Abstract
A characteristic feature of COVID‐19, the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection, is the dysregulated immune response with impaired type I and III interferon (IFN) expression and an overwhelming inflammatory cytokine storm. RIG‐I‐like receptors (RLRs) and cGAS–STING signaling pathways are responsible for sensing viral infection and inducing IFN production to combat invading viruses. Multiple proteins of SARS‐CoV‐2 have been reported to modulate the RLR signaling pathways to achieve immune evasion. Although SARS‐CoV‐2 infection also activates the cGAS–STING signaling by stimulating micronuclei formation during the process of syncytia, whether SARS‐CoV‐2 modulates the cGAS–STING pathway requires further investigation. Here, we screened 29 SARS‐CoV‐2‐encoded viral proteins to explore the viral proteins that affect the cGAS–STING signaling pathway and found that SARS‐CoV‐2 open reading frame 10 (ORF10) targets STING to antagonize IFN activation. Overexpression of ORF10 inhibits cGAS–STING‐induced interferon regulatory factor 3 phosphorylation, translocation, and subsequent IFN induction. Mechanistically, ORF10 interacts with STING, attenuates the STING–TBK1 association, and impairs STING oligomerization and aggregation and STING‐mediated autophagy; ORF10 also prevents the endoplasmic reticulum (ER)‐to‐Golgi trafficking of STING by anchoring STING in the ER. Taken together, these findings suggest that SARS‐CoV‐2 ORF10 impairs the cGAS–STING signaling by blocking the translocation of STING and the interaction between STING and TBK1 to antagonize innate antiviral immunity.
Keywords: autophagy, COVID‐19, IFN, ORF10, SARS‐CoV‐2, STING
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has resulted in a worldwide pandemic that has caused a vast number of infections and fatalities. The SARS‐CoV‐2 genome is a positive‐sense, nonsegmented, single‐stranded RNA with a length of 29.9 kb. The SARS‐CoV‐2 genome encodes a 5′ frameshifted polyprotein (open reading frame a [ORF1a]/ORF1ab), which is further processed by virally encoded proteinases and produces 16 nonstructural proteins (NSP1–16), 4 structural proteins, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, and approximately 9 accessory proteins: ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c, and ORF10. 1 , 2 The NSPs and structural proteins are common and well conserved in all coronaviruses; however, the accessory proteins encoded by distinct coronaviruses vary in number, location, and size, which are usually not required for virus replication but are often important for viral pathogenesis. 1 Compared with the SARS‐CoV‐1 and other coronavirus genomes, the ORF10 protein, which contains only 38 amino acids, is a unique protein within the SARS‐CoV‐2 genome. 1 A previous proteomics study reported that ORF10 interacts with an E3 ubiquitin ligase complex containing Cullin‐2, Rbx1, Elongin B, Elongin C, and ZYG11B, which are responsible for degrading proteins through ubiquitination. 3 In addition to interacting with the E3 ligase complex, ORF10 also interacts with the depalmitoylation enzyme PPT1. 3 The protein‐protein interaction map suggests that ORF10 may modulate the cellular ubiquitination or palmitoylation system to facilitate viral replication. A recent study suggests that ZYG11B is dispensable for SARS‐CoV‐2 infection, and the interaction between ORF10 and ZYG11B is not relevant for SARS‐CoV‐2 infection. 4 However, whether ORF10 facilitates viral replication by modulating other cellular processes remains unknown.
The cyclic GMP–AMP (cGAMP) synthase (cGAS) is the sensor of cytosolic double‐stranded DNA from DNA viruses, retroviruses, or extracellular, mitochondrial, and nuclear DNA that gain access to the cytosol. 5 Upon DNA binding, cGAS catalyzes the synthesis of the second messenger cGAMP from ATP and GTP. cGAMP binds to the endoplasmic reticulum (ER)‐resident adaptor protein STING and triggers STING oligomerization. Upon oligomerization, STING translocates from the ER to the Golgi. On traveling through the ER–Golgi intermediate compartment (ERGIC) and Golgi, STING recruits and activates TBK1. 6 TBK1 then phosphorylates itself, STING, and interferon regulatory factor 3 (IRF3), resulting in IRF3 entering the nucleus and initiating the transcription of type I and type III interferon (IFN), IFN‐stimulated gene (ISG), and other immune‐response genes to orchestrate host antiviral immunity. 7 During this process, the trafficking of STING from the ER to the Golgi is critical for activating the downstream signaling cascade. In the Golgi, STING interacts with TBK1 to be phosphorylated and aggregated. After trafficking to the Golgi apparatus, STING then relocates to the endosomes and binds with IRF3 in endosomes. 8 Treatment of cells with brefeldin A (BFA), an inhibitor of protein trafficking between the ER and the Golgi apparatus, inhibits IFN production induced by STING agonists. 7 In addition to activation of IRF3, STING also induces noncanonical autophagy through its direct interaction with light chain 3 (LC3). 9 STING‐containing ERGIC has been shown to serve as a membrane source for LC3 lipidation, which is an essential step in autophagosome biogenesis. Autophagy induction via STING trafficking plays an important role in the clearance of DNA and viruses. 10 The activation of autophagy by STING is independent of its TBK1 activity. Ultimately, STING within autophagosomes and STING from the Golgi are both delivered to the lysosome for degradation. 6
The RIG‐I‐like receptors (RLRs), consisting of RIG‐I, MDA5, and LGP2, play a major role in sensing RNA virus infection to establish host antiviral immunity. The evasion of antiviral innate immunity by SARS‐CoV‐2 mainly focuses on the antagonism of the RLR signaling pathway since RIG‐I/MDA5 are responsible for sensing the viral genome of RNA viruses, including coronaviruses. 11 , 12 Recent studies have shown that SARS‐CoV‐2 infection also activates the cGAS–STING signaling pathway through the sensing of released mitochondrial DNA and chromatin DNA shuttled from the nucleus as a result of cell‐to‐cell fusion, a widespread phenomenon in SARS‐CoV‐2‐infected cells and COVID‐19 patients. 13 , 14 Therefore, there is an urgent need to investigate whether SARS‐CoV‐2 proteins can counteract the cGAS–STING signaling pathway and achieve efficient immune evasion. Here, we screened 29 SARS‐CoV‐2‐encoded viral proteins and discovered that ORF10 of SARS‐CoV‐2 was a potent antagonist of the cGAS–STING signaling pathway by targeting STING. Overexpression of ORF10 facilitated viral infection by blocking STING‐induced IFN production and autophagy. Mechanistically, ORF10 interacted with STING and impaired the STING‐TBK1 interaction, prevented STING translocation from the ER to the Golgi, and suppressed STING oligomerization and aggregation. This study reveals the function of ORF10 in facilitating viral immune evasion and provides insights into the pathogenesis and medical treatment of SARS‐CoV‐2.
2. MATERIALS AND METHODS
2.1. Cell culture, plasmids, and transfection
HEK293T HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) with 10% heat‐inactivated fetal bovine serum (FBS; Gibco). HK‐2 cells were cultured by DMEM/F12 (1:1) complete medium with 10% FBS. All cells were cultured at 37°C in a humidified incubator with 5% CO2. Plasmids expressing cGAS, STING, TBK1, or various SARA‐CoV‐2 proteins were constructed in our previous publications. 15 , 16 , 17 , 18 IFN‐β luciferase reporter plasmid pGL3‐IFN‐β‐Luc, IFN‐λ1 luciferase reporter plasmid pGL3‐IFN‐λ1‐Luc, and ISG luciferase reporter plasmid pISRE‐Luc have been described previously. 19 , 20 , 21 SARS‐CoV‐2 ORF10 gene (Gene ID: 43740576) was synthesized according to the genome sequence of the SARA‐CoV‐2 Wuhan‐Hu‐1 strain (NC_045512.2) (GENERAL BIOL) and cloned into pCAG expression vector. Plasmids were transfected into HEK293T and HeLa cells by Lipofectamine 3000 (Thermo Fisher Scientific). cGAMP (Invivogen) was transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific) as described in a previous reference. 22
2.2. Antibodies and reagents
Rabbit anti‐DYKDDDDK (D6W5B), rabbit anti‐IRF3 (D83B9), rabbit anti‐pIRF3 (4D46), rabbit anti‐TBK1 (3031S), rabbit anti‐pTBK1 (D52C2), and rabbit anti‐pSTING (E9A9K) antibodies were from Cell Signaling Technology; rabbit anti‐STING, mouse anti‐β‐actin, and rabbit anti‐calnexin antibodies were from Proteintech; rabbit anti‐hemagglutinin (HA) (H6908) and mouse anti‐Flag M2 antibodies were from Sigma‐Aldrich; mouse anti‐Myc (9E10) antibody was from Origene; rabbit anti‐GM130 antibody was from Abcam; and mouse anti‐HA antibody was from MDL Biotech. Protein A/G beads were from Santa Cruz Biotechnology, and the anti‐Myc and anti‐HA magnetic beads were from Bimake. Alexa Fluor 488 goat anti‐rabbit immunoglobulin G (IgG), Alexa Fluor 568 goat anti‐mouse IgG, Alexa Fluor 488 goat anti‐mouse IgG, and Alexa Fluor 568 goat anti‐rabbit IgG secondary antibodies were from Thermo Fisher Scientific.
2.3. Real‐time quantitative polymerase chain reaction
Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse‐transcribed into first‐strand complementary DNA (cDNA) using HiScript III First Strand cDNA Synthesis Kit with gDNA wiper (Vazyme). Real‐time quantitative polymerase chain reaction (RT‐qPCR) assays were performed on a LightCycler (LC480; Roche) using SYBR Green Kit UltraSYBR Mixture (CWBIO) following the manufacturer's instructions. The relative abundance of all indicated genes was normalized to the messenger RNA (mRNA) level of GAPDH. The comparative C T method (ΔΔC T method) was used for the calculation of fold change in gene expression as described in a previous reference. 19 The primer sequences of target genes are described in Table 1.
Table 1.
Primers were used in this study.
| Primer name | Sequence (5′–3′) | Usage |
|---|---|---|
| GAPDH‐F | GGAGCGAGATCCCTCCAAAAT | RT‐qPCR |
| GAPDH‐R | GGCTGTTGTCATACTTCTCATGG | |
| IFN‐β‐F | TTGCTCTCCTGTTGTGCTTC | RT‐qPCR |
| IFN‐β‐R | AAGCCTCCCATTCAATTGCC | |
| IFN‐λ1‐F | GAGGCCCCCAAAAAGGAGTC | RT‐qPCR |
| IFN‐λ1‐R | AGGTTCCCATCGGCCACATA | |
| ISG56‐F | CTAAGCAAAACCCTGCAGAAC | RT‐qPCR |
| ISG56‐R | TCAGGCATTTCATCGTCATC | |
| CXCL10‐F | GTGGCATTCAAGGAGTACCTC | RT‐qPCR |
| CXCL10‐R | GACCTTTCCTTGCTAACTGCT | |
| ORF10‐F | GGGGTACCGCCACCATGGGCTACATCAACGTGTTCG | Expression plasmid |
| ORF10‐R | GCTCTAGAGGTCAGATTGAAGTTCACCACATC |
Abbreviations: F, forward primer; R, reverse primer; RT‐qPCR, real‐time quantitative polymerase chain reaction.
2.4. Luciferase reporter assays
HEK293T cells cultured in 48 well plates were transiently transfected with luciferase reporter plasmids (IFN‐β‐Luc, IFN‐λ1‐Luc, or ISRE‐Luc) together with plasmids expressing the indicated protein in each experiment. The pRL‐TK plasmid (Promega) was cotransfected to serve as an internal control. Thirty‐six hours after transfection, the Dual‐Luciferase Reporter Assay Kit (Vazymea) was used to measure the luciferase activity according to the manufacturer's instructions as described in our previous studies. 19 , 20
2.5. Viruses and infection
Herpes simplex virus type 1–green fluorescent protein (HSV‐1–GFP), ICP0‐null HSV‐1 virus (HSV‐1‐∆ICP0), and the transcription and replication‐competent SARS‐CoV‐2 virus‐like particles (trVLPs) were used to infect human cells as described previously. 15 , 17 , 23 Briefly, the virus was diluted into a prewarmed serum‐free DMEM medium at 37°C before infection and incubated with target cells for 1–2 h for viral entry. At the end of the infection, the supernatant was discarded, and fresh DMEM containing 10% FBS was added back.
2.6. Enzyme‐linked immunosorbent assay
The concentration of secreted IFN‐β was detected using an Enzyme‐Linked Immunosorbent Assay (ELISA) Kit (Beyotime) according to the manufacturer's instructions.
2.7. Nuclear and cytoplasmic fractionation
The cytoplasmic and nuclear extracts of HEK293T cells were fractionated with a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime) according to the manufacturer's protocol as described. 20
2.8. Co‐immunoprecipitation and immunoblotting
Co‐immunoprecipitation (Co‐IP) assays were performed as described in our previous publications. 20 , 24 In brief, HEK293T cells were collected and lysed with less stringent lysis buffer (1.0% [vol/vol] NP‐40, 50 mM Tris‐HCl [pH 7.4], 50 mM ethylenediaminetetraacetic acid [EDTA], and 150 mM NaCl) or more stringent radioimmunoprecipitation assay (RIPA) buffer (1.0% [vol/vol] NP‐40, 50 mM Tris‐HCl [pH 7.4], 50 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) according to the requirements of the experiments. The cell lysates were centrifuged at 14 000g for 10 min, then the supernatants were collected and incubated with the indicated antibodies followed by the addition of protein A/G beads (Santa Cruz Biotechnology) or with corresponding antibody‐conjugated magnetic beads (Bimake). After incubation overnight at 4°C, beads were washed and then boiled with 2× SDS loading buffer to elute the immunoprecipitants. Protein samples separated using SDS‐polyacrylamide gel electrophoresis (PAGE), semi‐denaturating detergent‐agarose gel electrophoresis (SDD‐AGE), and native PAGE were performed as described previously. 17 , 20 , 24 The samples on gels were then transferred onto a polyvinylidene difluoride membrane (Millipore), blocked with 3% (wt/vol) bovine serum albumin (BSA), and probed with indicated primary antibodies and corresponding secondary antibodies. Immunoreactive bands were visualized by ECL Western blotting detection reagent (Pierce).
2.9. Immunostaining and confocal microscopy
Confocal immunofluorescence microscopy studies were performed as described in our previous publications. 20 , 24 Briefly, 5 × 104 HeLa cells or 1 × 105 HeLa cells were grown on 12‐well slides one day before transfection with the indicated plasmids. For immunostaining, plasmid transfected or virus‐infected HeLa cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X‐100, and blocked with 5% BSA. The cells were then reacted with indicated primary antibodies at 4°C overnight followed by a fluorescent secondary antibody (Invitrogen). Nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole in the mounting medium (Abcam). Images of the cells were taken with a Zeiss LSM880 laser scanning confocal microscope and processed using the Zeiss LSM image browser.
2.10. Statistics
Data are presented as the mean ± SEM. unless indicated otherwise. All statistical calculations were carried out with Microsoft Excel and GraphPad Prism 8 software. For comparisons between two groups, the two‐tailed unpaired Student's t‐test was conducted to determine the significance. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. ORF10 inhibits STING‐induced IFN activation
SARS‐CoV‐2 is a single‐stranded RNA virus that has been shown to be sensed by RLRs after entering host cells. However, recent studies indicate that SARS‐CoV‐2 infection also activates the cGAS–STING signaling pathway through chromatin DNA and mitochondrial DNA release. 13 , 14 The antagonism of the RLR signaling pathway by SARS‐CoV‐2 proteins has been studied by several groups, and the effect of these viral proteins on the cGAS–STING signaling pathway requires further investigation. Studies have also demonstrated that activation of STING by its agonist can effectively control SARS‐CoV‐2 replication and infection. 25 , 26 Given the important role of STING in defense against SARS‐CoV‐2 infection, we propose that SARS‐CoV‐2 may encode viral proteins to attenuate its activation. We have constructed the plasmids that can well express 29 SARS‐CoV‐2‐encoded viral proteins in our previous studies. 18 , 27 To screen which of these viral proteins may play a role in dampening the STING signaling, we transfected the STING expression vector together with plasmids expressing empty vector (control) or individual SARS‐CoV‐2 proteins into HEK293T cells. Thirty‐six hours later, the cells were harvested for the detection of IFN‐β mRNA expression using RT‐qPCR. Interestingly, we found that NSP1, NSP3C (containing the papain‐like protease domain), NSP4, NSP5, NSP10, NSP11, NSP12, NSP13, NSP14, NSP15, ORF10, and S have some effects on the inhibition of STING‐induced IFN‐β mRNA transcription (Figure 1A). Our previous study observed that among the SARS‐CoV‐2 proteins, only ORF7b, ORF8, NSP6, and ORF10 are localized in the ER. 27 Thus, our further study focused on the interaction between ORF10 and STING, which is an ER‐localized protein. We found that ORF10 also inhibits cGAS–STING‐induced expression of IFN‐β, IFN‐λ1, CXCL10, and ISG56 (Figure 1B). Luciferase reporter assays revealed that ORF10 suppressed STING‐induced activation of IFN‐β, IFN‐λ1, and ISG (ISRE‐Luc) reporters (Figure 1C). In A549 cells, overexpression of ORF10 suppresses STING‐induced IFN‐β production (Figure 1D). In HK‐2 cells, which are highly susceptible to SARS‐CoV‐2, ORF10 impairs SARS‐CoV‐2 trVLP induced IFN‐β upregulation (Figure 1E). The HSV‐1 virus is a model DNA virus that activates the cGAS‐STING signaling pathway. We use HSV‐1‐∆ICP0, which induces higher levels of type I IFNs than wild‐type HSV‐1, to activate the cGAS‐STING signaling pathway in HK‐2 cells. We observed that ORF10 inhibited IFN‐β induction by HSV‐1‐∆ICP0 (Figure 1F). The ELISA assays showed that less IFN‐β is released into the culture supernatant from HK‐2 cells expressing ORF10 than in the control cells (Figure 1G). These results indicate that SARS‐COV‐2 ORF10 inhibits activation of the cGAS–STING signaling pathway.
Figure 1.
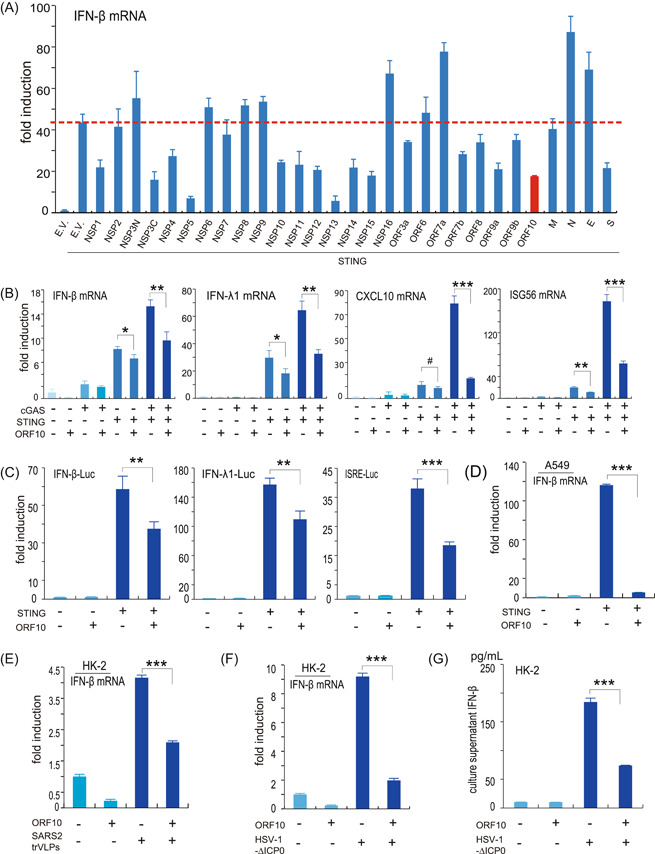
SARS‐CoV‐2 ORF10 suppresses IFN production induced by the cGAS–STING signaling pathway. (A) Identification of SARS‐CoV‐2 proteins perturbing STING‐induced IFN‐β activation. HEK293T cells were transfected with empty vector (E.V.) or STING expression vectors in the presence or absence of various SARS‐CoV‐2 protein expression vectors as indicated; 36 h later, the cells were harvested for total RNA extraction and subsequent RT‐qPCR examination of IFN‐β mRNA level. (B) SARS‐CoV‐2 ORF10 inhibits the expression of IFN‐β, IFN‐λ1, CXCL10, and ISG56 induced by the cGAS–STING signaling pathway. HEK293T cells were transfected with protein expression vectors as indicated. Thirty‐six hours posttransfection, total RNA was isolated and used to analyze the transcriptional level of the indicated genes by RT‐qPCR. (C) SARS‐CoV‐2 ORF10 inhibits STING‐induced activation of IFN‐β, IFN‐λ1, or ISG luciferase reporters. Luciferase reporter plasmids of IFN‐β (IFN‐β‐Luc), IFN‐λ1 (IFN‐λ1‐Luc), or ISGs (ISRE‐Luc) were cotransfected with STING‐ and/or ORF10‐expressing plasmids into HEK293T cells. pRL‐TK Renilla was transfected as an internal control. Thirty‐six hours later, transactivation of the luciferase reporters was determined using dual‐luciferase reporter assays. (D) SARS‐CoV‐2 ORF10 inhibits STING‐induced IFN‐β production in A549 cells. Protein expression plasmids were transfected into A549 cells as indicated. Thirty‐six hours later, A549 cells were harvested and subjected to RNA isolation and RT‐qPCR analysis of IFN‐β expression. (E) SARS‐CoV‐2 ORF10 inhibits IFN‐β expression induced by SARS‐CoV‐2 trVLP infection. HK‐2 cells were transfected with ORF10 expression plasmid, 36 h later, the cells were infected with SARS‐CoV‐2 trVLP for 12 h and then the cells were harvested for RNA isolation and RT‐qPCR analysis. (F, G) SARS‐CoV‐2 ORF10 inhibits IFN‐β expression and secretion in HK‐2 cells. ORF10 expression plasmid was transfected into HK‐2 cells, 36 h posttransfection, HSV‐1‐∆ICP0 was used to infect the cells as indicated, 12 h postinfection, the cells and culture supernatant were harvested for RT‐qPCR and ELISA analysis, respectively. cGAS, cyclic GMP–AMP synthase; ELISA, enzyme‐linked immunosorbent assay; HSV, herpes simplex virus type 1; IFN, interferon; ISG, IFN‐stimulated gene; mRNA, messenger RNA; NSP, nonstructural protein; ORF10, open reading frame 10; RT‐qPCR, real‐time quantitative polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; trVLP, transcription‐ and replication‐competent virus‐like particle. *p < 0.05; **p < 0.01; and ***p < 0.001.
3.2. ORF10 interacts with STING
Given that ORF10 efficiently dampens the STING‐mediated signaling pathway, we next investigated the molecular mechanism by which ORF10 counteracts the cGAS–STING signaling pathway. We first examined the interaction between ORF10 with the signaling molecules in the cGAS–STING signaling pathway. We found that ORF10 interacted with STING and TBK1 (Figure 2A,B) with a reciprocal Co‐IP assay when using a less stringent lysis buffer (1.0% [vol/vol] NP‐40, 50 mM Tris‐HCl [pH 7.4], 50 mM EDTA, 150 mM NaCl). To exclude the nonspecific bindings, we used a quite stringent RIPA buffer (1.0% [vol/vol] NP‐40, 50 mM Tris‐HCl [pH 7.4], 50 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS) to perform Co‐IP assays. The results indicated that ORF10 could be immunoprecipitated by STING but not by TBK1. Thus, ORF10 can specifically bind with STING but not TBK1 (Figure 2C). Confocal microscopy analysis showed that STING was colocalized with ORF10 (Figure 2D). On the basis of these observations, we reasoned that ORF10 might have a potential role in regulating the STING‐mediated antiviral innate immune pathway by interacting with STING. To investigate whether ORF10 affects TBK1 activity on IFN induction, RT‐qPCR and luciferase reporter assays were performed. The results indicated that ORF10 showed little effect on the expression of IFN‐β, IFN‐λ1, CXCL10, and ISG56 induced by TBK1 (Figure 2E); ORF10 has no effect on TBK1‐induced activation of IFN‐β, IFN‐λ1, and ISG luciferase reporters (Figure 2F). These results suggest that ORF10 specifically interacts with STING to affect STING‐induced IFN activation.
Figure 2.
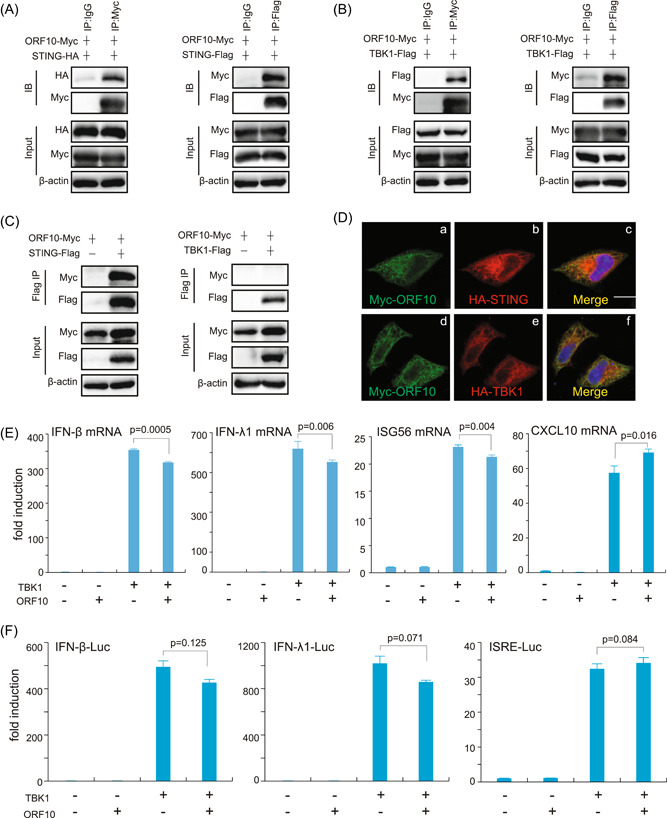
SARS‐CoV‐2 ORF10 interacts with STING. (A–C) Co‐IP analysis of the binding of ORF10 with STING and TBK1. HEK293T cells were transfected with the indicated plasmids for 24 h before Co‐IP assays. The cells were lysed with less stringent lysis buffer (1.0% [vol/vol] NP‐40, 50 mM Tris‐HCl [pH 7.4], 50 mM EDTA, 150 mM NaCl) (A, B) or more stringent RIPA buffer (1.0% [vol/vol] NP‐40, 50 mM Tris‐HCl [pH 7.4], 50 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS) (C) supplemented with a protease inhibitor cocktail (Sigma‐Aldrich) and a phosphatase inhibitor cocktail (Sigma‐Aldrich). The input and immunoprecipitates were immunoblotted (IB) with the indicated antibodies. (D) Representative confocal images of immunofluorescence staining for ORF10 with STING and TBK1 in HeLa cells. (E, F) ORF10 had no effect on TBK1‐induced IFN activation. Plasmids expressing ORF10 or TBK1 were transfected into HEK293T cells. Thirty‐six hours later, the cells were harvested for total RNA extraction and RT‐qPCR analysis (E). IFN‐β‐Luc, IFN‐λ1‐Luc, or ISRE‐Luc plasmids were transfected into HEK293T cells with ORF10‐ and/or TBK1‐expression plasmids. pRL‐TK Renilla was used as an internal control. Thirty‐six hours after transfection, the induction of luciferase reporter activities was analyzed by dual‐luciferase reporter assays (F). Co‐IP, co‐immunoprecipitation; EDTA, ethylenediaminetetraacetic acid; IFN, interferon; IgG, immunoglobulin G; mRNA, messenger RNA; ORF10, open reading frame 10; RIPA, radioimmunoprecipitation assay; RT‐qPCR, real‐time quantitative polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SDS, sodium dodecyl sulfate.
3.3. ORF10 impairs STING‐induced IRF3 phosphorylation and nuclear translocation
Next, we investigated whether ORF10 affected the signaling transduction events downstream of STING. To test this hypothesis, we examined whether ORF10 affected the activation of TBK1 and IRF3 by STING overexpression. We observed that STING alone efficiently induced the phosphorylation of TBK1 and IRF3, while when cotransfected with ORF10, the phosphorylation of TBK1 and IRF3 elicited by STING was notably attenuated (Figure 3A,B). In addition, we also found that the phosphorylation of STING, a marker of STING activation, was also attenuated by the overexpression of ORF10 (Figure 3A,B). Activated IRF3 enters the nucleus and initiates the transcription of target genes such as type I and type III IFNs. Thus, we studied the effect of ORF10 on cGAS–STING‐induced IRF3 translocation. The cytoplasmic and nuclear proteins were fractionated and then subjected to immunoblotting analysis. We observed that the nuclear translocation of IRF3 triggered by cGAS–STING signaling was prevented in cells expressing ORF10 compared with the corresponding control cells (Figure 3C,D). These results suggest that ORF10 suppressed IRF3 phosphorylation and nuclear translocation activated by the cGAS–STING signaling pathway.
Figure 3.
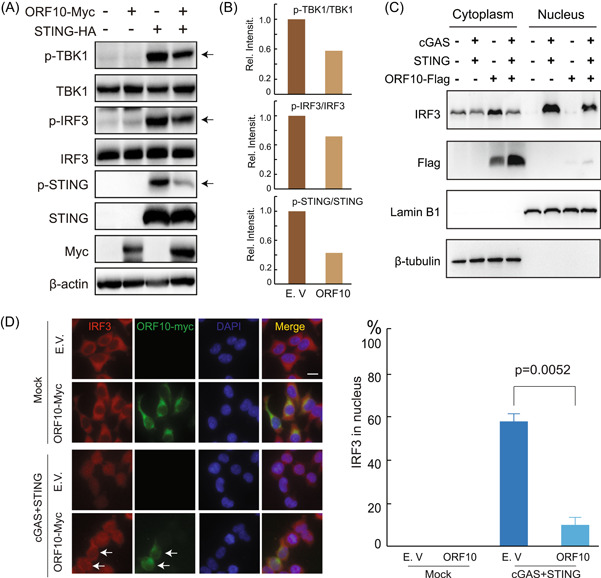
SARS‐CoV‐2 ORF10 specifically inhibits cGAS–STING signaling activation. (A) The plasmids expressing STING or SARS‐CoV‐2 ORF10 were transfected into HEK293T cells and cultured in six‐well plates as indicated. Twenty‐four hours after transfection, phosphorylated TBK1 (p‐TBK1), total TBK1, p‐IRF3, total IRF3, and STING were detected by immunoblotting. (B) Band intensity of the immunoblotting result from (A) was quantified by ImageJ and illustrated by Microsoft Excel. (C, D) SARS‐CoV‐2 ORF10 prevented cGAS–STING‐induced nuclear translocation of IRF3. (C) HEK293T cells were transfected with plasmids as indicated. Twenty‐four hours later, the cells were harvested and the cytoplasmic and nuclear proteins were fractionated. IRF3, ORF10, Lamin B1 (nuclear marker), and β‐tubulin (cytoplasmic marker) in the fractions were detected using indicated antibodies. (D) Confocal microscopy analysis of the nuclear translocation of IRF3 induced by cotransfection of cGAS and STING plasmids in HEK293T cells expressing an empty vector (E.V.) or ORF10. cGAS, cyclic GMP–AMP synthase; IRF3, interferon regulatory factor 3; ORF10, open reading frame 10; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.4. ORF10 inhibits the STING–TBK1 interaction along with STING oligomerization and aggregation
Since ORF10 interacted with both STING and TBK1 and affected the STING signaling, we next investigated whether ORF10 inhibited the oligomerization and aggregation of STING, the hallmark of STING activation. We first transfected HA‐tagged STING and Flag‐tagged STING in the absence or presence of ORF10 and found that ORF10 did not affect the interaction of HA‐STING and Flag‐STING (Figure 4A); however, overexpression of ORF10 significantly inhibited the interaction of STING and TBK1 (Figure 4B). In addition, native PAGE analysis indicated that ORF10 decreased the oligomerization of STING (Figure 4C). Then, we used SDD‐AGE to detect TBK1‐induced STING aggregation. The SDD‐AGE assay showed that STING aggregated upon coexpression with TBK1, while STING aggregation was attenuated by the overexpression of ORF10 (Figure 4D). A previous study suggested that STING oligomers formed aggregates after translocation from the ER to the Golgi, and this process was enhanced by TBK1 through its interaction with, and phosphorylation of, STING. 6 , 28 We have already observed that ORF10 expression efficiently affected the interaction between STING and TBK1 (Figure 4B). Consistent with a previous study, 28 we observed that TBK1 induced STING aggregation, while the aggregation was significantly decreased upon the ORF10 expression (Figure 4D). Overall, these results indicate that ORF10 inhibits STING activation by perturbing the STING–TBK1 interaction and decreasing STING oligomerization and aggregation.
Figure 4.
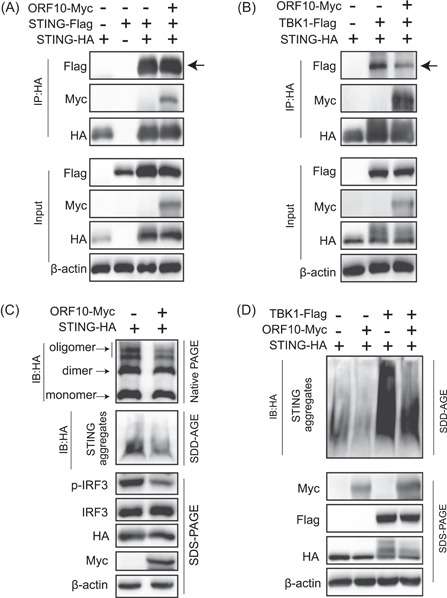
SARS‐CoV‐2 ORF10 abrogates the STING–TBK1 interaction and impairs STING activation. (A, B) ORF10 blocks STING–TBK1 interaction but does not affect STING–STING interaction. HEK293T cells were transfected with protein expression plasmids as indicated. Twenty‐four hours later, cell lysates were subjected to Co‐IP analysis with anti‐HA antibody followed by immunoblotting analysis with the indicated antibodies. (C) ORF10 inhibits STING oligomerization and aggregation. HEK293T cells were transfected with HA‐STING with or without Myc‐ORF10 expression plasmids for 24 h, and then the cells were lysed for native PAGE (top), SDD‐AGE (middle), and SDS‐PAGE (bottom) analysis of STING activation as indicated. (D) ORF10 inhibits TBK1‐induced STING aggregation. HEK293T cells were transfected with the plasmids expressing STING, TBK1, or ORF10 as shown. Twenty‐four hours later, the cells were lysed for SDD‐AGE (top) and SDS‐PAGE (bottom) analysis. HA, hemagglutinin; IB, immunoblotted; IP, immunoprecipitation; IRF3, interferon regulatory factor 3; ORF10, open reading frame 10; PAGE, polyacrylamide gel electrophoresis; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SDD‐AGE, semi‐denaturating detergent‐agarose gel electrophoresis; SDS‐PAGE, sodium dodecyl sulfate‐polyacrylamide gel electrophoresis.
3.5. ORF10 prevents STING translocation from the ER to the Golgi
Given that ORF10 dampens STING activation, there are two possible scenarios. First, ORF10 is localized in the ER and anchors STING in the ER to prevent its translocation. Anchoring of STING in the ER subsequently affects the downstream signaling. Second, ORF10 does not affect the translocation step; rather, it prevents the aggregation of STING by perturbing the interaction between STING and TBK1. To investigate the first scenario, we employed two strategies: First, we transfected a plasmid expressing mouse STING in the presence or absence of ORF10. In the absence of ORF10, we observed that mouse STING was efficiently translocated from the ER to the Golgi and colocalized with the Golgi marker GM130 upon stimulation with the mouse STING‐specific agonist DMXAA (Figure 5A, middle panel compared with the top panel). However, this translocation process was significantly impaired in the presence of ORF10 (Figure 5A, bottom panel compared with the middle panel). Consistently, we found that ORF10 efficiently attenuated the DMXAA‐induced phosphorylation of mouse STING, human TBK1, and human IRF3, which are the downstream steps of STING translocation (Figure 5B). Second, to further validate whether ORF10 affects the translocation of STING, we constructed the STING‐associated vasculopathy with onset in infancy (SAVI) mutant V147L. The SAVI mutant of STING is directly localized to the Golgi and constitutively activates the downstream signaling events. 29 If ORF10 blocks the translocation of wild‐type STING, the effect of STING V147L on downstream activation will not be affected. We found that overexpression of ORF10 did not affect the phosphorylation of TBK1 and IRF3 caused by STING V147L, while overexpression of ORF10 efficiently decreased the phosphorylation of TBK1 and IRF3 induced by wild‐type STING (Figure 5C). In addition, we observed that STING colocalized with ORF10, while STING V147L did not show colocalization with ORF10 through confocal microscopy (Figure 5D). Overall, ORF10 inhibits the cGAS‐STING signaling pathway by impairing the translocation of STING.
Figure 5.
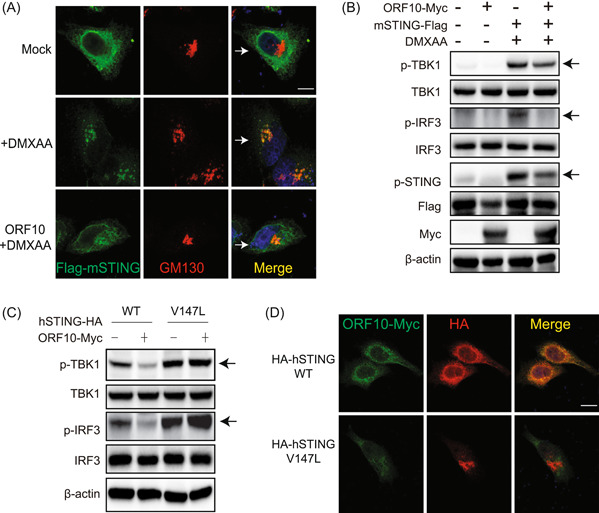
SARS‐CoV‐2 ORF10 prevents the translocation of STING from the endoplasmic reticulum to the Golgi. (A) Confocal microscopy analysis of STING translocation in HeLa cells transfected with the plasmid expressing mouse STING (mSTING) with or without ORF10 followed by stimulation with DMXAA. GM130 was used as a Golgi marker. Scale bars = 10 μm. (B) Immunoblotting analysis of the effect of ORF10 on DMXAA‐induced activation of mouse STING, human TBK1, and human IRF3. HeLa cells were transfected with protein expression plasmids followed by DMXAA stimulation as indicated for 24 h before immunoblotting analysis. (C) ORF10 inhibits TBK1 and IRF3 phosphorylation induced by wild‐type (WT) STING but not mutant STING (V147L). HeLa cells were transfected with protein expression plasmids for 24 h before immunoblot analysis as indicated. (D) ORF10 colocalizes with WT STING but not mutant STING (V147L). HeLa cells were transfected with plasmids expressing ORF10 and WT STING or STING (V147L). Twenty‐four hours later, cells were subjected to immunofluorescence staining with indicated antibodies. Scale bars = 10 μm. HA, hemagglutinin; IRF3, interferon regulatory factor 3; ORF10, open reading frame 10; p‐IRF3, phospho‐IRF‐3; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.6. ORF10 attenuates STING‐induced autophagy and facilitates HSV‐1 replication
Previous studies have suggested that STING activation stimulates the autophagy process independent of TBK1 activation and IFN induction, which is also important for clearing viruses. 9 , 10 Since ORF10 affects STING activation, we investigated whether ORF10 also affects STING‐induced autophagy. Overexpression of STING triggered strong LC3‐I to LC3‐II conversion, while this conversion was efficiently blocked by ORF10 expression (Figure 6A). Consistently, we observed that STING expression led to strong activation of LC3 puncta in the presence of STING, while coexpression of ORF10 together with STING affected LC3 puncta formation (Figure 6B). Given that ORF10 efficiently blocked STING‐induced autophagy and IFN production, we consider that ORF10 likely facilitates the replication of the HSV‐1 virus, which is a model DNA virus, in studying the cGAS–STING signaling pathway. As expected, we observed that overexpression of STING decreased HSV‐1–GFP replication, while overexpression of ORF10 increased viral replication with fluorescence analysis, viral plaque assays, and immunoblotting analysis of HSV‐1–GFP and ICP8 (Figure 6C–E).
Figure 6.
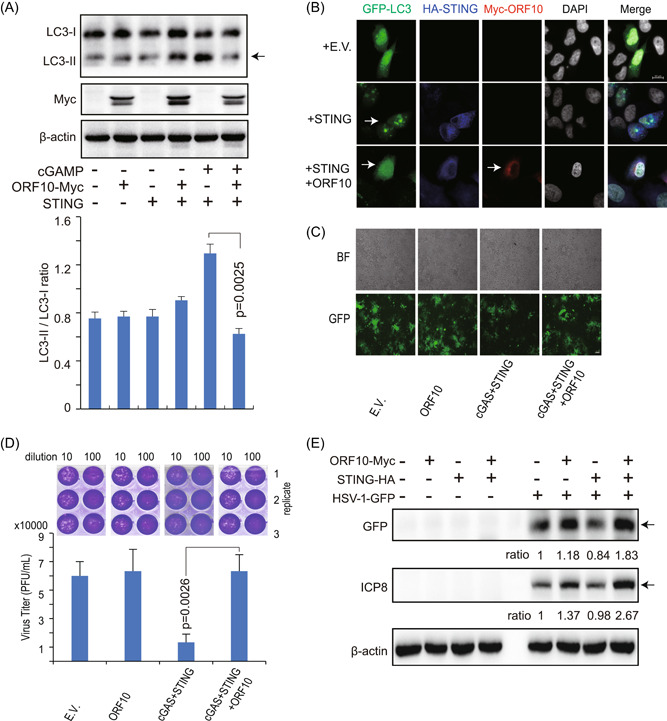
SARS‐CoV‐2 ORF10 inhibits STING‐induced autophagy and facilitates HSV‐1 replication. (A) Immunoblotting analysis of LC3‐II, a marker of autophagic activity. HEK293T cells were transfected with the plasmids as indicated. Twenty‐four hours later, cGAMP was transfected into the indicated cells for 6 h and then the cells were collected for immunoblotting analysis (top). The signals from the corresponding protein bands were quantitated using ImageJ and the ratio of LC3‐II over LC3‐I was determined (bottom). (B) Confocal microscopy analysis of LC3 in HeLa cells transfected with indicated plasmids for 24 h. (C) Fluorescence microscopy analysis of HSV‐1–GFP in the HEK293T cells transfected with the indicated plasmids for 24 h followed by HSV‐1–GFP infection (MOI = 0.01) for 24 h. (D) The culture supernatant in (C) was collected for plaque assays. (E) Immunoblotting analysis of HSV‐1–GFP and HSV‐1 ICP8 in the HEK293T cells transfected with indicated plasmids for 24 h followed by HSV‐1–GFP infection (MOI = 0.01) for 6 h. Relative ratios of GFP and ICP8 expressions in experimental groups (Lanes 7–9) to HSV‐1–GFP only group (Lane 6) were determined by ImageJ and indicated below. BF, bright field; cGAMP, cyclic GMP–AMP; cGAS, cyclic GMP–AMP synthase; DAPI, 4′,6‐diamidino‐2‐phenylindole; E.V., empty vector; GFP, green fluorescent protein; HA, hemagglutinin; HSV‐1, herpes simplex virus type 1; LC3, light chain 3; MOI, multiplicity of infection; ORF10, open reading frame 10; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
4. DISCUSSION
A typical feature of severe COVID‐19 patients is the imbalanced immune response with an impaired type I IFN response and overwhelming inflammatory cytokine storm, suggesting that SARS‐CoV‐2 encodes a series of antiviral signaling antagonists. 30 Recent studies have demonstrated that a list of structural, nonstructural, and accessory proteins of SARS‐CoV‐2 play roles in diminishing innate immune signaling. 31 , 32 , 33 For example, the SARS‐CoV‐2 M protein blocks the RIG‐I–MAVS–TRFA3–TBK1 complex formation or affects the aggregation of MAVS to impair downstream signaling. 21 , 34 N protein restrains the stress granule formation to affect the recognition of viral RNA by RIG‐I. 20 NSP5 affects immune signaling in a bifunctional manner either by impairing the K63‐linked polyubiquitination of RIG‐I or decreasing the translocation of IRF3 and STAT1. 20 , 35 , 36 ORF9b interacts with TOM70 to affect RLR‐MAVS signaling, 37 while another study indicated that ORF9b can affect the TBK1 activation and subsequently impair the RLR, cGAS‐STING, and TLR3 signaling pathways. 19 These studies focused on investigating the function of viral proteins in counteracting RLR‐MAVS, downstream kinases TBK1, and transcription factor IRF3. Recent studies have indicated that SARS‐CoV‐2 infection leads to mitochondrial DNA release and chromatin DNA shuttling from the nucleus to the cytosol to activate the cGAS‐STING signaling pathway. 13 , 14 STING agonists can efficiently restrict the SARS‐CoV‐2 infection in vivo and in vitro. 25 , 26 , 38 These studies suggest that cGAS‐STING signaling plays an essential role in counteracting SARS‐CoV‐2 infection.
STING is an essential modulator of antiviral innate immune signaling. Therefore, many viruses, including DNA and RNA viruses, have evolved a list of proteins to impair STING signaling through multiple mechanisms. For example, HSV‐1 encodes the ICP27 protein, which interacts with TBK1 and STING to affect their association. 39 HCMV possesses at least two proteins to affect STING activation. UL82 impairs the STING trafficking by disrupting the STING‐iRhom2‐TRAPβ translocation complex, 40 while US9 disrupts STING oligomerization and STING‐TBK1 association through competitive interaction. 41 The inhibition of STING signaling is not restricted to DNA viruses. Many RNA viruses also encode STING antagonists, such as NS4B from hepatitis C virus and NS2B from dengue virus. 42 , 43 These studies underline the supreme importance of STING in counteracting viral infection. Although SARS‐CoV‐2 ORF3a and NSP5 (3C‐like protease, 3CLpro or MPro) have been shown to inhibit STING activity, 44 whether SARS‐CoV‐2 encodes other proteins to counteract cGAS‐STING signaling remains largely unknown. In this study, we screened 29 SARS‐CoV‐2 proteins to identify which viral proteins affect STING‐induced IFN‐β induction and identified ORF10, which is specifically localized in the ER, as a STING antagonist. Furthermore, we found that ORF10 not only diminishes the interaction between STING and TBK1 but also anchors STING to the ER to prevent its Golgi trafficking. Blocking these processes eventually results in the impairment of STING oligomerization and aggregation, leading to the deficiency of downstream signaling activation.
STING trafficking plays an important role in activating the downstream type I IFN signaling and autophagy. However, the dysregulated trafficking of STING also leads to overwhelming autoimmunity. Multiple clinical autoinflammatory syndromes, including SAVI and coatomer protein subunit α (COPA), are closely associated with the abnormal localization of STING. 45 The SAVI mutant of STING is a gain‐of‐function mutation, 45 in which the amino acid changes (such as V155M and N154S) facilitate the aggregation and activation of STING due to conformational changes. 46 SAVI mutants autonomously localize to the Golgi instead of the ER to induce IFN and inflammatory cytokine production. 29 COPA syndrome is due to the COPA mutation, instead of STING itself. COPA is essential for sorting protein retrieval from the Golgi to the ER, while mutation of COPA leads to failure of the retrograde transport of STING from the Golgi to the ER and constitutive activation of downstream signaling. 47 The eventual result of these syndromes is the incorrect localization of STING. To further dissect the mechanism of ORF10‐mediated inhibition of STING signaling, we investigated whether ORF10 affected the signaling transduction events induced by STING V147L, one of the STING SAVI mutants. We found that ORF10 potently inhibited the downstream signaling induced by wild‐type STING but not V147L, revealing that ORF10 exerts its function mainly by blocking the translocation of STING from the ER to the Golgi (Figure 7).
Figure 7.
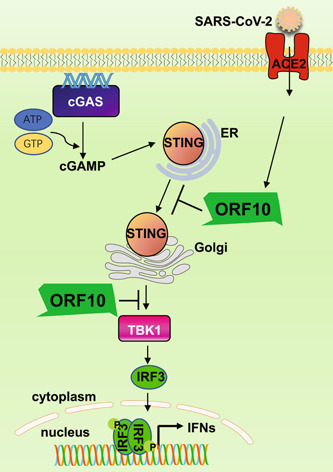
A schematic diagram describing the molecular mechanisms of SARS‐CoV‐2 ORF10‐mediated inhibition of the cGAS–STING signaling pathway. SARS‐CoV‐2 infection causes cGAS activation, which produces cGAMP and induces STING translocation from the ER to the Golgi. SARS‐CoV‐2 ORF10 prevents STING translocation and inhibits STING–TBK1 interaction to impair STING activation, leading to the suppression of STING‐mediated IRF3 activation and IFN induction. ACE2, angiotensin‐converting enzyme 2; cGAMP, cyclic GMP–AMP; cGAS, cyclic GMP–AMP synthase; ER, endoplasmic reticulum; IFN, interferon; IRF3, interferon regulatory factor 3; ORF10,open reading frame 10; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Autophagy has been reported to play an important role in controlling SARS‐CoV‐2 infection, and SARS‐CoV‐2 also manipulates autophagy to facilitate its infection and replication. 48 , 49 , 50 , 51 For example, ORF3a stops the fusion of lysosomes and blocks the HOPS complex‐mediated assembly of the SNARE complex required for autolysosome formation, while ORF7a reduces the acidity of lysosomes to block autophagy. SARS‐CoV‐2 was reported to promote autophagy to suppress IFN induction to antagonize antiviral innate immunity. 51 STING trafficking is also important for autophagy induction; thus, we studied the effect of ORF10 on STING‐induced autophagy. We found that ORF10 could prevent STING‐induced autophagy and virus clearance. Further studies are required to understand how ORF10 modulates STING‐induced autophagy.
In summary, our results extend the understanding of the interactions between SARS‐CoV‐2 and antiviral innate immunity and revealed the molecular mechanisms underlying ORF10 blunting of STING‐induced type I and type III IFN expression by impeding STING‐TBK1 interaction and STING translocation from the ER to the Golgi (Figure 7).
AUTHOR CONTRIBUTIONS
Chengjiang Gao and Pei‐Hui Wang conceptualized the study. Lulu Han, Yi Zheng, Jian Deng, Mei‐Ling Nan, Yang Xiao, Meng‐Wei Zhuang, Jing Zhang, and Wei Wang performed the experiments. Lulu Han, Yi Zheng, and Pei‐Hui Wang wrote the first draft of the manuscript. All the authors contributed to revising the manuscript and approved the final version for publication.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
The authors would like to thank the Translational Medicine Core Facility of Shandong University for the consultation and instrument availability that supported this study. The authors are grateful to Prof. Jian Li (Shandong University) for kindly providing the GFP‐LC3 plasmids and Prof. Qiang Ding (Tsinghua University) for providing SARS‐CoV‐2 trVLP. This study was supported by grants from the National Key R&D Program of China (2021YFC2701203 to Pei‐Hui Wang), grants from the Natural Science Foundation of China (82101856 to Pei‐Hui Wang), grants from the Key Research and Development Program of Shandong Province (2020CXGC011305 to Pei‐Hui Wang), grants from the Natural Science Foundation of Shandong Province (ZR2020QC085 to Pei‐Hui Wang), grants from the Natural Science Foundation of Jiangsu Province (BK20200225 to Pei‐Hui Wang), grants from the Natural Science Foundation of China (81930039, 31730026, 81525012) awarded to Chengjiang Gao, and Future Scholar Program of Shandong University and the Natural Science Foundation of China (81901604) awarded to Yi Zheng.
Han L, Zheng Y, Deng J, et al. SARS‐CoV‐2 ORF10 antagonizes STING‐dependent interferon activation and autophagy. J Med Virol. 2022;94:5174‐5188. 10.1002/jmv.27965
Lulu Han, Yi Zheng, and Jian Deng contributed equally to this study.
Contributor Information
Chengjiang Gao, Email: cgao@sdu.edu.cn.
Pei‐Hui Wang, Email: pei-hui.wang@connect.hku.hk.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus genomes. Virol J. 2020;17(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS‐CoV‐2 transcriptome. Cell. 2020;181(4):914‐921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mena EL, Donahue CJ, Vaites LP, et al. ORF10‐cullin‐2‐ZYG11B complex is not required for SARS‐CoV‐2 infection. Proc Natl Acad Sci USA. 2021;118(17):e2023157118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS‐STING signalling. Nat Rev Mol Cell Biol. 2020;21(9):501‐521. [DOI] [PubMed] [Google Scholar]
- 6. Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS‐STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21(9):548‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18(2):157‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu D, Wu H, Wang C, et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2019;26(9):1735‐1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gui X, Yang H, Li T, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu J, Shi Y, Pan X, et al. SARS‐CoV‐2 ORF9b inhibits RIG‐I‐MAVS antiviral signaling by interrupting K63‐linked ubiquitination of NEMO. Cell Rep. 2021;34(7):108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin X, Riva L, Pu Y, et al. MDA5 governs the innate immune response to SARS‐CoV‐2 in lung epithelial cells. Cell Rep. 2021;34(2):108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Domizio JD, Gulen MF, Saidoune F, et al. The cGAS‐STING pathway drives type I IFN immunopathology in COVID‐19. Nature. 2022;603:145‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Z, Zhang X, Lei X, et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS‐CoV‐2 infection. Signal Transduct Target Ther. 2021;6(1):382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu B, Zhang M, Chu H, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63‐linked polyubiquitination. Nat Immunol. 2017;18(2):214‐224. [DOI] [PubMed] [Google Scholar]
- 16. Song G, Liu B, Li Z, et al. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63‐linked ubiquitination of TBK1. Nat Immunol. 2016;17(12):1342‐1351. [DOI] [PubMed] [Google Scholar]
- 17. Wang PH, Fung SY, Gao WW, et al. A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing. Nucleic Acids Res. 2018;46(8):4054‐4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Shang L, Zhang J, et al. An antibody‐based proximity labeling map reveals mechanisms of SARS‐CoV‐2 inhibition of antiviral immunity. Cell Chem Biol. 2022;29(1):5‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han L, Zhuang MW, Deng J, et al. SARS‐CoV‐2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG‐I/MDA‐5‐MAVS, TLR3‐TRIF, and cGAS‐STING signaling pathways. J Med Virol. 2021;93(9):5376‐5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng Y, Deng J, Han L, et al. SARS‐CoV‐2 NSP5 and N protein counteract the RIG‐I signaling pathway by suppressing the formation of stress granules. Signal Transduct Target Ther. 2022;7(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng Y, Zhuang MW, Han L, et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) membrane (M) protein inhibits type I and III interferon production by targeting RIG‐I/MDA‐5 signaling. Signal Transduct Target Ther. 2020;5(1):299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhuang MW, Cheng Y, Zhang J, et al. Increasing host cellular receptor‐angiotensin‐converting enzyme 2 expression by coronavirus may facilitate 2019‐nCoV (or SARS‐CoV‐2) infection. J Med Virol. 2020;92(11):2693‐2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ju X, Zhu Y, Wang Y, et al. A novel cell culture system modeling the SARS‐CoV‐2 life cycle. PLoS Pathog. 2021;17(3):e1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou J, Han L, Zhao Z, et al. USP18 positively regulates innate antiviral immunity by promoting K63‐linked polyubiquitination of MAVS. Nat Commun. 2021;12(1):2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Humphries F, Shmuel‐Galia L, Jiang Z, et al. A diamidobenzimidazole STING agonist protects against SARS‐CoV‐2 infection. Sci Immunol. 2021;6(59):eabi9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M, Ferretti M, Ying B, et al. Pharmacological activation of STING blocks SARS‐CoV‐2 infection. Sci Immunol. 2021;6(59):eabi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J, Cruz‐Cosme R, Zhuang MW, et al. A systemic and molecular study of subcellular localization of SARS‐CoV‐2 proteins. Signal Transduct Target Ther. 2020;5(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Z, Liu G, Sun L, et al. PPM1A regulates antiviral signaling by antagonizing TBK1‐mediated STING phosphorylation and aggregation. PLoS Pathog. 2015;11(3):e1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mukai K, Konno H, Akiba T, et al. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7:11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuen CK, Lam JY, Wong WM, et al. SARS‐CoV‐2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9(1):1418‐1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lei X, Dong X, Ma R, et al. Activation and evasion of type I interferon responses by SARS‐CoV‐2. Nat Commun. 2020;11(1):3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia H, Cao Z, Xie X, et al. Evasion of type I interferon by SARS‐CoV‐2. Cell Rep. 2020;33(1):108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fu YZ, Wang SY, Zheng ZQ, et al. SARS‐CoV‐2 membrane glycoprotein M antagonizes the MAVS‐mediated innate antiviral response. Cell Mol Immunol. 2021;18(3):613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fung SY, Siu KL, Lin H, Yeung ML, Jin DY. SARS‐CoV‐2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3. Int J Biol Sci. 2021;17(6):1547‐1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Y, Ma L, Zhuang Z, et al. Main protease of SARS‐CoV‐2 serves as a bifunctional molecule in restricting type I interferon antiviral signaling. Signal Transduct Target Ther. 2020;5(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang HW, Zhang HN, Meng QF, et al. SARS‐CoV‐2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol. 2020;17(9):998‐1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bernard NJ. A STING in the tail for SARS‐CoV‐2. Nat Immunol. 2021;22(7):800. [DOI] [PubMed] [Google Scholar]
- 39. Christensen MH, Jensen SB, Miettinen JJ, et al. HSV‐1 ICP27 targets the TBK1‐activated STING signalsome to inhibit virus‐induced type I IFN expression. EMBO J. 2016;35(13):1385‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu YZ, Su S, Gao YQ, et al. Human cytomegalovirus tegument protein UL82 inhibits STING‐Mediated signaling to evade antiviral immunity. Cell Host Microbe. 2017;21(2):231‐243. [DOI] [PubMed] [Google Scholar]
- 41. Choi HJ, Park A, Kang S, et al. Human cytomegalovirus‐encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat Commun. 2018;9(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ding Q, Cao X, Lu J, et al. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59(1):52‐58. [DOI] [PubMed] [Google Scholar]
- 43. Wrighton K. The STING behind dengue virus infection. Nat Rev Microbiol. 2018;16(6):330. [DOI] [PubMed] [Google Scholar]
- 44. Rui Y, Su J, Shen S, et al. Unique and complementary suppression of cGAS‐STING and RNA sensing‐ triggered innate immune responses by SARS‐CoV‐2 proteins. Signal Transduct Target Ther. 2021;6(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fremond ML, Crow YJ. STING‐Mediated lung inflammation and beyond. J Clin Immunol. 2021;41(3):501‐514. [DOI] [PubMed] [Google Scholar]
- 46. Ergun SL, Fernandez D, Weiss TM, Li L. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell. 2019;178(2):290‐301. [DOI] [PubMed] [Google Scholar]
- 47. Lepelley A, Martin‐Niclós MJ, Le Bihan M, et al. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J Exp Med. 2020;217(11):e20200600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sui C, Xiao T, Zhang S, et al. SARS‐CoV‐2 NSP13 inhibits type I IFN production by degradation of TBK1 via p62‐dependent selective autophagy. J Immunol. 2022;208(3):753‐761. [DOI] [PubMed] [Google Scholar]
- 49. Hayn M, Hirschenberger M, Koepke L, et al. Systematic functional analysis of SARS‐CoV‐2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021;35(7):109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miao G, Zhao H, Li Y, et al. ORF3a of the COVID‐19 virus SARS‐CoV‐2 blocks HOPS complex‐mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56(4):427‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hui X, Zhang L, Cao L, et al. SARS‐CoV‐2 promote autophagy to suppress type I interferon response. Signal Transduct Target Ther. 2021;6(1):180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


