Abstract
By its functional interaction with a RecA polymer, the mutagenic UmuD′C complex possesses an antirecombination activity. We show here that MucA′B, a functional homolog of the UmuD′C complex, inhibits homologous recombination as well. In F− recipients expressing MucA′B from a Ptac promoter, Hfr × F− recombination decreased with increasing MucA′B concentrations down to 50-fold. In damage-induced pKM101-containing cells expressing MucA′B from the native promoter, recombination between a UV-damaged F lac plasmid and homologous chromosomal DNA decreased 10-fold. Overexpression of MucA′B together with UmuD′C resulted in a synergistic inhibition of recombination. RecA[UmuR] proteins, which are resistant to UmuD′C inhibition of recombination, are inhibited by MucA′B while promoting MucA′B-promoted mutagenesis efficiently. The data suggest that MucA′B and UmuD′C contact a RecA polymer at distinct sites. The MucA′B complex was more active than UmuD′C in promoting UV mutagenesis, yet it did not inhibit recombination more than UmuD′C does. The enhanced mutagenic potential of MucA′B may result from its inherent superior capacity to assist DNA polymerase in trans-lesion synthesis. In the course of this work, we found that the natural plasmid pKM101 expresses around 45,000 MucA and 13,000 MucB molecules per lexA(Def) cell devoid of LexA. These molecular Muc concentrations are far above those of the chromosomally encoded Umu counterparts. Plasmid pKM101 belongs to a family of broad-host-range conjugative plasmids. The elevated levels of the Muc proteins might be required for successful installation of pKM101-like plasmids into a variety of host cells.
SOS mutagenesis results from inaccurate trans-lesion DNA synthesis carried out by a DNA polymerase, probably DNA polymerase III, in conjunction with RecA and a mutagenic complex formed by the two SOS proteins UmuD′ and UmuC (for a review, see reference 44). The UmuD′C complex is thought to assist DNA polymerase in DNA synthesis across a lesion, while a RecA polymer acts as a directional chaperone to position the UmuD′C complex right at a lesion (6, 11, 38). A RecA polymer also acts as a recombinase in homologous recombination (for a review, see reference 22) and as a coprotease in the cleavage of LexA repressor and UmuD, the native form of the mutagenically active UmuD′ (for reviews, see references 21 and 44).
In addition to being an essential component of trans-lesion DNA synthesis, the UmuD′C complex is an antagonist of RecA-mediated recombination. When prematurely expressed at elevated intracellular levels, the UmuD′C complex prevents recombinational repair of UV-damaged DNA as well as recombination of undamaged DNA (37). The antirecombination activity of the UmuD′C complex correlates with its interaction with a RecA polymer (6, 38).
Here, we sought to determine if the property of the UmuD′C complex of inhibiting homologous recombination is shared by other mutagenic complexes. We tested the MucA′B complex, which is a member of a family of mutagenesis proteins structurally and functionally related to the UmuD′C complex (23, 46).
The MucA′B complex originates from plasmid pKM101, a variant of the multidrug resistance plasmid R46 (27). Like the chromosomal umuDC genes, the mucAB genes are organized in an operon and are regulated by LexA (12, 29). Like UmuD, the native MucA protein is processed into an active form, MucA′ (17, 35). Among the mutagenic repair genes, the mucAB genes from pKM101 appear to be the most efficient in promoting mutagenesis (5, 24). This property has led to their inclusion in the Ames tester strains, which detect environmental mutagens (25).
We found that the MucA′B complex inhibits homologous recombination as does its cellular UmuD′C counterpart. In F− recipients expressing MucA′B from a Ptac promoter, conjugational recombination decreased with increasing MucA′B concentrations down to 50-fold. In damage-induced pKM101-containing cells expressing MucA′B from the native promoter, recombination between a UV-damaged F lac plasmid and homologous chromosomal DNA also decreased markedly. Overexpression of MucA′B with UmuD′C results in a synergistic inhibition of recombination. Interestingly, the mutant RecA[UmuR] proteins, which are resistant to UmuD′C (38), remain sensitive to MucA′B. These data suggest that MucA′B and UmuD′C interact with a RecA polymer at distinct sites.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture, and media.
The bacterial strains and plasmids used are described in Tables 1 and 2, respectively. Plasmid pGY9411 was constructed by inserting the mucA′B NcoI-BamHI fragment of pRW294 into the vector pTrc99A under control of the Ptac promoter and the associated Shine-Dalgarno sequence. Plasmid pGY9463 (mucA′) was derived from pGY9411 by truncating the mucA′B operon at the BglII site, thus deleting 95% of mucB. Plasmid pGY9863 was derived from plasmid pGY9738 (o1cumuD′C) by replacing the mutant o1c with the wild-type operator. The three constructs were verified by sequencing through the modified regions.
TABLE 1.
Bacterial strains
| Strain | Sex and relevant markers | Source or reference |
|---|---|---|
| AB1157 (GY752) | F−leuB6 argE3 Strr | Laboratory collection |
| DM1420 (GY2901) | F−; as AB1157 but lexA51(Def) sfiA11 | Laboratory collection |
| Hfr3000 (GY306) | HfrH | Laboratory collection |
| GY4587 | F−; as AB1157 but sfiA11 | 38 |
| GY7962 | F−recA99 Δ(lac-pro) Spcr | 37 |
| GY8500 | F+ | 37 |
| GY8630 | F42-10 recA13 uvrB501 | 37 |
| GY9054 | F−; as GY9743 but Δ(srl-recA)306::Tn10 | This work |
| GY9427 | F−; as GY9743 but Δara leu::Tn10 | This work |
| GY9461 | F−; as GY9427 but carries pGY9738 (o1cumuD′C) | This work |
| GY9743 | F−; as GY4587 but ΔumuDC595::cat | This work |
| GY10229 | As Hfr3000 but ΔumuDC595::cat | This work |
| GY10419 | F−; as GY9743 but lacZ::kan | This work |
TABLE 2.
Plasmids
| Name | Vector plasmid | Descriptiona | Refer-ence(s) |
|---|---|---|---|
| F42-10 | Derivative of F lac, F42; carries a Tn10 insertion located outside the chromo-somal DNA; lac+ Tetr | 9, 37 | |
| pGB2 | Low-copy-number vector; Spcr | 8 | |
| pGY9738 | pGB2 | o1cumuD′C Spcr | 38 |
| pGY9863 | pGB2 | umuD′C Spcr | This work |
| pRW144 | pGB2 | mucAB Spcr | 18 |
| pRW294 | pGB2 | mucA′B Spcr | 41 |
| pTrc99A | Derivative of pBR322; Ptac Ampr | 1 | |
| pGY9411 | pTrc99A | Ptac::mucA′B Ampr | This work |
| pGY9463 | pTrc99A | Ptac::mucA′ Ampr | This work |
| pKM101 | mucAB Ampr | 27 | |
| pGW16 | Derivative of pKM101; o1cmucAB Ampr | 26 | |
| pGW208 | Derivative of pKM101; mucA421::Tn5 mucB+ Ampr Kanr | 34 | |
| pGY5887 | Mini-F | recA+ Kanr | 11 |
| pGY7104 | Mini-F | recA1730 Kanr | 11 |
| pGY9735 | Mini-F | recA410 Kanr | 38 |
| pGY9753 | Mini-F | recA411 Kanr | 38 |
| pGY9771 | Mini-F | recA415 Kanr | 38 |
Bacteria were grown at 37°C. The growth of the cultures was monitored by absorbance at 650 nm. Minimal medium was M9 supplemented with 0.2% (wt/vol) glucose, 30 μg of thiamine per ml, and the required amino acids. The rich media LBT and LAT, enriched minimal medium M9C, and low-arginine solid medium were as described previously (3, 6). Bacteria containing plasmids were grown in media supplemented with the appropriate antibiotics. Antibiotics and chemicals were used at the following final concentrations: streptomycin, 200 μg/ml; kanamycin, 50 μg/ml; tetracycline 10 μg/ml; ampicillin, 15 μg/ml for pKM101 or 100 μg/ml for derivatives of pTrc99A; chloramphenicol, 10 μg/ml; spectinomycin, 100 μg/ml for chromosomal spectinomycin resistance (Spcr) mutations or 50 μg/ml for derivatives of pGB2; rifampin, 100 μg/ml; 5-bromo-4-chloro-5-indolyl-β-d-galactoside (X-Gal), 40 μg/ml; and isopropyl-β-d-thiogalactopyranoside (IPTG), 0.5 mM if not otherwise stated.
Conjugational crosses.
As a rule, donor and recipient bacteria were grown in LBT to 2 × 108 ml−1 and mated on a filter at a 1.5:1 donor/recipient ratio. After 60 min, the mating mixture was vigorously shaken in M9, and diluted samples were plated on selective plates.
In conjugation experiments in which zygotic induction of prophage λ was determined, HfrH (λind553) donors and F− recipients were mated in liquid at a 1:10 donor/recipient ratio for 40 min. Phages resulting from zygotic induction were plated on a lawn of GY8500 (F+) indicator bacteria (6).
Rescue of a chromosomal lac marker by a UV-damaged F42-10 plasmid was measured by using a double mating procedure (16). The UV-damaged F42-10 plasmid was first transferred to an F− recA+ recipient, where recombination took place. The surviving plasmids were then transferred to a second indicator recipient that was recombination deficient and had the lac region deleted to determine their lac genotype.
Assay of UV mutagenesis.
Derivatives of AB1157 [argE3(Oc) Rifs] were grown in enriched minimal medium M9C to an optical density at 650 nm (OD650) of 0.3, centrifuged, resuspended in M9 buffer, and exposed to UV light. To measure argE3→arg+ mutagenesis, the UV-irradiated cells were plated on minimal medium containing a trace amount of arginine to select for the induced Arg+ revertants and on minimal medium fully supplemented with arginine for counting of the total number of surviving cells. To measure mutagenesis to rifampin resistance (Rifr), the UV-irradiated cells were diluted fivefold in LBT medium, incubated overnight, and plated on LAT plates with and without rifampin to determine the frequency of the induced Rifr mutants.
Quantitation of MucA, MucA′, and MucB.
Whole-cell extracts were prepared from a known amount of cells as described by Nastri et al. (28). Cell concentrations were determined by counting bacteria in a Malassez chamber with a microscope. Protein concentrations were measured with a protein assay kit (Bio-Rad) by the manufacturer’s microassay procedure. Portions of the cell extracts were run on a sodium dodecyl sulfate–15% polyacrylamide gel along with increasing amounts of purified MucA, MucA′, and MucB proteins (31) as standards. The proteins were transferred to a nitrocellulose membrane and treated with a 1:5,000 dilution of anti-MucA and -A′ antibodies (17) or with a 1:25,000 dilution of anti-MucB antibodies (43). MucA, MucA′, and MucB proteins were visualized by using 125I-labeled protein A, and the corresponding bands were scanned with a PhosphorImager and quantified with Image-Quant software (Molecular Dynamics).
RESULTS
A low level of MucA′ is expressed from plasmid pRW294.
In an effort to construct a mutant constitutively expressing the MucA′B complex, we found an unexpectedly low steady-state level of MucA′ in lexA(Def) bacteria carrying plasmid pRW294, which carries an engineered mucA′B operon (Table 3). In comparison, lexA(Def) bacteria carrying plasmid pRW144, a corresponding mucAB plasmid, had a 15-fold higher MucA concentration (11,000 MucA molecules versus 700 MucA′ molecules per cell) (Table 3). The steady-state level of MucB expressed from pRW294 was also reduced compared to that from pRW144 but not as dramatically (Table 3). MucA′ was efficiently produced from MucA in mitomycin-treated lexA(Def) cells, suggesting that the difference between MucA and MucA′ expression (Table 3) was not due to a high rate of degradation of MucA′ by a cellular protease. We also checked by sequencing that our isolate of plasmid pRW294 did not carry a mutation in the promoter or the Shine-Dalgarno sequence that could account for the poor MucA′ expression. We suggest that the deletion of the first 26 N-terminal codons of mucA, giving rise to mucA′ (36), might cause an unfavorable change in the mRNA secondary structure that might impair transcription or translation of the engineered mucA′B operon.
TABLE 3.
Concentrations of MucA, MucA′, and MucB proteins with expression from the native promotera
| Plasmid | Host genotype | Mitomycin (μg/ml) | Concn (molecules/cell)b
|
||
|---|---|---|---|---|---|
| MucA | MucA′ | MucB | |||
| pKM101 | lexA+ | 0 | <500 | <500 | <350 |
| 2.0 | 8,000 | 20,000 | 7,000 | ||
| lexA(Def) | 0 | 45,000 | 4,500 | 13,000 | |
| 2.5 | 22,500 | 48,000 | 13,500 | ||
| pRW144 | lexA(Def) | 0 | 11,000 | 2,000 | 2,500 |
| 2.5 | 700 | 16,000 | NDc | ||
| pRW294 | lexA(Def) | 0 | NAd | 700 | 500 |
| 2.5 | NA | 1,400 | 800 | ||
Strains GY10419 (lexA+) and DM1421 [lexA51(Def)] carrying the indicated plasmids were grown in LBT to an OD650 of 0.4, diluted twofold in the same medium, and incubated for 90 min with or without mitomycin.
Calculated from the MucA, -A′, and -B concentrations in cell extracts, the molecular masses of MucA (16,527 Da), MucA′ (13,570 Da), and MucB (46,321 Da), and the bacterial cell concentration from which cell extracts were prepared.
ND, not done.
NA, not applicable.
Elevated levels of Muc proteins are expressed from plasmid pKM101.
The data in Table 3 also show that lexA(Def) bacteria carrying plasmid pKM101 had around 45,000 MucA, 48,000 MucA′ (after mitomycin treatment), and 13,000 MucB molecules per cell. The concentration of RecA in the same cells, which provided an internal standard, was approximately 76,000 monomers per cell, a value in agreement with previous evaluations (6, 32).
The levels of the Muc proteins expressed from pKM101 appear to be surprisingly elevated, considering the low-level production of their chromosomally encoded Umu counterparts [2,500 UmuD and 200 UmuC molecules per lexA(Def) cell (45)]. Interestingly, the Muc levels expressed from pKM101 were also markedly higher than those expressed from plasmid pRW144, in which the mucAB operon has been subcloned (Table 3). This suggests that plasmid pKM101 may carry a regulatory element enhancing the expression of the mucAB operon upon SOS induction. In the leading region of pKM101 there are several conserved elements possessing a strong promoter and an SOS box-like sequence (10); however, none of these elements is in the mucAB orientation so as to play a regulatory role in MucAB expression. Alternatively, expression of MucAB may be sensitive to the level of supercoiling, which may differ between the large pKM101 plasmid and the small pRW144 plasmid. Finally, it should be noted that the levels of the Muc proteins expressed from pKM101 in an SOS-induced cell are far above those required for efficient mutagenesis (see below). This raises the question whether a high concentration of the Muc proteins is required for an additional plasmid pKM101 function.
Enhanced mutagenic activity of the MucA′B complex.
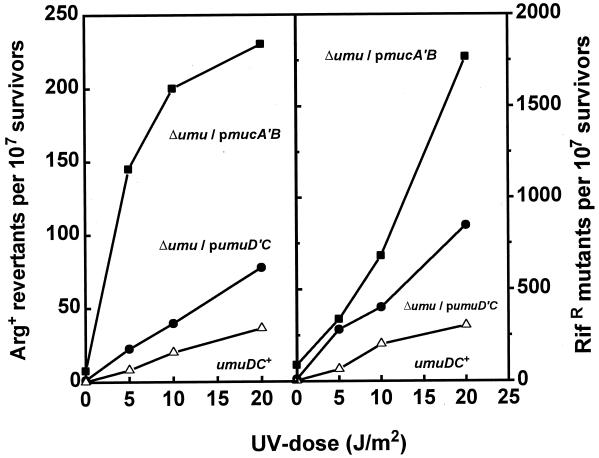
The poor expression of MucA′B from plasmid pRW294 nevertheless entailed high levels of SOS mutagenesis. Indeed, ΔumuDC bacteria carrying plasmid pRW294 were threefold more UV mutable than corresponding cells carrying a umuD′C plasmid and sixfold more UV mutable than wild-type umuDC+ bacteria (Fig. 1). These results are in line with those of Lawrence et al. (24) for single-stranded phage mutagenesis and indicate that the MucA′B complex is mutagenically more active than the UmuD′C complex. Thus, two factors appear to contribute to the elevated mutability of mucAB bacteria: speed of MucA processing (17) and efficient mutagenic activity of MucA′B.
FIG. 1.
The MucA′B complex is more mutagenic than UmuD′C. Derivatives of GY9743 (ΔumuDC) carrying plasmid pRW294 (mucA′B) (filled squares) or plasmid pGY9863 (umuD′C) (filled circles) and GY4587 (umuDC+) (open triangles) were assayed for UV-induced Arg+ (left panel) or Rifr (right panel) mutants after exposure to the UV doses indicated on the abscissa.
Inhibition of Hfr × F− recombination as a function of the MucA′B concentration in recipient bacteria.
We wanted to assess whether the MucA′B complex behaves as an inhibitor of homologous recombination. For this purpose, we measured Hfr × F− recombination in recipient bacteria in which the concentration of the MucA′B complex was made to vary by having the mucA′B genes placed under the control of a tac promoter. Expression of MucA′B from the hybrid operon was induced with increasing IPTG concentrations and monitored by measuring the levels of MucA′ and MucB. The concentration of the MucA′B complex was equated to the concentration of MucB that was produced in a limited amount relative to its MucA′ partner (Table 4).
TABLE 4.
Concentrations of MucA′ and MucB proteins with expression from a Ptac::mucA′B hybrid operona
| IPTG (μM) | Concn (molecules/cell)b
|
|
|---|---|---|
| MucA′ | MucB | |
| 0 | 1,300 | 350 |
| 10 | 6,000 | 1,600 |
| 50 | 51,000 | 14,000 |
| 100 | 90,000 | 19,500 |
| 500 | 110,000 | 21,000 |
Strain GY9427 (lexA+ lacY) carrying plasmid pGY9411 (Ptac::mucA′B) was grown in LBT to an OD650 of 0.2 and incubated for an additional 90 min in the same medium supplemented with IPTG at the indicated concentrations.
See Table 3, footnote b.
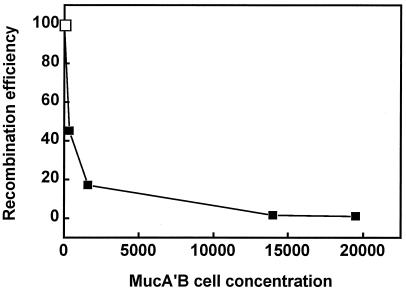
The efficiency of conjugational recombination decreased sharply when the concentration of the MucA′B complex increased (Fig. 2). A basal level of MucA′B, around 350 MucA′B complexes per cell, reduced the yield of recombinants twofold as compared to that of a corresponding recipient devoid of MucA′B. When the MucA′B concentration increased to around 1,600 complexes per cell, recombination decreased sixfold. A 10-fold-higher MucA′B concentration inhibited recombination more than 50-fold. However, a greater increase in the MucA′B concentration did not reduce further the recombination efficiency. We checked that overexpression of the MucA′B complex did not prevent the transmission of the Hfr DNA by measuring zygotic induction of prophage λ in parallel crosses with a lysogenic derivative of the HfrH donor (6) (data not shown).
FIG. 2.
Inhibition of Hfr × F− recombination as a function of increasing MucA′B cell concentration. GY9427 (F− leu::Tn10 ΔumuDC) (open symbol) and its derivative carrying plasmid pGY9411 (Ptac::mucA′B) (filled symbols) were mated with GY306 (HfrH), and selection was for Leu+ Strr recombinants. Prior to mating, expression of MucA′B in the recipient bacteria was induced with IPTG as described in Table 4, footnote a, so as to attain the MucA′B cell concentrations indicated on the abscissa.
Both the MucA′ and MucB proteins are required to inhibit homologous recombination. Indeed, there was no reduction of recombination in the absence of MucB in recipients producing MucA′ alone (data not shown). Furthermore, the native MucAB complex produced at an elevated concentration from plasmid pGW16, an operator-constitutive derivative of pKM101 (26), inhibited recombination only twofold (data not shown).
We conclude that the mutagenically active MucA′B complex, like the UmuD′C complex, possesses an antirecombination activity; however, the present results, compared with the data of Sommer et al. (37) and of Boudsocq et al. (6), suggest that MucA′B is rather less efficient than UmuD′C in inhibiting homologous recombination. Indeed, compared to UmuD′C, a higher concentration of the MucA′B complex is required for inhibiting recombination. In our system, about 1,600 MucA′B complexes per cell caused a sixfold drop in recombination, whereas a concentration of 700 UmuD′C complexes per cell induced from a PBAD promoter or 440 UmuD′C complexes per cell constitutively expressed from a mutant operator produced a similar inhibition (6, 37).
pKM101-promoted inhibition of recombination between an incoming UV-damaged F lac plasmid and homologous chromosomal DNA in SOS-induced recipient cells.
Does the MucA′B complex expressed from its natural promoter under physiological conditions, i.e., in damage-induced pKM101-containing cells, inhibit homologous recombination? To answer this question, we tested whether pKM101 would inhibit recombination between an incoming UV-damaged F lac plasmid and homologous chromosomal DNA in recipients that had been induced with mitomycin before the F conjugal transfer.
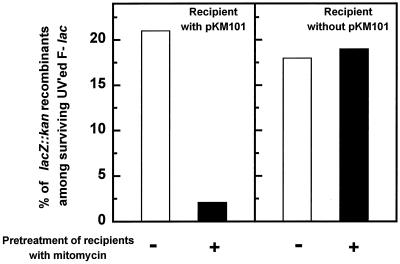
Recombination of the UV-damaged F lac plasmid occurred at a high frequency in a recipient devoid of pKM101. Indeed, about 20% of the surviving plasmids were found to have incorporated the chromosomal lacZ::kan allele (Fig. 3, right panel).
FIG. 3.
pKM101-promoted inhibition of recombination between an incoming UV-damaged (UV’ed) F lac plasmid and homologous chromosomal DNA in recipients induced with mitomycin. GY8630 (recA13 uvrB501/F42-10) donors exposed to 25 J of UV light m2 were mated with GY10419 (F− lacZ::kan ΔumuDC Strr) (right panel) or its derivative carrying plasmid pKM101 (left panel) pretreated for 90 min with 0 (open bars) or 2 (filled bars) μg of mitomycin per ml. After this first mating, the exconjugants were mated again with the indicator recipient GY7962 [F− recA99(Am) Δlac Spcr] and plated on X-Gal plates supplemented with tetracycline and spectinomycin and with or without kanamycin to determine the frequencies of lacZ::kan recombinants as indicated on the ordinate.
An elevated recombination frequency was also observed in a recipient carrying pKM101 in the absence of an SOS-inducing pretreatment (Fig. 3, left panel). In contrast, when these cells were treated with mitomycin, the yield of F lacZ::kan recombinants decreased 10-fold (Fig. 3, left panel). This drop in recombination is correlated with the enhanced production of the Muc proteins, which were found to increase from very low basal levels (below our detection limits) to around 8,000 MucA, 20,000 MucA′, and 7,000 MucB molecules per cell after mitomycin induction (Table 3). Furthermore, no recombination inhibition occurred when pKM101 carried a mucA421::Tn5 mutation (data not shown).
Conjugal transfer of UV-damaged single-stranded DNA generates discontinuities (DNA gaps) in the complementary strand synthesized in the recipient (19). Similar gaps arise after replication of damaged chromosomal DNA (30). Our results suggest that the MucA′B complex, when it reaches a high enough concentration after DNA damage, inhibits the recombinational filling of unresolved postreplicative gaps.
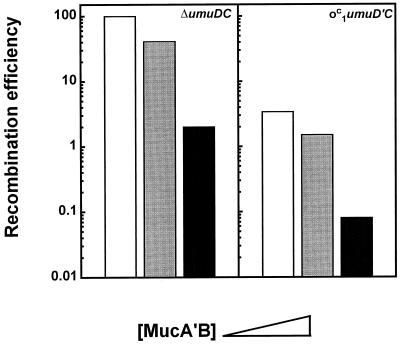
Synergism of MucA′B and UmuD′C in inhibiting recombination.
The kinetics of recombination inhibition by MucA′B shown in Fig. 2 indicates that the target sites become saturated at high MucA′B concentrations. We asked whether recombination could be further inhibited if MucA′B was expressed in a recipient that also overproduces the UmuD′C complex. For this purpose, the Ptac::mucA′B construct was introduced into GY9461, an o1c-umuD′C recipient constitutively expressing around 1,000 UmuD′C complexes per cell (37) (data not shown). In the absence of MucA′B, GY9461 bacteria showed a 30-fold decrease in recombination proficiency (Fig. 4, right panel) (37). However, when MucA′B was also expressed in the same cells, recombination was further inhibited (Fig. 4, right panel). At a MucA′B concentration of around 20,000 complexes per cell, recombination was decreased more than 1,000-fold, while it decreased only 50-fold in the absence of UmuD′C (Fig. 4, left panel).
FIG. 4.
Synergistic inhibitory action of MucA′B and UmuD′C on recombination. GY9427 (F− leu::Tn10 ΔumuDC) (left panel) and GY9461 (F− leu::Tn10 ΔumuDC/o1cumuD′C) (right panel) and their derivatives carrying plasmid pGY9411 (Ptac::mucA′B) were mated with GY306 (HfrH). Prior to mating, expression of MucA′B in the recipients was induced with 0 or 500 μM IPTG. Open bars, recipients without MucA′B; gray and dark bars, recipients expressing MucA′B from a Ptac promoter at basal or IPTG-induced levels, respectively.
These data suggest that MucA′B and UmuD′C may act synergistically in inhibiting recombination. MucAB and UmuDC have diverged during evolution and do not form mixed complexes (29, 33). The observed synergism of MucA′B and UmuD′C can be accounted for if each complex contacts a RecA polymer at distinct sites. Indeed, if each of the two mutagenic complexes binds independently to a RecA polymer at distinct sites, one can explain why when the interaction sites of one complex are saturated, the other complex can still bind to a RecA polymer, inhibiting further recombination.
RecA[UmuR] proteins are sensitive to recombination inhibition by MucA′B.
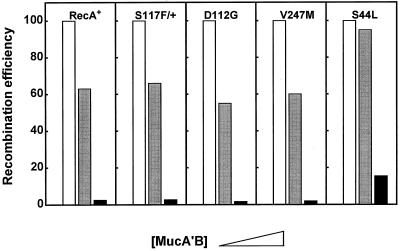
We asked whether the recently isolated mutant RecA[UmuR] proteins, which overcome UmuD′C recombination inhibition (38), are also sensitive to recombination inhibition by MucA′B. We tested mutants RecA D112G, S117F, V247L, and S44L, which have mutations located at the three sites where the [UmuR] amino acid changes have been shown to occur: the tail domain (D112G and S117F), the head domain (V247L), and the core of a RecA monomer (S44L) (38). RecA S117F, which is dominant over RecA+ in conferring Umu resistance (38), was used in a diploid combination with RecA+.
The sensitivity of the recA[UmuR] bacteria to MucA′B was assessed by measuring the efficiency of conjugational recombination in derivatives expressing the MucA′B complex from a Ptac promoter either at a low basal level or at an elevated IPTG-induced concentration (Fig. 5). As seen in Fig. 5, the RecA[UmuR] proteins were sensitive to MucA′B recombination inhibition. RecA mutants D112G, S117F (in the diploid combination with RecA+), and V247L behaved like wild-type RecA+. RecA S44L partially escaped recombination inhibition, yet recombination was still decreased 10-fold at an elevated MucA′B concentration (Fig. 5).
FIG. 5.
RecA[UmuR] proteins are sensitive to recombination inhibition by MucA′B. Derivatives of GY9054 (F− leuB6 ΔrecA ΔumuDC/mini-F-recA+, -recA410, -recA411, or -recA415) and of GY9743 (F− leuB6 recA+ ΔumuDC/mini-F-recA1730) carrying plasmid GY9411 (Ptac::mucA′B) (gray and dark bars) or the vector pTrc99A (open bars) were crossed with GY10229 (HfrH). Prior to mating, expression of MucA′B in the recipients was induced with 0 (gray bars) or 500 (dark bars) μM IPTG.
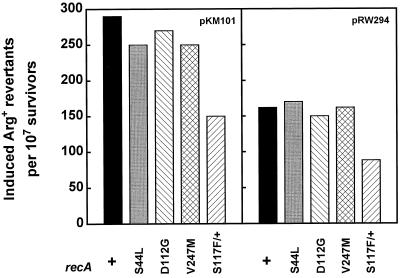
The MucA′B complex promotes efficient mutagenesis in recA[UmuR] bacteria.
It has been shown that the recA[UmuR] bacteria have a reduced UV mutability, a defect that has been attributed to a poor interaction of a RecA[UmuR] polymer with the UmuD′C complex (38). If the MucA′B complex interacts rather efficiently with a RecA[UmuR] protein, one might expect that it would alleviate the mutability defect of a recA[UmuR] strain. Indeed, recA[UmuR] bacteria carrying either pKM101, the natural mucAB plasmid, or plasmid pRW294, which expresses the MucA′B complex at a low level, promoted UV mutagenesis efficiently (Fig. 6). Mutant recA S44L, D112G, and V247L displayed the same level of Muc-promoted mutability as recA+. The recA+/recA S117F heterogenote, which is the most affected in Umu-promoted mutagenesis (38), also recovered about 60% of the wild-type mutability in the presence of MucA′B.
FIG. 6.
Efficient Muc-promoted mutagenesis in recA[UmuR] bacteria. Derivatives of GY9054 (ΔrecA ΔumuDC/mini-F-recA+, -recA410, -recA411, or -recA415) and of GY9743 (recA+ ΔumuDC/mini-F-recA1730) carrying plasmid pKM101 (left panel) or plasmid pRW294 (right panel) were assayed for Arg+ reversion after exposure to 20 J of UV light m2.
DISCUSSION
Antirecombination activity of mutagenic complexes.
We demonstrated here that the MucA′B complex from plasmid pKM101 inhibits homologous recombination as its cellular homolog UmuD′C does. An antirecombination activity might be a general property shared by other members of the family of Umu-like mutagenesis proteins and might be related to their common ability to interact with a RecA polymer (7, 13, 23).
Recombination and mutagenesis are two repair processes that take place sequentially on the single-stranded discontinuities (DNA gaps) arising after replication of damaged DNA (30). The two processes require single-stranded DNA around which a RecA polymer wraps. Most RecA-coated gaps are first mended by recombinational repair. At the remaining gaps, the binding of a mutagenic complex to RecA prevents recombination from occurring and switches repair into a mutagenic pathway. The induced mutability might be the product of the capacity of a mutagenic complex to assist DNA polymerase in trans-lesion synthesis coupled with its antagonistic action on recombination.
The enhanced mutagenic activity of the MucA′B complex is not correlated with an enhanced antirecombination activity.
The MucAB proteins are more proficient in mutagenesis than the related UmuDC proteins (5, 23). This difference has been attributed to more efficient processing of MucA than of UmuD (17). Our data demonstrate that the MucA′B complex possesses an inherent superior capacity to stimulate cellular mutagenesis.
Two sets of interactions might be specifically relevant to the MucA′B mutagenic activity: (i) an interaction of MucA′B with a RecA polymer (13), which might target the MucA′B complex at a DNA lesion while inhibiting recombination, and (ii) an interaction of MucA′B with subunits of DNA polymerase III and with single-stranded-DNA-binding protein (31, 42), which might account for the function of the MucA′B complex to stimulate trans-lesion synthesis. A comparison of our present data with those of Sommer et al. (37) and of Boudsocq et al. (6) indicates that quantitatively MucA′B does not inhibit recombination more than UmuD′C does. Thus, it is unlikely that the MucA′B complex binds a RecA polymer with a greater affinity. The enhanced mutagenic potential of the MucA′B complex seems to be more correlated with an increased efficiency in helping DNA polymerase to synthesize across a large range of DNA lesions. Such a view is supported by the work of Lawrence et al. (24), who showed that the MucA′B complex per se is more active than UmuD′C in promoting replication past a defined abasic site carried by a single-stranded DNA vector.
MucA′B and UmuD′C may interact with a RecA polymer at different sites.
The MucA′B complex also relies on the RecA chaperoning function to be active in mutagenesis (reference 36 and unpublished results). It is thought that, like UmuD′C, the MucA′B complex is targeted at a DNA lesion through its interaction with a RecA polymer (3, 13). Our data suggest that the two mutagenic complexes might contact a RecA polymer at distinct sites.
We observed, indeed, that the recently isolated RecA[UmuR] proteins, which are resistant to UmuD′C inhibition of recombination (38), are sensitive to recombinational inhibition by MucA′B and support MucA′B-promoted mutagenesis efficiently. The mutant RecA[UmuR] proteins seem to interact efficiently with MucA′B. An interaction of MucA′ with a RecA[UmuR] polymer, namely, RecA S117F, has been demonstrated in vitro (13).
Two hypotheses may account for the interaction of MucA′B with a mutant RecA[UmuR] polymer: (i) MucA′B proteins may have evolved to be functional in bacteria divergent from Escherichia coli and may bind RecA with a relaxed specificity, and (ii) the binding of MucA′B to RecA may involve specific interactions, with MucA′B and UmuD′C contacting a RecA polymer at distinct sites. Our findings that MucA′B and UmuD′C act synergistically in inhibiting recombination support the second hypothesis. If each of the two mutagenic complexes binds a RecA polymer independently at distinct sites, then one can explain why when the interaction sites of one complex are saturated, the other complex can still bind to a RecA polymer, inhibiting further recombination.
A test of these models awaits the isolation of recA mutations specifically altering RecA-MucA′B interactions. It is interesting that RecA S44L was less sensitive to recombination inhibition by MucA′B than the other RecA[UmuR] proteins. The S44 amino acid lies in the core of a RecA monomer (39). Should the S44L amino acid change have a far-reaching effect on the overall structure of a RecA monomer, it might partially alter a putative MucA′B binding site.
Elevated levels of Muc proteins are expressed from the plasmid pKM101.
In the course of this work, we found that the concentrations of the Muc proteins expressed from the natural plasmid pKM101 in an SOS-induced cell are particularly elevated, taking into account the low-level production of their chromosomally encoded Umu counterparts. For instance, the level of MucA expressed from pKM101 in lexA(Def) bacteria, devoid of LexA, was 18 times higher than that of UmuD (45,000 MucA molecules per cell versus 2,500 UmuD molecules per cell) (Table 3) (45).
We speculate that an elevated level of the plasmid-encoded mutagenic proteins might be required for an additional plasmid function. pKM101 belongs to the IncN group of broad-host-range conjugative plasmids. We suggest that the muc genes may become transiently induced during conjugation and may facilitate the installation of pKM101 into a variety of host cells. Although they are not essential for conjugal transfer under laboratory conditions, the Muc proteins may enhance the fitness of plasmid transmission if they speed up replication of the transferred plasmid single-stranded DNA by interacting with the host DNA polymerase. Interestingly, in pKM101 the muc operon is located in the leading region, the first segment that enters the recipient cell during conjugation (4). The leading regions of many conjugative plasmids carry installation genes that are thought to promote the establishment of an immigrant plasmid into a new host cell (2, 14, 20).
Bacterial conjugation mediates genetic exchange not only between cells of the same species but also between members of distantly related and even unrelated genera. This promiscuous gene transfer provides a mechanism for making available a huge pool of genes for bacterial evolution (for a review, see reference 15). In this regard, it is significant that half of the mutagenic repair genes are carried by conjugative plasmids (40).
ACKNOWLEDGMENTS
We benefited from the qualified assistance of D. Bouillon and G. Coste in genetic and molecular biology experiments. We thank M. Pierre for her help in preparing the manuscript. We are grateful to R. Woodgate for providing anti-MucA and -A′ and anti-MucB antibodies as well as valuable strains and to Z. Livneh for the generous gift of purified MucA, MucA′, and MucB proteins. We thank R. Woodgate, A. Roca, and P. Stragier for helpful discussions and comments and R. Devoret for his constant advice.
C. Venderbure is the recipient of a fellowship from the Ministère de l’Education Nationale de la Recherche et de la Technologie. F. Boudsocq benefited from fellowships from the Association pour la Recherche sur le Cancer and from the Fondation pour la Recherche Médicale. This work was supported by grants to A. Bailone from the Ligue Nationale contre le Cancer, from the Association pour la Recherche sur le Cancer (no. 9601), and from the Ministère de l’Education Nationale de la Recherche et de la Technologie (AS Radiobiologie 98-15) and by funds to UMR 216 from the Institut Curie and from the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 2.Bagdasarian M M, Bailone A, Angulo J, Scholz P, Bagdasarian M, Devoret R. PsiB, an anti-SOS protein, is transiently expressed by the F sex factor during its transmission to an Escherichia coli K12 recipient. Mol Microbiol. 1992;6:885–893. doi: 10.1111/j.1365-2958.1992.tb01539.x. [DOI] [PubMed] [Google Scholar]
- 3.Bailone A, Sommer S, Knezevic J, Dutreix M, Devoret R. A RecA protein mutant deficient in its interaction with the UmuDC complex. Biochimie. 1991;73:479–484. doi: 10.1016/0300-9084(91)90115-h. [DOI] [PubMed] [Google Scholar]
- 4.Belogurov A A, Delver E P, Rodzevich O V. IncN plasmid pKM101 and IncI1 plasmid ColIb-P9 encode homologous antirestriction proteins in their leading regions. J Bacteriol. 1992;174:5079–5085. doi: 10.1128/jb.174.15.5079-5085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco M, Herrera G, Aleixandre V. Different efficiency of UmuDC and MucAB proteins in UV light induced mutagenesis in Escherichia coli. Mol Gen Genet. 1986;205:234–239. doi: 10.1007/BF00430433. [DOI] [PubMed] [Google Scholar]
- 6.Boudsocq F, Campbell M, Devoret R, Bailone A. Quantitation of the inhibition of Hfr × F− recombination by the mutagenesis complex UmuD′C. J Mol Biol. 1997;270:201–211. doi: 10.1006/jmbi.1997.1098. [DOI] [PubMed] [Google Scholar]
- 7.Bruck I, Woodgate R, McEntee K, Goodman M F. Purification of a soluble UmuD′C complex from Escherichia coli. Cooperative binding of UmuD′C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 8.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 9.Clark A J, Low K B. Pathways and systems of homologous recombination in Escherichia coli. In: Low K B, editor. The recombination of genetic material. San Diego, Calif: Academic Press, Inc.; 1988. pp. 155–215. [Google Scholar]
- 10.Delver E P, Belogurov A A. Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J Mol Biol. 1997;271:13–30. doi: 10.1006/jmbi.1997.1124. [DOI] [PubMed] [Google Scholar]
- 11.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J R, Walker G C, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elledge S J, Walker G C. The muc genes of pKM101 are induced by DNA damage. J Bacteriol. 1983;155:1306–1315. doi: 10.1128/jb.155.3.1306-1315.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank E G, Hauser J, Levine A S, Woodgate R. Targeting of the UmuD, UmuD′ and MucA′ mutagenesis proteins to DNA by RecA protein. Proc Natl Acad Sci USA. 1993;90:8169–8173. doi: 10.1073/pnas.90.17.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golub E, Bailone A, Devoret R. A gene encoding an SOS inhibitor is present in different conjugative plasmids. J Bacteriol. 1988;170:4392–4394. doi: 10.1128/jb.170.9.4392-4394.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiney D G. Broad host range conjugative and mobilizable plasmids in gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 75–103. [Google Scholar]
- 16.Hall J D, Howard-Flanders P. Recombinant F′ factors from Escherichia coli K-12 strains carrying recB or recC. J Bacteriol. 1972;110:578–584. doi: 10.1128/jb.110.2.578-584.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser J, Levine A S, Ennis D G, Chumakov K M, Woodgate R. The enhanced mutagenic potential of the MucAB proteins correlates with the highly efficient processing of the MucA protein. J Bacteriol. 1992;174:6844–6851. doi: 10.1128/jb.174.21.6844-6851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho C, Kulaeva O I, Levine A S, Woodgate R. A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446B, and R471a. J Bacteriol. 1993;175:5411–5419. doi: 10.1128/jb.175.17.5411-5419.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard-Flanders P, Rupp W D, Wilkins B M, Cole R S. DNA replication and recombination after UV-irradiation. Cold Spring Harbor Symp Quant Biol. 1968;33:195–305. doi: 10.1101/sqb.1968.033.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Jones A L, Barth P T, Wilkins B M. Zygotic induction of plasmid ssb and psiB genes following conjugative transfer of IncI1 plasmid ColIb-P9. Mol Microbiol. 1992;6:605–613. doi: 10.1111/j.1365-2958.1992.tb01507.x. [DOI] [PubMed] [Google Scholar]
- 21.Koch W H, Woodgate R. The SOS response. In: Nickoloff J A, Hoeskstra M F, editors. DNA damage and repair. Vol. 1. Totowa, N.J: Humana Press, Inc.; 1997. pp. 107–134. [Google Scholar]
- 22.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence C W, Borden A, Woodgate R. Analysis of the mutagenic properties of the UmuDC, MucAB and RumAB proteins, using a site-specific abasic lesion. Mol Gen Genet. 1996;251:493–498. doi: 10.1007/BF02172378. [DOI] [PubMed] [Google Scholar]
- 25.McCann J, Choi E, Yamazaki E, Ames B N. Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals. Proc Natl Acad Sci USA. 1975;72:5135–5139. doi: 10.1073/pnas.72.12.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNally K P, Freitag N E, Walker G C. LexA-independent expression of a mutant mucAB operon. J Bacteriol. 1990;172:6223–6231. doi: 10.1128/jb.172.11.6223-6231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortelmans K E, Stocker B A D. Segregation of the mutator property of plasmid R46 from its ultraviolet-protecting property. Mol Gen Genet. 1979;167:317–327. doi: 10.1007/BF00267425. [DOI] [PubMed] [Google Scholar]
- 28.Nastri H G, Guzzo A, Lange C S, Walker G C, Knight K L. Mutational analysis of the RecA protein L1 region identifies this area as a probable part of the co-protease substrate binding site. Mol Microbiol. 1997;25:967–978. doi: 10.1111/j.1365-2958.1997.mmi533.x. [DOI] [PubMed] [Google Scholar]
- 29.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rupp W D, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 31.Sarov-Blat L, Livneh Z. The mutagenesis protein MucB interacts with single strand DNA binding protein and induces a major conformational change in its complex with single-stranded DNA. J Biol Chemistry. 1998;273:5520–5527. doi: 10.1074/jbc.273.10.5520. [DOI] [PubMed] [Google Scholar]
- 32.Sassanfar M, Roberts J W. Nature of the SOS inducing signal in E. coli: the involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 33.Sedgwick S G, Lodwick D, Doyle N, Crowne H, Strike P. Functional complementation between chromosomal and plasmid mutagenic DNA repair genes in bacteria. Mol Gen Genet. 1991;229:428–436. doi: 10.1007/BF00267466. [DOI] [PubMed] [Google Scholar]
- 34.Shanabruck W G, Walker G C. Localization of the plasmid (pKM101) gene(s) involved in recA+ lexA+-dependent mutagenesis. Mol Gen Genet. 1980;179:289–297. doi: 10.1007/BF00425456. [DOI] [PubMed] [Google Scholar]
- 35.Shiba T, Iwasaki H, Nakata A, Shinagawa H. Proteolytic processing of MucA protein in SOS mutagenesis: both processed and unprocessed MucA may be active in the mutagenesis. Mol Gen Genet. 1990;224:169–176. doi: 10.1007/BF00271549. [DOI] [PubMed] [Google Scholar]
- 36.Slater S C, Maurer R. Requirement for bypass of UV-induced lesions in single-stranded DNA of bacteriophage ΦX174 in Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:1251–1255. doi: 10.1073/pnas.88.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer S, Bailone A, Devoret R. The appearance of the UmuD′C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 38.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol Microbiol. 1998;28:281–292. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 39.Story R M, Weber I T, Steitz T A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 40.Strike P, Lodwick D. Plasmid genes affecting DNA repair and mutation. J Cell Sci (Suppl) 1987;6:303–321. doi: 10.1242/jcs.1984.supplement_6.20. [DOI] [PubMed] [Google Scholar]
- 41.Szekeres E S, Woodgate R, Lawrence C W. Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis. J Bacteriol. 1996;178:2559–2563. doi: 10.1128/jb.178.9.2559-2563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tadmor Y, Ascarelli-Goell R, Skaliter R, Livneh Z. Overproduction of the beta subunit of DNA polymerase III holoenzyme reduces UV mutagenesis in Escherichia coli. J Bacteriol. 1992;174:2517–2524. doi: 10.1128/jb.174.8.2517-2524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanooka H, Tanaka K, Shinozaki K. Heterospecific expression of misrepair-enhancing activity of mucAB in Escherichia coli and Bacillus subtilis. J Bacteriol. 1991;173:2906–2914. doi: 10.1128/jb.173.9.2906-2914.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker G C. SOS-regulated proteins in translesion DNA synthesis and mutagenesis. Trends Biochem Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 45.Woodgate R, Ennis D G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 46.Woodgate R, Sedgwick S G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]