VI, et al. Massive plasmablast response elicited in the acute phase of hantavirus pulmonary syndrome. Immunology. 2017;151(1):122‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wrammert J, Onlamoon N, Akondy RS, et al. Rapid and massive virus‐specific plasmablast responses during acute dengue virus infection in humans. J Virol. 2012;86(6):2911‐2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varnaite R, Garcia M, Glans H, et al. Expansion of SARS‐CoV‐2‐specific antibody‐secreting cells and generation of neutralizing antibodies in hospitalized COVID‐19 patients. J Immunol. 2020;205(9):2437‐2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee FE, Falsey AR, Halliley JL, Sanz I, Walsh EE. Circulating antibody‐secreting cells during acute respiratory syncytial virus infection in adults. J Infect Dis. 2010;202(11):1659‐1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fink K. Origin and function of circulating Plasmablasts during acute viral infections. Front Immunol. 2012;3:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wrammert J, Smith K, Miller J, et al. Rapid cloning of high‐affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Halliley JL, Kyu S, Kobie JJ, et al. Peak frequencies of circulating human influenza‐specific antibody secreting cells correlate with serum antibody response after immunization. Vaccine. 2010;28(20):3582‐3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee FE, Halliley JL, Walsh EE, et al. Circulating human antibody‐secreting cells during vaccinations and respiratory viral infections are characterized by high specificity and lack of bystander effect. J Immunol. 2011;186(9):5514‐5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sze DM, Toellner KM, Garcia de Vinuesa C, Taylor DR, IC ML. Intrinsic constraint on plasmablast growth and extrinsic limits of plasma cell survival. J Exp Med. 2000;192(6):813‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammarlund E, Thomas A, Amanna IJ, et al. Plasma cell survival in the absence of B cell memory. Nat Commun. 2017;8(1):1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357(19):1903‐1915. [DOI] [PubMed] [Google Scholar]
- 28. Benner R, Hijmans W, Haaijman JJ. The bone marrow: the major source of serum immunoglobulins, but still a neglected site of antibody formation. Clin Exp Immunol. 1981;46(1):1‐8. [PMC free article] [PubMed] [Google Scholar]
- 29. Manz RA, Thiel A, Radbruch A. Lifetime of plasma cells in the bone marrow. Nature. 1997;388(6638):133‐134. [DOI] [PubMed] [Google Scholar]
- 30. Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long‐lived plasma cells. Immunity. 1998;8(3):363‐372. [DOI] [PubMed] [Google Scholar]
- 31. Weisel FJ, Zuccarino‐Catania GV, Chikina M, Shlomchik MJ. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity. 2016;44(1):116‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halliley JL, Tipton CM, Liesveld J, et al. Long‐lived plasma cells are contained within the CD19(−)CD38(hi)CD138(+) subset in human bone marrow. Immunity. 2015;43(1):132‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nguyen DC, Garimalla S, Xiao H, et al. Factors of the bone marrow microniche that support human plasma cell survival and immunoglobulin secretion. Nat Commun. 2018;9(1):3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joyner CJ, Ley AM, Nguyen DC, et al. Generation of human long‐lived plasma cells by developmentally regulated epigenetic imprinting. Life Sci Alliance. 2022;5(3):e202101285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakharkar M, Rappazzo CG, Wieland‐Alter WF, et al. Prolonged evolution of the human B cell response to SARS‐CoV‐2 infection. Sci Immunol. 2021;6(56):eabg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaebler C, Wang Z, JCC L, et al. Evolution of antibody immunity to SARS‐CoV‐2. Nature. 2021;591(7851):639‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science. 2021;371(6529):eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klein SL, Pekosz A, Park HS, et al. Sex, age, and hospitalization drive antibody responses in a COVID‐19 convalescent plasma donor population. J Clin Invest. 2020;130(11):6141‐6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science. 2020;370(6521):1227‐1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haddad NS, Nguyen DC, Kuruvilla ME, et al. One‐stop serum assay identifies COVID‐19 disease severity and vaccination responses. Immunohorizons. 2021;5(5):322‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodda LB, Netland J, Shehata L, et al. Functional SARS‐CoV‐2‐specific immune memory persists after mild COVID‐19. Cell. 2021;184(1):169‐183 e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Z, Muecksch F, Schaefer‐Babajew D, et al. Naturally enhanced neutralizing breadth against SARS‐CoV‐2 one year after infection. Nature. 2021;595(7867):426‐431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anand SP, Prevost J, Nayrac M, et al. Longitudinal analysis of humoral immunity against SARS‐CoV‐2 spike in convalescent individuals up to 8 months post‐symptom onset. bioRxiv. 2021. https://www.biorxiv.org/content/10.1101/2021.01.25.428097v1. Accessed January 07, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Egbert ER, Xiao S, Colantuoni E, et al. Durability of spike Immunoglobin G antibodies to SARS‐CoV‐2 among health care workers with prior infection. JAMA Netw Open. 2021;4(8):e2123256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turner JS, Kim W, Kalaidina E, et al. SARS‐CoV‐2 infection induces long‐lived bone marrow plasma cells in humans. Nature. 2021;595(7867):421‐425. [DOI] [PubMed] [Google Scholar]
- 46. Gallais F, Gantner P, Bruel T, et al. Evolution of antibody responses up to 13 months after SARS‐CoV‐2 infection and risk of reinfection. EBioMedicine. 2021;71:103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dehgani‐Mobaraki P, Zaidi AK, Yadav N, Floridi A, Floridi E. Longitudinal observation of antibody responses for 14 months after SARS‐CoV‐2 infection. Clin Immunol. 2021;230:108814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perreault J, Tremblay T, Fournier MJ, et al. Waning of SARS‐CoV‐2 RBD antibodies in longitudinal convalescent plasma samples within 4 months after symptom onset. Blood. 2020;136(22):2588‐2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marot S, Malet I, Leducq V, et al. Rapid decline of neutralizing antibodies against SARS‐CoV‐2 among infected healthcare workers. Nat Commun. 2021;12(1):844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roltgen K, Powell AE, Wirz OF, et al. Defining the features and duration of antibody responses to SARS‐CoV‐2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54):eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS‐CoV‐2 in Iceland. N Engl J Med. 2020;383(18):1724‐1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wheatley AK, Juno JA, Wang JJ, et al. Evolution of immune responses to SARS‐CoV‐2 in mild‐moderate COVID‐19. Nat Commun. 2021;12(1):1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haveri A, Ekstrom N, Solastie A, et al. Persistence of neutralizing antibodies a year after SARS‐CoV‐2 infection in humans. Eur J Immunol. 2021;51(12):3202‐3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He Z, Ren L, Yang J, et al. Seroprevalence and humoral immune durability of anti‐SARS‐CoV‐2 antibodies in Wuhan, China: a longitudinal, population‐level, cross‐sectional study. Lancet. 2021;397(10279):1075‐1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alfego D, Sullivan A, Poirier B, Williams J, Adcock D, Letovsky S. A population‐based analysis of the longevity of SARS‐CoV‐2 antibody seropositivity in the United States. EClinicalMedicine. 2021;36:100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lumley SF, Wei J, O'Donnell D, et al. The duration, dynamics, and determinants of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) antibody responses in individual healthcare workers. Clin Infect Dis. 2021;73(3):e699‐e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Krutikov M, Palmer T, Tut G, et al. Prevalence and duration of detectable SARS‐CoV‐2 nucleocapsid antibodies in staff and residents of long‐term care facilities over the first year of the pandemic (VIVALDI study): prospective cohort study in England. Lancet Healthy Longev. 2022;3(1):e13‐e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oved K, Olmer L, Shemer‐Avni Y, et al. Multi‐center nationwide comparison of seven serology assays reveals a SARS‐CoV‐2 non‐responding seronegative subpopulation. EClinicalMedicine. 2020;29:100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marklund E, Leach S, Axelsson H, et al. Serum‐IgG responses to SARS‐CoV‐2 after mild and severe COVID‐19 infection and analysis of IgG non‐responders. PLoS One. 2020;15(10):e0241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu W, Russell RM, Bibollet‐Ruche F, et al. Predictors of nonseroconversion after SARS‐CoV‐2 infection. Emerg Infect Dis. 2021;27(9):2454‐2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suthar MS, Zimmerman MG, Kauffman RC, et al. Rapid generation of neutralizing antibody responses in COVID‐19 patients. Cell Rep Med. 2020;1(3):100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beaudoin‐Bussieres G, Laumaea A, Anand SP, et al. Decline of humoral responses against SARS‐CoV‐2 spike in convalescent individuals. MBio. 2020;11(5):e02590‐e02520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ibarrondo FJ, Fulcher JA, Goodman‐Meza D, et al. Rapid decay of anti‐SARS‐CoV‐2 antibodies in persons with mild Covid‐19. N Engl J Med. 2020;383(11):1085‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu J, Liang B, Chen C, et al. SARS‐CoV‐2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID‐19. Nat Commun. 2021;12(1):1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Crawford KHD, Dingens AS, Eguia R, et al. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2021;223(2):197‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Markmann AJ, Giallourou N, Bhowmik DR, et al. Sex disparities and neutralizing‐antibody durability to SARS‐CoV‐2 infection in convalescent individuals. mSphere. 2021;6(4):e0027521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pilz S, Chakeri A, Ioannidis JP, et al. Allerberger F SARS‐CoV‐2 re‐infection risk in Austria. Eur J Clin Invest. 2021;51(4):e13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Townsend JP, Hassler HB, Wang Z, et al. The durability of immunity against reinfection by SARS‐CoV‐2: a comparative evolutionary study. Lancet Microbe. 2021;2(12):e666‐e675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Abu‐Raddad LJ, Chemaitelly H, Coyle P, et al. SARS‐CoV‐2 antibody‐positivity protects against reinfection for at least seven months with 95% efficacy. EClinicalMedicine. 2021;35:100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gazit S, Shlezinger R, Perez G, et al. Comparing SARS‐CoV‐2 natural immunity to vaccine‐induced immunity: reinfections versus breakthrough infections. 2021. https://www.medrxiv.org/content/10.1101/2021.08.24.21262415v1. Accessed January 25, 2022. [DOI] [PMC free article] [PubMed]
- 71. Isho B, Abe KT, Zuo M, et al. Persistence of serum and saliva antibody responses to SARS‐CoV‐2 spike antigens in COVID‐19 patients. Sci Immunol. 2020;5(52):eabe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS‐CoV‐2 spike protein in COVID‐19 patients. Sci Immunol. 2020;5(52):eabe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ripperger TJ, Uhrlaub JL, Watanabe M, et al. Orthogonal SARS‐CoV‐2 serological assays enable surveillance of low‐prevalence communities and reveal durable humoral immunity. Immunity. 2020;53(5):925‐933 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS‐CoV‐2 infection. Nat Med. 2021;27(7):1205‐1211. [DOI] [PubMed] [Google Scholar]
- 75. Irsara C, Egger AE, Prokop W, et al. Clinical validation of the Siemens quantitative SARS‐CoV‐2 spike IgG assay (sCOVG) reveals improved sensitivity and a good correlation with virus neutralization titers. Clin Chem Lab Med. 2021;59(8):1453‐1462. [DOI] [PubMed] [Google Scholar]
- 76. Brochot E, Demey B, Touze A, et al. Anti‐spike, anti‐nucleocapsid and neutralizing antibodies in SARS‐CoV‐2 inpatients and asymptomatic individuals. Front Microbiol. 2020;11:584251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020;26(6):845‐848. [DOI] [PubMed] [Google Scholar]
- 78. Ogega CO, Skinner NE, Blair PW, et al. Durable SARS‐CoV‐2 B cell immunity after mild or severe disease. J Clin Invest. 2021;131(7):e145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat Med. 2020;26(8):1200‐1204. [DOI] [PubMed] [Google Scholar]
- 80. Peluso MJ, Takahashi S, Hakim J, et al. SARS‐CoV‐2 antibody magnitude and detectability are driven by disease severity, timing, and assay. Sci Adv. 2021;7(31):eabh3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Woodruff MC, Ramonell RP, Nguyen DC, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID‐19. Nat Immunol. 2020;21(12):1506‐1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Woodruff MC, Ramonell RP, Saini AS, et al. Relaxed peripheral tolerance drives broad de novo autoreactivity in severe COVID‐19. medRxiv. 2021. https://www.medrxiv.org/content/10.1101/2020.10.21.20216192v3. Accessed January 15, 2022. [Google Scholar]
- 83. Seydoux E, Homad LJ, MacCamy AJ, et al. Analysis of a SARS‐CoV‐2‐infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53(1):98‐105 e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaneko N, Kuo HH, Boucau J, et al. Loss of Bcl‐6‐expressing T follicular helper cells and germinal centers in COVID‐19. Cell. 2020;183(1):143‐157 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Duan YQ, Xia MH, Ren L, et al. Deficiency of Tfh cells and germinal center in deceased COVID‐19 patients. Curr Med Sci. 2020;40(4):618‐624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kuri‐Cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID‐19. Sci Immunol. 2020;5(49):eabd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kreer C, Zehner M, Weber T, et al. Longitudinal isolation of potent near‐germline SARS‐CoV‐2‐neutralizing antibodies from COVID‐19 patients. Cell. 2020;182(4):843‐854 e812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Juno JA, Tan HX, Lee WS, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID‐19. Nat Med. 2020;26(9):1428‐1434. [DOI] [PubMed] [Google Scholar]
- 89. Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS‐CoV‐2. Nature 2020;584(7821):443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Adamo S, Chevrier S, Cervia C, et al. Profound dysregulation of T cell homeostasis and function in patients with severe COVID‐19. Allergy. 2021;76(9):2866‐2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang J, Wu Q, Liu Z, et al. Spike‐specific circulating T follicular helper cell and cross‐neutralizing antibody responses in COVID‐19‐convalescent individuals. Nat Microbiol. 2021;6(1):51‐58. [DOI] [PubMed] [Google Scholar]
- 92. Fenoglio D, Dentone C, Parodi A, et al. Characterization of T lymphocytes in severe COVID‐19 patients. J Med Virol. 2021;93(9):5608‐5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Seow J, Graham C, Merrick B, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS‐CoV‐2 infection in humans. Nat Microbiol. 2020;5(12):1598‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS‐CoV‐2: a case study. Lancet Infect Dis. 2021;21(1):52‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sterlin D, Mathian A, Miyara M, et al. IgA dominates the early neutralizing antibody response to SARS‐CoV‐2. Sci Transl Med. 2021;13(577):eabd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Li GM, Chiu C, Wrammert J, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross‐reactive memory B cells. Proc Natl Acad Sci USA. 2012;109(23):9047‐9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Garimalla S, Nguyen DC, Halliley JL, et al. Differential transcriptome and development of human peripheral plasma cell subsets. JCI Insight. 2019;4(9):e126732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Garcia‐Bates TM, Cordeiro MT, Nascimento EJ, et al. Association between magnitude of the virus‐specific plasmablast response and disease severity in dengue patients. J Immunol. 2013;190(1):80‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bernardes JP, Mishra N, Tran F, et al. Longitudinal multi‐omics analyses identify responses of megakaryocytes, erythroid cells, and Plasmablasts as hallmarks of severe COVID‐19. Immunity. 2020;53(6):1296‐1314 e1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Boulanger M, Molina E, Wang K, Kickler T, Xu Y, Garibaldi BT. Peripheral plasma cells associated with mortality benefit in severe COVID‐19: a marker of disease resolution. Am J Med. 2021;134(8):1029‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620‐2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jego G, Bataille R, Pellat‐Deceunynck C. Interleukin‐6 is a growth factor for nonmalignant human plasmablasts. Blood. 2001;97(6):1817‐1822. [DOI] [PubMed] [Google Scholar]
- 103. Nguyen DC, Joyner CJ, Sanz I, Lee FE. Factors affecting early antibody secreting cell maturation into Long‐lived plasma cells. Front Immunol. 2019;10:2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cervia C, Nilsson J, Zurbuchen Y, et al. Systemic and mucosal antibody responses specific to SARS‐CoV‐2 during mild versus severe COVID‐19. J Allergy Clin Immunol. 2021;147(2):545‐557 e549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang Z, Lorenzi JCC, Muecksch F, et al. Enhanced SARS‐CoV‐2 neutralization by dimeric IgA. Sci Transl Med. 2021;13(577):eabf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Byazrova M, Yusubalieva G, Spiridonova A, et al. Pattern of circulating SARS‐CoV‐2‐specific antibody‐secreting and memory B‐cell generation in patients with acute COVID‐19. Clin Transl Immunology. 2021;10(2):e1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sokal A, Chappert P, Barba‐Spaeth G, et al. Maturation and persistence of the anti‐SARS‐CoV‐2 memory B cell response. Cell. 2021;184(5):1201‐1213 e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Winklmeier S, Eisenhut K, Taskin D, et al. Persistence of functional memory B cells recognizing SARS‐CoV‐2 variants despite loss of specific IgG. iScience. 2022;25(1):103659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hartley GE, Edwards ESJ, Aui PM, et al. Rapid generation of durable B cell memory to SARS‐CoV‐2 spike and nucleocapsid proteins in COVID‐19 and convalescence. Sci Immunol. 2020;5(54):eabf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Nielsen SCA, Yang F, Jackson KJL, et al. Human B cell clonal expansion and convergent antibody responses to SARS‐CoV‐2. Cell Host Microbe. 2020;28(4):516‐525 e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cho A, Muecksch F, Schaefer‐Babajew D, et al. Anti‐SARS‐CoV‐2 receptor‐binding domain antibody evolution after mRNA vaccination. Nature. 2021;600(7889):517‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hoehn KB, Ramanathan P, Unterman A, et al. Cutting edge: distinct B cell repertoires characterize patients with mild and severe COVID‐19. J Immunol. 2021;206(12):2785‐2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pape KA, Dileepan T, Kabage AJ, et al. High‐affinity memory B cells induced by SARS‐CoV‐2 infection produce more plasmablasts and atypical memory B cells than those primed by mRNA vaccines. Cell Rep. 2021;37(2):109823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nguyen DC, Duan M, Ali M, Ley A, Sanz I, Lee FE. Plasma cell survival: the intrinsic drivers, migratory signals, and extrinsic regulators. Immunol Rev. 2021;303(1):138‐153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wen W, Su W, Tang H, et al. Immune cell profiling of COVID‐19 patients in the recovery stage by single‐cell sequencing. Cell Discov. 2020;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhu L, Yang P, Zhao Y, et al. Single‐cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID‐19 and influenza patients. Immunity. 2020;53(3):685‐696 e683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fan X, Chi X, Ma W, et al. Single‐cell RNA‐seq and V (D) J profiling of immune cells in COVID‐19 patients. MedRxiv. 2020. https://www.medrxiv.org/content/10.1101/2020.05.24.20101238v1. Accessed 01/28/2022. [Google Scholar]
- 118. Jenks SA, Cashman KS, Zumaquero E, et al. Distinct effector B cells induced by unregulated toll‐like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity. 2018;49(4):725‐739 e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Qi F, Zhang W, Huang J, Fu L, Zhao J. Single‐cell RNA sequencing analysis of the Immunometabolic rewiring and Immunopathogenesis of coronavirus disease 2019. Front Immunol. 2021;12:651656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shapiro‐Shelef M, Calame K. Regulation of plasma‐cell development. Nat Rev Immunol. 2005;5(3):230‐242. [DOI] [PubMed] [Google Scholar]
- 121. Price MJ, Scharer CD, Kania AK, Randall TD, Boss JM. Conserved epigenetic programming and enhanced Heme metabolism drive memory B cell reactivation. J Immunol. 2021;206(7):1493‐1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Niu L, Wittrock KN, Clabaugh GC, Srivastava V, Cho MW. A structural landscape of neutralizing antibodies against SARS‐CoV‐2 receptor binding domain. Front Immunol. 2021;12:647934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Piccoli L, Park YJ, Tortorici MA, et al. Mapping neutralizing and Immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell. 2020;183(4):1024‐1042 e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Boonyaratanakornkit J, Morishima C, Selke S, et al. Clinical, laboratory, and temporal predictors of neutralizing antibodies against SARS‐CoV‐2 among COVID‐19 convalescent plasma donor candidates. J Clin Invest. 2021;131(3):e144930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Salazar E, Kuchipudi SV, Christensen PA, et al. Convalescent plasma anti‐SARS‐CoV‐2 spike protein ectodomain and receptor‐binding domain IgG correlate with virus neutralization. J Clin Invest. 2020;130(12):6728‐6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Garcia‐Beltran WF, Lam EC, Astudillo MG, et al. COVID‐19‐neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476‐488. e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Voss C, Esmail S, Liu X, et al. Epitope‐specific antibody responses differentiate COVID‐19 outcomes and variants of concern. JCI Insight. 2021;6(13):e148855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Amanat F, Thapa M, Lei T, et al. SARS‐CoV‐2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. 2021;184(15):3936‐3948 e3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Arvin AM, Fink K, Schmid MA, et al. A perspective on potential antibody‐dependent enhancement of SARS‐CoV‐2. Nature. 2020;584(7821):353‐363. [DOI] [PubMed] [Google Scholar]
- 130. Cloutier M, Nandi M, Ihsan AU, Chamard HA, Ilangumaran S, Ramanathan S. ADE and hyperinflammation in SARS‐CoV2 infection‐ comparison with dengue hemorrhagic fever and feline infectious peritonitis. Cytokine. 2020;136:155256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Fleming AB, Raabe V. Current studies of convalescent plasma therapy for COVID‐19 may underestimate risk of antibody‐dependent enhancement. J Clin Virol. 2020;127:104388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hotez PJ, Corry DB, Bottazzi ME. COVID‐19 vaccine design: the Janus face of immune enhancement. Nat Rev Immunol. 2020;20(6):347‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jiang S. Don't rush to deploy COVID‐19 vaccines and drugs without sufficient safety guarantees. Nature. 2020;579:321. [DOI] [PubMed] [Google Scholar]
- 134. Sanchez‐Zuno GA, Matuz‐Flores MG, Gonzalez‐Estevez G, et al. A review: antibody‐dependent enhancement in COVID‐19: the not so friendly side of antibodies. Int J Immunopathol Pharmacol. 2021;35:20587384211050199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wen J, Cheng Y, Ling R, et al. Antibody‐dependent enhancement of coronavirus. Int J Infect Dis. 2020;100:483‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Corapi WV, Olsen CW, Scott FW. Monoclonal antibody analysis of neutralization and antibody‐dependent enhancement of feline infectious peritonitis virus. J Virol. 1992;66(11):6695‐6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Jaume M, Yip MS, Cheung CY, et al. Anti‐severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH‐ and cysteine protease‐independent FcgammaR pathway. J Virol. 2011;85(20):10582‐10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kam YW, Kien F, Roberts A, et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS‐CoV challenge despite their capacity to mediate FcgammaRII‐dependent entry into B cells in vitro. Vaccine. 2007;25(4):729‐740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Luo F, Liao FL, Wang H, Tang HB, Yang ZQ, Hou W. Evaluation of antibody‐dependent enhancement of SARS‐CoV infection in rhesus macaques immunized with an inactivated SARS‐CoV vaccine. Virol Sin. 2018;33(2):201‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yang ZY, Werner HC, Kong WP, et al. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA. 2005;102(3):797‐801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yip MS, Leung NHL, Cheung CY, et al. Antibody‐dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Yip MS, Leung HL, Li PH, et al. Antibody‐dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J. 2016;22(3):25‐31. [PubMed] [Google Scholar]
- 143. Wan Y, Shang J, Sun S, et al. Molecular mechanism for antibody‐dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liu L, Wei Q, Lin Q, et al. Anti‐spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS‐CoV infection. JCI Insight. 2019;4(4):e123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Li D, Edwards RJ, Manne K, et al. In vitro and in vivo functions of SARS‐CoV‐2 infection‐enhancing and neutralizing antibodies. Cell. 2021;184(16):4203‐4219 e4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhou Y, Liu Z, Li S, et al. Enhancement versus neutralization by SARS‐CoV‐2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021;34(5):108699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chu H, Chan JF, Wang Y, et al. Comparative replication and immune activation profiles of SARS‐CoV‐2 and SARS‐CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID‐19. Clin Infect Dis. 2020;71(6):1400‐1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Grant RA, Morales‐Nebreda L, Markov NS, et al. Circuits between infected macrophages and T cells in SARS‐CoV‐2 pneumonia. Nature. 2021;590(7847):635‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bost P, Giladi A, Liu Y, et al. Host‐viral infection maps reveal signatures of severe COVID‐19 patients. Cell. 2020;181(7):1475‐1488 e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Knoll R, Schultze JL, Schulte‐Schrepping J. Monocytes and macrophages in COVID‐19. Front Immunol. 2021;12:720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lu Q, Liu J, Zhao S, et al. SARS‐CoV‐2 exacerbates proinflammatory responses in myeloid cells through C‐type lectin receptors and Tweety family member 2. Immunity. 2021;54(6):1304‐1319 e1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Bahnan W, Wrighton S, Sundwall M, et al. Spike‐dependent Opsonization indicates both dose‐dependent inhibition of phagocytosis and that non‐neutralizing antibodies can confer protection to SARS‐CoV‐2. Front Immunol. 2021;12:808932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Beaudoin‐Bussieres G, Chen Y, Ullah I, et al. A fc‐enhanced NTD‐binding non‐neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS‐CoV‐2 infection. Cell Rep. 2022;38:110368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Schafer A, Muecksch F, Lorenzi JCC, et al. Antibody potency, effector function, and combinations in protection and therapy for SARS‐CoV‐2 infection in vivo. J Exp Med. 2021;218(3):e20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ullah I, Prevost J, Ladinsky MS, et al. Live imaging of SARS‐CoV‐2 infection in mice reveals that neutralizing antibodies require fc function for optimal efficacy. Immunity. 2021;54(9):2143‐2158 e2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yu J, Tostanoski Lisa H, Peter L, et al. DNA vaccine protection against SARS‐CoV‐2 in rhesus macaques. Science. 2020;369(6505):806‐811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Gorman MJ, Patel N, Guebre‐Xabier M, et al. Fab and fc contribute to maximal protection against SARS‐CoV‐2 following NVX‐CoV2373 subunit vaccine with matrix‐M vaccination. Cell Rep Med. 2021;2(9):100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Chakraborty S, Gonzalez J, Edwards K, et al. Proinflammatory IgG fc structures in patients with severe COVID‐19. Nat Immunol. 2021;22(1):67‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Chakraborty S, Gonzalez JC, Sievers BL, et al. Early non‐neutralizing, afucosylated antibody responses are associated with COVID‐19 severity. Sci Transl Med. 2022;14:eabm7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tso FY, Lidenge SJ, Poppe LK, et al. Presence of antibody‐dependent cellular cytotoxicity (ADCC) against SARS‐CoV‐2 in COVID‐19 plasma. PloS One. 2021;16(3):e0247640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Chen X, Rostad CA, Anderson LJ, et al. The development and kinetics of functional antibody‐dependent cell‐mediated cytotoxicity (ADCC) to SARS‐CoV‐2 spike protein. Virology. 2021;559:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Adeniji Opeyemi S, Giron Leila B, Purwar M, et al. COVID‐19 severity is associated with differential antibody fc‐mediated innate immune functions. MBio. 2021;12(2):e00281‐00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Farkash I, Feferman T, Cohen‐Saban N, et al. Anti‐SARS‐CoV‐2 antibodies elicited by COVID‐19 mRNA vaccine exhibit a unique glycosylation pattern. Cell Rep. 2021;37(11):110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zohar T, Loos C, Fischinger S, et al. Compromised humoral functional evolution tracks with SARS‐CoV‐2 mortality. Cell. 2020;183(6):1508‐1519 e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kobayashi K, Tachibana M, Tsutsumi Y. Neglected roles of IgG fc‐binding protein secreted from airway mucin‐producing cells in protecting against SARS‐CoV‐2 infection. Innate Immun. 2021;27(6):423‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Teijaro JR, Farber DL. COVID‐19 vaccines: modes of immune activation and future challenges. Nat Rev Immunol. 2021;21(4):195‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383(27):2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Haas EJ, Angulo FJ, McLaughlin JM, et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS‐CoV‐2 infections and COVID‐19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397(10287):1819‐1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Angel Y, Spitzer A, Henig O, et al. Association between vaccination with BNT162b2 and incidence of symptomatic and asymptomatic SARS‐CoV‐2 infections among health care workers. JAMA. 2021;325(24):2457‐2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid‐19 vaccine in a Nationwide mass vaccination setting. N Engl J Med. 2021;384(15):1412‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Thompson MG, Burgess JL, Naleway AL, et al. Prevention and attenuation of Covid‐19 with the BNT162b2 and mRNA‐1273 vaccines. N Engl J Med. 2021;385(4):320‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Rosenberg ES, Holtgrave DR, Dorabawila V, et al. New COVID‐19 cases and hospitalizations among adults, by vaccination status ‐ New York, may 3‐July 25, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(37):1306‐1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Rosenberg ES, Dorabawila V, Easton D, et al. Covid‐19 vaccine effectiveness in New York state. N Engl J Med. 2022;386(2):116‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single‐dose Ad26.COV2.S vaccine against Covid‐19. N Engl J Med. 2021;384(23):2187‐2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Milman O, Yelin I, Aharony N, et al. Community‐level evidence for SARS‐CoV‐2 vaccine protection of unvaccinated individuals. Nat Med. 2021;27(8):1367‐1369. [DOI] [PubMed] [Google Scholar]
- 177. Piano Mortari E, Russo C, Vinci MR, et al. Highly specific memory B cells generation after the 2nd dose of BNT162b2 vaccine compensate for the decline of serum antibodies and absence of mucosal IgA. Cell. 2021;10(10):2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Azzi L, Dalla Gasperina D, Veronesi G, et al. Mucosal immune response in BNT162b2 COVID‐19 vaccine recipients. EBioMedicine. 2022;75:103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Widge AT, Rouphael NG, Jackson LA, et al. Durability of responses after SARS‐CoV‐2 mRNA‐1273 vaccination. N Engl J Med. 2021;384(1):80‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Pegu A, O'Connell SE, Schmidt SD, et al. Durability of mRNA‐1273 vaccine‐induced antibodies against SARS‐CoV‐2 variants. Science. 2021;373(6561):1372‐1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature. 2021;592(7855):616‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Wisnewski AV, Campillo Luna J, Redlich CA. Human IgG and IgA responses to COVID‐19 mRNA vaccines. PLoS One. 2021;16(6):e0249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Turner JS, O'Halloran JA, Kalaidina E, et al. SARS‐CoV‐2 mRNA vaccines induce persistent human germinal Centre responses. Nature. 2021;596(7870):109‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Walsh EE, Frenck RW Jr, Falsey AR, et al. Safety and immunogenicity of two RNA‐based Covid‐19 vaccine candidates. N Engl J Med. 2020;383(25):2439‐2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS‐CoV‐2 ‐ preliminary report. N Engl J Med. 2020;383(20):1920‐1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Israel A, Shenhar Y, Green I, et al. Large‐scale study of antibody titer decay following BNT162b2 mRNA vaccine or SARS‐CoV‐2 infection. medRxiv. 2021. https://www.medrxiv.org/content/10.1101/2021.08.19.21262111v1. Accessed January 15, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Krammer F, Srivastava K, Alshammary H, et al. Antibody responses in seropositive persons after a single dose of SARS‐CoV‐2 mRNA vaccine. N Engl J Med. 2021;384(14):1372‐1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Saadat S, Rikhtegaran Tehrani Z, Logue J, et al. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS‐CoV‐2. JAMA. 2021;325(14):1467‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Goel RR, Apostolidis SA, Painter MM, et al. Distinct antibody and memory B cell responses in SARS‐CoV‐2 naive and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6(58):eabi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Collier AY, Yu J, McMahan K, et al. Differential kinetics of immune responses elicited by Covid‐19 vaccines. N Engl J Med. 2021;385(21):2010‐2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. van Gils MJ, Ayesha Lavall AH, van der Straten K, et al. Four SARS‐CoV‐2 vaccines induce quantitatively different antibody responses against SARS‐CoV‐2 variants. 2021. https://www.medrxiv.org/content/10.1101/2021.09.27.21264163v1. Accessed January 25, 2022.
- 192. Barouch DH, Stephenson KE, Sadoff J, et al. Durable humoral and cellular immune responses 8 months after Ad26.COV2.S vaccination. N Engl J Med. 2021;385(10):951‐953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Earle KA, Ambrosino DM, Fiore‐Gartland A, et al. Evidence for antibody as a protective correlate for COVID‐19 vaccines. Vaccine. 2021;39(32):4423‐4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Lustig Y, Sapir E, Regev‐Yochay G, et al. BNT162b2 COVID‐19 vaccine and correlates of humoral immune responses and dynamics: a prospective, single‐Centre, longitudinal cohort study in health‐care workers. Lancet Respir Med. 2021;9(9):999‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA‐1273 COVID‐19 vaccine efficacy clinical trial. Science. 2022;375(6576):43‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Frenck RW Jr, Klein NP, Kitchin N, et al. Safety, immunogenicity, and efficacy of the BNT162b2 Covid‐19 vaccine in adolescents. N Engl J Med. 2021;385(3):239‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Goldberg Y, Mandel M, Bar‐On YM, et al. Waning immunity after the BNT162b2 vaccine in Israel. N Engl J Med. 2021;385(24):e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Wall EC, Wu M, Harvey R, et al. Neutralising antibody activity against SARS‐CoV‐2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397(10292):2331‐2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Naaber P, Tserel L, Kangro K, et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet Reg Health Eur. 2021;10:100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Thakkar A, Gonzalez‐Lugo JD, Goradia N, et al. Seroconversion rates following COVID‐19 vaccination among patients with cancer. Cancer Cell. 2021;39(8):1081‐1090 e1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Canaday DH, Carias L, Oyebanji OA, et al. Reduced BNT162b2 messenger RNA vaccine response in severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐naive nursing home residents. Clin Infect Dis. 2021;73(11):2112‐2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Muller L, Andree M, Moskorz W, et al. Age‐dependent immune response to the Biontech/Pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin Infect Dis. 2021;73(11):2065‐2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Abu Jabal K, Ben‐Amram H, Beiruti K, et al. Impact of age, ethnicity, sex and prior infection status on immunogenicity following a single dose of the BNT162b2 mRNA COVID‐19 vaccine: real‐world evidence from healthcare workers, Israel, December 2020 to January 2021. Euro Surveill. 2021;26(6):2100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Doria‐Rose N, Suthar MS, Makowski M, et al. Antibody persistence through 6 months after the second dose of mRNA‐1273 vaccine for Covid‐19. N Engl J Med. 2021;384(23):2259‐2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Mor O, Zuckerman NS, Hazan I, et al. BNT162b2 vaccine effectiveness was marginally affected by the SARS‐CoV‐2 beta variant in fully vaccinated individuals. J Clin Epidemiol. 2021;142:38‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Abu‐Raddad LJ, Chemaitelly H, Butt AA, National Study Group for COVID‐19 Vaccination . Effectiveness of the BNT162b2 Covid‐19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med. 2021;385(2):187‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Pilishvili T, Gierke R, Fleming‐Dutra KE, et al. Effectiveness of mRNA Covid‐19 vaccine among U.S. health care personnel. N Engl J Med. 2021;385(25):e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Brown CM, Vostok J, Johnson H, et al. Outbreak of SARS‐CoV‐2 infections, including COVID‐19 vaccine breakthrough infections, associated with large public gatherings ‐ Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2021;70(31):1059‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Bergwerk M, Gonen T, Lustig Y, et al. Covid‐19 breakthrough infections in vaccinated health care workers. N Engl J Med. 2021;385(16):1474‐1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Levine‐Tiefenbrun M, Yelin I, Alapi H, et al. Viral loads of Delta‐variant SARS‐CoV‐2 breakthrough infections after vaccination and booster with BNT162b2. Nat Med. 2021;27(12):2108‐2110. [DOI] [PubMed] [Google Scholar]
- 211. Chia PY, Ong SWX, Chiew CJ, et al. Virological and serological kinetics of SARS‐CoV‐2 Delta variant vaccine breakthrough infections: a multicentre cohort study. Clin Microbiol Infect. 2021;28(4):612.e1‐612.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Hetemaki I, Kaariainen S, Alho P, et al. An outbreak caused by the SARS‐CoV‐2 Delta variant (B.1.617.2) in a secondary care hospital in Finland, May 2021. Euro Surveill. 2021;26(30):2100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Dougherty K, Mannell M, Naqvi O, Matson D, Stone J. SARS‐CoV‐2 B.1.617.2 (Delta) variant COVID‐19 outbreak associated with a gymnastics facility ‐ Oklahoma, April‐may 2021. MMWR Morb Mortal Wkly Rep. 2021;70(28):1004‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Shitrit P, Zuckerman NS, Mor O, Gottesman BS, Chowers M. Nosocomial outbreak caused by the SARS‐CoV‐2 Delta variant in a highly vaccinated population, Israel, July 2021. Euro Surveill. 2021;26(39):2100822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Puranik A, Lenehan PJ, Silvert E, et al. Comparison of two highly‐effective mRNA vaccines for COVID‐19 during periods of alpha and Delta variant prevalence. medRxiv. 2021. https://www.medrxiv.org/content/10.1101/2021.08.06.21261707v2. Accessed February 21, 2022. [Google Scholar]
- 216. Sheikh A, McMenamin J, Taylor B, Robertson C, Public health S, the EIIC . SARS‐CoV‐2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397(10293):2461‐2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Pouwels KB, Pritchard E, Matthews PC, et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS‐CoV‐2 infections in the UK. Nat Med. 2021;27(12):2127‐2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Tenforde MW, Self WH, Naioti EA, et al. Sustained effectiveness of Pfizer‐BioNTech and Moderna vaccines against COVID‐19 associated hospitalizations among adults ‐ United States, march‐July 2021. MMWR Morb Mortal Wkly Rep. 2021;70(34):1156‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Fowlkes A, Gaglani M, Groover K, et al. Effectiveness of COVID‐19 vaccines in preventing SARS‐CoV‐2 infection among frontline workers before and during B.1.617.2 (Delta) variant predominance ‐ eight U.S. locations, December 2020‐august 2021. MMWR Morb Mortal Wkly Rep. 2021;70(34):1167‐1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Tang P, Hasan MR, Chemaitelly H, et al. BNT162b2 and mRNA‐1273 COVID‐19 vaccine effectiveness against the SARS‐CoV‐2 Delta variant in Qatar. Nat Med. 2021;27(12):2136‐2143. [DOI] [PubMed] [Google Scholar]
- 221. León MT, Dorabawila V, Nelson L, et al. COVID‐19 Cases and Hospitalizations by COVID‐19 Vaccination Status and Previous COVID‐19 Diagnosis — California and New York, May–November 2021. Morbi Mortal Wkly Rep. 2022;71(4):125‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. NVV C, Ngoc NM, Nguyet LA, et al. An observational study of breakthrough SARS‐CoV‐2 Delta variant infections among vaccinated healthcare workers in Vietnam. EClinicalMedicine. 2021;41:101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Antonelli M, Penfold RS, Merino J, et al. Risk factors and disease profile of post‐vaccination SARS‐CoV‐2 infection in UKusers of the COVID symptom study app: a prospective, community‐based, nested, case‐control study. Lancet Infect Dis. 2022;22(1):43‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Grannis SJ, Rowley EA, Ong TC, et al. Interim estimates of COVID‐19 vaccine effectiveness against COVID‐19‐associated emergency department or urgent care clinic encounters and hospitalizations among adults during SARS‐CoV‐2 B.1.617.2 (Delta) variant predominance ‐ nine states, June‐august 2021. MMWR Morb Mortal Wkly Rep. 2021;70(37):1291‐1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell. 2020;183(4):996‐1012 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Goel RR, Painter MM, Apostolidis SA, et al. mRNA vaccines induce durable immune memory to SARS‐CoV‐2 and variants of concern. Science. 2021;374(6572):abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Geers D, Shamier MC, Bogers S, et al. SARS‐CoV‐2 variants of concern partially escape humoral but not T‐cell responses in COVID‐19 convalescent donors and vaccinees. Sci Immunol. 2021;6(59):eabj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Tarke A, Sidney J, Methot N, et al. Impact of SARS‐CoV‐2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2(7):100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS‐CoV‐2 in rhesus macaques. Nature. 2021;590(7847):630‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Zuo J, Dowell AC, Pearce H, et al. Robust SARS‐CoV‐2‐specific T cell immunity is maintained at 6 months following primary infection. Nat Immunol. 2021;22(5):620‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Cromer D, Steain M, Reynaldi A, et al. Neutralising antibody titres as predictors of protection against SARS‐CoV‐2 variants and the impact of boosting: a meta‐analysis. Lancet Microbe. 2022;3(1):e52‐e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Garcia‐Beltran WF, Lam EC, St Denis K, et al. Multiple SARS‐CoV‐2 variants escape neutralization by vaccine‐induced humoral immunity. Cell. 2021;184(9):2372‐2383 e2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Pradenas E, Trinite B, Urrea V, et al. Clinical course impacts early kinetics,magnitude, and amplitude of SARS‐CoV‐2 neutralizing antibodies beyond 1 year after infection. Cell Rep Med. 2022;3(2):100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Hacisuleyman E, Hale C, Saito Y, et al. Vaccine breakthrough infections with SARS‐CoV‐2 variants. N Engl J Med. 2021;384(23):2212‐2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Lumley SF, O'Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS‐CoV‐2 infection in health care workers. N Engl J Med. 2021;384(6):533‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Mannar D, Saville JW, Zhu X, et al. SARS‐CoV‐2 omicron variant: antibody evasion and cryo‐EM structure of spike protein‐ACE2 complex. Science. 2022;375:eabn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. He X, Hong W, Pan X, Lu G, Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm (2020). 2021;2:838‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Accorsi EK, Britton A, Fleming‐Dutra KE, et al. Association between 3 doses of mRNA COVID‐19 vaccine and symptomatic infection caused by the SARS‐CoV‐2 omicron and Delta variants. JAMA. 2022;327:639‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Wald A. Booster vaccination to reduce SARS‐CoV‐2 transmission and infection. JAMA. 2022;327(4):327‐328. [DOI] [PubMed] [Google Scholar]
- 240. Collie S, Champion J, Moultrie H, Bekker LG, Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N Engl J Med. 2021;386(5):494‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature. 2021;602:657‐663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Spitzer A, Angel Y, Marudi O, et al. Association of a third dose of BNT162b2 vaccine with incidence of SARS‐CoV‐2 infection among health Care Workers in Israel. JAMA. 2022;327:341‐349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Bar‐On YM, Goldberg Y, Mandel M, et al. Protection of BNT162b2 vaccine booster against Covid‐19 in Israel. N Engl J Med. 2021;385(15):1393‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Arbel R, Hammerman A, Sergienko R, et al. BNT162b2 vaccine booster and mortality due to Covid‐19. N Engl J Med. 2021;385(26):2413‐2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Barda N, Dagan N, Cohen C, et al. Effectiveness of a third dose of the BNT162b2 mRNA COVID‐19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet. 2021;398(10316):2093‐2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Hall VG, Ferreira VH, Ku T, et al. Randomized trial of a third dose of mRNA‐1273 vaccine in transplant recipients. N Engl J Med. 2021;385(13):1244‐1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Choi A, Koch M, Wu K, et al. Safety and immunogenicity of SARS‐CoV‐2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med. 2021;27(11):2025‐2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Atmar RL, Lyke KE, Deming ME, et al. Homologous and heterologous Covid‐19 booster vaccinations. N Engl J Med. 2022;386:1046‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Falsey AR, Frenck RW Jr, Walsh EE, et al. SARS‐CoV‐2 neutralization with BNT162b2 vaccine dose 3. N Engl J Med. 2021;385(17):1627‐1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Garcia‐Beltran WF, St Denis KJ, Hoelzemer A, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 omicron variant. Cell. 2022;185:457‐466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Dejnirattisai W, Huo J, Zhou D, et al. SARS‐CoV‐2 omicron‐B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022;185(3):467–484 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Cele S, Gazy I, Jackson L, et al. Escape of SARS‐CoV‐2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593(7857):142‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Carreno JM, Alshammary H, Tcheou J, et al. Activity of convalescent and vaccine serum against SARS‐CoV‐2 omicron. Nature. 2021;602(7898):682‐688. [DOI] [PubMed] [Google Scholar]
- 254. Kim W, Zhou JQ, Horvath SC, et al. Germinal Centre‐driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Hammarlund E, Thomas A, Poore EA, et al. Durability of vaccine‐induced immunity against tetanus and diphtheria toxins: a cross‐sectional analysis. Clin Infect Dis. 2016;62(9):1111‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Amanna IJ, Hammarlund E, Lewis MW, Slifka MK. Impact of infection or vaccination on pre‐existing serological memory. Hum Immunol. 2012;73(11):1082‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Slifka MK, Amanna IJ. Role of Multivalency and antigenic threshold in generating protective antibody responses. Front Immunol. 2019;10:956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Sings HL. Pneumococcal conjugate vaccine use in adults ‐ addressing an unmet medical need for non‐bacteremic pneumococcal pneumonia. Vaccine. 2017;35(40):5406‐5417. [DOI] [PubMed] [Google Scholar]
- 259. Quast I, Tarlinton D. B cell memory: understanding COVID‐19. Immunity. 2021;54(2):205‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Giannotta G, Giannotta N. mRNA COVID‐19 vaccines and Long‐lived plasma cells: a complicated relationship. Vaccines (Basel). 2021;9(12):1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Milne G, Hames T, Scotton C, et al. Does infection with or vaccination against SARS‐CoV‐2 lead to lasting immunity? Lancet Respir Med. 2021;9(12):1450‐1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Ferdinands MJ, Rao S, Dixon EB, et al. Waning 2‐dose and 3‐dose effectiveness of mRNA vaccines against COVID‐19–associated emergency department and urgent care encounters and hospitalizations among adults during periods of delta and omicron variant predominance — VISION Network, 10 States, August 2021–January 2022. Morb Mortal Wkly Rep. 2022;71:255‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Song G, He WT, Callaghan S, et al. Cross‐reactive serum and memory B‐cell responses to spike protein in SARS‐CoV‐2 and endemic coronavirus infection. Nat Commun. 2021;12(1):2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Brewer RC, Ramadoss NS, Lahey LJ, Jahanbani S, Robinson WH, Lanz TV. BNT162b2 vaccine induces divergent B cell responses to SARS‐CoV‐2 S1 and S2. Nat Immunol. 2022;23(1):33‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Samanovic MI, Cornelius AR, Gray‐Gaillard SL, et al. Robust immune responses are observed after one dose of BNT162b2 mRNA vaccine dose in SARS‐CoV‐2‐experienced individuals. Sci Transl Med. 2022;14(631):eabi8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Kramer KJ, Wilfong EM, Voss K, et al. Single‐cell profiling of the antigen‐specific response to BNT162b2 SARS‐CoV‐2 RNA vaccine. bioRxiv. 2021. https://www.biorxiv.org/content/10.1101/2021.07.28.453981v1. Accessed January 07, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Wang Y, Wang X, Luu LDW, et al. Single‐cell transcriptomic atlas of individuals receiving inactivated COVID‐19 vaccines reveals distinct immunological responses between vaccine and natural SARS‐CoV‐2 infection. medRxiv. 2021. https://www.medrxiv.org/content/10.1101/2021.08.30.21262863v1. Accessed January 22, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Ivanova EN, Devlin JC, Buus TB, et al. SARS‐CoV‐2 mRNA vaccine elicits a potent adaptive immune response in the absence of IFN‐mediated inflammation observed in COVID‐19. medRxiv. 2021. https://www.medrxiv.org/content/10.1101/2021.04.20.21255677v2. Accessed January 22, 2022. [Google Scholar]
- 269. Mades A, Chellamathu P, Kojima N, et al. Detection of persistent SARS‐CoV‐2 IgG antibodies in oral mucosal fluid and upper respiratory tract specimens following COVID‐19 mRNA vaccination. Sci Rep. 2021;11(1):24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Hassan AO, Kafai NM, Dmitriev IP, et al. A single‐dose intranasal Chad vaccine protects upper and lower respiratory tracts against SARS‐CoV‐2. Cell. 2020;183(1):169‐184 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Burton DR, Topol EJ. Toward superhuman SARS‐CoV‐2 immunity? Nat Med. 2021;27(1):5‐6. [DOI] [PubMed] [Google Scholar]
- 272. Ebinger JE, Fert‐Bober J, Printsev I, et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS‐CoV‐2. Nat Med. 2021;27(6):981‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Couch RB, Kasel JA. Immunity to influenza in man. Annu Rev Microbiol. 1983;37:529‐549. [DOI] [PubMed] [Google Scholar]
- 274. Abu‐Raddad LJ, Chemaitelly H, Ayoub HH, et al. Association of Prior SARS‐CoV‐2 infection with risk of breakthrough infection following mRNA vaccination in Qatar. JAMA. 2021;326(19):1930‐1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Murugesan M, Mathews P, Paul H, et al. Protective effect conferred by prior infection and vaccination on COVID‐19 in a healthcare worker cohort in South India. Preprints with The Lancet. 2021. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3914633. Accessed January 25, 2022. [DOI] [PMC free article] [PubMed]
- 276. Vitale J, Mumoli N, Clerici P, et al. Assessment of SARS‐CoV‐2 reinfection 1 year after primary infection in a population in Lombardy, Italy. JAMA Intern Med. 2021;181(10):1407‐1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Hall VJ, Foulkes S, Charlett A, et al. SARS‐CoV‐2 infection rates of antibody‐positive compared with antibody‐negative health‐care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. 2021;397(10283):1459‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Hansen CH, Michlmayr D, Gubbels SM, Molbak K, Ethelberg S. Assessment of protection against reinfection with SARS‐CoV‐2 among 4 million PCR‐tested individuals in Denmark in 2020: a population‐level observational study. Lancet. 2021;397(10280):1204‐1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Sheehan MM, Reddy AJ, Rothberg MB. Reinfection rates among patients who previously tested positive for coronavirus disease 2019: a retrospective cohort study. Clin Infect Dis. 2021;73(10):1882‐1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Abu‐Raddad LJ, Chemaitelly H, Malek JA, et al. Assessment of the risk of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) reinfection in an intense Reexposure setting. Clin Infect Dis. 2021;73(7):e1830‐e1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Stamatatos L, Czartoski J, Wan YH, et al. mRNA vaccination boosts cross‐variant neutralizing antibodies elicited by SARS‐CoV‐2 infection. Science. 2021;372:1413‐1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Deng X, Garcia‐Knight MA, Khalid MM, et al. Transmission, infectivity, and neutralization of a spike L452R SARS‐CoV‐2 variant. Cell. 2021;184(13):3426‐3437 e3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Bozio CH, Grannis SJ, Naleway AL, et al. Laboratory‐confirmed COVID‐19 among adults hospitalized with COVID‐19‐like illness with infection‐induced or mRNA vaccine‐induced SARS‐CoV‐2 immunity ‐ nine states, January‐September 2021. MMWR Morb Mortal Wkly Rep. 2021;70(44):1539‐1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Pardi N, Tuyishime S, Muramatsu H, et al. Expression kinetics of nucleoside‐modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release. 2015;217:345‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Shenai MB, Rahme R, Noorchashm H. Equivalency of protection from natural immunity in COVID‐19 recovered versus fully vaccinated persons: a systematic review and pooled analysis. Cureus. 2021;13(10):e19102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Cavanaugh AM, Spicer KB, Thoroughman D, Glick C, Winter K. Reduced risk of reinfection with SARS‐CoV‐2 after COVID‐19 vaccination ‐ Kentucky, may‐June 2021. MMWR Morb Mortal Wkly Rep. 2021;70(32):1081‐1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Chen Y, Tong P, Whiteman NB, et al.. Differential antibody dynamics to SARS‐CoV‐2 infection and vaccination. 2021. https://www.biorxiv.org/content/10.1101/2021.09.09.459504v1. Accessed January 19, 2022.
- 288. Lozano‐Ojalvo D, Camara C, Lopez‐Granados E, et al. Differential effects of the second SARS‐CoV‐2 mRNA vaccine dose on T cell immunity in naive and COVID‐19 recovered individuals. Cell Rep. 2021;36(8):109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Manisty C, Otter AD, Treibel TA, et al. Antibody response to first BNT162b2 dose in previously SARS‐CoV‐2‐infected individuals. Lancet. 2021;397(10279):1057‐1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Prendecki M, Clarke C, Brown J, et al. Effect of previous SARS‐CoV‐2 infection on humoral and T‐cell responses to single‐dose BNT162b2 vaccine. Lancet. 2021;397(10280):1178‐1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Bates TA, McBride SK, Winders B, et al. Antibody response and variant cross‐neutralization after SARS‐CoV‐2 breakthrough infection. JAMA. 2022;327(2):179‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Bates TA, McBride SK, Leier HC, et al. Vaccination before or after SARS‐CoV‐2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol. 2022;7:eabn8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Hall V, Foulkes S, Insalata F, et al. Effectiveness and durability of protection against future SARS‐CoV‐2 infection conferred by COVID‐19 vaccination and previous infection; findings from the UK SIREN prospective cohort study of healthcare workers March 2020 to September 2021. 2021. https://www.medrxiv.org/content/10.1101/2021.11.29.21267006v1. Accessed January 19, 2022.
- 294. Deng W, Bao L, Liu J, et al. Primary exposure to SARS‐CoV‐2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818‐823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Chandrashekar A, Liu J, Martinot AJ. SARS‐CoV‐2 infection protects against rechallenge in rhesus macaques. Science. et al., 2020;369(6505):812‐817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Gundlapalli AV, Salerno RM, Brooks JT, et al. SARS‐CoV‐2 serologic assay needs for the next phase of the US COVID‐19 pandemic response. Open Forum Infect Dis. 2021;8(1):ofaa555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Feng S, Phillips DJ, White T, et al. Correlates of protection against symptomatic and asymptomatic SARS‐CoV‐2 infection. Nat Med. 2021;27(11):2032‐2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Barnes CO, Jette CA, Abernathy ME, et al. SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS‐CoV‐2 spike. Nature. 2020;584(7821):450‐456. [DOI] [PubMed] [Google Scholar]
- 300. Zost SJ, Gilchuk P, Chen RE, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS‐CoV‐2 spike protein. Nat Med. 2020;26(9):1422‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN‐COV2, a neutralizing antibody cocktail, in outpatients with Covid‐19. N Engl J Med. 2021;384(3):238‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Lucas C, Klein J, Sundaram ME, et al. Delayed production of neutralizing antibodies correlates with fatal COVID‐19. Nat Med. 2021;27(7):1178‐1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Baumgarth N. The shaping of a B cell Pool maximally responsive to infections. Annu Rev Immunol. 2021;39:103‐129. [DOI] [PubMed] [Google Scholar]
- 304. Abayasingam A, Balachandran H, Agapiou D, et al. COSIN Study Group Long‐term persistence of RBD(+) memory B cells encoding neutralizing antibodies in SARS‐CoV‐2 infection. Cell Rep Med. 2021;2(4):100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Zhou W, Wang W, Wang H, Lu R, Tan W. First infection by all four non‐severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect Dis. 2013;13:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short‐lasting. Nat Med. 2020;26(11):1691‐1693. [DOI] [PubMed] [Google Scholar]
- 307. Galanti M, Shaman J. Direct observation of repeated infections with endemic coronaviruses. J Infect Dis. 2021;223(3):409‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105(2):435‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Sariol A, Perlman S. Lessons for COVID‐19 immunity from other coronavirus infections. Immunity. 2020;53(2):248‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Huang AT, Garcia‐Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11(1):4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Alshukairi AN, Khalid I, Ahmed WA, et al. Antibody response and disease severity in healthcare worker MERS survivors. Emerg Infect Dis. 2016;22(6):1113‐1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS‐associated coronavirus after recovery. N Engl J Med. 2007;357(11):1162‐1163. [DOI] [PubMed] [Google Scholar]
- 313. Guo X, Guo Z, Duan C, et al. Long‐term persistence of IgG antibodies in SARS‐CoV infected healthcare workers. 2020. https://www.medrxiv.org/content/10.1101/2020.02.12.20021386v1. Accessed January 07, 2022.
- 314. Nie Y, Wang G, Shi X, et al. Neutralizing antibodies in patients with severe acute respiratory syndrome‐associated coronavirus infection. J Infect Dis. 2004;190(6):1119‐1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Liu W, Fontanet A, Zhang PH, et al. Two‐year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193(6):792‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Wei J, Matthews PC, Stoesser N, et al. Anti‐spike antibody response to natural SARS‐CoV‐2 infection in the general population. Nat Commun. 2021;12(1):6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Monto AS, Malosh RE, Petrie JG, Martin ET. The doctrine of original antigenic sin: separating good from evil. J Infect Dis. 2017;215(12):1782‐1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Zhang A, Stacey HD, Mullarkey CE, Miller MS. Original antigenic sin: how first exposure shapes lifelong anti‐influenza virus immune responses. J Immunol. 2019;202(2):335‐340. [DOI] [PubMed] [Google Scholar]
- 319. Fonville JM, Wilks SH, James SL, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346(6212):996‐1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Lin CY, Wolf J, Brice DC, et al. Pre‐existing humoral immunity to human common cold coronaviruses negatively impacts the protective SARS‐CoV‐2 antibody response. Cell Host Microbe. 2022;30(1):83‐96 e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Anderson EM, Goodwin EC, Verma A, et al. Seasonal human coronavirus antibodies are boosted upon SARS‐CoV‐2 infection but not associated with protection. Cell. 2021;184(7):1858‐1864 e1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Sagar M, Reifler K, Rossi M, et al. Recent endemic coronavirus infection is associated with less‐severe COVID‐19. J Clin Invest. 2021;131(1):e143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Aguilar‐Bretones M, Westerhuis BM, Raadsen MP, et al. Seasonal coronavirus‐specific B cells with limited SARS‐CoV‐2 cross‐reactivity dominate the IgG response in severe COVID‐19. J Clin Invest. 2021;131(21):e150613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Aydillo T, Rombauts A, Stadlbauer D, et al. Immunological imprinting of the antibody response in COVID‐19 patients. Nat Commun. 2021;12(1):3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Maemura T, Kuroda M, Armbrust T, Yamayoshi S, Halfmann PJ, Kawaoka Y. Antibody‐dependent enhancement of SARS‐CoV‐2 infection is mediated by the IgG receptors FcgammaRIIA and FcgammaRIIIA but does not contribute to aberrant cytokine production by macrophages. MBio. 2021;12(5):e0198721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Gouma S, Weirick ME, Bolton MJ, et al. Health care worker seromonitoring reveals complex relationships between common coronavirus antibodies and COVID‐19 symptom duration. JCI Insight. 2021;6(16):e150449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Dowell AC, Butler MS, Jinks E, et al. Children develop robust and sustained cross‐reactive spike‐specific immune responses to SARS‐CoV‐2 infection. Nat Immunol. 2022;23(1):40‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Shen XR, Li Q, Li HL, et al. Antibody‐dependent enhancement of SARS‐CoV‐2 infection of human immune cells: in vitro assessment provides insight in COVID‐19 pathogenesis. Viruses. 2021;13(12):2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Jenks SA, Cashman KS, Woodruff MC, Lee FE, Sanz I. Extrafollicular responses in humans and SLE. Immunol Rev. 2019;288(1):136‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Rothaeusler K, Baumgarth N. B‐cell fate decisions following influenza virus infection. Eur J Immunol. 2010;40(2):366‐377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Di Niro R, Lee SJ, Vander Heiden JA, et al. Salmonella infection drives promiscuous B cell activation followed by Extrafollicular affinity maturation. Immunity. 2015;43(1):120‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Tipton CM, Fucile CF, Darce J, et al. Diversity, cellular origin and autoreactivity of antibody‐secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16(7):755‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Mathew D, Giles JR, Baxter AE, et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Zhang JY, Wang XM, Xing X, et al. Single‐cell landscape of immunological responses in patients with COVID‐19. Nat Immunol. 2020;21(9):1107‐1118. [DOI] [PubMed] [Google Scholar]
- 335. Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS‐CoV‐2 in a COVID‐19 recovered patient cohort and their implications. medRxiv 2020. https://www.medrxiv.org/content/10.1101/2020.03.30.20047365v2. Accessed January 07, 2022. [Google Scholar]
- 336. Richardson C, Chida AS, Adlowitz D, et al. Sanz I Molecular basis of 9G4 B cell autoreactivity in human systemic lupus erythematosus. J Immunol. 2013;191(10):4926‐4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Reed JH, Jackson J, Christ D, Goodnow CC. Clonal redemption of autoantibodies by somatic hypermutation away from self‐reactivity during human immunization. J Exp Med. 2016;213(7):1255‐1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Zhang Y, Xiao M, Zhang S, et al. Coagulopathy and antiphospholipid antibodies in patients with Covid‐19. N Engl J Med. 2020;382(17):e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life‐threatening COVID‐19. Science. 2020;370(6515):eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Nalbandian A, Sehgal K, Gupta A, et al. Post‐acute COVID‐19 syndrome. Nat Med. 2021;27(4):601‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Al‐Aly Z, Xie Y, Bowe B. High‐dimensional characterization of post‐acute sequelae of COVID‐19. Nature. 2021;594(7862):259‐264. [DOI] [PubMed] [Google Scholar]
- 342. Woodruff MC, Walker TA, Truong AD, et al. Evidence of persisting autoreactivity in post‐acute sequelae of SARS‐CoV‐2 infection. medRxiv 2021. https://www.medrxiv.org/content/10.1101/2021.09.21.21263845v1. Accessed January 07, 2022. [Google Scholar]
- 343. Chang SE, Feng A, Meng W, et al. New‐onset IgG autoantibodies in hospitalized patients with COVID‐19. Nat Commun. 2021;12(1):5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Su Y, Yuan D, Chen DG, et al. Multiple early factors anticipate post‐acute COVID‐19 sequelae. Cell. 2022;185:881‐895.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Wang EY, Mao T, Klein J, et al. Diverse functional autoantibodies in patients with COVID‐19. Nature. 2021;595(7866):283‐288. [DOI] [PubMed] [Google Scholar]
- 346. Liu Y, Ebinger JE, Mostafa R, et al. Paradoxical sex‐specific patterns of autoantibody response to SARS‐CoV‐2 infection. J Transl Med. 2021;19(1):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Bertin D, Kaphan E, Weber S, et al. Persistent IgG anticardiolipin autoantibodies are associated with post‐COVID syndrome. Int J Infect Dis. 2021;113:23‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Bowles L, Platton S, Yartey N, et al. Lupus anticoagulant and abnormal coagulation tests in patients with Covid‐19. N Engl J Med. 2020;383(3):288‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Zuo Y, Estes SK, Ali RA, et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID‐19. Sci Transl Med. 2020;12(570):eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Bastard P, Gervais A, Le Voyer T, et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID‐19 deaths. Sci Immunol. 2021;6(62):eabl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Murphy WJ, Longo DL. A possible role for anti‐idiotype antibodies in SARS‐CoV‐2 infection and vaccination. N Engl J Med. 2021;386(4):394‐396. [DOI] [PubMed] [Google Scholar]
- 353. Ryan FJ, Hope CM, Masavuli MG, et al. Long‐term perturbation of the peripheral immune system months after SARS‐CoV‐2 infection. BMC Med. 2022;20(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Arunachalam PS, Scott MKD, Hagan T, et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596(7872):410‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Sudre CH, Murray B, Varsavsky T, et al. Attributes and predictors of long COVID. Nat Med. 2021;27(4):626‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Cervia C, Zurbuchen Y, Taeschler P, et al. Immunoglobulin signature predicts risk of post‐acute COVID‐19 syndrome. Nat Commun. 2022;13(1):446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


