Abstract
The dcuB gene of Escherichia coli encodes an anaerobic C4-dicarboxylate transporter that is induced anaerobically by FNR, activated by the cyclic AMP receptor protein, and repressed in the presence of nitrate by NarL. In addition, dcuB expression is strongly induced by C4-dicarboxylates, suggesting the presence of a novel C4-dicarboxylate-responsive regulator in E. coli. This paper describes the isolation of a Tn10 mutant in which the 160-fold induction of dcuB expression by C4-dicarboxylates is absent. The corresponding Tn10 mutation resides in the yjdH gene, which is adjacent to the yjdG gene and close to the dcuB gene at ∼93.5 min in the E. coli chromosome. The yjdHG genes (redesignated dcuSR) appear to constitute an operon encoding a two-component sensor-regulator system (DcuS-DcuR). A plasmid carrying the dcuSR operon restored the C4-dicarboxylate inducibility of dcuB expression in the dcuS mutant to levels exceeding those of the dcuS+ strain by approximately 1.8-fold. The dcuS mutation affected the expression of other genes with roles in C4-dicarboxylate transport or metabolism. Expression of the fumarate reductase (frdABCD) operon and the aerobic C4-dicarboxylate transporter (dctA) gene were induced 22- and 4-fold, respectively, by the DcuS-DcuR system in the presence of C4-dicarboxylates. Surprisingly, anaerobic fumarate respiratory growth of the dcuS mutant was normal. However, under aerobic conditions with C4-dicarboxylates as sole carbon sources, the mutant exhibited a growth defect resembling that of a dctA mutant. Studies employing a dcuA dcuB dcuC triple mutant unable to transport C4-dicarboxylates anaerobically revealed that C4-dicarboxylate transport is not required for C4-dicarboxylate-responsive gene regulation. This suggests that the DcuS-DcuR system responds to external substrates. Accordingly, topology studies using 14 DcuS-BlaM fusions showed that DcuS contains two putative transmembrane helices flanking a ∼140-residue N-terminal domain apparently located in the periplasm. This topology strongly suggests that the periplasmic loop of DcuS serves as a C4-dicarboxylate sensor. The cytosolic region of DcuS (residues 203 to 543) contains two domains: a central PAS domain possibly acting as a second sensory domain and a C-terminal transmitter domain. Database searches showed that DcuS and DcuR are closely related to a subgroup of two-component sensor-regulators that includes the citrate-responsive CitA-CitB system of Klebsiella pneumoniae. DcuS is not closely related to the C4-dicarboxylate-sensing DctS or DctB protein of Rhodobacter capsulatus or rhizobial species, respectively. Although all three proteins have similar topologies and functions, and all are members of the two-component sensor-kinase family, their periplasmic domains appear to have evolved independently.
Escherichia coli can utilize C4-dicarboxylates (aspartate, fumarate, malate, and succinate) as energy sources during both aerobic and anaerobic growth (8). Uptake of C4-dicarboxylates is achieved by the aerobic DctA system and by the anaerobic DcuA, DcuB, and DcuC systems. DcuA and DcuB are homologous proteins (36% identical), and studies with corresponding dcuA and dcuB mutants suggested that they perform similar roles in C4-dicarboxylate transport (29). A more recent study on the expression of the dcuA and dcuB genes (12) indicated that DcuA has a general function in C4-dicarboxylate transport whereas DcuB primarily mediates C4-dicarboxylate transport during anaerobic fumarate respiration (12). These studies further showed that dcuA is constitutively expressed whereas dcuB expression is highly regulated. The dcuB gene is strongly induced anaerobically by FNR, repressed in the presence of nitrate by NarL, and is subject to cyclic AMP receptor protein (CRP)-mediated catabolite repression. In addition, dcuB transcription is strongly induced (up to 70-fold) by C4-dicarboxylates (aspartate, fumarate, malate, maleate, and succinate) (12). The mechanism of the C4-dicarboxylate-dependent induction of dcuB is unknown. However, the frd and nuo operons of E. coli have also been shown to be regulated by C4-dicarboxylates, albeit weakly, via an undefined mechanism (2, 16a, 18). Together, these findings suggest that E. coli possesses an uncharacterized C4-dicarboxylate-responsive transcriptional regulator controlling the expression of at least three genes or operons (12).
Although nothing is known of the putative C4-dicarboxylate-responsive transcriptional regulator of E. coli, such systems have been identified in other bacteria. Rhizobium meliloti and Rhizobium leguminosarum each contain two-component sensor-regulators, DctB and DctD encoded by dctBD, that activate transcription of the adjacent dctA genes (specifying the C4-dicarboxylate transporter) in response to C4-dicarboxylates (38). Rhodobacter capsulatus also contains a two-component sensor-regulator, DctS and DctR encoded by dctSR, which is required for high-affinity C4-dicarboxylate transport mediated by the products of the adjacent dctPQM genes (9a, 14). It is therefore assumed that DctS and DctR are involved in the C4-dicarboxylate-dependent induction of dctPQM. The DctB and DctS proteins are thought to be membrane-bound sensor-kinases containing a periplasmic C4-dicarboxylate sensing domain in their N-terminal segments and a cytosolic histidine-kinase domain in the C-terminal regions. Although the DctB and DctS proteins appear to have similar sensory functions and are both members of the two-component sensor-kinase family, they are not otherwise closely related, and surprisingly, their N-terminal domains exhibit no apparent sequence similarity. The DctR and DctD proteins are members of different subfamilies of the response-regulators. DctR contains two domains: an N-terminal acceptor domain and a C-terminal DNA-binding domain. DctD contains three domains: an N-terminal acceptor domain, a centrally located domain that mediates ς54-dependent transcriptional activation of dctA, and a C-terminal domain that is responsible for binding to the upstream activator sequence of the dctA gene (17).
The E. coli genome does not contain genes encoding close homologs of the DctB-DctD or DctS-DctR pairs, showing that the putative C4-dicarboxylate-responsive transcriptional regulatory system of E. coli is not closely related to those of Rhizobium and Rhodobacter. Indeed, the studies described here reveal that E. coli contains a new two-component regulatory system, designated DcuS-DcuR, that regulates the expression of dcuB and other genes in response to external C4-dicarboxylates. Furthermore, the DcuS-DcuR proteins are not closely related to the DctB-DctD or DctS-DctR proteins but instead are members of the CitA-CitB sub-family of two-component sensor-regulators. While this paper was under review, Zientz et al., published a paper that also identifies the role of DcuS-DcuR in the transcriptional regulation of gene expression in E. coli (41). The results described here are largely in agreement with those of Zientz et al. (41).
MATERIALS AND METHODS
Media, growth conditions, strains, and general methods.
All strains of E. coli used in this study are listed in Table 1. For growth studies, strains were generally grown aerobically or anaerobically in M9 minimal salts (Sigma) with either glucose (0.4%) or glycerol (0.4%) as the carbon source, supplemented with 1 mM MgSO4, 0.1 mM CaCl2, and 0.5-mg/ml vitamin B1. Where used, fumarate, nitrate, or trimethylamine N oxide (TMAO) was present at 50 mM. Unless otherwise stated, cultures were grown at 37°C either aerobically in 250-ml conical flasks with shaking or anaerobically in stationary 10-ml bijou bottles. Standard genetic procedures were performed as described by Sambrook et al. (26) with DH5α grown aerobically at 37°C in L-broth supplemented, as required, with 15 μg of tetracycline/ml or with 10 or 50 μg of chloramphenicol/ml. DNA labeling was achieved with the Ready to Go DNA Labelling Kit (Pharmacia) and [α-32P]dCTP.
TABLE 1.
Strains of E. coli and phages and plasmids used
| Strain, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | Δ(argF-lac)U169 (φ80ΔlacZM15) recA | 26 |
| IMW157 | AN387 dcuC::mini-Tn10(Camr) | 37 |
| JRG1788 | MC1000 [λAΦZ.4(frdA′-lacZ)] | 30 |
| JRG1938 | RK4353 [λRZ5::fumA′-lacZ] | 35 |
| JRG2814 | AN387 dcuA::spc dcuB::kan | 29 |
| JRG3351 | MC4100 [λRS45::dctA′-lacZ] | 6 |
| JRG3835 | MC4100 [λRS45::dcuB′-lacZ] | 12 |
| JRG3983 | JRG3835 dcuS::mini-Tn10 | This work |
| JRG3984 | JRG3351 dcuS::mini-Tn10 | This work |
| JRG3985 | JRG1788 dcuS::mini-Tn10 | This work |
| JRG3986 | JRG1938 dcuS::mini-Tn10 | This work |
| MC4100 | Δ(argF-lac)U169 rpsL | 28 |
| TG1 | Δ(lac-pro) lacIq lacZΔM15 | 10 |
| SCA1 | MC4100 [λRS45::dcuB′-lacZ] dcuA::spc dcuB::kan | This work |
| SCA2 | MC4100 [λRS45::dcuB′-lacZ] dcuA::spc dcuB::kan dcuC::mini-Tn10(Camr) | This work |
| SCA3 | MC4100 dcuS::mini-Tn10 | This work |
| Phages | ||
| P1vir1 | 24 | |
| λNK1098 | 34 | |
| Plasmids | ||
| pGS78 | dcuSR | 13 |
| pGS1179 | pSU19 + 1.1-kb SalI dcuS′ fragment | This work |
| pGS1180 | pGS1179 + 2.5-kb SphI-HindIII ′dcuS-dcuR fragment | This work |
| pHSG576 | 33 | |
| pLH21 | J. K. Broome-Smith | |
| pPG1 | pSU18 + 4.3-kb PstI dcuS::mini-Tn10 fragment | This work |
| pPG2 | pHSG576 + 3.5-kb EcoRI-HindIII dcuS-dcuR fragment | This work |
| pSU18/19 | 22 | |
| pYZ4 | 36 |
To investigate the effects of the dcuS mutation on the expression of the dctA, fumA, and frdA genes, the dcuS::Tn10 mutation of strain JRG3983 was transferred via P1-mediated transduction to the corresponding lacZ fusion strains JRG1788, JRG1938, and JRG3351 (see Table 1). Strain SCA2 (dcuA dcuB dcuC) was constructed in two steps: first, the dcuA dcuB double mutation of JRG2814 was transferred via P1-mediated transduction to strain JRG3835 to generate strain SCA1, and second, the dcuC mutation of strain IMW157 (37) was similarly transferred to SCA1 to generate strain SCA2.
Transposon mutagenesis and isolation of a dcuS::Tn10 mutant.
Transposon mutagenesis was performed with the mini-Tn10 carrying phage λNK1098 and the procedure described by Way et al. (34). JRG3835 (dcuB-lacZ) was grown aerobically in 50 ml of λym (1% tryptone, 0.25% sodium chloride, 0.2% maltose, and 0.1% yeast extract) liquid medium at 37°C to an optical density at 650 nm (OD650) of approximately 0.5. Cells were then harvested by centrifugation, resuspended in 5 ml of λym (1 mM isopropyl-β-d-thiogalactopyranoside [IPTG]) and infected with λNK1098 at a multiplicity of infection of 0.3. After incubation at 21°C for 30 min to allow phage adsorption, the culture was incubated for 90 min at 37°C to allow expression of the antibiotic resistance genes. Cells were then pelleted, washed in 10 ml of L-broth containing 50 mM sodium citrate, and resuspended in 1 ml of L-broth containing sodium citrate. Aliquots of 0.1 ml of the resuspended cells were spread onto agar plates containing M9 minimal salts, 0.4% glycerol, 50 mM TMAO, 50 mM fumarate, 1.25 mM sodium pyrophosphate, 20 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) ml−1, and tetracycline and then incubated aerobically at 42°C for 36 h. Approximately 10% of the 8 × 103 Tcr mutants obtained were Lac−, possibly due to instability of the dcuB-lacZ bearing prophage. Most of the rest were strongly Lac+, except for 14 weakly Lac+ Tcr mutants, which were further screened by aerobic propagation at 37°C for 24 h on M9 minimal medium plates containing glycerol, TMAO, X-Gal, and tetracycline with and without fumarate. Tcr mutants exhibiting a weak Lac+ phenotype in both the presence and absence of fumarate were selected as potential C4-dicarboxylate regulatory mutants.
Southern hybridization.
Chromosomal DNA was isolated from strain JRG3983 with the Wizard Genomic DNA Purification kit (Promega). Aliquots of approximately 10 μg of chromosomal DNA were digested with restriction enzymes, electrophoresed, denatured, transferred to a nylon membrane, and hybridized at 65°C with an [α-32P]-labeled 0.85-kb EcoRI-HindIII fragment of the mini-Tn10 transposon.
Recovery of the transposon and flanking DNA from JRG3983 and construction of pPG2.
A 4.2-kb chromosomal fragment containing the Tn10 insertion (together with ∼1 kb of flanking chromosomal DNA) of JRG3983 was PCR amplified with Pfu Turbo DNA polymerase (Stratagene) and two primers: DcuS-f, 5′-ccctgcagATTGCGTCGTCATCGATAATTAATACA-3′; and DcuS-r, 5′-ccctgcagACAAGAATTGCTGAATTACCGTAAGTC-3′ (mismatches are shown in small capitals, PstI sites are in boldface, and the corresponding annealing sites are indicated in Fig. 2). The 4.2-kb PCR product was purified, digested with PstI, and cloned into pSU18 to generate plasmid pPG1. The nucleotide sequence of one of the regions flanking the Tn10 fragment of pPG1 was determined with the Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and two primers (Tn10-A, 5′-TTCAGTGATCCATTGCTG-3′; and Tn10-B, 5′-CAAAGGGAATCATAGATC-3′) designed to anneal to adjacent regions (40 bp apart) of the downstream segment of the tetR gene of Tn10.
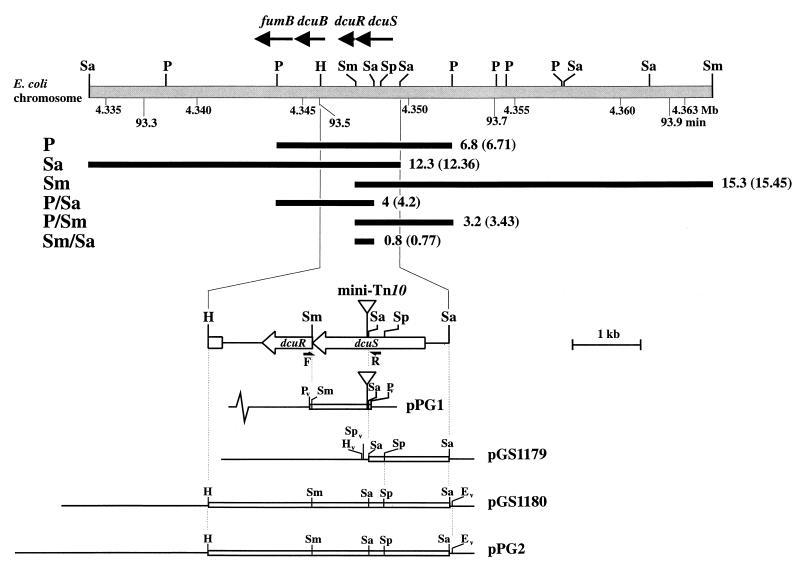
FIG. 2.
Restriction map of the dcuSR region of the E. coli chromosome and relevant plasmids. The lightly shaded bar represents chromosomal DNA and the solid bars directly below indicate the positions of the mini-Tn10-hybridizing restriction fragments detected by Southern blot analysis of chromosomal DNA from JRG3983. The observed (and expected) sizes (in kilobases), corrected for the size of mini-Tn10 (3.2 kb), are shown for each restriction fragment. Vector and insert DNA of plasmids are represented by thin lines and open bars, respectively. F, priming site for the DcuS-forward primer; R, priming site for the DcuS reverse primer. The position of the mini-Tn10 insertion is shown. Restriction sites are as follows: E, EcoRI; H, HindIII; P, PstI; Sa, SalI; Sm, SmaI; and Sp, SphI. The subscript v denotes vector restriction sites used in subcloning. The coordinates (in megabases [Mb]) and chromosomal restriction map (minutes [min]) are from Burland et al. (5) and Blattner et al. (1), respectively.
The dcuSR genes were cloned in two steps: first, the 1.1-kb SalI partial-dcuS fragment of pGS78 (13) was inserted into plasmid pSU19 to generate pGS1179, and second, the 0.24-kb HindIII-SphI fragment of pGS1179 was replaced with the 2.5-kb HindIII-SphI ′dcuB-dcuR-dcuS′-containing fragment of pGS78 to produce pGS1180 (Table 1 and Fig. 2). Plasmid pPG2 was constructed by cloning the 3.5-kb HindIII-EcoRI dcuSR-containing fragment of pGS1180 into the vector pHSG576 (33) (Table 1 and Fig. 2).
Construction and analysis of dcuS-blaM fusions.
Fourteen site-directed dcuS-blaM fusions were created by PCR with the dcuS-containing plasmid pGS1180 as template, Pfu DNA polymerase (Stratagene), DcuS-F (5′-gggccATGgGACATTCATTGCCCTAC-3′ (start codon underlined, mismatches in small capitals, and NcoI site in boldface) as the forward primer, and 14 codon-specific reverse primers (26-mers, with the EcoRV-site-containing sequence CCGATATC at the 5′ termini and 18 homologous bases at the 3′ termini). The 0.09 to 1.5-kb NcoI- and EcoRV-treated PCR fragments and the 0.85-kb SmaI-SacI blaM cassette of pLH21 were coligated into the corresponding sites of the Knr plasmid pYZ4 (36) to give dcuS-blaM fusions appropriately positioned downstream of the IPTG-inducible lacUV5 promoter. E. coli TG1 was transformed with the dcuS-blaM-containing plasmids and propagated on solid medium containing M9 minimal salts, 0.4% glucose, and kanamycin (50 μg ml−1). Knr transformants were tested for growth when inoculated at low or high density on solid glucose minimal medium containing kanamycin and ampicillin (35 μg ml−1). The MICs of ampicillin (AP) for transformants carrying dcuS-blaM fusions were determined as described by Golby et al. (11).
The dcuS-blaM fusion points were determined by nucleotide sequencing with the Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and a primer (BLAM1, 5′-CTCGTGCACCCAACTGA-3′) complementary to codons 14 to 18 of blaM (11).
β-Galactosidase measurements.
Preparation of cell extracts and measurements of β-galactosidase activity and protein content were performed on samples taken during the mid- to late-log phase, as described by Golby et al. (12), except that β-galactosidase activities were measured with a Biolumin 960 microtiter plate spectrophotometer (Molecular Dynamics) and protein content was determined with a Dynatech MRX microtiter plate spectrophotometer (Dynatech Laboratories).
Specific β-galactosidase activities (micromoles of o-nitrophenyl-β-d-galactopyranoside per minute per milligram of protein) were averaged from samples taken from two independent cultures. Each of the two samples was assayed in duplicate. Standard deviations were generally within 10%.
RESULTS
Isolation of a C4-dicarboxylate regulation mutant.
In order to identify the regulatory system responsible for mediating the C4-dicarboxylate-dependent induction of dcuB expression, we sought to generate mutants of JRG3835 (dcuB-lacZ) in which the fumarate-dependent expression of dcuB is perturbed. The chosen method exploited the observation that the dcuB gene is strongly induced by fumarate when JRG3835 is grown aerobically on minimal agar containing glycerol, TMAO, and X-Gal. By using such indicator plates, dcuB regulation mutants could be detected by their weak Lac+ phenotype in the presence of fumarate. The C4-dicarboxylate induction of dcuB-lacZ expression in strain JRG3835 was previously shown, in liquid medium, to be strictly dependent on the absence of oxygen (12). The aerobic induction on agar plates is presumably due to low-oxygen tensions at the centers of colonies.
JRG3835 was subjected to random mutagenesis with the mini-Tn10 transposon carried by phage λNK1098 according to the protocol described in Materials and Methods. Approximately 8,000 Tcr mutants were screened by growth on indicator plates. After 36 h of growth at 42°C under aerobic conditions, 14 weakly Lac+ Tcr mutants were selected and further screened by aerobic growth at 37°C for 24 h on indicator plates with and without fumarate. One mutant, designated JRG3983, was found to be weakly Lac+ in both the presence and absence of fumarate. The P1-mediated transfer of the Tn10(Tcr) mutation of JRG3983 to strain JRG3835 resulted in the 100% cotransfer of the regulatory defect. P1-mediated transfer to MC4100 revealed that the Tn10(Tcr) mutation is not linked to the dcuB-lacZ fusion. Therefore, JRG3983 appears to possess a Tn10-induced mutation, located outside the dcuB-lacZ promoter-operator region, which results in the loss of fumarate-dependent induction of dcuB expression.
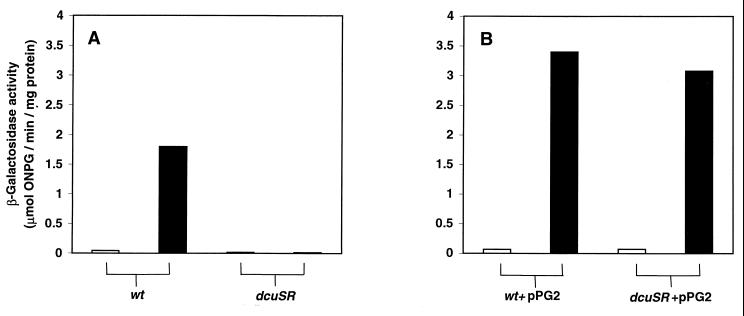
The regulatory defect of the mutant was further explored by comparing the β-galactosidase specific activities of JRG3983 with those of the parent strain, JRG3835, after anaerobic growth in liquid minimal medium containing glycerol and TMAO, with and without fumarate (Fig. 1A). In the parental strain, expression of dcuB was increased 43-fold by fumarate (from 0.042 to 1.8 μmol/min/mg). However, in the mutant, dcuB expression was unaffected by fumarate (0.013 μmol/min/mg without fumarate compared to 0.011 with fumarate). Indeed, dcuB expression was threefold lower than that of the parent in the absence of fumarate and 160-fold lower in the presence of fumarate (Fig. 1A). These findings indicate that the Tn10 insertion of JRG3983 completely inactivates the C4-dicarboxylate regulator controlling dcuB expression.
FIG. 1.
Expression of the dcuB-lacZ transcriptional fusion in the uncomplemented (A) and complemented (B) dcuB regulatory mutant (JRG3983) and parent (JRG3835) strains. Growth was performed under anaerobic conditions in M9 minimal medium containing 0.4% glycerol, 50 mM TMAO with (closed bars) or without (open bars) 50 mM fumarate. β-Galactosidase activities were assayed in mid- to late-logarithmic cultures of JRG3835 (wt) and JRG3983 (dcuSR) (A) and JRG3835(pPG2) (wt+pPG2) and JRG3983(pPG2) (dcuSR+pPG2) (B).
Identification of the genes encoding the C4-dicarboxylate regulator.
The location of the mini-Tn10 insertion in the chromosome of JRG3938 was determined by Southern blot analysis (data summarized in Fig. 2). Chromosomal DNA from JRG3983 was digested with restriction enzymes known not to possess recognition sites within the mini-Tn10 transposon. The resulting fragments were separated by gel electrophoresis, blotted, and hybridized with a labeled mini-Tn10 fragment (data not shown). The sizes of the hybridizing bands, corrected for the presence of the transposon (3.2 kb), match the 93.5-min region of the E. coli physical map only, and locate the mini-Tn10 insertion between the SalI and SmaI sites of the yjdH (dcuS) gene (Fig. 2). The precise location of the transposon was determined by cloning the PCR-amplified 4.2-kb SalI-SmaI mini-Tn10-containing fragment of JRG3983 into pSU18 to generate pPG1 and then sequencing across the Tn10-yjdH fusion site (see Materials and Methods and Fig. 2). In this way the mini-Tn10 transposon was shown to be inserted 810 bp downstream of the translational start point of yjdH, between the first and second bases of codon 271 (specifying a histidine residue), such that the Tn10 tetA gene is copolar with yjdH. The yjdH gene, together with yjdG, form the predicted yjdHG (or f543-f239) operon. These genes are just 570 bp upstream of dcuB (5) and they encode a putative two-component regulatory system that had previously been predicted to function as the C4-dicarboxylate regulatory system of dcuB (12). Therefore, the yjdHG genes were redesignated dcuSR.
Complementation of the dcuS mutation of JRG3983.
It is likely that the mini-Tn10 insertion of JRG3983 inactivated the downstream dcuR gene, as well as dcuS, due to polar effects on transcription. Therefore, in order to complement the C4-dicarboxylate regulatory defect of JRG3983, the 3.6-kb HindIII-SalI fragment of pGS78 (35), containing the entire dcuSR operon, was cloned into the medium-copy-number plasmid, pSU19 to generate pGS1180 (see Fig. 2 and Materials and Methods). However, the corresponding transformant, JRG3983(pGS1180), was unable to grow anaerobically in minimal medium containing glycerol, TMAO, and fumarate, suggesting that multiple copies of the dcuRS genes are deleterious under these growth conditions. To circumvent this problem, the 3.6-kb HindIII-EcoRI dcuSR-containing fragment of pGS1180 was cloned into the low-copy-number plasmid pHSG576 (33) to generate pPG2 (Fig. 2). The introduction of pPG2 into the mutant strain JRG3983 restored the fumarate-dependent induction of dcuB expression, resulting in a 280-fold higher expression of dcuB in JRG3983(pPG2) than in JRG3983 (3.1 versus 0.011 μmol/min/mg) (Fig. 1) and a 44-fold increase in the induction by fumarate (from 0.07 to 3.1 μmol/min/mg) (Fig. 1A) in JRG3983(pPG2). Transformation of JRG3835 or JRG3983 with pPG2 increased dcuB expression, with respect to the wild type (JRG3835), to similar levels (Fig. 1B). In the absence of fumarate, dcuB expression in the transformant was 1.5- to 1.7-fold higher than that of the wild type, and in the presence of fumarate expression was 1.7- to 1.9-fold higher than that of the wild type. These data confirm that the C4-dicarboxylate regulatory defect is indeed due to inactivation of dcuS (and/or dcuR) and further show that hyperexpression of dcuB is achieved by provision of multicopy dcuSR. This indicates that fumarate induction of dcuB expression is dependent on the concentration of the dcuSR products, DcuS and DcuR.
Role of the DcuS-DcuR system in the regulation of the dctA, frdABCD, and fumA genes.
The possibility that the DcuS-DcuR system regulates the expression of other genes involved in C4-dicarboxylate transport or metabolism was investigated by transferring the dcuS mutation of JRG3983 to strains containing the appropriate single-copy lacZ gene fusions. Expression of the dctA gene, encoding the aerobic C4-dicarboxylate transporter (DctA), was investigated by using a dctA-lacZ transcriptional fusion (6). The expression of dctA in JRG3351 (dctA-lacZ) during aerobic growth in minimal medium containing glycerol was ∼twofold increased by succinate (from 0.80 to 1.6 μmol/min/mg) whereas in the dcuS mutant (JRG3984) succinate caused an ∼twofold decrease in expression (from 0.79 to 0.43 μmol/min/mg) (Table 2). Thus, the dcuS mutation resulted in an ∼fourfold decrease in dctA expression in the presence of succinate (from 1.6 to 0.43 μmol/min/mg) (Table 2), indicating that dctA expression is under the direct or indirect control of DcuS-DcuR.
TABLE 2.
Effects of dcuS on dctA-lacZ and frdA-lacZ expression
| Fusiona | Substratesb | β-Galactosidase activityd (μmol of ONPG/min/mg of protein)
|
|
|---|---|---|---|
| dcuS+ | dcuS::Tn10 | ||
| dctA′-lacZ | Glycerol + O2 | 0.80 ± 0.08 | 0.79 ± 0.05 |
| Glycerol + O2 + succinate | 1.56 ± 0.08 | 0.43 ± 0.02 | |
| frdA′-lacZc | Glycerol + TMAO | 0.096 ± 0.005 | 0.022 ± 0.003 |
| Glycerol + TMAO + fumarate | 0.56 ± 0.05 | 0.025 ± 0.0003 | |
Strains were JRG3351, JRG3984, JRG1788, and JRG3985 (Table 1).
M9 salts medium was used with 0.4% glycerol, with or without 50 mM succinate or fumarate, and with or without 50 mM TMAO.
Casamino Acids (0.05%) was included in the growth medium, and growth was at 30°C to prevent induction of the temperature-sensitive prophage.
Values are means ± standard deviations from four measurements.
The expression of the frdABCD operon, encoding the subunits of fumarate reductase, was studied with an frdA-lacZ translational fusion (30). The expression of the frdA-lacZ fusion in the dcuS mutant, JRG3985, and its parent, JRG1788, was determined after anaerobic growth in minimal medium containing glycerol, TMAO, and 0.05% Casamino Acids with and without fumarate (Table 2). The expression of frdA in JRG1788 was increased sixfold by the presence of fumarate (from 0.096 to 0.56 μmol/min/mg) (Table 2). However, the dcuS mutation of JRG3985 abolished frdA induction by fumarate (Table 2). Expression of frdA in the dcuS mutant was ∼4-fold lower than that of the wild type in the absence of fumarate (0.022 versus 0.096 μmol/min/mg) and 22-fold lower in the presence of fumarate (0.025 versus 0.56 μmol/min/mg) (Table 2). These findings indicate that, as for dcuB, expression of frdA is strongly induced by the DcuS-DcuR system in response to C4-dicarboxylates. Surprisingly, previous studies indicated that the frdABCD operon is only 1.5-fold induced by fumarate (18). This discrepancy probably arises from differences in the growth conditions employed, in particular the presence of glucose in the medium used by Jones and Gunsalus (18).
The expression of the fumA gene, encoding the aerobic fumarase A, was studied by using a fumA-lacZ translational fusion (35). The β-galactosidase activity of this strain was found to be undetectable after growth in minimal medium, necessitating the use of L-broth (containing glycerol with or without succinate) as the growth medium for measurement of aerobic fumA-lacZ expression. The expression of fumA in JRG1938 (fumA-lacZ) was not affected by succinate, and the dcuS mutation did not affect expression in either the presence or absence of succinate (expression levels remained at ∼0.08 μmol/min/mg in all cases). Thus, it appears that neither C4-dicarboxylates nor the DcuS-DcuR system regulates the fumA gene.
The above studies establish that three E. coli genes or operons (dcuB, frdABCD, and dctA), having functions in the transport or metabolism of C4-dicarboxylates, are activated by the DcuS-DcuR system in response to C4-dicarboxylates. However, the fumA gene is not DcuS-DcuR regulated. The dcuB, frdABCD, and dctA genes (and probably the fumarase B-encoding fumB gene, since fumB is at least partly cotranscribed with dcuB [12]) appear to be members of a new regulon, designated the DcuSR regulon. It is likely that DcuS acts as a C4-dicarboxylate-sensing histidine kinase that reports C4-dicarboxylate concentration to DcuR, which in turn directly activates the transcription of genes in the DcuSR regulon. Appropriately, the DcuS-DcuR system allows the anaerobically expressed members of the DcuSR regulon (dcuB-fumB and frdABCD) to be coordinately up regulated by C4-dicarboxylates, thus ensuring that the transport (DcuB), production (fumarase B), and utilization (fumarate reductase) of fumarate are jointly induced during anaerobic fumarate respiration. Induction of the aerobic C4-dicarboxylate transporter (DctA) by DcuS-DcuR is also appropriate and is consistent with previous studies showing that C4-dicarboxylates increase Dct transport activity in E. coli (19, 21).
Growth properties of the dcuS mutant.
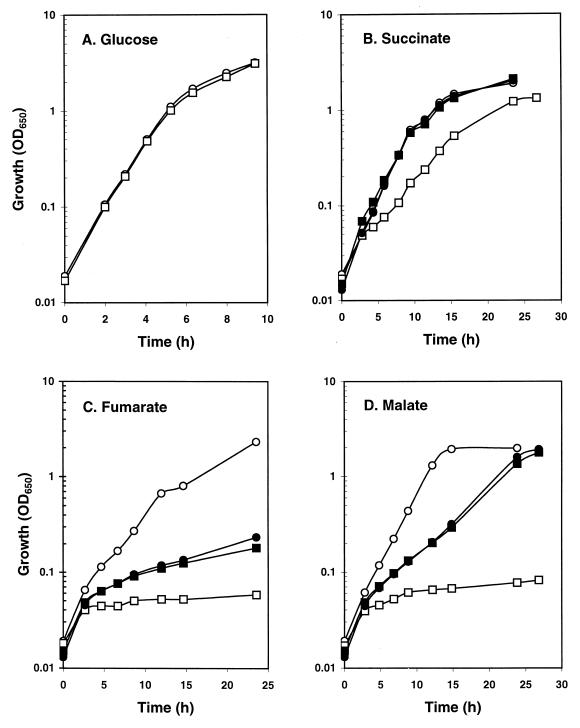
The possibility that the regulatory defect of the dcuS mutant leads to an associated growth deficiency was tested with a dcuS mutant, SCA3 (MC4100 dcuS). The parental (MC4100) and mutant (SCA3) strains grew identically under aerobic conditions in minimal medium containing glucose (Fig. 3A), glycerol, pyruvate, acetate, or lactate as the sole carbon source (data not shown). However, under aerobic conditions with C4-dicarboxylates as sole carbon sources, the dcuS mutation significantly lowered the growth rates (Fig. 3B to D). With succinate as sole carbon source, the log-phase growth rate of dcuS mutant (SCA3) was approximately 1.8-fold lower than that of the parental strain MC4100 (Fig. 3B). This growth difference was fully reversed by complementation with the dcuSR-containing plasmid, pPG2 (Fig. 3B). With fumarate or malate as sole carbon source, growth of the dcuS mutant was negligible (Fig. 3C and D, respectively). Complementation with pPG2 restored the ability of the dcuS mutant to grow with fumarate and malate, but growth was not as strong as that of the parent (Fig. 3C and D). However, MC4100(pPG2) grew at the same rate as SCA3(pPG2) in fumarate and malate minimal medium (Fig. 3C and D), showing that pPG2 reduces the fumarate- and malate-dependent growth of the parental strain. This provides further evidence that multiple copies of the dcuSR operon can be deleterious, but it is unclear why pPG2 affects growth with fumarate and malate but not with succinate.
FIG. 3.
Growth of the dcuS mutant SCA3 and the parent MC4100 under aerobic conditions in M9 minimal salts medium containing 0.4% glucose (A), 50 mM succinate (B), 50 mM fumarate (C), or 50 mM malate (D). The strains used were MC4100 (○), SCA3 (□), MC4100(pPG2) (●), and SCA3(pPG2) (■). The plasmid pPG2 had no effect on the growth of MC4100 or SCA3 during growth with glucose (data not shown).
It was somewhat surprising to find that the dcuS mutation has a greater effect on aerobic growth with fumarate and malate than with succinate. This dcuS-associated phenotype closely matches that of a dctA mutant (6), which suggests that the aerobic growth defects of the dcuS mutant are a consequence of the fourfold lower dctA expression revealed earlier (Table 2). However, it is not clear why a relatively small reduction in dctA transcription causes such a dramatic growth defect with fumarate and malate. Presumably, the dcuS mutation affects aspects of aerobic C4-dicarboxylate metabolism or transport that have not been revealed here. The better growth of the dcuS mutant in succinate relative to that in fumarate or malate is probably due to the presence of an uncharacterized and putative succinate-specific transporter (designated SucT) that enables good aerobic growth of dctA mutants on succinate (6) and is probably not strongly regulated by the DcuS-DcuR system.
Surprisingly, the dcuS mutation had no significant effect on growth under anaerobic conditions in minimal medium containing glycerol and fumarate (data not shown). This indicates that the weak expression of the frdABCD genes in the dcuS mutant is sufficient to provide adequate fumarate reductase activities for fumarate respiration under the conditions used. It is also consistent with the observation that dcuB mutants are not growth impaired when cultured under conditions of anaerobic fumarate respiration (29).
DcuS responds to external C4-dicarboxylates.
The studies described above show that the DcuS-DcuR system regulates gene expression in response to the addition of C4-dicarboxylates to the culture medium. Previous studies showed that the expression of dcuB is dependent on the concentration of C4-dicarboxylates in the medium, suggesting that the regulatory response of the DcuS-DcuR system is quantitative with respect to C4-dicarboxylate concentration (12). However, it is not clear whether the signal sensed by the DcuS-DcuR system is intra- or extracellularly located. Clearly, if intracellular C4-dicarboxylates are sensed by the DcuS-DcuR system, then mutants unable to transport C4-dicarboxylates into the cell would display a regulatory phenotype similar to that of the dcuS mutant. For this reason, the C4-dicarboxylate-dependent induction of dcuB was tested in a dcuA dcuB dcuC mutant, SCA2, which is unable to grow anaerobically on glycerol and fumarate because of its deficient anaerobic C4-dicarboxylate transport activity (37). The expression of the dcuB gene during anaerobic growth in minimal medium containing glycerol and TMAO with and without fumarate was not affected by the transport defect of SCA2, although the transport mutant did show a threefold increase in dcuB expression in the absence of fumarate (Table 3). These findings suggest that internalization of C4-dicarboxylates is not a requirement for DcuS-DcuR-dependent transcriptional activation, and this in turn suggests that DcuS senses external (periplasmic) rather than internal (cytosolic) substrate.
TABLE 3.
Effects of the dcuA dcuB dcuC triple mutation on dcuB′-lacZ expressiona
| Substratesb | β-Galactosidase activity (μmol of ONPG/min/mg of protein)
|
|
|---|---|---|
| wtb | dcuA dcuB dcuCb | |
| Glycerol + TMAO | 0.069 ± 0.03 | 0.24 ± 0.03 |
| Glycerol + TMAO + fumarate | 1.75 ± 0.07 | 1.79 ± 0.05 |
The strains used were JRG3835 (wt) and SCA2 (dcuA dcuB dcuC).
Growth conditions were as described in the legend to Fig. 1.
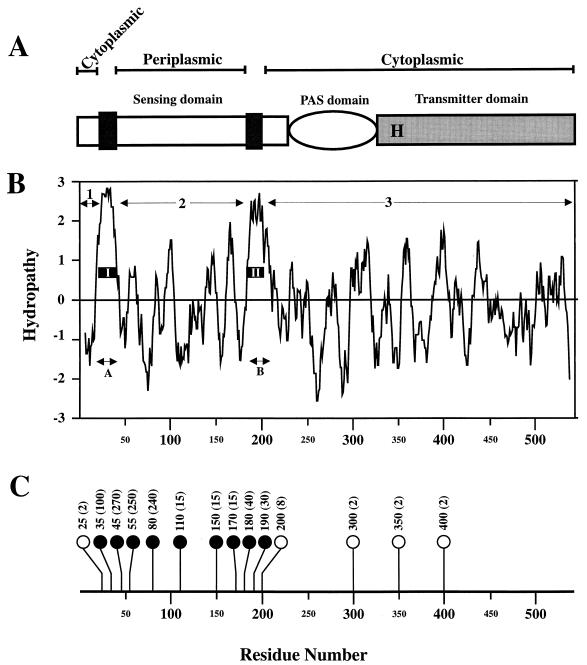
Topological analysis of DcuS.
Inspection of the hydropathy profile of the 543-amino-acid-residue DcuS protein revealed two highly hydrophobic segments (A and B) of sufficient length to span the membrane and three hydrophilic regions (1, 2, and 3) likely to be extramembranous (Fig. 4B). An analysis of the DcuS sequence using the TMpred program (16) predicted that DcuS is an integral membrane protein having the following structural features: two membrane spanning α-helices (I and II) corresponding to hydrophobic segments A and B, a 21-residue N-terminal cytosolic region (region 1), a 140-residue periplasmic domain incorporating residues 43 to 182 (region 2), and a 341-residue cytosolic domain (region 3) (Fig. 4A and B). This topological prediction was tested by creating fusions between a series of truncated dcuS derivatives and a blaM gene encoding a leaderless β-lactamase (see Materials and Methods). Fourteen in-phase dcuS′-blaM fusions were produced and tested with the corresponding transformants. Transformants for which the MIC was 2 to 10 μg of AP ml−1 were presumed to encode fusion proteins in which the BlaM region remains in the cytoplasm or membrane, whereas those for which the MIC was ≥10 μg of Ap ml−1 were considered to encode fusion proteins in which the BlaM segment is directed to the periplasm (4).
FIG. 4.
Topological organization of the DcuS protein. (A) Schematic representation of the structure of DcuS. The black boxes denote the predicted transmembrane helices and the letter H indicates the position of the putative autophosphorylated His residue. (B) The hydropathy profile of DcuS, generated with a window of 11 residues (20). The two regions of hydrophobicity (A and B) and the three regions of hydrophilicity (1, 2, and 3) are indicated, as are two predicted membrane-spanning helices (I and II). (C) Resistance to AP of strains expressing DcuS′-BlaM fusion proteins. The positions of the fusions in each of the 14 DcuS′-BlaM proteins are shown. Solid circles represent periplasmic fusion proteins, and open circles represent cytoplasmic fusion proteins. The MICs (μg of AP ml−1) for E. coli strains expressing the corresponding fusion proteins are shown in brackets.
The fusion positions of the fourteen dcuS′-blaM fusions, and the corresponding AP MICs, are shown in Fig. 4C. Fusions within hydrophilic region 2 gave MICs of 15 to 270 μg of AP ml−1, confirming that this region is indeed located in the periplasm. The reason for the relatively low MICs (≤30 μg of AP ml−1) for fusions in the 110- to 180-residue region of the periplasmic loop is unknown. However, it is possible that fusions in this region generate toxic and/or unstable BlaM fusions, as observed previously for fusions between membrane proteins and BlaM (15, 27). Fusions in hydrophilic region 3 gave MICs of 2 μg of AP ml−1, supporting the prediction that this region is cytoplasmic. Therefore, the MIC data are entirely in agreement with the topological model (Fig. 4A) showing that DcuS contains an ∼140-residue periplasmic domain.
Structure-function relationships between DcuS-DcuR and other two-component sensor-regulators.
Database searches show that the DcuS protein is closely related (24 to 35% identical) to six other proteins from the two-component sensor-kinase/transmitter family: CitA from E. coli and K. pneumoniae, CitS from Bacillus subtilis and Streptomyces coelicolor, and YufL and YdbF from B. subtilis. Together with DcuS, these six proteins form a subgroup (the CitA-like proteins) of the transmitter family in which the sequence similarities are evenly distributed along the entire lengths of their aligned polypeptides (data not shown). Thus, the predicted transmembrane helices and periplasmic domain of DcuS are conserved throughout the subgroup, suggesting that they all sense extracytoplasmic signals. No other proteins in the databases have sequences that significantly resemble the periplasmic-sensing domain of the CitA-like proteins (other than the weakly related putative periplasmic domain of the HydH sensor-kinase of E. coli). This is consistent with the observation that the N-terminal sensing domains of two-component sensor-kinases are poorly conserved (25, 31, 32). It is of particular interest to note that the periplasmic domains of the respective C4-dicarboxylate-sensing DctS and DctB proteins of R. capsulatus and rhizobial species do not possess any apparent sequence similarities with that of DcuS. This is surprising given that DctS, DctB, and DcuS appear to have similar topological organizations and are all members of the histidine-kinase two-component sensor-regulator family.
The only other member of the CitA group that has been characterized is the CitA protein of K. pneumoniae (3). This protein was identified as a citrate-sensing histidine kinase having a predicted topology analogous to that reported here for DcuS (3). The periplasmic location of the N-terminal domain of CitA was supported by results obtained with a single citA-phoA fusion (3a). Together with the cognate receiver protein CitB, CitA is involved in activating the transcription of the citAB genes (encoding CitA and CitB) and other genes required for citrate fermentation (citS, oadGAB, and citDEF). However, the location of the signal (intra- or extracellular) sensed by CitA was not established (3). Interestingly, it appears that CitA and DcuS both sense carboxylic acids, which raises the possibility that all members of the CitA-like group are carboxylic-acid sensors.
The C-terminal region of DcuS (residues 331 to 543) and the other CitA-like proteins bears strong sequence similarity to the highly conserved histidine-kinase domain of the transmitters. The five sequence motifs (the H, N, G1, F, and G2 regions [25]) that are characteristic of the kinase domain are present in the CitA-like proteins (data not shown). In particular, the histidine residue that is phosphorylated in other transmitters is conserved in all seven CitA-like proteins (data not shown). Thus it would appear that DcuS possesses all the sequence features expected of a functional transmitter protein. In addition to the N-terminal sensor domain and the C-terminal histidine-kinase domain, the CitA-like proteins possess a central region of ∼130 residues that is well conserved within the CitA family. BLASTP searches (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast) of the nonredundant database show that an ∼100-residue segment of this central region resembles the PAS (or S [sensory]) domains at the central or N-terminal regions of more than 50 proteins in the non-CitA-like sensor kinase or ς54-dependent transcriptional activator families (selected sequences are shown in Fig. 5). Good sequence similarity is restricted to two parts of the PAS domain, the S1 box and the S2 box, which are 50 to 60 residues apart (Fig. 5). PAS domains were first found in proteins associated with light and clock regulation in eukaryotes, where the domains are normally organized in pairs and function in protein-protein interactions (39). More recently, PAS domains have been identified in a large family of prokaryotic and eukaryotic sensor proteins involved in sensing light, oxygen, redox status, and other signals (38, 39). So far, four different redox-responsive cofactors (flavin adenine dinucleotide, heme, [2Fe-2S], and 4-hydroxy-cinnamoyl), presumed to provide specific sensing capabilities, have been found associated with PAS domains (38, 39). It appears likely that the PAS domains of prokaryotic sensor proteins are involved in redox or oxygen sensing (39).
FIG. 5.
Alignment of the central PAS domain of the CitA-like proteins with homologous regions from eight representative proteins. Highly conserved (>70%) residues are boxed and are listed in the consensus sequence in uppercase letters, the absolutely conserved Asn residue is in boldface, relevant well-conserved (33 to 70%) residues are listed in lowercase letters. U, conserved bulky hydrophobic residues; o, conserved nonbulky hydrophobic residues. The PAS consensus sequence of Zhulin et al. (39) is shown for comparison. The S1 and S2 box regions are indicated by black lines (the broken lines indicate regions of conservation that extend beyond those regions reported by Zhulin et al. [38]). The residue number of the first amino acid displayed is shown for each sequence. The proteins are from the following organisms: Ec, E. coli; Kp, K. pneumoniae; Bs, B. subtilis; Sc, S. coelicolor; LL, Lactococcus lactis; and Av, Azotobacter vinelandii. Aer, NifL, AtoS, PhoR, and RseE are sensor histidine-kinases, YqiR and RocR are members of the ς54-dependent transcriptional activator family, and the other proteins are members of the CitA family.
The presence of PAS domains in the CitA-like proteins has, apparently, not been reported previously (see the “complete multiple alignment of PAS domains” referred to in Zhulin and Taylor [39]). This may be because the pattern of residue conservation for the CitA PAS domains differs somewhat from that of other PAS domains (Fig. 5). In particular, the S1 box on the CitA-like proteins is extended by 10 residues relative to that described by Zhulin and Taylor (38) and the C-terminal portion is poorly conserved (Fig. 5). Also, many of the residues conserved in the S2 box of the CitA-like proteins are not conserved in the PAS domains of other proteins (39). This suggests that the PAS domain of the CitA-like proteins may function differently from other PAS domains. Although the function of the PAS domain of the CitA-like proteins is unknown, it is likely to act either as a sensor (e.g., for oxygen or redox status) or in transmitting the signal of the sensor periplasmic domain to the C-terminal transmitter domain. Sensitivity of the DcuS-PAS domain to oxygen, nitrate, redox status, or metabolic status might partly explain the FNR-, NarL-, and CRP-independent regulation of dcuB expression by oxygen, nitrate, and glucose, respectively (12).
The 239-residue DcuR protein closely resembles (29 to 49% identity) the CitB-like proteins of the two-component regulator-receiver family (CitB of E. coli and K. pneumoniae, YdbG, YufM, and CitR/YflQ of B. subtilis; “CitR” of S. coelicolor; and Ygd1 of Bacillus megaterium). Since the sensor and regulators of the CitA- and CitB-like proteins form distinct groups it is likely that the CitB-like proteins act as the partner proteins for the corresponding CitA-like proteins, as for CitA-CitB of K. pneumoniae and DcuS-DcuR of E. coli. This conclusion is supported by the close juxtapositions of the corresponding genes on the chromosomes of the host organisms. The N-terminal region (residues 1 to 127) of the DcuR protein (and the other CitB-like proteins) is very similar to the histidine-kinase receiver module of other two-component regulator proteins. This region includes the aspartate residue (Asp-56 in DcuR) that is the site for phosphorylation in other receiver proteins (25). However, the C-terminal region (residues 128 to 239 for DcuR) appears to be unique to the CitB-like proteins. The C-terminal or “output” domains of response regulators are highly variable and normally function as DNA-binding domains. An analysis of the C-terminal domain of DcuR using the MOTIF program (http://www.genome.ad.jp/SIT/SIT.html) revealed a probable DNA-binding helix-turn-helix motif (residues 177 to 218) resembling that of the GntR and DeoR families of bacterial gene regulators. By analogy with other response regulators, it is likely that the C-terminal domain or output domain of DcuR is a DNA-binding domain enabling specific interaction with the promoter-operator regions of the genes in the DcuSR regulon. The DNA-binding activity of the output domain is likely to be regulated in response to the phosphorylation state of the receiver domain. The receiver domain would accept phosphate from the phosphorylated form of DcuS, which would autophosphorylate in response to the presence of external C4-dicarboxylates sensed by the periplasmic domain.
The CitB protein of K. pneumoniae is the only CitB-like protein that has been characterized (23). Gel retardation and DNase I footprinting studies showed that phospho-CitB binds with high affinity to multiple A+T-rich sites between the divergent promoters of the citC-citS intergenic region. However, no consensus sequence for the CitB-binding site has been established (23). The low DNA-binding affinities of the isolated CitB C-terminal domain and the unphosphorylated CitB protein indicated that CitB belongs to class I of the two-domain response regulators (23), in which interaction between the receiver and output domains inhibits receiver domain dimerization (9). It is therefore likely that DcuR and the other CitB-like proteins are also class I response regulators.
DISCUSSION
The studies described here reveal that E. coli contains a CitA-CitB-like two-component sensor-regulator system, designated DcuS-DcuR, that activates the transcription of the dcuB, frdABCD, and dctA genes in response to the presence of external C4-dicarboxylates. The DcuS protein contains a periplasmic input domain near the N-terminus that is presumed to sense C4-dicarboxylates (aspartate, fumarate, malate, maleate, and succinate), a central PAS domain of uncertain function, and a C-terminal transmitter domain. The DcuR protein contains an N-terminal receiver domain and a C-terminal output domain containing a potential DNA-binding helix-turn-helix motif. Thus, the DcuS-DcuR system possesses all the features required to function as a classical two-component response-regulator.
Many of the results of Zientz et al., reported while this paper was under review, are similar to those obtained here (41). Zientz et al. found that the DcuS-DcuR system activates frdA and dcuB expression in response to C4-dicarboxylates (41). However, the degrees of regulation reported by Zientz et al. (41) were just 80- and 2.5-fold inductions, respectively, whereas our data indicate 160- and 22-fold inductions, respectively. These differences could reflect the different lacZ fusions and growth media employed (such as the use of dimethyl sulfoxide rather than TMAO). Furthermore, Zientz et al. found that dctA expression is ∼threefold induced by succinate in both the wild-type and dcuR strains and that the dcuR mutation results in an ∼threefold lower expression for dctA (41). Consequently, it was concluded that dctA is not regulated by the DcuS-DcuR system in response to C4-dicarboxylates. Our results clearly show that dctA is regulated by DcuS-DcuR in response to C4-dicarboxylates. This discrepancy could once again be explained by the different growth media used (i.e., glycerol or succinate versus glycerol with or without succinate) or the use of different fusions. Zientz et al. showed that neither the nuo operon, the dcuC gene, nor the sdhC gene are DcuS-DcuR or C4-dicarboxylate regulated, and we show that the fumA gene is also not affected by these factors (41). The data presented here extends those of Zientz et al. (41) by revealing a growth defect for the dcuS mutant and by showing that a low-copy-number plasmid carrying the dcuSR genes complements the regulatory and growth defects of the dcuS mutant. In addition, we have experimentally determined the membrane topology of DcuS and shown that DcuS (and other CitA-like proteins) contains a central PAS domain. Also, our studies with a mutant deficient in anaerobic transport of C4-dicarboxylates show that the DcuS-DcuR system responds to external C4-dicarboxylates. A similar conclusion was made by Zientz et al. based on the finding that maleate (not transported by the Dcu systems) induces the DcuS-DcuR system anaerobically (41).
Some interesting questions remain to be answered concerning the nature of the DcuS-DcuR system: what factors influence dcuSR expression and how do these affect regulation of the DcuSR regulon; what is the nature of the DcuR DNA-binding site(s); which other genes (if any) are members of the DcuSR regulon; how does the DcuS-DcuR regulatory system regulate expression jointly with other regulators such as FNR, NarL, and CRP; does the DcuS-DcuR system contribute to the FNR-, NarL-, and CRP-independent regulation of dcuB by oxygen, nitrate, and glucose, respectively (12); and what is the function of the central PAS domain of DcuS? The answers to these questions await further investigation.
ACKNOWLEDGMENTS
We thank P. Poole for helpful comments on the manuscript and the BBSRC for a project grant (S.C.A. and J.R.G.), an Advanced Fellowship (S.C.A.) and a Special Studentship award (S.D.).
REFERENCES
- 1.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;6:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 3.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 3a.Bott, M., M. Meyer, and P. Dimroth. 1995. Unpublished data.
- 4.Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM β-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 5.Burland V, Plunkett G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. 6. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, S. J., P. Golby, D. Omrani, J. R. Guest, D. J. Kelly, and S. C. Andrews. Unpublished observations.
- 7.Devereux J. The GCG sequence analysis software package, version 6.0. Madison, Wis: Genetics Computer Group, Inc.; 1989. [Google Scholar]
- 8.Engel P, Kramer R, Unden G. Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from aerobic dicarboxylate uptake. J Bacteriol. 1992;174:5533–5539. doi: 10.1128/jb.174.17.5533-5539.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler U, Weiss V. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induced dimerization of receiver modules. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Forward J A, Behrendt M C, Wyborn N R, Cross R, Kelly D J. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctQMP genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J Bacteriol. 1997;179:5482–5493. doi: 10.1128/jb.179.17.5482-5493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. England: Cambridge University; 1984. [Google Scholar]
- 11.Golby P, Kelly D J, Guest J R, Andrews S. Topological analysis of DcuA, an anaerobic C4-dicarboxylate transporter of Escherichia coli. J Bacteriol. 1998;180:4821–4827. doi: 10.1128/jb.180.18.4821-4827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golby P, Kelly D J, Guest J R, Andrews S C. Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J Bacteriol. 1998;180:6586–6596. doi: 10.1128/jb.180.24.6586-6596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guest J R, Miles J S, Roberts R E, Woods S A. The fumarase genes of Escherichia coli: location of the fumB gene and discovery of a new gene (fumC) J Gen Microbiol. 1985;131:2971–2984. doi: 10.1099/00221287-131-11-2971. [DOI] [PubMed] [Google Scholar]
- 14.Hamblin M J, Shaw J G, Kelly D J. Sequence analysis and interposon mutagenesis of a sensor-kinase (DctS) and response-regulator (DctR) controlling synthesis of the high affinity C4-dicarboxylate transport system in Rhodobacter capsulatus. Mol Gen Genet. 1993;237:215–224. doi: 10.1007/BF00282803. [DOI] [PubMed] [Google Scholar]
- 15.Henderson P J F. The 12-transmembrane helix transporters. Curr Opin Cell Biol. 1993;5:708–721. doi: 10.1016/0955-0674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 16.Hofman K, Stoffel W. TMbase—a database of membrane spanning protein segments. J Biol Chem. 1993;347:166. [Google Scholar]
- 16a.Iuchi S, Kuritzkes D R, Lin E C C. Escherichia coli mutant with altered respiratory control of the frd operon. J Bacteriol. 1985;161:1023–1028. doi: 10.1128/jb.161.3.1023-1028.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Gu B, Albright L M, Nixon B T. Conservation between coding and regulatory elements of Rhizobium meliloti and Rhizobium leguminoserum dct genes. J Bacteriol. 1989;171:5244–5253. doi: 10.1128/jb.171.10.5244-5253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones H M, Gunsalus R P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J Bacteriol. 1987;169:3340–3349. doi: 10.1128/jb.169.7.3340-3349.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kay W W, Kornberg H L. The uptake of C4 dicarboxylic acids by Escherichia coli. Eur J Biochem. 1971;18:274–281. doi: 10.1111/j.1432-1033.1971.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Lo T C Y, Rayman K, Sanwal B D. Transport of succinate in Escherichia coli K12. J Biol Chem. 1972;247:6323–6331. [PubMed] [Google Scholar]
- 22.Martinez E, Bartolomé B, De La Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 23.Meyer M, Dimroth P, Bott M. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A + T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J Mol Biol. 1997;269:719–731. doi: 10.1006/jmbi.1997.1076. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for E. coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 25.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sarsaro J P, Pittard A J. Membrane topology analysis of Escherichia coli K-12 Mtr permease by alkaline phosphatase and β-galactosidase fusions. J Bacteriol. 1995;177:297–306. doi: 10.1128/jb.177.2.297-306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silhavy T J, Barman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 29.Six S, Andrews S C, Unden G, Guest J R. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct) J Bacteriol. 1994;176:6470–6478. doi: 10.1128/jb.176.21.6470-6478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiro S, Roberts R E, Guest J R. FNR-dependent repression of the ndh gene of Escherichia coli and metal ion requirement for FNR-regulated gene expression. Mol Microbiol. 1989;3:601–608. doi: 10.1111/j.1365-2958.1989.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 31.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1995;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson R V, Alex L A, Simon M I. Histidine and aspartate phosphorylation: two-component systems and the limits of homology. Trends Biochem Sci. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 33.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 34.Way J C, Davies M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 35.Woods S A, Guest J R. Differential roles of the Escherichia coli fumarases and fnr-dependent expression of fumarase B and aspartase. FEMS Microbiol Lett. 1987;48:219–224. [Google Scholar]
- 36.Yarosh O K, Charles T C, Finan T M. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Broome-Smith J K. Correct insertion of a simple eukaryotic plasma-membrane protein into the cytoplasmic membrane of Escherichia coli. Gene. 1990;96:51–57. doi: 10.1016/0378-1119(90)90340-w. [DOI] [PubMed] [Google Scholar]
- 38.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhulin I B, Taylor B L. Correlation of PAS domains with electron-transport associated proteins in completely sequenced microbial genomes. Mol Microbiol. 1998;29:1522–1523. [PubMed] [Google Scholar]
- 40.Zientz E, Six S, Unden G. Identification of a third secondary carrier (DcuC) for anaerobic C4-dicarboxylate transport in Escherichia coli: roles of the three Dcu carriers in uptake and exchange. J Bacteriol. 1996;178:7241–7247. doi: 10.1128/jb.178.24.7241-7247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zientz E, Bongaerts J, Unden G. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J Bacteriol. 1998;178:7241–7247. doi: 10.1128/jb.180.20.5421-5425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]