Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the seventh member of the coronavirus family that can infect humans. Recently, more contagious and pathogenic variants of SARS‐CoV‐2 have been continuously emerging. Clinical candidates with high efficacy and ready availability are still in urgent need. To identify potent anti‐SARS‐CoV‐2 repurposing drugs, we evaluated the antiviral efficacy of 18 selective estrogen receptor modulators (SERMs) against SARS‐CoV‐2 infection. Six SERMs exhibited excellent anti‐SARS‐CoV‐2 effects in Vero E6 cells and three human cell lines. Clomifene citrate, tamoxifen, toremifene citrate, and bazedoxifene acetate reduced the weight loss of hamsters challenged with SARS‐CoV‐2, and reduced hamster pulmonary viral load and interleukin‐6 expression when assayed at 4 days postinfection. In particular, bazedoxifene acetate was identified to act on the penetration stage of the postattachment step via altering cholesterol distribution and endosome acidification. And, bazedoxifene acetate inhibited pseudoviruses infection of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV. These results offer critical information supporting bazedoxifene acetate as a promising agent against coronaviruses.
Keywords: bazedoxifene acetate, cholesterol distribution, endosome acidification, selective estrogen receptor modulator, severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is yet far from being under control. 1 At present, SARS‐CoV‐2 variants continuously arise and the variants of concern include Alpha, Beta, Gamma, Delta, and Omicron. 2 Although various types of SARS‐CoV‐2 vaccines have been used for vaccinations in a large scale, recent studies indicate that existing vaccines are less effective in protecting against SARS‐CoV‐2 variants. 3 , 4 , 5 And there are only a few drugs approved for clinical treatment of COVID‐19. 6 , 7 Currently, the severity of the ongoing global pandemic still urges the efforts to discover new antiviral compounds. 7
Compared with novel drug development requiring both large budgets and long lead times, drug repurposing is a faster way to discern new treatment candidates for emerging infectious diseases. In our previous screening work, five selective estrogen receptor modulators (SERMs) were identified to inhibit the infection of SARS‐CoV‐2. 8 To further evaluate the anti‐SARS‐CoV‐2 efficacy of SERMs, 18 SERMs available in the market were selected for further evaluation of the antiviral activity in vitro and in vivo. Six SERMs potently inhibited SARS‐CoV‐2 infection in four cell lines. And, four SERMs showed prophylactic anti‐SARS‐CoV‐2 efficacy in vivo using Syrian hamsters challenged with SARS‐CoV‐2. The role of the four SERMs in the SARS‐CoV‐2 life cycle was subjected to further mechanism investigation. The results showed that bazedoxifene acetate (Baze‐A) acted on the penetration stage of the post‐attachment step via altering the cholesterol distribution and endosome acidification. Furthermore, Baze‐A inhibited the pseudoviruses infection of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV, indicating that Baze‐A is a promising antiviral candidate for coronavirus infection.
2. MATERIALS AND METHODS
2.1. Cells, viruses, and compounds
The African green monkey kidney cell line Vero E6 (kindly provided by Dr. Rong Zhang, Fudan University), human epithelial colorectal adenocarcinoma cell line Caco‐2, and human cervical cancer cell lines overexpressing human ACE2 (Hela‐ACE2) 8 were maintained in Gibco Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids, 1% l‐glutamine and 1% penicillin/streptomycin at 37℃ and 5% CO2. The human lung adenocarcinoma cell line Calu‐3 was cultured in Gibco DMEM: F‐12 supplemented with 15% FBS and 1% penicillin/streptomycin. The SARS‐CoV‐2 strain (GenBank accession number: 622319) was isolated from a laboratory‐confirmed COVID‐19 patient. 9 All experiments involving live SARS‐CoV‐2 were performed in a Biosafety Level 3 (BLS‐3) laboratory of the Navy Medical University. All compounds used in the experiments were purchased from Selleck Chemicals.
2.2. Dose‐response analysis and cytotoxicity assay
Cell lines were plated in a 96‐well plate (4 × 104 cells/well). Then, cells were infected with SARS‐CoV‐2 in the presence of the indicated compounds with serial dilutions (30 μM–0.9375 μM). DMSO was the negative control and remdesivir (Rem) was considered as the positive control. Vero E6, Caco‐2, Hela‐ACE2, and Calu‐3 cells were infected with SARS‐CoV‐2 at MOI of 0.1, 0.1, 0.1, and 1, respectively. SARS‐CoV‐2 infection was detected using immunofluorescence at 24 h postinfection (hpi). Simultaneously, cells were plated in parallel and continuously exposed to various concentrations of the compounds for 48h. Compound cytotoxicity was determined using a Cell Counting Kit‐8 (Beyotime). The results were detected using the Synergy H1 microplate reader (BioTek Instruments) with absorbance at 450nm. The EC50 and CC50 values were calculated using GraphPad Prism 9.
2.3. Immunofluorescence assay
Infected cells were fixed with methanol. After blocking with 3% bovine serum albumin for 2 h, the cells were incubated overnight at 4℃ with a rabbit‐anti‐SARS‐CoV‐2 NP antibody (Sino Biological). Then, the cells were incubated with the Alexa Fluor 488‐conjugated secondary antibody (Thermo Fisher Scientific) for 2h at room temperature. Finally, phosphate‐buffered saline (PBS) supplemented with 0.1 μg/ml DAPI (Sigma‐Aldrich) was added to the cells for at least 15 min before imaging. Images were acquired using Cytation 5 (Biotek). All of the acquired data was analyzed by GraphPad Prism 9.
2.4. Antiviral evaluation in a SARS‐CoV‐2 infected hamster model
Male Syrian hamsters (Beijing Vital River Laboratory Animal Technology Co.), aged 6 weeks, were kept in BSL‐3 housing and fed with standard diet and water. All experimental protocols were approved by the Institutional Committee for Animal Care and Biosafety of the Navy Medical University. Hamsters were randomly distributed into eight groups (6/group). Each hamster was intranasally inoculated with 105 TCID50 of SARS‐CoV‐2. Drugs were delivered intraperitoneally at the indicated doses (Figure 1A). DMSO was considered as vehicle control. To evaluate the viral load and histopathological changes, three hamsters were euthanized and killed at 4 days postinfection (dpi) to collect the lungs. The others were observed daily to record body weight until 14 dpi. The viral load and inflammation in the lung tissue homogenates were detected using quantitative real‐time reverse‐transcription polymerase chain reaction (qRT‐PCR) methods. Tissue pathology of infected animals was examined and scored.
Figure 1.
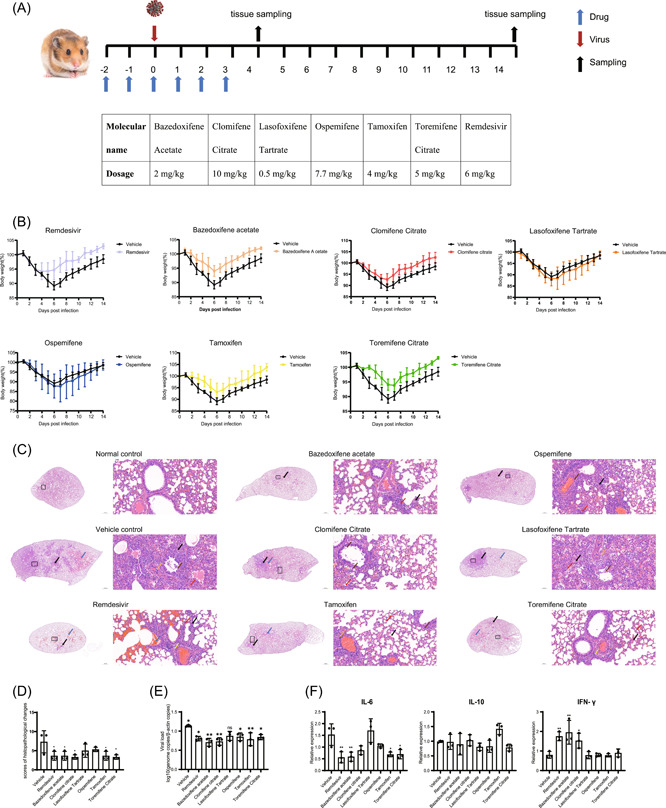
Antiviral evaluation in a SARS‐CoV‐2 infected hamster model. (A) Scheme and drug dosage of the animal experiment. (B) Body weights changes are shown with standard errors of the means of the three hamsters. Weight change is demonstrated as the percent of the initial body weight of day 0 for all animals. (C) Representative images of H&E‐stained lung tissue section from hamsters treated with different drugs. Black arrows: sites of alveolar wall thicken or congestion; red arrows: sites of alveolar wall inflammatory infiltration; blue arrows: sites of alveolar space infiltration or exudation or hemorrhage; brown arrows: sites of epithelium desquamation with inflammatory infiltration; orange arrows: sites of vessel wall inflammatory infiltration; yellow arrows: sites of peribronchiolar or perivascular inflammatory infiltration. (D) Histological analysis of lung pathology. To distinguish comprehensive lung pathological changes, semiquantitative histology scores were given to each lung tissue. Lung inflammation score taking into account (i) the severity of bronchitis; (ii) the severity of alveolar space infiltration or exudation or hemorrhage; (iii) the severity of vascular edema or infiltration. (E) Viral loads in the lung tissue homogenates at 4 dpi was measured by determining the genome copies/β‐actin copies by qRT‐PCR methods. (F) Relative mRNA expression of IL‐6, IL‐10, and IFN‐γ in the lungs of the indicated groups (n = 3), as detected in the hamster lung tissue homogenate at 4 dpi. Data is represented as mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. H&E, hematoxylin and eosin; IFN‐γ, interferon gamma; IL, interleukin; ns, not significant; qRT‐PCR, quantitative reverse‐transcription polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation
2.5. Time‐of‐drug‐addition assay
A time‐of‐drug‐addition assay was performed to investigate which stages of the SARS‐CoV‐2 life cycle antiviral drugs interfered with, as previously described. 8 , 10
2.6. Temperature shift assay
The protocol used for the temperature shift assay has been previously reported. 11 , 12 Cells were seeded in 96‐well plates (4 × 104 cells/well). Cells were pre‐cooled at 4℃ for 1 h. For the attachment assay, compounds diluted in viruses co‐treated cells for 1 h at 4℃ (final concentration 10 μM, MOI = 2). Then the medium was replaced with fresh medium and incubation continued at 37℃. For the penetration assay, cells were incubated with virus (MOI = 2) for 1 h at 4℃ and then fresh medium with diluted compounds (final concentration 10 μM) at 37℃ for 1h. Then plate was replaced with fresh medium and incubation continued at 37℃. At 10 hpi, the cells were fixed and immunofluorescence images were acquired.
2.7. Pseudovirus‐based inhibition assay
The pseudovirus of SARS‐CoV, original SARS‐CoV‐2, Delta variant, and Omicron variant of SARS‐CoV‐2 were harvested as previously reported. 8 Hela‐ACE2 cells were seeded in 96‐well plates (2 × 104 cells/well). Then, cells treated with compounds were infected with the pseudovirus (MOI = 0.5). At 72 hpi, the cells were imaged and EC50 values of selected compounds were analyzed.
2.8. Membrane fusion assay
HEK293T cells, used as effector cells, were transfected with the S protein expression vector and lenti‐EGFP (293 T/S/GFP) or empty plasmid lenti‐EGFP (293 T/GFP). For S‐mediated cell–cell fusion assays, 293 T/S/GFP or 293T/EGFP were digested without trypsin and then seeded onto Hela‐ACE2 cells. After a 6‐h coculture at 37℃ and 5% CO2, fused cells were then imaged using Cytation 5 (Biotek).
2.9. Filipin staining
Cells were plated and treated with selected compounds at the indicated concentrations (5 μM) for 24 h. And then cells were fixed with 4% paraformaldehyde and washed twice with PBS followed by incubation with 50 μg/ml filipin (Sigma‐Aldrich) in PBS for 1 h at room temperature. Next, the cells were washed again with PBS, after which fluorescence images were obtained using confocal fluorescence microscope. The images were analyzed by ImageJ software. All of the acquired data was analyzed by GraphPad Prism 9.
2.10. Endosome acidification assay
Cells were treated with either DMSO, positive control NH4Cl (10 mM), or selected compounds (5 μM) for 2 h and then low endosomal pH was detected by incubating cells with Lysotracker Red (Beyotime) for an additional 30min. Cells were fixed with 4% paraformaldehyde and imaged using Cytation 5.
2.11. qRT‐PCR
For animal experiments, to evaluate the viral loads and relative expression of IL‐6, IL‐10, and IFN‐γ, qRT‐PCR was performed using a TB Green Fast qPCR Mix (Takara Bio). The primer sequences were listed in Table 1.
Table 1.
The primer sequences
| Gene | Forward primer | Reverse primer |
|---|---|---|
| SARS‐CoV‐2 N | 5′‐GGGGAACTTCTCCTGCTAGAAT‐3′ | 5′‐CAGACATTTTGCTCTCAAGCTG‐3′ |
| Hamster β‐actin | 5′‐AGCAGTCTGTTGGAGCAAGC‐3′ | 5′‐ TCTAGGGAATTGGGGTGGCT‐3′ |
| Hamster IL‐6 | 5′‐GAGACTGGGGATGTCTGTAGC‐3′ | 5′‐ GGATGGAAGTCTCTTGCGGAG‐3′ |
| Hamster IL‐10 | 5′‐GGTTGCCAAACCTTATCAGAAATG‐3′ | 5′‐ TTCACCTGTTCCACAGCCTTG‐3′ |
| Hamster IFN‐γ | 5′‐TGTTGCTCTGCCTCACTCAGG‐3′ | 5′‐ AAGACGAGGTCCCCTCCATTC‐3′ |
Abbreviations: IFN‐γ, interferon gamma; IL, interleukin; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
2.12. Statistical analysis
Data analysis was performed using GraphPad Prism 9. A paired two‐tailed Student's t test was used to determine statistically significant differences between two groups while one‐ and two‐way analysis of variance was applied for multiple comparisons. Values are presented as the mean ± SEM. p values are represented as follows *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
3. RESULTS
3.1. Evaluation of the anti‐SARS‐CoV‐2 activity of SERMs in vitro
To evaluate the anti‐SARS‐CoV‐2 efficacy of 18 SERMs, Vero E6 cell line, a commonly used and robust cell model of SARS‐CoV‐2 infection, was first utilized. The screening procedure was conducted as previously described (Online Supporting Information Methods). Seven SERMs (final concentration 10 μM), raloxifene hydrochloride (Ralo‐H), tamoxifen (Tamo), clomifene citrate (Clom‐C), lasofoxifene tartrate (Laso‐T), Baze‐A, toremifene citrate (Tore‐C), and ospemifene (Ospe), exhibited potent anti‐SARS‐CoV‐2 activity (Data not shown). The 50% effective concentration (EC50) and 50% cytotoxic concentration (CC50) values of the seven SERMs were further determined. Remarkably, Baze‐A, Laso‐T, and Ospe showed the strongest antiviral activity in Vero E6 cells with the selective index (SI) of 13.11, 10.94, and 9.66, respectively (Table 2).
Table 2.
Cytotoxicity and antiviral activity of the SERMs
| Vero E6 (MOI = 0.1) | Caco‐2 (MOI = 0.1) | Calu‐3 (MOI = 1) | Hela‐ACE2 (MOI = 0.1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC50 | EC50 | SI | CC50 | EC50 | SI | CC50 | EC50 | SI | CC50 | EC50 | SI | |
| (μM) | (μM) | (μM) | (μM) | (μM) | (μM) | (μM) | (μM) | |||||
| Bazedoxifene acetate | 63.46 | 4.84 | 13.11 | 23.63 | 1.81 | 13.06 | 107.10 | 5.29 | 20.25 | 69.37 | 1.64 | 42.30 |
| Clomifene citrate | 19.78 | 2.84 | 6.96 | 13.69 | 0.42 | 32.60 | 110.00 | 5.93 | 18.55 | 19.84 | 2.49 | 7.97 |
| Lasofoxifene tartrate | 25.90 | 2.58 | 10.94 | 23.73 | 1.84 | 12.90 | 39.81 | 6.03 | 6.60 | 18.15 | 0.64 | 28.36 |
| Ospemifene | 81.28 | 8.41 | 9.66 | 62.71 | 0.71 | 88.32 | 93.83 | 8.32 | 11.28 | 92.90 | 3.10 | 29.97 |
| Tamoxifen | 17.05 | 4.51 | 3.78 | 15.63 | 0.22 | 71.05 | 60.64 | 5.65 | 10.73 | 33.32 | 2.13 | 15.65 |
| Toremifene citrate | 24.25 | 5.33 | 4.55 | 12.74 | 3.37 | 3.78 | 113.70 | 5.81 | 19.57 | 71.94 | 2.70 | 26.64 |
| Raloxifene HCl | 46.51 | 5.16 | 9.01 | 55.71 | 1.68 | 33.16 | 22.00 | 11.53 | 1.91 | 59.80 | 3.10 | 19.29 |
Abbreviations: CC50, 50% cytotoxic concentration; EC50, 50% effective concentration; MOI, multiplicity of infection; SERMs, selective estrogen receptor modulators.
To further validate the antiviral efficacy of the seven selected SERMs in human cell lines, Caco‐2, Calu‐3, and Hela‐ACE2 cells were utilized. Among the seven SERMs, Tamo, Clom‐C, Ospe, and Ralo‐H showed much higher SI in Caco‐2 cells. All the selected SERMs except for Ralo‐H and Laso‐T showed a SI of > 10 in Calu‐3 cells (Table 2). Remarkably, Baze‐A exhibited a SI > 10 in the four cell lines. Ralo‐H was excluded from further study for showing high cytotoxicity in Calu‐3 cells. Encouragingly, all of the other six SERMs exhibit potent antiviral effects in the four cell lines, indicating that the antiviral activity of these drugs is not achieved by affecting cell viability but by affecting virus infection.
3.2. Evaluation of the antiviral efficacy of six SERMs against SARS‐CoV‐2 in vivo
Syrian hamsters were treated with different drugs via intraperitoneal injection at the indicated dosages, which were transferred from already approved human dosing when treating other diseases (Figure 1A). The administration of Baze‐A, Clom‐C, Tamo, Tore‐C, and Rem resulted in excellent protection against hamster weight change accompanied by rescuing hamsters from moderate clinical signs such as lethargy, ruffled fur, and hunched back posture (Figure 1B). Three hamsters per group were killed at 4 dpi, when the viral loads and histopathological changes were expected to be the most prominent, to determine the viral load and inflammatory factors. 13 Histological examination showed the severity of the inflammation in hamsters that received Baze‐A, Clom‐C, Rem, Tamo, and Tore‐C was significantly alleviated (Figure 1C,D). And, compared with vehicle group, treatments with five SERMs except for Laso‐T significantly decreased the viral load in hamster lungs (Figure 1E). Concurrently, messenger RNA (mRNA) expression levels of interleukin‐6 (IL‐6), interleukin‐10 (IL‐10), and interferon‐γ (IFN‐γ) of hamster lungs were determined. The IL‐6 mRNA expression levels were remarkably diminished in the hamsters treated with Baze‐A, Tamo, and Tore‐C, consistent with those of hamsters treated with Rem, while there was no significant difference in the IL‐10 levels. Intriguingly, the mRNA expression of IFN‐γ was elevated in hamsters treated with Baze‐A, Clom‐C, and Rem (Figure 1F). This may be ascribable to an immunomodulatory effect of SERMs on the pathogen‐disturbed inflammatory response. Taken together, Baze‐A, Clom‐C, Tamo, and Tore‐C altered the course of the SARS‐COV‐2 infection in hamsters.
3.3. Effects of the selected SERMs on the SARS‐CoV‐2 life cycle
To investigate which steps of the SARS‐CoV‐2 life cycle were interrupted by Baze‐A, Clom‐C, Tamo, and Tore‐C that inhibited SARS‐CoV‐2 both in vitro and in vivo, time‐of‐drug‐addition assays were conducted in Vero E6 cells (Figure 2A). Clom‐C and Tamo displayed antiviral activity at 0 hpi while Tore‐C inhibited SARS‐CoV‐2 infection at all the indicated time points, suggesting Clom‐C and Tamo acted on the virus entry step while Tore‐C were multiple‐target drugs. Intriguingly, Baze‐A significantly suppressed virus replication at 0 and 2 hpi while no detectable effect was found when Baze‐A was maintained at 5 and 8 hpi, similar to Chloroquine (Chlo), a lysosomal inhibitor, strongly indicating that Baze‐A specifically targeted virus entry and internalization (Figure 2B). These results indicated that all of the four SERMs disturbed the early stage of the SARS‐CoV‐2 life cycle.
Figure 2.
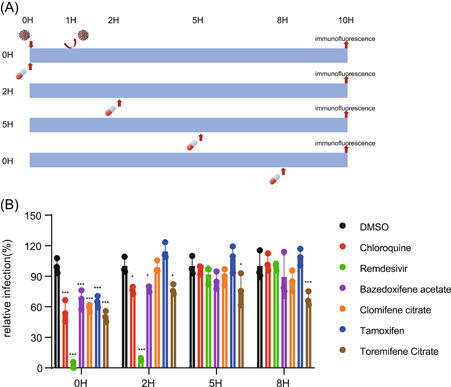
Effects of SERMs on SARS‐CoV‐2 life cycle. (A) The schedule of the time‐of‐drug‐addition asssay. Vero E6 cells were incubated with SARS‐CoV‐2 for 1 h to synchronize the assay, the culture medium was then removed. Cells were treated with the selected compounds at a final concentration of 5 μM at 0, 2, 5, and 8 h after the supernatants were removed. And, the infection was qualified at 10 h postinoculation after fixation and staining for SARS‐CoV‐2 NP. (B) Infection was calculated and quantificated. Data is represented as mean± SEM of n = 3 independent experiments. Statistical analysis is compared with the DMSO‐treated group. (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). DMSO, dimethyl sulfoxide; MOI, multiplicity of infection; ns, not significant; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of mean; SERMs, selective estrogen receptor modulators
3.4. Baze‐A inhibited SARS‐CoV‐2 entry at the post‐attachment step
To delineate the early step of the virus life cycle that is interfered by the four SERMs, temperature shift experiments were performed to distinguish between the viral attachment and post‐attachment (penetration) steps (Figure 3A). The SARS‐CoV‐2 infectivity was not affected by the four SERMs at the attachment step in both Vero E6 and Calu‐3 cells. By contrast, Baze‐A treatment led to a significant reduction of virus infection at the penetration step, similar to the positive control Chlo (Figure 3B). The results suggested that Baze‐A targeted the post‐attachment (penetration) step of entry, which is consistent with the results of the time‐of‐drug‐addition assay.
Figure 3.
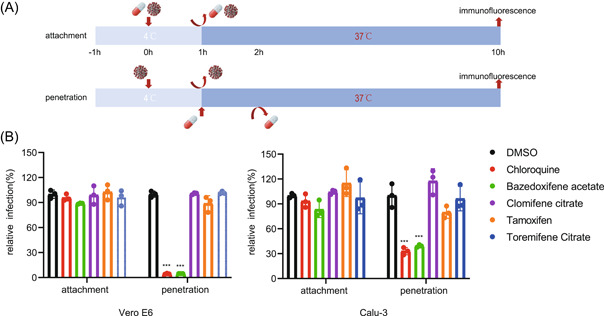
Baze‐A inhibited SARS‐CoV‐2 entry on postattachment step. (A) Scheme of temperature shift experiment. Vero E6 and Calu‐3 cells were pre‐cooled at 4℃ for 1 h. For the attachment assay, cells were treated with compounds diluted in viruses for 1 h at 4℃ (10 μM, MOI = 2). For the penetration assay, virus without drugs was added to cells for 1 h at 4℃ (MOI = 2). After washed off with PBS, warm medium with diluted compounds (10 μM) were added to cells at 37℃ and incubated for 1 h. Chlo was included as a positive control. (B) Quantification of temperature shift assay. Relative infection was measured by immunofluorescence assay. Data is represented as mean ± SEM of n = 3 independent experiments. Statistical analysis is compared with the DMSO‐treated group. (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). DMSO, dimethyl sulfoxide; MOI, multiplicity of infection; ns, not significant; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of mean
3.5. Baze‐A inhibited SARS‐CoV‐2 S‐mediated membrane fusion
To further explore the role of Baze‐A in SARS‐CoV‐2 cell entry, membrane fusion assays were performed. Baze‐A exhibited inhibitory effect in full‐time treatment, pretreatment to Hela‐ACE2 cells and pretreatment to 293T cells groups, demonstrating that the targets of Baze‐A were related to host cell factors and possibly entangled with membrane fusion step (Figure 4).
Figure 4.
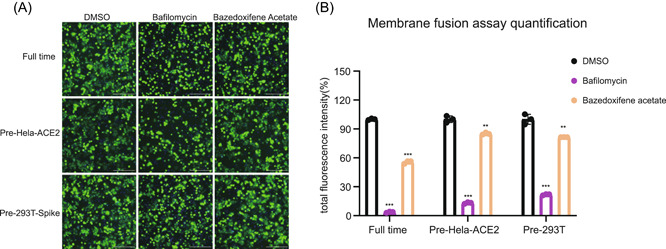
Baze‐A inhibited SARS‐CoV‐2 S‐mediated membrane fusion. For pre‐Hela‐ACE2 group, Hela‐ACE2 were pretreated in the presence of Baze‐A or positive control bafilomycin at the indicated concentration (10 μM) for 2 h. For pre‐293T‐Spike group, 293T/S/GFP cells were pretreated at the indicated concentration (10 μM) for 2 h. For full‐time treatment, HEK293T and Hela‐ACE2 were incubated with the Baze‐A (10 μM) all the time. Representative immunofluorescence images (A) and quantification of fused cells (B). Data is represented as mean ± SEM of n = 3 independent experiments. Statistical analysis is compared with the DMSO‐treated group. (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). DMSO, dimethyl sulfoxide; ns, not significant; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of mean
3.6. Baze‐A altered the cholesterol distribution and endosome acidification
Cholesterol‐rich lipid rafts and acidic endosomal pH is reported to be important for SARS‐CoV‐2 entry and virus–cell fusion. 14 , 15 , 16 , 17 Next, a cholesterol accumulation assay and an endosome acidification assay were conducted to further investigate the key steps that were interfered by Baze‐A. Compared with the DMSO‐treated group, the pattern of cholesterol distribution in the cells treated with Baze‐A was obviously altered and larger filipin‐positive vesicles in the cytoplasm were formed (Figure 5A–C). In addition, Baze‐A obviously inhibited endosome acidification, which is similar to NH4Cl, the positive control (Figure 5D–F). The above results suggested that Baze‐A participate in the process of cholesterol distribution and endosome acidification of target cells, thus inhibiting the entry of SARS‐CoV‐2.
Figure 5.
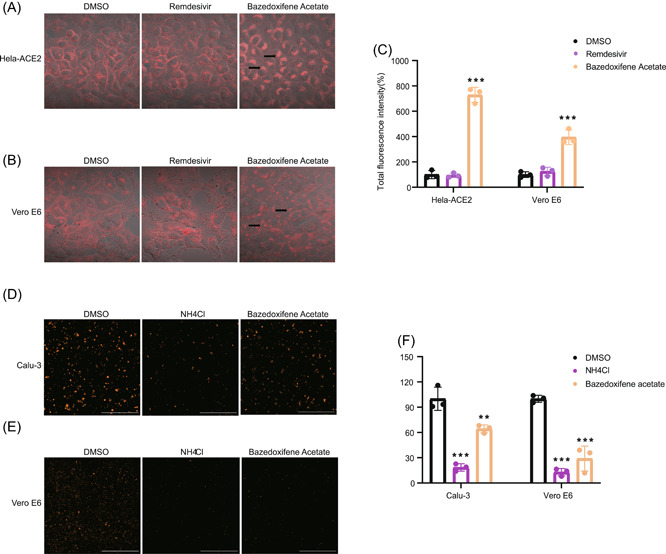
Baze‐A altered the cholesterol distribution and endosome acidification. Hela‐ACE2 (A) and Vero‐E6 (B) cells were treated for 24 h with either DMSO or drugs (5uM). Cells were then fixed, stained with filipin, and imaged on a confocal fluorescence microscope. Representative images are shown. Arrows indicate sites of cholesterol accumulation. (C) Quantification of cholesterol accumulation assay was performed by calculating the total fluorescence intensity. Calu‐3 (D) and Vero E6 (E) cells were treated for 2 h with either DMSO or compounds (5 μM) and then low endosomal pH was detected by incubating with Lysotracker Red; 10 mM NH4Cl was used as the positive control. (F) Quantification of endosome acidification assay was performed by calculating the total fluorescence intensity. Data is represented as mean ± SEM of n = 3 independent experiments. Statistical analysis is compared with the DMSO‐treated group. (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). DMSO, dimethyl sulfoxide; ns, not significant; SEM, standard error of mean
3.7. Baze‐A exhibited antiviral activity on pseudovirus infection of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV
A pseudovirus of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV were utilized to explore the antiviral effect of Baze‐A. The results showed that Baze‐A significantly inhibited the pseudovirus infection of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV with EC50 values of 7.306 μM, 1.716 μM, 3.446 μM, and 3.039 μM, respectively (Figure 6). The results offered critical information supporting Baze‐A is a promising antiviral candidate for coronaviruses infection.
Figure 6.
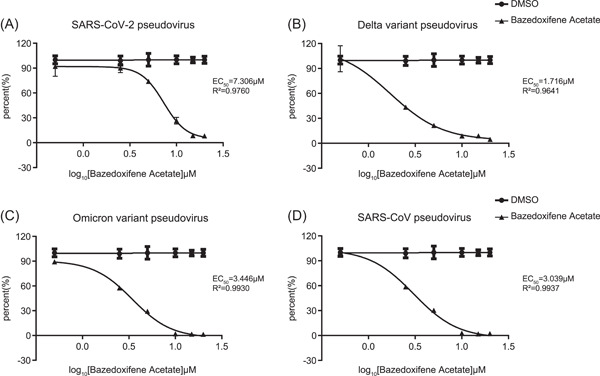
Baze‐A exhibited antiviral activity on pseudovirus infection of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV. Cells were treated with a series concentration of Baze‐A and then infected with pseudovirus of original SARS‐CoV‐2 (A), Delta variant of SARS‐CoV‐2 (B), Omicron variant of SARS‐CoV‐2 (C), and SARS‐CoV (D). DMSO, dimethyl sulfoxide; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
4. DISCUSSION
SERMs combine with and selectively act on the estrogen receptors (ERs) and produce hormone‐like or antiestrogen effects in different target tissues. To date, SERMs including fulvestrant (Fulv), Ralo‐H, Tamo‐C, Tore‐C, Baze‐A, and Ospe have already been approved by the Food and Drug Administration for the treatment of cancer or osteoporosis. Moreover, emerging reports have indicated the pleiotropic effect of SERMs, such as weight loss, neuroprotection, cardio‐protection, and antimicrobial activity. Interestingly, some SERMs have shown good antiviral activity (Table S1). And some SERMs have been reported to exhibit inhibitory activity against SARS‐CoV‐2 infection, whereas the underlying mechanism and their antiviral activity in vivo are poorly defined as the existing studies are cell‐based or in silico screening. 18 , 19
In this study, we evaluated the antiviral efficacies of 18 SERMs against SARS‐CoV‐2 infection across four cell lines and six SERMs performed excellent anti‐SARS‐CoV‐2 effects. Furthermore, the antiviral efficacy of six SERMs for SARS‐CoV‐2 was analyzed in vivo using hamster model. Then the mechanisms of the selected four SERMs against SARS‐CoV‐2 were explored. First, the possibility that the SERMs acted through the classical estrogen signaling pathway was ruled out (Figure S1), given the abortive effects of the estradiol and Fulv against SARS‐CoV‐2 infection and various expression of ERα and ERβ in different cell lines. Then Clom‐C, Tamo, and Baze‐A were proved to specifically participate in the early step of viral infection. The possibility that the selected SERMs may play a role via pathway including ACE2 expression, the binding affinity between ACE2 and SARS‐CoV‐2 Spike protein and the enzyme activity of TMPRSS2 was also excluded (Supplementary figure S2). While in temperature shift assay, Baze‐A was screened to act on the penetration of post‐attachment step, which was reinforced in the experiment of membrane fusion, possibly contributed to changed cholesterol distribution and endosome acidification.
Bazedoxifene (Baze) is a third‐generation SERM. The main indication for Baze, not affecting the stimulation of the female breast and uterus, is the treatment of postmenopausal osteoporosis. 20 Recent research shows that Baze inhibits Mycobacterium tuberculosis in macrophages by promoting autophagy. 21 Furthermore, Baze exhibited anti‐inflammatory and anti‐atherosclerotic effects via the inhibition of IL‐6/IL‐6R/STAT3 signaling. 22 Our study showed that Baze‐A inhibits SARS‐CoV‐2 infection in vitro and in vivo, the mechanism of which is impeding the postattachment step of SARS‐CoV‐2 entry. Interestingly, the lungs of hamsters treated with Baze‐A exhibited relatively lower IL‐6 mRNA levels in comparison with those of the vehicle group, consistent with a previous study. 23
Cholesterol is a prerequisite for viral endocytosis, micropinocytosis, and membrane fusion. 24 , 25 Moreover, increasing evidence suggests that lipid rafts participate in mediating the cell entry of enveloped viruses, including influenza virus, African swine fever virions, and Ebola virus. 26 , 27 Therefore, the role of cholesterol in understanding the SARS‐CoV‐2 infection process has attracted increasing attention. Recently, the major role of cholesterol‐rich membrane lipid rafts in SARS‐CoV‐2 pseudovirus infection and authentic SARS‐CoV‐2 infection, and depletion of accessible cholesterol on the plasma membrane leading to inhibition of virus–cell fusion have been reported. 17 , 28 The broad antiviral effect of Baze‐A, which was evaluated on authentic SARS‐CoV‐2 infection and pseudovirus infection of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV, may be partially explained by the effect of Baze‐A on rewiring the host cell factors.
Clom is used to treat female infertility due to anovulation, and is an efficient drug for reversing impotence in men with hypogonadism due to low testosterone secretion. 29 Clom has been reported to show antiviral effects against SARS‐CoV‐2 via several lipid‐dependent mechanisms. 18 , 30 Our assays showed that Clom inhibited the entry stage of SARS‐CoV‐2 using an authentic isolated strain while it did not act either on the attachment or post‐attachment step during the temperature shift assay. The exact mechanism underlying the activity of Clom against SARS‐CoV‐2 needs further study.
Tamo has been reported to show broad‐spectrum antiviral activity against Ebola virus, Zika virus, vesicular stomatitis virus, and herpes simplex virus type 1 infection. 31 However, Hulya and his colleagues proposed that Tamo may increase the COVID‐19 risk due to its antiestrogen and P‐glycoprotein inhibitory effects. 32 Tore destabilizes the Ebola virus glycoprotein to inhibit Ebola virus infection. Martin et al. suggested the repurposing of the FDA‐approved Tore to treat COVID‐19 by blocking the spike glycoprotein and NSP14 of SARS‐CoV‐2 based on in silico analysis. 33 The time‐of‐drug‐addition analysis showed that Tamo disturbed the early stage of SARS‐CoV‐2 infection, and Tore is a multiple target drug, while the detailed mechanisms need further exploration.
In summary, we screened the inhibitory efficacy of 18 SERMs and identified Baze‐A as a promising drug against SARS‐CoV‐2 in vitro and in vivo, the mechanism of which was likely contributed to the changed cholesterol distribution and endosome acidification. Notably, Baze‐A showed antiviral activity on pseudovirus of original SARS‐CoV‐2, Delta variant, Omicron variant, and SARS‐CoV. Moreover, Clom‐C, Tamo, and Tore‐C also showed good anti‐SARS‐CoV‐2 efficacy in different cell lines and hamster infection model. As novel variants of SARS‐CoV‐2 become prevalent, more studies will be focused on the emerging prevalent variants and evaluating therapeutic cocktails to optimize therapeutic options.
AUTHOR CONTRIBUTIONS
Cuiling Ding, Zhongtian Qi, and Ping Zhao designed experiments. Gen Miao, Haoran Peng, Hailin Tang, Yangang Liu, Xu Zheng, Bin Liu, Liangliang Jiang, Wanda Tang, Yanhua He, Yan Liu, and Hao Ren performed experiments and analyzed data. Gen Miao and Cuiling Ding wrote the manuscript. Cuiling Ding, Zhongtian Qi, and Ping Zhao reviewed and edited the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Miao G, Peng H, Tang H, et al. Antiviral efficacy of selective estrogen receptor modulators against SARS‐CoV‐2 infection in vitro and in vivo reveals bazedoxifene acetate as an entry inhibitor. J Med Virol. 2022;94:4809‐4819. 10.1002/jmv.27951
Gen Miao, Haoran Peng and Hailin Tang contributed equally to this study
Contributor Information
Ping Zhao, Email: pnzhao@163.com.
Zhongtian Qi, Email: qizt@smmu.edu.cn.
Cuiling Ding, Email: cuilingding@smmu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Dhar MS, Marwal R, Vs R, et al. Genomic characterization and epidemiology of an emerging SARS‐CoV‐2 variant in Delhi, India. Science (New York, NY). 2021;374:eabj9932‐eabj9999. 10.1126/science.abj9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Araf Y, Akter F, Tang YD, et al. Omicron variant of SARS‐CoV‐2: genomics, transmissibility, and responses to current COVID‐19 vaccines. J Med Virol. 2022;94:1825‐1832. 10.1002/jmv.27588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zinatizadeh MR, Zarandi PK, Zinatizadeh M, Yousefi MH, Amani J, Rezaei N. Efficacy of mRNA, adenoviral vector, and perfusion protein COVID‐19 vaccines. Biomed Pharmacother. 2021;146:112527. 10.1016/j.biopha.2021.112527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pérez‐Then E, Lucas C, Monteiro VS, et al. Immunogenicity of heterologous BNT162b2 booster in fully vaccinated individuals with CoronaVac against SARS‐CoV‐2 variants Delta and Omicron: the Dominican Republic experience. medRxiv . 2021. 10.1101/2021.12.27.21268459 [DOI]
- 5. Liu J, Liu Y, Xia H, et al. BNT162b2‐elicited neutralization of B.1.617 and other SARS‐CoV‐2 variants. Nature. 2021;596(7871):273‐275. 10.1038/s41586-021-03693-y [DOI] [PubMed] [Google Scholar]
- 6. Fernandes Q, Inchakalody VP, Merhi M, et al. Emerging COVID‐19 variants and their impact on SARS‐CoV‐2 diagnosis, therapeutics and vaccines. Ann Med. 2022; 54(1):524‐540. 10.1080/07853890.2022.2031274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scavone C, Mascolo A, Rafaniello C, et al. Therapeutic strategies to fight COVID‐19: which is the status artis? Br J Pharmacol. 2022;179(10):2128‐2148. 10.1111/bph.15452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng H, Ding C, Jiang L, et al. Discovery of potential anti‐SARS‐CoV‐2 drugs based on large‐scale screening in vitro and effect evaluation in vivo. Sci China Life Sci. 2021;65:1181‐1197. 10.1007/s11427-021-2031-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hao‐ran P, Cheng‐zhong L, Hai‐lin T, et al. Isolation and identification of two strains of severe acute respiratory syndrome coronavirus 2 from coronavirus disease 2019 patients in shanghai. Acad J Second Mil Med Univ. 2020;41:365‐370. [Google Scholar]
- 10. Yuan S, Yin X, Meng X, et al. Clofazimine broadly inhibits coronaviruses including SARS‐CoV‐2. Nature. 2021;593(7859):418‐423. 10.1038/s41586-021-03431-4 [DOI] [PubMed] [Google Scholar]
- 11. Yuan S, Wang R, Chan JF, et al. Metallodrug ranitidine bismuth citrate suppresses SARS‐CoV‐2 replication and relieves virus‐associated pneumonia in Syrian hamsters. Nat Microbiol. 2020;5(11):1439‐1448. 10.1038/s41564-020-00802-x [DOI] [PubMed] [Google Scholar]
- 12. Chan S‐W, Shafi T, Ford RC. Kite‐shaped molecules block SARS‐CoV‐2 cell entry at a post‐attachment step. Viruses. 2021;13(11):2306. 10.3390/v13112306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imai M, Iwatsuki‐Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS‐CoV‐2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117(28):16587‐16595. 10.1073/pnas.2009799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu Y, Feng F, Hu G, et al. A genome‐wide CRISPR screen identifies host factors that regulate SARS‐CoV‐2 entry. Nat Commun. 2021;12(1):961. 10.1038/s41467-021-21213-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. e8 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu Y, Liu DX, Tam JP. Lipid rafts are involved in SARS‐CoV entry into Vero E6 cells. Biochem Biophys Res Commun. 2008;369(2):344‐349. 10.1016/j.bbrc.2008.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Zhu W, Fan M, et al. Dependence of SARS‐CoV‐2 infection on cholesterol‐rich lipid raft and endosomal acidification. Comput Struct Biotechnol J. 2021;19:1933‐1943. 10.1016/j.csbj.2021.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong HL, Cao JL, Shen CG, et al. Several FDA‐Approved drugs effectively inhibit SARS‐CoV‐2 infection in vitro. Front Pharmacol. 2020;11:609592. 10.3389/fphar.2020.609592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smetana K, Jr. , Rosel D. Brábek J. raloxifene and bazedoxifene could be promising candidates for preventing the COVID‐19 related cytokine storm, ARDS and mortality. In Vivo. 2020;34(5):3027‐3028. 10.21873/invivo.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yavropoulou MP, Makras P, Anastasilakis AD. Bazedoxifene for the treatment of osteoporosis. Expert Opin Pharmacother. 2019;20(10):1201‐1210. 10.1080/14656566.2019.1615882 [DOI] [PubMed] [Google Scholar]
- 21. Ouyang Q, Zhang K, Lin D, Feng CG, Cai Y, Chen X. Bazedoxifene suppresses intracellular mycobacterium tuberculosis growth by enhancing autophagy. mSphere. 2020;5(2):e00124‐20, 10.1128/mSphere.00124-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo P, Wang Y, Zhao C, et al. Bazedoxifene exhibits anti‐inflammation and anti‐atherosclerotic effects via inhibition of IL‐6/IL‐6R/STAT3 signaling. Eur J Pharmacol. 2021;893:173822. 10.1016/j.ejphar.2020.173822 [DOI] [PubMed] [Google Scholar]
- 23. Ma H, Yan D, Wang Y, et al. Bazedoxifene exhibits growth suppressive activity by targeting interleukin‐6/glycoprotein 130/signal transducer and activator of transcription 3 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110(3):950‐961. 10.1111/cas.13940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sandvig K, Kavaliauskiene S, Skotland T. Clathrin‐independent endocytosis: an increasing degree of complexity. Histochem Cell Biol. 2018;150(2):107‐118. 10.1007/s00418-018-1678-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mesquita FS, Abrami L, Sergeeva O, et al. S‐acylation controls SARS‐CoV‐2 membrane lipid organization and enhances infectivity. Dev Cell. 2021;56(20):2790‐2807. e8 10.1016/j.devcel.2021.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bavari S, Bosio CM, Wiegand E, et al. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 2002;195(5):593‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cuesta‐Geijo MÁ, Chiappi M, Galindo I, et al. Cholesterol flux is required for endosomal progression of African swine fever virions during the initial establishment of infection. J Virol. 2016;90(3):1534‐1543. 10.1128/JVI.02694-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang S, Li W, Hui H, et al. Cholesterol 25‐Hydroxylase inhibits SARS‐CoV‐2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39(21):e106057. 10.15252/embj.2020106057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scovell JM, Khera M. Testosterone replacement therapy versus clomiphene citrate in the young hypogonadal Male. Eur Urol Focus. 2018;4(3):321‐323. 10.1016/j.euf.2018.07.033 [DOI] [PubMed] [Google Scholar]
- 30. Ghasemnejad‐Berenji M, Pashapour S, Ghasemnejad‐Berenji H. Therapeutic potential for clomiphene, a selective estrogen receptor modulator, in the treatment of COVID‐19. Med Hypotheses. 2020;145:110354. 10.1016/j.mehy.2020.110354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montoya MC, Krysan DJ. Repurposing estrogen receptor antagonists for the treatment of infectious disease. mBio. 2018;9(6):e02272‐18, 10.1128/mBio.02272-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vatansev H, Kadiyoran C, Cumhur Cure M, Cure E. COVID‐19 infection can cause chemotherapy resistance development in patients with breast cancer and tamoxifen May cause susceptibility to COVID‐19 infection. Med Hypotheses. 2020;143:110091. 10.1016/j.mehy.2020.110091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin WR, Cheng F. Repurposing of FDA‐approved toremifene to treat COVID‐19 by blocking the spike glycoprotein and NSP14 of SARS‐CoV‐2. J Proteome Res. 2020;19(11):4670‐4677. 10.1021/acs.jproteome.0c00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


